The Small Nuclear Genomes of Selaginella Are Associated with a Low Rate of Genome Size Evolution (original) (raw)
Abstract
The haploid nuclear genome size (1C DNA) of vascular land plants varies over several orders of magnitude. Much of this observed diversity in genome size is due to the proliferation and deletion of transposable elements. To date, all vascular land plant lineages with extremely small nuclear genomes represent recently derived states, having ancestors with much larger genome sizes. The Selaginellaceae represent an ancient lineage with extremely small genomes. It is unclear how small nuclear genomes evolved in Selaginella. We compared the rates of nuclear genome size evolution in Selaginella and major vascular plant clades in a comparative phylogenetic framework. For the analyses, we collected 29 new flow cytometry estimates of haploid genome size in Selaginella to augment publicly available data. Selaginella possess some of the smallest known haploid nuclear genome sizes, as well as the lowest rate of genome size evolution observed across all vascular land plants included in our analyses. Additionally, our analyses provide strong support for a history of haploid nuclear genome size stasis in Selaginella. Our results indicate that Selaginella, similar to other early diverging lineages of vascular land plants, has relatively low rates of genome size evolution. Further, our analyses highlight that a rapid transition to a small genome size is only one route to an extremely small genome.
Keywords: evolution, flow cytometry, genome size, lycophytes, Selaginella, Selaginellaceae
Introduction
Genome size exhibits an extraordinary amount of variation in vascular plants. This variation ranges from the extremely small genomes of Genlisea tuberosa (61.1 Mb; Lentibulariaceae; [Fleischmann et al. 2014]) to the extremely large genomes of Paris japonica (150 Gb; Melanthiaceae; [Pellicer et al. 2010]). Substantial genome size variation has also been found within single genera such as Genlisea (Fleischmann et al. 2014) and Eleocharis (Zedek et al. 2010), as well as across populations of a single species such as teosinte in Central America (Díez et al. 2013) or Arabidopsis thaliana in Sweden (Long et al. 2013). However, relatively slow rates of genome size evolution may characterize many ferns (Nakazato et al. 2008; Barker and Wolf 2010; Barker 2013; Bomfleur et al. 2014; Clark et al. 2016) and gymnosperms (Morse et al. 2009; Nystedt et al. 2013).
Genome sizes may increase through two chief mechanisms: polyploidy or transposable element (TE) expansion. Whole genome duplications (WGDs) are a common source of genome size variation among closely related species. Nearly 25% of vascular plant speciation events are associated with a shift to a higher ploidal level (Wood et al. 2009; Mayrose et al. 2011; Barker et al. 2015). All seed plants (Jiao et al. 2011; Li et al. 2015) and flowering plants have also experienced at least one round of ancient polyploidy (Schleuter et al. 2004; Cui et al. 2006; Jaillon et al. 2007; Barker et al. 2008, 2009, 2016; Schmutz et al. 2010; Shi et al. 2010; D’Hont et al. 2012; Tomato Genome Consortium 2012; Ibarra-Laclette et al. 2013; Jiao et al. 2014; Kagale et al. 2014; Edger et al. 2015). However, most of the observed variation in vascular plant genome size is attributed to the differential accumulation of TEs such as long terminal repeat (LTRs) retrotransposons (SanMiguel et al. 1996; Hill et al. 2005; Neumann et al. 2006; Vitte and Bennetzen 2006; Hawkins et al. 2009; Schnable et al. 2009; Willing et al. 2015). Rapid bursts of TE activity and proliferation are common in many plant nuclear genomes (Ungerer et al. 2006; Wicker and Keller 2007; Baucom et al. 2009; Baidouri and El Panaud 2013), and stress the ongoing evolutionary arms-race between the host genome and “parasitic” TEs (Kato et al. 2003; Slotkin and Martienssen 2007; Hollister and Gaut 2009; Hollister et al. 2011; Kim and Zilberman 2014).
Expansions in nuclear genome size do not proceed unchecked (Bennetzen and Kellogg 1997). Instead, genome size increases are frequently offset by deletion mechanisms. These include unequal and illegitimate homologous recombination which can remove portions of repeated LTRs (Devos et al. 2002; Ma et al. 2004; Hawkins et al. 2009; Lee and Kim 2014), as well as nonhomologous end joining following double-stranded DNA breaks which can delete large portions of the genome (Fawcett et al. 2012; Chen et al. 2013). Indeed, the extremely small genomes of Genlisea spp. and Utricularia spp. [Lentibulariaceae] result from recent TE deletion from ancestors with larger genomes (Hu et al. 2011; Ibarra-Laclette et al. 2013), and the extremely large genomes of Fritillaria [Liliaceae] reflect an absence of non-coding DNA removal and the slow accumulation of genome size (Kelly et al. 2015). Thus, recent research suggests that extremely small plant nuclear genomes evolve from the rapid reduction of TEs, whereas extremely large genomes result from the accumulation of TEs with little deletion (Barker 2013; Nystedt et al. 2013; Kelly et al. 2015).
An alternative hypothesis is that extremely small plant nuclear genomes may be ancestral and experienced relatively little expansion over time. One potential example of this pattern of genome size evolution is the cosmopolitan lycophyte genus Selaginella (Selaginellaceae). Selaginella is the largest genus of heterosporous nonseed plants with over 700 spp. (Jermy 1956, 1990) found in a diversity of ecological niches ranging from warm humid tropics, arid deserts, alpine mountain tops, and cold dry tundra (Tryon 1955; Valdespino 1993; Mickel et al. 2004; Arrigo et al. 2013). Besides their unique phylogenetic position in the evolutionary history of vascular land plants, Selaginella are also distinguished as the only clade of vascular land plants that lack a shared WGD event in their ancient history (Banks et al. 2011; Jiao et al. 2011; Li et al. 2015). Despite a crown group age minimum of ∼310 Ma and deep divergences among extant subgenera of ∼250 Ma (Kenrick and Crane 1997; Korall and Kenrick 2004; Arrigo et al. 2013), their estimated genome sizes are not only extremely small (1C DNA = 84–156 Mb; [Obermayer et al. 2002; Little et al. 2007]) but show incredibly little variation considering the ample time for genome size expansion and contraction.
