Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and CNS inflammation via the aryl hydrocarbon receptor (original) (raw)
. Author manuscript; available in PMC: 2016 Nov 9.
Published in final edited form as: Nat Med. 2016 May 9;22(6):586–597. doi: 10.1038/nm.4106
Abstract
Astrocytes play important roles in the central nervous system (CNS) during health and disease. Through genome-wide analyses we detected a transcriptional response to type I interferons (IFN-I) in astrocytes during experimental CNS autoimmunity and also in CNS lesions from multiple sclerosis (MS) patients. IFN-I signaling in astrocytes reduces inflammation and experimental autoimmune encephalomyelitis (EAE) disease scores via the ligand-activated transcription factor aryl hydrocarbon receptor (AhR) and suppressor of cytokine signaling 2 (SOCS2). The anti-inflammatory effects of nasally administered IFN-β are partly mediated by AhR. Dietary tryptophan is metabolized by the gut microbiota into AhR agonists that act on astrocytes to limit CNS inflammation. EAE scores were increased following ampicillin treatment during the recovery phase, and CNS inflammation was reduced in antibiotic-treated mice by supplementation with the tryptophan metabolites indole, indoxyl-3-sulfate (I3S), indole-3-propionic acid (IPA) and indole-3-aldehyde (IAld), or the bacterial enzyme tryptophanase. In individuals with MS, the circulating levels of AhR agonists were decreased. These findings suggest that IFN-I produced in the CNS act in combination with metabolites derived from dietary tryptophan by the gut flora to activate AhR signaling in astrocytes and suppress CNS inflammation.
Astrocytes are the most abundant cell population in the central nervous system (CNS). They participate in diverse functions including control of the blood-brain barrier (BBB), the regulation of metabolism, the modulation of neuronal transmission and CNS development and repair1–9. Astrocytes also play important roles during CNS injury and disease, and are thought to participate in the pathogenesis of multiple sclerosis (MS) and its animal model experimental autoimmune encephalomyelitis (EAE)10–12. Astrocyte activity is affected by factors produced within and outside the CNS, therefore, the study of these factors may shed light on the regulation of astrocyte function in health and disease and identify new therapeutic approaches for human neurologic disorders.
The microbial flora and its products have been shown to control T cell-dependent inflammation through several mechanisms including the conversion of precursors provided by the diet into immune regulatory metabolites13–15. However, less is known about the effects of the diet and microbial products on the inflammatory response of resident cells in the CNS. Here we identify an IFN-I and AhR axis that integrates immunologic, metabolic and environmental cues to regulate astrocyte activity and CNS inflammation.
Results
Astrocytes show a transcriptional response to IFN-I during EAE
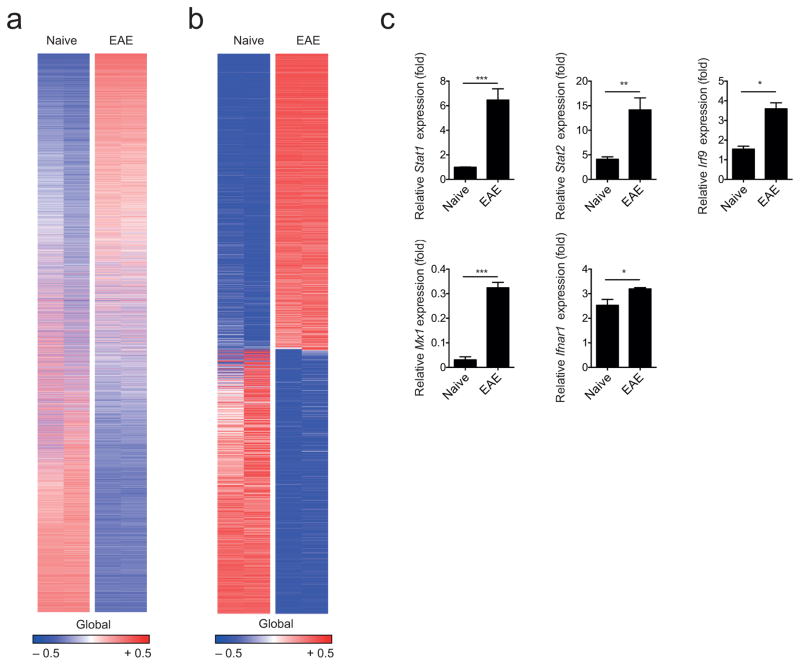
To study the regulation of astrocyte function during autoimmune CNS inflammation, we induced EAE in C57Bl/6 mice by immunization with myelin oligodendrocyte glycoprotein 35–55 (MOG35-55) in Complete Freund’s Adjuvant (CFA) and analyzed mRNA expression in astrocytes by RNA-sequencing (Supplementary Figs. 1a,b). We detected 17,964 expressed genes (Fig. 1a), and found 1,879 transcripts that were differentially regulated in astrocytes during EAE compared to astrocytes from naive mice (Fig. 1b). Although these transcripts were associated with different functional families, ingenuity pathway analysis and functional gene clustering revealed that most genes were linked to IFN-I signaling (Supplementary Table 1). Upregulation of genes associated with IFN-I signaling genes during EAE was validated in an independent set of astrocyte samples by qPCR (Fig. 1c).
Figure 1. CNS inflammation induces a type I IFN signature in astrocytes.
(a) Heatmap of all 17,964 expressed genes (detected at level 0.1 in at least half of the samples) sorted by their differential expression (signal to noise ratio) between naive and EAE (peak disease) astrocytes; representatives out of two independent experiments (n = 2 per group). Gene expression levels are row centered and log2 transformed and saturated at − 0.5 and + 0.5 for visualization. (b) Heatmap of 1,869 differentially expressed genes sorted by their differential expression (signal to noise ratio) between naive and EAE (peak disease) astrocytes; representatives out of two independent experiments (n = 2 per group). Gene expression levels are row centered and log2 transformed and saturated at − 0.5 and + 0.5 for visualization. (c) qPCR analysis of transcription factors and relevant genes involved in type I IFN signaling from FACS-sorted naive and EAE astrocytes (n = 3; mean + SEM, Student’s _t_-test; normalized to Naive Stat1). Significance levels: *P < 0.05, **P < 0.01, ***P < 0.001, n.s.: not statistically significant.
We also validated the upregulation of genes previously associated with EAE, including Ccl2, Nos2 and Csf24,8,11 (Supplementary Fig. 1c). When adjusted for the number of cells, astrocytes were a dominant source of Ccl2, Nos2 and Csf2 expression in the inflamed CNS (Supplementary Figs. 1d,e). The expression of these genes in astrocytes was more strongly induced by immunization with MOG35-55 in CFA than with CFA alone, suggesting that it is mostly triggered by immune cell infiltration into the CNS (Supplementary Fig. 2).
IFN-I signaling in astrocytes limits CNS inflammation
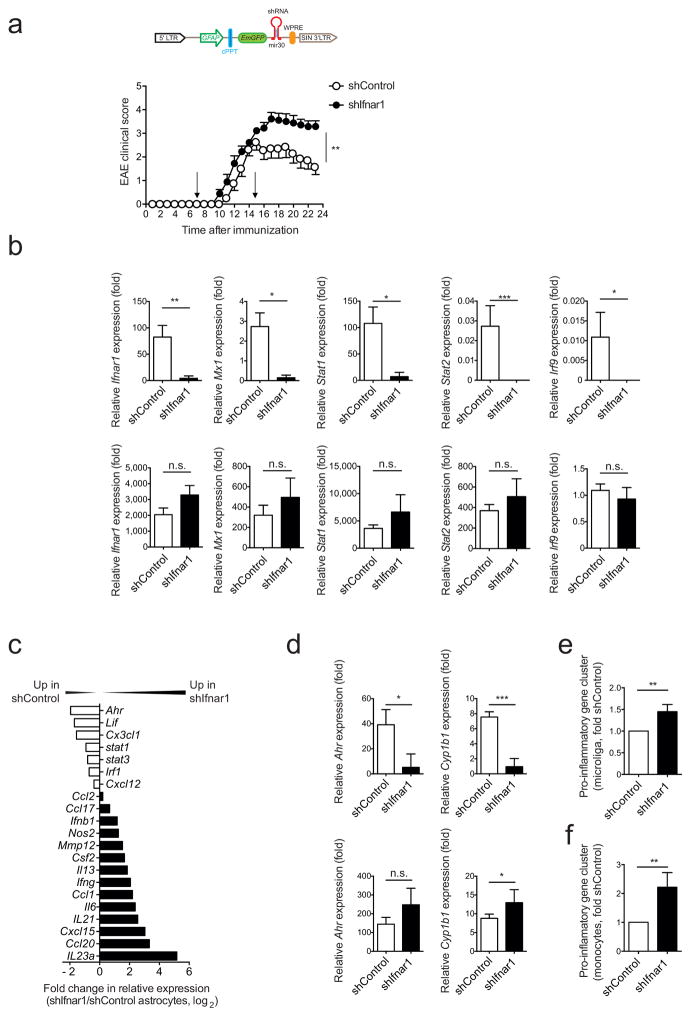
IFN-I are important regulators of inflammation in the context of infections, autoimmunity and other physiological processes16–18. To investigate the role of IFN-I signaling in astrocytes during EAE we knocked-down the interferon alpha/beta receptor 1 (Ifnar1) using a lentivirus-delivered shRNA (shIfnar1) expressed under the control of the GFAP promoter, which in this vector also controls the expression of green fluorescent protein (GFP)11,19 (Fig. 2a). The lentivirus was administered intracerebroventricularily into the CNS 7 and 15 days after EAE induction; a lentivirus carrying a non-targeting shRNA was used as a control (shControl). IFNAR1 silencing worsened EAE, as indicated by higher disease scores and failure to recover (Fig. 2a).
Figure 2. Type I IFN signaling in astrocytes limits CNS inflammation.
EAE was induced by active immunization with MOG35-55 in C57Bl/6 WT mice, which were injected intracerebroventricularly at days 7 and 15 after disease induction with Ifnar1 targeting (shIfnar1) or control (shControl) lentiviruses. (a) Top: Schematic of the astrocyte-specific shRNA targeting lentiviral vector. cPPT, central polypurine tract termination; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; mir30, micro-RNA30. Bottom: Clinical scores of shControl or shIfnar1-injected mice (mean ± s.e.m., representative out of two independent experiments with n = 10 mice per group; Two-way ANOVA). (b) Astrocytes infected with shControl or shIfnar1 were identified by GFP expression at the peak of EAE and isolated by FACS-sorting. qPCR analysis of indicated genes from GFP+ astrocytes (upper row) and the entire CD11b+CD45lo microglia population (lower row); n = 4 mice per group, representative of two independent experiments; Student’s _t_-test; normalized to shControl Microglia Irf9). (c) Fold change in mRNA expression of the indicated genes from sorted astrocytes of shIfnar1 and shControl mice during peak disease as determined by Nanostring analysis (fold change in relative expression as determined by log2(shIfnar1/shControl)). Representative out of two independent experiments of pooled astrocytes of n = 3 mice per group. (d) qPCR analysis of Ahr and Cyp1b1 expression in astrocytes and microglia sorted as in (b); n = 4 mice per group, representative of two independent experiments; Student’s _t_-test; normalized to astrocytes shIfnar1 Cyp1b1. (e,f) Nanostring analysis of pro-inflammatory gene clusters (Supplementary Table 3) from sorted microglia (e) and Ly-6Chi pro-inflammatory monocytes (f); ratio of count numbers of shIfnar1 to shControl; representative out of two independent experiments of pooled microglia and macrophages with n = 3 mice per group. Significance levels: *P < 0.05, **P < 0.01, ***P < 0.001), n.s.: not statistically significant.
