Comparison of Diversities and Compositions of Bacterial Populations Inhabiting Natural Forest Soils (original) (raw)
Abstract
The diversity and composition of soil bacterial communities were compared among six Austrian natural forests, including oak-hornbeam, spruce-fir-beech, and Austrian pine forests, using terminal restriction fragment length polymorphism (T-RFLP, or TRF) analysis and sequence analysis of 16S rRNA genes. The forests studied differ greatly in soil chemical characteristics, microbial biomass, and nutrient turnover rates. The aim of this study was to relate these differences to the composition of the bacterial communities inhabiting the individual forest soils. Both TRF profiling and clone sequence analysis revealed that the bacterial communities in soils under Austrian pine forests, representing azonal forest types, were distinct from those in soils under zonal oak-hornbeam and spruce-fir-beech forests, which were more similar in community composition. Clones derived from an Austrian pine forest soil were mostly affiliated with high-G+C gram-positive bacteria (49%), followed by members of the α_-Proteobacteria_ (20%) and the Holophaga/Acidobacterium group (12%). Clones in libraries from oak-hornbeam and spruce-fir-beech forest soils were mainly related to the Holophaga/Acidobacterium group (28 and 35%), followed by members of the Verrucomicrobia (24%) and the α_-Proteobacteria_ (27%), respectively. The soil bacterial communities in forests with distinct vegetational and soil chemical properties appeared to be well differentiated based on 16S rRNA gene phylogeny. In particular, the outstanding position of the Austrian pine forests, which are determined by specific soil conditions, was reflected in the bacterial community composition.
Forests represent the natural vegetation cover in the majority of landscapes in Central Europe (11). Different types of forest exist which, at their natural sites, are distributed according to regional climate conditions, if they are situated on zonal soils, or otherwise respond to specific, azonal soil conditions (11). In Austria, 47% of the total area presently consists of forested land (12). Although forests have widely been used for wood production, involving selective promotion of singular tree species or replacement of natural species by planted tree species, some remnants of forest still exist which have a natural vegetation composition and hence have been designated as forest reserves. These natural forests are of great value in silviculture, where they represent model systems for studying processes which are important for the implementation of sustainable forest management. Natural forests have also gained recognition as sites of high biodiversity, where complex relationships between fauna, flora, and microflora are maintained due to the structural richness of the habitat and because anthropogenic disturbance by management is restricted.
A range of Austrian natural forests have been surveyed with respect to plant species composition, soil chemical parameters, microbial biomass, and microbially mediated nutrient transformation. Thereby, distinct soil chemical and microbiological conditions have been found to prevail in contrasting forest types (13, 14, 29; E. Hackl, M. Pfeffer, C. Donat, G. Bachmann, and S. Zechmeister-Boltenstern, submitted for publication). However, these findings have not yet been set in relation to the key players in soil nutrient turnover, i.e., the microorganisms inhabiting the different environments. The present study is focused on the bacterial communities of the forest soils and their interrelationships with vegetation and soil environmental conditions.
Soil bacteria are an essential component of the biotic community in natural forests and they are largely responsible for ecosystem functioning, because they participate in most nutrient transformations. Although the main diversity of life has been proven to be microbial, the vast majority of soil bacteria still remain unknown due to the fact that only a minor percentage of naturally occurring microorganisms can be cultured (23). To survey the phylogenetic diversity present in bacterial communities, 16S rRNA gene-based molecular techniques have commonly been applied, which circumvent limitations of culture-based studies. Forest soil bacterial diversity has been characterized by DNA sequence analysis of 16S rRNA genes, derived for instance from soils under Amazonian forest (3), under pinyon-juniper woodlands (7), and under lodgepole pine trees (4). While sequence analysis of clones from 16S rRNA gene clone libraries provides the most detailed, reliable information about the composition of microbial communities, this method is time-consuming and expensive. Terminal restriction fragment length polymorphism (TRF, or T-RFLP) analysis, in contrast, is a DNA-based method which allows rapid comparison of complex bacterial communities. TRF analysis has been applied for a wide range of environmental samples, including agricultural soils (19, 25), grassland (18), and forest soils (7) and biological soil crusts (24). Comparison of bacterial communities by TRF analysis has proven to provide information consistent with analysis of clone libraries (7).
In the present study, 16S rRNA gene-based community analysis has been applied to compare the diversity and composition of the bacterial communities in soils under six natural forest stands which differ in soil and vegetation characteristics, soil microbial biomass, and microbial nutrient turnover rates (13, 14). These include two oak-hornbeam and two spruce-fir-beech forests, which represent zonal vegetation types of the region, as well as two pine forests, which are found on azonal soils that are typically stony and water and nutrient limited. The aim of the study was to relate the bacterial community composition to the already-known characteristics of these forest soils.
MATERIALS AND METHODS
Sampling sites.
Six forest stands were selected which were all situated within the eastern part of Austria and featured two oak-hornbeam, two spruce-fir-beech, and two Austrian pine forests. These forest types are common throughout Europe. Oak-hornbeam and spruce-fir-beech forests comprise zonal vegetation types found in the region and therefore reflect the regional climate (11). Austrian pine (Pinus nigra Arnold) forests are azonal vegetation communities, which may occur in various climatic regions because they are associated with specific soil conditions (11). They have developed on soils over dolomite stone that are restricted to a small layer and are nutrient limited and prone to desiccation (14). Soil and vegetation characteristics of the study sites as well as soil chemical properties have been described by Hackl et al. (14). Site characteristics are presented in Tables 1 and 2.
TABLE 1.