Given the dynamics of extremely small, derived genomes in angiosperms, we tested whether the small nuclear genomes of Selaginella result from abrupt reductions or if they have ancestrally small genomes with little expansion over time. To address this question, we collected genome size estimates for 31 species of Selaginella. Combined with haploid nuclear genome size and plastid sequence data for Selaginella and other vascular plants available in public databases, we used a comparative phylogenetic approach to analyze the rates of genome size across >1,160 representative vascular plant species. We tested whether Selaginella and other vascular plant genome sizes evolved stochastically under a Brownian motion (BM) model or drift around a long-term mean under Ornstein–Uhlenbeck (OU) models. From the best fitting models, we compared the estimated rates of genome size evolution among plant clades to assess the relative rate of Selaginella genome size evolution. Our assembled data and analyses provide new insight into the evolutionary dynamics of small vascular plant nuclear genomes.
Materials and Methods
Flow Cytometry
Specimens of 31 Selaginella taxa across the Selaginellaceae were collected from the field and the University of Arizona Herbarium (Tucson, AZ). Fresh specimens were air dried for 1 week at 21 °C, then stored in plastic bags in the dark, and later rehydrated with distilled water for 24–36 h at 21 °C prior to use. Many Selaginella possess a unique metabolism that permits desiccation to extremely low water potentials and resurrection from metabolic dormancy following the availability of moisture while keeping their nuclei intact. Herbarium specimens were rehydrated for 12–18 h in PBS buffer with 0.1% v/v Triton X-100. Voucher specimens for flow cytometry are deposited at the University of Arizona Herbarium (table 1).
Table 1.
New Flow Cytometry Estimates of 1C Haploid Nuclear Genome Size in Selaginella
| Taxon | Mean Haploid DNA Content (pg/1C) | Mean Haploid DNA Content (Mb/1C) | Coefficient of Variation (%) | Voucher |
|---|---|---|---|---|
| Selaginella arenicola Underw. | 0.1 | 95.96 | 2.8 | ARIZ361220 |
| Selaginella arizonica Maxon | 0.09 | 92.64 | 2.6 | A. Baniaga 604 |
| Selaginella arizonica Maxon | 0.09 | 90.36 | 3.53 | ARIZ357741 |
| Selaginella asprella Maxon | 0.1 | 96.17 | 5.61 | A. Baniaga 617 |
| Selaginella bigelovii Underw. | 0.15 | 146.87 | 3.66 | A. Baniaga 625 |
| Selaginella cinerascens A.A. Eaton | 0.13 | 124.34 | 4.6 | A. Baniaga 664 |
| Selaginella densa Rydb. | 0.12 | 117.8 | 3.74 | ARIZ231165 |
| Selaginella eremophila Maxon | 0.09 | 91.48 | 1.34 | A. Baniaga 622 |
| Selaginella exaltata (Kunze) Spring | 0.11 | 106.53 | 2.06 | ARIZ380655 |
| Selaginella extensa Underw. | 0.13 | 130.37 | 1.36 | ARIZ250761 |
| Selaginella flabellata (L.) Spring | 0.12 | 112.53 | 2.23 | ARIZ224742 |
| Selaginella hansenii Hieron. | 0.11 | 110.5 | 2.38 | ARIZ187140 |
| Selaginella landii Greenm. & N. Pfeiff. | 0.11 | 107.68 | 1.96 | ARIZ007154 |
| Selaginella lepidophylla (Hook. & Grev.) Spring | 0.17 | 166.41 | 9.86 | A. Baniaga 584 |
| Selaginella leucobryoides Maxon | 0.12 | 114.89 | 2.12 | ARIZ210058 |
| Selaginella martensii Spring | 0.1 | 96.63 | 2.43 | ARIZ259786 |
| Selaginella mutica D.C. Eat. ex Underw. | 0.13 | 130.06 | 2.31 | A. Baniaga 595 |
| Selaginella oregana D.C. Eaton | 0.13 | 129.23 | 0.86 | ARIZ393369 |
| Selaginella peruviana (Milde) Hieron. | 0.13 | 123.6 | 5.07 | A. Baniaga 588 |
| Selaginella pilifera A. Braun | 0.11 | 106.48 | 1.35 | ARIZ409958 |
| Selaginella pulcherrima Liebm. | 0.12 | 113.48 | 2.35 | ARIZ292925 |
| Selaginella rupestris (L.) Spring | 0.11 | 109.04 | 3.83 | ARIZ180913 |
| Selaginella rupincola Underw. | 0.14 | 134.23 | 4.31 | A. Baniaga 618 |
| Selaginella selaginoides (L.) P. Beauv. ex Mart. & Schrank | 0.08 | 81.45 | 2.76 | ARIZ32203 |
| Selaginella sellowi Hieron. | 0.12 | 121.66 | 2.39 | ARIZ238573 |
| Selaginella tortipila A. Braun | 0.12 | 121.99 | 1.7 | ARIZ341819 |
| Selaginella underwoodii Hieron. | 0.08 | 82.21 | 1.24 | MD Windham 4148 |
| Selaginella wallacei Hieron. | 0.13 | 125.5 | 5.47 | A. Baniaga 624 |
| Selaginella watsonii Underw. | 0.19 | 182.4 | 3.3 | A. Baniaga 625 |
| Selaginella weatherbiana R.M. Tryon | 0.12 | 118.97 | 4.87 | MD Windham 4147 |
| Selaginella willdenowii (Desv. ex Poir.) Baker | 0.09 | 91.34 | 3.25 | ARIZ146729 |
| Selaginella wrightii Hieron. | 0.11 | 106.17 | 5.67 | A. Baniaga 592 |
A modified procedure based on Arumuganathan and Earle (1991) and Little et al. (2007) was used for the nuclei isolation and staining procedure. In a cold room, ∼50 mg of fresh mature Arabidopsis thaliana “Columbia-0” rosette leaf tissue, 50 mg of fresh Selaginella shoot tips, or 5 mg of dried herbarium sample were chopped in chilled 800 µl of buffer (9.6 mmol/l MgSO4, 48 mmol/l KCl, 4.8 mmol/l HEPES, 1 mmol/l dithiothreitol, 0.25% v/v Triton X-100, pH 8.0) in a glass plate resting on a ceramic tile in an ice bucket. The homogenate was filtered through a gauze mesh and then filtered through a 40-µm nylon mesh. An additional 800 µl of buffer was added to the chopped tissue and filtered with gauze and nylon mesh and then combined with the previous homogenate. Then 400 µg of RNAse solution was added to each solution followed by 200 µl of a propidium iodide solution. Samples were then incubated at 37°C for 15 min and then kept in the dark at 4 °C prior to flow cytometry estimation. This procedure was replicated 3–5 times per taxon on the same individual.