As assessed by qPCR, Ifnar1 expression was efficiently knocked down in GFP+ astrocytes sorted from shIfnar1-treated mice, but not in microglia (Fig. 2b). Transcripts associated with the response to IFN-I such as Mx1 and other genes of the IFN-I signaling pathway (Stat1, Stat2, Irf9) were down-regulated in shIfnar1-treated astrocytes but not in microglia (Fig. 2b). In agreement with the worsening of EAE, using custom-made NanoString nCounter arrays (Supplementary Table 2) we detected upregulation of pro-inflammatory genes (Ifng, Il6, Nos2, Ccl2 and Il23a) in astrocytes from shIfnar1-treated mice (Fig. 2c). Moreover, Ifnar1 knock-down reduced the expression of the immunomodulatory transcription factor aryl hydrocarbon receptor (Ahr) and its target gene Cyp1b1 in astrocytes (Figs. 2c,d). Although IFNAR1 knock-down was restricted to astrocytes, it was also associated with the increased expression of pro-inflammatory transcripts in microglia and Ly-6Chi monocytes (Figs. 2e,f and Supplementary Table 3) suggesting that IFN-I signaling modulates functional interactions between astrocytes and myeloid cells. Taken together, these data show that IFN-I signaling in astrocytes limits CNS inflammation and induces AhR expression.
IFN-I induces AhR expression in astrocytes
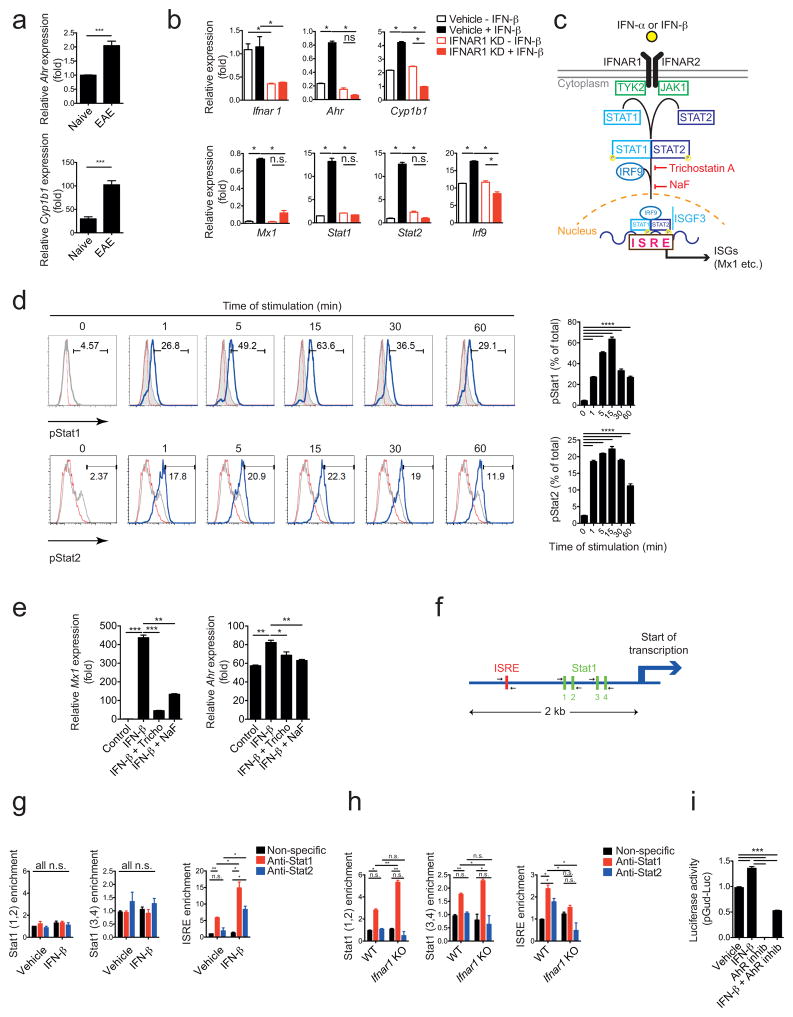
AhR regulates inflammation through its effects on several components of the immune system20,21. We first investigated whether AhR expression is controlled by IFN-I signaling in astrocytes. Similar to what has been previously reported for interferon-β IFN-β22, expression of Ahr and its target gene Cyp1b1 was increased in astrocytes during EAE (Fig. 3a). In agreement with our in vivo findings, treatment with IFN-β up-regulated Ahr expression in primary mouse astrocytes in culture in an IFNAR1-dependent manner (Fig. 3b).
Figure 3. Interferon-β induces AhR expression in astrocytes.
(a) qPCR analysis of Ahr and Cyp1b1 expression in astrocytes sorted from naive mice and mice with peak EAE scores (mean+s.e.m., n = 3, Student’s _t_-test; normalized to Naive Ahr). (b) mRNA expression of the indicated genes in in vitro cultured astrocytes transduced with shControl or shIfnar1 lentivirus and treated with IFN-β or vehicle (n = 3, representative of three independent experiments; one-way ANOVA followed by Tukey’s multiple comparisons test; normalized to Vehicle – IFN-β Ifnar1). (c) Schematic of type I interferon signaling pathway17. (d) FACS analysis of pStat1 expression in WT astrocytes stimulated with IFN-β or vehicle; left in histograms: red line: isotype, grey line: unstimulated; blue line: anti-pStat1; right in bar graph: quantification of GFP+; representative of two independent experiments; one-way ANOVA followed by Tukey’s multiple comparisons test; (e) mRNA expression of interferon response gene (ISG) Mx1 and Ahr as determined by qPCR in astrocytes activated with IFN-β and inhibitors of the type I IFN pathway (n = 3, representative of two independent experiments,; one-way ANOVA followed by Tukey’s multiple comparisons test; normalized to Control Mx1). (f) Schematic of predicted Stat1 and ISRE binding sites in AhR promoter as determined by bioinformatics analysis. (g) ChIP from WT astrocytes incubated with IFN-β or control in vitro using anti-STAT1 or non-specific antibodies to determine binding of Stat1 to its binding sites in the AhR promoter (n = 3, representative of two independent experiments, one-way ANOVA followed by Tukey’s multiple comparisons test; g and h normalized to Vehicle non-specific stat1(1,2)). (h) EAE was induced in WT and Ifnar1 KO animals, astrocytes were purified at peak of disease by FACS sorting and ChIP analysis for STAT1 binding in the AhR promoter was performed as in (g). (i) HEK293 cells were transfected with an AhR-responsive reporter (pGud-Luc) and treated as indicated with IFN-β or AhR Inhibitor CH-223191 (n = 3, representative of three independent experiments, one-way ANOVA followed by Tukey’s multiple comparisons test; normalized to Vehicle). Significance levels: * P<0.05, ** P<0.01, *** P<0.001.
Astrocytes produce IL-2723 and IL-27 is thought to contribute to the therapeutic effects of IFN-β in MS24. Although IL-27 up-regulates AhR expression in DCs and CD4+ Tr1 regulatory cells25–27, IFN-β treatment also up-regulated AhR expression in IL-27 receptor deficient astrocytes, indicating that IFN-β induces AhR expression independently of IL-27 (Supplementary Fig. 3a).
We then analyzed the direct regulation of AhR expression by IFN-β. IFNAR1 activation triggers JAK1/TYK2-dependent phosphorylation of STAT1, which dimerizes with phosphorylated STAT217. The STAT1/STAT2 heterodimer assembles with IRF9 to form a trimolecular complex called IFN-stimulated gene factor 3 (ISGF3), which translocates to the nucleus and binds specific IFN-response elements (ISREs) to control the expression of interferon-stimulated genes (ISGs) (Fig. 3c). In line with these findings, exposure of primary cultures of astrocytes to IFN-β resulted in the phosphorylation of both STAT1 and STAT2, which peaked after 15 minutes (Fig. 3d). Inhibition of ISGF3 assembly by trichostatin A or impairment of ISGF3 translocation to the nucleus by sodium fluoride (NaF) inhibited the up-regulation of both the ISGF3 target genes Mx1 and Ahr (Fig. 3e).
A bioinformatic analysis of the AhR promoter identified an ISRE that could be targeted by STAT1/STAT2-containing ISGF3 complexes formed in response to IFNAR1 activation, and also 4 STAT1 binding sites that could be recognized following STAT1 activation by other stimuli such as IFN-γ (Fig. 3f). In vitro activation of astrocytes with IFN-β resulted in the recruitment of STAT1 and STAT2 to the ISRE in the Ahr promoter, but not to the STAT1 binding sites, as determined in chromatin immunoprecipitation (ChIP) assays (Fig. 3g). These data suggest that STAT1 is recruited to the ISRE as part of an IFN-I dependent ISGF3 complex containing STAT2. STAT1 and STAT2 were also recruited to the ISRE in the Ahr promoter in astrocytes during EAE in an IFNAR1-dependent manner (Fig. 3h). Considering up-regulation of Ahr expression in astrocytes dependent on IFNAR1 during EAE (Figs. 2c,d), these data suggest that ISGF3 drives Ahr expression in astrocytes during CNS inflammation. Moreover, its increased recruitment to STAT1 binding sites following Ifnar1 knock-down suggest that IFN-I limits the accessibility of STAT1 to those sites in vivo, probably through the formation of STAT1/STAT2 heterodimers and/or epigenetic mechanisms.
To further investigate the effects of IFN-β on AhR-dependent transcriptional regulation we performed reporter assays using a construct in which an AhR-responsive promoter controls luciferase expression28. Exposure of HEK293 cells to IFN-β transactivated the AhR-responsive promoter (Fig. 3i). This transactivation was mediated by AhR, as evidenced by its inhibition by the AhR-specific antagonist CH-223191 (Fig. 3i). Collectively, these findings suggest that ISGF3 assembled in astrocytes in response to IFN-β induces AhR-dependent transcriptional programs.
