A Concentration-Dependent Liquid Phase Separation Can Cause Toxicity upon Increased Protein Expression (original) (raw)
Summary
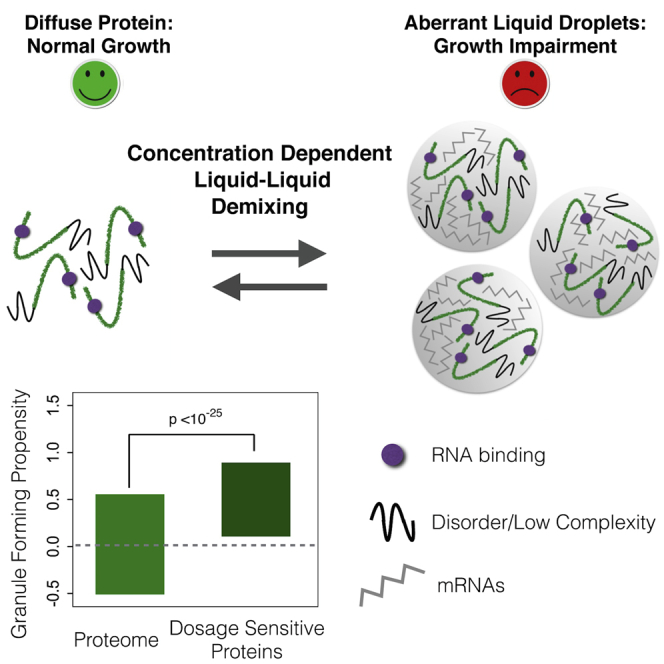
Multiple human diseases are associated with a liquid-to-solid phase transition resulting in the formation of amyloid fibers or protein aggregates. Here, we present an alternative mechanism for cellular toxicity based on a concentration-dependent liquid-liquid demixing. Analyzing proteins that are toxic when their concentration is increased in yeast reveals that they share physicochemical properties with proteins that participate in physiological liquid-liquid demixing in the cell. Increasing the concentration of one of these proteins indeed results in the formation of cytoplasmic foci with liquid properties. Demixing occurs at the onset of toxicity and titrates proteins and mRNAs from the cytoplasm. Focus formation is reversible, and resumption of growth occurs as the foci dissolve as protein concentration falls. Preventing demixing abolishes the dosage sensitivity of the protein. We propose that triggering inappropriate liquid phase separation may be an important cause of dosage sensitivity and a determinant of human disease.
Graphical Abstract
Highlights
- •
Dosage-sensitive proteins in yeast have a high propensity for liquid-liquid demixing - •
Increased protein concentration can force a liquid phase separation, titrating proteins and RNAs from the cytoplasm - •
Preventing liquid-liquid demixing averts dosage sensitivity - •
Inappropriate liquid phase separation may be a determinant of human genetic disease
Bolognesi et al. report that proteins that are harmful when overexpressed have properties associated with liquid-liquid demixing and that increased protein concentration can force a liquid phase transition causing cellular toxicity.
Introduction
A subset of the proteins encoded in any genome is toxic when their expression level is increased (Gelperin et al., 2005, Sopko et al., 2006). Even small increases in dosage can be detrimental (Tomala et al., 2014) with dosage sensitivity causing a large number of human diseases, ranging from developmental defects to psychiatric disorders (Girirajan et al., 2011, Veitia and Birchler, 2010).
Whether an individual protein is toxic or not when overexpressed may depend both on its specific functions and on its intrinsic physicochemical properties. For example, imbalance in regulatory networks or in the assembly of protein complexes (Papp et al., 2003, Veitia, 2003), aggregation (Geiler-Samerotte et al., 2011, Tartaglia et al., 2007), and mass-action driven promiscuous molecular interactions (Vavouri et al., 2009) have all been suggested to cause dosage sensitivity. However, the precise molecular mechanism by which each individual protein becomes harmful when overexpressed is normally unknown.
The cytoplasm and nuclei of cells are crowded environments containing very high concentrations of macromolecules. One principle that is becoming increasingly appreciated as a means for how cells organize and compartmentalize their contents is liquid-liquid phase separation (Brangwynne et al., 2009, Hyman et al., 2014, Jain et al., 2016). Liquid demixing creates non-membrane bound organelles that rapidly exchange molecules with the surrounding cytoplasm and increases the concentration of particular macromolecules within the separated phase. Examples include germ granules (Brangwynne et al., 2009), the nucleolus (Brangwynne et al., 2011, Weber and Brangwynne, 2015), and other ribonucleoprotein (RNP) assemblies (Lin et al., 2015, Mitchell et al., 2013). Although the precise molecular details about how liquid-liquid demixing is initiated are still unknown, the process is tightly controlled (Wippich et al., 2013).
Several proteins involved in physiological liquid-liquid demixing are prone to form protein aggregates or amyloids when they carry disease-causing mutations (Hyman et al., 2014, Kim et al., 2013, Patel et al., 2015) and indeed these mutations can promote a transition from a liquid droplet to a solid phase in vitro (Patel et al., 2015). This has led to the proposal that a liquid-to-solid phase transition is a mechanism of cellular toxicity (Patel et al., 2015).
Here, based on a proteome-wide analysis in yeast, we report that dosage-sensitive proteins share characteristics with proteins known to undergo physiological liquid-liquid demixing. Overexpressing one of these proteins revealed that it is indeed the induction of a liquid-liquid phase separation that correlates with toxicity. The condensation of this concentration-dependent liquid phase requires the RNA-binding domains of the protein and decondensation occurs as protein concentrations drop, reversing the growth impairment. Genetically preventing the liquid phase separation prevents the dosage sensitivity of this protein. This establishes that inappropriate liquid-liquid demixing can be a cause of concentration-dependent dosage sensitivity. Thus, both liquid-to-solid and liquid-to-liquid phase transitions can underlie cellular toxicity and may contribute to human genetic disease.
Results
Dosage-Sensitive Proteins Have an Intrinsic Propensity for Liquid-Liquid Phase Separation
To better understand concentration-dependent cellular toxicity, we analyzed the properties of dosage-sensitive genes identified in two genome-wide screens in yeast (Makanae et al., 2013, Sopko et al., 2006). In addition to the previously reported high intrinsic protein disorder content (Vavouri et al., 2009), we found that dosage-sensitive proteins have a strong propensity to bind nucleic acids (Figure 1A). Recent studies have highlighted the potential for disordered and low complexity sequences to bind RNA (Castello et al., 2012), so we reasoned that these two features might be related. Consistent with this, we found that dosage-sensitive proteins are enriched for nucleic acid binding-propensity even after removing all known transcription factors (Figure S1A).
