SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells (original) (raw)
Significance
Long noncoding RNAs (lncRNAs) are important regulators of gene expression, but their structural features are largely unknown. We used structure-selective chemical probing to examine the structure of the Xist lncRNA in living cells and found that the RNA adopts well-defined and complex structures throughout its entire 18-kb length. By looking for changes in reactivity induced by the cellular environment, we were able to identify numerous previously unknown hubs of protein interaction. We also found that the Xist structure governs specific protein interactions in multiple distinct ways. Our results provide a detailed structural context for Xist function and lay a foundation for understanding structure–function relationships in all lncRNAs.
Keywords: RNA structure, RNA–protein interaction, SHAPE-MaP, X-inactivation
Abstract
The 18-kb Xist long noncoding RNA (lncRNA) is essential for X-chromosome inactivation during female eutherian mammalian development. Global structural architecture, cell-induced conformational changes, and protein–RNA interactions within Xist are poorly understood. We used selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) to examine these features of Xist at single-nucleotide resolution both in living cells and ex vivo_._ The Xist RNA forms complex well-defined secondary structure domains and the cellular environment strongly modulates the RNA structure, via motifs spanning one-half of all Xist nucleotides. The Xist RNA structure modulates protein interactions in cells via multiple mechanisms. For example, repeat-containing elements adopt accessible and dynamic structures that function as landing pads for protein cofactors. Structured RNA motifs create interaction domains for specific proteins and also sequester other motifs, such that only a subset of potential binding sites forms stable interactions. This work creates a broad quantitative framework for understanding structure–function interrelationships for Xist and other lncRNAs in cells.
Long noncoding RNAs (lncRNAs) play central roles in the regulation of gene expression through interactions with numerous protein partners (1) and are necessary for normal health and development (2, 3). The 18-kb Xist lncRNA is essential for X-chromosome inactivation during female eutherian mammalian development and is an archetype of gene-silencing lncRNAs. During the early stages of X inactivation, Xist accumulates in cis around the future inactive X chromosome and recruits protein complexes that apply repressive chromatin modifications, leading to stable gene silencing (3, 4).
Genetic deletion studies have demarcated several broad regions of function within Xist. Several tandem repeat regions (labeled A–F in the mouse) show moderate conservation (5–7), and at least two of these, repeat A and the rodent-specific repeat C, are implicated in silencing and localization to the inactive X. Deletion of the final 7.5-kb exon of Xist causes a defect in its localization (8), and the 1.5-kb region encompassing repeats F and B is required for accumulation of heterochromatic marks over the inactive X (4); however, beyond these initial characterizations, the mechanisms by which gene silencing, heterochromatinization, and localization of Xist on the X chromosome occur are not well understood. In particular, the role of RNA structure in orchestrating these distinct functions remains unclear.
Several previous studies have suggested the importance of RNA structures in specific regions of Xist (9–12), but overall, the locations and structures of functional domains within Xist are poorly defined. Detailed structural maps of other functional RNAs, such as ribosomal RNAs (13) and the HIV RNA genome (14–16), have been fundamental to understanding the mechanisms by which individual domains within large RNAs execute discrete cellular functions. A detailed and quantitative structural map of Xist would be expected to have a similar transformative impact.
Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) provides a biophysically rigorous measurement of local nucleotide flexibility that is independent of base identity (15). SHAPE-MaP readily detects modifications in highly complex environments, including in the cell nucleus (17), and, unlike alternative RNA-probing methods with deep sequencing readout, is unaffected by biases introduced during complex ligation-based library preparation steps (15). SHAPE data are sufficient for distinguishing between structural models (18) and detecting distinct modes of protein binding in cells (17). SHAPE-informed structural models have consistently yielded rich insights into the biological functions of diverse RNAs (14, 15, 18–21) and, in many cases, uncovered novel functional elements (14–17, 20).
Using SHAPE-MaP, we examined full-length, authentic transcripts of mouse Xist at single-nucleotide resolution in mouse trophoblast stem cells (TSCs) and under protein-free conditions (ex vivo). TSCs demonstrate prototypical epigenetic patterns over the inactive X chromosome (22) and require Xist for continued silencing (23). SHAPE data identified 33 regions in Xist that form well-defined structures with complexities comparable to those of functional elements within RNA viruses and ribosomal RNAs (15, 21). We found extensive significant differences between in-cell SHAPE reactivities and those obtained ex vivo, indicating that many nucleotides of Xist interact with proteins or have different conformations in cells vs. a cell-free state. The perspective obtained here supports novel and specific models of the complex interrelationships among lncRNA sequence, structure, and function.
Results
Ex Vivo Structure Probing.
We probed full-length Xist, after gentle and nondenaturing extraction from cells, using the SHAPE reagents 1-methyl-7-nitroisatoic anhydride (1M7), 1-methyl-6-nitroisatoic anhydride (1M6), and _N_-methyl-isatoic anhydride (NMIA) (24, 25) and obtained ex vivo SHAPE reactivities for 86% of nucleotides in Xist (Fig. 1_A_). We also probed the Xist structure in living cells with 1M7. Full biological replicates of 1M7 probing, performed more than 1 y apart, showed good agreement over thousands of nucleotides under ex vivo conditions (Spearman’s ρ = 0.65; SI Appendix, Fig. S1_A_). In-cell replicates exhibited a more modest correlation (Spearman’s ρ = 0.50; SI Appendix, Fig. S1_B_). Critically, however, the in-cell replicates yielded highly similar outcomes in subsequent analyses (SI Appendix, Figs. S2 and S3 and Supporting Text).
