Genetic Interactions Between Transcription Factors Cause Natural Variation in Yeast (original) (raw)
. Author manuscript; available in PMC: 2016 Aug 15.
Published in final edited form as: Science. 2009 Jan 23;323(5913):498–501. doi: 10.1126/science.1166426
Abstract
Our understanding of the genetic basis of phenotypic diversity is limited by the paucity of examples in which multiple, interacting loci have been identified. We show that natural variation in the efficiency of sporulation, the program in yeast that initiates the sexual phase of the life cycle, between oak tree and vineyard strains is due to allelic variation between four nucleotide changes in three transcription factors: IME1, RME1, and RSF1. Furthermore, we identified that selection has shaped quantitative variation in yeast sporulation between strains. These results illustrate how genetic interactions between transcription factors are a major source of phenotypic diversity within species.
Understanding the molecular basis of natural phenotypic diversity is a major challenge in modern genetics (1–6). Knowing how individual genetic polymorphisms combine to produce phenotypic change could strengthen evolutionary theory and advance applications such as personalized medicine (7, 8). Many loci that contribute to variation have been identified across taxa, but only a small fraction has been resolved to the nucleotide level (9, 10). Examples of complex traits in which causative polymorphisms have been identified at multiple contributing loci are even rarer (11). As a result, the interactions between nucleotide changes in nature, and thus the genetic mechanisms of phenotypic change, are largely unknown.
Crosses of laboratory strains of the yeast Saccharomyces cerevisiae have identified genes and polymorphisms governing complex traits (12–21). However, these lines may harbor laboratory-engineered gene deletions and deleterious mutations that are pleiotropic for multiple traits which may obscure the natural genetic architecture. The natural diversity of this species, which includes isolates from clinical, vineyard, and oak tree environments, remains largely untapped (22–24).
Sporulation efficiency is a highly heritable, complex trait that varies among natural populations of S. cerevisiae (25). Sporulation is a core developmental program that initiates the sexual phase of the yeast life cycle and promotes long-term survival during dessication or starvation (26). Sporulation is triggered as a response to environmental change (27), and is hypothesized to be under different selective pressures in different habitats (28). Accordingly, wild isolates from North American oak trees and associated soil samples sporulate with efficiencies approaching 100%, but strains isolated from naturally occurring vineyard fermentations sporulate at lower rates (25). Genetic dissection of the underlying natural variation in sporulation efficiency provides an opportunity to uncover the nucleotide changes that govern ecologically driven variation.
To identify quantitative trait loci (QTL) governing natural variation in sporulation, we crossed YPS606, a strain isolated from the bark of an oak tree in Pennsylvania that sporulates at 99% efficiency, and BC187, a strain originating from a California wine barrel that sporulates at only 3.5% (25). We developed genetic markers by shotgun sequencing these parent strains, and produced a genetic linkage map of 225 loci typed in 374 recombinant segregants. This map covers the yeast genome at an average of 11 centimorgan intervals (Table S1). Composite interval mapping (29) was used to identify five QTL on four chromosomes (Fig. S1, Table 1) that significantly co-segregated with variation in sporulation efficiency with an experiment-wide error rate of P = 0.05 (LOD > 3.1, which is significant by permutation analysis). Alleles from the oak parent at four of the QTL were linked to an increase in sporulation efficiency (markers L7.9, L7.17, L10.14, L13.6). At the locus on chromosome 11 (marker L11.2), an allele from the poorly sporulating vineyard parent promotes more efficient sporulation in segregating progeny. The presence of a vineyard allele promoting higher sporulation was expected based on previously observed transgressive segregants that sporulate more efficiently than the oak parent (25).
Table 1.
Significant QTL for sporulation efficiency.
