Protein–Protein Interactions Governing Septin Heteropentamer Assembly and Septin Filament Organization in Saccharomyces cerevisiae (original) (raw)
Abstract
Mitotic yeast (Saccharomyces cerevisiae) cells express five related septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) that form a cortical filamentous collar at the mother-bud neck necessary for normal morphogenesis and cytokinesis. All five possess an N-terminal GTPase domain and, except for Cdc10, a C-terminal extension (CTE) containing a predicted coiled coil. Here, we show that the CTEs of Cdc3 and Cdc12 are essential for their association and for the function of both septins in vivo. Cdc10 interacts with a Cdc3–Cdc12 complex independently of the CTE of either protein. In contrast to Cdc3 and Cdc12, the Cdc11 CTE, which recruits the nonessential septin Shs1, is dispensable for its function in vivo. In addition, Cdc11 forms a stoichiometric complex with Cdc12, independent of its CTE. Reconstitution of various multiseptin complexes and electron microscopic analysis reveal that Cdc3, Cdc11, and Cdc12 are all necessary and sufficient for septin filament formation, and presence of Cdc10 causes filament pairing. These data provide novel insights about the connectivity among the five individual septins in functional septin heteropentamers and the organization of septin filaments.
INTRODUCTION
Septins play crucial but ill defined roles in many cellular processes, such as cytokinesis, cellularization, and exocytosis in yeast, insects, and mammals (Kinoshita, 2003). Genes encoding septins were first identified in budding yeast via temperature-sensitive mutations that prevent cytokinesis at the restrictive temperature and cause accumulation of multibudded and multinucleated cells, with markedly elongated buds (Hartwell, 1971). Five septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) are expressed in mitotic yeast and are structural components of a cortical collar of highly ordered filaments at the mother-bud neck (Byers and Goetsch, 1976; Frazier et al., 1998). On dialysis into low-salt buffer, septin-enriched fractions isolated from yeast polymerize into paired filaments of 7–9 nm in diameter (Frazier et al., 1998).
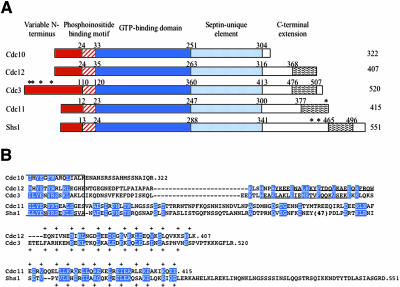
All septins contain a conserved core comprising a GTP-binding domain and, to its C-terminal side, a region unique to septin family members of as yet unknown function (Figure 1). The N terminus is variable, except for a tract of basic residues that may represent a phosphoinositide-binding region (Zhang et al., 1999; Casamayor and Snyder, 2003). Most, but not all, septins have a prominent C-terminal extension (CTE). Available algorithms (Woolfson and Alber, 1995; Wolf et al., 1997) predict that a segment with coiled coil-forming capacity might be present within each CTE.
Figure 1.
Primary structure of S. cerevisiae mitotic septins. (A) Schematic depiction of the domain structure of yeast septins. Characteristic features include N termini of variable length and sequence (red box), tract of basic and hydrophobic residues implicated in septin-phosphoinositide interaction (hatched box), globular GTPase domain (dark blue box), a sequence element diagnostic of septins (light blue box), and the variable C-terminal extensions (white box) containing, where indicated, a predicted coiled coil segment (wavy box). The number of residues in each protein is indicated to the right, and the residue numbers above demarcate the boundaries of the indicated domains. Asterisks denote known sites of covalent attachment of Smt3 (yeast SUMO). (B) Amino acid sequence of the C-termini beyond the last 15 residues of the septin-unique element (boxed). In each of the indicated septins, the CTEs (number of residues) are as follows: Cdc10 (18), Cdc12 (91), Cdc3 (107), Cdc11 (115), and Shs1 (210). Identities and conservative substitutions shared by all five septins, or by the indicated septin pairs, are shown as white letters on blue boxes. Period indicates the C-terminal end of each polypeptide; dashes indicate gaps introduced to maximize the alignment, or to position the predicted coiled coil forming segments in the same approximate register. Strongly predicted α-helical segments (in Cdc3 and Cdc12 only) are underlined; residues constituting the diagnostic 4-3 repeat of primarily hydrophobic residues characteristic of coiled coils are indicated by the plus sign. Number in parentheses indicates a large insert present in Shs1.
In yeast, septins congregate early in the cell cycle as a patch at the incipient bud site, which transforms into a cylindrical collar during bud emergence. This patch-to-collar transition correlates with a sharp decrease in septin mobility, as determined by fluorescence recovery after photobleaching (Caviston et al., 2003; Dobbelaere et al., 2003). Correspondingly, well-ordered septin arrays have been observed via electron microscopy (EM) only in budded cells (Byers and Goetsch, 1976; Frazier et al., 1998). We have demonstrated recently that GTP binding to Cdc10 and Cdc12 is required for collar assembly in vivo and for septin polymerization in vitro (Versele and Thorner, 2004). In addition, phosphorylation of septins by Cla4, a p21-activated kinase (Dobbelaere et al., 2003; Versele and Thorner, 2004), and by Gin4, a microtubule-associated protein/microtubule affinity-regulating kinase family member (Longtine et al., 1998; Mortensen et al., 2002), is necessary for proper septin collar assembly. Time-lapse confocal microscopy of septin structures in certain mutants of Cdc42, the physiological activator of Cla4, or in mutants of Cdc42 GAPs (Rga1, Rga2, and Bem3), indicates that the initial patch at the bud site is a flat ring or disk, which subsequently expands into the collar as the bud emerges (Caviston et al., 2003). The collar remains at the bud neck throughout the cell cycle until it is split at cytokinesis and disassembles upon completion of cell division. Dephosphorylation of septins by the phosphatase PP2A (bound to a regulatory subunit, Rts1) may be required for the disassembly process (Dobbelaere et al., 2003).
Aside from their vital function in cytokinesis, septins play important roles in bud-site selection (Zahner et al., 1996), bud morphology (Barral et al., 1999; Shulewitz et al., 1999), and maintenance of both asymmetric bud-specific localization of integral membrane proteins and polarization of actin (Barral et al., 2000; Takizawa et al., 2000). The septin collar also serves as a structural scaffold at the bud neck that recruits different components at specific stages during the cell cycle (Gladfelter et al., 2001; Sakchaisri et al., 2004). Some bud neck-associated proteins localize only to the daughter side, whereas others localize to the mother side, suggesting that the septin filaments in the collar have some sort of inherent polarity or asymmetry.
In all cell types studied, different individual septin monomers assemble into heteromeric multiseptin complexes—at the cleavage furrow of dividing cells or elsewhere at the leading edges of membrane extensions (Fares et al., 1995; Field et al., 1996; Frazier et al., 1998; Kinoshita et al., 1998; Beites et al., 1999). Heterodimerization of mammalian Sept6 and Sept7 is required for their association with the septin regulator Borg3 (Sheffield et al., 2003). Purified or recombinant septins assemble into higher-order structures, such as linear filaments, rings, and spirals (Field et al., 1996; Kinoshita et al., 1997; Frazier et al., 1998; Kinoshita et al., 2002). The nature of these multiseptin complexes and the rules governing their formation have not been elucidated, nor do we know how these complexes order themselves into filaments. To date, no crystal structure for any individual septin, septin complex, or septin filament has been reported.
Given the remarkable diversity among septins in the lengths and features of their CTEs, and given the known role of coiled-coils in mediating protein–protein associations, we investigated the role of the CTE in each septin with regard, first, to the effect of its absence in vivo. We then examined in vitro the role of the CTE in each septin with regard to its ability to mediate association with each of the other septins, individually and in reconstituted heteromeric complexes prepared from purified recombinant septins. Finally, we examined the competence of those complexes, if formed, to assemble into filaments and the ultrastructural properties of those filaments.
MATERIALS AND METHODS
Strains and Growth Conditions
Yeast strains used in this study are listed in Table 1. Standard rich (YP) and defined minimal (SC) media (Sherman et al., 1986), containing either 2% glucose (Glc), 2% raffinose (Raf), or 2% galactose (Gal) as the carbon source and supplemented with appropriate nutrients to maintain selection for plasmids, were used for yeast cultivation. Cells were routinely grown at 30°C, except where otherwise indicated. Standard yeast genetic techniques were performed according to Sherman et al. (1986).
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source/reference |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Brachmann et al., 1998 |
| BY4743 | MATa/MATαhis3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0 | Brachmann et al., 1998 |
| Y21935 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0 leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0 cdc12Δ::KANMX4/CDC12 | Invitrogen, Carlsbad, CA |
| BJ2168 | MATaleu2 trp1 ura3-52 prbI-I122 pep4-3 prc1-407 gal2 | Jones, 1991 |
| YMVB17a | BJ2168 CDC12(Δ337-407)(URA3) | This study |
| Y25223 | BY4743 CDC3/cdc3Δ(::KANMX4) | Invitrogen, Carlsbad, CA |
| YMVB32b | BY4743 CDC11/cdc11Δ(::HIS3) | This study |
| YMVB5b | BY4741 cdc10Δ(::KANMX4) | This study |
Plasmids and Recombinant DNA Methods
Plasmids used in this study are listed in Table 2. Plasmids were constructed using standard procedures (Sambrook et al., 1989) in Escherichia coli strain DH5α (Hanahan, 1983). The lack of errors in constructs prepared by polymerase chain reaction (PCR), correct in-frame junctions, and site-directed mutations were verified by nucleotide sequence analysis (Sanger et al., 1977). Genomic DNA that served as template for PCR amplifications was prepared from strain BY4741 (Table 1).
Table 2.