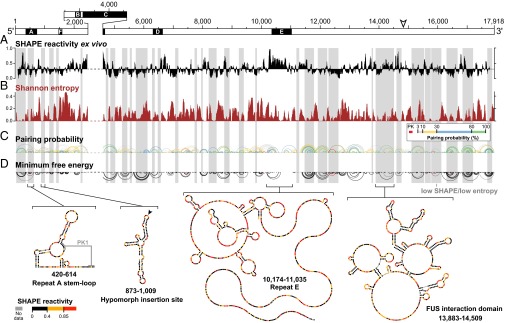
Fig. 1.
Structural architecture of the Xist lncRNA. (A) Ex vivo 1M7 reactivities shown as the median reactivity over 55-nt sliding windows relative to the global median. Values above or below the line are more or less flexible than the median, respectively. Repeats B and C were excluded from analysis due to lack of uniquely aligning reads. (B) Shannon entropy values for the ex vivo secondary structure model, smoothed over 55-nt sliding windows. High values indicate many possible structures, and low values indicate a single well-defined structure. Gray shading marks well-defined structures with low SHAPE reactivity and low Shannon entropy. (C) Base-pairing probabilities in Xist. Arcs represent base pairs and are color-coded by probability. Pseudoknotted helices are shown in red. (D) Minimum free energy secondary structure model of Xist. Arcs are inverted relative to C. Secondary structure models for selected domains are color-coded according to SHAPE reactivity. Secondary structures of all low-SHAPE/low-entropy regions are shown in SI Appendix, Fig. S5.
We used the cell-free ex vivo data to guide initial RNA structure modeling. We searched for and identified 10 potential pseudoknots (26) and modeled the secondary structure of Xist using the three-reagent differential SHAPE strategy, which yields highly accurate RNA structure models (15, 25). The structure was also modeled without SHAPE data and with only 1M7 data (SI Appendix, Fig. S4 A and B).
To assess our models, we examined the structural context of 105 single-nucleotide variants (SNVs) within mouse Xist (27). For each structural model (no data, 1M7 only, or three-reagent differential), we counted the SNVs that disrupted structure by creating base pair mismatches. With increasing data quality, the probability that SNVs are structurally disruptive by chance decreases significantly (SI Appendix, Fig. S4_C_; P = 0.35, 0.15, and 0.027 for the no data, 1M7 only, and three-reagent models, respectively). Just as lack of selective pressure leads to increased SNV abundance in genetic elements with low functional potential (28), we infer that SNVs occur predominantly in unstructured regions in our model because many RNA structures within Xist are important for function.
We further assessed how well each secondary structure element was defined by its sequence and the experimental SHAPE data by calculating Shannon entropies at nucleotide resolution (15) (Fig. 1_B_). Previous work with large viral RNAs has shown that functional elements are overrepresented in regions with both low SHAPE reactivity (indicating a high degree of structure) and low Shannon entropy (indicating well-defined structure) (15, 21). We identified 33 regions with low SHAPE reactivity and Shannon entropy in the Xist RNA (Fig. 1 C and D, gray shading and SI Appendix, Fig. S5). Of the well-defined domains, three-fourths have not been described previously (SI Appendix, Supporting Text).
Many of the well-defined structural elements in Xist are located within the final 4,000 nucleotides (Fig. 1 C and D). Much of this region was missing from the original Xist annotation in mouse (7) and is dispensable for gene silencing in transgenic settings (29, 30). Nevertheless, the extent of defined structures within the region suggested functional roles. To test this possibility, we induced expression of full-length Xist or a 14.8-kb transcript lacking the 3′ end from an isogenic site within the β-globin gene locus of a male mouse embryonic stem cell line (31). We found that the half-life of full-length Xist was threefold longer than that of the truncated version (SI Appendix, Fig. S6), consistent with a role for 3′ structured elements in maintaining Xist stability in cells.