AhR expression in astrocytes limits CNS inflammation
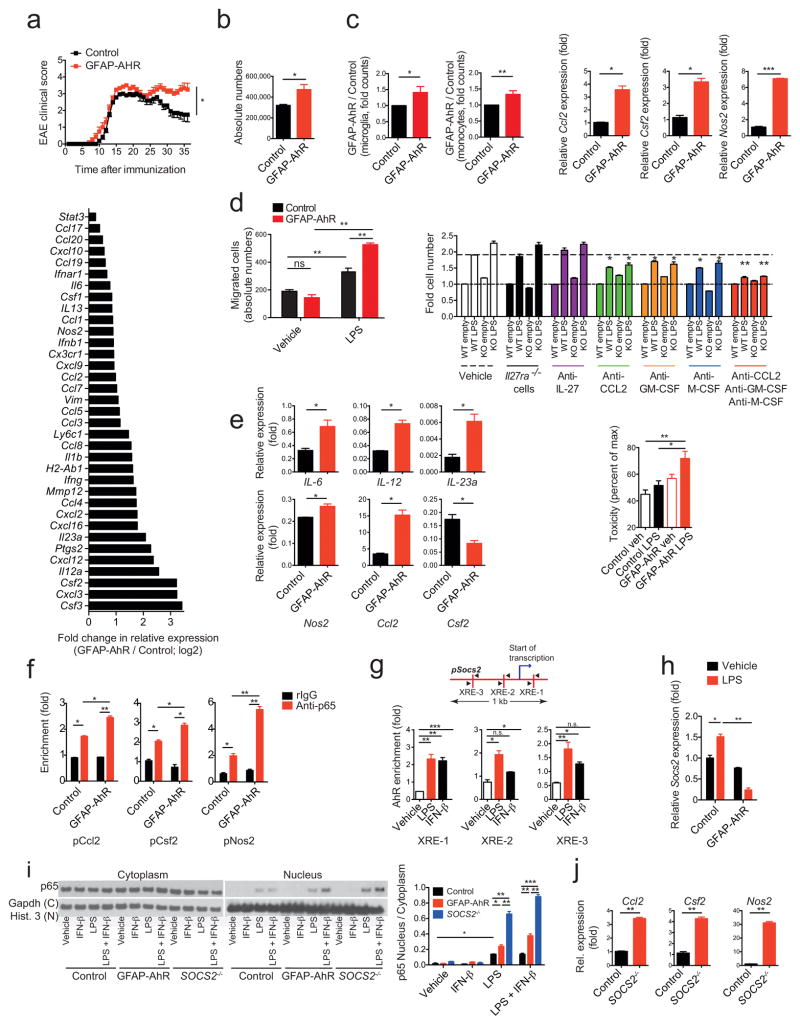
To study the role of AhR expressed in astrocytes on the regulation of CNS inflammation we deleted AhR in astrocytes by crossing AhRfl/fl mice with mice expressing the Cre recombinase under the control of the GFAP promoter29. EAE was induced in GFAP-Crepos;AhRfl/fl (GFAP-AhR deficient) and GFAP-Creneg;AHRfl/fl (control) mice and disease course was monitored. EAE disease scores were worse in GFAP-AhR deficient mice, mostly characterized by failure to recover during the chronic phase of the disease (Fig. 4a and Supplementary Table 4).
Figure 4. AhR in astrocytes limits CNS inflammation.
EAE in GFAP-AhR-deficient or control mice. (a) Top: Clinical scores (mean ± s.e.m.; representative out of five independent experiments with n = 10 mice per group; Two-way ANOVA). Bottom: Ratio of RNA abundances in pro-inflammatory cluster from sorted GFAP-AhR-deficient and control astrocytes at the peak of disease (fold change in relative expression as determined by log2(GFAP-AhR/Control). Representative of two independent experiments of pooled astrocytes of n = 3 mice per group. (b) Absolute number of CNS infiltrating CD11b+Ly-6Chi monocytes as assessed by FACS analysis. n = 5 per group, representative of five independent experiments, Student’s _t_-test. (c) Left two figures: Nanostring analysis of pro-inflammatory gene clusters (Supplementary Table 3) from sorted CD11b+CD45lo microglia (left) and CD11b+Ly-6Chi monocytes (right); numbers of GFAP-AhR divided by Control; representative out of two independent experiments of pooled microglia and macrophages of n = 3 mice per group; Student’s _t_-test. Right three figures: RNA expression of indicated genes in astrocytes sorted from WT and GFAP-AhR mice at peak of disease. (n = 3, Student’s _t_-test; normalized to Control Ccl2) (d) Left: Supernatants of LPS or vehicle stimulated WT or GFAP-AhR-deficient astrocytes were investigated in migration assays using CD11b+Ly6Chi WT monocytes as migrating cells (absolute cell numbers; n = 3; representative of three independent experiments; one-way ANOVA, Tukey’s multiple comparisons test). Right: Migration assay using blocking antibodies as indicated or IL-27R KO macrophages (fold cell numbers; n = 3; representative of three independent experiments; one-way ANOVA within treatment groups, Turkey’s multiple comparisons test). (e) Left panel: Sorted CD11b+Ly6Chi monocytes were co-cultured with activated control or GFAP-AhR-deficient astrocytes, re-isolated and gene-expression analyzed by qPCR (n = 3, representative of two independent experiments; Student’s _t_-test; normalized to Control Ccl2 in c). Right graph: Neurotoxicity assay with supernatants from control or GFAP-AhR-deficient astrocytes after activation with LPS or vehicle n = 3, representative of two independent experiments, one-way ANOVA, Tukey’s multiple comparisons test). (f) ChIP analysis of binding of NF-kB (p65) to the promoters of Ccl2, Csf2 and Nos2 in Control or GFAP-AhR-deficient astrocytes after activation with LPS. (n = 3, representative of two independent experiments, one-way ANOVA, Tukey’s multiple comparisons test). (g) Schematic of predicted AhR binding sites (XREs) in the SOCS2 promoter (upper graph) and ChIP analysis of AhR binding to the promoter of SOCS2 in astrocytes after stimulation with indicated conditions (lower bar graphs). (n = 3, representative of two independent experiments, one-way ANOVA, Tukey’s multiple comparisons test; f, g normalized to pCcl2 Control rIgG). (h) Relative expression of Socs2 in WT or GFAP-AhR astrocytes after stimulation with LPS (representative out of two independent experiments; one-way ANOVA, Tukey’s multiple comparisons test; normalized to Control Vehicle). (i) Western blot detecting NF-kB (p65; left) and quantification (right) of the ratio of nuclear to cytoplasmatic fraction of WT, GFAP-AhR, and SOCS2−/− astrocytes stimulated with indicated conditions (representative out of three independent experiments, one-way ANOVA, Tukey’s multiple comparisons test). (j) qPCR of expression levels of Ccl2, Csf2, and Nos2 in Control and SOCS2−/− astrocytes after stimulation with LPS (representative out of two independent experiments; Student’s _t_-test; normalized to Control Ccl2). Significance levels: * P<0.05, ** P<0.01, *** P<0.001.
GFAP is also expressed outside the CNS, including cells of the enteric nervous system30. Thus, it is possible that GFAP-driven AhR deletion in these cells and not astrocytes is responsible for the worsening of EAE in GFAP-AhR mice. To investigate the role of AhR in GFAP+ astrocytes during EAE, we constructed a lentivirus in which an AhR-targeting shRNA was expressed under the control of the GFAP promoter (shAhR-LV) and used it to knock down Ahr expression in GFAP+ astrocytes. The administration of shAhR-LV resulted in a significant worsening of EAE (Supplementary Figs. 3b,c), suggesting that disease worsening in GFAP-AhR mice results from AhR deficiency in GFAP+ astrocytes and not in GFAP+ cells outside the CNS.
AhR deficiency in astrocytes did not affect recall T cell responses in terms of proliferation or cytokine production in the periphery nor the recruitment of effector and regulatory T cells to the CNS (Supplementary Figs. 3d–f). However, expression of chemokines (for instance, Ccl2, Ccl20 and Cxcl10), cytokines (e.g. Il6, Il12, Il23) and pro-inflammatory markers such as Vim, Nos2, and Csf2 was increased in AhR-deficient astrocytes during EAE (Fig. 4a, lower panel). In agreement with the increased production of chemokines, AhR deletion in astrocytes was associated with an increase in the number of CD11b+CD45+Ly-6Chi CNS-infiltrating monocytes (Fig. 4b and Supplementary Fig. 3f). In addition, expression of markers associated with inflammation and neurodegeneration were increased in microglia and monocytes from GFAP-AhR mice (Supplementary Table 3 and Fig. 4c).
To investigate the role of AhR in astrocytes we studied WT and AhR-deficient primary mouse astrocytes in culture. Expression of Ccl2, Csf2 and Nos2 was increased upon activation of AhR-deficient astrocytes with LPS (Fig. 4c). Astrocyte-produced factors have been linked to the recruitment of inflammatory monocytes to the CNS31. Supernatants from stimulated astrocytes promoted migration of CD11b+Ly-6chi monocytes in transwell assays, and this migration-promoting activity was increased in supernatants from AhR-deficient astrocytes (Fig. 4d). Blocking antibodies to CCL-2, GM-CSF and/or M-CSF interfered with the migration of monocytes induced by astrocyte supernatants, suggesting that the increased expression of these molecules in AhR-deficient astrocytes is responsible for their increased chemotactic activity (Fig. 4d).
Astrocyte-produced factors modulate the polarization of microglia and monocytes, and may also have direct neurotoxic effects8,11. Co-culture of activated AhR-deficient astrocytes with monocytes increased expression of pro-inflammatory markers (Il6, Il12, Il23a, Nos2, Ccl2) and decreased the expression of IL-10 in re-isolated monocytes (Fig. 4e). Moreover, culture supernatants from AhR-deficient astrocytes were more neurotoxic in vitro as compared to supernatants from WT astrocytes (Fig. 4e). Taken together, these data show that AhR signaling in astrocytes controls transcriptional programs that regulate the recruitment of Ly-6Chi inflammatory monocytes to the CNS, the activation of microglia and monocytes and astrocytic neurotoxic activities during EAE.
SOCS2 induced by AhR in astrocytes limits NF-κB activation
The transcription factor NF-κB regulates the response of astrocytes to activation and their potential pathogenic activities during CNS inflammation32,33. To study the mechanisms used by AhR to regulate astrocyte function, we analyzed by ChIP the recruitment of NF-κB to the Ccl2, Csf2 and Nos2 promoters, because these genes have been linked to the recruitment of inflammatory monocytes to the CNS, the induction of neurotoxic activities in monocytes and microglia, and direct neurotoxic activities in astrocytes4,8,11. Following in vitro activation, binding of NF-κB to the Ccl2, Csf2 and Nos2 promoters was increased in AhR-deficient astrocytes, suggesting that AhR limits NF-κB activation in astrocytes (Fig. 4f).
AhR is reported to induce the expression of SOCS2 in B cells34, which interferes with NF-κB activation35. Activation of astrocytes in vitro with LPS or IFN-β induces recruitment of AhR to target sites in the promoter of Socs2, and the AhR-dependent upregulation of Socs2 expression (Figs. 4g,h). Activation of AhR- or SOCS2-deficient astrocytes with LPS in vitro resulted in increased NF-κB activation, supporting a role for SOCS2 in the suppression of astrocyte activation by AhR (Fig. 4i). Expression of Ccl2, Csf2 and Nos2 was increased in in vitro activated SOCS2-deficient astrocytes (Fig. 4j). Thus, these data suggest that AhR controls the pathogenic activities of astrocytes during EAE by limiting NF-κB activation in a SOCS2-dependent manner.