Plasmids used in this study
| Plasmid | Description | Source/reference |
|---|---|---|
| pMVB12 | _CDC12_-His6 | Versele and Thorner, 2004 |
| pMVB13 | _CDC12ΔC_-His6 | Versele and Thorner, 2004 |
| pMVB120 | _CDC11_-His6 | Versele and Thorner, 2004 |
| pBEG2 | His6-CDC3 | Versele and Thorner, 2004 |
| pBEG2 | His6-CDC10 | Versele and Thorner, 2004 |
| pGST-CDC11 | GST-CDC11 | D. Kellogg (University of California, Santa Cruz, CA) |
| pMVB29a | GST-CDC11ΔC | This study |
| pMVB24 | GST-CDC10 | Versele and Thorner, 2004 |
| pMVB25 | GST-CDC12 | Versele and Thorner, 2004 |
| pMVB26a | GST-CDC12ΔC | This study |
| pMVB27 | GST-CDC3 | Versele and Thorner, 2004 |
| pMVB30a | GST-CDC3ΔC | This study |
| pBEG4b | GST-SHS1 | This study |
| pBEG24b | GST-SHS1ΔC | This study |
| pMVB172c | His6-GST-CTECdc12 | This study |
| pMVB121 | pETDuet-His6_CDC12_ | Versele and Thorner, 2004 |
| pMVB128d | pETDuet-His6_CDC12-CDC10_ | This study |
| pMVB134d | pETDuet-His6_CDC10_ | This study |
| pMVB132d | pACYDuet-CDC3 | This study |
| pMVB133d | pACYDuet-CDC3-CDC11 | This study |
| pMVB127d | pACYDuet-CDC11 | This study |
| pMVB122 | pETDuet-His6_CDC12-CDC3_ | Versele and Thorner, 2004 |
| pMVB129d | pETDuet-His6_CDC12ΔC_ | This study |
| pMVB130d | pETDuet-His6_CDC12ΔC-CDC10_ | This study |
| pMVB135d | pETDuet-His6_CDC12ΔC-CDC3_ | This study |
| pMVB45c | CEN, LEU2, CDC12 | This study |
| pMVB48e | CEN, LEU2, CDC12ΔC | This study |
| pMVB52e | CEN, LEU2, CDC12ΔC(S43V) | This study |
| pMVB53e | CEN, LEU2, CDC12ΔC(T48N) | This study |
| pLP17 | CEN, LEU2, CDC12-GFP | Lippincott and Li, 1998 |
| pMVB33f | CEN, LEU2, CDC12ΔC-GFP | This study |
| pMVB62g | 2μ, LEU2, CDC12-GFP | This study |
| pMVB63g | 2μ, LEU2, CDC12ΔC-GFP | This study |
| pMVB23h | URA3, CDC12ΔC | This study |
| pMVB54i | 2μm, LEU2, CDC12ΔC | This study |
| pMVB2j | CEN, URA3, GAL1-CDC12-GFP | This study |
| pMVB160j | CEN, URA3, GAL1-CDC12ΔC-GFP | This study |
| pMVB162k | CEN, LEU2, GAL1-CDC12 | This study |
| pMVB164k | CEN, LEU2, GAL1-CDC12ΔC | This study |
| pSB5 | CEN, URA3, CDC11-GFP | Versele and Thorner, 2004 |
| CDC3-GFP | CEN, URA3, CDC3-GFP | B. Haarer (SUNY Upstate Med. Ctr., Syracuse, NY) |
| pLA10 | CEN, URA3, CDC10-GFP | Cid et al., 1998 |
| pMVB38l | 2μm, LEU2, P(CDC12)-GFP-CTECdc12 | This study |
| pMVB100m | CEN, URA3, CDC3 | This study |
| pMVB102m | CEN, URA3, CDC3ΔC | This study |
| pSB1n | CEN, URA3, CDC11 | This study |
| pSB2n | CEN, URA3, CDC11ΔC | This study |
| YCpUG-CDC11o | CEN, URA3, GAL1-CDC11 | This study |
| YCpUG-CDC11ΔCo | CEN, URA3, GAL1-CDC11ΔC | This study |
Protein Binding to Immobilized Glutathione S_-Transferase (GST) Fusions_
Freshly transformed BL21(DE3) cells carrying a plasmid expressing the indicated GST-fusion protein were grown, induced, and lysed as described in detail in Versele and Thorner (2004). The cleared lysates were incubated with a 50% slurry of Pharmacia glutathione-Sepharose (Pfizer, New York, NY) for 1 h at 4°C. The beads were washed three times with 1 ml of wash buffer (50 mM sodium phosphate, pH 7.5, 125 mM NaCl, 0.1% Tween 20), and finally, resuspended in binding buffer (50 mM sodium phosphate, pH 7.5, 125 mM NaCl, 0.05% Tween 20, 0.1 mM dithiothreitol [DTT]). Cdc12(His)6, Cdc12(Δ339–407)(His)6, (His)6-Cdc3, (His)6-Cdc10, or Cdc11(His)6 was prepared using Ni2+-nitrilotriacetate (NTA) affinity beads also as described in Versele and Thorner (2004). These proteins were diluted in binding buffer and incubated with GST-coated glutathione-Sepharose for 30 min at room temperature. The resulting precleared supernatant fractions were then incubated with glutathione-Sepharose beads coated with the GST-fusion protein to be tested or with GST alone (as a negative control) in binding buffer for 1 h at room temperature. The beads were then washed four times with ice-cold binding buffer. Proteins were solubilized by addition of 100 μl of two times-concentrated SDS-PAGE sample buffer and boiling for 5 min. Samples of the solubilized eluate were resolved by SDS-PAGE and analyzed using Coomassie Blue stain or immunoblotting by using anti-His(C-term) antibody (Invitrogen, Carlsbad, CA) for the detection of C-terminally tagged Cdc12, Cdc12ΔC, and Cdc11, or anti-His (Roche Diagnostics, Indianapolis, IN) for the detection of N-terminally tagged Cdc3 and Cdc10.
Preparation of Septin Complexes
Cultures (100 ml) of E. coli BL21(DE3) coexpressing His6Cdc12 or His6Cdc10, and untagged Cdc3, Cdc10, and Cdc11 from corresponding plasmids (Table 2) constructed from bicistronic vectors with compatible replication origins (pET-Duet and pACY-Duet; Novagen, Madison, WI) were grown to an A600 nm = 1, induced with isopropyl-β-d-thiogalactoside (1 mM final) for 3 h at 37°C, collected by centrifugation, and washed once in lysis buffer (250 mM NaCl, 2 mM MgCl2, 40 μM GDP, 1 mM EDTA, 5 mM β-mercapto-ethanol, 0.5% Tween 20, 12% glycerol, 50 mM Tris-HCl, pH 8.0). Washed cells were resuspended in lysis buffer (5 ml) containing protease inhibitor mix (Complete EDTA-free; Roche Diagnostics), and 0.2 mg/ml lysozyme, incubated on ice for 15 min, and then lysis was completed by two 15-s pulses of sonication (Branson sonifier 450). Lysates were clarified at 4°C by centrifugation at 12,000 × g. The resulting supernatant fraction (∼5 ml) was mixed with 200 μl of a 50:50 slurry of Ni2+-NTA agarose beads (QIAGEN, Valenica, CA) that had been preequilibrated in lysis buffer and incubated for 1 h at 4°C on a rollerdrum. Beads were collected by centrifugation for 1 min at 500 × g and washed with five 1-ml portions of rinse buffer (250 mM NaCl, 20 mM imidazole, 0.1% Tween 20, 50 mM Tris-HCl, pH 8.0). His6-tagged proteins and any interacting proteins were then eluted with 200 μl of 250 mM NaCl, 300 mM imidazole, 0.1% Tween 20, 12% glycerol, 50 mM Tris-HCl, pH 8.0. After elution, the eluate containing the resulting protein complexes was dialyzed against a low-salt buffer (50 mM KCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, 50 mM potassium phosphate, pH 7.2).
Antibodies
The polyclonal Cdc12-specific antiserum used was raised against purified Cdc12ΔC, as described in detail previously (Versele and Thorner, 2004). Other antibodies were obtained from the following sources: mouse monoclonal anti-His(C-term) (Invitrogen), mouse monoclonal anti-green fluorescent protein (GFP) (Roche Diagnostics), mouse monoclonal anti-His (Roche Diagnostics), and rabbit polyclonal anti-Cdc11 (gift from Doug Kellogg, University of California, Santa Cruz, Santa Cruz, CA).
Preparation of Yeast Extracts for Immunoblots
Protein extracts were prepared in lysis buffer (125 mM NaCl, 0.1% Tween 20, 1 mM DTT, 50 mM sodium phosphate buffer, pH 7.5), containing a mixture of protease inhibitors (Complete; Roche Diagnostics). The suspended cells were subjected to vigorous shaking with glass beads (0.5 mm in diameter) in an automated device (FastPrep 120; Bio 101, Vista, CA/Thermo Savant, Holbrook, NY), and the resulting lysates were clarified by centrifugation. A sample (30 μg of total protein) of these extracts was resolved by SDS-PAGE, blotted onto nitrocellulose filters, and analyzed using appropriate antibodies, which were detected using chemiluminescence (Super-Signal Pico West; Pierce Chemical, Rockford, IL).
Immuno-staining and Negative-Stain Electron Microscopy to Monitor Filament Formation
For immuno-staining, protein samples (1 μg) were applied onto 12-well polylysine-coated glass slides, blocked with 2% bovine serum albumin in PBS, incubated with polyclonal anti-Cdc12ΔC antibodies, washed, incubated with a TRITC-conjugated secondary anti-rabbit IgG antibody, and viewed in a BH-2 epifluorescence microscope (Olympus, Tokyo, Japan) using a 100× objective, equipped with a red (545 nm) band-pass filter (Chroma Technology, Brattleboro, VT). Images were collected using a charged-coupled device camera (Olympus), processed with Magnafire SP imaging software (Optronics, Goleta, CA) and Photoshop (Adobe Systems, Mountain View, CA). For negative-stain electron microscopy, protein samples (0.25 μg) were applied to polylysine-treated, glow-discharged, carbon-coated copper grids; stained with 1% uranyl acetate in 50% methanol; and viewed in a Philips Tecnai 12 transmission electron microscope (FEI Company, Eindhoven, The Netherlands). Measurements were carried out using the Digital Micrograph R01 tool (Gatan, Inc., Pleasanton, CA).
Microscopy
To visualize GFP-fusions in live cells in real time, yeast from culture samples were collected by filtration, washed once with PBS, and viewed immediately under an epifluorescence microscope (BH-2; Olympus) by using a 100× objective equipped with a GFP band-pass filter (Chroma Technology). Images were collected and processed as described for immunostaining. To stain nuclei, cells were fixed briefly in 70% ethanol, washed several times with PBS, and incubated with 4,6-diamidino-2-phenylindole (DAPI) (0.2 μg/ml final). To stain chitin, cells were incubated with 0.1 mg/ml calcofluor white (Fluorescent Brightener 28; Sigma-Aldrich, St. Louis, MO) for 10 min and washed several times with PBS. Chitin-stained cells were visualized upon excitation with UV light.
RESULTS
The CTE Is Necessary for Cdc12 Function
Cdc12 contains an apparent coiled coil-forming segment at its C terminus (Figure 1A), as predicted by algorithms like MultiCoil (Wolf et al., 1997) that recognize sequence covariation in 4-3 hydrophobic repeats compatible with the formation of coiled coils. In Cdc12, the predicted 40-residue coiled coil extends from Glu368 to the C-terminal residue (Lys407). In addition, programs for secondary structure prediction indicate that the immediately preceding sequence (Arg341-Trp367) has a striking propensity to form an extended α-helix. Both of these elements lie downstream of the septin-unique domain, which flanks the GTP-binding domain and is present in all septins (Figure 1B). The predicted α-helix is broken at Asn339-Pro340. Therefore, we consider the region from residue 339 to the C terminus to be the CTE.