Prior to flow cytometry estimation samples were mixed with the standard at a 1:1 ratio (sample:standard). Measurements of at least 20,000 events were collected with a FACscan flow cytometer (488 nm laser; BD Biosciences, San Jose, CA) at the University of Arizona ARL Cytometry Core Facility (Tucson, AZ). All flow cytometric data were collected within a 1-h time period. Flow cytometry data were analyzed with BD CellQuest Pro acquisition software and the mean value of each peak was used for further calculations. The internal standard A. thaliana “Columbia-0” diploid genome size was assumed to be 0.34 pg/2C which is the median of published values (Doležel et al. 1998; Bennett 2003; Schmuths et al. 2004), and it was also assumed that 1 pg of DNA equals 9.78×108 bp (Doležel et al. 1998, 2003).
Phylogenetic Comparative Analyses
Genome size data for all available vascular plant taxa were compiled from the Kew Plant DNA C-values database (Bennett and Leitch 2012) and those not yet listed in the database from two phylogenetically important surveys (Bai et al. 2012; Gorelick et al. 2014), as well as our new estimates for Selaginella taxa. When multiple haploid genome size estimates were available, we used the lowest reported cytotype and when multiple estimates were reported for the lowest cytotype we used the mean for further analyses. For all taxa with an estimated haploid genome size, we searched the PhyLoTA browser (phylota.net, last accessed May 29, 2015) and GenBank for the rbcL sequence. The intersection of these two databases resulted in a data set of 1,510 taxa that had both a 1C DNA genome size estimate and rbcL sequence, including 7 previously estimated Selaginella taxa, and 29 new flow cytometry estimates for Selaginella (supplementary file S1, Supplementary Material online). Sequences were aligned using MAFFT ver. 7.2 (Katoh and Standley 2013) and manually inspected and trimmed in JalView 2 (Waterhouse et al. 2009) to an aligned sequence length of 1,474 bp. A phylogeny for all taxa was inferred using RAxML ver. 8.1 specifying a GTR GAMMA substitution model with Physcomitrella patens for an outgroup. The RAxML phylogeny was congruent with published relationships of vascular plant taxa (Wickett et al. 2014). Using the highest scoring RAxML topology, we applied 38 age constraints (supplementary file S2, Supplementary Material online) to calibrate the phylogeny with the non-parametric dating method PATHd8 (Britton et al. 2007).
To test for differences in the rate of genome size evolution between Selaginella and other vascular plant clades, we compared the fit of two BM and four OU models. These models are used to evaluate hypotheses regarding the evolutionary rates of continuous traits, such as genome size, while accounting for the relationships of the taxa and the time they have been evolving. They may be used to compare observed differences across taxa with unique traits or between entire clades. The major difference between BM and OU is that BM models a random walk process with an equal probability of moving to any value in parameter space, while the OU process incorporates an attractor with a tendency to move back to an optimum value. This framework estimates key parameters of the model such as the rate of stochastic motion (σ2), and in the OU models the optimum (θ) and the strength of attraction towards that optimum (α) (Hansen 1997; Butler and King 2004; O’Meara et al. 2006; Beaulieu et al. 2012). These models were implemented in the R package OUwie (Beaulieu et al. 2012) on 14 vascular plant clades including Selaginella in a single analysis as well as 24 independent pairwise analyses between Selaginella and 24 vascular plant clades (see Supplementary Material online for pairwise comparisons). We used a sample size corrected Akaike Information Criterion (AICc) to select the best fit model, with the best supported model having a Δ AICc > 2. When a Δ AICc < 2 was found, the simpler model with fewer parameters was chosen. All genome size data were log10 transformed to comply with assumptions of BM in which a trait may equally increase or decrease in the same magnitude given its current state (O’Meara et al. 2006; Beaulieu et al. 2010). For the OU rate tests, θ0 was dropped from the model and assumed to be distributed according to a stationary distribution of the OU process. Confidence intervals for parameter estimates were obtained from 100 parametric bootstraps implemented in the R package OUwie (Beaulieu et al. 2012) for the best fit model in all analyses.
Results
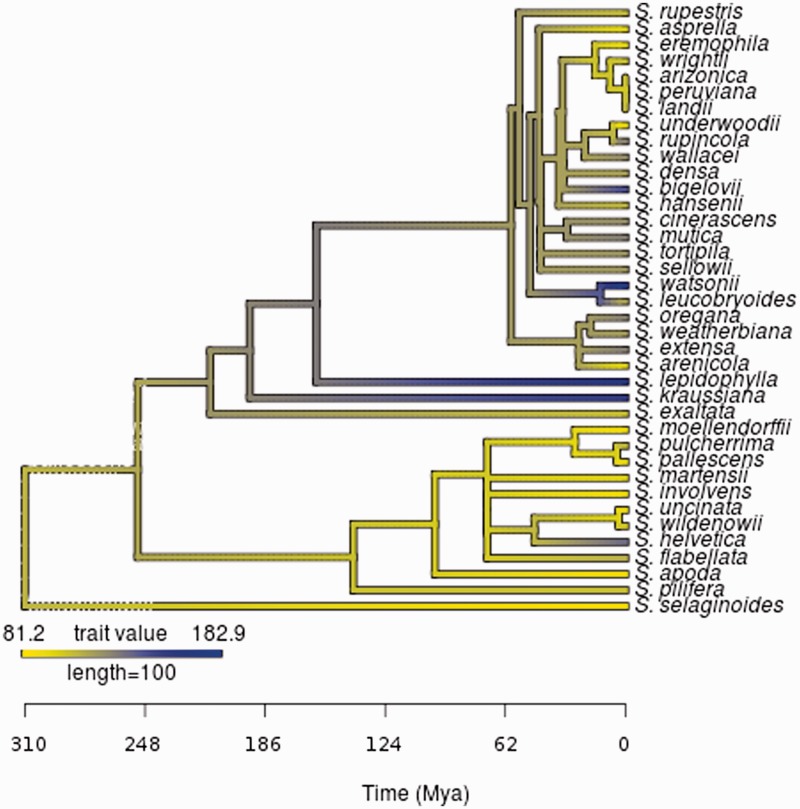
Haploid genome size (1C DNA) was estimated across the Selaginellaceae for 31 Selaginella taxa for a total of 29 new genome size estimates (table 1). No significant difference was observed between flow cytometry estimates from herbarium specimens and fresh collected specimens. In addition, low variation (CV = 1.94%) was found in our standard of A. thaliana “Col-0” when we randomly sampled ten replicates across all dates. The estimated haploid genome size in Selaginellaceae ranges 2.2-fold from 81.2 Mb in S. selaginoides to 182.4 Mb in S. watsonii (fig. 1). Within this range several taxa with relatively larger haploid genome sizes include S. helvetica (136.9 Mb), the diploid cytotype of S. kraussiana (156 Mb), as well as some members of subgenus Tetragonostachys such as S. bigelovii (146.8 Mb), S. lepidophylla (166.4 Mb), S. watsonii (182.4 Mb), and the only known tetraploid (Therrien 2004) in the analysis S. rupincola (134 Mb). Notably, these flow cytometry estimates are comparable to previously published values. For example, our estimates of S. flabellata (1C DNA = 0.115 pg) are nearly identical to those of Bouchard (1976), 1C DNA = 0.12 pg, who used light microscopy Feulgen staining. However, our genome size measurement for S. pulcherrima (1C DNA = 0.116 pg) is ∼20% greater than a previous estimate (1C DNA = 0.093 pg; [Little et al. 2007]) (table 1 and fig. 1).