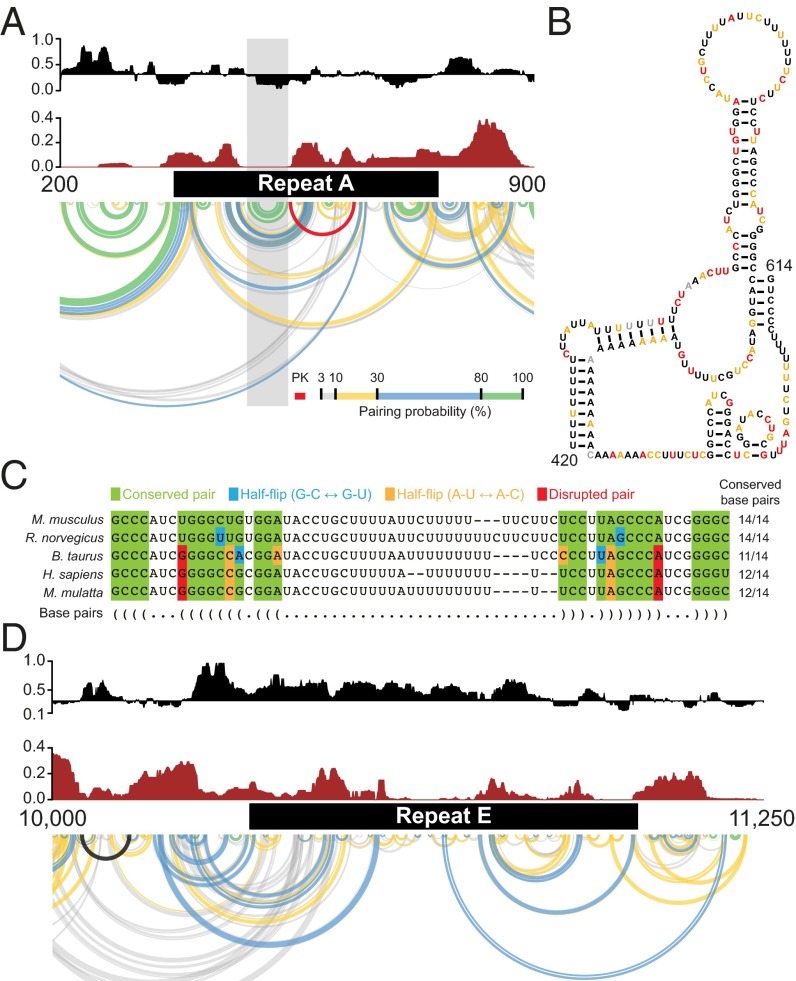
The 400-nt long repeat A region at the 5′ end of Xist is one of the most clearly conserved regions of the RNA (5–7). Repeat A is required for stable accumulation of spliced Xist in cells and for gene silencing (3, 4). In the mouse, repeat A includes seven and one-half copies of a 24-nt repeat unit separated by U-rich spacers of variable lengths. Prior models of this region have emphasized self-contained structures consisting of either small intrarepeat stem-loops (30), large interrepeat structures (12), or a combination of both (10). In contrast, SHAPE data obtained in the context of full-length native Xist indicate that the repeat A region has high Shannon entropy and likely exhibits significant structural variability (Fig. 2_A_). A single hairpin with a GC-rich stem and AU-rich loop that bridges repeats three and four is the only well-defined element in repeat A in our model (Fig. 2 A and B); these nucleotides exhibit high sequence conservation (Fig. 2_C_). Repeat A nucleotides also likely interact with adjacent segments of Xist in the full-length RNA (Fig. 2_A_), and the base of the repeat A stem loop may form a pseudoknot (Fig. 2_B_). Elements of prior repeat A models (10, 12, 30) occur among the structures generated by our ensemble analysis (SI Appendix, Supporting Text); however, high Shannon entropies support the model that this region is structurally dynamic, a feature that may facilitate accessible interaction with protein cofactors.
Fig. 2.
Ex vivo structural features of repeat regions A and E. (A) SHAPE reactivity (black), Shannon entropy (brown), and pairing probabilities (Bottom) for repeat A and the surrounding region. Arcs are color-coded as in Fig. 1. A single high-probability stem-loop structure is predicted to occur within repeat A (gray shading). Xist regions outside of repeat A may interact with this region. (B) Secondary structure of the repeat A stem-loop and predicted pseudoknot. (C) Comparative sequence alignment of the repeat A stem-loop element. Base pairs in mouse are indicated in dot-bracket notation. Nucleotides are color-coded to indicate conservation of base pairing. (D) SHAPE reactivity (black) and Shannon entropy (brown) for the repeat E region. SHAPE reactivities are high and Shannon entropies are low in this region, indicating a high probability of lack of defined structure as illustrated by base pairing probability arcs (Bottom).
High probability pairing regions are predicted to exist in well-defined motifs just upstream (nucleotides 49–352) and downstream (nucleotides ∼850–1,300) of repeat A. These regions have not been genetically disrupted in isolation of repeat A and their role in Xist function is not known. However, these regions bracket the essential repeat A element in Xist and may cooperate with the repeat to encode function in the 5′ end of the lncRNA.
Repeat E, which has no known function, also forms a dynamic and flexible structure. This region spans roughly 1 kb at the beginning of exon 7 and consists of U-rich repeats of 20–25 nt (5). Repeat E exhibits low Shannon entropy and high SHAPE reactivity, indicating that this region is unstructured (Fig. 2_D_). Nucleotides in repeat E are accessible for unencumbered interaction with RNA binding proteins and we will show below that proteins extensively target this element.
Our model also provides structural context for previously characterized Xist mutant phenotypes. For example, a 16-nt insertion located 3′ of repeat A causes a hypomorphic phenotype (32). The insertion falls in the middle of a well-defined hairpin structure with low Shannon entropy (Fig. 1_D_, filled arrowhead). The insertion likely leads to a rearrangement of local structure that affects the biological activity of the repeat A region or attenuates a function of the hairpin itself. A 4-kb inversion of nucleotides 5,984–9,954 leads to a similar hypomorphic phenotype with incomplete silencing (33). This inversion overlaps 14 structural elements in the Xist RNA model (SI Appendix, Fig. S5).
