A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1 (original) (raw)
Abstract
Lv1 restriction of HIV-1 in the cells of Old World monkeys is associated with the expression of the Trim5 gene. Uniquely, in owl monkey kidney cells, HIV-1 restriction is dependent on the ability of incoming viral capsid protein to bind cyclophilin A (CypA). Cloning of the owl monkey Trim5 gene now reveals the presence of an inserted CypA pseudogene within intron 7 of the Trim5 gene. This insertion results in the formation of a chimeric Trim5-CypA transcript. Transfer of a cDNA corresponding to this transcript into human cells confers cyclosporin A-sensitive resistance to HIV-1 infection. The restriction factor appears to be a chimeric protein created by retrotransposon-mediated exon shuffling.
There is increasing evidence that mammalian cells possess a variety of innate barriers to retroviral infection. These include receptor variation (1) or blockade (2), enzymes that degrade retroviral nucleic acids (3), and a class of so-called restriction factors that block one or more postentry, preintegration steps in the viral life cycle (4). To infect a cell, a retrovirus must either sidestep the defense mechanisms or encode activities to overcome them (5).
Retroviral restriction factors interact with the retroviral capsid protein (CA) in a specific and saturable manner and appear to prevent reverse transcription and/or transit of newly synthesized DNA to the cell nucleus (4). The mouse Fv1 gene was the first of these factors to be described and has two alleles, Fv1n and Fv1b, targeting B- and N-tropic murine leukemia viruses (MLV), respectively (6-8). Viral tropism is determined mainly by MLV CA amino acid 110 (9). Subsequently, it was shown that a number of other species, including humans, possess an activity, Ref1, which specifically restricts N-tropic but not B-tropic MLV (10).
At the same time, studies of lentivirus replication provided evidence for the role of CA in determining replication potential in cells derived from a variety of primates (11). Further studies provided evidence for the presence of a dominant factor, resembling Ref1, showing restriction of a variety of lentiviruses (12-14). This factor was termed Lv1. This activity shows species specificity in the profiles of viruses restricted (4, 15). Lv1 from different species can also be distinguished by the effects of treatments influencing the interaction between HIV-1 and the cellular protein cyclophilin A (CypA). CA from HIV-1, but not simian immunodeficiency virus, specifically binds CypA (16), an interaction that can be prevented by the drug cyclosporin A (CsA) or by specific mutations in CA such as G89V (17-20). The roles of CypA binding in HIV-1 assembly and entry remain poorly defined, but one attractive possibility is that CypA binding to CA prevents Lv1 restriction in human cells (21). By contrast, HIV-1 restriction in owl monkey kidney (OMK) cells appears to be absolutely dependent on CypA binding because CsA treatment or introduction of the G89V mutation abolishes restriction in these cells (21).
Recently, Sodroski et al. (22) demonstrated that the α isoform of the Trim5 gene from rhesus monkey, but not human, could restrict HIV-1, thereby suggesting Trim5α as a candidate for Lv1 activity. Subsequently, three groups showed that Trim5α from various species inhibited lentivirus and N-tropic MLV replication, with the expected virus restriction profiles (23-25). However, Trim5 from owl monkey was not cloned in these studies. Here we report experiments to examine the owl monkey gene. We show that the owl monkey Trim5 locus encodes a Trim5-CypA fusion protein, thereby providing a rationale for the absolute requirement for CypA binding in HIV-1 restriction by OMK cells.
Materials and Methods
Restriction Assays. Assays for restriction activity were carried out as described by performing two-color fluorescence-activated cell sorting (FACS) analysis (25-27).
Cells and Viruses. Cells, culture conditions, and viruses were as described in ref. 25.