| Chromosome | Nearest Marker | LOD score | Variance Explained (%) | Additive Effect (%) |
|---|---|---|---|---|
| 7 | L7-9 | 86.42 | 41 | 20 |
| 7 | L7-17 | 4.7 | 2 | 4 |
| 10 | L10-14 | 68.2 | 29 | 16 |
| 11 | L11-2 | 3.9 | 1 | -3 |
| 13 | L13-6 | 28.7 | 10 | 10 |
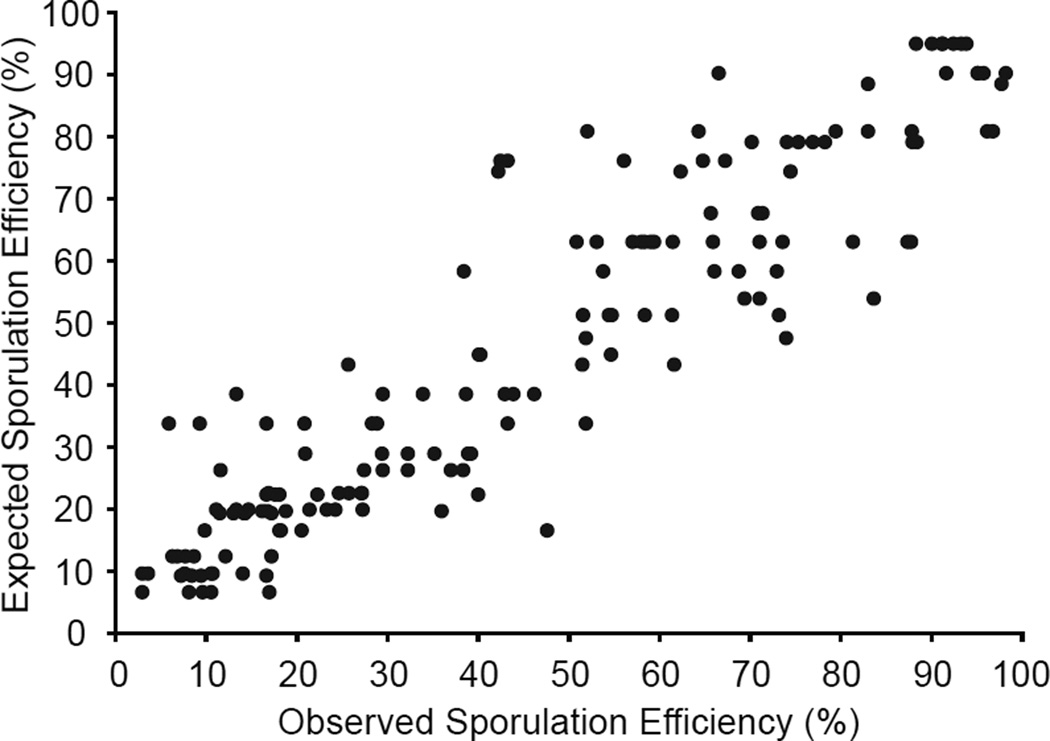
We quantified the total amount of variation explained by these five QTL using a linear model trained on 200 segregants (Table S2) (29). In an independent test set of 155 segregants, the model explains 88% of the phenotypic variation (squared correlation coefficient (R2) = 0.88), with an average prediction error of ±8% (Fig. 1). This indicates that the five QTL explain most of the variation in sporulation efficiency in this cross. To obtain the best fit, the model must incorporate two and three-way interactions between loci (F-test, P < 0.001 ) (29), indicating that the genetic architecture of sporulation efficiency is non-additive and complex. Although incorporating the effects of all five loci produces the best-fit model (F-test, P < 0.001), only three of the loci have large effects. When the minor QTL (markers L7.17, L11.2) are ignored, the results are virtually identical (R2 = 0.87, prediction error ± 8%).
Fig. 1. A linear model of the effects of five QTL on sporulation efficiency.
Expected values of sporulation efficiency plotted as a function of observed values for 155 segregants. Expected values are derived from a linear model based from 200 independent segregants (Table S2).