Broad Effects of the Cellular Environment on Xist Structure.
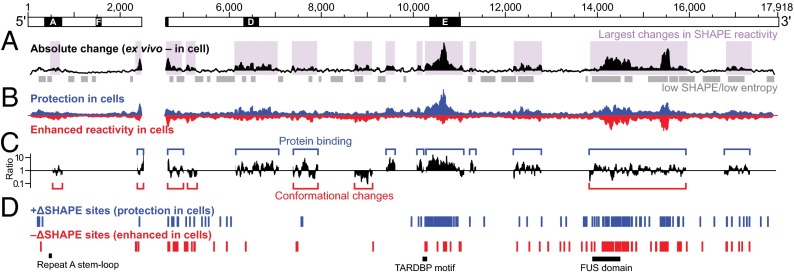
To assess the impact of the cellular environment upon Xist, we probed Xist structure in living cells in biological replicate experiments using the 1M7 SHAPE reagent (SI Appendix, Figs. S1 and S2) and evaluated reactivity changes relative to ex vivo measurements in two complementary ways. First, by searching for regions with an average absolute change greater than the global median, we identified 13–15 regions in each replicate that are strongly affected by the cellular environment (Fig. 3_A_, purple shading). These regions overlap well-defined RNA secondary structure domains and structurally variable regions, and are highly similar between biological replicates (SI Appendix, Fig. S3_A_). Reduced in-cell SHAPE reactivities, relative to the ex vivo state, tend to report direct protein–RNA interactions, whereas increased reactivity in cells are often reflective of RNA conformational changes (17). On this basis, we identified regions of Xist that likely interact with proteins and those that have different structures ex vivo and in cells (Fig. 3 B and C and SI Appendix, Fig. S3 A and B).
Fig. 3.
Effects of the cellular environment on Xist lncRNA structure. (A) Absolute difference between ex vivo and in-cell SHAPE reactivities in 50-nt sliding windows. Purple shading indicates regions with the strongest differences between ex vivo and in-cell reactivity. Repeat E is characterized by a large absolute change, as are regions spanning 6,000–8,000, 12,100–13,400, 13,800–16,000 and 16,700–17,400. Regions with low SHAPE reactivity and low Shannon entropy are indicated with gray bars. (B) Contributions of positive (blue) and negative (red) reactivity differences to the total absolute change. In-cell values were subtracted from ex vivo values, such that positive differences represent reduced reactivity in cells. The sum of the blue and red areas equals the height of the black histogram in A. (C) Ratio between positive and negative reactivity differences within regions of substantial reactivity change. Blue and red brackets indicate regions where protections or enhancements (or both) are most abundant. (D) Positive and negative ΔSHAPE sites. Blue and red sites exhibit protection vs. enhancement in cells and are generally consistent with protein binding and conformational changes, respectively. ΔSHAPE analyses of biological replicates yield similar patterns (SI Appendix, Fig. S3).
Nucleotides in repeat E underwent striking changes in SHAPE reactivity; this region was largely unstructured ex vivo but was very unreactive (and thus structurally constrained) in cells (Fig. 3 A–C and SI Appendix, Fig. S3 A–C). There also appeared to be extensive protein binding to repeat D in cells. There were notable changes in absolute SHAPE reactivity in repeat A, but these were not as strong as those within other regions in Xist, suggesting that repeat A participates in RNA–protein interactions in cells but that these interactions are less stable than those with other Xist motifs. The lack of predicted RNA structure in these repeat regions ex vivo suggests they present relatively unhindered access to proteins.
We also observed large differences between ex vivo and in-cell SHAPE reactivities in regions that span many of the low SHAPE/low Shannon entropy domains in the ex vivo model (Fig. 3 A and B), for example, positions 12,100–13,300, 13,700–16,000, and 16,700–17,300. Regions that exhibit large changes in SHAPE reactivity in cells span nearly the entirety of the Xist RNA, are characterized by multiple distinct features, and are comprised of both structurally variable elements (repeats A, D, and E), and large, structurally well-defined RNA domains.
Localized Cellular Effects on Xist Structure.
Each individual reactivity measurement in a SHAPE-MaP experiment includes an error estimate (15), thus allowing for statistically rigorous analysis of local changes in RNA structure. We have developed a comparison framework (termed ΔSHAPE) that incorporates these error estimates and identifies specific compact sites within an RNA likely to be bound by protein or likely to have distinct conformations under two conditions (17). Thus, ∆SHAPE analysis complements the identification of large-scale structural changes identified above.
In side-by-side analyses of in-cell and ex vivo 1M7 SHAPE probing replicates performed >1 y apart, we identified roughly 200 ΔSHAPE sites at which Xist is strongly impacted by the cellular environment (SI Appendix, Supporting Text). Owing to the stringency of the ∆SHAPE framework, these sites are expected to represent a subset of the strongest Xist interaction sites. Of the ∼200 ΔSHAPE sites identified in each replicate, 43 are shared, and these likely represent extremely stable interactions. We analyzed the global sequence and structural context of ΔSHAPE sites within each replicate in parallel, and observed highly similar overall profiles.