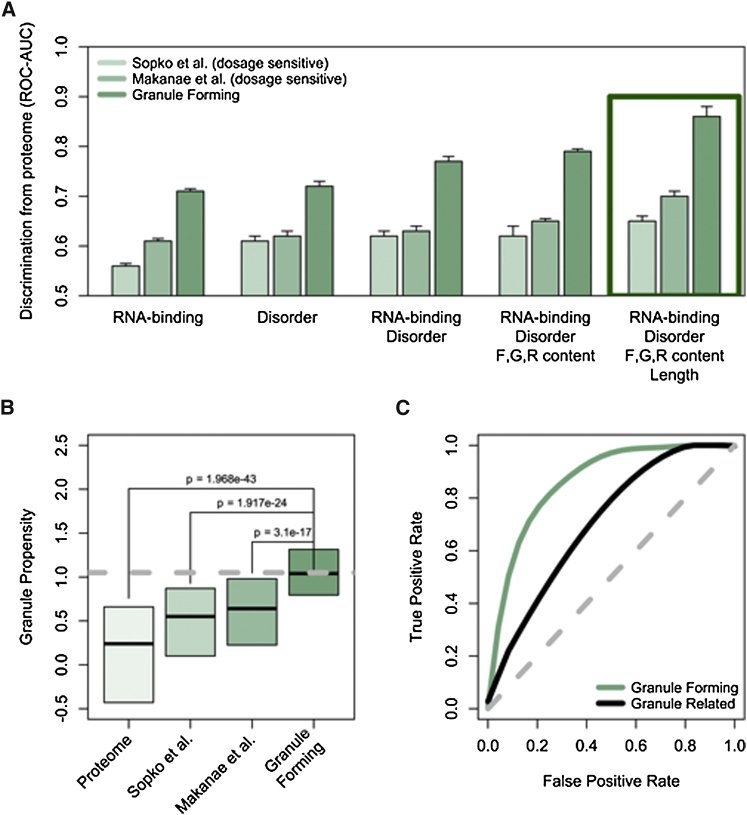
Figure 1.
Dosage-Sensitive Proteins Have an Intrinsic Propensity for Liquid-Liquid Phase Separation
(A) Ability of physico-chemical features to discriminate dosage-sensitive and granule forming sets from the rest of the yeast proteome. The area under the receiver operating characteristic curve (AUC) is used to measure performances of individual properties and combinations of them. Performances of the _cat_GRANULE algorithm (Experimental Procedures) are highlighted with a green square.
(B) Distribution of granule propensities calculated with _ca_tGRANULE (Experimental Procedures) for annotated granule forming proteins (n = 120), dosage-sensitive proteins as reported in Sopko et al. (2006) (n = 770) and Makanae et al. (2013) (n = 777) and the rest of the S. cerevisiae proteome (n = 3,726). Boxes represent the range between the 25th and 75th percentile. Grey dashed line indicates the propensity value for Mip6p.
(C) Performance of the model on “granule forming” genes (AUC: 0.86) and on the independent test set “granule related” genes (AUC: 0.72). See also Table S1 for full table of ORFs.
See also Figure S1.
Structural disorder and nucleic acid-binding are properties of proteins that assemble into RNP foci. The formation of these foci occurs by liquid-liquid phase separation (Brangwynne et al., 2009, Hyman et al., 2014, Weber and Brangwynne, 2012) and it is normally a highly regulated process (Malinovska et al., 2013). Based on the enrichment of the same properties among dosage-sensitive proteins, we hypothesized that some dosage-sensitive proteins might impair cellular function by triggering an inappropriate liquid phase separation as their concentration increases.
A Computational Method for Predicting Liquid Demixing also Distinguishes Dosage-Sensitive Genes
To evaluate the potential of proteins to participate in phase separation, we trained an algorithm to distinguish between proteins known to localize to foci within the cytoplasm or nucleus and the rest of the proteome (Experimental Procedures; Table S1, the _cat_GRANULE algorithm) (Mitchell et al., 2013). Our approach predicts whether a protein takes part in foci-formation by considering the contributions of structural disorder and nucleic acid binding propensities and, to a lesser degree, sequence length and arginine, glycine, and phenylalanine content (R, G, F), which are enriched in granule-forming proteins (Kato et al., 2012, Thandapani et al., 2013) (Figure 1A; Experimental Procedures). The algorithm showed good performance with an area under the receiver operating characteristic curve, an area under the curve (AUC), of 0.86 (Figures 1A and 1C, on an independent test set the AUC is 0.72; Experimental Procedures).
Applying the approach to the entire yeast proteome revealed that dosage-sensitive proteins (Makanae et al., 2013, Sopko et al., 2006) have a propensity to assemble into foci that is intermediate between known granule-associated proteins and the rest of the proteome (Figure 1B). In addition, known granule-associated proteins are enriched among dosage-sensitive proteins (Figure S1B). Indeed, our algorithm is a better predictor of dosage sensitivity than intrinsic disorder content, nucleic acid binding propensity, or a linear combination of the two (Figure 1A).