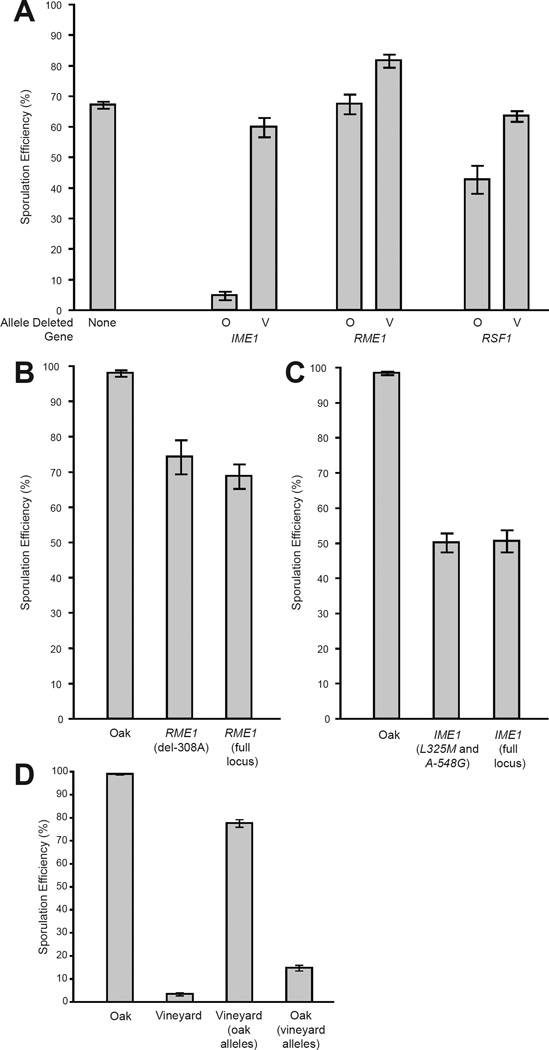
One major QTL (marker L7.9) covers a 100kb confidence interval on chromosome 7. RME1, a transcription factor that suppresses sporulation in specific cell types (30), resides in this peak (Fig. S1A). To test if allelic variation in RME1 produces variation in sporulation efficiency, we deleted each parental allele of RME1 in a hybrid background [through reciprocal hemizygosity analysis (18)]. This showed that the allelic contributions of RME1 from each parent were different, confirming that variation in RME1 affects sporulation efficiency (Fig. 2A). The coding region of RME1 contains no amino acid substitutions between the oak and vineyard parents suggesting that the allelic difference is regulatory. By replacement (29), we confirmed that a single nucleotide insertion/deletion 308bp upstream of the initiation codon (Fig. S2A) accounts for the effect of the RME1 locus on sporulation efficiency (Fig. 2B). The vineyard strain allele [RME1(del-308A)], which has a deletion of a single adenine relative to the oak strain, also reduces sporulation efficiency in laboratory strains (21). This nucleotide change presumably increases the expression of RME1, which represses sporulation, as the RME1(del-308A) allele is expressed at higher levels than the oak allele (25, 31).
Fig. 2.
A. Reciprocal hemizygosity analysis of RME1, IME1, and RSF1 in an oak/vineyard hybrid background_._ O = oak parent allele, V = vineyard parent allele. B. Sporulation efficiency in oak, in oak with the vineyard parent allele (RME1(del-308A), and in oak with the entire vineyard parent locus including all the coding and non-coding polymorphisms. Variation among multiple replicate clones of the experiment (F test, P < 0.001) explains the discrepancy between the RME1(del-308A) and full allele in the oak background and this is not an effect of the two replacement types (F test, P = 0.13). C. Replacement of both the oak IME1 causative nucleotides with the vineyard parent alleles (L325M and A-548G) in the oak parent is equivalent to replacing the entire IME1 vineyard parent locus, coding and non-coding, in the oak parent (full locus). D. Replacements of all four alleles in the vineyard parent strain (oak alleles) compared to placing the vineyard alleles in the oak parent. Error bars denote one standard deviation.
A second major QTL (L10.14) located in a 50kb confidence interval on chromosome 10 also contained a strong candidate gene, IME1 (Fig. S1B). IME1 is a transcriptional activator and master regulator that initiates yeast sporulation (32). Reciprocal hemizygosity analysis confirmed that IME1 quantitatively controls sporulation efficiency (Fig. 2A). We identified 8 polymorphisms in the coding regions, both synonymous and non-synonymous, and 39 polymorphisms in the non-coding regions of IME1 between the oak and vineyard strain. Allele replacements demonstrated that two of these polymorphisms account for the full effect of the IME1 locus on sporulation efficiency (Fig. 2C). Although other polymorphisms in this region may affect sporulation efficiency, they must be redundant with the two causative alleles we identified. We identified a causative non-synonymous substitution in the vineyard strain, IME1(L325M) (Fig. S2B), which resides in a Ime1 domain essential for protein-protein interactions with Rim11 and Ume6, which also regulate the initiation of sporulation (33). Mutation of this leucine reduces the ability of Ime1 to activate transcription (34). We also identified a non-coding IME1(A-548G) polymorphism in an 11bp sequence that is conserved among three yeast species closely related to S. cerevisiae (Fig. S2C).