Fig. 1.—
Chronogram of 1C haploid nuclear genome size across members of the Selaginellaceae. Color shading indicates relative genome size change in the phylogeny from small (yellow) to large (blue). The estimated haploid nuclear genome size in Selaginellaceae ranges 2.2-fold from 81.2 Mb in S. selaginoides (yellow) to 182.4 Mb in S. watsonii (blue).
Across both the single and pairwise rate test comparisons between Selaginella and other vascular plant clades, the best fit model was consistently an OU model. This best fit OU model was either the OUMV or OUMVA model (table 2 and supplementary file S3, Supplementary Material online). The OUMV model, which infers a different rate parameter (σ2), and optimum value (θ) for Selaginella and other plant clades, better fit rates of genome size evolution for the single rate test comparison and a majority (20/24) of the pairwise rate test comparisons. The OUMVA model, which infers a different optimum value (θ), different stochastic rate parameter (σ2), and different attraction parameter (α), for Selaginella and other plant clades, better fit rates of genome size evolution for four (4/24) pairwise rate test comparisons. The inferred optimum value for Selaginella across both the single and 24 pairwise rate tests ranged from 1C haploid nuclear DNA content of 111–113 Mb (i.e., 0.113–0.115 pg). Notably, this was the smallest optimum value for haploid nuclear genome size among the tested clades (tables 2 and 3).
Table 2.
Top Model Parameter Estimates from the Single OUwie Rate Analysis
| Clade | OUMV α | OUMV σ2 | OUMV θ | OUMV θ SE |
|---|---|---|---|---|
| Selaginella | 0.03647364 | 0.000656334 | −0.94285954 | 0.01949149 |
| Monilophytes | 0.03647364 | 0.01043217 | 0.90984577 | 0.06124471 |
| Gymnosperms | 0.03647364 | 0.00200809 | 1.09681094 | 0.03357669 |
| Asparagales | 0.03647364 | 0.01256434 | 0.85979987 | 0.07296574 |
| Arecales | 0.03647364 | 0.007693061 | 0.43315083 | 0.08922684 |
| Poales | 0.03647364 | 0.01430594 | 0.10831398 | 0.07829409 |
| Fabales | 0.03647364 | 0.00675 | −0.05193303 | 0.04325768 |
| Rosales | 0.03647364 | 0.00653 | −0.08601322 | 0.05287082 |
| Brassicales | 0.03647364 | 0.01002 | −0.25069191 | 0.04577896 |
| Myrtales | 0.03647364 | 0.00805 | −0.11469388 | 0.09157143 |
| Caryophyllales | 0.03647364 | 0.01068326 | −0.02380324 | 0.03871096 |
| Lamiales | 0.03647364 | 0.01433344 | 0.04439613 | 0.08218401 |
| Solanales | 0.03647364 | 0.007225038 | 0.3528727 | 0.1703527 |
| Asterales | 0.03647364 | 0.01228269 | 0.3901742 | 0.1326304 |
Table 3.
Summary of Alternative Models of Haploid Nuclear Genome Size Evolution across Vascular Land Plants in a Single Rate Analysis
| Model | n | −lnL | AIC | |ΔAICc| | | ----- | ---- | -------- | -------- | ---------- | | BM1 | 1163 | −356.432 | 716.8751 | 535.3402 | | BMS | 1163 | −220.997 | 470.3597 | 288.8248 | | OUM | 1163 | −171.827 | 374.0727 | 192.5378 | | OU1 | 1163 | −296.722 | 599.4638 | 417.9289 | | OUMV | 1163 | −63.1014 | 181.5349 | 0 | | OUMVA | 1163 | 27249.77 | −54418.8 | 54600.3049 |
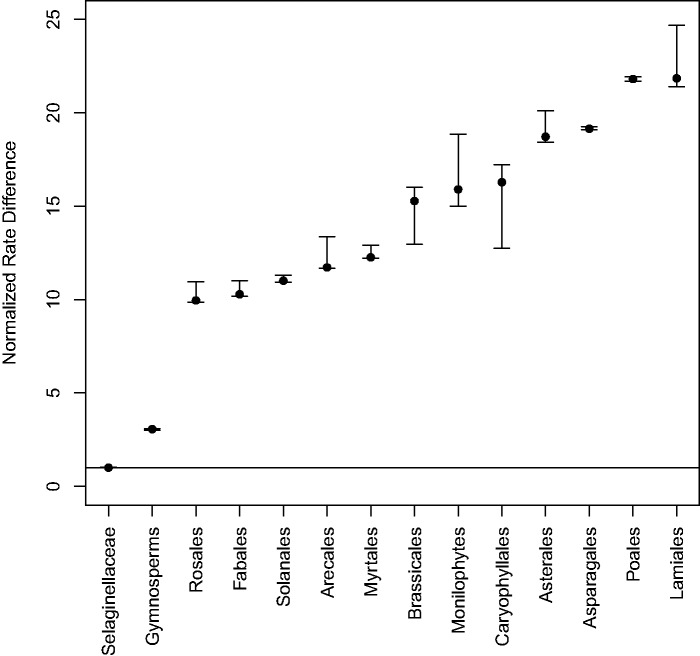
Over both the single and 24 pairwise rate tests Selaginella was inferred to have a lower rate of genome size evolution (σ2) than all other vascular plant clades included in the study (fig. 2 and supplementary file S3, Supplementary Material online). In the single rate test analysis, gymnosperms had the second lowest rate of genome size evolution inferred, and was only 3.1 times faster than Selaginella. The vascular plant clades Rosales, Fabales, and Solanales, had rates of genome size evolution that were 10–12 times greater than Selaginella, but relatively lower compared with other euphyllophyte taxa. In addition, the clades with the highest rates of genome size evolution were the Asterales, Asparagales, Poales, and Lamiales (fig. 2).