The Dosage-Sensitive Protein Mip6p Changes Localization when Overexpressed
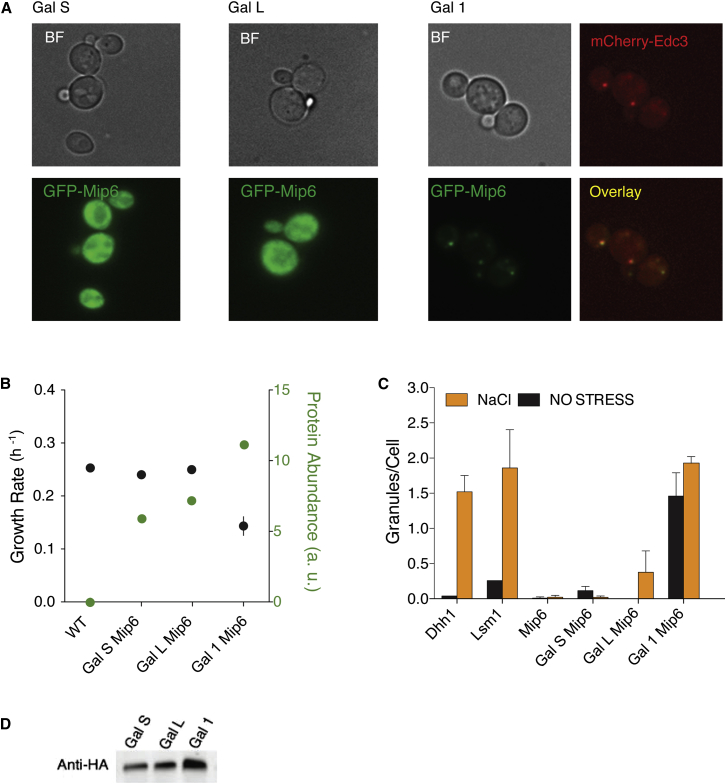
Using three inducible promoters of increasing strength, we overexpressed ten dosage-sensitive proteins with intermediate to high granule propensity and identified one protein, Mip6p, whose sub-cellular localization was altered when overexpressed (Table S2) (Figures 2A and 2D). Mip6p is a lowly expressed (Wang et al., 2015) protein containing four RNA recognition motif (RRM) domains and two short low-complexity regions. It has a granule-propensity that deviates above 1 SD from the proteome mean (granule propensity = 1.05). Moderate overexpression of Mip6p is not toxic (Figure 2B). However, strong overexpression inhibits growth (Figure 2B). When moderately overexpressed, Mip6p has a mostly diffuse cytoplasmic localization (Figure 2A), while at concentrations that impair growth, the protein re-localizes to cytoplasmic foci (Figures 2A and 2B).
Figure 2.
Mip6p Changes Localization when Overexpressed
(A) Cellular localization of Mip6-GFP at different levels of overexpression. Co-localization with Edc3p is observed when Mip6 is expressed from the Gal1 promoter.
(B) Growth rates of strains expressing Mip6 under the control of different promoters, error bars represent SDs of three independent replicates.
(C) Number of foci per cell as counted by monitoring GFP-tagged endogenous levels of Dhh1, Lsm1, and Mip6 in BY4741 (left), or counted in the same strains as in A); 50–100 cells for each condition were counted.
(D) Western blot where an antibody against HA was used to detect different amounts of Mip6p upon expression under weak (GalS), medium (GalL), and strong (Gal1) promoters.
See also Figure S2.
Mip6p Foci Associate with P-Body Components
To characterize the assemblies formed by Mip6p, we co-expressed it with markers for various cellular compartments and structures and found that the Mip6p foci partially co-localize with Edc3p (Figure 2A). Edc3p is a known component of cytoplasmic ribonucleoprotein granules referred to as P-bodies (Kedersha et al., 2005) (Figure 2A). The Mip6p foci also partially co-localize with another P-body component, Dcp1p (Figure S2). P-bodies constitute sites of mRNA storage and turnover and, in wild-type cells, their formation can be stimulated in response to osmotic stress (Decker and Parker, 2012). However, P-bodies induced by osmotic stress do not contain Mip6p when it is expressed at endogenous levels (Figure 2C). Rather, Mip6p only co-localizes with P-body components when overexpressed (Figure 2C) and in both the presence and absence of osmotic stress (Figure 2C). Thus, Mip6p both induces the formation of P-body-like assemblies and partially localizes to these foci when overexpressed.
Mip6p Foci Have Liquid Properties
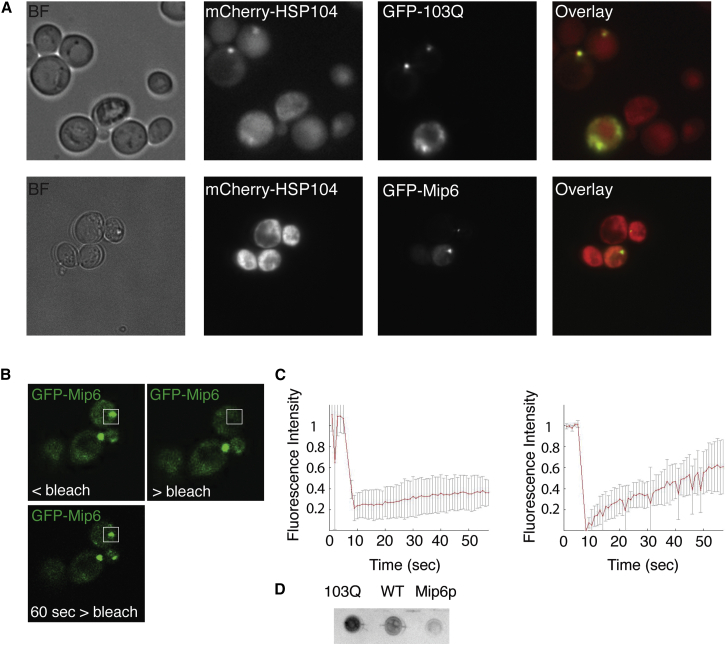
Next, we further characterized the biophysical properties of the Mip6p foci. We found that overexpression of Mip6p did not result in the formation of large insoluble protein aggregates similar to those of a poly-glutamine peptide (103Q) (Figure 3A), which we overexpressed with a GFP tag. The Mip6p foci also did not contain the chaperone Hsp104p (Figure 3A), a marker of intra-cellular aggregation compartments in yeast (Kaganovich et al., 2008). Indeed, the conformational antibody OC that specifically binds to fibrillar species (Kayed et al., 2007), showed no binding to whole cell protein extracts from Mip6p overexpressing cells but showed preferential interaction with 103Q aggregates (Figure 3D). The Mip6p foci, unlike 103Q aggregates, showed rapid fluorescence recovery after photo-bleaching (Figures 3B and 3C). We also observed that the Mip6p foci disassemble when cells are treated with the aliphatic alcohol 1,6-hexanediol, which is able to disrupt weak hydrophobic interactions (Kroschwald et al., 2015, Patel et al., 2007). This effect is reversible, as Mip6p foci reassemble within 30 min after removal of the alcohol (Figure S3A). Moreover, we found that the Mip6p foci are often inherited by daughter cells (Figure S3B), further distinguishing them from misfolded protein compartments that are asymmetrically inherited by mother cells (Ogrodnik et al., 2014). Occasionally, we observed fusion of two foci in the cytoplasm, as well as splitting of one focus into two (Figure S3C). Taken together, our results show that the Mip6p foci have a dynamic structure and liquid-like properties, rapidly exchanging molecules with the free cytoplasmic protein pool. This set of properties recapitulates the behavior of a known liquid phase-separating protein from Caenorhabditis elegans, PGL-3 (Brangwynne et al., 2009), when we overexpressed it in yeast (Figure S4).