The third major QTL affecting sporulation occurs in a 100kb region of chromosome 13 (L13.6, Fig. S1D). Reciprocal hemizygosity analysis of genes in this region identified RSF1 as a candidate gene affecting sporulation efficiency (Fig. 2A). Allele replacements (Fig. S3) confirmed that a single derived polymorphism in the vineyard strain—coding for a substitution of a conserved glutamic acid with a glycine RSF1(_D181G)_—is responsible for the allelic effect of RSF1 (Fig. S2D). RSF1 encodes a transcriptional activator of mitochondrial genes critical for cellular respiration (35). Since a respiratory signal promotes IME1 expression and sporulation (36), the vineyard allele likely reduces the function of Rsf1.
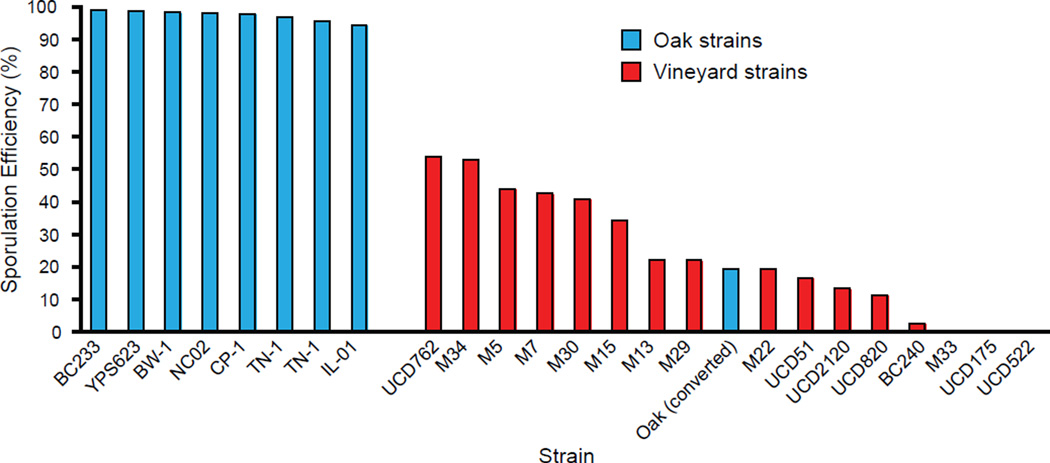
In total, we identified four nucleotide changes causing variation in sporulation efficiency by directly affecting three transcription factors governing pathways that regulate the initiation of sporulation. Our QTL model (Table S2) includes, however, potentially undiscovered linked alleles that could account for some of the variation. Therefore, we engineered yeast strains isogenic to each parent, but carrying the causative alleles from the opposite parent (Fig. 2D) (29). The vineyard parental strain, which sporulates at 3.5 ± 0.1%, increases to 78 ± 2% when carrying all four oak alleles. In the oak parent, the replacement of these four nucleotides reduced sporulation efficiency from 99 ± 0.2% to 14.9 ± 1% and places the phenotype of the oak strain background within the range normally seen only among vineyard strains (Fig. 3).
Fig. 3.
Variation among oak strains relative to the oak parental strain (BC233) and vineyard strains relative to the vineyard parental strain (BC240) show fixed and variable sporulation efficiencies, respectively. Oak(converted) is the parental oak strain transformed with the 4 causative nucleotides from the vineyard strain.
Our QTL model also predicts that that the four causative nucleotides will interact. We therefore compared the phenotypes of strains isogenic to the oak background carrying all possible permutations of the four oak and vineyard alleles. We chose the oak strain background for this experiment because isolates of other species of yeast sporulate efficiently, supporting that the oak strain resembles the ancestral state of sporulation efficiency from which we hypothesize the vineyard parent alleles arose. This is further supported by the fact that three of the four vineyard alleles reducing sporulation [_IME1(L325M), IME1(A-548G), RSF1(D181G)_] are derived (Fig. S2).