In both replicates, the first 2.5 kb of Xist exhibited very few ΔSHAPE sites, consistent with the occurrence of dynamic or SHAPE-invisible protein interactions within the region, whereas ΔSHAPE sites were abundant in other regions (Fig. 3_D_ and SI Appendix, Fig. S3_C_). We hypothesized that sequences critical to _Xist_–protein interactions may be overrepresented among +ΔSHAPE sites (in which reactivity is lower in cells than ex vivo). We searched these sites for sequence motifs and identified two U-rich sequence motifs, E1 and E2 (SI Appendix, Fig. S2), located in repeat E. No other significant sequence motifs spanning ∆SHAPE sites were identified.
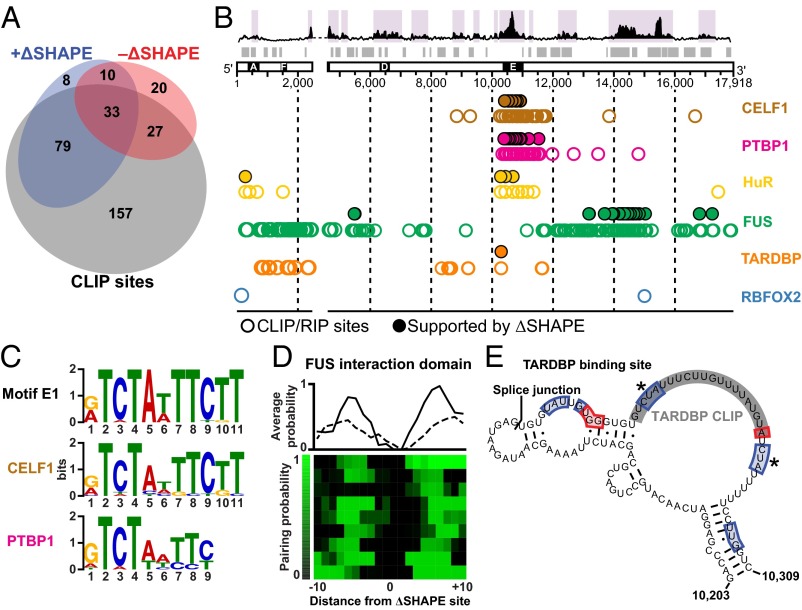
To identify sites in Xist where specific protein interactions occur, we searched for proteins both previously identified as Xist partners in TSCs (34) and present in the CLIPdb protein cross-linking and immunoprecipitation database (35) and identified CELF1, PTBP1, TARDBP, FUS, and RBFOX2 (34, 36, 37). We also performed digestion-optimized RIP-seq experiments in TSCs to identify binding sites for HuR, another _Xist_-interacting protein (34). We expected to find that proteins that bound stably to Xist during our 2-min probing period would perturb the RNA structure and yield clear ∆SHAPE signals. For all proteins except RBFOX2, we identified CLIP or RIP sites that overlapped with positive and negative ΔSHAPE sites in each replicate. We found that, on average, 76% of ΔSHAPE sites overlapped with CLIP or RIP sites, whereas only 53% of the total reported CLIP sites coincided with ΔSHAPE signals (Fig. 4_A_ and SI Appendix, Fig. S3_D_). This latter low number likely reflects differences between cell types, the high stringency used in the ΔSHAPE analysis (17), and the high background of CLIP experiments (38).
Fig. 4.
Distinct classes of _Xist_-protein interactions. (A) Overlap between ΔSHAPE sites and CLIP- or RIP-identified sites; 79% of ΔSHAPE sites overlap CLIP or RIP sites. (B) Locations of CLIP or RIP protein-binding sites that overlap +ΔSHAPE sites. Regions of large absolute change (purple shading) and low SHAPE/low Shannon entropy (gray bars) are highlighted. CLIP- or RIP-defined protein binding sites are shown as open circles; sites confirmed by ∆SHAPE measurements, as filled circles. (C) Sequence motifs identified in CELF1 (Middle) and PTBP1 (Bottom) ΔSHAPE-confirmed sites are similar to the E1 motif (Top). (D) Clustering of pairing probabilities from CLIP-confirmed +ΔSHAPE sites reveal a structure-based preference for FUS binding. Average base-pairing probabilities for the major cluster of +ΔSHAPE-overlapping FUS sites are shown for each replicate (solid and dashed lines). (E) Structural context of the single ΔSHAPE-confirmed TARDBP binding site. The CLIP site is shaded gray. ΔSHAPE sites of in-cell protection and enhancement are boxed in blue and red, respectively. ΔSHAPE sites identified in both replicates are marked with an asterisk. The splice junction between Xist exons 6 and 7 is highlighted.