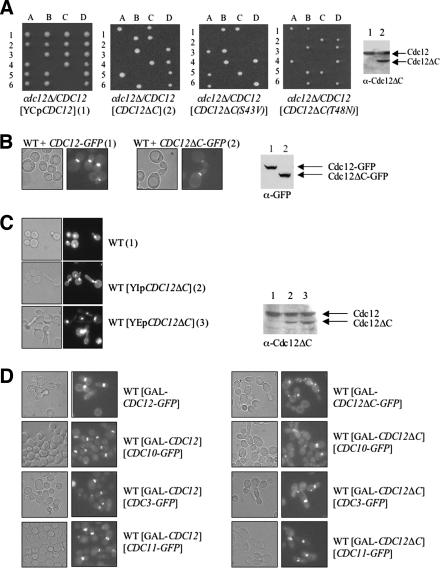
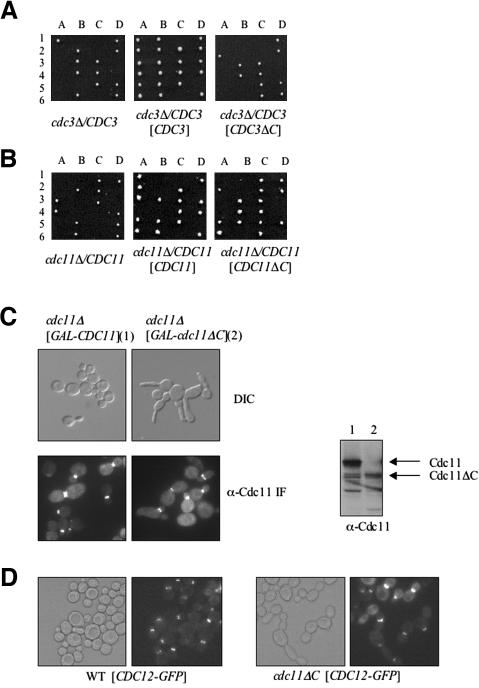
Cdc12, Cdc3, and Cdc11 are required for viability, whereas Cdc10 is essential only at 37°C, and Shs1 is dispensable even at elevated temperature. Therefore, we first used a genetic approach to examine whether the CTE is necessary for Cdc12 function in vivo. For this purpose, we constructed a truncated allele, CDC12(Δ_339–407) (hereafter Cdc12ΔC) and tested whether it was able to support growth when present as the sole source of Cdc12. Either full-length CDC12 or the truncated allele, under control of the authentic CDC12 promoter, was introduced on a low-copy-number (CEN) plasmid into a heterozygous cdc12_Δ/CDC12 diploid. After sporulation and tetrad dissection, the ability of normal Cdc12 and Cdc12ΔC to rescue the inviability of the _cdc12_Δ spores was compared (Figure 2A). For diploids transformed with the plasmid expressing normal Cdc12, all of the tetrads (≥20 analyzed) displayed three or four viable spores, showing that plasmid-borne CDC12 readily rescued the inviability of _cdc12_Δ spores (which also were marked by the insertion of a resistance marker, kanMX, to the antibiotic G418). By contrast, for the diploids transformed with the plasmid expressing Cdc12ΔC, only two viable spores were obtained in every tetrad (≥20 analyzed), and those spores were always G418 sensitive. In the heterozygous diploids, and when produced from a CEN plasmid, the truncated protein was stably expressed at a level equivalent to full-length Cdc12 (Figure 2A, far right). Hence, Cdc12ΔC could not support cell viability. Cdc12ΔC was unable to rescue viability of _cdc12_Δ cells even when overexpressed from a multicopy (2 μm DNA) plasmid or from the GAL1 promoter (unpublished data). Hence, the inability of Cdc12ΔC to rescue Cdc12-deficient cells was not due to its lack of expression or instability. Also, the truncated protein is folded because Cdc12ΔC binds GTP as tightly as normal Cdc12 and has a GTPase activity that is at least 5 times higher than wild-type Cdc12 (Versele and Thorner, 2004). These properties of the mutant protein suggested that perhaps its elevated GTPase activity might account for its deleterious function in vivo. Consistent with this possibility, full-length Cdc12 mutants compromised for either GTP binding or GTP hydrolysis are able to rescue the lethality of _cdc12_Δ cells (Versele and Thorner, 2004). However, when mutations (T48N and S43V, respectively) that abrogate, respectively, GTP binding and GTP hydrolysis were introduced into Cdc12ΔC (Versele and Thorner, 2004), neither of the corresponding Cdc12ΔC derivatives could support the growth of _cdc12_Δ spores (Figure 2A). Thus, the CTE of Cdc12 is necessary for the essential function of Cdc12 in vivo and that function is independent of the GTP-dependent activities of Cdc12.
Figure 2.
The essential function of Cdc12 requires its C-terminal extension. (A) A cdc12_Δ/CDC12_ heterozygous diploid (Y21935) was transformed with a CEN plasmid carrying either wild-type CDC12 (pMVB45), CDC12_Δ_C (pMVB48), _CDC12(S43V)Δ_C (pMVB52), or _CDC12(T48N)Δ_C (pMVB53), sporulated and individual tetrads (1–6) were dissected and the resulting spore clones (A–D) were germinated on YPD plates. Samples (30 μg of total protein) of extracts were prepared and analyzed by SDS-PAGE and immunoblotting with an anti-Cdc12ΔC antibody [lane 1, Y21935 transformed with a CEN plasmid expressing wild-type CDC12; lane 2, Y21935 transformed with the same CEN vector expressing CDC12_Δ_C (right-most panel)]. (B) Wild-type cells (BY4741) were transformed with a CEN plasmid expressing either _CDC12_-GFP (pLP17) or _CDC12_Δ_C_-GFP (pMVB33), grown at 26°C to midexponential phase on SCGlc(-Leu), and samples of each culture were analyzed by differential interference contrast (DIC) optics (left) or fluorescence microscopy (right). Samples (30 μg of total protein) of extracts were prepared from the same cultures and analyzed by SDS-PAGE and immunoblotting with an anti-GFP monoclonal antibody (lane 1, BY4741 expressing _CDC12_-GFP; lane 2, BY4741 expressing _CDC12_Δ_C_-GFP; right-most panel). (C) Cdc12ΔC interferes with septin collar assembly. A wild-type strain (BJ2168) was transformed with an empty vector or with either an integrating vector (YMVB17) or a multi-copy (2 μm DNA) vector expressing CDC12_Δ_C, and samples of each were examined by bright field (left) or stained with DAPI to reveal the location of nuclei and examined by fluorescence microscopy (right). Samples (30 μg of total protein) of extracts were prepared from the same cultures and analyzed by SDS-PAGE and immunoblotting with polyclonal anti-Cdc12ΔC antibody (lane 1, BJ2168 transformed with empty vector; lane 2, BJ2168 transformed with the integrating vector expressing CDC12_Δ_C; lane 3, BJ2168 transformed with the multi-copy vector expressing CDC12_Δ_C; right-most panel). (D) A wild-type strain (BY4741) expressing either _GAL1_-_CDC12_-GFP (pMVB2) (left column) or _GAL1_-CDC12_Δ_C-GFP (pMVB160) (right column), or the same strains coexpressing either _GAL1_-CDC12 (pMVB162) or _GAL1_-CDC12_Δ_C (pMVB164) along with each of the indicated plasmids [_CDC3_-_GFP, CDC10_-GFP (pLA10) or _CDC11_-GFP (pSB5)], were grown to midexponential phase on SCGlc(-Leu,-Ura) at 30°C, and samples were analyzed by DIC optics (left) or fluorescence microscopy to visualize GFP-tagged septins (right).
Another potential explanation for the inability of Cdc12ΔC to substitute for normal Cdc12 is that it may not localize properly. Because Cdc12 is essential and Cdc12ΔC is nonfunctional, we could only test whether the CTE is required to target Cdc12 to the bud neck by expressing full-length Cdc12-GFP or Cdc12ΔC-GFP in otherwise wild-type cells. Both Cdc12-GFP and Cdc12ΔC-GFP were expressed at the same level and were recruited to the bud neck (Figure 2B). However, we consistently noted that Cdc12ΔC-GFP was incorporated less efficiently into the collar than its wild-type counterpart, despite its equivalent expression, as judged by the reproducibly dimmer signal at the bud neck in every cell (Figure 7A). Moreover, heterozygous cdc12/CDC12 diploids transformed with the plasmid expressing CDC12_Δ_C displayed a mild elongated-bud morphology. This dominant effect suggested that Cdc12ΔC interferes with the function of Cdc12 in the same cell.
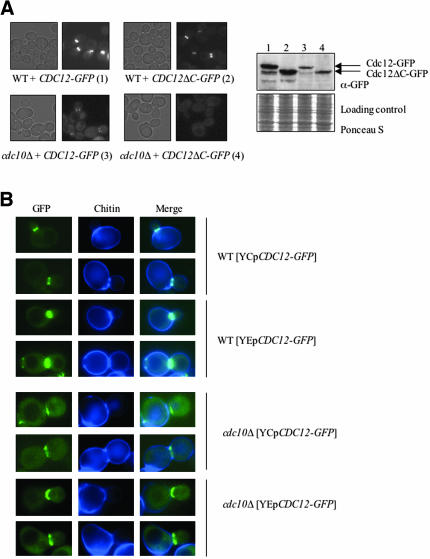
Figure 7.
Cdc10 is essential for normal septin collar organization at the bud neck. (A) Wild-type strain (BY4741) and a _cdc10_Δ mutant (YMVB5) were transformed with plasmids expressing Cdc12-GFP (pLP17) or Cdc12ΔC-GFP (pMVB33), grown at 26°C to midexponential phase on SCGlc(-Leu), and samples of each culture were viewed by differential interference contrast optics (left) or fluorescence (right) microscopy (GFP). Samples (30 μg of total protein) of extracts prepared from the same cultures was analyzed by SDS-PAGE and immunoblotting by using an anti-GFP monoclonal antibody. The blot was stained with Ponceau S as a loading control. (B) Wild-type strain (BY4741) and _cdc10_Δ mutant (YMVB5) were transformed with either a CEN plasmid (pLP17) or a multi-copy (2 μm DNA) vector (pMVB62) expressing Cdc12-GFP, grown to midexponential phase at 26°C on SCGlc(-Leu), and samples of each culture were incubated with calcofluor white to stain chitin and then viewed by fluorescence microscopy, by using appropriate cut-off filters to view GFP and chitin staining, as indicated.
To confirm this conclusion, a haploid strain was constructed that contained a single copy of the normal CDC12 gene and a single copy of the truncated allele (also under control of a CDC12 promoter) integrated into the genome at the CDC12 locus. We found that even one copy of Cdc12ΔC in otherwise wild-type cells was sufficient to disrupt normal cell morphology, as indicated by the markedly elongated buds (Figure 2C). When the ratio of Cdc12ΔC to Cdc12 was further elevated by expression of CDC12_Δ_C from a multicopy plasmid, morphological perturbation of the cells was even more pronounced, even though the steady-state level of Cdc12ΔC was still somewhat less than that of Cdc12 (Figure 2C). Expression of Cdc12ΔC (from its own promoter) on either a CEN or 2 μm DNA plasmid was sufficient to noticeably perturb the bud neck localization of C-terminally GFP-tagged versions of Cdc3, Cdc10, Cdc11, and Cdc12 itself (unpublished data). Prolonged overexpression of Cdc12ΔC driven by the _GAL1_-promoter caused even more severe defects in cell morphology and grossly disrupted the organization of the septin collar, as judged by the mislocalization and aberrant deposition of three other septins, Cdc3, Cdc10, and Cdc11 (each marked with a C-terminal GFP tag) or of Cdc12ΔC-GFP itself (Figure 2D). Overexpression of normal Cdc12 in the same manner had no such effects. Thus, the CTE of Cdc12 is critically important for the proper organization of septins in vivo.