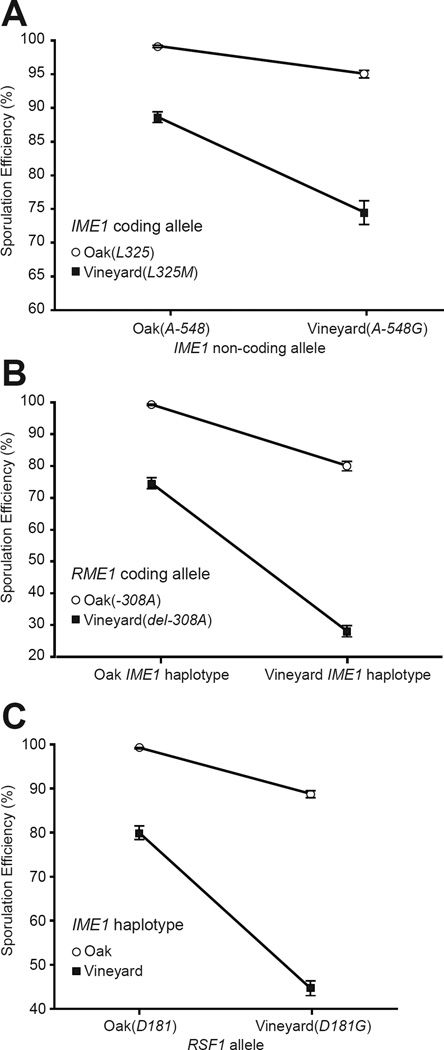
Analysis of the allele replacement strains revealed extensive interactions between the four nucleotides as all possible two, three, and four way interactions are statistically significant in an analysis of variance (Table S3). The interactions indicate that the vineyard alleles work synergistically to reduce sporulation efficiency. For example, the IME1 coding and non-coding polymorphisms interact and double the sum of their individual effects (Fig. 4A). The strongest interactions observed occurred between RME1(del-308A) and the two polymorphisms at IME1 (Fig. 4B). The RSF1 vineyard allele, which causes less than a 3% change on its own, also was found to interact synergistically with the other vineyard alleles (Fig. 4C) and was responsible for 11%, 13%, and 16% drops in sporulation efficiency through its two-way interactions (Table S3). These results illustrate mechanisms by which combinations of alleles can produce a phenotypic change larger than expected from individual effects.
Fig. 4.
Single nucleotide interactions governing sporulation efficiency as shown by replacing single nucleotides in the oak parent with vineyard parent alleles. A. The effect of placing the vineyard allele IME1(A-548G) in the oak background is greater when the vineyard coding allele IME1(L325M) is also present. B. The effect of the two causative nucleotides in IME1 (IME1 haplotype) is greater when the vineyard RME1(del-308A) allele is also present. C. The effect of the RSF1(D181G) allele is greater when the vineyard alleles occur at IME1 (IME1 haplotype). A full list of interactions is available in Table S3. Error bars denote one standard deviation.
Genetic interactions (epistasis) are often seen between genomic regions affecting quantitative traits (37), and this study demonstrates how a small number of nucleotides can create complex, quantitative variation in phenotype highlighting the importance of single nucleotides on epistasis. This emphasizes the need to incorporate genetic interactions into models that seek to accurately predict phenotype from genotype. If prevalent, genetic interactions between nucleotides will be a major hurdle in the endeavor to connect genetic and phenotypic variation in humans (38). Our identification of epistasis between RME1 and IME1 supports the idea that a search for epistasis should incorporate prior knowledge of functional relationships between genes and proteins (39).
We found that alleles reducing sporulation efficiency were at varying frequencies in additional vineyard strains (Table S4). The RME1 polymorphism was found to be ubiquitous, whereas the IME1 polymorphisms were the rarest. Over half of the vineyard strains harbor at least two of the alleles reducing sporulation suggesting that the two-way interactions we identified may be producing phenotypic change in nature. However, the four nucleotides we identified are not sufficient to predict all of the differences in sporulation efficiency among vineyard strains. For example, strain UCD51 sporulates less efficiently than M34 and M15 (Fig. 3) despite carrying fewer of the alleles reducing sporulation that we identified (Table S4). This suggests that additional alleles reducing sporulation efficiency remain to be identified in the vineyard population.