Site characteristics (14) and TRF diversity data from six natural forest soils
| Forest type | Site | Eleva- tion (mi)b | Annual climate data | Soil type | Geology | TRF diversity statisticsa | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Temp (°C) | Pptc (mm) | Phylotype richness (S) | Shannon- Weiner (H) | Simpson (D) | Evenness (E) | |||||
| Oakhornbeam | Johannser Kogel | 325 | 8.8 | 643 | Dystric planosol | Laab formation | 91 (3), bc | 3.82 (0.05), abc | 0.969 (0.003), b | 0.588 (0.055), a |
| Kolmberg | 270 | 8.7 | 593 | Calcaric planosol | Micashist/limestone | 82 (2), c | 3.73 (0.04), c | 0.968 (0.002), b | 0.587 (0.033), a | |
| Spruce-fir-beech | Rothwald | 1,035 | 5.5 | 1,759 | Chromic cambisol | Dolomite | 90 (2), b | 3.82 (0.04), bc | 0.968 (0.001), b | 0.588 (0.036), a |
| Neuwald | 995 | 5.8 | 1,262 | Stagnic luvisol | Sandstone | 96 (4), ab | 3.86 (0.06), abc | 0.971 (0.002), b | 0.587 (0.061), a | |
| Austrian pine | Stampfltal | 640 | 7.0 | 668 | Rendzic leptosol | Dolomite | 98 (4), a | 3.95 (0.06), a | 0.978 (0.002), a | 0.597 (0.068), a |
| Merkenstein | 475 | 8.2 | 554 | Rendzic leptosol | Dolomite | 101 (3), a | 3.89 (0.05), ab | 0.967 (0.004), b | 0.585 (0.042), a |
TABLE 2.
Soil chemical and microbiological characteristics of six natural forest soilsc
| Forest type | Site | pH (CaCl2)d | Organic C (%) | Total soil N (%) | C/N ratio | Microbial biomass (μg/g of organic C) | PLFAs | ||
|---|---|---|---|---|---|---|---|---|---|
| FE-Na | SIR-Cb | Total nmol/g of organic C | Bacterial/ fungal | ||||||
| Oak-hornbeam | Johannser Kogel | 5.0 (0.1) | 5.60 (0.56) | 0.34 (0.03) | 16.07 (0.54) | 1.27 (0.12) | 10.66 (0.86) | 5.11 (0.36) | 17.66 (1.42) |
| Kolmberg | 5.7 (0.3) | 4.93 (0.28) | 0.26 (0.02) | 18.92 (0.52) | 1.60 (0.40) | 15.88 (2.70) | 4.98 (0.36) | 18.42 (1.39) | |
| Spruce-fir-beech | Rothwald | 5.2 (0.3) | 21.44 (2.98) | 0.88 (0.12) | 24.55 (1.58) | 1.90 (0.20) | 14.98 (2.38) | 4.46 (0.46) | 26.99 (1.89) |
| Neuwald | 4.1 (0.2) | 9.90 (0.84) | 0.42 (0.03) | 23.44 (0.72) | 1.21 (0.16) | 12.02 (1.10) | 4.64 (0.27) | 33.17 (2.69) | |
| Austrian pine | Stampfltal | 7.5 (0.0) | 17.73 (1.37) | 0.43 (0.08) | 57.07 (10.64) | 0.56 (0.12) | 3.10 (0.30) | 1.74 (0.21) | 7.99 (0.73) |
| Merkenstein | 7.6 (0.0) | 10.67 (0.70) | 0.21 (0.02) | 52.84 (4.47) | 0.57 (0.07) | 3.42 (0.45) | 2.41 (0.18) | 5.73 (0.35) |
All stands studied have in common that the vegetation composition is considered to be natural in terms of not having been changed by human activities. The Rothwald and Neuwald sites are virgin spruce-fir-beech forests that have never been used for wood production. Both oak-hornbeam forests and both Austrian pine forest stands have similar plant communities as well as similar soil types, but the two spruce-fir-beech forests differ in soil type and geology (Table 1). While the Rothwald forest is situated on nutrient-rich cambisol over dolomite stone, the Neuwald forest soil is classified as leptosol, which is situated on a sandstone substrate with slow nutrient release (14).
Soil sample collection.
The mineral soils under the six forest stands were sampled in Autumn 2002. Ten individual samples (0- to 10-cm depth) were collected from each forest stand at intervals of 5 m along transects of 50 m in length, which had been established during earlier studies (13, 14). The soil samples were taken to the laboratory in cooling boxes. They were sieved (2-mm mesh) and then kept at −20°C until analysis.
TRF community profiles of soil samples.
The individual soil samples were well homogenized, and DNA was extracted from approximately 0.5 g of each sample by using the UltraClean soil DNA isolation kit (MoBio Laboratories, Inc.). This DNA was then used as template for PCR with 16S rRNA gene primers 8f (10) labeled with 6-carboxyfluorescein (MWG) at the 5′ end and 926r (22). Each 50-μl mixture contained 10 to 20 ng of extracted DNA, 1× reaction buffer (Gibco BRL), 200 μM (each) dATP, dCTP, dGTP, and dTTP, 3 mM MgCl2, a 0.2 μM concentration of the primers, and 2.5 U of Taq DNA polymerase (Gibco BRL). Amplifications were performed in a PTC-100 thermocycler (MJ Research, Inc.) with an initial denaturing step of 5 min at 95°C followed by 30 cycles of 30 s at 95°C, 1 min of annealing at 53°C, and 10 min of extension at 72°C. Three amplification products were prepared from each DNA sample and were then pooled to reduce PCR bias. Approximately 200 ng of fluorescently labeled PCR amplification products was digested with the restriction enzyme AluI (Gibco BRL) and subsequently purified by passage through Sephadex G-200 columns. Aliquots of 5 μl were mixed with 2 μl of loading buffer (diluted five times in deionized formamide; Fluka) and 0.3 μl of the DNA fragment length standard (Rox 500; PE Applied Biosystems Inc., Foster City, Calif.). Mixtures were denatured for 2 min at 92°C, and fluorescently labeled TRFs were then detected using an ABI 373A automated DNA sequencer (PE Applied Biosystems, Inc.) in the GeneScan mode. Lengths of labeled fragments were determined by comparison with the internal standard using the GeneScan 2.5 software package (PE Applied Biosystems Inc.).