Molecular Cloning. cDNAs corresponding to differently spliced variants of transcripts from the OMK Trim5 locus were cloned by using RT-PCR. mRNA was isolated from OMK cells by using the Oligotex direct mRNA minikit (Qiagen, Crawley, U.K.), reverse-transcribed, and PCR-amplified by using the primers described below. PCR products were cloned into the pENTR/TOPO vector (Invitrogen), sequenced, and transferred to pLGatewayIY (25). Trim5-V1, -V2, and -V3 were isolated after nested PCR by using a variation of the 3′ rapid amplification of cDNA ends (3′ RACE) technique (28); 100 ng of OMK mRNA was reverse-transcribed by using 3′ CDS primer A (Smart RACE cDNA amplification kit, Clontech) and AMV RT (Roche Applied Science). A 3′ RACE reaction was performed by using the Expand High Fidelity PCR system (Roche Applied Science), the F-RACE primer (CACCATGGCTTCTGGAATCCTGGTTAATGTAAAGG), and the Nested Universal Primer A (Clontech). One microliter of the PCR product was reamplified by using PfuUltra DNA polymerase (Stratagene) and the Trim5α-F (CACCATGGCTTCTGGAATCCTGGTTAATG) and Trim5α-R (TCAAGAGCT TGGTGAGCACAGAGTCATGGG) primers. Trim5-V4 and Trim5-V5 were synthesized with Trim5α-F in combination with CypA-R (T TAT TCGAGT TGTCCACAGTCAGC) (based on the human CypA sequence), using PfuUltra DNA polymerase. Trim5-V6 was derived from Trim5-V2 by deletion of CypA sequences in two rounds of PCR. First, with Trim5-V2 as a template, the 5′ and 3′ ends were synthesized with Trim5α-F plus Trim5α-R1 (GACAGA AGTCCA ACGCTACTGGGTGCAGCTCATGTGACACTGGTTCCAAG) and Trim5α-F1 (GACAGAAGTCCA ACGCTACTGGGCAGCTCATGTGACACTGGT TCCAAG) plus TRIM5α-R primers, respectively. The two DNA fragments were then used to prime a joining reaction by amplification with Trim5α-F and Trim5α-R.
Owl monkey genomic DNA prepared from OMK cells and a sample of whole blood (Aotus trivirgatus, cryopreserved since 1994) was purified by using Qiagen kits. A genomic DNA clone containing the 3′ end of Trim5 exon 7, the 5′ end of exon 8, and intervening sequences was obtained by PCR using F3gen (CCGACAGAAGTCCAACGCTACTGG) and R3gen (TGACAGTACATGAAGGGTGACTTGG) primers and KOD HiFi polymerase (Novagen) (30 cycles; 15 sec at 95°C, 30 sec at 62°C, and 30 sec at 72°C). Other genomic PCRs used F3gen with CypA-R (ACCCAAAGCGCTCCATGG-CCTCCAC) and CypA-F5 (GAGAACTTCATCCTAAAGCATACAGG) with R3gen. Sequences have been deposited in GenBank.