All four sporulation-reducing alleles were absent from all oak isolates, consistent with the possibility that selection for high sporulation efficiency occurs in woodland environments. This hypothesis was supported by the spectrum of polymorphism in IME1, a positive regulator of sporulation (Table S5). In oak strains, we observed a slower rate of non-synonymous substitution relative to synonymous substitutions—which is consistent with purifying selection (Z test_, P_ < 0.01) (29). In vineyard strains, the rate of non-synonymous substitution was not lower than the rate of synonymous substitution, suggesting relaxed selection in vineyard strains (Z test, P = 0.99).
Variation at RSF1 is consistent with selection reducing sporulation efficiency in the vineyard strains. A McDonald-Kreitman test (40) reveals an excess of non-synonymous polymorphisms in the S. cerevisiae RSF1 gene relative to its ortholog in Saccharomyces paradoxus (Fisher’s exact test, P < 0.001). These substitutions occurred almost entirely in the vineyard strains, as eight of nine oak isolates sequenced show no polymorphism at this locus (Table S6). This result alone is consistent with relaxed selection on sporulation efficiency but relaxed selection should still result in a rate of non-synonymous substitutions lower or equal to the synonymous rate. Instead, the rate of non-synonymous substitution at RSF1 in the vineyard strains is significantly greater than the synonymous rate (Z test, P < 0.02) (29). This result cannot be explained by relaxed selection and instead suggests selective pressure favoring derived alleles of RSF1 in vineyard strains.
Selection is hypothesized to favor genetic changes with limited pleiotropy (8). Deletion of RSF1 causes a severe growth defect when yeast are raised on glycerol (35), and some vineyard strains harbor frame-shifts and premature termination codons in RSF1 that likely affect both sporulation efficiency and growth rate on glycerol (Table S6). We tested if the D181G polymorphism affected growth rate in glycerol and found no effect in the oak parent (t test, P = 0.99, N = 3) or the vineyard parent (t test, P = 0.60, N = 3). This suggests that the D181G allele is not a complete loss-of-function allele and may alter only a subset of RSF1 functions.
A major challenge in studying phenotypic change is to find predictable patterns among the genetic differences that create diversity. In this particular case, the causative polymorphisms included both rare and common alleles, and occur in both coding and non-coding sequences. One commonality underlying these effects, however, is that all of the changes occur in transcription factors, and thus exert their effects through the differential expression of downstream regulated genes. We argue that change in these transcription factor genes was predictable. IME1 and RME1 regulate a battery of functionally related genes whose expression is specific to a single cell type: sporulating cells (41, 42). Therefore the roles of IME1 and RME1 are analogous to those of transcription factors controlling “differentiation gene batteries” that drive terminal cell differentiation in animal development (43). Because these transcription factors coordinately regulate the expression of functionally related genes, changes in their function have the potential for large phenotypic effects (“coordinated pleiotropy”) (2). Furthermore, evolution is hypothesized to favor changes in this type of transcription factor, as their role in a specific cell type limits the potential for deleterious consequences (8, 43, 44). Our results point to transcription factor polymorphism as a major source of natural phenotypic diversity within species.
Supplementary Material
01
Acknowledgments
We thank Laura Kyro for technical assistance, and Justin Fay, Betsy Engle, Hyun-Seok Kim and Mark Johnston for sharing reagents. We also thank Justin Fay, Scott Doniger, Chris Hittinger and members of the Cohen Lab for helpful advice and discussions. This research was funded by NSF-MCB-0543156. J.P.G. has recieved support from NHGRI-T32HG000045.