Astrocytic AhR mediates the suppressive effects of IFN-β
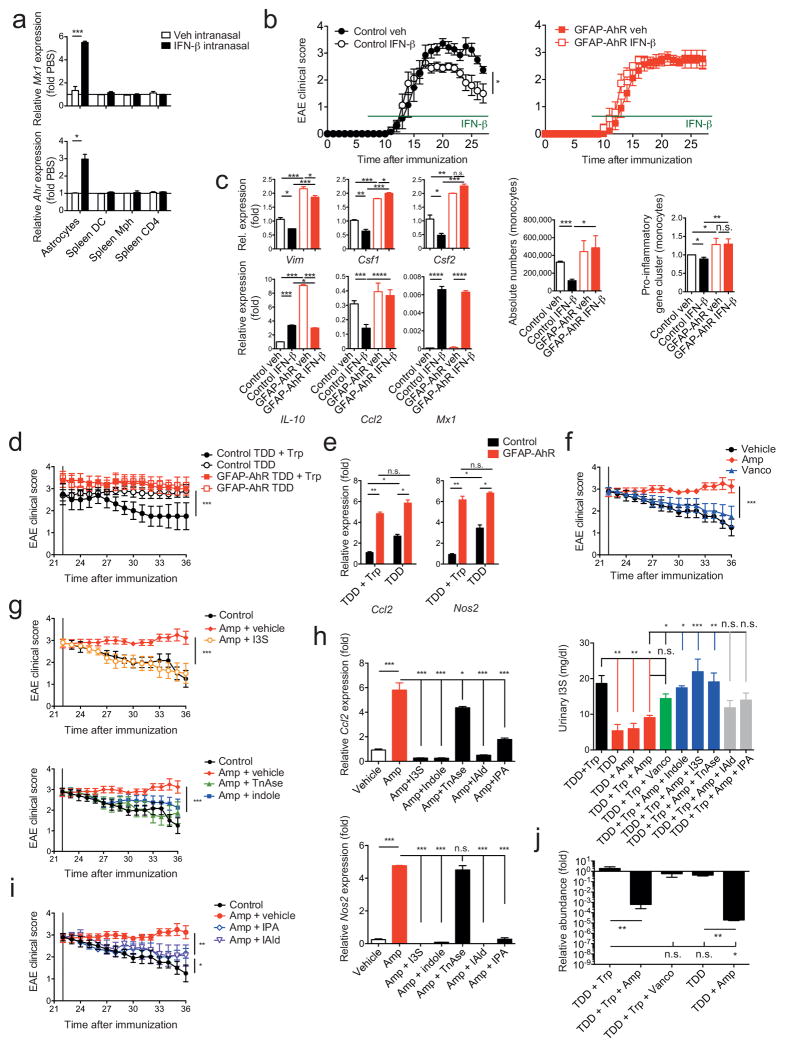
IFN-β is a first line therapy for MS and suppresses EAE36,37. Previous studies in mice have focused mainly on the effects of peripherally administered IFN-β on T cells and monocytes, because IFN-β does not cross the blood brain barrier37. To investigate the role of AhR on the effects of IFN-β on astrocytes during EAE we administered IFN-β intranasally (i.n.) because this route of administration results in accumulation of IFN-β in the CNS38. In support of a CNS-restricted biodistribution of i.n. administered IFN-β, we detected the up-regulation of Mx1 expression in astrocytes, but not in splenic cells from WT mice (Fig. 5a). I.n. administration of IFN-β started 7 days after disease induction reduced EAE scores in WT but not in GFAP-AhR deficient mice (Fig. 5b). Conversely, the specific deletion of AhR in microglia in CX3CR1-CreERT2;AhRfl/fl mice, one month after treatment with tamoxifen39, did not alter the effects of i.n. IFN-β (Supplementary Figs. 4a,b). Expression of pro-inflammatory cytokines and activation markers in astrocytes were decreased in WT mice treated with i.n. IFN-β. This was not observed in astrocytes from GFAP-AhR-deficient mice (Fig. 5c left). Of note, the expression of the IFN-I responsive gene Mx1 was not altered in GFAP-AhR as compared to control mice, indicating the co-existence of AhR-dependent and -independent IFN-I regulated genes. Il10 expression was up-regulated in AhR-deficient astrocytes (Fig. 5c left), suggesting that AhR does not drive Il10 expression in astrocytes as reported for T cells25,27,40,41. The number of CNS-infiltrating monocytes (Fig. 5c middle) and expression of pro-inflammatory molecules (Fig. 5c right) was decreased in WT but not GFAP- AhR deficient mice after i.n. IFN-β treatment. Hence, IFN-β signaling in astrocytes limits CNS inflammation in an AhR dependent manner.
Figure 5. Microbial metabolites of tryptophan and IFN-β suppress CNS inflammation via AhR in astrocytes.
(a) qPCR from sorted astrocytes and splenic DCs, macrophages or T cells from naive WT mice treated intranasally with 5.000 IU hIFN-β or PBS daily for 2 days (n = 3, Student’s _t_-test; normalized to Astrocytes Ahr). (b) EAE in control or GFAP-AhR mice under intranasal IFN-β treatment. Clinical scores of control (left) or GFAP-AhR-deficient (right) mice (mean ± s.e.m. in left graph; representative out of three independent experiments with n = 10 mice per group; Two-way ANOVA). (c) Left panel: RNA abundances from Control and GFAP-AhR astrocytes of indicated genes (n = 3, representative of two independent experiments, one-way ANOVA, Tukey’s multiple comparison test; normalized to Control Veh Vim). Middle graph: Quantification of CNS infiltrating CD11b+Ly6Chi inflammatory monocytes; representative out of three independent experiments with n = 10 mice per group; one-way ANOVA, Tukey’s multiple comparison test. Right graph: RNA analysis of pro-inflammatory gene cluster from sorted monocytes; ratio of count numbers of specific treatment group to Control veh; representative out of two independent experiments of pooled monocytes of n = 3 mice per group; one-way ANOVA, Tukey’s multiple comparison test. (d) GFAP-AhR-deficient and control animals under indicated treatment starting from day 22 after EAE induction (TDD: Tryptophan depleted diet; Trp: Tryptophan; Clinical scores, mean ± s.e.m.; representative out of two independent experiments with n = 10 mice per group; Two-way ANOVA; Tukey’s multiple comparison test). (e) qPCR of Ccl2 and Nos2 in indicated treatment conditions as in (c; normalized to Control TDD+Trp Ccl2, one-way ANOVA, Tukey’s multiple comparison test) (f, g, i) Clinical scores of WT mice treated with indicated conditions starting from day 22 after EAE induction (mean ± s.e.m.; representative out of two independent experiments with n = 5 mice per group; Two-way ANOVA; Tukey’s multiple comparisons test) (h) Left: qPCR of relative mRNA abundances for Ccl2 and Nos2 from astrocytes sorted at day 36 from experimental groups as in f, g, and i; (n = 3, representative of two independent experiments; one-way ANOVA followed by Tukey’s multiple comparisons test normalized to Vehicle Ccl2). Right: Measurement of I3S in urine samples collected at day 36 of experimental groups as in f, g, i (n = 3; representative of two independent experiments; one-way ANOVA followed by Tukey’s multiple comparison test). (j) SYBR Green qPCR of Lactobacillus reuteri bacterial DNA isolated from fecal samples of indicated groups at day 36 after EAE induction (n = 4 per group, representative of two independent experiments; one-way ANOVA followed by Tukey’s multiple comparisons test normalized to TDD+Trp). Significance levels: * P<0.05, ** P<0.01, *** P<0.001.
Dietary tryptophan limits CNS inflammation via AhR
The activity of AhR is regulated by small molecules, some of which are provided by the diet and the commensal flora20. The AhR-dependent anti-inflammatory effects of IFN-β suggest that dietary and microbial AhR ligands might regulate IFN-I signaling in astrocytes and CNS inflammation. Tryptophan (Trp) is an essential amino acid provided by the diet that is metabolized to into several AhR ligands20. To study the AhR-dependent effects of the diet on the regulation of CNS inflammation by astrocytes we induced EAE in WT and GFAP AhR-deficient mice and, starting on day 22 we fed the mice a Trp-depleted diet (TDD) or TDD supplemented with Trp (TDD+Trp) as described42. Lack of dietary Trp worsened EAE scores (Supplementary Fig. 5a), without affecting the levels of Trp or its catabolites serotonin and kynurenic acid (Supplementary Fig. 5b). The worsening of EAE scores could be reverted by Trp supplementation in control but not GFAP-AhR-deficient mice (Fig. 5d), suggesting that Trp-derived ligands of AhR act through astrocytes. Dietary Trp deficiency was associated with increased Ccl2 and Nos2 expression in astrocytes, which could not be reverted by Trp supplementation in GFAP-AhR deficient mice (Fig. 5e).
Trp-derived AhR agonists are generated through complex biochemical pathways catalyzed by commensal and host enzymes42–44. Indeed, Ampicillin (Amp)-sensitive Vancomycin (Vanco)-resistant bacteria generate AhR agonists from dietary Trp44. Thus, we tested the effects of antibiotic oral administration on late stage EAE and found that Amp, but not Vanco, interfered with disease recovery (Fig. 5f).
Bacterial tryptophanase (TnAse) catalyzes the conversion of dietary Trp to indole, which is used in the liver as a precursor for the synthesis of the AhR agonist indoxyl-3-sulfate (I3S)43,45. I3S is undetectable in germ free mice, indicating that its generation is dependent on the commensal flora43. Indeed, Amp administration decreased I3S urinary levels (Fig. 5h right). Moreover, peripherally administered I3S crosses the BBB and activates AhR in astrocytes (Supplementary Fig. 5c). Collectively, these data suggest that I3S produced by Amp-sensitive commensal bacteria participates in the regulation of astrocyte activity by AhR.
I3S or indole supplementation reduced EAE disease scores in Amp-treated mice (Fig. 5g) and increased I3S to levels (Fig. 5h right) to a similar extent to those detected in non-Amp treated mice. Moreover, I3S or indole supplementation reduced Ccl2 and Nos2 expression in astrocytes in an AhR-dependent manner (Fig. 5h and Supplementary Figs. 5e,f). Similar results were obtained when recombinant TnAse was administered by gavage to Amp-treated mice (Figs. 5g and Supplementary Figs. 5e,f).
TnAse-independent enzymatic reactions in commensal bacteria can also result in the synthesis of AhR agonists different from I3S such as indole-3-propionic acid (IPA) and indole-3-aldehyde (IAld) 44. Supplementation with IPA or IAld also reduced EAE scores in Amp-treated mice while reducing Ccl2 and Nos2 expression in astrocytes (Fig. 5h,i). IPA or IAld administration, however, did not restore peripheral I3S concentrations (Fig. 5h right).