Analysis of TRF profiles.
The DNA quantity analyzed for each of the 60 TRF community profiles was compared and standardized to the lowest quantity, according to the method of Dunbar et al. (8). TRFs of 39 to 500 bp in length and with heights of ≥50 fluorescence units were included in the analysis.
TRF data from individual soil samples were subjected to principal component analysis (PCA) to elucidate major variation patterns. Both fragment length and peak height were used as parameters for profile comparison. The scores of the first two components were subsequently used to compare differences between the forest stands in the TRF patterns with differences in soil chemical and microbiological properties by calculating Spearman's correlation coefficients (P ≤ 0.05). Average TRF data from 10 community profiles per forest site were subjected to cluster analysis using the Euclidean distance measure. A dendrogram was constructed using an unweighted pair-group method with arithmetic mean (UPGMA). All statistical procedures were performed using Statistica version 5.1 (1997 ed.).
Diversity statistics were calculated from standardized profiles of individual soil samples by using the number and height of peaks in each profile as representations of the number and relative abundance of phylotypes, as defined by Dunbar et al. (7). Phylotype richness (S) was calculated as the total number of distinct TRF sizes (between 39 and 496 bp) in a profile. The Shannon-Weiner diversity index (26) was calculated as follows: H = −Σ(pi) (log2_pi_), where p is the proportion of an individual peak height relative to the sum of all peak heights. Simpson's diversity index (27) was calculated as follows: D = 1 − [−Σ(pi)2]. Evenness (26) was calculated from the Shannon-Weiner diversity function as follows: E = _H/H_max, where _H_max = log2(S). Diversity data were tested for significant differences among forest sites by using the nonparametric Mann-Whitney U-test (Statistica 5, version 97).
16S rRNA gene clone libraries.
Clone libraries were constructed for the forest sites Kolmberg (oak-hornbeam forest), Rothwald (spruce-fir-beech forest), and Stampfltal (Austrian pine forest). The DNA templates used for construction of clone libraries were the same DNA preparations from which 16S rRNA genes for TRF analysis were amplified. Twenty independent PCRs (10 subsamples in duplicate) were performed for each sampling site by using the primers 8f (10) and 1520r (20) and the conditions described above. 16S rRNA gene amplicons were pooled and purified by gel extraction using the Concert rapid gel extraction system (Gibco BRL). Amplicons were ligated into the TpCR 4-TOPO vector (Invitrogen), and ligation products were transformed into Escherichia coli DH5α-T1 (Invitrogen) according to the manufacturer's instructions. Approximately 50 colonies were picked for each study site, each suspended in 80 μl of Tris-EDTA buffer (pH 8.0), and incubated at 100°C for 10 min. The lysis product (1.5 μl) was added to PCR mixtures using the primers M13f (5′-GTAAAACGAGGCCAG-3′) and M13r (5′-CAGGAAACAGCTATGAC-3′) and the conditions described above to amplify cloned inserts. PCR products were purified with the QIAquick PCR purification system (QIAGEN) according to the manufacturer's instructions and used as templates in sequencing reactions.
DNA sequencing.
Partial 16S rRNA gene sequencing was performed by applying the primer 518r (21) by the dideoxy chain termination method (24a) by using an ABI 373A automated DNA sequencer and the ABI Prism Big Dye terminator cycle sequencing kit (PE Applied Biosystems Inc.). Sequences were subjected to BLAST analysis (1) with the National Center for Biotechnology Information (NCBI) database. Sequences were checked for the presence of chimeric artifacts using the Chimera Check program of the Ribosomal Database Project (5).
TRF analysis of 16S rRNA gene clones.
16S rRNA genes of the clones were amplified by PCR using the M13f and M13r primers (Invitrogen) and the conditions described above. Each reaction mixture (1.5 μl) was used as template in a second PCR containing forward primer 8f (10) labeled with 6-carboxyfluorescein (MWG) at the 5′ end and 926r (22). Reaction conditions were the same as described above except that only 16 cycles of PCR were used instead of 30. Fluorescently labeled PCR amplification products from four to six clones (differing in TRF size) were mixed and collectively digested with the restriction enzyme AluI (Gibco BRL) and subsequently purified by passage through Sephadex G-200 columns. Aliquots of 1 μl were mixed with 3 μl of loading buffer (diluted five times in deionized formamide [Fluka]) and 0.3 μl of the DNA fragment length standard (Rox 500; PE Applied Biosystems Inc.). Mixtures were denatured for 2 min at 92°C, and fluorescently labeled TRFs were then detected using an ABI 373A automated DNA sequencer (PE Applied Biosystems Inc.) in the GeneScan mode. Lengths of labeled fragments were determined by comparison with the internal standard by using the GeneScan 2.5 software package (PE Applied Biosystems Inc.).
Nucleotide sequence accession number.
The nucleotide sequences determined in this study have been deposited in the NCBI database under accession numbers AY395085 to AY395222.
RESULTS
TRF community profiles.
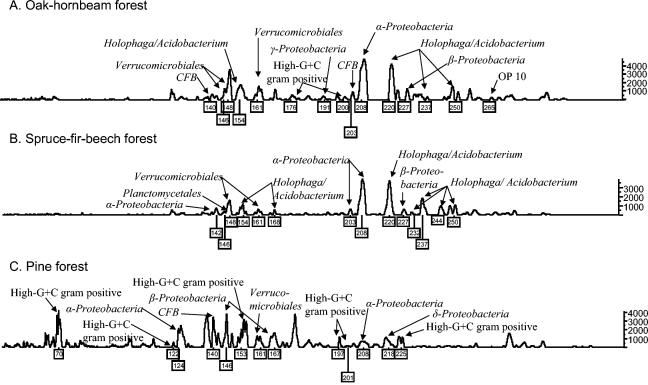
Soil samples were collected from six different natural forests at 10 sampling locations each, and bacterial 16S rRNA genes were amplified and analyzed by TRF community profiling, resulting in 60 individual TRF community profiles. Figure 1 depicts typical TRF community profiles derived from soils of an oak-hornbeam, a spruce-fir-beech, and an Austrian pine forest. Fragments which were found to be represented by bacterial groups identified in clone libraries are labeled with fragment sizes, and the respective phylogenetic groups are indicated.