Footnotes
References
- 1.Wagner GP, Lynch VJ. Trends Ecol Evol. 2008 Jul;23:377. doi: 10.1016/j.tree.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Wray GA, et al. Mol Biol Evol. 2003 Sep;20:1377. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 3.Stern DL. Evolution. 2000 Aug;54:1079. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 4.Hoekstra HE, Coyne JA. Evolution. 2007 May;61:995. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 5.Carroll SB. PLoS Biol. 2005 Jul;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackay TF. Annu Rev Genet. 2001;35:303. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- 7.Kruglyak L. Nat Rev Genet. 2008 Apr;9:314. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 8.Stern DL, Orgogozo V. Evolution. 2008 Aug 14; [Google Scholar]
- 9.Flint J, Mott R. Nat Rev Genet. 2001 Jun;2:437. doi: 10.1038/35076585. [DOI] [PubMed] [Google Scholar]
- 10.Glazier AM, Nadeau JH, Aitman TJ. Science. 2002 Dec 20;298:2345. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- 11.Steiner CC, Weber JN, Hoekstra HE. PLoS Biol. 2007 Sep;5:e219. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demogines A, Smith E, Kruglyak L, Alani E. PLoS Genet. 2008 Jul;4:e1000123. doi: 10.1371/journal.pgen.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demogines A, Wong A, Aquadro C, Alani E. PLoS Genet. 2008 Jun;4:e1000103. doi: 10.1371/journal.pgen.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heck JA, et al. Proc Natl Acad Sci U S A. 2006 Feb 28;103:3256. doi: 10.1073/pnas.0510998103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha H, Nicholson BP, Steinmetz LM, McCusker JH. PLoS Genet. 2006 Feb;2:e13. doi: 10.1371/journal.pgen.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yvert G, et al. Nat Genet. 2003 Sep;35:57. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 17.Brem RB, Storey JD, Whittle J, Kruglyak L. Nature. 2005 Aug 4;436:701. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinmetz LM, et al. Nature. 2002 Mar 21;416:326. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Ari G, et al. PLoS Genet. 2006 Nov 17;2:e195. doi: 10.1371/journal.pgen.0020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HS, Fay JC. Proc Natl Acad Sci U S A. 2007 Dec 4;104:19387. doi: 10.1073/pnas.0708194104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deutschbauer AM, Davis RW. Nat Genet. 2005 Dec;37:1333. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- 22.Naumov GI, Naumova ES, Sniegowski PD. Can J Microbiol. 1998 Nov;44:1045. [PubMed] [Google Scholar]
- 23.Marullo P, et al. FEMS Yeast Res. 2007 Sep;7:941. doi: 10.1111/j.1567-1364.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- 24.McCullough MJ, Clemons KV, Farina C, McCusker JH, Stevens DA. J Clin Microbiol. 1998 Feb;36:557. doi: 10.1128/jcm.36.2.557-562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerke JP, Chen CT, Cohen BA. Genetics. 2006 Oct;174:985. doi: 10.1534/genetics.106.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kupiec M, Byers B, Espositio RE, Mitchell AP. In: The molecular and cellular biology of the yeast Saccharmoyces: Cell cycle and cell biology. Pringle JR, Broach JR, Jones EW, editors. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- 27.Honigberg SM, Purnapatre K. J Cell Sci. 2003 Jun 1;116:2137. doi: 10.1242/jcs.00460. [DOI] [PubMed] [Google Scholar]
- 28.Nachman I, Regev A, Ramanathan S. Cell. 2007 Nov 2;131:544. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 29.Materials and Methods are available in supplementary materials
- 30.Mitchell AP, Herskowitz I. Nature. 1986;319:738. doi: 10.1038/319738a0. Feb 27-Mar 5. [DOI] [PubMed] [Google Scholar]
- 31.Doniger SW, et al. PLoS Genet. 2008 Aug;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kassir Y, Granot D, Simchen G. Cell. 1988 Mar 25;52:853. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 33.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Mol Cell Biol. 1996 May;16:2518. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith HE, Driscoll SE, Sia RA, Yuan HE, Mitchell AP. Genetics. 1993 Apr;133:775. doi: 10.1093/genetics/133.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu L, Roberts G, Simon K, Yu J, Hudson AP. Curr Genet. 2003 Jul;43:263. doi: 10.1007/s00294-003-0398-z. [DOI] [PubMed] [Google Scholar]
- 36.Jambhekar A, Amon A. Curr Biol. 2008 Jul 8;18:969. doi: 10.1016/j.cub.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlborg O, Haley CS. Nat Rev Genet. 2004 Aug;5:618. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- 38.Moore JH. Hum Hered. 2003;56:73. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- 39.Pattin KA, Moore JH. Hum Genet. 2008 Aug;124:19. doi: 10.1007/s00439-008-0522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonald JH, Kreitman M. Nature. 1991 Jun 20;351:652. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 41.Chu S, et al. Science. 1998 Oct 23;282:699. [Google Scholar]
- 42.Primig M, et al. Nat Genet. 2000 Dec;26:415. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 43.Davidson EH, Erwin DH. Science. 2006 Feb 10;311:796. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 44.Lynch VJ, Wagner GP. Evolution. 2008 Sep;62:2131. doi: 10.1111/j.1558-5646.2008.00440.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01