Figure 3.
Mip6p Foci Have Liquid Properties
(A) Localization of the chaperone Hsp104 in cells showing either 103Q assemblies or Mip6p cytoplasmic foci.
(B) An example of fluorescence recovery after photobleaching for Mip6p foci.
(C) Average trends of fluorescence recovery after photo-bleaching for 103Q (left) and Mip6p (right) cytoplasmic assemblies.
(D) Dot blot assay showing differential binding of the OC conformational antibody to protein extracts from wild-type (WT) cells or cells expressing 103Q or Mip6p.
See also Figures S3 and S4.
Focus Formation Is Reversible with Growth Resuming upon Dissolution
Using single-cell imaging, we investigated the behavior of Mip6p assemblies when the Mip6p protein concentration was reduced in live cells. We expressed Mip6p from an inducible promoter until foci formed and then changed the media to repress further protein production (Experimental Procedures). Whereas the cytoplasmic foci formed by 103Q remained stable, the assemblies formed by Mip6p and PGL-3 dissolved with the redistribution of the fluorescent signal across the whole cytoplasm (Movies S1, S2, and S3). The maintenance of the Mip6p foci therefore requires a high protein concentration; as the concentration drops below a critical level, the foci dissolve.
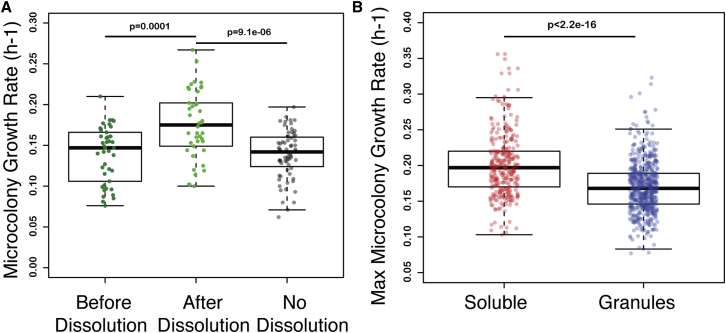
We used automated microscopy to follow the growth dynamics of individual micro-colonies following the inhibition of Mip6p production. We found a strong correlation between the dissolution of foci and the resumption of rapid colony growth, consistent with Mip6p foci being the cause of impaired growth (Figure 4A). When the foci disappear, cells resume growth. These findings are in line with what we observe for PGL-3 overexpressing cells in the same single colony assay (Figure S4A). By means of fluorescence-activated cell sorting (FACS) we could also separate cells with higher fluorescence intensity and containing cytoplasmic assemblies from cells with lower protein concentrations and mostly diffuse fluorescence. Following sorting, we allowed the two populations to grow in a micro-colony growth assay and confirmed higher growth rates for cells with diffuse Mip6p compared to cells with Mip6p foci (Figure 4B).
Figure 4.
Growth Resumes after Dissolution of Foci
(A) Average growth rates quantified by automated micro-colony growth assay before dissolution of cytoplasmic Mip6p assemblies (dark green), after their dissolution (light green), or for the colonies in which dissolution was not observed (gray). Boxes represent the 95% confidence interval (CI), while points correspond to the growth rate measurements of all individual micro-colonies.
(B) Average growth rate during re-growth after FACS sorting for cells displaying cytoplasmic assemblies (blue) and cells displaying soluble fluorescence (red).
See also Figures S3 and S4 and Movies S1, S2, and S3.
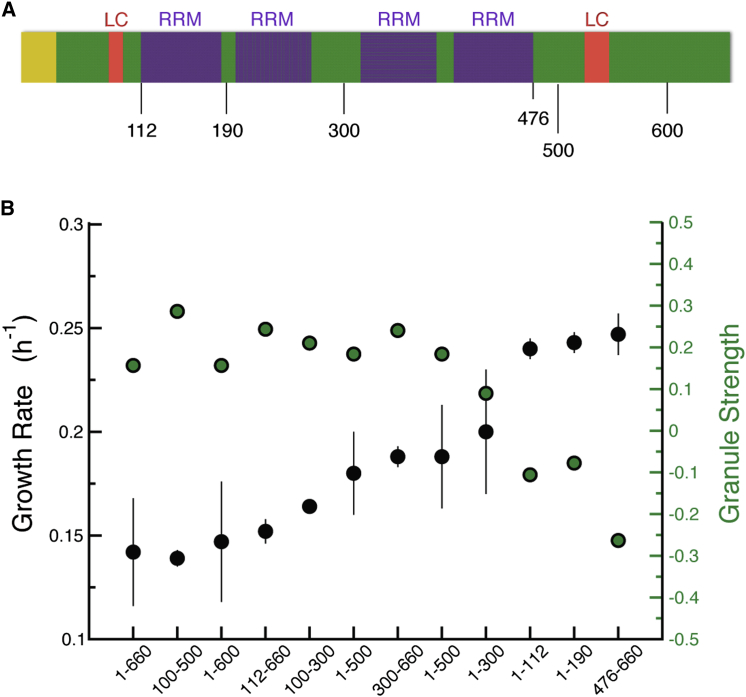
Focus Formation and Toxicity Require RNA Binding Domains
To identify the regions of Mip6p required for focus formation, we overexpressed a series of protein fragments and monitored their subcellular localization by fluorescence microscopy. Mip6p contains an N-terminal disordered region (aa 1–32), two short low complexity regions (aa 82–93 and aa 525–546), and four RRMs (aa 112–185, aa 200–270, aa 314–385, and aa 403–476) (Figure 5A). Whereas overexpression of the two N-terminal fragments (aa 1–112 and aa 1–190) or the C terminus (aa 476–660) did not result in the formation of foci, overexpression of other truncated variants did (Figure S5). The protein fragments leading to formation of cytoplasmic assemblies vary in length, but all contain at least two RRMs (Figure S5). The N-terminal disordered region and the low complexity regions are not required for triggering focus assembly, but two RRMs alone are sufficient, suggesting that focus formation requires Mip6p to interact with RNA. Using our computational method to score each fragment on the basis of its intrinsic physicochemical properties (Granule Strength, Equation 3), we find that the variants with a value >0 are associated with foci formation (Figures 5B and S5).