Cdc12 Interacts with Cdc3 via the CTE and with Cdc10 Independently of the CTE
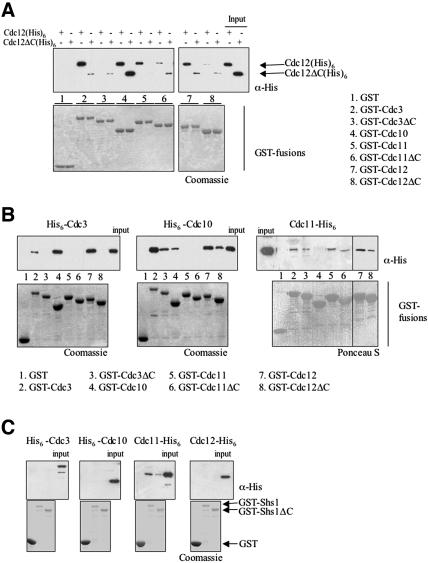
To determine whether the CTE of Cdc12 participates in direct septin–septin interactions, an in vitro assay was used to test the binding of any given septin (with or without its CTE) to every other septin (with or without its CTE). First, glutathione-agarose beads were coated with GST fusions either to full-length septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) or to derivatives of each septin lacking its cognate CTE, namely, Cdc3(Δ427–520), Cdc11(Δ357–415), Cdc12(Δ339–407), and Shs1(Δ418–551). Cdc10 is the only yeast septin that lacks a CTE (Figure 1). Second, each septin to be tested (with or without its CTE) was tagged with a (His)6 tract (at either its N or C terminus), expressed in bacteria, purified by metal-chelate affinity chromatography, and then incubated in solution with the bead-bound GST-fusions. After washing the beads, bound proteins were eluted and analyzed by SDS-PAGE. Neither Cdc12-His6 nor Cdc12ΔC-His6 bound to beads coated with GST alone (Figure 3A). Full-length Cdc12-His6 interacted strongly with three other septins (GST-Cdc3, GST-Cdc10 and GST-Cdc11) and also with itself (GST-Cdc12), but not with GST-Shs1 (Figure 3C). By comparison, binding of Cdc12ΔC-His6 to GST-Cdc3 and GST-Cdc11, and to GST-Cdc12 itself, was markedly reduced (Figure 3A). In contrast, Cdc12 and Cdc12ΔC interacted with GST-Cdc10 with apparently equal affinity. Conversely, a GST-fusion to Cdc3ΔC interacted poorly with either Cdc12-His6 or Cdc12ΔC-His6 (Figure 3A). Also, the interactions between Cdc12-His6 and Cdc12ΔC-His6 with either GST-Cdc11ΔC or GST-Cdc12ΔC were reduced compared with their interactions with full-length GST-Cdc11 and GST-Cdc12 (Figure 3A).
Figure 3.
Role of septin CTEs in pairwise septin–septin interactions. (A) Equivalent amounts of the indicated GST-septin fusions [Cdc3, Cdc3(Δ427–521), Cdc10, Cdc11, Cdc11(Δ357–416), Cdc12, and Cdc12(Δ339–407)] were immobilized on glutathione-agarose beads, incubated with either Cdc12(His)6 or Cdc12ΔC(His)6, and washed extensively. Bound proteins were eluted with SDS-PAGE sample buffer, resolved by SDS-PAGE, and analyzed either by Coomassie Blue (to verify equal loading of the GST-fusion proteins) or by immunoblotting by using anti-His(C-term) antibody to detect the input and bound (His)6-tagged proteins. Input, 20% of the amount of Cdc12-His6 or Cdc12ΔC-His6 added in each binding reaction. (B) The same GST-septin fusions as in A, immobilized on glutathione-agarose, were incubated with His6-Cdc3, His6-Cdc10, and Cdc11-His6, as indicated, and analyzed as described in A, except either anti-His antibody or anti-His(C-term) was used, as appropriate. C. GST-Shs1 and GST-Shs1(Δ418–551) were immobilized on glutathione-agarose and incubated with the indicated His6-tagged septins and analyzed as described in B.
To corroborate and extend these conclusions, the ability of His6-Cdc3, His6-Cdc10, and Cdc11-His6 to associate with the panel of GST-fusions was assessed. His6-Cdc3 did not bind detectably to GST alone (Figure 3B, left). His6-Cdc3 bound well to GST-Cdc12, yet not to GST-Cdc12ΔC, confirming that the CTE of Cdc12 is essential for Cdc3–Cdc12 association (Figure 3B). His6-Cdc3 also bound to Cdc10, but not to GST-Cdc11 (Figure 3B) or GST-Shs1 (Figure 3C). Like Cdc12-His6, His6-Cdc3 displayed some self-association that was dependent on its CTE.
Consistent with the preceding results, His6-Cdc10 bound well to both GST-Cdc12 and GST-Cdc12ΔC (Figure 3B, middle). Likewise, in accord with the binding of His6-Cdc3 to GST-Cdc10, His6-Cdc10 bound to both GST-Cdc3 and GST-Cdc3ΔC. His6-Cdc10 did not bind detectably to either GST-Cdc11 (Figure 3B, middle) or to GST-Shs1 (Figure 3C). Like Cdc12 and Cdc3, Cdc10 displayed a modest degree of self-association.
In agreement with the lack of interaction between His6-Cdc10 and GST-Cdc11, Cdc11-His6 did not interact detectably with GST-Cdc10 (Figure 3B, right). The strongest interaction observed for Cdc11-His6 was with GST-Shs1 (Figure 3C). In fact, the only septin able to associate with Shs1 was Cdc11. The binding of Cdc11-His6 to GST-Shs1 was markedly and reproducibly reduced when the CTE of Shs1 was absent. Cdc11-His6 also interacted with GST-Cdc3, GST-Cdc3ΔC, GST-Cdc12, GST-Cdc12ΔC, and with itself (either GST-Cdc11 or GST-Cdc11ΔC) (Figure 3B, right). Thus, all of these interactions of Cdc11 were largely independent of the CTEs of any of the septins.
The results of these binding interactions are compiled in Table 3. Briefly, Cdc3–Cdc12 interaction requires the CTE of each protein; both Cdc3 and Cdc12 associate with Cdc10, and do so independently of their CTEs. Cdc10 does not associate at all with Cdc11 or Shs1. The only septin that binds Shs1 is Cdc11, and this interaction is largely dependent on the CTE of Shs1. Finally, Cdc3, Cdc10, Cdc11, and Cdc12, all self-associate to at least some degree (Shs1 was not tested). However, this analysis of the direct pairwise interactions between the septins, although very informative, left a few ambiguities. Cdc11 bound to Cdc12, but it was unclear from these particular experiments whether this interaction is influenced by the CTE of either protein. It also was unresolved whether Cdc11 binds to Cdc3.
Table 3.
Summary of direct septin–septin interactions
| His6a | |||||
|---|---|---|---|---|---|
| GSTb | Cdc3 | Cdc10 | Cdc12 | Cdc12ΔC | Cdc11 |
| Cdc3 | + | +++ | +++ | + | + |
| Cdc3ΔC | – | ++ | – | + | +/– |
| Cdc10 | +++ | + | +++ | +++ | – |
| Cdc11 | – | – | ++ | + | + |
| Cdc11ΔC | – | – | + | + | +/– |
| Cdc12 | +++ | +++ | ++ | + | + |
| Cdc12ΔC | – | ++ | + | + | +/– |
| Shs1 | – | – | – | – | ++ |
| Shs1ΔC | – | – | – | – | –/+ |
Requirements for Septin Complex Formation
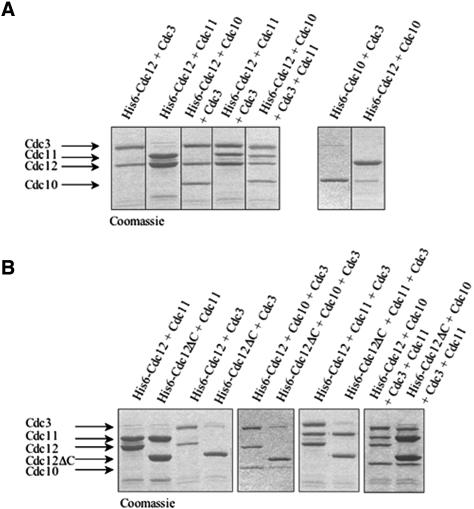
In the cell, septins are all present together, and multivalent contacts among them may contribute to higher order interactions that mediate formation of the multiseptin complexes found in vivo. Therefore, we coexpressed the core septins Cdc3, Cdc10, Cdc11, and Cdc12 (with or without their CTE) in all possible binary, tertiary, and quaternary combinations in E. coli by using compatible bicistronic vectors. (Shs1 was not included in this analysis because, unlike the core septins, it is not an essential protein in vivo and because our binding assays indicated that it only associates with Cdc11 and does so via its CTE.) In each case, only a single septin (usually Cdc12) carried a (His)6 tag and was the septin expressed from the vector and promoter that ensured that it would be the protein expressed in the most limiting amount (unpublished data). The His6-tagged protein and any associated polypeptides were purified by Ni2+-chelate affinity chromatography and the composition of the resulting complexes was analyzed by SDS-PAGE.
As judged by densitometry of the Coomassie Blue-stained protein bands, formation of stoichiometric complexes between Cdc3 and His6-Cdc12 and between His6-Cdc12 and Cdc11 was observed reproducibly (Figure 4A). No complexes between His6-Cdc3 and Cdc11, or between His6-Cdc10 and Cdc11, were detected (unpublished data). Revealingly, the amount of Cdc3 bound in complexes with His6-Cdc10, and the amount of Cdc10 bound in complexes with His6-Cdc12, was strikingly substoichiometric (Figure 4A, right). In marked contrast, Cdc10 was incorporated stoichiometrically into complexes that contained both Cdc3 and His6-Cdc12 (Figure 4A, left), suggesting that efficient incorporation of Cdc10 into multiseptin complexes requires its simultaneous association with both Cdc3 and Cdc12.
Figure 4.
Reconstitution of stoichiometric septin complexes requires the CTE of Cdc12. (A) His6-Cdc12 or His6-Cdc10, as indicated, was coexpressed in E. coli with the indicated untagged septin(s), and any resulting complexes were purified using Ni2+-NTA-agarose. After washing, bound proteins were eluted with imidazole, resolved by SDS-PAGE, and stained with Coomassie Blue. (B) His6-Cdc12 or His6-Cdc12ΔC, as indicated, was coexpressed with the other indicated untagged septin(s), and analyzed as described in A.