Given the low false-positive detection rate of protein binding when considering only +ΔSHAPE sites (17), we focused on CLIP sites corroborated by +ΔSHAPE values. We identified sites likely bound by CELF1, PTBP1, and HuR in repeat E, showed that sites for FUS are concentrated in the well-folded RNA domains spanning positions 13,900–15,000, and defined a single site strongly bound by TARDBP at position 10,285 (Fig. 4_B_, filled circles and SI Appendix, Fig. S3_E_). These results were observed independently in both biological replicates. Despite the relatively small number of proteins in our analysis, the data indicate that the 3′ end of Xist is extensively involved in in-cell interactions. This analysis also confirms that repeat E is a major protein-binding platform (Fig. 4_B_).
ΔSHAPE-confirmed CELF1 and PTBP1 CLIP sites are located almost exclusively in repeat E (Fig. 4_B_). These proteins function in RNA processing (39, 40) and may regulate Xist splicing or editing. We used sequence clustering to define consensus motifs from +ΔSHAPE-supported CLIP sites for CELF1 and PTBP1 and found that both overlap with motif E1 (Fig. 4_C_ and SI Appendix, Fig. S3_F_). No strong consensus sequence was identified among non-∆SHAPE-validated CLIP sites, although many fall within repeat E. Thus, CELF1 and PTBP1 likely interact with repeat E in a sequence-specific manner.
We identified HuR-binding sites throughout repeat E (Fig. 4_B_ and SI Appendix, Fig. S3_E_). HuR promotes mRNA stability through interactions with AU-rich elements (AREs) (41). Consistent with an affinity for ARE motifs, HuR was widely detected throughout the U-rich repeat E (SI Appendix, Fig. S7_A_). Searching over subsequences corresponding to +ΔSHAPE in-cell protections returned a U-rich consensus containing elements from motifs E1 and E2 (SI Appendix, Fig. S7_B_). Repeat E may be particularly susceptible to ARE-mediated degradation, and coating this region with proteins, especially HuR, may inhibit RNA decay.
FUS is an abundant, nuclear-enriched protein involved in the regulation of transcription, RNA processing, and DNA damage repair. FUS binds to many RNAs, and its binding has been characterized as promiscuous (42). In contrast to this view, in the context of full-length Xist RNA, +ΔSHAPE signals in CLIP sites indicative of FUS binding cluster strongly at nucleotides 13,000–15,000 in each replicate (Fig. 4_B_ and SI Appendix, Fig. S3_E_). This region has a well-defined RNA structure (Fig. 1 and SI Appendix, Fig. S8) and a mixture of positive and negative ΔSHAPE sites (Fig. 3 C and D and SI Appendix, Fig. S3_C_). We analyzed the pairing probabilities over FUS-associated +ΔSHAPE sites within this region and identified a structural context for FUS binding; FUS-protected nucleotides occur in single-stranded motifs flanked by base-paired structures (Fig. 4_D_ and SI Appendix, Figs. S3_G_ and S8). FUS undergoes RNA-induced multimerization (43), an observation consistent with the complex structural rearrangements detected within the FUS interaction domain when comparing ex vivo and in-cell data. A local increase in FUS concentration via multimerization may lead to cooperative binding, which protects some regions from in-cell SHAPE modification while making others more accessible.
ΔSHAPE analyses support a single major CLIP-identified binding site for the TARDBP protein (Fig. 4_E_ and SI Appendix, Fig. S3_H_). TARDBP is an RNA- and DNA-binding protein with a reported preference for UG-rich sequences; it is both a transcription repressor and a splicing regulator (44). The single TARDBP-binding site in Xist detected by our analyses of both replicate experiments is part of a UG-rich structural motif (positions 10,203–10,309) encompassing the splice junction between exons 6 and 7 (Fig. 4_E_). A threefold reduction in Xist transcript levels has been reported in adult mouse brains depleted of TARDBP via antisense knockdown (45); our analysis of these data further show that levels of incorrectly spliced Xist transcripts increased by twofold (SI Appendix, Fig. S9 A and B), suggesting that TARDBP controls the amount of Xist present in a cell. Although in principle many of the reported CLIP sites for TARDBP are detectable by ∆SHAPE (SI Appendix, Fig. S9), only a single site overlapped with a strong +∆SHAPE signal. The median SHAPE reactivity of this site was much higher than that of any other reported TARDBP CLIP site. These data suggest that the remaining TARDBP sites are occluded by RNA structure or are not sufficiently stable to cause a detectable reduction in SHAPE reactivity when in-cell data and ex vivo data are compared. Most broadly, this analysis indicates that Xist RNA structure can specify a unique accessible protein-binding site.
It is intriguing that the 5′ end of Xist lacks ΔSHAPE sites. The regions near and including repeat A are important for Xist silencing activity (30, 34). We hypothesize that RNA–protein interactions may be less stable here than in other regions, and reanalyzed the ΔSHAPE data with reduced stringency in an attempt to identify potential weaker sites. With these criteria, we identified only four to six additional interaction sites in the first 1,000 nucleotides of Xist (SI Appendix, Fig. S10), suggesting that proteins interact transiently with a dynamic 5′ end or bind to double-stranded elements in such a way as to not exhibit SHAPE reactivity changes.