Figure 5.
Protein Domains Able to Form Foci Impair Growth when Overexpressed
(A) A scheme of MIP6p sequence as annotated by ELM (Dinkel et al., 2016) (yellow, disordered region; purple, RNA recognition motifs; red, low complexity sequences.
(B) Growth rate (black) of strains overexpressing Mip6p truncated variants with corresponding granule strength (green). Error bars represent SD of three independent replicates. Granule strength is calculated on fragments extracted from the Mip6p profile (Figure S7B) as explained in the Experimental Procedures.
See also Figure S5.
Next, we tested whether overexpression of the different protein fragments affected growth. Only overexpression of the fragments that formed cytoplasmic assemblies caused a growth defect (Figure 5B) with growth rates anti-correlated with the score assigned to the sequence by our computational model (Figure 5B). We conclude that both foci formation and cellular toxicity are primarily driven by the RNA-binding domains of Mip6p.
Foci Contain RNA
The presence of RNA in the Mip6p foci was confirmed by their co-localization with the nucleic acid dye Syto14 (Schisa et al., 2001) (Figure S6A). To further understand the role of RNA in the formation of the Mip6p foci, we treated cells overexpressing Mip6p with cycloheximide, an inhibitor of translation elongation that traps mRNAs on polysomes and depletes them from the cytoplasm (Teixeira et al., 2005). Consistent with a role for mRNA in their stability, cycloheximide treatment dissolved foci formed upon Mip6p overexpression (Figure S6B).
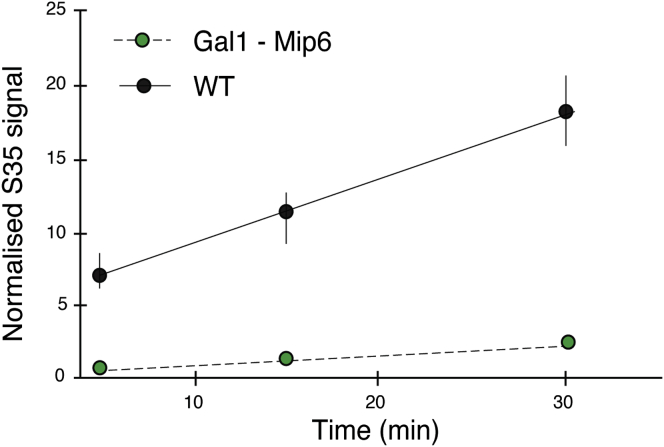
Mip6p Foci Are Associated with Translation Inhibition
How does Mip6p focus formation trigger toxicity? Physiological P-bodies contain protein translation factors and are associated with translation slow down (Parker and Sheth, 2007, Walters et al., 2015). A simple hypothesis is therefore that Mip6p foci, by titrating proteins and mRNAs from the cytoplasm, impair the translation capacity of the cell. Consistent with this, we found that global translation rates were severely reduced in cells overexpressing Mip6p (Figure 6).
Figure 6.
Translation Rates Are Reduced in Cells Overexpressing Mip6p
S35Met incorporation over time as measured in WT cells and in cells overexpressing Mip6p, where cytoplasmic foci are evident and fitness is impaired.
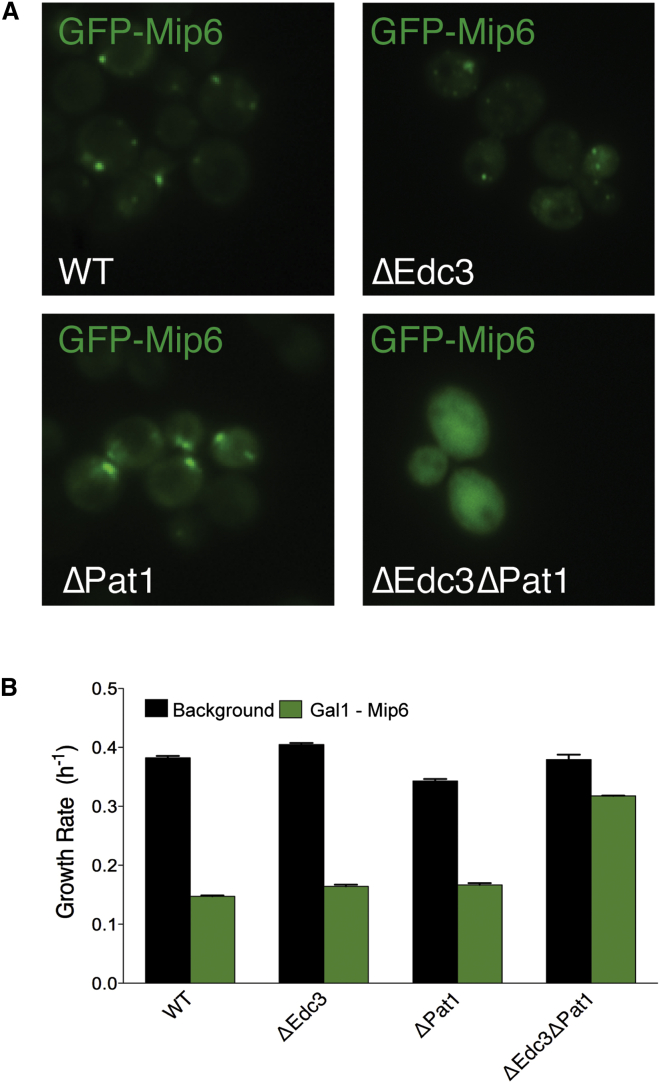
Preventing Liquid-Liquid Demixing Prevents Dosage Sensitivity
The correlations between formation of foci and growth impairment when Mip6p (Figure 2B) or its fragments (Figure 5B) are overexpressed and direct comparison across micro-colonies (Figure 4A) strongly suggest that foci formation is required for growth impairment. To provide an additional test of this, we reasoned that deletion of genes encoding P-body components might prevent the formation of Mip6p foci. Deleting both edc3 and pat1 has been previously shown to impair P-body formation (Lavut and Raveh, 2012), and indeed deletion of these two genes abolished the formation of Mip6p foci (Figures 7A and S6C). Edc3p and Pat1p are therefore required for the formation of the foci when Mip6p is overexpressed, allowing us to test whether focus formation is required for toxicity. Deletion of edc3 and pat1 suppresses the growth impairment caused by overexpression of Mip6p (Figure 7B).