Fig. 2.—
Results of OUwie rate comparisons across all 14 vascular plant clades included in our analyses normalized by the stochastic rate (σ2) of genome size evolution found in Selaginella. The horizontal line represents the rate of genome size evolution in the Selaginellaceae. Clades are organized on the _x_-axis according to their relative rate difference to Selaginella. The 95% confidence intervals were calculated from 100 parametric bootstraps implemented in OUwie.
Discussion
Our analyses find consistent evidence that Selaginella possess extremely small genomes (1C DNA = 81.2–182.4 Mb) and have some of the lowest rates of plant nuclear genome size evolution. Selaginella were 502.2–603.4 megabases smaller than the modal angiosperm nuclear genome size of 684.6 megabases (Leitch et al. 1998). This places them in the lower 0.05–0.12% of vascular plant nuclear genome sizes (Bennett and Leitch 2012). Our phylogenetic comparative analyses indicate that the small genomes of Selaginella are a product of low rates of genome size evolution rather than recent reductions in genome size. Relatively low rates of genome size evolution have been previously observed in other lineages of vascular plants such as monilophytes (Barker and Wolf 2010; Bomfleur et al. 2014; Clark et al. 2016) and gymnosperms (Morse et al. 2009; Nystedt et al. 2013). Our analyses indicate that Selaginella nuclear genome sizes evolve an order of magnitude slower (σ2_Selaginella_ = 0.00066) than both of these lineages; ferns (σMonilophytes2=0.010)andgymnosperms(σ2Gymnosperms = 0.002). Notably, our comparative ranking of genome size evolution across the vascular plants support previous analyses that nonflowering plants have lower rates of genome size evolution than many angiosperm lineages (Nakazato et al. 2008; Barker and Wolf 2010; Leitch and Leitch 2012; Clark et al. 2016). A history of paleopolyploidy in ferns (Barker and Yatskievych 2009; Barker and Wolf 2010; Barker 2013; Vanneste et al. 2015) and gymnosperms (Li et al. 2015) has likely contributed to their higher rates of genome size evolution. In contrast, low rates of genome size evolution in Selaginella may be an order of magnitude lower because of the absence of paleopolyploidy in the Selaginellaceae (Banks et al. 2011).
Unlike the significantly larger nuclear genomes of most ferns, gymnosperms, and other lycopsids (Bainard et al. 2011; Barker 2013; Lomax et al. 2014), Selaginella have extremely small genomes and relatively little variation in genome size. The low size and variation are maintained across deep divergences among extant Selaginella subgenera dating back to the Carboniferous and Permian-Triassic boundary (Kenrick and Crane 1997; Korall and Kenrick 2004; Arrigo et al. 2013). Our rate analyses are compatible with this pattern and suggest a process of genome size stasis as evident by the strong support for OU models and the large attraction parameter estimates. Across all of our rate analyses, unambiguous support was found for OU models over BM models. The Selaginellaceae consistently had a slower rate of genome size evolution (OUMV) than all other vascular plants. This result contrasts with other extremely small genomes of vascular plants. For example, the genomes of Lentibulariaceae represent relatively recent derived states from ancestors with larger genome sizes (Leushkin et al. 2013; Fleischmann et al. 2014; Veleba et al. 2014). The Utricularia gibba [Lentibulariaceae] genome contains not only smaller amounts of noncoding DNA, but also the presence of solo LTRs which is suggestive of similar mechanisms of large scale genome size reduction through TE deletion as found in A. thaliana (Hu et al. 2011; Ibarra-Laclette et al. 2013).
The smaller genome sizes of Selaginella cannot be fully explained by similar mechanisms. Although the S. moellendorffii genome has slightly smaller introns on an average and ∼15% fewer protein coding genes than A. thaliana, the S. moellendorffii genome is comprised of a greater proportion of TEs with LTRs comprising roughly a third of the genome (Banks et al. 2011). TEs are concentrated in centromeric and pericentromeric regions in other extremely small plant genomes (Arabidopsis Genome Initiative 2000; Hu et al. 2011). In contrast, TEs are evenly distributed throughout Selaginella genomes (Brandes et al. 1997; Banks et al. 2011). Thus, Selaginella genomes have a more typical TE distribution in their genomes suggesting that they have not experienced recent, sharp reductions in their TE concentrations.
Ultimately changes in genome size reflect genetic variation introduced by mutation, and the subsequent rates of genome size evolution observed are driven by the underlying population level processes of selection and drift (Lynch and Conery 2003; Lynch 2007). Despite changes in genome size having phenotypic consequences for both cellular energetics (Lane and Martin 2010; Lynch and Marinov 2015), and important functional traits in plants such as minimum generation time, cell size, stomatal density, stomatal guard cell size, and seed mass (Bennett 1972; Knight and Ackerly 2002; Knight et al. 2005; Knight and Beaulieu 2008; Beaulieu et al. 2010). Currently, the relative roles of selection and drift on the small nuclear genome sizes of Selaginella are not yet clear.