FIG. 1.
Representative TRF profiles of bacterial communities from mineral soils under three Austrian natural forests: oak-hornbeam forest (A), spruce-fir-beech forest (B), and Austrian pine forest (C). Fragments corresponding to dominant phylogenetic groups represented by 16S rRNA gene clones derived from the same soils are indicated and labeled with the respective peak heights.
Both fragment length and signal intensity were considered as parameters for profile comparison after standardization of TRF data according to the lowest DNA quantity, as described by Dunbar et al. (8). TRF analysis after standardization yielded between 67 and 113 TRFs in individual community profiles which had intensities of at least 50 fluorescence units (FU). The number of TRFs with intensities higher than 500 FU ranged between 13 and 22 for the individual profiles. For the different forest sites, however, the number of peaks with intensities of ≥50 FU averaged between 82 (Kolmberg oak-hornbeam forest) and 101 (Merkenstein pine forest). The term phylotype richness was applied to the number of peaks with intensities of ≥50 FU and among all soils showed highest values for the Merkenstein pine forest soils. The Shannon-Weiner index and the Simpson index were calculated as a measure of diversity, which showed only a few differences among the forest soils. However, values of the Simpson index were significantly higher for the Stampfltal pine forest. No significant differences were found in the evenness of distribution of TRFs among the forest sites studied (Table 1).
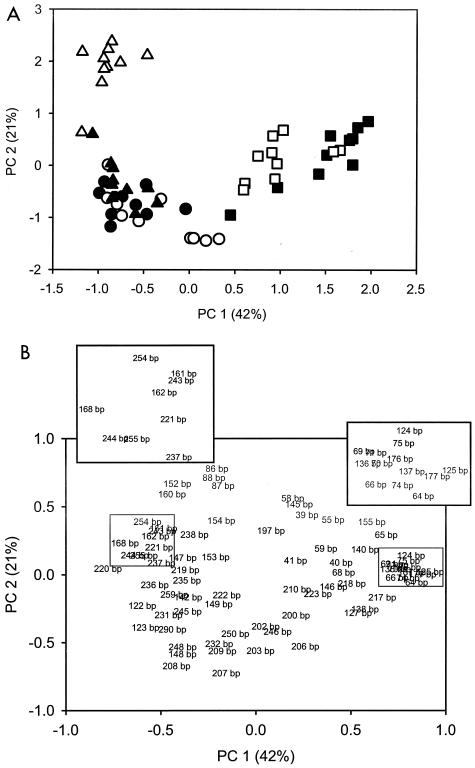
To elucidate major distribution patterns, a PCA of TRF data was performed (Fig. 2). In the score plot of the PCA, the soils of the Austrian pine forests were separated from the other forest soils along the first principal component (PC 1). TRF profiles of the pine forest soils contained fragments of 64, 65, 66, 69, 70, 71,74, 75, 124, 125, 136, 137, 140, 176, 177, and 217 bp in high abundances (>300 FU), which were heavily loaded on PC 1. Some of these fragment lengths were found to be represented by 16S rRNA gene clones from the Stampfltal pine forest soil, for instance, members of the high-G+C gram positives (70 bp), α_-Proteobacteria_ (124 bp), and the Cytophaga/Flexibacter/Bacteroides group (140 bp), as illustrated in Fig. 1. Compared with the pine forest profiles, the profiles of the oak-hornbeam and spruce-fir-beech forest soils showed higher intensities in the fragments of 168, 220, 236, 237, and 244 bp, which had negative loadings along PC 1. 16S rRNA gene clones from the Kolmberg oak-hornbeam forest and the Rothwald spruce-fir-beech forest which corresponded with these fragment lengths were all affiliated with the Holophaga/Acidobacterium division (Fig. 1). The soils of both oak-hornbeam forests as well as the Rothwald spruce-fir-beech forest clustered together, whereas the Neuwald spruce-fir-beech forest soils were set apart from these soils along the second principal component (PC 2). Compared with profiles from the other forest soils, TRF profiles from the Neuwald forest had higher intensities of fragments with 86, 87, 88, 152, 160, 161, and 254 bp, which had positive loadings on PC 2. Differences between the forest stands in the TRF patterns corresponded with differences in the soils' chemical and microbiological properties, presented in Table 2. Significant correlations were found between PC 1 and soil pH (r = 0.76; P = 0.0001), soil C-to-N ratio (r = 0.79; P = 0.0000), the ratio of fungal to bacterial phospholipid fatty acids (PLFAs) (r = 0.83, P = 0.0000), and microbial biomass measured as SIR-C g of organic C−1 (r = 0.56; P = 0.0079), FE-N g of organic C−1 (r = 0.50; P = 0.0219), and total PLFAs g of organic C−1 (r = 0.72; P = 0.0002). No significant correlations have been found between PC 2 and soil chemical and microbiological properties.
FIG. 2.
Score plot (A) and loading plot (B) of PCA of TRF data. In the loading plot, only fragments with heights of ≥300 FU are shown. •, oak-hornbeam forest (Johannser Kogel); ○, oak-hornbeam forest (Kolmberg); ▴, spruce-fir-beech forest (Rothwald); ▵, spruce-fir-beech forest (Neuwald); ▪, Austrian pine forest (Stampfltal); □, Austrian pine forest (Merkenstein).