Lactobacillus reuteri (L. reuteri) has been described to participate in the transformation of dietary Trp into AhR agonists 44. Interestingly, treatment with Amp but not with Vanco was associated with a reduction in L. reuteri 16S-RNA levels in the feces of treated mice (Fig. 5j). Taken together, these data suggest that L. reuteri and other Amp-sensitive Vanco-resistant commensal bacteria catalyze via TnAse dependent and independent pathways the conversion of dietary Trp into AhR agonists that modulate astrocyte function during EAE.
IFN-I/AhR signaling modulates human astrocyte activation
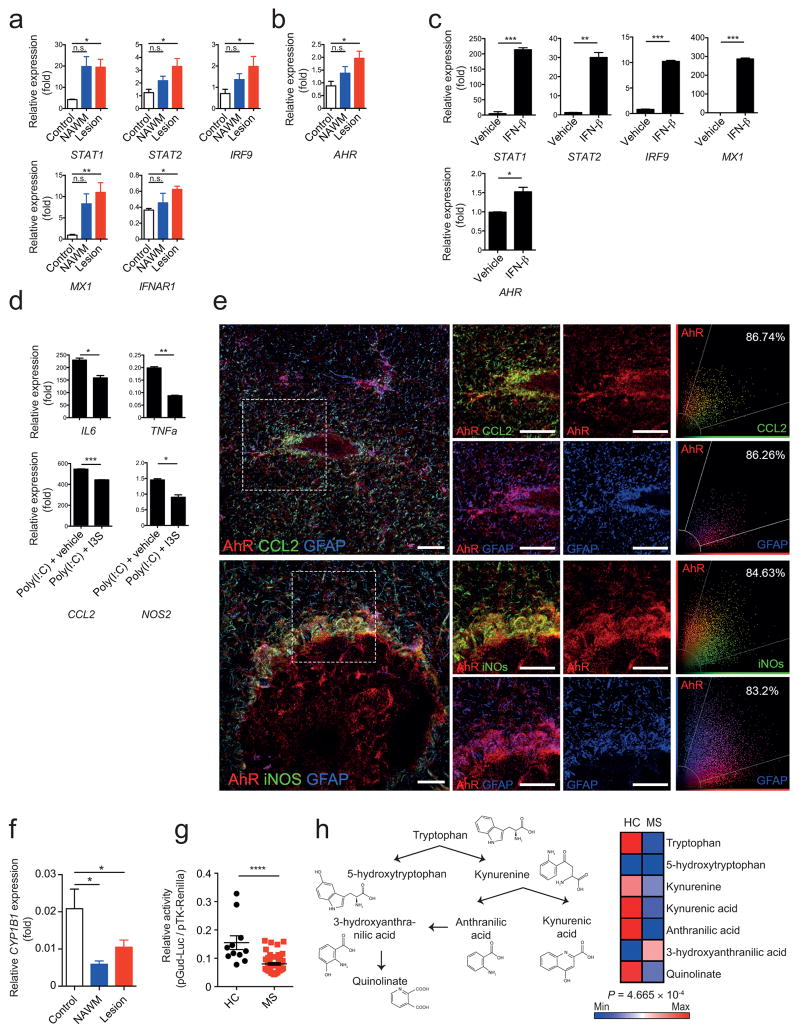
To examine the clinical relevance of our findings we analyzed the expression of genes involved in IFN-I signaling in brain samples taken from MS lesions and normal appearing white matter (NAWM) from individuals with MS, as well as controls. Expression of MX1 and the IFN-I pathway genes STAT1, STAT2 and IRF9 was upregulated in brain samples from individuals with MS (Fig. 6a). AHR expression was also upregulated in MS lesions (Fig. 6b).
Figure 6. Human astrocyte activation is controlled by IFN-β and AhR signaling.
(a,b) qPCR analysis of indicated mRNA expression in samples from lesions and normal appearing white matter (NAWM) from individuals with MS, or healthy controls relative to GAPDH (n = 4 Controls, n = 5 MS NAWM, n = 10 MS Lesion; one-way ANOVA, Tukey’s multiple comparison test; normalized to Control STAT2). (c) qPCR of RNA levels from human fetal astrocytes treated with IFN-β or vehicle in vitro (n = 3, representative of two independent experiments, Student’s _t_-test; normalized to Vehicle AHR). (d) qPCR of pro-inflammatory genes from human fetal astrocytes activated with Poly(I:C) and treated with 3-Indoxylsulfate (I3S) or vehicle. (n = 3, representative of two independent experiments; Student’s _t_-test; normalized to Poly(I:C)+I3S NOS2). (e) Immunofluorescence staining of human white matter brain tissue of active MS lesions for AhR (red), CCL2 (green, left), iNOS (green, right) and GFAP (blue) (Data shown are representative of n = 12 fields from three distinct MS brains) and co-expression of AhR and CCL2, AhR and iNOS, and AhR and GFAP. Scatter graphs (right panel) show the distribution of pixels and extent of colocalization in percentage. Scale bar: 20 μm. (f) qPCR analysis of CYP1B1 expression as in a,b. (g) Luciferase assay for the determination of absolute amount of AhR ligands in human serum (representative of two independent experiments with 11 Healthy controls, 49 MS; student’s _t_-test) (h) Schematic of tryptophan metabolism (left), and heatmap of median abundances of tryptophan metabolites in serum of healthy controls (HC) and multiple sclerosis patients (right, n = 11 HC, n = 49 MS; Hotelling’s _T_2-test). Significance levels: * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001, n.s. not statistically significant.
In MS brain samples pSTAT1, MX1 and AhR co-localized with GFAP+ astrocytes in active and chronic MS lesions, as assessed by immunofluorescence (Supplementary Fig. 6), suggesting that IFN-I signaling promotes AhR expression in human astrocytes. Incubation of human fetal astrocytes in vitro with IFN-β up-regulated the expression of AHR (Fig. 6c). Activation of AhR by I3S decreased the expression of the pro-inflammatory genes NOS2, TNFa, IL6 and CCL2 in human astrocytes (Fig. 6d). In co-staining studies, however we detected the co-expression of AHR with CCL2 and iNos in GFAP+ cells (Fig. 6e and Supplementary Fig. 7), suggesting that although AhR expression is induced by IFN-I signaling during MS, the AhR-dependent regulation of astrocyte function may be impaired in MS.
Expression of the AhR transcriptional target CYP1B1 is decreased in MS lesions and NAWM (Fig. 6f). Using an AhR-responsive reporter to quantify AhR agonistic activity in MS and healthy control sera, decreased AhR agonistic activity was detected in sera from individuals with MS as compared to controls (Fig. 6g). Moreover, using a metabolomic approach we detected decreased levels of Trp-derived AhR-activating molecules and related metabolites in MS samples (Fig. 6h). Taken together, these data support a role for the IFN-β/AhR axis in the regulation of human astrocytes, and suggest that deficits in AhR agonists provided by the metabolism, the diet, the commensal flora or the environment may contribute to the pathogenesis of MS.
Discussion
In this work we describe a novel IFN-I/AhR axis that limits pathogenic astrocyte functions in MS and potentially, other neurologic disorders. Beneficial and deleterious effects have been assigned to IFN-β in CNS inflammation. Peripheral IFN-β administration is first line disease modifying therapy for MS, thought to act through its effects on dendritic cells (DCs), B and T cells24,37. IFN-β, however, boosts the production of pathogenic antibodies and exacerbates immunopathology in neuromyelitis optica46. These opposing effects of IFN-β on CNS inflammation in MS and neuromyelitis optica highlight the need to study its cell-specific effects on CNS immunopathology.
Peripherally administered IFN-β does not cross the BBB, but substantial amounts of IFN-β are produced by astrocytes, microglia and CNS-infiltrating plasmacytoid DCs during EAE47. Microglia, infiltrating monocytes and neurons are cell populations directly targeted by IFN-β to arrest CNS inflammation and neurodegeneration22,48,49. We found significant anti-inflammatory and neurodegeneration-arresting effects of IFNAR1 signaling in astrocytes mediated by AhR-driven transcriptional programs. Thus, our data support targeting IFNAR1 signaling within the CNS for the therapeutic control of inflammation and astrocyte-driven neurodegeneration. The translational value of IFNAR1 activation in astrocytes, however, should be evaluated in the context of the deleterious effects of excessive IFN-I signaling in the CNS50.
The study of AhR signaling in EAE has been focused on its role in the regulation of effector and regulatory T cells, either directly or through the modulation of DC function27,40,41,51–53. AhR agonists with physiologically relevant important immunomodulatory activities are provided by environmental sources and also by cellular metabolism54,55. Together with recent reports of decreased levels of AhR agonists in inflammatory bowel disease56, our data suggest that imbalances in the uptake, production and/or degradation of AhR agonists may contribute to the pathogenesis of MS and other immune-mediated diseases, establishing a new link between the environment, metabolism and inflammation.
Environmental factors are known to contribute to the development of MS, but limited information is available about the identity of most of those factors and their mechanisms of action. Besides cellular metabolism, the diet and the commensal flora are abundant physiological sources of AhR agonists20. Depending on the experimental model, pro- and anti-inflammatory effects of the commensal flora have been recently reported57,58,14,15,59 highlighting the need to identify the molecular mechanisms by which the commensal flora regulates the immune response. In this context, our work contributes to the understanding of the anti-inflammatory effects of AhR agonists generated from dietary Trp by TnAse dependent and independent pathways in commensal bacteria44. Moreover, our results identify a molecular pathway through which the diet in cooperation with the commensal flora modulate the activity of CNS resident cells and neuroinflammation. In addition, the regulation of AhR expression by IFN-β suggest that the microbiome may affect not only the development of CNS autoimmunity, but also its response to disease modifying therapies60. Thus, the study of the metabolic and environmental factors that regulate astrocyte activity may shed light on CNS physiology, identify mechanisms of disease pathogenesis and drive the development of more efficacious therapeutic interventions for MS and other neurologic diseases.
Online Methods
Animals
C57BL/6J, Ifnar1−/−, IL-27ra−/−, GFAP-Cre, and AhRfl/fl mice were obtained from the Jackson Laboratory and were all female. Mice with a specific deletion of AhR in astrocytes were generated by crossing GFAP-Cre and AhRfl/fl mice, efficient deletion of AhR in astrocytes was verified by PCR and Western blotting (data not shown). CX3CR1-CreERT2 mice39 were a kind gift from Steffen Jung (Weizmann Institute of science) and were bred to AhRfl/fl mice. For activation of Cre recombinase activity, 5 week old mice were injected subcutaneously with 4 mg tamoxifen (Sigma) in 200 μl warm corn oil at two time points 48 hours apart. Four weeks later, the expression of AhR in microglia and macrophages was determined by qPCR and mice subjected to EAE induction. All mice were on the C57Bl/6 background and were kept in a pathogen-free facility at the Harvard Institutes of Medicine. All experiments were carried out in accordance with guidelines prescribed by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School.