Interestingly, many Selaginella species possess a unique ability to resurrect from metabolic dormancy following soil moisture availability, and they have independently evolved this trait multiple times (Korall and Kenrick 2004; Arrigo et al. 2013). The lifestyle of some Selaginella taxa is consistent with other extremely small genomes found in both plant and animal extremophile taxa. Relevant examples in photosynthetic organisms include the ephemeral pond endemic Genlisea tuberosa with the smallest measured vascular plant genome (61.1 Mb; [Rivadavia et al. 2013; Fleischmann et al. 2014]), the desiccation-tolerant grass Oropetium thomaeum (245 Mb; [VanBuren et al. 2015]), as well as Ostreococcus tauri a unicellular green alga of oligotrophic waters known for its extremely rapid growth rates and smallest measured genome of photosynthetic eukaryotes (12.6 Mb; [Derelle et al. 2006]). In addition, the smallest known insect genome, Belgica antarctica (99 Mb), is the only insect species endemic to Antarctica that survives through a combination of cold and desiccation tolerance (Kelley et al. 2014). However, desiccation tolerance is a complex integrated physiological process and not all desiccation tolerant organisms have extremely small genomes. For example, the vascular plant Boea hygrometrica [Gesneriaceae] does not have an extremely small genome (1.7 Gb; [Xiao et al. 2015]), nor do animals well known to withstand centuries of desiccation such as tardigrades or (78.2–802 Mb; [Garagna et al. 1996]) or bdelloid rotifers (489 Mb–2.34 Gb; [Welch and Meselson 2003]). Future research should clarify these peculiar observations.
Among our other results, we find that ferns had higher than expected rates of genome size evolution. Several flowering plant orders such as the Rosales, Fabales, and Brassicales had lower rates of genome size evolution than the ferns. Previous analyses suggested that ferns may have relatively slow rates of genome size evolution (Barker and Wolf 2010; Barker 2013; Bomfleur et al. 2014; Schneider et al. 2015; Wolf et al. 2015; Clark et al. 2016). However, none of these studies provided a direct comparison of the rates of monilophyte genome size evolution relative to other vascular plant clades. It may be that a greater frequency of polyploid speciation events among ferns (Wood et al. 2009), and a tendency for ferns to retain DNA following WGD (Nakazato et al. 2008; Bainard et al. 2011; Clark et al. 2016) explains their higher rates of genome size evolution.
Our study highlights Selaginella as an important clade of vascular plants that has not only extremely small genome sizes, but also the lowest relative rate of genome size evolution in vascular land plants. Our analyses support the hypothesis that Selaginella has ancestrally small genomes with few stochastic changes and consistent selection for a smaller genome size. This stands in contrast to the derived small genomes of other vascular plants that have close relatives with much larger genomes. Previous research suggests that ancestral genome sizes in the flowering plants were small (Soltis et al. 2003), but the dynamics of polyploidy and TE evolution have led to increases in many lineages. Even in families with consistently small genome sizes, such as the Brassicaceae, analyses find evidence of dynamic genome size evolution (Lysak et al. 2009). Unlike all other vascular plant lineages (Barker et al. 2008, 2009; Jiao et al. 2011, 2012; McKain et al. 2012; Jiao and Paterson 2014; Soltis et al. 2014; Cannon et al. 2015; Edger et al. 2015; Li et al. 2015), Selaginella genomes have not duplicated in the past. Future analyses of Selaginella genomes are needed to understand if they represent a model of vascular plant genome evolution in the absence of paleopolyploidy. Regardless, our results suggest that there are many ways to a small genome, and Selaginella provides a unique example of genome size evolution among vascular plants.
Supplementary Material
Supplementary files S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Supplementary Data
Acknowledgments
We thank the University of Arizona Herbarium staff and volunteers as well as the University of Arizona ARL Cytometry Core Facility. We also thank M.D. Windham, R. E. Baniaga, A. Wighton, A. Zimmerman, B. Penton, K.K. Fujimoto, and S. Hawthorne for assistance with Selaginella collections. Material and seeds of A. thaliana were provided by J. Brock, E. Forsythe, A. Gloss, and T. O’Connor. Finally, we thank S. M. Lambert, Z. Li, S. Jorgensen, and X. Qi for helpful discussion and comments on the manuscript. A.E.B. was supported by a National Science Foundation IGERT Fellowship in Functional and Evolutionary Genomics and a Ginny Saylor Research Grant from the Tucson Chapter of the Arizona Native Plant Society. N.A. was funded by SNSF Ambizione research grant (PZ00P3 148224). M.S.B. was supported by NSF-IOS-1339156.
Literature Cited
- Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant _Arabidopsis thaliana._Nature 408:796–815. [DOI] [PubMed] [Google Scholar]
- Arrigo N, et al. 2013. A total evidence approach to understanding phylogenetic relationships and ecological diversity in Selaginella subg. _Tetragonostachys._Am J Bot. 100:1672–1682. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. 1991. Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep. 9:229–241. [Google Scholar]
- Bai C, Alverson WS, Follansbee A, Waller DM. 2012. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann Bot. 110:1623–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidouri M, El Panaud O. 2013. Comparative genomic paleontology across plant kingdom reveals the dynamics of TE-driven genome evolution. Genome Biol Evol. 5:954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainard JD, Henry TA, Bainard LD, Newmaster SG. 2011. DNA content variation in monilophytes and lycophytes: large genomes that are not endopolyploid. Chromosome Res. 19:763–775. [DOI] [PubMed] [Google Scholar]
- Banks JA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332:960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS. 2013. Karyotype and genome evolution in Pteridophytes In: Leitch IJ, editor. Plant genome diversity. Vol. 2. Vienna, Springer; p. 245–253. [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. 2015. On the relative abundance of autopolyploids and allopolyploids. New Phytol (in press) doi:10.1111/nph.13698. [DOI] [PubMed] [Google Scholar]
- Barker MS, et al. 2008. Multiple paleopolyploidizations during the evolution of the Compositae reveal patterns of duplicate gene retention after millions of years. Mol Biol Evol. 25:2445–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, et al. Forthcoming 2016. Most Compositae are descendants of a paleohexaploid and all share a paleotetraploid ancestor with the Calyceraceae. Am J Bot. [DOI] [PubMed]
- Barker MS, Vogel H, Schranz ME. 2009. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol. 1:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Wolf PG. 2010. Unfurling fern biology in the genomics age. Bioscience 60:177–185. [Google Scholar]
- Barker MS, Yatskievych G. 2009. Summary of the 2008 AFS Symposium: from gels to genomics: the evolving landscape of pteridology. A celebration of Gerald Gastony’s contributions to fern evolutionary biology. Am Fern J. 99:117–141. [Google Scholar]