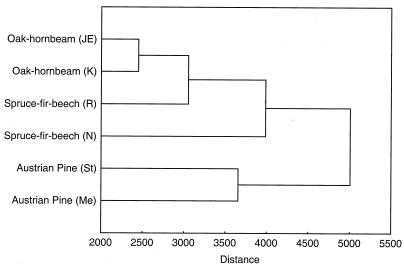
The dendrogram of the cluster analysis which was based on average TRF values from each forest site illustrated the differences between the bacterial communities inhabiting the different soil environments (Fig. 3). Corresponding with results of the PCA, the communities in the two pine forest soils clustered and were distinct from the communities of the other forest soils, which clustered separately. Within this second cluster, the communities of the two oak-hornbeam forests grouped with a low distance value, while major differences were apparent among the communities of the Rothwald and the Neuwald spruce-fir-beech stands.
FIG. 3.
Dendrogram of TRF profiles from six natural forest soils generated from Euclidean distance values, using the UPGMA method. Cluster analysis was performed with average TRF values of 10 individual subsamples from the study sites. JE, Johannser Kogel; K, Kolmberg; R, Rothwald; N, Neuwald; St, Stampfltal; Me, Merkenstein.
16S rRNA gene clone sequences.
Soils of the Kolmberg, Rothwald, and Stampfltal forest sites, featuring oak-hornbeam, spruce-fir-beech, and Austrian pine forests, respectively, were surveyed by constructing 16S rRNA gene libraries. Partial (nonchimeric) sequence information for 46 (Kolmberg), 51 (Rothwald), and 41 (Stampfltal) clones was obtained, covering approximately 500 bp. The majority of these sequences (82%) showed at least 95% similarity to sequences deposited in the NCBI database, while the remaining sequences were only moderately related (92 to 94%) to known genes.
The relative abundances of clones affiliated to various bacterial groups differed for the three forest soils. In the clone library derived from the Kolmberg oak-hornbeam forest, clones affiliated with the Holophaga/Acidobacterium division were most abundant, comprising 28% of total clones. Clones representing members of the Verrucomicrobia division were the second most abundant (24%), and those affiliated with the Cytophaga/Flexibacter/Bacteroides group were third most abundant (11%). Members of the α_-Proteobacteria_ accounted for 9% of the sequences. The remaining clones were affiliated with high-G+C gram positives, Planctomycetes, β_-,γ_-, and δ_-Proteobacteria_, and candidate divisions BD and OP10. One clone (clone L13K) could not be classified into the known bacterial divisions. In the clone library from the Rothwald spruce-fir-beech forest, the majority of clones were also classified as Holophaga/Acidobacterium (35%). Members of the α_-Proteobacteria_ accounted for 27%. Minor parts of the library belonged to the Verrucomicrobiales (10%) and Planctomycetales (10%) groups as well as to members of the β_-, γ-, and δ_-Proteobacteria (each 6%). The clone library from the Stampfltal Austrian pine forest differed from the other libraries in that the majority of clones was classified as high-G+C gram positives (49%). Again, sequences that affiliated with the α_-Proteobacteria_ were highly abundant (20%). Twelve percent of the clones were represented by Holophaga/Acidobacterium, and the remaining 16S rRNA genes showed the highest similarity to the β- and δ_-Proteobacteria_, the Cytophaga/Flexibacter/Bacteroides group, and to the Verrucomicrobia.
In Table 3, the phylogenetic assignments are given for clones of the three libraries which corresponded to dominant TRFs in the community profiles. TRF lengths observed in profiles have been reported to differ from sequence-determined TRF lengths, depending on subtle differences in molecular weight, either from purine content or dye label (16). To allow comparison of TRF lengths of the clones with TRFs in community profiles, the true fragment lengths of the clones were therefore determined by subjecting them to TRF analysis. In addition to the observed TRF sizes of the clones, the TRF sizes predicted from sequence information are also shown in Table 3, providing information on TRF drift.
TABLE 3.
Phylogenetic assignment of clones from mineral soil from three different natural forests corresponding to highly abundant TRFs in community profilesa
| Site and TRF size (bp) | Predicted TRF size (bp) | Mean peak height (FU) | Corres- ponding clone | Closest NCBI match/% homology | Phylogenetic group |
|---|---|---|---|---|---|
| Oak-hornbeam forest (Kolmberg) | |||||
| 146 | 146 | 872 | N08K | Bacterium Ellin428 (AF432252)/95 | Verrucomicrobia |
| 148 | 148 | 1,843 | M18K | Uncultured cupper smeltery bacterium D69 (AF337838)/94 | Verrucomicrobia |
| 148 | 148 | 1,843 | J02K | Uncultured bacterial clone Ac57 (AF388362)/98 | Verrucomicrobia |
| 148 | 148 | 1,843 | M05K | Uncultured bacterial clone Ac57 (AF388362)/98 | Verrucomicrobia |
| 154 | 155 | 703 | L03K | Uncultured acidobacterium UA1 (AF200696)/98 | Holophaga/Acidobacterium |
| 161 | 162 | 642 | M02K | Verrucomicrobial clone N42.61PG (AF431600)/99 | Verrucomicrobia |
| 161 | 162 | 642 | L16K | Agricultural soil bacterial clone SC-1-74 (AJ252653)/96 | Verrucomicrobia |
| 162 | 162 | 442 | G05k | Uncultured bacterial clone Ac49 (AF388313)/97 | Verrucomicrobia |
| 203 | 204 | 765 | H01K | Uncultured bacterium GKS2-164 (AJ290028)/96 | Cytophaga/Flexibacter/Bacteroides |