GST Pull-Down. The CA protein of HIV-1 was PCR-amplified by using pNL4.3 with primers CA-F (GGGAATTCATGCCTATAGTGCAGAACCTCCAGGGG) and CA-R (GGCTCGAGTCACAAAACTCTTGCTTTATGGGCCGGG) and cloned into pGEX6P1 (Amersham Biosciences, Chalfont St. Giles, U.K.) between the _Eco_RI and _Xho_I restriction sites to generate a GST-CA fusion. The resulting plasmid was introduced into competent BL21(DE3) Escherichia coli (One Shot, Invitrogen). Two-hundred-milliliter cultures were induced for 3 h with 1 mM isopropyl β-D-thiogalactoside, and cell extracts were prepared by resuspending cell pellets in 5 ml of Yeast Protein Extraction Reagent (Pierce), 5 mM MgCl2, and 125 units of Benzonase nuclease (Novagen), and incubating for 5 min at room temperature and 30 min on a spinning wheel at 4°C, followed by centrifugation at 30,000 × g for 30 min. Trim-V2, -V4, and -V6 were subcloned into pEXP2-DEST Gateway (Invitrogen) from the pENTR constructs by recombination using Gateway LR Clonase (Invitrogen). Two micrograms of plasmid DNA was used for in vitro transcription/translation with the TNT coupled reticulocyte lysate system (Promega) labeling the protein product with Redivue L-[35S]methionine (Amersham Biosciences). One milliliter of glutathione Sepharose 4B beads (Amersham Biosciences) was incubated overnight at 4°C with 500 μl of crude bacterial extract (corresponding to ≈500 μg of GST-CA). The beads were washed three times with 10 ml of GST pull-down buffer (20 mM Tris, pH 8.0/100 mM NaCl/5 mM MgCl2/0.5% Nonidet P-40/5% glycerol/1 mM EDTA/10 μg/ml BSA). Fifteen microliters of _in vitro-_translated TRIM5-V2, -V4, or -V6 was incubated with 50 μl of beads in a final volume of 1.5 ml of GST pull-down buffer for 1 h at 4°C. Beads were then washed five times in GST pull-down buffer and resuspended in 30 μl of SDS/PAGE loading buffer for 10 min at 95°C. Eluates were run on a 10% polyacrylamide gel. Before autoradiography, overnight at -80°C, gels were incubated for 30 min in 50% methanol/10% acetic acid, for 30 min in Amplify reagent (Amersham Biosciences), and for 5 min in 7% methanol/7% acetic acid/1% glycerol, and dried for 1 h at 80°C.
Results
Presence of a CypA Pseudogene in the Owl Monkey Trim5 Locus. There is now good evidence that the Trim5 gene encodes Lv1 activity in a variety of Old World monkeys (22-25). However, the OMK restriction factor shows a unique pattern of HIV-1 restriction. It does not affect HIV-1 modified to prevent CypA binding, and restriction is abolished by the treatment of cells with CsA (21). We therefore asked whether Trim5 was also responsible for the restriction activity of OMK cells. Our previous attempts to clone the OMK Trim5 gene by using RT-PCR were unsuccessful (25). Therefore, we used a variant of the 3′ RACE technique for this purpose. Products from this reaction were cloned and sequenced. Among the first clones screened, we identified one product showing a fusion between Trim5 and CypA sequences (Trim5-V1; see below).
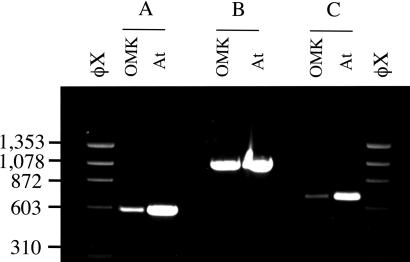
Using the cDNA sequence, we designed primers to OMK DNA corresponding to exons 7 and 8 of the human Trim5α locus. PCRs with genomic DNA isolated from OMK cells and from a sample of A. trivirgatus blood revealed a single product of 1.1 kb (Fig. 1), ≈0.55 kb longer than the predicted corresponding product with human DNA. Reactions with the exon 7 primer plus a CypA reverse primer, and a CypA forward primer with the exon 8 primer both gave bands of about 0.6 kb. Sequencing of the 1.1-kb product revealed the presence of an intact CypA ORF (Fig. 2). This ORF has all of the hallmarks of a processed pseudogene (29). An intact, intronless CypA ORF is followed by a 3′ untranslated region with a short A-rich sequence located 18 nt after a potential polyadenylation signal. The CypA sequences are flanked by a 15-nt duplication that appears only once in the corresponding human Trim5 intron. Potential splice-acceptor and splice-donor sequences were also identified within the pseudogene. Together, these observations suggested the possibility of a Trim5-CypA fusion protein that might provide an elegant explanation for the unique CypA dependence of HIV-1 restriction in OMK cells.
Fig. 1.