- Baucom RS, et al. 2009. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5:e1000732.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Jhueng D-J, Boettiger C, O’Meara BC. 2012. Modeling stabilizing selection: expanding the Ornstein-Uhlenbeck model of adaptive evolution. Evolution 66:2369–2383. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Smith SA, Leitch IJ. 2010. On the tempo of genome size evolution in angiosperms. J Bot. 2010:1–8. [Google Scholar]
- Bennett MD. 1972. Nuclear DNA content and minimum generation time in herbaceous plants. P Roy Soc Lond B Biol. 181:109–135. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 2003. Comparisons with Caenorhabditis (100 Mb) and Drosophila (175 Mb) using flow cytometry show genome size in Arabidopsis to be 157 Mb and thus 25% larger than the Arabidopsis Genome Initiative estimate of 125 Mb. Ann Bot. 91:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. 2012. Plant DNA C-values database. Available from: http://www.kew.org/cvalues. [DOI] [PubMed]
- Bennetzen JL, Kellogg EA. 1997. Do plants have a one-way ticket to genomic obesity? Plant Cell 9:1–7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomfleur B, McLoughlin S, Vajda V. 2014. Fossilized nuclei and chromosomes reveal 180 million years of genomic stasis in royal ferns. Science 343:1376–1377. [DOI] [PubMed] [Google Scholar]
- Bouchard RA. 1976. DNA amount and organization in some lower vascular plants. University of Chicago. [Google Scholar]
- Brandes A, et al. 1997. Comparative analysis of the chromosomal and genomic organization of Ty1-copia-like retrotransposons in pteridophytes, gymnosperms and angiosperms. Plant Mol Biol. 33:11–21. [DOI] [PubMed] [Google Scholar]
- Britton T, Anderson CL, Jacquet D, Lundqvist S, Bremer K. 2007. Estimating divergence times in large phylogenetic trees. Syst Biol. 56:741–752. [DOI] [PubMed] [Google Scholar]
- Butler MA, King AA. 2004. Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am Nat. 164:683–695. [DOI] [PubMed] [Google Scholar]
- Cannon SB, et al. 2015. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol. 32:193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, et al. 2013. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat Commun. 4:1595.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, et al. 2016. Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytol. 210:1072–1082. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. 2006. Widespread genome duplications throughout the history of flowering plants. Genome Res. 16:738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hont A, et al. 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217. [DOI] [PubMed] [Google Scholar]
- Derelle E, et al. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A. 103:11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in _Arabidopsis._Genome Res. 12:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez CM, et al. 2013. Genome size variation in wild and cultivated maize along altitudinal gradients. New Phytol. 199:264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Bartos J, Voglmayr H, Greilhuber J. 2003. Nuclear DNA content and genome size of trout and human. Cytometry A. 51:127–128. [DOI] [PubMed] [Google Scholar]
- Doležel J, et al. 1998. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot. 82:17–26. [Google Scholar]
- Edger PE, et al. 2015. The butterfly plant arms-race escalated by gene and genome duplications. Proc Natl Acad Sci U S A. 112:8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Rouze P, Van de Peer Y. 2012. Higher intron loss rate in Arabidopsis thaliana than A. lyrata is consistent with stronger selection for a smaller genome. Mol Biol Evol. 29:849–859. [DOI] [PubMed] [Google Scholar]
- Fleischmann A, et al. 2014. Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperms. Ann Bot. 114:1651–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagna S, Rebecchi L, Guidi A. 1996. Genome size variation in tardigrada. Zoo J Linn Soc Lond. 116:115–121. [Google Scholar]
- Gorelick R, Fraser D, Zonneveld BJM, Little DP. 2014. Cycad (Cycadales) chromosome numbers are not correlated with genome size. Int J Plant Sci. 175:986–997. [Google Scholar]
- Hansen TF. 1997. Stabilizing selection and the comparative analysis of adaptation. Evolution 51:1341–1351. [DOI] [PubMed] [Google Scholar]
- Hawkins JS, Proulx SR, Rapp RA, Wendel JF. 2009. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc Natl Acad Sci U S A. 42:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill P, Burford D, Martin DMA, Flavell AJ. 2005. Retrotransposon populations of Vicia species with varying genome size. Mol Genet Genomics. 273:371–381. [DOI] [PubMed] [Google Scholar]
- Hollister JD, et al. 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and _Arabidopsis lyrata._Proc Natl Acad Sci U S A. 108:2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19:1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 43:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E, et al. 2013. Architecture and evolution of a minute plant genome. Nature 498:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, et al. 2007. The grapevine genome suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467. [DOI] [PubMed] [Google Scholar]
- Jermy A. 1956. Subgeneric names in _Selaginella._Fern Gaz. 13:117–118. [Google Scholar]
- Jermy A. 1990. Selaginellaceae In: Kramer K, Green P, editors. The families and genera of vascular plants. vol. 1. Pteridophytes and gymnosperms. New York: Springer; p. 39–45. [Google Scholar]
- Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. 2012. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13:R3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Li J, Tang H, Paterson AH. 2014. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26:2792–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Paterson AH. 2014. Polyploidy-associated genome modifications during land plant evolution. Philos T Roy Soc B. 369:20130355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, et al. 2014. Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26:2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T. 2003. Role of CG and non-CG methylation in immobilization of transposons in _Arabidopsis._Curr Biol. 13:421–426. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, et al. 2014. Compact genome of the Antarctic midge is likely an adaptation to an extreme environment. Nat Commun. 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LJ, et al. 2015. Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytol. 208:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. 1997. The origin and early diversification of land plants a cladistic study. Washington: Smithsonian Institution Press. [Google Scholar]
- Kim MY, Zilberman D. 2014. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 19:320–326. [DOI] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD. 2002. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecol Lett. 5:66–76. [Google Scholar]
- Knight CA, Beaulieu JM. 2008. Genome size scaling through phenotype space. Ann Bot. 101:759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. 2005. The large genome constraint hypothesis: evolution, ecology and phenotype. Ann Bot. 95:177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korall P, Kenrick P. 2004. The phylogenetic history of Selaginellaceae based on DNA sequences from the plastid and nucleus: extreme substitution rates and rate heterogeneity. Mol Phylogenet Evol. 31:852–864. [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467:929–934. [DOI] [PubMed] [Google Scholar]