Figure 7.
Mip6p Cytoplasmic Foci Are Causative of Fitness Impairment
(A) Cellular localization of Mip6p when overexpressed in different genetic deletion backgrounds.
(B) Growth rate of strains overexpressing Mip6p in different genetic deletion backgrounds. The Gal1 promoter is integrated at the endogenous Mip6 locus. Error bars represent SD of three independent replicates.
See also Figure S6.
Discussion
We have shown here that dosage-sensitive proteins in yeast share physicochemical properties with proteins known to undergo liquid phase separation and that, for at least one protein, liquid-liquid demixing is associated with the growth impairment when the protein is overexpressed. The physicochemical properties associated with liquid phase separation are more common in the proteins of eukaryotes (Figure S7A). For example, whereas disordered regions make up ∼5% of the proteome in bacteria, they compose >30% in humans (van der Lee et al., 2014). Thus, many human proteins may have the potential to induce liquid phase separations when their concentration is increased. We propose therefore that the mechanism of dosage sensitivity that we have demonstrated here for Mip6p—a concentration-dependent triggering of liquid phase separation—may also occur in human disease.
Most recent studies have focused on liquid-to-solid phase changes as a mechanism of cellular pathology (Jain et al., 2016, Kato et al., 2012, Kim et al., 2013, Patel et al., 2015). For example, the protein FUS participates in physiological liquid-liquid phase separation in vivo and both disease mutations and increased concentration can accelerate the conversion of FUS liquid compartments to solid aggregates in vitro (Patel et al., 2015). Indeed, given that both liquid-to-liquid and liquid-to-solid phase transitions are intrinsically concentration-dependent (Lee et al., 2013, Weber and Brangwynne, 2012) it is reasonable to propose that both types of phase transition can cause dosage sensitivity, cellular toxicity, and disease. Thus, whereas for some proteins overexpression or mutation will promote an inappropriate liquid-to-solid phase transition, for other proteins an inappropriate liquid-to-liquid transition may be the cause of concentration- or mutation-dependent pathology.
Finally, we note that Mip6p foci—that have a substantial influence on growth rate—are often inherited by both the mother and daughter cells after cell division. Two important remaining questions are therefore whether and how such assemblies could act as epigenetic elements of inheritance in an isogenic population.
Experimental Procedures
Yeast Strains and Plasmids
Overexpression was performed using promoter swapping in a derivative of Saccharomyces cerevisiae S288C (MATa ura3-52 his3Δ200 leu2 lys2-801 ade2-101 trp1Δ63), or S288C BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) in the case of the experiments on the double deletion strain (_edc3_Δ, _pat1_Δ) reported in Figure 7. The Gal-1, Gal-L, and Gal-S promoters were amplified via tool-box PCR amplification (Janke et al., 2004). For localization and immunochemistry analysis, the same genes were tagged with either GFP or hemagglutinin (HA) by means of the same method. Amplified fragments were integrated via a standard lithium acetate transformation protocol and the correct insertion of the promoter was verified by PCR on extracted genomic DNA. 103Q and PGL-3 were expressed via cloning into p426-Gal1 (Addgene). For the characterization of Mip6p deletion variants, fragments of the coding sequence were amplified and cloned into p426-Gal1 vector.
Fitness Assays
Strains were grown to saturation in either Synthetic Complete or YPDA (Raffinose 2%) medium and diluted to ∼0.15 optical density (OD) 600 nm for a second growth of ∼6–8 hr in the same medium. At this stage, strains were diluted again at ∼0.15 OD 600 nm in Raffinose or Galactose medium to assess growth in non-inducing versus inducing conditions. Growth was monitored by measuring OD 600 nm in a 96-well plate incubated at 30°C inside the Infinite M200 microplate reader (Tecan). Growth curves were fitted and growth rate was calculated implementing the approach described by Hall et al. (2014).
Localization Screen
Strains were grown in inducing conditions until exponential phase and imaged under 100× magnification on a DMRE fluorescence microscope with PRIOR Lumen 200 light (Leica).
Flow Cytometry
Cells were analyzed at a medium flow rate in a FACSaria flow cytometer. The signal coming from cell aggregates was eliminated on the basis of the forward scatter area and height. Populations were separated on the basis of fluorescence intensity. We collected ∼106 events for each sample. Immediately after sorting, cells were imaged under fluorescence light or incubated at 30°C for fitness assays, either in an Infinite M200 microplate reader (Tecan) or in an Image Xpress Microscope (Molecular Devices).
Automated Colony Growth Assays
S. cerevisiae cells grown in different conditions were diluted and attached to a 96-well plate by Concanavalin-A-mediated cell adhesion. Imaging was automatically performed every 60 min on an Image Xpress Microscope (Molecular Devices). Colonies were identified in a bright field image in two steps. First, we identified clusters of pixels where the bright pixels of the image were juxtaposed with the dark pixels (that occur in the case of yeast cells). To assign the bright and dark pixels in each image, mean and SD of pixel intensities were calculated. Bright pixels had intensities greater than mean + 2.2 × SD, whereas dark ones corresponded to intensities lower than mean − 2.2 × SD. In addition, Sobel edge detection (Sobel, 1968) was used to identify sharp changes in intensity, which happen in the case of yeast cells at the cell boundary. After classification of all the clusters in an image, the centroid position for each cluster was calculated. In the second step, the centroids of the clusters in the series of images taken over time were computationally aligned. Clusters that showed at least doubling in area over the entire period of observation were considered in our analysis. Growth rate was calculated as change in the natural log-transformed area over time. To identify fluorescent cytoplasmic assemblies in each image, mean fluorescence intensity and SD of all pixels were calculated. Only pixels with intensities greater than mean + 3 × SD were considered as cytoplasmic assemblies. To avoid spurious inference on dissolution of foci, only the clusters with observations of foci in at least five time points and observations for dissolved protein on at least five time points were considered in our analysis.