Detection of CypA sequences within the OMK Trim5 gene. PCR products with genomic DNA from OMK cells and A. trivirgatus whole blood (At). (A) Primers, Trim5-F3gen (exon 7) plus CypA-R1. (B) Primers, Trim5-F3gen (exon 7) plus Trim5-R3gen (exon 8). (C) Primers, CypA-F1 plus Trim5-R3gen (exon 8). Markers, _Hae_III-digested φX174 DNA.
Fig. 2.
Sequence of the CypA insert. The genomic sequence of OMK between exons 7 and 8 of the Trim5 gene is shown. The CypA insert is in capitals, with the normal CypA ORF underlined and translation start and stop sites in bold. Dotted underlining indicates the sequences corresponding to the 11 additional amino acids found in Trim-V2 and -V4. Sequences homologous to the human intronic sequences are in lowercase, with the insertion-associated duplication boxed. The ends of Trim5 exons 7 and 8 are shown in bold capitals. Potential splice-donor (SD) and splice-acceptor (SA) sites are indicated.
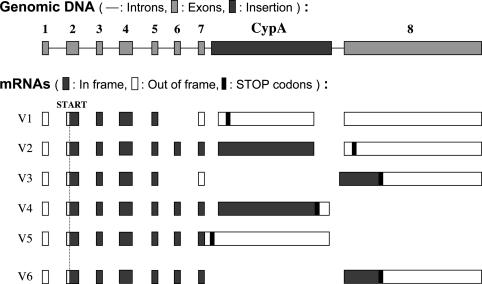
Analysis of Trim5-CypA Fusion Transcripts. The first Trim5-CypA mRNA cloned, Trim5-V1 (Fig. 3) would not be predicted to yield an in-frame fusion protein because of the absence of Trim5 exon 6. We therefore screened our collection of RACE products further. This screen led to the identification of two further variants, Trim5-V2 and -V3. V2 would be predicted to yield a fusion protein containing amino acids 1-299 of Trim5α, followed by 11 aa derived from the 5′ untranslated region of CypA plus amino acids 1-147 of CypA. However, it lacks the last 17 aa of CypA; these are replaced by 16 aa derived from exon 8 of Trim5 but in a different reading frame. V3 has exons 1-5 of Trim5, exon 7 read in a different frame, five nucleotides of unknown origin, followed by the Trim5 exon 8.
Fig. 3.
Structure and transcripts from the OMK Trim5 gene. The structure of the Trim5 locus of OMK cells is illustrated at the top of the figure (not to scale). Below are the deduced structures of five naturally occurring spliced processed RNAs (V1-V5) and one potential transcript generated by recombinant techniques (V6). In frame, predicted translation in the normal Trim5 or CypA reading frame; Out of frame, use of an alternative frame.
To clone a variant containing all of the CypA coding sequences, we used RT-PCR with primers directed to the 5′ end of Trim5 and the 3′ end of the CypA ORF. Two products, Trim-V4 and -V5, were obtained in this way. V4 would be predicted to yield a fusion protein analogous to V2 but with the CypA C-terminal residues. A comparison of the predicted amino acid sequences of proteins encoded by Trim-V4 and -V2 is shown in Fig. 4. V5 is an incompletely spliced version of V4. Whether other spliced variants are made in vivo is not clear. One possibility is that the CypA sequences are removed by normal splicing between exons 7 and 8 of the Trim5 gene. We have been unable to detect such a product, but for further functional analyses, we generated such a cDNA in vitro, Trim5-V6, by PCR-mediated deletion of CypA-derived sequences from Trim5-V2.
Fig. 4.
Predicted sequence of Trim5-CypA fusion proteins. The predicted amino acid sequence of Trim4-V4 is shown with CypA-derived sequences underlined. Trim5-V2 is identical to Trim5-V4 for amino acids 1-457; the final 16 residues are indicated in the bottom line in lowercase.