- Lee S-I, Kim N-S. 2014. Transposable elements and genome size variations in plants. Genomics Inform. 12:87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. 2012. Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol. 194:629–646. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD. 1998. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Ann Bot. 82:85–94. [Google Scholar]
- Leushkin EV, et al. 2013. The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences. BMC Genomics. 14:476.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. 2015. Early genome duplications in conifers and other seed plants. Sci Adv. 1:e1501084.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DP, Moran RC, Brenner ED, Stevenson DW. 2007. Nuclear genome size in _Selaginella._Genome 50:351–356. [DOI] [PubMed] [Google Scholar]
- Lomax BH, et al. 2014. Reconstructing relative genome size of vascular plants through geological time. New Phytol. 201:636–644. [DOI] [PubMed] [Google Scholar]
- Long Q, et al. 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 45:884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sunderland, MA: Sinauer. [Google Scholar]
- Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302:1401–1404. [DOI] [PubMed] [Google Scholar]
- Lynch M, Marinov GK. 2015. The bioenergetic costs of a gene. Proc Natl Acad Sci U S A. 112:15690–15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Beaulieu JM, Meister A, Leitch IJ. 2009. The dynamic ups and downs of genome size evolution in Brassicaceae. Mol Biol Evol. 26:85–98. [DOI] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL. 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, et al. 2011. Recently-formed polyploids diversify at lower rates. Science 333:1257.. [DOI] [PubMed] [Google Scholar]
- McKain MR, et al. 2012. Phylogenomic analysis of transcriptome data elucidates co-occurence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae). Am J Bot. 99:397-406. [DOI] [PubMed] [Google Scholar]
- Mickel JT, Smith AR, Valdespino IA. 2004. Selaginella In: Mickel JT, Smith AR, editors. The pteridophytes of Mexico. New York: New York Botanical Garden; p. 550–602. [Google Scholar]
- Morse AM, et al. 2009. Evolution of genome size and complexity in _Pinus._PLoS One 4:e4332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato T, Barker MS, Rieseberg LH, Gastony GJ. 2008. Evolution of the nuclear genome of ferns and lycophytes In: The biology and evolution of ferns and lycophytes, Ranker TA, Haufler CH, editors. Oxford: Cambridge University Press; p.177-200. [Google Scholar]
- Neumann P, Koblízková A, Navrátilová A, Macas J. 2006. Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 173:1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. [DOI] [PubMed] [Google Scholar]
- O’Meara BC, Ane C, Sanderson MJ, Wainwright PC. 2006. Testing for different rates of continuous trait evolution using likelihood. Evolution 60:922–933. [PubMed] [Google Scholar]
- Obermayer R, Leitch IJ, Hanson L, Bennett MD. 2002. Nuclear DNA C-values in 30 species doubles the familial representation in pteridophytes. Ann Bot. 90:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Fay MF, Leitch IJ. 2010. The largest eukaryotic genome of them all? Bot J Linn Soc. 164:10–15. [Google Scholar]
- Rivadavia F, Gonella PM, Fleischmann A. 2013. A new and tuberous species of Genlisea (Lentibulariaceae) from the Campos Rupestres of Brazil. Syst Bot. 38:464–470. [Google Scholar]
- SanMiguel P, et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768. [DOI] [PubMed] [Google Scholar]
- Schleuter JA, et al. 2004. Mining EST databases to resolve evolutionary events in major crop species. Genome 47:868–876. [DOI] [PubMed] [Google Scholar]
- Schmuths H, Meister A, Horres R, Bachmann K. 2004. Genome size variation among accessions of _Arabidopsis thaliana._Ann Bot. 93:317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463:178–183. [DOI] [PubMed] [Google Scholar]
- Schnable PS, et al. 2009. The B73 maize genome: complexity, diversity, dynamics. Science 326:1112–1115. [DOI] [PubMed] [Google Scholar]
- Schneider H, et al. 2015. Are the genomes of royal ferns really frozen in time? Evidence for coinciding genome stability and limited evolvability in the royal ferns. New Phytol. 207:10–13. [DOI] [PubMed] [Google Scholar]
- Shi T, Huang H, Barker MS. 2010. Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Ann. Bot. 106:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 8:272–285. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Bennett MD, Leitch IJ. 2003. Evolution of genome size in the angiosperms. Am J Bot. 90:1596–1603. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS. 2014. The polyploidy revolution then…and now: Stebbins revisited. Am J Bot. 101:1057–1078. [DOI] [PubMed] [Google Scholar]
- Therrien JP. 2004. Phylogeny of Selaginella subgenus Tetragonostachys based on nuclear and chloroplast DNA sequence data [dissertation thesis]. University of Kansas.
- Tomato Genome Consortium 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryon RM., Jr. 1955. Selaginella Rupestris and its allies. Ann Mo Bot Gard. 42:1–99. [Google Scholar]
- Ungerer MC, Strakosh SC, Zhen Y. 2006. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr Biol. 16:R872–R873. [DOI] [PubMed] [Google Scholar]
- Valdespino IA. 1993. Selaginellaceae In: Flora of North America Editorial Committee, editors. Flora of North America. Vol. 2. Pteridophytes and gymnosperms. New York: Oxford University Press; p. 38–63. [Google Scholar]
- VanBuren R, et al. 2015. Single-molecule sequencing of the desiccation-tolerant grass _Oropetium thomaeum._Nature doi:10.1038/nature15714. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Sterck L, Myburg AA, Van de Peer Y, Mizrachi E. 2015. Horsetails are ancient polyploids: evidence from _Equisetum giganteum._Plant Cell 6:1567–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleba A, et al. 2014. Genome size and genomic GC content evolution in the miniature genome-sized family Lentibulariaceae. New Phytol. 203:22–28. [DOI] [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL. 2006. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci U S A. 103:17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch DMB, Meselson M. 2003. Oocyte nuclear DNA content and GC proportion in rotifers of the anciently asexual Class Bdelloidea. Bio J Linn Soc. 79:85–91. [Google Scholar]
- Wicker T, Keller B. 2007. Genome-wide comparative analysis of Copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual Copia families. Genome Res. 17:1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, et al. 2014. A phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 111:E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing E-M, et al. 2015. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat Plants. 1:14023.. [DOI] [PubMed] [Google Scholar]
- Wolf PG, et al. 2015. An exploration into fern genome space. Genome Biol Evol. 7:2533–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T, et al. 2009. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 106:13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao LH, et al. 2015. The resurrection genome of Boea hygrometrica: a blueprint for survival of dehydration. Proc Natl Acad Sci U S A. 112:5833–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Šmerda J, Šmarda P, Bureš P. 2010. Correlated evolution of LTR retrotransposons and genome size in the genus _Eleocharis._BMC Plant Biol. 10:265.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data