EAE induction and treatments
EAE was induced in eight to ten weeks old mice by subcutaneous immunization with 200 μg MOG35–55 peptide emulsified in complete Freund’s adjuvant (CFA, Difco Laboratories) per mouse, followed by administration of 200 ng pertussis toxin (PTX, List biological laboratories, Inc.) on days 0 and 2 as described26. Clinical signs of EAE were assessed according to the following score: 0, no signs of disease; 1, loss of tone in the tail; 2, hind limb paresis; 3, hind limb paralysis; 4, tetraplegia; 5, moribund. Human Interferon-beta 1a (Rebif, Merck Serono) or vehicle control was administered daily at a dose of 5.000 IU intranasally or intraperitoneally as outlined in the specific experiments. Antibiotics, Trp-indoles, and TnAse were administered daily by oral gavage starting from day 22 after EAE induction at the following doses: Ampicillin 6 mg/20 g body weight (BW), vancomycin 3 mg/20 g BW; indole, indole-3-propionic acid, indole-3-aldehyde at 400 μg/20g BW, TnAse at 200 μg/20 g BW. I3S was administered daily intraperitoneally at a dose of 200 μg/20g BW. All agents were purchased from Sigma Aldrich.
Isolation of cells from adult mouse CNS
Mononuclear cells were isolated from the CNS as previously described11. Astrocytes, monocytes, and microglia were sorted as described before11 and outlined in Supplementary Fig. 1. Isolated CNS cells were stained with fluorochrome-conjugated antibody to CD11b (M1/70, 1:50), CD45 (90, 1:50), Ly6C (HK1.4. 1:100), CD105 (N418, 1:100), CD140a (APA5, 1:100), CD11c (N418, 1:100), F4/80 (BM8, 1:50), O4 (O4, Miltenyi Biotec, 1:10), and CD19 (eBio1D3, 1:100). All antibodies were from eBioscience or BD Pharmingen unless otherwise mentioned. Microglia were sorted as CD11b+ cells with low CD45 expression and low Ly6C (CD11b+CD45lowLy6Clow), inflammatory monocytes were considered as CD45hiCD11b+Ly6Chi61. Astrocytes were sorted as CD11blowCD45low Ly6ClowCD105lowCD140alowCD11blowF4/80lowO4lowCD19low after the exclusion of lymphocytes, microglia, oligodendrocytes, monocytes (Supplementary Fig. 1). Sorted cells were found to be >85% GFAP+ by FACS analysis (Supplementary Fig. 1). We confirmed that we had isolated a relatively pure population of astrocytes by qPCR analysis of the expression of the astrocyte markers gfap, aldh1l1 and aqp4.
Flow cytometry staining and acquisition
Mononuclear cell suspensions were prepared as previously described11. Antibodies for flow cytometry were purchased from eBioscience or BD Pharmingen and used at a concentration of 1:100 unless recommended otherwise by the manufacturer. Cells were then analyzed on a LSRII or MACSQuant flow cytometer (BD Biosciences and Miltenyi Biotec, respectively). As outlined in the individual figures, Th1 cells were defined as CD3+CD4+IFN-g+IL-17−IL-10−Foxp3−, Th17 cells as CD3+CD4+IFN-g−IL-17+IL-10−Foxp3−, Treg cells as CD3+CD4+IFN-g−IL-17−IL-10−/+Foxp3+, microglia as CD11b+CD45lowLy6Clow, and pro-inflammatory monocytes as CD45hiCD11b+Ly6Chi.
RNA-sequencing
Mice were sacrificed at day 28 after disease induction and astrocytes isolated as described above. RNA was sequenced using the strand-specific TruSeq protocol. High coverage (>50M) strand-specific paired-end 76bp reads were aligned to the mm10/GRCm38 mouse reference genome using TopHat v2.0.1162. Gene expression levels were estimated for 38922 GenCode Release M2 (GRCm38.p2) mouse gene annotations using Cuffquant and Cuffnorm v2.2.1 quartile normalized FPKMs and differential expression was assessed using Cuffdiff v2.2.162.
nCounter gene expression
100 ng of total RNA was hybridized with reporter and capture probes in custom-made astrocyte-targeted nCounter Gene Expression code sets (Supplementary Table 1) according to manufacturer’s instructions (NanoString Technologies). Data were analyzed using nSolver Analysis software.
qPCR
RNA was extracted with RNAeasy kit (Qiagen), cDNA was prepared and used for qPCR with the results normalized to Gapdh. All primers and probes were from Applied Biosystems. Mouse: Ahr Mm00478932_m1, Ccl20 Mm01268754_m1, Ccl2 Mm00441242_m1, Ccl8 Mm01297183_m1, Cxcl3 Mm01701838_m1, Cyp1a1 Mm00487218_m1, Cyp1b1 Mm00487229_m1, Gapdh Mm99999915_g1, Ido1 Mm00492590_m1, Ido2 Mm00524210_m1, Ifnar1 Mm00439544_m1, Ifnb1 Mm00439552_s1, Il10 Mm00439614_m1, Il12a Mm00434165_m1, Il23a Mm01160011_g1, Il27 Mm00461162_m1, Il6 Mm00446190_m1, Irf9 Mm00492679_m1, Mx1 Mm00487796_m1, Nos2 Mm00440502_m1, Stat1 Mm00439531_m1, Stat2 Mm00490880_m1, Tdo2 Mm00451266_m1, Tgfb1 Mm01178820_m1, Tnfa Mm00443258_m1, Vim Mm01333430_m1. Human: AHR Hs00169233_m1, CCL2 Hs00234140_m1, CYP1B1 Hs02382916_s1, IFNAR1 Hs01066118_m1, IL6 Hs00985639_m1, IRF9 Hs00196051_m1, MX1 Hs00895608_m1, NOS2 Hs01075529_m1, STAT1 Hs01013996_m1, STAT2 Hs01013123_m1, TNFA Hs01113624_g1.
T cell proliferation
Splenocytes were cultured in X-VIVO 15 medium (Lonza) and were plated for 72 h at a density of 5 × 105 cells per well with increasing concentrations of MOG35–55 peptide. During the final 16 h, cells were pulsed with 1 Ci [3H]thymidine (PerkinElmer), followed by collection on glass fiber filters and analysis of incorporated [3H]thymidine in a beta-counter (1450 MicroBeta TriLux; PerkinElmer). For intracellular cytokine staining, cells were stimulated for 4 h with PMA (phorbol 12-myristate 13-acetate; 50 ng/ml; Sigma), ionomycin (1 μg/ml; Sigma) and monensin (GolgiStop; 2 μM BD Biosciences). After staining of surface markers, cells were fixed and made permeable according to the manufacturer’s instructions (BD Cytofix/Cytoperm Kit (BD Biosciences) or Foxp3 Fixation/Permeabilization (eBioscience).
Primary astrocyte cultures
Cerebral cortices from neonatal mice aged 1–3 d were dissected, carefully stripped of their meninges, digested with 0.25% trypsin-EDTA and DNAse I (1 mg/ml) for 15 mins, and dispersed to single-cell level by passing through a cell strainer (70 μm). The cell suspension was then cultured at 37 °C in humidified 5% CO2, 95% air on poly-L-Lysin (Sigma) precoated 175 cm2 cell culture flasks. Medium was replaced every 4–5 d. After 7–10 d cells reached confluence and astrocytes were isolated by mild trypsinization with Trypsin-EDTA (0.06%) as previously described11. Cells were >95% astrocytes as determined by staining with GFAP or GLAST, with less than 5% contamination of CD11b+ microglia cells (not shown). After the isolation procedure, cells were further plated as required for the specific experiments. Concentrations of agents were 100 ng/ml for LPS (Sigma), 500 IU/ml for mIFN-β (R&D Systems), or 50 μg/ml 3-Indoxyl-sulfate (Sigma). Unless otherwise indicated, RNA analysis was done 24 h after start of treatment. For Western Blot, cells were pretreated with IFN-β or vehicle for 24 h, thereafter LPS was added and protein prepared after 2 h.
Plasmids
Constructs encoding STAT1 and IRF-1 as well as pTK-Renilla were from Addgene. Reporter plasmids pAhR-Luc and pGud-Luc have been described before28. The pLenti-GFAP-EGFP-mir30-shAct1 vector25 was a gift from Guang-Xian Zhang19.
Transfections and luciferase assays
HEK293 cells were grown in DMEM supplemented with 10% FBS and transfected with Fugene-HD Transfection Reagent (Roche), reporter constructs (pAhR-Luc/pTK-Renilla or pGud-Luc/pTK Renilla) as well transcription factor expression constructs (pStat1, pIrf9) as indicated. In some experiments, transfected cells were exposed to 500 IU/ml of mIFN-β (Bio X cell) or AhR inhibitor CH223191 (Sigma). For the measurement of AhR ligands in human serum, 15.000 HEK293 cells were plated in 96 well plates 24 h before transfection with pGud-Luc and pTK-Renilla. After 24 h transfected cells were incubated with DMEM supplemented with 10% of patient serum. Luciferase activity was analyzed 24 h later using Dual Luciferase Reporter System (Promega) and normalized to Renilla luciferase activity.
In vitro knockdown with shRNA
Ifnar1 expression was knocked down in mouse primary astrocytes in culture using a lentivirus vector carrying an _Ifnar1_-targeting shRNA (TRCN0000374694), a lentivirus carrying a non-targeting sequence was used as a control (TRCN0000018782) (Sigma). Astrocytes were incubated with lentiviruses and 8 μg/ml polybrene (both from Sigma-Aldrich) for 24 h and thereafter incubated with mIFN-β (Bio-X-cell) or vehicle. GFP+ astrocytes were sorted, and gene knockdown was verified by qPCR. Transduction efficiency as determined by GFP-expression in transduced astrocytes was around 20% (Fig. 2b and not shown).
In vivo astrocyte-specific knockdown with shRNA lentivirus
shRNA sequences against Ifnar1 or Ahr and a non-targeting control shRNA were cloned into the pLenti-GFAP-EGFP-mir30-shAct1 vector19 backbone by replacement of the shAct1 with the above-mentioned in vitro validated shRNA sequence against Ifnar1 (5′-CCGGGAATGAGGTTGATCCGTTTATCTCGAGATAAACGGATCAACCTCATTCTTTTTG-3′) or Ahr (5′-CCGGCATCGACATAACGGACGAAATCTCGAGATTTCGTCCGTTATGTCGATGTTTTTG-3′) as previously described11. Lentivirus particles were generated by transfecting HEK293FT cells (Invitrogen) the newly generated pLenti-GFAP-EGFP-mir30-shRNA vectors and the ViraPower Packaging mix (helper plasmids pLP1, pLP2, pLP/VSV-G, Invitrogen). Supernatants were collected, filtered through a 0.45-μm PVDF filter, and concentrated overnight with the Lenti-X concentrator kit (Clontech). The viral titer was determined using the Lenti-X qRT-PCR titration kit (Clontech). For in vivo knockdown immunized mice were anesthetized at indicated time points, positioned in a Kopf Stereotaxic Alignment System and injected with 107 IU of shIfnar1 or shControl virus using a Hamilton syringe 0.44 mm posterior to the bregma, 1.0 mm lateral to it and 2.2 mm below the skull surface. The injection system was retracted slowly, skin incisions closed carefully by surgical sutures, and mice allowed to awake in a specific cage pre-warmed by red light and checked thrice daily thereafter.