The only other ternary complex observed was between Cdc3, Cdc11, and His6-Cdc12 (Figure 4A, left), in keeping with the fact that Cdc12 associated in stoichiometric binary complexes with both Cdc3 and Cdc11. Presence of all three of these septins in stoichiometric amounts in the ternary complex suggests that Cdc3 and Cdc11 associate with different sites on Cdc12. This conclusion is also in accord with the GST-pull down assays, in which strong Cdc3–Cdc12 association required the CTE of each protein (Figure 3A), whereas Cdc11 was able to associate with either full-length Cdc12 or Cdc12 lacking its CTE (Figure 3B).
When Cdc10, Cdc11, and His6-Cdc12 were coexpressed, His6-Cdc12-Cdc11 binary complexes were recovered, but no significant amount of Cdc10 was incorporated (unpublished data). Moreover, when His6-Cdc3, Cdc10, and Cdc11 were coexpressed, no protein other than His6-Cdc3 was recovered efficiently; likewise, when Cdc3, His6-Cdc10, and Cdc11 were coexpressed, no other protein copurified with Cdc10 (unpublished data). However, when all four proteins were coexpressed, stoichiometric heterotetrameric complexes of Cdc3, Cdc10, Cdc11, and His6-Cdc12 were readily and reproducibly recovered (Figure 4A). These results reinforce the conclusion that presence of Cdc12 is critical to assembly of heteromeric septin complexes.
Finally, this coexpression approach was used as an independent means to confirm the role of the Cdc12 CTE in Cdc3–Cdc12 interaction, but not in Cdc12–Cdc11 interaction. Indeed, as expected, Cdc11 was copurified in stoichiometric complexes with either His6-Cdc12 or His6-Cdc12ΔC, whereas Cdc3 formed stoichiometric complexes with His6-Cdc12, but copurified in only trace amounts with His6-Cdc12ΔC (Figure 4B, left). Likewise, stoichiometric ternary complexes containing Cdc3, Cdc10, and His6-Cdc12 were readily purified, but neither Cdc3 nor Cdc10 were efficiently recovered with His6-Cdc12ΔC (Figure 4B, middle), in agreement with the fact that the CTE of Cdc12 is required for Cdc3–Cdc12 association and that incorporation of Cdc10 into complexes requires the presence of both Cdc3 and Cdc12 (Figure 4A).
Similarly, stoichiometric ternary complexes containing Cdc3, Cdc11, and His6-Cdc12 can be readily purified, but Cdc3 is very inefficiently recovered with His6-Cdc12ΔC, even though Cdc11 binding to His6-Cdc12ΔC is undiminished (Figure 4B, middle). Finally, heterotetrameric complexes prepared with His6-Cdc12 contain stoichiometric amounts of each of the other three core septins; but, in the absence of the Cdc12 CTE, only Cdc11 is recovered in a stoichiometric amount with His6-Cdc12Δ and both Cdc3 and Cdc10 are greatly underrepresented (Figure 4B, right). These results confirm that the CTE of Cdc12 is required for its association with Cdc3, but not with Cdc11.
The Cdc12 CTE, but Not Its Coiled Coil Element, Is Sufficient for Interaction with Cdc3
Because Cdc12 and Cdc3 association requires their CTEs (and self-association of each of these two septins is also dependent on their CTEs), the simplest interpretation of these findings is that the CTEs physically interact. Moreover, the presence of predicted coiled coil sequences within the CTEs raised the possibility that CTE–CTE interaction is mediated by coiled coil formation. To determine whether the predicted coiled coil sequences are, by themselves, sufficient to interact, synthetic peptides corresponding to Cdc12(369–407), Cdc3(459–503) and, as a control, Cdc11(369–415), were prepared and their circular dichroism signal was monitored at 222 nm (diagnostic of helix formation) as a function of temperature (unpublished data). No individual peptide adopted a stable α-helical conformation at physiological pH, temperature, and salt concentration. At high peptide concentration (400 μM), Cdc12(369–407) formed an unstable homo-oligomer under physiological conditions. Unexpectedly, mixing Cdc12(369–407) and Cdc3(459–503) produced no synergistic effects, and no stable interactions were detected in any two-way or three-way combinations of the three peptides. Thus, the predictions of MultiCoil (Wolf et al., 1997) notwithstanding, these isolated polypeptide segments were unable to form stable autonomous coiled coils.
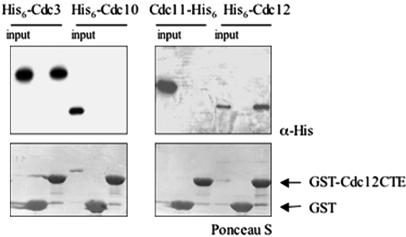
Given that the predicted coiled coil sequences in both Cdc3 and Cdc12 are preceded by an additional segment predicted to be α-helical that could buttress the helix-forming propensity of a juxtaposed coiled coil, we examined whether a longer portion of the Cdc12 CTE that included the additional predicted α-helix would be sufficient to mediate association of Cdc12 CTE with other septins. Using purified bacterially expressed proteins, only His6-Cdc3 and His6-Cdc12, and not His6-Cdc10 or Cdc11-His6, bound to beads coated with purified GST-CTECdc12 (residues 339–407) (Figure 5). Thus, the entire CTE of Cdc12 is both necessary and sufficient for mediating the interaction between Cdc12 and Cdc3 and also contributes to Cdc12–Cdc12 self-association. In vivo a GFP-CTECdc12 chimera was not stably recruited to the bud neck (unpublished data); however, when extracts of such cells were passed over glutathione-agarose beads coated with the panel of GST-septin fusions, GFP-CTECdc12 bound avidly to GST-Cdc3, but not to GST-Cdc3ΔC (unpublished data), consistent with the binding observed between the highly purified proteins (Figure 5). The fact that GFP-CTECdc12 was not stably incorporated into the septin collar, whereas Cdc12ΔC-GFP is (Figure 2B), indicates that stable filament assembly requires the globular, N-terminal GTP-binding domain of Cdc12. This conclusion is in accord with our finding that GTP binding to Cdc12 is indeed essential for septin collar formation in vivo and for septin filament polymerization in vitro (Versele and Thorner, 2004).
Figure 5.
The Cdc12 CTE is sufficient for interaction with Cdc3. GST-Cdc12(339–407) [GST-CT-ECdc12] expressed in bacteria from pMVB172 was immobilized on glutathione-agarose and incubated with the indicated purified His6-tagged septins. After washing, bound proteins were eluted, resolved by SDS-PAGE, and analyzed either by Ponceau S dye (to verify equal loading of the GST-fusion proteins) or by immunoblotting by using anti-His monoclonal antibody to detect the input and bound (His)6-tagged proteins. Input, 20% of the total amount of the indicated His6-tagged protein that was added in each binding reaction.
Roles for the CTE of Cdc3 and Cdc11
The pairwise interaction assays, the analysis of multiseptin complex formation, and the binding properties of the isolated CTECdc12 described above all demonstrate that Cdc12 interacts via its CTE with Cdc3, and vice versa. These two core septins are both essential proteins in yeast. By contrast, the only septin that requires its CTE to interact with Cdc11 is the nonessential septin Shs1. If the reason that the CTE of Cdc12 is required for viability (Figure 2A) is that it is essential for mediating association with the CTE of Cdc3, the Cdc3 CTE should therefore also be necessary for cell viability. By the same logic, if the only function of the CTE of Cdc11 is to recruit the nonessential septin Shs1, a Cdc11ΔC mutant should be viable.
To test these ideas, truncations that removed the CTEs of Cdc3 and Cdc11, namely, Cdc3(Δ440–520) and Cdc11(Δ372–415), respectively, were constructed and introduced on plasmids into the corresponding heterozygous cdc3_Δ/CDC3_ or cdc11_Δ/CDC11_ diploids. After sporulation, the resulting tetrads were analyzed (in each case, at least 20 tetrads were examined). In our hands, deletion of CDC11 is lethal in BY4741 (a derivative of S288c), as has been observed for other S288c derivatives (Casamayor and Snyder, 2003), and in W303 (unpublished data), in contrast to what has been reported for one strain background (YEF473), in which viable _cdc11_Δ haploids purportedly were recovered (Frazier et al., 1998). As anticipated, Cdc3ΔC was unable to rescue the viability of _cdc3_Δ spores (Figure 6A), whereas Cdc11ΔC was able to maintain the viability of _cdc11_Δ spores (Figure 6B). Even when overexpressed from a multi-copy plasmid, Cdc3ΔC was unable to support growth when present as the sole source of Cdc3 (unpublished data). Likewise, _cdc3_Δ haploids carrying a _URA3_-marked plasmid CDC3 (to maintain viability) and _LEU2_-based plasmids (either centromeric or multi-copy) expressing CDC3_Δ_C were unable to grow when plated on medium containing 5-fluoro-orotic acid, which selects for cells that have lost the URA3 plasmid. As observed for overexpression of Cdc12ΔC (Figure 2, C and D), overexpression of Cdc3ΔC in otherwise wild-type cells caused elongated bud morphology and cytokinesis defects, and perturbed the localization of Cdc3-GFP, as well as that of Cdc10-GFP, Cdc11-GFP, and Cdc12-GFP (unpublished data). Thus, like the Cdc12 CTE, the Cdc3 CTE is critically important for proper organization of septins in vivo.
Figure 6.
The Cdc3 CTE is essential, but the Cdc11 CTE is not. (A) A cdc3_Δ/CDC3_ heterozygous diploid (Y25223) was transformed with empty vector, or a CEN plasmid carrying either CDC3 (pMVB100) or CDC3_Δ_C (pMVB102), sporulated and the meiotic products analyzed described in the legend of Figure 2A. (B) A cdc11_Δ/CDC11_ heterozygous diploid (YMVB32) was transformed with empty vector, or a CEN plasmid expressing CDC11 (pSB1) or CDC11_Δ_C (pSB2), and analyzed as in A. C. A _cdc11_Δ mutant expressing either CDC11 or CDC11_Δ_C from the GAL1 promoter on CEN vectors (YCpUG-CDC11 and YCpUG-CDC11ΔC, respectively) were grown to midexponential phase and analyzed by differential interference contrast (top) or indirect immunofluorescence by using rabbit polyclonal anti-Cdc11 as the primary antibody (bottom). Samples (30 μg total protein) of extracts prepared from the same cultures were analyzed by SDS-PAGE and immunoblotting by using the anti-Cdc11 antibody. D. A _cdc11_Δ mutant expressing either CDC11 (WT) from pSB1 or CDC11_Δ_C (cdc11_Δ_C) from pSB2 was transformed with a plasmid (pLP17) expressing Cdc12-GFP, grown to midexponential phase and viewed by DIC (left) or fluorescence (right) microscopy (GFP).