FRAP
S. cerevisiae cells grown until exponential phase in inducing conditions were immobilized to an 8-well cover slide by Concanavalin-A-mediated cell adhesion. Cells were then imaged under a Confocal TCS SP5 microscope (Leica) where bleaching was achieved with 488 Laser Power at 50% for three frames (1.3 s/frame) while recovery was recorded for 50 frames. The curves were then fitted to a single exponential, following normalization, with the EasyFrap package (Rapsomaniki et al., 2012).
Immunochemistry
Samples were run in precast NOVEX NuPAGE 4%–12% gels in denaturing conditions. The Invitrogen iBlot system was used to transfer proteins to PVDF membranes. After blocking, membranes were incubated overnight at 4°C with anti-GFP rabbit antibody (Santa Cruz sc-8334), anti-HA 3F10 rat antibody (Roche 11867431001), or anti-PGKD1 mouse antibody (Novex 459250) diluted 1:1,000, 1:4,000, and 1:10,000, respectively. Secondary incubation with anti-Protein G HRP conjugated (Millipore 18-161) at 1:10,000 was performed at room temperature (RT) during 1 hr. ImageJ 1.49v software was used to quantify protein bands. Dot blots were performed by microfiltration where lysate samples passed through a 0.1 nm nitrocellulose membrane. The membrane was then blocked and incubated with the anti-amyloid fibrils OC antibody (Millipore AB2286) in 1:1,000 dilution.
Cycloheximide Treatment
Strains were grown until exponential phase in inducing conditions followed by 1 hr incubation with 0.25% CHX before observation under the microscope.
Translation Efficiency
Ten hours after induction with galactose, cells were incubated with 35S-methionine (NEG709A001MC) for 5, 15, or 30 min. A standard ethanol/glass beads protocol was used for whole protein extraction. Samples were loaded in a denaturing acrylamide gel and blue colloidal was applied for visualizing the total amount of loaded protein. A CL-Xposure Film was then employed to detect radioactivity on a Typhoon Trio imager (GE Healthcare). Quantification was performed using ImageQuant (GE Healthcare), followed by normalization of the 35S signal to Colloidal Blue intensity.
The _cat_GRANULE Algorithm
The tendency to assemble into foci is predicted using RNA binding and structural disordered propensities identified through a computational screening (Table S3) (Klus et al., 2014), as well as amino acid patterns and the polypeptide length. We selected the combination of RNA-binding and structural disordered predictors that best discriminates proteins with potential for foci formation (120 genes “Granule Forming” from Mitchell et al. [2013]) (Table S1) from the proteome (4,145 proteins excluding 770 genes from Sopko et al. [2006] and 777 genes from Makanae et al. [2013]). To cross-validate our predictions, we used an independent set of genes identified through a QuickGO search with query “granules” (16 proteins in common with “granule forming” and 64 “granule related”; Table S1).
The granule propensity (g) of an amino acid s at position i is defined as
| gi(s)=aR·Ri(s)+aD·Di(s)+aP·Pi(s), | (Equation 1) |
|---|
where R i = (R C,R N)i are the amino acid propensities of classical R C and recently discovered R N RNA-binding proteins (Castello et al., 2012), D i = (D C,D B)i estimates the structural disorder content based on coil D C and bending D B features (Deléage and Roux, 1987, Isogai et al., 1980), P i = (P RG,P FG)i takes into account arginine-glycine and phenylalanine-glycine content (Kato et al., 2012, Thandapani et al., 2013) (Table S4). Each property is calculated on a heptapeptide centered at position i in the sequence.
The overall granule propensity is calculated as
| granulepropensity=1ℓ∑i=1ℓgi(s)+aℓlog(ℓ) | (Equation 2) |
|---|
where ℓ is the sequence length.
The coefficients aR=(0.487.24), aD=(0.2611.54), aP=(1.981.42), and aℓ=0.25 were determined using a Monte Carlo search. The granule propensity shown in Figure 1C is _Z_-normalized (mean 0 and SD 1 on the yeast proteome).
The performances of the algorithm was assessed using a 5-fold cross-validation (area under the receiver operating characteristic (ROC) curve, AUC, of 0.86 for “granule forming” genes; Figure 1A) and on an additional set (AUC of 0.72 for “granule related” genes; Figure 1A). With respect to dosage-sensitive genes identified by Sopko et al. (2006) and Makanae et al. (2013), AUCs are 0.65 and 0.69 (Figure 1B). Gene groups are discriminated even when DNA-binding proteins are removed from original sets (Sopko et al. [2006]: AUC = 0.64; Makanae et al. [2013]: AUC = 0.67). Elimination of sequence length contribution from Equation 2 (aℓ=0) marginally reduces overall performances (“granule forming”: AUC = 0.80; “granule related”: AUC = 0.70).
To visualize the granule propensity g˜i of each amino acid i we average g i(s) on a sliding window of 50 amino acids and _Z_-normalize the score (mean 0 and SD of 1 on the yeast proteome). The granule strength (Figure 4B) is calculated using the fraction of amino acids with g˜i>0:
| granulestrength=∑fragg˜iϑ(g˜i)∑fragϑ(g˜i) | (Equation 3) |
|---|
where ϑ(x) is the Heaviside function that is 1 if x > 0 and zero otherwise on the fragment of interest (frag). In the case of full-length Mip6, the granule strength of the entire sequence can be visualized in Figure S7B.
All protein sequences were retrieved from the UniProt database (Uniprot Consortium, 2013). The _cat_GRANULE algorithm is freely available at http://service.tartaglialab.com/grant_submission/catGRANULE.
Author Contributions
B.B., N.L., and R.D. performed all the experiments. D.C. and G.G.T. built the computational model. B.L. conceived the hypothesis. B.B, G.G.T., and B.L. designed the experiments and wrote the manuscript.
Acknowledgments
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (BFU2011-26206 and BFU2014-55054-P), a European Research Council Consolidator grant IR-DC (616434), a European Research Council Starting Grant (Ribomylome 309545), Agencia de Gestio d’Ajuts Universitaris i de Recerca (AGAUR), the EMBO Young Investigator Program, the EMBL-CRG Systems Biology Program, the AXA Research Fund, and by the FP7 Marie Curie COFUND action. We also acknowledge support of the Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa 2013-2017,” SEV-2012-0208. All confocal and automated imaging was performed at the CRG Advanced Light Microscopy Facility. All sorting experiments were performed at the CRG/UPF FACS facility. We are grateful to Dr. Juana Diez at the Virology Unit of the UPF, Barcelona, for providing plasmids and discussing the results of this study.