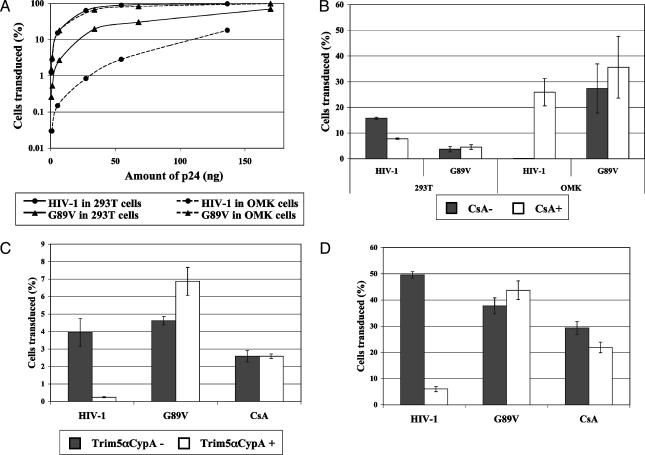
Trim5-CypA Restriction of HIV-1. We next set out to examine the restriction properties of the various cloned Trim5 cDNAs from OMK cells. For comparison, the patterns of natural restriction in OMK cells were also examined. Virus titration on human 293T and OMK cells showed that HIV-1 is significantly restricted in OMK, compared with human cells, an inhibitory effect that can be overcome by introduction of the G89V mutation (Fig. 5_A_). CsA treatment shows slight inhibition of HIV-1 on human cells but abolishes restriction in OMK cells (Fig. 5_B_). CsA has little effect on the virus with G89V. These results confirm those of earlier reports (13, 21).
Fig. 5.
Restriction activity of Trim5-CypA. (A) Titration of wild-type HIV-1 and G89V mutant on human 293T cells and OMK cells. HIV-1 and G89V titers are represented by circles and triangles, respectively, and titers in 293T and OMK cells are represented by smooth and dashed lines, respectively. (B) Effect of CsA on HIV-1 titers. Human 293T cells and OMK cells were transduced with HIV-1 or G89V (5 ng of P24) carrying the enhanced GFP marker, and the percentage of transduced cells was determined by FACS 48 h posttransduction. CsA (3 μg/ml) was added 2 h before transduction. White bars, CsA-treated cells; gray bars, untreated cells. Shown is the average of two different experiments. (C) HT1080 cells transduced with Trim5α-CypA were challenged with 1 × 104 infectious units of tester virus carrying enhanced GFP 2 days posttransduction. The percentage of Trim5-CypA-positive cells that was transduced by the tester virus is compared with the percentage in Trim5-CypA-negative cells. Means and standard deviation of four different experiments are shown. White bars, results with Trim5-CypA-positive cells; gray bars, Trim5-CypA-negative cells. (D) As in C, but with 1 × 105 infectious units of tester virus.
To test the cloned cDNAs, we used a two-color FACS assay originally developed for the study of Fv1 (26, 27) and adapted for HIV-1 (25). Human HT1080 cells were transduced with an MLV-based vector encoding the gene to be examined and yellow fluorescent protein, then challenged with tester viruses encoding enhanced GFP. The effect of the introduced gene can be assessed by comparing the proportion of GFP positive cells in the cell populations with and without yellow fluorescence.
The introduction of Trim5-V4, the variant carrying exons 1-7 of Trim5 fused to the intact CypA ORF, into human HT1080 cells resulted in the inhibition of both low and high doses of HIV-1 tester virus (Fig. 5 C and D). No restriction was seen of G89V or of wild-type HIV-1 in the presence of CsA. Trim5-V4 expression thereby recapitulates the restriction properties of Lv1 in OMK cells. By contrast, Trim5-V2, which differs only in the very C-terminal region of the predicted protein, showed no effect on HIV-1 replication (data not shown). Interestingly, Trim5-V6, the equivalent of the Trim5α gene present in other species of primate, also showed no restriction activity against either HIV-1 or MLV (data not shown), presumably as a result of sequence divergence.