| 207 | 208 | 1,536 | M01K | Uncultured alphaproteobacterial clone N27.63SM (AF431139)/99 | Alphaproteobacteria |
| 208 | 208 | 1,772 | H29K | Uncultured gold mine bacterium D17 (AF337873)/95 | Cytophaga/Flexibacter/Bacteroides |
| 208 | 210 | 1,772 | H20K | Uncultured bacterial clone BCM3S-34B (AY102909)/99 | Alphaproteobacteria |
| 208 | 210 | 1,772 | L08K | Uncultured alphaproteobacterial clone glen99_6 (AY150881)/99 | Alphaproteobacteria |
| 220 | 222 | 1,539 | H24K | Uncultured acidobacterium clone SMS9 (AY043842)/99 | Holophaga/Acidobacterium |
| 245 | 249 | 635 | J01K | Uncultured bacterium clone CCM3a (AY221061)/98 | Holophaga/Acidobacterium |
| Spruce-fir-beech forest (Rothwald) | |||||
| 153 | 157 | 727 | A05R | Uncultured Acidobacteriales clone WS044 (AY174199)/98 | Holophaga/Acidobacterium |
| 154 | 155 | 1,003 | A16R | Uncultured eubacterium WR896 (AJ292801)/99 | Holophaga/Acidobacterium |
| 154 | 155 | 1,003 | C11R | Uncultured acidobacterium UA1 (AF200696)/99 | Holophaga/Acidobacterium |
| 154 | 155 | 1,003 | B25R | Agricultural soil bacterial clone SC-1-45 (AJ252636)/99 | Betaproteobacteria |
| 154 | 157 | 1,003 | A03R | Uncultured bacterial clone N12.55WL (AF432710)/99 | Deltaproteobacteria |
| 154 | 157 | 1,003 | A15R | Uncultured bacterial clone N26.112SM (AF432735)/99 | Deltaproteobacteria |
| 154 | 158 | 1,003 | Roth04 | Uncultured bacterial clone C46 (AF432770)/99 | Deltaproteobacteria |
| 203 | 206 | 628 | C07R | Uncultured bacterial clone CCU18 (AY221076)/98 | Alphaproteobacteria |
| 208 | 210 | 2,503 | B06R | Unidentified eubacterial clone DA122 (Y12598)/98 | Alphaproteobacteria |
| 208 | 210 | 2,503 | B26R | Uncultured eubacterium WR8100 (AJ292844)/99 | Alphaproteobacteria |
| 208 | 210 | 2,503 | Roth06 | Uncultured eubacterial clone IAFR109 (AF270944)/99 | Alphaproteobacteria |
| 209 | 206 | 1,808 | A09R | Uncultured bacterial clone CCU18 (AY221076)/99 | Alphaproteobacteria |
| 220 | 222 | 2,334 | C15R | Uncultured earthworm cast bacterial clone c234 (AY154590)/99 | Holophaga/Acidobacterium |
| 235 | 236 | 1,070 | B29R | Uncultured eubacterium WD235 (AJ292680)/98 | Gammaproteobacteria |
| 235 | 236 | 1,070 | C19R | Uncultured eubacterium WD235 (AJ292680)/100 | Gammaproteobacteria |
| 236 | 237 | 1,045 | A04R | Uncultured acidobacterial clone NOS7.146WL (AY043714)/96 | Holophaga/Acidobacterium |
| 237 | 238 | 785 | C16R | Uncultured acidobacterial clone NOS7.146WL (AY043714)/97 | Holophaga/Acidobacterium |
| 237 | 237 | 785 | A14R | Uncultured planctomycete YNPRH54A (AF465657)/92 | Planctomycetales |
| 244 | 246 | 1,010 | B11R | Uncultured bacterial clone BCTP21 (AF485788)/94 | Holophaga/Acidobacterium |
| Austrian pine forest (Stampfltal) | |||||
| 69 | 70 | 1,313 | F10ST | Actinobacterium dtb36 (AJ309984)/96 | High-G+C gram positive |
| 70 | 73 | 1,601 | E07ST | Unidentified actinobacterium d13 (AJ292034)/97 | High-G+C gram positive |
| 124 | 127 | 371 | D02ST | Pedomicrobium manganicum (X97691)/97 | Alphaproteobacteria |
| 140 | 142 | 654 | D07ST | Rhizosphere soil bacterial clone RSC-II-66 (AJ252690)/96 | Cytophaga/Flexibacter/Bacteroides |
| 146 | 147 | 1,111 | D22ST | Uncultured gold mine bacterium D (AF337867)/99 | Betaproteobacteria |
| 153 | 156 | 621 | D11ST | Uncultured soil bacterial clone Tc127-88 (AY242751)/94 | High-G+C gram positive |
| 153 | 156 | 621 | D31ST | Unidentified eubacterium (AF010117)/93 | High-G+C gram positive |
| 161 | 162 | 368 | F13ST | Unidentified eubacterium (AF010030)/98 | Verrucomicrobia |
| 200 | 204 | 337 | E01ST | Uncultured actinobacterial clone GCPF6 (AY129782)/96 | High-G+C gram positive |
| 200 | 204 | 337 | D12ST | Uncultured soil bacterial clone Tc120-G03 (Ay242750)/97 | High-G+C gram positive |
| 208 | 210 | 728 | D10ST | Uncultured gold mine bacterium D23 (AF337879)/95 | Alphaproteobacteria |
DISCUSSION
Distinct bacterial communities in pine forest soils.
By comparison of TRF profiles, compositional differences were detected in the bacterial communities inhabiting a variety of natural forest soils. The bacterial communities from Austrian pine forest soils clustered separately from communities from oak-hornbeam and spruce-fir-beech forests, indicating that they were compositionally distinct. These results were consistent with expectations based on the vegetation and soil chemical characteristics. The two pine forest soils were set apart from the other four forest soils by contrasting soil conditions, such as higher soil pH, higher C/N ratio, lower amounts of microbial biomass g of organic C−1, and a higher ratio of fungal to bacterial biomass (14; Hackl et al., submitted). Our findings are generally in agreement with PFLA analyses of the same forest sites (Hackl et al., submitted). PLFA analysis discriminated among the microbial communities of various soils by assigning them to few functional groups but gave little information about their phylogenetic affiliations. TRF analysis showed that several bacterial groups (represented by specific TRFs) were highly abundant in the pine forests but were less prevalent or missing in the other soils. Cloning and sequencing of 16S rRNA genes indicated that these bacterial groups were mainly composed of high-G+C gram positives.