Published: June 16, 2016
Footnotes
Contributor Information
Gian Gaetano Tartaglia, Email: gian.tartaglia@crg.eu.
Ben Lehner, Email: ben.lehner@crg.eu.
Supplemental Information
Document S1. Figures S1–S7, Tables S2 and S4, and Movies S1–S3
Table S1. Granule Forming Proteins, Related to the Experimental Procedures
Table S3. List of Physicochemical Properties Evaluated to Build the _cat_GRANULE Algorithm, Related to the Experimental Procedures
Movie S1. Dissolution of MIP6p Cytoplasmic Assemblies followed by Microscopy, Related to Figure 4
Movie S2. Dissolution of PGL-3 Cytoplasmic Assemblies followed by Microscopy, Related to Figure 4
Movie S3. Dissolution of 103Q cytoplasmic Assemblies followed by Microscopy, Related to Figure 3
Document S2. Article plus Supplemental Information
References
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Decker C.J., Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deléage G., Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- Dinkel H., Van Roey K., Michael S., Kumar M., Uyar B., Altenberg B., Milchevskaya V., Schneider M., Kühn H., Behrendt A. ELM 2016--data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016;44(D1):D294–D300. doi: 10.1093/nar/gkv1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte K.A., Dion M.F., Budnik B.A., Wang S.M., Hartl D.L., Drummond D.A. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. USA. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D.M., White M.A., Wilkinson M.L., Kon Y., Kung L.A., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S., Campbell C.D., Eichler E.E. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B.G., Acar H., Nandipati A., Barlow M. Growth rates made easy. Mol. Biol. Evol. 2014;31:232–238. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Isogai Y., Némethy G., Rackovsky S., Leach S.J., Scheraga H.A. Characterization of multiple bends in proteins. Biopolymers. 1980;19:1183–1210. doi: 10.1002/bip.1980.360190607. [DOI] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R., Head E., Sarsoza F., Saing T., Cotman C.W., Necula M., Margol L., Wu J., Breydo L., Thompson J.L. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim N.C., Wang Y.-D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klus P., Bolognesi B., Agostini F., Marchese D., Zanzoni A., Tartaglia G.G. The cleverSuite approach for protein characterization: predictions of structural properties, solubility, chaperone requirements and RNA-binding abilities. Bioinformatics. 2014;30:1601–1608. doi: 10.1093/bioinformatics/btu074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavut A., Raveh D. Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet. 2012;8:e1002527. doi: 10.1371/journal.pgen.1002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.F., Brangwynne C.P., Gharakhani J., Hyman A.A., Jülicher F. Spatial organization of the cell cytoplasm by position-dependent phase separation. Phys. Rev. Lett. 2013;111:088101. doi: 10.1103/PhysRevLett.111.088101. [DOI] [PubMed] [Google Scholar]
- Lin Y., Protter D.S.W., Rosen M.K., Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanae K., Kintaka R., Makino T., Kitano H., Moriya H. Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Res. 2013;23:300–311. doi: 10.1101/gr.146662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim. Biophys. Acta. 2013;1834:918–931. doi: 10.1016/j.bbapap.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Mitchell S.F., Jain S., She M., Parker R. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrodnik M., Salmonowicz H., Brown R., Turkowska J., Średniawa W., Pattabiraman S., Amen T., Abraham A.C., Eichler N., Lyakhovetsky R., Kaganovich D. Dynamic JUNQ inclusion bodies are asymmetrically inherited in mammalian cell lines through the asymmetric partitioning of vimentin. Proc. Natl. Acad. Sci. USA. 2014;111:8049–8054. doi: 10.1073/pnas.1324035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Pál C., Hurst L.D. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Patel S.S., Belmont B.J., Sante J.M., Rexach M.F. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Rapsomaniki M.A., Kotsantis P., Symeonidou I.-E., Giakoumakis N.-N., Taraviras S., Lygerou Z. easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics. 2012;28:1800–1801. doi: 10.1093/bioinformatics/bts241. [DOI] [PubMed] [Google Scholar]
- Schisa J.A., Pitt J.N., Priess J.R. P granules and mRNA. Development. 2001;128:1–12. doi: 10.1242/dev.128.8.1287. [DOI] [PubMed] [Google Scholar]
- Sobel I. SAIL; 1968. A 3x3 Isotropic Gradient Operator for Image Processing. Stanford Artificial Intelligence Project. [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Tartaglia G.G., Pechmann S., Dobson C.M., Vendruscolo M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 2007;32:204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thandapani P., O’Connor T.R., Bailey T.L., Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- Tomala K., Pogoda E., Jakubowska A., Korona R. Fitness costs of minimal sequence alterations causing protein instability and toxicity. Mol. Biol. Evol. 2014;31:703–707. doi: 10.1093/molbev/mst264. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T., Semple J.I., Garcia-Verdugo R., Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Veitia R.A. Nonlinear effects in macromolecular assembly and dosage sensitivity. J. Theor. Biol. 2003;220:19–25. doi: 10.1006/jtbi.2003.3105. [DOI] [PubMed] [Google Scholar]
- Veitia R.A., Birchler J.A. Dominance and gene dosage balance in health and disease: why levels matter! J. Pathol. 2010;220:174–185. doi: 10.1002/path.2623. [DOI] [PubMed] [Google Scholar]
- Walters R.W., Muhlrad D., Garcia J., Parker R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA. 2015;21:1660–1671. doi: 10.1261/rna.053116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Herrmann C.J., Simonovic M., Szklarczyk D., von Mering C. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S.C., Brangwynne C.P. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Weber S.C., Brangwynne C.P. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F., Bodenmiller B., Trajkovska M.G., Wanka S., Aebersold R., Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S7, Tables S2 and S4, and Movies S1–S3
Table S1. Granule Forming Proteins, Related to the Experimental Procedures
Table S3. List of Physicochemical Properties Evaluated to Build the _cat_GRANULE Algorithm, Related to the Experimental Procedures
Movie S1. Dissolution of MIP6p Cytoplasmic Assemblies followed by Microscopy, Related to Figure 4
Movie S2. Dissolution of PGL-3 Cytoplasmic Assemblies followed by Microscopy, Related to Figure 4
Movie S3. Dissolution of 103Q cytoplasmic Assemblies followed by Microscopy, Related to Figure 3
Document S2. Article plus Supplemental Information