Subcellular fractionation and immunoblot analysis
In vitro astrocyte cultures were treated as indicated in specific experiments, subcellular fractions generated using Cell Fractionation kit (Cell Signaling) and 10 μg of nuclear and cytoplasmic fractions were separated by 4–12% Bis-Tris Nupage gels (Invitrogen, USA) and transferred onto PVDF membranes (Millipore). As primary antibodies we used rabbit anti- GAPDH mAb (14C10, Cell Signaling), anti-Histone H3 rabbit polyclonal Ab (EMD Millipore), anti-NF-κB p65 rabbit mAb (D14E12, Cell Signaling), followed by goat anti-rabbit IgG HRP linked AB (7074S, Cell Signaling). All antibodies were used at a dilution of 1:1.000. Blots were developed using the SuperSignal West Femto Maximum sensitivity kit (Thermo Scientific/Life Technologies). Data quantification was done using Image J software 1.48v (NIH) and specific signals normalized to GAPDH (cytoplasm) or Histone 3 (nucleus).
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1% paraformaldehyde and lysed with 350 μl lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) containing 1× protease inhibitor cocktail (Roche Molecular Biochemicals, USA). Chromatin was sheared by sonication and supernatants were collected after centrifugation and diluted in ChIP incubation buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.0). 10 μg of antibody was prebound for 6 h to protein A– and protein G–Dynal magnetic beads (Invitrogen, USA) and washed three times with ice-cold PBS plus 1% BSA, and then added to the diluted chromatin and immunoprecipitated rotating overnight. The magnetic bead-chromatin complexes were then washed 3 times in RIPA buffer (50 mM HEPES (pH 7.6), 1 mM EDTA, 0.7% Na deoxycholate, 1% NP-40, 0.5 M LiCl) followed by 2 times with TE buffer. Immunoprecipitated chromatin was then extracted with 1% SDS, 0.1 M NaHCO3 and heated at 65 °C for 8 h to reverse the paraformaldehyde cross-linking. DNA fragments were purified with a QIAquick DNA purification kit (Qiagen, USA) and analyzed using the SYBR Green real-time PCR kit (Takara Bio Inc., USA). Anti-AhR (BML-SA210, Enzo Life Sciences, USA), anti-STAT1 (Cat. #9172, Cell Signaling Technology, USA), anti-STAT2 (Cat. #4597, Cell Signaling Technology, USA), and NF-κB p65 (D14E12) XP Rabbit mAb (Cat. #8242, Cell Signaling Technology, USA) were used as indicated in specific experiments. The following primer pairs were used: ccl2:p65 forward, 5′-CAGCTAAATATCTCTCCCGAAGG-3′, and reverse, 5′-CATAGATGCCCACAGCTCAT-3′; csf2:p65 forward, 5′-GACCAGATGGGTGGAGTGACC-3′, and reverse, 5′-AGCCACACGCTTCTGGTTCC-3′; nos2:p65(1) forward, 5′-CACAGACTAGGAGTGTCCATCA-3′, and reverse, 5′-GCAGCAGCCATCAGGTATTT-3′; nos2:p65(2) forward, 5′-ACCATGCGAAGATGAGTGGA-3′, and reverse, 5′-AGCCAGGAACACTACACAGAA-3′. pAhR:Stat1(1) forward, 5′-TGCGACTGGAGAGCATTC-3′, and reverse, 5′-TCCTGAAGTCTGATGGAGGA-3′; pAhR:Stat2(2) forward, 5′-TTGGGTCTCATTCTGCAGAC-3′, and reverse, 5′-CTTGCAGCTGGTCGATTTCA-3′; pAhR-ISRE forward, 5′-TGGCATGAATCACCAGAAAGA-3′, and reverse, 5′-CAGTTCTTTTGCAGTACCCACA-3′. pSOCS:AhR(1) forward, 5′-CCAGCTCCCTACCTGTTTGA-3′, and reverse, 5′-GGAATGGAGCGGACAGGA-3′; pSOCS2:AhR(2) forward, 5′-ATGAGTCAACACGTCCCAGA-3′, and reverse, 5′-CTGCACACTCTCGTTTTGGG-3′, pSOCS2:AhR(3), forward, 5′-TGGCAAAGTCTCTCGCAGA-3′, and reverse, 5′-TGCTCGGGGTTAAATGGTAC-3′.
Generation of astrocyte conditioned media for migration and neurotoxicity assays
In vitro astrocyte cultures from WT or AhR-GFAP-deficient pups were treated with LPS (100 ng/ml) or vehicle for 24 hours, extensively washed and supplemented with fresh culture medium. 48 h later, supernatants were spun down and kept for migration and neurotoxicity assays at −80°C.
Monocyte migration assay
Splenic monocytes from WT or IL-27R−/− mice were pre-enriched by CD11b beads (Miltenyi) and sorted for F4/80+SSClowLy-6Chi. These monocytes were seeded in the upper chamber of a 24-well cell culture insert with 5 μm pore-size (Corning) containing astrocyte preconditioned media (see above). In some experiments, the preconditioned media was supplemented with antibodies to IL-27 p28/IL-30 (Cat. AF1834), CCL2 (Clone 123616), M-CSF (Clone 131621), GM-CSF (Clone MP122E9, all R&D Systems), or combinations of the latter. Migrating monocytes were quantified in the lower chamber after 3 h.
Neurotoxicity assay
N2A Neuronal cells (ATCC CCL-131, ATCC, Manassas) were grown in 96 Well-plates and pre-activated with mIFN-γ (100 ng/ml, R&D Systems) for 24 h. Thereafter, medium was replaced after extensive washes with PBS with astrocyte-conditioned media. Cytotoxicity was measured using LDH release (CytoTox 96® Non-Radioactive Cytotoxicity Assay, Promega) after 24 h as suggested by manufacturer’s protocol.
Measurement of urinary I3S
Urinary indican (I3S) was measured using the Indican Assay Kit (MAK128, Sigma Aldrich) in undiluted urine samples.
Measurement of CNS serotonin levels
50 mg of minced CNS tissue was resuspended in 200 μl of PBS and homogenized by sonication. After spinning down the extract to clear cellular debris serotonin levels were measured in the supernatant using the Serotonin Research Elisa Kit (Cat. BA E-5900, LDN Immunoassays, Germany).
Quantification of Lactobacillus reuteri DNA in mouse feces
Bacterial DNA in 200 mg of fecal samples from EAE mice was purified using QIAamp DNA Stool Mini Kit (Qiagen) and subjected to SYBR Green qPCR using primers specific to Lactobacillus reuteri as previously published44. Lactobacillus reuteri primers were used as follows: Forward: 5′-ACCGAGAACACCGCGTTATTT-3′, Reverse: 5′-CATAACTTAACCAAACAATCAAAGATTGTCT-3′. Relative abundances were normalized to the control group (TDD+Trp).
Human primary astrocytes
Human fetal astrocytes were isolated as previously described11,63 from human CNS tissue from fetuses at 17–23 weeks of gestation obtained from the Human Fetal Tissue Repository (Albert Einstein College of Medicine) following Canadian Institutes of Health Research–approved guidelines. Primary human astrocytes were treated with hIFN-β (500 IU, Rebif, Merck Serono) or vehicle, poly(I:C) (10 mg/ml) with or without 3-indoxyl-sulfate (50 μM, Sigma) or untreated (control). After 24 h, total RNA was isolated, transcribed and subjected to qPCR.
MS tissue and Immunofluorescence
Brain tissue was obtained from untreated individuals with clinically diagnosed MS and neuropathologically confirmed MS, and healthy controls as previously described11,64. All MS individuals and controls, or their next of kin, had given informed consent for autopsy and use of their brain tissue for research purposes. Ethical approval was given before autopsy (CHUM ethical approval: SL05.022 and SL05.023 and BH07.001). MS samples were processed and immunostained as previously described2. Briefly, sections were thawed, fixed, washed and blocked with donkey serum 10%. Sections were then incubated overnight at 4oC with antibodies against AhR (rabbit anti AhR, Enzo Life Sciences) and CCL2 (mouse anti MCP-1, BD Biosciences) or iNOS (mouse anti iNOS – Abcam). After washes the samples were incubated at room temperature for 40 minutes with the secondary antibodies (Donkey anti rabbit RRX and donkey anti mouse Alexa 488, Jackson ImmunoResearch). Sections were then incubated with anti-GFAP antibody directly conjugated with Alexa 647 (BD Biosciences) for 1h. In other staining set, the sections were incubated overnight at 4°C with antibodies against AhR (rabbit anti AhR, Enzo Life Sciences) and Mx1 (mouse anti Mx1, Santa Cruz Biotechnology) or pSTAT1 (mouse anti pSTAT1 – BD Biosciences). Sections were then incubated with secondary antibodies and anti-GFAP-Alexa 647 as indicated above. Finally, all the sections were washed and mounted in gelvatol containing TOPRO-3 (Invitrogen). Imaging was performed using a Leica SP5 confocal microscope and the Leica LAS AF software. Images were processed using Adobe Photoshop CS2. For imaging analysis all the data were acquired using the same settings, which were originally standardized on NAWM sections. The degree of co-localization of AhR with CCL2, iNOS, Mx1, pSTAT1 and GFAP as well as GFAP with CCL2, iNOS, Mx1 and pSTAT1 was determined using the Leica LAS software. The overlap coefficient is expressed in percentage where 100% represents the maximum degree of colocalization and 0% denotes no colocalization.
Serum metabolite analysis
Serum samples were collected at the University of Sevilla from healthy controls (11) and individuals with MS (49 MS). The study was approved by the Institutional Review Board of Brigham and Women’s Hospital and all subjects provided written informed consent. Tryptophan and tryptophan metabolites were profiled using two targeted LC-MS-based metabolomics methods as described previously65. Briefly, tryptophan, kynurenine, kynurenic acid and anthranilic acid were measured in the positive ion mode using a 4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX, Framingham MA) coupled to a 1200 Series binary HPLC pump (Agilent, Santa Clara, CA) and an HTS PAL autosampler (Leap Technologies, Carrboro, NC). Serum samples (10μL) were extracted using nine volumes of 74.9:24.9:0.2 (v/v/v) acetonitrile/methanol/formic acid containing stable isotope-labeled internal standards (0.2 ng/μL valine-d8, Isotec; and 0.2 ng/μL phenylalanine-d8 (Cambridge Isotope Laboratories, Inc., Tewksbury MA)). The samples were centrifuged (10 min, 9,000 × g, 4°C) and the supernatants (10 μL) injected onto a 150 × 2.1 mm Atlantis HILIC column (Waters). The column was eluted isocratically at a flow rate of 250 μL/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 1 minute followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 minutes. The ion spray voltage was 4.5 kV and the source temperature was 450°C. Kynurenine was measured in the negative ion mode using an ACQUITY UPLC (Waters Corp, Milford MA) coupled to a 5500 QTRAP triple quadrupole mass spectrometer (AB SCIEX, Framingham MA). Serum samples (30μL) were extracted using 120 μL of 80% methanol containing 0.05 ng/μL inosine-15N4, 0.05 ng/μL thymine-d4, and 0.1 ng/μL glycocholate-d4 as internal standards (Cambridge Isotope Laboratories, Inc., Tewksbury MA). The samples were centrifuged (10 min, 9,000 × g, 4°C) and the supernatants (10 μL) were injected directly onto a 150 × 2.0 mm Luna NH2 column (Phenomenex, Torrance CA). The column was eluted at a flow rate of 400 μL/min with initial conditions of 10% mobile phase A (20 mM ammonium acetate and 20 mM ammonium hydroxide (Sigma-Aldrich) in water (VWR)) and 90% mobile phase B (10 mM ammonium hydroxide in 75:25 v/v acetonitrile/methanol (VWR)) followed by a 10 min linear gradient to 100% mobile phase A. The ion spray voltage was −4.5 kV and the source temperature was 500°C. Raw data were processed using MultiQuant 1.2 software (AB SCIEX, Framingham MA) for automated peak integration. Metabolite peaks were manually reviewed for quality of integration and compared against standard reference standards to confirm identities.