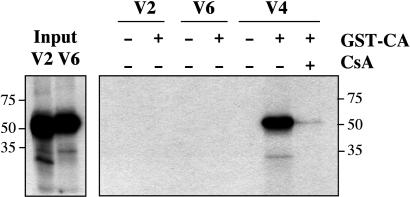
Trim5-CypA Interaction with CA. CypA binds to HIV-1 CA protein (16). To test whether the Trim5-CypA fusion proteins encoded by Trim5-V2 and -V4 were also capable of binding HIV-1 CA, we incubated GST-tagged CA with _in vitro_-synthesized 35S-labeled Trim5-CypA and tested for binding to glutathione-coated beads. Fig. 6 illustrates the result on one such experiment. Restriction-competent Trim5-V4 binds CA; binding is abolished by CsA addition. By contrast, Trim5-V2, which does not restrict HIV-1, showed no CA binding. Trim5-V6, equivalent to intact OMK Trim5α, also showed no CA binding.
Fig. 6.
Trim5-CypA binds HIV-1 CA. Autoradiograms of gels from a GST pull-down experiment with _in vitro-_transcribed/translated Trim5-V2, -V4, and -V6. (Left) Gel with the products of the in vitro transcription/translation reactions for Trim5-V2 and -V6. (Right) Products precipitated by incubation with GST-CA and glutathione-coated beads. CsA was added to 20 μg/ml where indicated.
Discussion
In this study, we show that a processed CypA pseudogene is present within the Trim5 locus of OMK cells and provide evidence that a chimeric Trim5-CypA protein is responsible for Lv1 restriction in OMK cells. Recently, others have reported similar findings (30). As is the case in OMK cells, Trim5-CypA-mediated restriction appears dependent on a CypA-CA interaction. An alternatively processed variant, encoded by Trim5-V2, which differs only at the extreme C terminus of the predicted protein, neither binds CA nor restricts HIV-1. The structure of CypA provides a plausible explanation for this finding; CypA appears to show an eight-stranded antiparallel β-barrel structure (31). Changes in the final β-sheet are therefore likely to disrupt the whole structure. The observed lack of restriction activity in Trim5-V6-transduced cells implies that even if full-length Trim5α protein is made in OMK cells, it does not contribute to HIV-1 restriction. In addition, siRNA to Trim5 has been reported to inhibit HIV-1 replication in OMK cells (23, 30). Taken together, these observations imply that Trim5-CypA is the primary factor responsible for HIV-1 restriction in OMK cells.
Intact Trim5α appears to be constructed in a modular fashion with the domains comprising the tripartite motif followed by a SPRY domain (32). In the Trim5-CypA chimera, the CypA residues almost precisely replace the SPRY domain. Restriction specificity of Trim5α from human, rhesus and African green monkey for HIV-1 and MLV maps to the SPRY domain (M.W.Y., S.N., and J.P.S., unpublished work). It therefore seems likely that the CypA insertion results in the exchange of one virus-binding domain with another. The Trim5-CypA fusion protein might therefore be valuable for studying the role in restriction of the remaining portion of the Trim protein, not least by providing an ideal unrestricted control, G89V, in studies designed to follow incoming virus.
The inserted CypA sequences present within intron 7 of the OMK Trim5 gene appear to represent a typical processed pseudogene, lacking introns but containing a 3′ poly(A) tract and generating a duplication in insertion site sequence (29). This pseudogene presumably resulted from L1-mediated retransposition of an authentic CypA transcript (33). This insertion does not appear to have occurred during the growth on OMK cells in culture because the pseudogene was also present in genomic DNA from a sample of frozen whole blood taken from an owl monkey. Both OMK cells and blood came from members of the A. trivirgatus species; it will be of interest to examine the distribution of the Trim5-CypA gene in more members of the Aotus genus from different geographic locations.
Owl monkeys are normally found in Central and South America. It therefore seems unlikely that they would have been exposed to primate lentiviruses, and it is unclear whether any selective advantage might be conferred by acquisition of the hybrid gene. However, even though CypA is a very common pseudogene (34), it seems improbable that the chimeric ORF would be maintained without selection unless it is a very recent insertion.