Conclusion
Comprehensive and quantitative nucleotide-resolution SHAPE-MaP structure probing revealed that Xist consists of multiple domains of well-defined secondary structure linked by structurally variable and dynamic regions (Fig. 1 and SI Appendix, Fig. S5), and supports existing domain-based models for lncRNA function (10, 46, 47). Fully one-half of the Xist lncRNA forms well-defined structure motifs, is significantly impacted by the cellular environment, or both. Structured elements at the 3′ end of Xist appear to function in part by increasing the cellular stability of the transcript. Repeat-containing regions are generally unstructured and are extensively bound by protein cofactors (Figs. 1–4).
We identified three distinct structure-based mechanisms by which protein cofactors form stable interactions with Xist. In each case, protein interactions corroborated by CLIP-seq or RIP-seq and ΔSHAPE data are focused within specific structural elements, and ΔSHAPE signals reveal specific details of these _Xist_–protein interactions. CELF1, PTBP1, and HuR exemplify widespread binding, likely with a degree of sequence specificity, to accessible, unstructured regions. FUS binding occurs in a region with a well-defined structure ex vivo that undergoes extensive rearrangement in cells. TARDBP appears to bind predominantly to a single site presented within a small structural domain. These findings highlight the impressive diversity of lncRNA–protein interactions and their distinct RNA structure-dependent interaction modes.
Cross-referencing of +ΔSHAPE sites with CLIP- and RIP-identified binding sites suggests that quantitative ΔSHAPE analysis is a rigorous approach for identifying stable RNA–protein interaction sites (Fig. 4 and SI Appendix, Fig. S3). Whereas CLIP studies often report binding across the entire transcript, our ΔSHAPE analysis revealed that stable binding sites tend to cluster within the RNA, as was observed for CELF1, PTBP1, and HuR within repeat E and for FUS within the FUS domain. In addition, only when our analysis was limited to +ΔSHAPE sites was a binding motif identified for HuR. ΔSHAPE can also detect site-specific interactions, as were observed for TARDBP. ΔSHAPE analyses of two independent replicates revealed similar overall patterns of protein interaction (SI Appendix, Fig. S3), despite relatively modest correlations for the in-cell experiments (SI Appendix, Fig. S1). Protein-binding events may simply vary between individual Xist ribonucleoprotein complexes, perhaps due to limited access to the lncRNA, low stability of subsets of lncRNA–protein interactions, or limited availability of protein-binding partners (SI Appendix, Supporting Text). ΔSHAPE analysis clearly enables characterization of RNA–protein interactions and examination of RNA structure-mediated recognition in a way that will be broadly useful in future studies of Xist and other lncRNAs as additional protein partners are identified.
This work embraces numerous innovations in quantitative RNA structure probing to define RNA structure, RNA–protein interactions, and the effects of the cellular environment on RNA architecture. Our approach deemphasizes the global minimum free energy structure in regions where multiple structures are likely to be sampled simultaneously and uses experimentally derived metrics to define structural domains. For individual regions with a high propensity to form well-determined stable motifs, we modeled Xist structures using the validated three-reagent differential SHAPE approach (15, 25). Differences between in-cell and ex vivo states were interpreted in the context of robust analysis of measurement errors (17) (SI Appendix, Supporting Text). Limitations are that RNA interactions were constrained to 600-nt windows and canonical base pairing, and there are uncertainties in the thermodynamic parameters used in modeling. Nevertheless, in-cell SHAPE-MaP represents a major advance in converting RNA structure probing from a qualitative tool to a quantitative and predictive tool for understanding RNA biology.
The structured and unstructured domains identified here define maps that are expected to be invaluable in guiding investigations into the mechanisms by which Xist elements contribute to X chromosome inactivation. Xist and other lncRNA transcripts may span kilobases to coordinate long-range protein and domain interactions (Figs. 3 and 4) that ultimately enable orchestration of epigenetic regulation on the kilobase to megabase scales (1, 3, 4). Many lncRNAs are likely to share features identified here for Xist, including densely arrayed secondary structural features, multiple distinctive modes of protein interaction, and the ability to serve as multidomain organizers of cellular function.
Methods
In-cell modification was carried out by treating mouse TSCs in fresh growth medium with 1M7 (10 mM final) and incubating at 37 °C for 5 min before RNA isolation. For ex vivo analyses, total cellular RNA was gently extracted from TSCs into RNA folding buffer (100 mM Hepes, pH 8.0, 100 mM NaCl, and 10 mM MgCl2), incubated at 37 °C for 20 min, and subjected to SHAPE modification with 1M7, 1M6, or NMIA. RNA was subjected to MaP reverse transcription (15) using _Xist_-specific primers, followed by _Xist_-specific PCR amplification and high-throughput sequencing library construction. SHAPE reactivities were calculated from raw sequencing reads using ShapeMapper, and secondary structures were modeled using SuperFold (15). Detailed descriptions of in-cell RNA probing, library construction, structure modeling, and bioinformatics analyses are provided in SI Appendix, Methods.