Analysis of 16S rRNA gene clone libraries allowed us to identify the most dominant members of the bacterial communities. In agreement with the results obtained by TRF analysis, clones contained in the library from the Stampfltal pine forest were affiliated with different bacterial groups than those in the libraries derived from the Kolmberg oak-hornbeam forest and the Rothwald spruce-fir-beech forest. Representing 49% of the total clones, the library from the pine forest was highly dominated by members of the high-G+C gram-positive bacteria. Similarly, in community fingerprints the corresponding TRFs were highly abundant. High-G+C gram positives were of minor importance in the Kolmberg oak-hornbeam forest and had no representatives in the library obtained from the Rothwald spruce-fir-beech forest. High-G+C gram positives were also most prevalent in 16S rRNA clone libraries from mineral forest soil samples in British Columbia, where lodgepole pine (Pinus contorta Douglas) is a major tree species (2). Nevertheless, clones representing this bacterial group were far less abundant in the British Columbian soil than in the Stampfltal samples. In clone libraries from organic forest soils (2) and from lodgepole pine rhizosphere soil (4) in British Columbia, however, α_-Proteobacteria_ were dominant. In the Stampfltal library clones which affiliated with the α_-Proteobacteria_ were second most abundant. High-G+C gram-positive bacteria were not detected or were of minor prevalence in clone libraries from forests other than pine forests (3, 4, 6, 9).
High-G+C gram-positive bacteria comprise a large variety of bacteria which have commonly been called actinomycetes because they include members which grow in a filamentous manner. The high prevalence of clones affiliated with this bacterial group in the Stampfltal clone library is in agreement with the fact that high concentrations of tuberculostearic acid (PLFA 10Me18:0) have previously been found in soils of the Stampfltal and Merkenstein pine forests (Hackl et al., submitted). This fatty acid is common in actinomycetes and has frequently been used to indicate actinomycetes in soils (17). Sequences retrieved from the Stampfltal pine forest soil were distributed among several phyla within the high-G+C gram positives, including filamentous actinomycetes. However, organisms related to _Streptomyces_—a common soil microorganism—were not detected. Filamentous growth may facilitate nutrient acquisition, which would be of advantage in the pine forest soils, which are prone to desiccation and nutrient imbalances (14). In addition, high quantities of PFLAs indicative of fungi have been reported in these soils (Hackl et al., submitted). Various Stampfltal 16S rRNA gene sequences grouped with clones obtained from Australian desert soils (15) and fell into a deeply branching line of actinobacteria which was most related, though moderately, to the genus Rubrobacter.
Pine forest soils represent habitats for soil bacteria which are prone to nutrient imbalances, and of all forest soils studied they are most limited in water availability (13, 14; Hackl et al., submitted). Nevertheless, by using TRF profiling high numbers of highly dominant bacterial groups were detected as phylotypes in these soils. In particular, the Stampfltal pine forest appeared to host a highly diverse soil bacterial community, as indicated by diversity indices calculated from TRF community data. In contrast, the Rothwald spruce-fir-beech forest soil, which is most nutrient rich and high in water content, exhibited only average values of phylotype richness and community diversity. It has to be considered that diversity measures based on TRFs only give information on the diversity of the numerically dominant bacterial groups and that minor but potentially highly diverse subsets of the soil bacterial communities may not have been detected by the TRF analysis. However, the bacterial communities of the pine forest soils are set apart from those in the other forest soils in that they are composed of a multitude of highly abundant bacterial groups. A possible explanation for this result may be that these soils host a huge variety of bacteria which are able to cope with a wide range of environmental conditions, including various levels of nutrient availability. A major portion of these bacteria might be active only in short periods after rainfall or when nutrients are episodically released. Indeed, the soil moisture content and the C/N ratio have been identified as the factors which are mainly responsible for within-site variability in microbial activity in pine forest soils (13).
Bacterial communities in oak-hornbeam and spruce-fir-beech forest soils.
The oak-hornbeam and the spruce-fir-beech forest soils contained several highly abundant bacterial groups which were less dominant in the pine forest soils. Comparison of community TRF profiles with the representative 16S rRNA gene clones implied that these bacterial groups are composed of Holophaga/Acidobacterium members. The view that members of the Holophaga/Acidobacterium group may constitute a major part of the bacterial communities in oak-hornbeam and spruce-fir-beech forest soils is further supported by the fact that 16S rRNA gene clones affiliated with this group were the most abundant in libraries from both the Kolmberg and the Rothwald forest soils. Among forest soils, soils under pinyon pine-juniper woodlands have been reported to contain acidobacteria in highest numbers (6). Sequences belonging to the Holophaga/Acidobacterium division have been retrieved from a multitude of habitats, including forest soils (e.g., references 2, 4, and 6) and have revealed extraordinary phylogenetic diversity within this group (9, 25, 28). So far, only a few cultivated representatives of the Holophaga/Acidobacterium division exist, and therefore ecological and physiological data are very limited. However, the phylogenetic diversity and ubiquity of the acidobacteria suggest that they show high metabolic versatility.
The bacterial communities from the two oak-hornbeam forest soils (Johannser Kogel and Kolmberg) clustered closely in comparisons of forest soil TRF profiles, indicating similarity in community composition. Conformities in the profiles included high intensities in fragments of 148 bp in length. In the library from the Kolmberg forest soil, several clones were present which corresponded to 148-bp fragments and which were affiliated with the Verrucomicrobia group. Clones representing Verrucomicrobia members were second most abundant in the Kolmberg clone library, suggesting that Verrucomicrobia organisms may occupy important roles in the oak-hornbeam forest soils.