Over the course of evolution, mammals have accumulated tens of thousands of pseudogenes (34, 35). Although the process of retrotransposition may have been necessary for genome evolution (36), the vast majority of existing pseudogenes are considered as “junk” DNA. Although some are transcribed, many have accumulated mutations that would preclude function, and only a very small number may play physiological roles (37-39). Indeed, even though exon shuffling may have been of great importance in the evolutionary past (40, 41), the Trim5-CypA protein appears to belong to a tiny handful of functional proteins unambiguously created in this manner.
Acknowledgments
We thank Graham Mitchell for the gift of owl monkey blood and Graham Preece and Chris Atkins for their assistance with FACS. This work was supported by the Medical Research Council and by EMBO Long-Term Fellowship ALTF 343-2001 (to S.N.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: OMK, owl monkey kidney; CsA, cyclosporin A; CypA, cyclophilin A; MLV, murine leukemia virus; FACS, fluorescence-activated cell sorter; 3′ RACE, rapid amplification of 3′ cDNA ends.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY684991-AY684995 and AY684997).
References
- 1.Marin, M., Tailor, C. S., Nouri, A., Kozak, S. L. & Kabat, D. (1999) J. Virol. 73**,** 9362-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner, M. B., Kozak, C. A. & O'Brien, S. J. (1991) Trends Genet. 7**,** 22-27. [DOI] [PubMed] [Google Scholar]
- 3.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. V., Watt, I. N., Neuberger, M. S. & Malim, M. H. (2003) Cell 116**,** 803-809. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. (2003) Trends Microbiol. 11**,** 286-291. [DOI] [PubMed] [Google Scholar]
- 5.Nisole, S. & Saib, A. (2004) Retrovirology 1**,** 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lilly, F. (1970) J. Natl. Cancer Inst. 45**,** 163-169. [PubMed] [Google Scholar]
- 7.Hartley, J. W., Rowe, W. P. & Huebner, R. J. (1970) J. Virol. 5**,** 221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Best, S., Le Tissier, P., Towers, G. & Stoye, J. P. (1996) Nature 382**,** 826-829. [DOI] [PubMed] [Google Scholar]
- 9.Kozak, C. A. & Chakraborti, A. (1996) Virology 225**,** 300-306. [DOI] [PubMed] [Google Scholar]
- 10.Towers, G., Bock, M., Martin, S., Takeuchi, Y., Stoye, J. P. & Danos, O. (2000) Proc. Natl. Acad. Sci. USA 97**,** 12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann, W., Schubert, D., LaBonte, J., Munson, L., Gibson, S., Scammell, J., Ferrigno, P. & Sodroski, J. (1999) J. Virol. 73**,** 10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Besnier, C., Takeuchi, Y. & Towers, G. (2002) Proc. Natl. Acad. Sci. USA 99**,** 11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowan, S., Hatziioannou, T., Cunningham, T., Musing, M. A., Gottlinger, H. G. & Bieniasz, P. D. (2002) Proc. Natl. Acad. Sci. USA 99**,** 11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Münk, C., Brandt, S. M., Lucero, G. & Landau, N. R. (2002) Proc. Natl. Acad. Sci. USA 99**,** 13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoye, J. P. (2002) Proc. Natl. Acad. Sci, USA 99**,** 11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luban, J., Bossolt, K. L., Franke, E. K., Kalpana, G. V. & Goff, S. P. (1993) Cell 73**,** 1067-1078. [DOI] [PubMed] [Google Scholar]