Supplementary Material
Supplementary File
Acknowledgments
We thank Dirk Schübeler and Oliver Bell for generously sharing the Hy/TK embryonic stem cell line and Kathrin Plath for sharing the pSM33 line. This work was supported by National Institutes of Health (NIH) Grant GM064803 and National Science Foundation (NSF) Grant MCB-1121024 (to K.M.W.), NSF Grant MCB-0842621 and NIH Grant CA157268 (to J.D.K.), and laboratory start-up funds provided by the Lineberger Comprehensive Cancer Center (to J.M.C.). M.J.S. is an NSF Graduate Research Fellow (Grant DGE-1144081) and was supported in part by an NIH training grant in molecular and cellular biophysics (Grant T32 GM08570). T.W.C. was supported in part by an NIH training grant in bioinformatics and computational biology (Grant T32 GM067553). D.M.L. was supported in part by an NIH training grant in genetics and molecular biology (Grant T32 GM007092).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequencing data have been deposited in the Sequence Read Archive (accession no. SRP074108). Processed data are available in the SI Appendix and at www.chem.unc.edu/rna.
References
- 1.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 3.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Gendrel A-V, Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol. 2014;30:561–580. doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- 5.Nesterova TB, et al. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001;11(5):833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown CJ, et al. The human XIST gene: Analysis of a 17-kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 7.Brockdorff N, et al. The product of the mouse Xist gene is a 15-kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 8.Yamada N, et al. Xist exon 7 contributes to the stable localization of Xist RNA on the inactive X chromosome. PLoS Genet. 2015;11(8):e1005430. doi: 10.1371/journal.pgen.1005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caparros M-L, Alexiou M, Webster Z, Brockdorff N. Functional analysis of the highly conserved exon IV of XIST RNA. Cytogenet Genome Res. 2002;99(1-4):99–105. doi: 10.1159/000071580. [DOI] [PubMed] [Google Scholar]
- 10.Fang R, Moss WN, Rutenberg-Schoenberg M, Simon MD. Probing Xist RNA structure in cells using targeted structure-seq. PLoS Genet. 2015;11(12):e1005668. doi: 10.1371/journal.pgen.1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duszczyk MM, Wutz A, Rybin V, Sattler M. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA. 2011;17(11):1973–1982. doi: 10.1261/rna.2747411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maenner S, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8(1):e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noller HF, Woese CR. Secondary structure of 16S ribosomal RNA. Science. 1981;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- 14.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460(7256):711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11(9):959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavender CA, Gorelick RJ, Weeks KM. Structure-based alignment and consensus secondary structures for three HIV-related RNA genomes. PLOS Comput Biol. 2015;11(5):e1004230. doi: 10.1371/journal.pcbi.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smola MJ, Calabrese JM, Weeks KM. Detection of RNA–protein interactions in living cells with SHAPE. Biochemistry. 2015;54(46):6867–6875. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGinnis JL, et al. In-cell SHAPE reveals that free 30S ribosome subunits are in the inactive state. Proc Natl Acad Sci USA. 2015;112(8):2425–2430. doi: 10.1073/pnas.1411514112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52(48):8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGinnis JL, Weeks KM. Ribosome RNA assembly intermediates visualized in living cells. Biochemistry. 2014;53(19):3237–3247. doi: 10.1021/bi500198b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mauger DM, et al. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci USA. 2015;112(12):3692–3697. doi: 10.1073/pnas.1416266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabrese JM, et al. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell. 2012;151(5):951–963. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mugford JW, Yee D, Magnuson T. Failure of extra-embryonic progenitor maintenance in the absence of dosage compensation. Development. 2012;139(12):2130–2138. doi: 10.1242/dev.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129(14):4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 25.Rice GM, Leonard CW, Weeks KM. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA. 2014;20(6):846–854. doi: 10.1261/rna.043323.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajdin CE, et al. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci USA. 2013;110(14):5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y, Jamison DC. The distribution of SNPs in human gene regulatory regions. BMC Genomics. 2005;6:140. doi: 10.1186/1471-2164-6-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5(4):695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 30.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30(2):167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 31.Lienert F, et al. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43(11):1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 32.Hoki Y, et al. Incomplete X-inactivation initiated by a hypomorphic Xist allele in the mouse. Development. 2011;138(13):2649–2659. doi: 10.1242/dev.061226. [DOI] [PubMed] [Google Scholar]
- 33.Senner CE, et al. Disruption of a conserved region of Xist exon 1 impairs Xist RNA localisation and X-linked gene silencing during random and imprinted X chromosome inactivation. Development. 2011;138(8):1541–1550. doi: 10.1242/dev.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161(2):404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y-CT, et al. CLIPdb: A CLIP-seq database for protein–RNA interactions. BMC Genomics. 2015;16:51. doi: 10.1186/s12864-015-1273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minajigi A, et al. Chromosomes: A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349(6245):aab2276. doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley KJ, Steitz JA. The “observer effect” in genome-wide surveys of protein–RNA interactions. Mol Cell. 2013;49(4):601–604. doi: 10.1016/j.molcel.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barreau C, Paillard L, Méreau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: Molecular characteristics and biological functions. Biochimie. 2006;88(5):515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21(10):3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65(20):3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Schwartz JC, Cech TR. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015;43(15):7535–7543. doi: 10.1093/nar/gkv679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Reports. 2013;5(4):918–925. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19(R1):R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14(4):459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40(11):5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Somarowthu S, et al. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58(2):353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File