Although absence of the Cdc11 CTE is not lethal, cells containing Cdc11ΔC as the only source of this septin displayed severe cytokinesis defects and elongated buds, indicating that Cdc11 CTE is required for proper septin collar function. These results with regard to the CTE of Cdc11 are in agreement with a previous report (Casamayor and Snyder, 2003), but at odds with another prior study (Lee et al., 2002). We found that Cdc11ΔC is more unstable than full-length Cdc11 (unpublished data), which suggested that the observed phenotypes of cells expressing Cdc11ΔC as the sole source of this septin might simply be due to the lower level of the mutant protein. However, overproduction of CDC11_Δ_C using the GAL1 promoter exacerbated, rather than ameliorated, the aberrant morphology of cells expressing Cdc11ΔC (Figure 6C). In contrast to Cdc12ΔC-GFP and Cdc3ΔC-GFP, Cdc11ΔC still localized normally to the bud neck, even in cells lacking normal Cdc11 (Figure 6C) and did not interfere with localization of Cdc12-GFP at the bud neck (Figure 6D). Thus, unlike the CTE of Cdc3 and Cdc12, the CTE of Cdc11 does not seem to play a major role in septin collar formation, but rather in septin collar function.
Roles of Cdc10 in Septin Collar Architecture
Cdc12 associates with Cdc3 via its CTE and with Cdc10 (and Cdc11) in a CTE-independent manner. To determine whether both of these modes of binding are necessary for incorporation of Cdc12 into the septin collar in vivo and to assess the specific role that Cdc10 may play in localizing Cdc12 to the bud neck, we examined the subcellular localization of Cdc12-GFP and Cdc12ΔC-GFP in normal and in _cdc10_Δ cells, which are viable at or below normal growth temperature (30°C). In wild-type cells, both Cdc12-GFP and Cdc12ΔC-GFP decorated the bud neck, but Cdc12ΔC seemed to be incorporated less efficiently, as expected, based on the reproducibly dimmer staining seen in every cell (Figures 2B and 7A). In cells lacking Cdc10, Cdc12-GFP still localized at the bud neck, but at a greatly reduced level. However, Cdc12-GFP is significantly less stable in cells lacking Cdc10 than in wild-type cells (Figure 7A, right), which presumably accounts for the reduction in signal observed. In marked contrast, no detectable Cdc12ΔC-GFP was present at the bud neck in cells lacking Cdc10, even though Cdc12ΔC-GFP is no less stable under these conditions than Cdc12-GFP (Figure 7A). Thus, in the absence of the Cdc3-Cdc12 interaction mediated by its CTE, recruitment of Cdc12 to the septin collar in vivo requires Cdc10. These data suggest that, in the absence of Cdc10, the residual interactions between Cdc12ΔC and other septins observed in vitro are not sufficient to permit its stable incorporation into the septin collar at the bud neck.
Close inspection revealed that, in cells lacking Cdc10, even normal Cdc12-GFP never localized to both sides of the septin collar, as it does in wild-type cells (Figure 7B). To mark the cell boundary so that we could discriminate septin recruitment to the mother and bud sides of the septin collar, the chitin deposited at the bud neck was stained with calcofluor white. Whether expressed in wild-type cells at a near normal level (from the CDC12 promoter on a CEN plasmid) or highly overexpressed (from a 2 μm DNA vector), Cdc12-GFP localization is centered around the chitin ring and seems to be symmetrically distributed on the mother and daughter sides of the septin collar (Figure 7B). In _cdc10_Δ cells, however, Cdc12-GFP localized only to the daughter-side of the bud neck, even when copiously overproduced (Figure 7B). Likewise, both Cdc3-GFP and Cdc11-GFP also localize asymmetrically to the bud side of the neck in _cdc10_Δ cells [unpublished data; see also Supplemental Material cited in Castillon _et al_. (2003)]. Thus, lack of Cdc10 grossly perturbs septin organization; therefore, Cdc10 plays an important role in dictating the arrangement of the septin filaments in the septin collar at the bud neck.
Cdc3, Cdc11, and Cdc12 Are Necessary and Sufficient for Septin Filament Formation
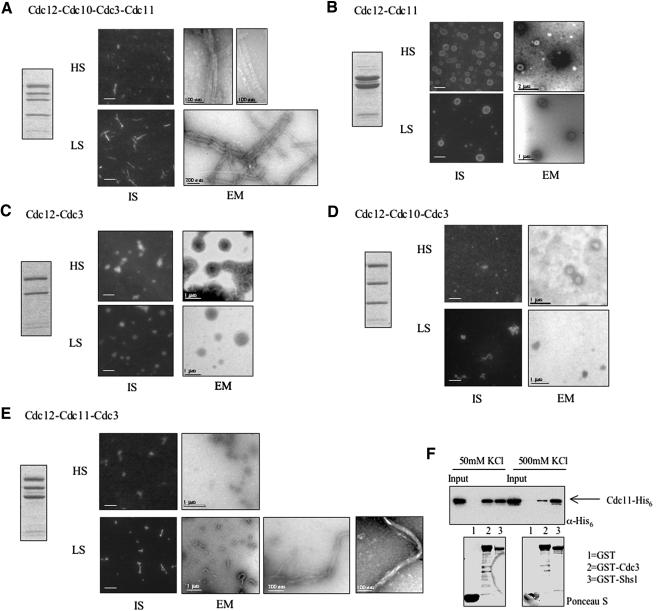
To gain insight about the contribution of individual septins to septin polymer assembly and the properties of septin filaments, each of the purified stoichiometric complexes reconstituted by coexpression of recombinant proteins was examined for its ability to form polymers by using both an immunostaining assay with anti-Cdc12ΔC antibodies and examination in the EM after negative staining (Figure 8). Because high ionic strength decreases the ability of heteromeric septin complexes to form septin filaments and low ionic strength promotes filament formation (Frazier et al., 1998; Kinoshita et al., 2002; Versele and Thorner, 2004), the filament-forming capacity of each complex was examined both in the high-salt (HS) elution buffer (containing 250 mM NaCl and 300 mM imidazole) and after dialysis into a low-salt (LS) buffer (50 mM KCl).
Figure 8.
Ultrastructural analysis of reconstituted septin filaments. (A–E) The ability of the indicated purified, recombinant, heteromeric septin complexes (left panel) to form filaments in the high-salt elution buffer used for purification (HS) or after dialysis against 50 mM KCl (LS) were analyzed, as indicate, either by immunostaining using anti-Cdc12ΔC antibodies (IS, bars represent 2% μm) or by EM after negative staining (bar represents scale indicated). (F) The interaction between Cdc3 and Cdc11 is weakened at high salt concentration. Binding assay was performed as described in the legend to Figure 3 at the salt concentrations indicated.
The His6Cdc12-Cdc3-Cdc10-Cdc11 heterotetrameric complexes formed filaments, even in HS buffer (Figure 8A). In the EM, the filaments are typically straight and almost invariably paired. The range of widths measured for individual filaments (n = 30) was 7–10 nm, and for paired filaments (n = 30) was 16–22 nm. Lengths varied greatly, from 50 nm to >1 μm. Occasionally, lateral striations occurring with a repeat length of 22–33 nm were visible (as measured on four different filament pairs, each containing >15 striations). In LS buffer, the filaments often associated laterally into bundles of up to 20 filaments. These bundles could be observed as straight, strikingly bright objects visible by light microscopy after immunostaining (Figure 8A).
In LS buffer (but not in HS), the His6Cdc12-Cdc3-Cdc11 ternary complex also formed filaments (Figure 8E). However, the appearance of these filaments was rather different from those generated from the heterotetrameric His6Cdc12–Cdc3–Cdc10–Cdc11 complexes. At the light microscope level, the filament bundles composed of the His6Cdc12–Cdc3–Cdc11 heterotrimeric complexes seemed shorter, more curved, and less bright than those formed from the His6Cdc12–Cdc3–Cdc10–Cdc11 heterotetrameric complexes. EM analysis confirmed that short, curved filaments were formed. Moreover, unlike the filaments made from the His6Cdc12–Cdc3-Cdc10–Cdc11 complex, these filaments were not paired in register, although many of the curved forms comprise apparently unorganized bundles of multiple filaments (Figure 8E).
His6Cdc12 alone did not polymerize into filaments, but sometimes small rings or disks (50–200 nm in diameter) could be observed by immunostaining and by EM (unpublished data). The His6Cdc12–Cdc11 complex nearly always formed rings or disks (200–500 nm in diameter) in either HS or LS buffers (Figure 8B). By contrast, the His6Cdc12–Cdc3 complex formed rather amorphous-looking blobs (400–500 nm in diameter) at the light microscope level, which, at the EM level, sometimes had a honeycomb-like appearance with knobby protrusions (Figure 8C). The His6Cdc12–Cdc3–Cdc10 ternary complex failed to form any regular structure that could be discerned by either immunostaining or EM (Figure 8D). Finally, the complexes that contain stoichiometric amounts of His6Cdc12ΔC and Cdc11, but substoichiometric amounts of Cdc3 and Cdc10, did not form filaments, but instead displayed rings (unpublished data), similar to those formed by the His6Cdc12–Cdc11 heterodimer complex (Figure 8B).
These data demonstrate that the three essential septins— Cdc3, Cdc11, and Cdc12—are both necessary and sufficient for polymerization of heteromeric septin complexes into filaments, whereas Cdc10 is required to permit those polymers to organize into straighter and more regular filament pairs. The simplest model compatible with all of these data (see Discussion) is that Cdc3–Cdc12–Cdc1l complexes polymerize end to end. We entertained the notion that the Cdc3–Cdc11 association observed under some circumstances (Figure 3B) might play a pivotal role in this polymerization. Therefore, and because polymerization is enhanced at low salt, we tested whether Cdc3–Cdc11 association is salt-sensitive. Indeed, binding of Cdc11-His6 to GST-Cdc3 was robust at low salt and decreased at high salt, whereas binding of Cdc11-His6 to GST-Shs1 (which is mediated by their CTEs) was not salt dependent (Figure 8F).
DISCUSSION
Formation of CTE-mediated Cdc3–Cdc12 Heterodimeric Complex Is Essential for Cell Viability
Two structural hallmarks of septins are a conserved GTPase domain, and in nearly all cases, a prominent CTE containing a predicted coiled coil segment. The essential yeast septin, Cdc12, is an intrinsically active GTPase, but mutations that abrogate its GTPase activity do not compromise the essential function of Cdc12 in vivo. A Cdc12 GTPase mutant, cdc12(S43V), and even a GTP binding-defective mutant, cdc12(T48N), rescue the inviability of _cdc12_Δ cells (Versele and Thorner, 2004). By contrast, we have shown here that absence of the CTE destroys the ability of Cdc12 to perform its essential function in vivo, but it does not compromise its ability to bind and hydrolyze GTP (Versele and Thorner, 2004).
The essential function of Cdc12 for which its CTE is required is formation of heterodimeric complexes with Cdc3 via interaction with the CTE of Cdc3. Without this critical pair, no stable multimeric septin complexes can be assembled and no filament formation can occur. Several observations support this conclusion. First, the pairwise association of Cdc12 with Cdc3, and vice versa, depends on an intact CTE in each protein (Figure 3). Second, even in the presence of other septins that might potentially stabilize higher oligomers, Cdc12ΔC is unable to form complexes that contain stoichiometric amounts of Cdc3, unlike full-length Cdc12 (Figure 4). Third, only complexes in which Cdc3 and Cdc12 are present stoichiometrically are competent to form filaments (Figure 8). Fourth, purified Cdc3 binds tightly to purified GST-CTECdc12 (Figure 5), and GFP-CTECdc12 in cell extracts associates avidly with GST-Cdc3, but not with GST-Cdc3ΔC (unpublished data). Fifth, like the Cdc12 CTE (but not the Cdc11 CTE or Cdc10 or Shs1), the Cdc3 CTE is essential for yeast cell viability. Sixth, elevated expression of either Cdc12ΔC or Cdc3ΔC (but not their full-length counterparts or Cdc11ΔC) interferes with proper septin collar assembly.