Similarities in bacterial community composition were observed within the two oak-hornbeam forests as well as within the two pine forests, which might reflect a selective influence of the dominant plant species at these sites on the soil bacteria. However, since the vegetation composition of the forests under study is supposed to be differentiated according to site conditions, its influence on the soil bacterial communities is closely connected with effects of other site conditions, such as climate, geological substrate, and soil type. In addition, it may be assumed that plant-specific effects through the input of litter may be more pronounced in the organic than in the mineral soil layer, which was studied. Thus, the community patterns observed in our study probably reflect the interplay of various factors, including plant species composition as well as soil type and other site conditions.
In contrast to the soil bacterial communities from the oak-hornbeam and pine forests, which seemed to be similar in composition within forest type, major differences were apparent when the soil bacterial communities from the Rothwald and the Neuwald spruce-fir-beech forests were compared. This may have been related to differences between the two spruce-fir-beech forest sites in soil type and geological substrate. While the Rothwald forest soil has been defined as planosol situated on dolomite and has been found to be especially nutrient rich with pH values of about 5, the Neuwald forest soil has been classified as stagnic luvisol situated on sandstone substrate with slow nutrient release and lower pH values (14). Compared with the Rothwald forest soil, considerably lower nutrient concentrations and slower nutrient turnover rates have been measured in the Neuwald forest soil, although both forests have a similar vegetation composition and similar topological characteristics (14). Results of the present study suggest that distinct bacterial communities are involved in nutrient cycling in the two soil environments.
Conclusions.
In the present study, 16S rRNA gene analysis proved appropriate for detecting compositional differences in the bacterial communities inhabiting various natural forest soils. Differences in bacterial community composition among forest soils were found to correspond with differences in other microbiological and soil chemistry characteristics. Our data suggest that the composition of the bacterial communities inhabiting natural forest soils is related to specific environmental conditions which go along with various types of natural forest vegetation. The comparative analysis of bacterial communities in soils under different types of natural forest has provided important baseline information about bacterial diversity and composition in undisturbed forest ecosystems. These data will allow us to go forward with studies in which the effects of management practices or environmental pollution on the soil bacterial communities and their functions will be evaluated.
Acknowledgments
This project was financed by the Austrian Ministry of Agriculture and Forestry, Environment and Water Management, and by the provincial government of Lower Austria.
We thank Alexandra Weilharter for her excellent assistance with the TRF and sequence analyses.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäfer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-PLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axelrood, P. E., M. L. Chow, C. C. Radomski, J. M. McDermott, and J. Davies. 2002. Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 48**:**655-674. [DOI] [PubMed] [Google Scholar]
- 3.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63**:**2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow, M. L., C. C. Radomski, J. M. McDermott, J. Davies, and P. E. Axelrood. 2002. Molecular characterisation of bacterial diversity in lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol. Ecol. 42**:**347-357. [DOI] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31**:**442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65**:**1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66**:**2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67**:**190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar, J., M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68**:**3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17**:**7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellenberg, H. 1996. Vegetation Mitteleuropas mit den Alpen. Eugen Ulmer Verlag, Stuttgart, Germany.
- 12.Frank, G., and G. Koch. 1999. Nationaler Bericht “Naturwaldreservate in Österreich.” Österreichischer Beitrag zur COST Aktion E4 Forest Reserves Research Network. [Online.] http://fbva.forvie.ac.at/inst1/publ/koch/naturwald98.html#2.
- 13.Hackl, E., G. Bachmann, and S. Zechmeister-Bolternstern. 2000. Soil microbial biomass and rhizosphere effects in natural forest stands. Phyton 40**:**83-90. [Google Scholar]
- 14.Hackl, E., G. Bachmann, and S. Zechmeister-Bolternstern. 2004. Microbial nitrogen turnover in soils under different types of natural forest. Forest Ecol. Manag. 188**:**101-112. [Google Scholar]
- 15.Holmes, A. J., J. Bowyer, M. P Holley, M. O'Donoghue, M. Montgomery, and M. R. Gillings. 2000. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 33**:**111-120. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan, C. W., and C. L. Kitts. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 54**:**121-125. [DOI] [PubMed] [Google Scholar]
- 17.Kroppenstedt, R. M. 1985. Fatty acid and menaquinon analysis of actinomycetes and related organisms, p. 173-199. In M. Goodfellow, and D. E. Minnikin (ed.), Chemical methods in bacterial systematics. Academic Press, London, United Kingdom.
- 18.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S. M. Barns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68**:**1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32**:**241-247. [DOI] [PubMed] [Google Scholar]
- 20.Massol-Deya, A. A., D. A. Odelson, R. F. Hickey, and J. M. Tiedje. 1995. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal gene sequences and restriction endonuclease analysis (ARDRA), p. 1-8. In Molecular microbial ecology manual 3.3.2. Kluwer, Dordrecht, The Netherlands.
- 21.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59**:**695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer, G., A. Teske, and C. O. Wirsen. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturating gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164**:**165-172. [DOI] [PubMed] [Google Scholar]
- 23.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276**:**734-740. [DOI] [PubMed] [Google Scholar]
- 24.Redfield, E., S. M. Barns, J. Belnap, L. L. Daane, and C. R. Kuske. 2002. Comparative diversity and composition of cyanobacteria in three predominant soil crusts of the Colorado Plateau. FEMS Microbiol. Ecol. 40**:**55-63. [DOI] [PubMed] [Google Scholar]
- 24a.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. USA 74**:**5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67**:**4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication, p. 125. University of Illinois Press, Urbana.
- 27.Simpson, E. H. 1949. Measurement of diversity. Nature 163**:**688. [Google Scholar]
- 28.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296**:**1064-1066. [DOI] [PubMed] [Google Scholar]
- 29.Zechmeister-Boltenstern, S., E. Hackl, G. Bachmann, C. Donat, and M. Pfeffer. 2000. Microbial nutrient turnover in forests with natural tree species composition, p. 296-297. In Proceedings of the International Conference on Forest Ecosystem Restoration. Universität für Bodenkultur, Vienna, Austria.