- 17.Franke, E. K., Yuan, H. E. H. & Luban, J. (1994) Nature 372**,** 359-362. [DOI] [PubMed] [Google Scholar]
- 18.Thali, M., Bukovsky, A., Kondo, E., Rosenwirth, B., Walsh, C. T., Sodroski, J. & Göttlinger, H. G. (1994) Nature 372**,** 363-365. [DOI] [PubMed] [Google Scholar]
- 19.Braaten, D., Franke, E. K. & Luban, J. (1996) J. Virol. 70**,** 4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo, S., Myszka, D. G., Yeh, C.-Y., McMurray, M., Hill, C. P. & Sundquist, W. I. (1997) J. Mol. Biol. 269**,** 780-795. [DOI] [PubMed] [Google Scholar]
- 21.Towers, G. J., Hatziioannou, T., Cowan, S., Goff, S. P., Luban, J. & Bieniasz, P. D. (2003) Nat. Med. 9**,** 1138-1143. [DOI] [PubMed] [Google Scholar]
- 22.Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissler, P. & Sodroski, J. (2004) Nature 427**,** 848-853. [DOI] [PubMed] [Google Scholar]
- 23.Hatziioannou, T., Perez-Caballero, D., Yang, A., Cowan, S. & Bieniasz, P. D. (2004) Proc. Natl. Acad. Sci. USA 101**,** 10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keckesova, Z., Ylinen, L. M. J. & Towers, G. J. (2004) Proc. Natl. Acad. Sci. USA 101**,** 10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yap, M. W., Nisole, S., Lynch, C. & Stoye, J. P. (2004) Proc. Natl. Acad. Sci. USA 101**,** 10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bock, M., Bishop, K. N., Towers, G. & Stoye, J. P. (2000) J. Virol. 74**,** 7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop, K. N., Bock, M., Towers, G. & Stoye, J. P. (2001) J. Virol. 75**,** 5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frohman, M. A., Dush, M. K. & Martin, G. R. (1988) Proc. Natl. Acad. Sci. USA 85**,** 8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiner, A. M., Deininger, P. L. & Efstradiadis, A. (1986) Annu. Rev. Biochem. 55**,** 631-661. [DOI] [PubMed] [Google Scholar]
- 30.Sayah, D. M., Sokolskaja, E., Berthoux, L. & Luban, J. (2004) Nature 430**,** 569-573. [DOI] [PubMed] [Google Scholar]
- 31.Kallen, J., Spitzfaden, C., Zurini, M. G. M., Wilder, G., Wilder, H., Wüthrich, K. & Walkinshaw, M. D. (1991) Nature 353**,** 276-279. [DOI] [PubMed] [Google Scholar]
- 32.Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., et al. (2001) EMBO J. 20**,** 2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esnault, C., Maestre, J. & Heidmann, T. (2000) Nat. Genet. 24**,** 363-367. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Z., Harrison, P. M., Liu, Y. & Gerstein, M. (2003) Genome Res. 13**,** 2541-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrents, D., Suyama, M., Zdobnov, E. & Bork, P. (2003) Genome Res. 13**,** 2559-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kazazian, H. H., Jr. (2004) Science 303**,** 1626-1632. [DOI] [PubMed] [Google Scholar]
- 37.Hirotsune, S., Yoshida, N., Chen., A., Garrett, L., Suglyama, F., Takahashi, S., Yagami, K.-I., Wynshaw-Boris, A. & Yoshiki, A. (2003) Nature 423**,** 91-100. [DOI] [PubMed] [Google Scholar]
- 38.Korneev, S. A., Park, J.-H. & O'Shea, M. (1999) J. Neurosci. 19**,** 7711-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long, M. & Langley, C. H. (1993) Science 260**,** 91-95. [DOI] [PubMed] [Google Scholar]
- 40.Long, M., de Souza, S. J. & Gilbert, W. (1995) Curr. Opin. Genet. Dev. 5**,** 774-778. [DOI] [PubMed] [Google Scholar]
- 41.Long, M., Deutsch, M., Wang, W., Betran, E., Brunet, F. G. & Zhang, J. (2003) Genetica (The Hague) 118**,** 171-182. [PubMed] [Google Scholar]