Wild-type yeast display circular, 10-nm profiles (in cross section) and 10-nm striations (in tangential/grazing sections) in negatively-stained EM thin sections of the bud neck (Byers and Goetsch, 1976). A lack of such objects at the neck in _cdc10_Δ cells was interpreted to mean that septins may be able to function in the absence of normal polymerization (Frazier et al., 1998). Our findings cast doubt on this interpretation. Rather, our findings suggest that septin filaments are present at the bud neck in _cdc10_Δ cells, but lack the normal organization seen in wild-type cells. First, we found that Cdc3–Cdc12–Cdc11 heterotrimeric complexes are competent to form filaments in vitro (Figure 8), and they seem to be competent to form filaments in vivo because septin-containing collars clearly assemble in _cdc10_Δ cells (Figure 7). Second, the filaments formed in vitro by Cdc3–Cdc12–Cdc11 complexes are strikingly different from those formed from Cdc3–Cdc10–Cdc12–Cdc11 complexes (Figure 8). Correspondingly, the collar architecture in cells lacking Cdc10 is clearly aberrant (Figure 7). Third, in keeping with the altered form of Cdc3–Cdc12–Cdc11-containing filaments, the septins themselves (Figure 7) and septin-associated proteins (unpublished data) are recruited to the bud neck in abnormal distributions in _cdc10_Δ mutants (Castillon et al., 2003). Finally, as we also have shown here, disruption of the filament-forming capacity of any of the three core septins, Cdc3, Cdc11, or Cdc12, is not compatible with yeast cell viability. Therefore, septin filament formation is essential for yeast cell survival.
Unlike the CTEs of Cdc3 and Cdc12, the CTE of Cdc11 is not essential for viability (although Cdc11 itself is). Thus, the globular, N-terminal GTP-binding domain of Cdc11 is sufficient for the essential function of Cdc11 (Figure 6A) and for its recruitment to the bud neck (Figure 6C). Nevertheless, cells containing Cdc11ΔC as the sole source of this septin displayed severe morphological and cytokinesis defects. The observed phenotype of the Cdc11ΔC mutant may be due, at least in part, to lack of recruitment of the nonessential septin Shs1 because we found that Cdc11 is the only septin that associates with Shs1 and that the CTE of Shs1 is important for this interaction (Figure 3C). However, the possibility cannot be excluded that the CTEs of septins serve other functions, in addition to their role in mediating septin–septin interactions. For example, it has been reported that Gin4, a protein kinase localized to the bud neck, binds to the region of Cdc3 that contains its presumptive coiled-coil (Longtine et al., 1998); however, a more recent study indicates that the septin to which Gin4 binds directly is Shs1 (Mortensen et al., 2002). Two-hybrid analysis indicates that the Cdc11 CTE may interact with Bem4, a protein implicated in bud emergence (Casamayor and Snyder, 2003), but this conclusion must be interpreted with caution because bridging by other yeast proteins could be responsible for the interaction observed in vivo. We found that another bud-neck associated protein kinase, Hsl1, binds directly to Cdc12 in vitro (Versele and Thorner, 2004); but, this interaction does not require the Cdc12 CTE (unpublished data). Similarly, in vitro binding of Bni5, another bud-neck–interacting protein, to Cdc11 does not require the CTE of this septin (Lee et al., 2002). It remains to be determined how septin filament-associated proteins are recruited to the bud neck, and whether the CTE of any septin participates in these interactions.
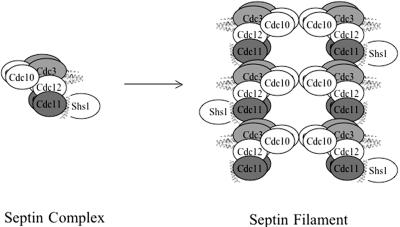
A Model for the Heteropentameric Septin Complex in Mitotic Cells
Based on the findings presented here, and taking into account prior analyses of the composition of septin complexes isolated from yeast extracts (Frazier et al., 1998; Mortensen et al., 2002), we propose a model for the arrangement of the subunits in the heteropentameric septin complex present in mitotic cells (Figure 9). Cdc12 is the linchpin of this assembly. Cdc12 interacts with Cdc3 via the CTE of each protein, and with Cdc11 via the N-terminal domain of each protein. Based on the lack of a CTE in Cdc10 (Figure 1), on the fact that the CTEs of neither Cdc3 nor Cdc12 are necessary for their interaction with Cdc10 (Figure 3), and on our observation that Cdc10 has a higher affinity for the Cdc3–Cdc12 complex than for either protein alone (Figure 4A), Cdc10 presumably interacts with an interface created by the globular N-terminal domains of Cdc3 and Cdc12.
Figure 9.
Model for septin heteropentamer organization and assembly into filaments. Together, the results obtained in this study indicate that septin heteropentamers are assembled with the interseptin contacts indicated and suggest that filaments are formed by the end-to-end polymerization of Cdc3–Cdc12–Cdc11 complexes, with Cdc10 serving as a bridge to bundle the polymers into paired filaments.
The C-terminal ends of the CTEs of Cdc3 and Cdc12 are predicted to form parallel coiled coils, but we have found no direct support for this structure, based on the lack of apparent interaction between the corresponding Cdc12 and Cdc3 peptides. Although coiled coil formation by the entire CTE of each protein is not ruled out, the absence of side chains to constitute a regular, 4-3 hydrophobic repeat in the N-terminal portion of each CTE suggests that a helical fold and novel contact surface distinct from the canonical coiled coil may mediate association between the CTEs of Cdc3 and Cdc12. We anticipate a parallel arrangement in the association of the Cdc3 and Cdc12 chains for three reasons: 1) the N-terminal domains of Cdc3 and Cdc12 (Cdc3ΔC and Cdc12ΔC) have detectable affinity for each other (Figure 3); 2) the properties of Cdc10 association with Cdc3–Cdc12 complexes (Figures 4 and 8) argues that the N-terminal domains of Cdc3 and Cdc12 must be in proximity; and 3) others have found that fluorescence energy resonance transfer only occurs between two mammalian septins, Sept6 and Sept7 (whose closest yeast orthologues are Cdc3 and Cdc12, respectively), when the fluorophores are both attached to the N termini of the proteins, but not when the probes are attached to opposite ends (C. Low and I. Macara, personal communication).
Cdc12 interacts strongly with Cdc11, and this association does not require the CTE of Cdc12 (Figures 3 and 4) and occurs in the presence of Cdc3 (Figure 4). Hence, in the heteropentameric unit, Cdc11 must be juxtaposed to Cdc12 without interfering with the interaction of Cdc12 with Cdc3 (Figure 9). Although a direct Cdc3-Cdc11 interaction can be detected (Figure 3), using the approach of bacterial coexpression to reconstitute septin complexes, Cdc11 had no impact on the recruitment of Cdc3 into the complex. Instead, we presented evidence (Figure 8F) for the idea that an electrostatic (salt-sensitive) interaction between Cdc3 and Cdc11 could play a key role in the polymerization of septin complexes into filaments (Figure 9). Finally, by analogy to the Cdc3–Cdc12 complex, the association between Cdc11 and Shs1 (Figure 3C) is presumably parallel and mediated via interaction of their respective CTEs.
In one study (Frazier et al., 1998), it was estimated that, in purified septin filaments recovered from yeast cells, Cdc3, Cdc10, and Cdc12 were present in approximately equal amounts, whereas Cdc11 was less abundant and Shs1 was not recovered. In a more recent analysis, Cdc11 and especially Shs1 were also substoichiometric to the other septins in complexes that copurified with Gin4 (Mortensen et al., 2002). The recombinant heterotetrameric complexes we prepared reproducibly contained stoichiometric amounts of all four input septins (Cdc3, Cdc10, Cdc11, and Cdc12) (Figure 4) and displayed robust filament formation (Figure 8). An important difference in our work, which may account for the observed difference in the efficiency of Cdc11 recovery in heteromeric septin complexes, is that we used only 250 mM salt during purification, whereas 0.5–1 M salt was used in the other studies cited.
Our model (Figure 9) depicts the core of the complex as comprising four sets of dimers. This proposed feature reflects our finding that all septins tested (Cdc3, Cdc10, Cdc11, and Cdc12) self-associated (Figure 2). The calculated M_r_ for this predicted 2:2:2:2 arrangement is 382 kDa, which is in remarkably good agreement with the apparent molecular weight (370 ± 60 kDa) of the native septin complex isolated from yeast (Frazier et al., 1998). Circumstantial evidence suggests that the (Cdc3)2–(Cdc12)2 sub-complex may involve a helical tetrameric assembly: Cdc3-Cdc3 and Cdc12-Cdc12 self-association requires the CTE of each protein; the interaction of Cdc3 and Cdc12 requires the CTE of each protein; and, the isolated CTE of Cdc12 associates both with itself and with Cdc3 (and the latter interaction depends on the CTE of Cdc3). The ability of the isolated Cdc12 CTE to interact with Cdc3 (Figure 5), but the inability of a synthetic peptide corresponding to just the predicted coiled coil-forming segment of Cdc12 to associate with the corresponding segment of Cdc3, argues that the entire CTE sequence (Figure 1) is crucial for stabilizing the CTE–CTE interaction.
Shs1 seems to be most peripheral component of the complex, in agreement with its substoichiometric presence in native septin complexes (Mortensen et al., 2002) and with the fact that it is not essential for cell viability. Despite its clear-cut, specific, and CTE-dependent binding to Cdc11 (Figure 2), we were unable to purify a stoichiometric complex between Cdc11 and Shs1 by coexpression in bacterial cells, suggesting that the competency of Cdc11 to recruit Shs1 in yeast may depend on its association with the other septins or on some other yeast-specific factor or posttranslational modification.
Polymerization of Septin Complexes into Filaments and Organization of the Septin Collar
Cdc3, Cdc11, and Cdc12 are all necessary, and the three together are sufficient, to form filaments in vitro, but only under low ionic strength conditions. Hence, we favor the view that the filaments arise from end-to-end polymerization of the Cdc3–Cdc12–Cdc11 complex via electrostatic contacts between the globular N-terminal heads of Cdc3 and Cdc11 (Figure 9). Our finding that septin polymerization occurs in the absence of Cdc10 is at odds with a prior report (Frazier et al., 1998). The following factors may account for the failure of others to observe such filaments. First, Cdc3–Cdc12–Cdc11 filaments are less stable than filaments formed from Cdc3–Cdc12–Cdc11–Cdc10 heterotetrameric complexes. Moreover, the latter polymerize even in high salt, the former do not. Second, Cdc11 is essential for filament formation (Figure 8), yet Cdc11 was reportedly substoichiometric in the native septin complexes purified in 1 M salt (Frazier et al., 1998). In contrast, Cdc11 was present in stoichiometric amounts in our recombinant complexes. Third, in the EM after negative staining with uranyl acetate, septin filaments lacking Cdc10 show weaker contrast and are harder to resolve from the background than filaments containing all four core septins, perhaps because filaments generated from Cdc3–Cdc12–Cdc11 ternary complexes are not paired in register, like the filaments generated from Cdc3–Cdc12–Cdc11–Cdc10 quaternary complexes.