Statistical analysis
Statistical analyses were performed with Prism software (GraphPad) using the statistical tests indicated in the individual figure legends. No sample size estimate was performed, but samples size was selected based on previous experiments. No samples were excluded. The investigators were blinded as to the treatment of mice in individual experiments. P values of <0.05 were considered significant. All error bars represent s.e.m. or SD as noted in the individual figure legends. Unless otherwise stated, ≥3 independent experiments were used for all assays and displayed figures are representative.
Data deposition
RNA-Sequencing data, Nanostring data, and human metabolomics data have been deposited at http://figshare.com/s/e99974829c4d11e5a57b06ec4b8d1f61.
Supplementary Material
1
2
Acknowledgments
This work was supported by grants AI075285, and AI093903 from the National Institutes of Health, RG4111A1 from the National Multiple Sclerosis Society and PA0069 and PA-1501-02847 from the International Progressive MS Alliance to FJQ. V.R. received support from an educational grant from Mallinkrodt Pharmaceuticals (A219074, V.R.) and by a fellowship from the German Research Foundation (DFG RO4866 1/1, V.R.), I.D.M. was supported by a postdoctoral fellowship (FG 2036-A1/1, I.D.M) from the National Multiple Sclerosis Society, L.B. was supported by a fellowship from the International Academy of Life Sciences, L.M. was supported by a postdoctoral fellowship (FG1941A1/2, L.M.) from the National Multiple Sclerosis Society, C-C.C. was supported by a postdoctoral research abroad program (104-2917-I-564 -024, C-C.C.) from the Ministry of Science and Technology, Taiwan, and M.C.T. was supported by a fellowship (BEX 0571/15-6) from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil.
Footnotes
Authors’ contributions:
V.R., I.D.M., L.B., M.C.T., J.E.K., L.M., C-C.C., H.K., J.I.A., M.B. and C.B.C. performed in vitro and in vivo experiments, B.P., R.Y., N.O. and N.P. performed bioinformatics analysis, N.A., G.I., C.B.C., A.P., S.J., M.P. and J.A. provided unique reagents, discussed and/or interpreted findings, V.R. and F.J.Q. wrote the manuscript and F.J.Q. designed and supervised the study and edited the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Allen NJ, et al. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez JI, et al. The Hedgehog pathway promotes blood-brain barrier integrity and CNS immune quiescence. Science. 2011;334:1727–1731. doi: 10.1126/science.1206936. [DOI] [PubMed] [Google Scholar]
- 3.Chung WS, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khakh BS, Sofroniew MV. Diversity of astrocyte functions and phenotypes in neural circuits. Nature neuroscience. 2015;18:942–952. doi: 10.1038/nn.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molofsky AV, et al. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature. 2014;509:189–194. doi: 10.1038/nature13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nature medicine. 2013;19:1584–1596. doi: 10.1038/nm.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieckmann P, Engelhardt B. Building up the blood-brain barrier. Nature medicine. 2003;9:828–829. doi: 10.1038/nm0703-828. [DOI] [PubMed] [Google Scholar]
- 8.Sofroniew MV. Astrocyte barriers to neurotoxic inflammation. Nature reviews. Neuroscience. 2015;16:249–263. doi: 10.1038/nrn3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai HH, et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science. 2012;337:358–362. doi: 10.1126/science.1222381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lassmann H. Mechanisms of white matter damage in multiple sclerosis. Glia. 2014;62:1816–1830. doi: 10.1002/glia.22597. [DOI] [PubMed] [Google Scholar]
- 11.Mayo L, et al. Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nature medicine. 2014;20:1147–1156. doi: 10.1038/nm.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao W, et al. Suppression of neuroinflammation by astrocytic dopamine D2 receptors via alphaB-crystallin. Nature. 2013;494:90–94. doi: 10.1038/nature11748. [DOI] [PubMed] [Google Scholar]
- 13.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 14.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 15.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baruch K, et al. Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science. 2014;346:89–93. doi: 10.1126/science.1252945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandler NG, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Y, et al. CNS-specific therapy for ongoing EAE by silencing IL-17 pathway in astrocytes. Molecular therapy: the journal of the American Society of Gene Therapy. 2012;20:1338–1348. doi: 10.1038/mt.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacological reviews. 2013;65:1148–1161. doi: 10.1124/pr.113.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 22.Prinz M, et al. Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity. 2008;28:675–686. doi: 10.1016/j.immuni.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald DC, et al. Suppressive effect of IL-27 on encephalitogenic Th17 cells and the effector phase of experimental autoimmune encephalomyelitis. Journal of immunology (Baltimore, Md : 1950) 2007;179:3268–3275. doi: 10.4049/jimmunol.179.5.3268. [DOI] [PubMed] [Google Scholar]
- 24.Mitsdoerffer M, Kuchroo V. New pieces in the puzzle: how does interferon-beta really work in multiple sclerosis? Ann Neurol. 2009;65:487–488. doi: 10.1002/ana.21722. [DOI] [PubMed] [Google Scholar]
- 25.Mascanfroni ID, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nature medicine. 2015;21:638–646. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascanfroni ID, et al. IL-27 acts on DCs to suppress the T cell response and autoimmunity by inducing expression of the immunoregulatory molecule CD39. Nat Immunol. 2013;14:1054–1063. doi: 10.1038/ni.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apetoh L, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol. 2010;11:854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeste A, Nadeau M, Burns EJ, Weiner HL, Quintana FJ. Nanoparticle-mediated codelivery of myelin antigen and a tolerogenic small molecule suppresses experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2012;109:11270–11275. doi: 10.1073/pnas.1120611109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weidemann A, et al. The glial cell response is an essential component of hypoxia-induced erythropoiesis in mice. J Clin Invest. 2009;119:3373–3383. doi: 10.1172/JCI39378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jessen KR, Mirsky R. Glial cells in the enteric nervous system contain glial fibrillary acidic protein. Nature. 1980;286:736–737. doi: 10.1038/286736a0. [DOI] [PubMed] [Google Scholar]
- 31.Kim RY, et al. Astrocyte CCL2 sustains immune cell infiltration in chronic experimental autoimmune encephalomyelitis. J Neuroimmunol. 2014;274:53–61. doi: 10.1016/j.jneuroim.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mc Guire C, Prinz M, Beyaert R, van Loo G. Nuclear factor kappa B (NF-kappaB) in multiple sclerosis pathology. Trends Mol Med. 2013;19:604–613. doi: 10.1016/j.molmed.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 33.van Loo G, et al. Inhibition of transcription factor NF-kappaB in the central nervous system ameliorates autoimmune encephalomyelitis in mice. Nat Immunol. 2006;7:954–961. doi: 10.1038/ni1372. [DOI] [PubMed] [Google Scholar]
- 34.Boverhof DR, et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces suppressor of cytokine signaling 2 in murine B cells. Molecular pharmacology. 2004;66:1662–1670. doi: 10.1124/mol.104.002915. [DOI] [PubMed] [Google Scholar]
- 35.Zadjali F, et al. SOCS2 deletion protects against hepatic steatosis but worsens insulin resistance in high-fat-diet-fed mice. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2012;26:3282–3291. doi: 10.1096/fj.12-205583. [DOI] [PubMed] [Google Scholar]
- 36.Axtell RC, et al. T helper type 1 and 17 cells determine efficacy of interferon-beta in multiple sclerosis and experimental encephalomyelitis. Nature medicine. 2010;16:406–412. doi: 10.1038/nm.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borden EC, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross TM, et al. Intranasal administration of interferon beta bypasses the blood-brain barrier to target the central nervous system and cervical lymph nodes: a non-invasive treatment strategy for multiple sclerosis. J Neuroimmunol. 2004;151:66–77. doi: 10.1016/j.jneuroim.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farez MF, et al. Melatonin Contributes to the Seasonality of Multiple Sclerosis Relapses. Cell. 2015;162:1338–1352. doi: 10.1016/j.cell.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gandhi R, et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nature immunology. 2010;11:846–853. doi: 10.1038/ni.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 43.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Schroeder JC, et al. The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry. 2010;49:393–400. doi: 10.1021/bi901786x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palace J, Leite MI, Nairne A, Vincent A. Interferon Beta treatment in neuromyelitis optica: increase in relapses and aquaporin 4 antibody titers. Arch Neurol. 2010;67:1016–1017. doi: 10.1001/archneurol.2010.188. [DOI] [PubMed] [Google Scholar]
- 47.Khorooshi R, et al. Induction of endogenous Type I interferon within the central nervous system plays a protective role in experimental autoimmune encephalomyelitis. Acta Neuropathol. 2015;130:107–118. doi: 10.1007/s00401-015-1418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dann A, et al. Cytosolic RIG-I-like helicases act as negative regulators of sterile inflammation in the CNS. Nature neuroscience. 2012;15:98–106. doi: 10.1038/nn.2964. [DOI] [PubMed] [Google Scholar]
- 49.Ejlerskov P, et al. Lack of Neuronal IFN-beta-IFNAR Causes Lewy Body- and Parkinson’s Disease-like Dementia. Cell. 2015;163:324–339. doi: 10.1016/j.cell.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldmann T, et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34:1612–1629. doi: 10.15252/embj.201490791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 52.Quintana FJ, et al. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2010;107:20768–20773. doi: 10.1073/pnas.1009201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 54.Bessede A, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 56.Monteleone I, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248. 248e231. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Atarashi K, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ochoa-Reparaz J, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. Journal of immunology (Baltimore, Md : 1950) 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 60.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nature neuroscience. 2011;14:1227–1235. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- 62.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jack CS, et al. TLR signaling tailors innate immune responses in human microglia and astrocytes. Journal of immunology (Baltimore, Md : 1950) 2005;175:4320–4330. doi: 10.4049/jimmunol.175.7.4320. [DOI] [PubMed] [Google Scholar]
- 64.Alvarez JI, et al. Focal disturbances in the blood-brain barrier are associated with formation of neuroinflammatory lesions. Neurobiology of disease. 2015;74:14–24. doi: 10.1016/j.nbd.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 65.Townsend MK, et al. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clinical chemistry. 2013;59:1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2