Although Cdc10 apparently is dispensable for septin polymerization in vitro, it is required for the lateral pairing of septin filaments. The paired filaments are straight and stable under high ionic strength conditions. Because Cdc10 self-associates, it is possible that pairing occurs via homomeric interactions of Cdc10 (Figure 9). Its mutual binding to Cdc3 and Cdc12, documented here, presumably allows Cdc10 to act as a linker to strengthen the interaction between these two other core septins (and perhaps to optimize their mutual orientation). These roles for Cdc10 —as both a brace within an individual filament and as a bridge to connect filaments laterally—fit with the observed phenotypes of a _cdc10_Δ mutant. First, a _cdc10_Δ cell is inviable above 30°C. Thus, although Cdc10 is not essential for cell viability under normal growth conditions, it does become essential under conditions where septin filament structure is under greater thermal stress. Second, even at lower temperatures, a _cdc10_Δ mutant displays obvious defects in morphology and cytokinesis, suggesting that, when Cdc10 is absent, there are abnormalities in the structure of the septin filaments and/or in their organization at the septin collar. Indeed, in thin-section EM, a _cdc10_Δ mutant does not show profiles that resemble the normal neck filaments seen in wild-type cells (Frazier et al., 1998). Reduced stability may make the Cdc12–Cdc3–Cdc11 filaments harder to visualize and/or unable to withstand the preparation and fixation conditions. Finally, we have shown here that, in cells lacking Cdc10, GFP-tagged septins never decorate the collar in the “double ring” typically seen in wild-type cells. Rather, in a _cdc10_Δ mutant, septins are predominantly localized to the daughter side of the bud neck. Thus, Cdc10-mediated filament pairing seems essential for proper organization of the septin filaments in the collar at the bud neck.
On a more speculative note, different models of septin filament organization at the bud neck have been proposed. In one, septin polymers are associated into filaments that are perpendicular to the mother-bud axis and organized circumferentially around the collar, like the coils of a spring. Impetus for this “spiral model” first came from EM images of grazing sections of the bud neck (Byers and Goetsch, 1976), but the idea is supported by the dramatic septin-containing coils seen in the elongated buds elicited when Candida albicans Int1 is overexpressed in S. cerevisiae (Gale et al., 2001) and by the finding that certain mammalian septin complexes self-assemble into spirals (Kinoshita et al., 2002). The alternative “picket fence” model is that, in the collar, septin filaments organize into lateral bundles aligned parallel to the motherbud axis. This idea was first suggested on the basis of the bar-like septin-containing aggregates seen in a _gin4_Δ mutant (Longtine et al., 1998).
Either model is consistent with the extensive lateral bundling of filaments we observed in our recombinant filament preparations. In the picket fence model, assuming that all of the lateral filament bundles are organized in the same direction, the intrinsic polarity of a filament comprised of nonsymmetrical subunits (Figure 9) could provide an explanation for how proteins can be recruited to one side of the collar or the other. Proteins that interact with Cdc3 or another Cdc3-binding protein would be associated with one side of the collar, and those that interact with Cdc11 or another Cdc11-binding protein would associate with the other side of the collar. Attractive as this idea is, there is little or no direct support for it. Instead, in freeze-fracture and scanning EM images of the cortical surface of yeast plasma membranes, those septin-containing structures that can be visualized have a “gauze-like” appearance (A. Rodal and J. Hartwig, personal communication), where the filaments lie across each other in a cross-hatched arrangement. This observation might suggest a third model for the organization of the septin filaments at the bud neck, in which the collar expands, like the slats in a child's safety gate, due to the pivoting of the filaments or filament bundles against each other. We are currently applying various strategies to help distinguish between these models.
Acknowledgments
We thank Doug Kellogg, Brian Haarer, Tim Stearns, and Rong Li for the generous gift of research materials; David S. King for the preparation of synthetic peptides; Erin Scott and Lisa Valdin for early technical help; Dagmar Truckses for vector construction; Françoise Roelants for critical reading of the manuscript; and other Thorner laboratory members for advice. This work was supported by a long-term fellowship from the Human Frontier Science Program Organization and by a North Atlantic Treaty Organisation Advanced Fellowship to M.V., by a fellowship from the Blanceflor Foundation to B.G., by fellowships from the Fulbright and Del Amo Foundations to V.J.C., by National Institutes of Health Predoctoral Traineeship GM-07232 to R.E.C., by National Institutes of Health Research Grant GM-48958 to T.A., and by National Institutes of Health research grant GM-21841 and facilities provided by the Berkeley campus Cancer Research Laboratory to J.T.
Abbreviations used: CTE, C-terminal extension; NTA, nitrilotriacetate.
References
- Bardwell, L., Cook, J.G., Zhu-Shimoni, J.X., Voora, D., and Thorner, J. (1998). Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase kss1 requires the dig1 and dig2 proteins. Proc. Natl. Acad. Sci. USA 95, 15400-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, Y., Mermali, V., Mooseker, M.S., and Snyder, M. (2000). Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5, 841-851. [DOI] [PubMed] [Google Scholar]
- Barral, Y., Parra, M., Bidlingmaier, S., and Snyder, M. (1999). Nim1-related kinases coordinate cell-cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13, 176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beites, C.L., Xie, H., Bowser, R., and Trimble, W.S. (1999). The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 2, 434-439. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- Byers, B., and Goetsch, L. (1976). A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69, 717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor, A., and Snyder, M. (2003). Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol. Cell. Biol. 23, 2762-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon, G.A., Adames, N.R., Rosello, C.H., Seidel, H.S., Longtine, M.S., Cooper, J.A., and Heil-Chapdelaine, R.A. (2003). Septins have a dual role in controlling mitotic exit in budding yeast. Curr. Biol. 13, 654-658. [DOI] [PubMed] [Google Scholar]
- Caviston, J.P., Longtine, M., Pringle, J.R., and Bi, E. (2003). The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid, V.J. Adamikova, L, Cenamor, R. Molina, M, Sanchez, M., Nombela, C. (1998). Cell integrity and morphogenesis in a budding yeast septin mutant. Microbiology 144, 3463-3474. [DOI] [PubMed] [Google Scholar]
- Dobbelaere, J., Gentry, M.S., Hallberg, R.L., and Barral, Y. (2003). Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4, 345-357. [DOI] [PubMed] [Google Scholar]
- Fares, H., Peifer, M., and Pringle, J.R. (1995). Localization and possible functions of Drosophila septins. Mol. Biol. Cell 6, 1843-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, C.M., al-Awar, O., Rosenblatt, J., Wong, M.L., Alberts, B., and Mitchinson, T.J. (1996). A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133, 605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier, J.A., Wong, M.L., Longtine, M.S., Pringle, J.R., Mann, M., Mitchison, T.J., and Field, C. (1998). Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143, 737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, C., Gerami-Nejad, M., McClellan, M., Vandoninck, S., Longtine, M.S., and Berman, J. (2001). Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell 12, 3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527-534. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A.S., Pringle, J.R., and Lew, D.J. (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681-689. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557-580. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. (1971). Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell. Res. 69, 265-276. [DOI] [PubMed] [Google Scholar]
- Jones, E.W. (1991). Tackling the protease problem in yeast. Methods Enzymol. 194, 428-453. [DOI] [PubMed] [Google Scholar]
- Jones, J.S., and Prakash, L. (1990). Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6, 363-366. [DOI] [PubMed] [Google Scholar]
- Kinoshita, M. (2003). The septins. Genome Biol. 4, 236.1-236.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, A., Kinoshita, M., Akiyama, H., Tomimoto, H., Akiguchi, I., Kumar, S., Noda, M., and Kimura, J. (1998). Identification of septins in neurofibrillary tangles in Alzheimer's disease. Am. J. Pathol. 153, 1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, M., Field, C.M., Coughlin, M.L., Straight, A.F., and Mitchison, T.J. (2002). Self- and actin-templated assembly of mammalian septins. Dev. Cell, 3, 791-802. [DOI] [PubMed] [Google Scholar]
- Kinoshita, M., Kumar, S., Mizoguchi, A., Ide, C., Kinoshita, A., Haraguchi, T., Hiraoka, Y., and Noda, M. (1997). Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 11, 1535-1547. [DOI] [PubMed] [Google Scholar]
- Lee, P.R., Song, S., Ro, H.S., Park, C.J., Lippincott, J., Li, R., Pringle, J.R., De Virgilio, C., Longtine, M.S., and Lee, K.S. (2002). Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 6906-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and Li, R. (1998). Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143, 1947-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., Fares, H., and Pringle, J.R. (1998). Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143, 719-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, K. (2000). A modular set of prokaryotic and eukaryotic expression vectors. Analyt. Biochem. 277, 109-120. [DOI] [PubMed] [Google Scholar]
- Mortensen, E.M., McDonald, H., Yates J. 3rd, Kellogg, D.R. (2002) Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell 13, 2091-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakchaisri, K., Asano, S., Yu, L.R., Shulewitz, M.J., Park, C.J., Park, J.E., Cho, Y.W., Veenstra, T.D., Thorner, J., and Lee, K.S. (2004). Coupling morphogenesis to mitotic entry. Proc. Natl. Acad. Sci. USA 101, 4124-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sanger, F., Nicklen, S., and Coulson, A.R. (1977). DNA sequencing with chain-termination inhibitor. Proc. Natl. Acad. Sci. USA 74, 5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield, P.J., Oliver, C.J., Kremer, B.E., Sheng, S., Shao, Z., and Macara, I.G. (2003). Borg/Septin interactions and the assembly of mammalian septin heterodimers, trimers and filaments. J. Biol. Chem., 278, 3483-3488. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Hicks, J.B. (1986). Laboratory Course Manual for Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Shulewitz, M.J., Inouye, C.J., and Thorner, J. (1999). Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7123-7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P.A., DeRisi, J.L., Wilhelm, J.E., and Vale, R.D. (2000). Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341-344. [DOI] [PubMed] [Google Scholar]
- Versele, M., and Thorner, J. (2004). Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol., 164, 701-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsen, P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793-1808. [DOI] [PubMed] [Google Scholar]
- Wolf, E., Kim, P.S., and Berger, B. (1997). MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6, 1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfson, D.N., and Alber, T. (1995). Predicting oligomerization states of coiled coils. Protein Sci. 4, 1596-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner, J.E., Harkins, H.A., and Pringle, J.R. (1996). Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 1857-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Kong, C., Xie, H., McPherson, P.S., Grinstein, S., and Trimble, W.S. (1999). Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr. Biol. 9, 1458-1467. [DOI] [PubMed] [Google Scholar]