RNA controls PolyQ protein phase transitions (original) (raw)
. Author manuscript; available in PMC: 2017 Jan 9.
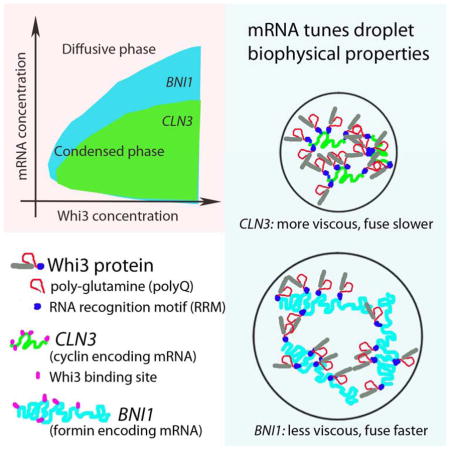
Summary
Compartmentalization in cells is central to the spatial and temporal control of biochemistry. In addition to membrane-bound organelles, membrane-less compartments form partitions in cells. Increasing evidence suggests that these compartments assemble through liquid-liquid phase separation. However the spatiotemporal control of their assembly, and how they maintain distinct functional and physical identities is poorly understood. We have previously shown an RNA-binding protein with a polyQ-expansion called Whi3 is essential for the spatial patterning of cyclin and formin transcripts in cytosol. Here, we show that specific mRNAs that are known physiological targets of Whi3 drive phase separation. mRNA can alter the viscosity of droplets, their propensity to fuse, and the exchange rates of components with bulk solution. Different mRNAs impart distinct biophysical properties of droplets indicating mRNA can bring individuality to assemblies. Our findings suggest that mRNAs can encode not only genetic information, but also the biophysical properties of phase-separated compartments.
Graphical abstract
Introduction
Compartmentalization of the cytoplasm into various organelles is a key aspect of intracellular organization, and is essential for accomplishing diverse and simultaneous biomolecular interactions. Membrane-less RNA/Protein (RNP) bodies are an important class of organelles that bind and regulate RNA, and thus play a central role in regulating the flow of genetic information (Anderson and Kedersha, 2006). These structures include processing bodies and stress granules in the cytoplasm, and nucleoli, Cajal bodies and PML bodies in the nucleus. It is becoming clear that RNP bodies are not only involved in numerous physiological processes, but also that dysregulated bodies are associated with a variety of diseases (Ramaswami et al., 2013).
Increasing evidence suggests that the assembly of RNP bodies is driven by a concentration-dependent phase transition, including: localization of P granules (Brangwynne et al., 2009; Elbaum-Garfinkle et al., 2015; Hubstenberger et al., 2013; Lee et al., 2013a), cell-size dependent assembly of the nucleolus (Berry, 2015; Weber and Brangwynne, 2015) phosphorylation-regulated assembly of RNA granules and stress granules (Wang et al., 2014; Wippich et al., 2013). Consistent with intracellular liquid-liquid phase separation, these bodies exhibit liquid-like behaviors such as wetting, dripping and relaxation to spherical structures upon fusion (Brangwynne et al., 2009; Brangwynne et al., 2011; Feric and Brangwynne, 2013; Wippich et al., 2013). The liquid-like properties of these droplets could facilitate their function, by allowing for high concentration of molecular components that nevertheless remain dynamic within the droplet.
The need for mechanisms to spatially and temporally organize the cytoplasm is particularly acute in large cells of multinucleated organisms such as the fungus Ashbya gossypii, where many nuclei share a common cytoplasm (Dietrich et al., 2004). Our work with this model organism has shown that asynchronous nuclear division is achieved by spatial organization of cyclin transcripts (CLN3) through the assembly of an RNA binding protein called Whi3. The Whi3 homolog in Saccharomyces cerevisiae also forms super-assemblies in this case that segregate to mother cells to memorize past events (Caudron and Barral, 2013; Gladfelter et al., 2006; Lee et al., 2013a; Lee et al., 2015). Interestingly, Whi3 assembly is also essential for organizing formin transcripts (BNI1) at polarity sites where Ashbya growth tips are located or new branches are generated (Lee et al., 2015) This suggests that the assembly of one protein can spatially organize transcripts for both nuclear autonomy and symmetry breaking in the same cell. However, the mechanism of Whi3 assembly and how this assembly might allow it to achieve these dual functions remains unclear.
Whi3 is predicted to be largely disordered, containing a long polyQ tract adjacent to an RNA recognition motif (RRM). Intrinsically disordered protein (IDP) motifs are commonly found in RNP bodies (Uversky et al., 2015; Wang et al., 2014) and Q/N rich sequences are particularly implicated in RNP assembly (Reijns et al., 2008). Whi3 also interacts via multivalent binding sites on its mRNA binding partners, CLN3 and BNI1, in the regulation of nuclear division and polarity, respectively (Lee et al., 2015; Lee et al., 2013b). These structural features of Whi3 and its partners are particularly interesting, since both IDPs (Elbaum-Garfinkle et al., 2015; Nott et al., 2015) and multivalent binding interactions (Li et al., 2012) can promote liquid-liquid phase separation; closely related ‘low complexity sequences’ (LCS) (Han et al., 2012; Kato et al., 2012) also can form hydrogels in vitro. Moreover, RNA has been shown to nucleate RNP assembly in vivo (Shevtsov and Dundr, 2011), and can induce preferential condensation by locally shifting the droplet phase boundary at transcription sites (Berry, 2015). Moreover, RNA can alter droplet properties in vitro (Elbaum-Garfinkle et al., 2015). Thus we hypothesized that Whi3 may form assemblies at different locations and for distinct functions in a single cytoplasm based on the specific mRNAs with which it is in complex.
In this work, we show that specific mRNAs drive Whi3 assembly into dynamic liquid-like droplets with distinct biophysical properties. These droplets are spherical, exhibit rapid fluorescence recovery after photobleaching (FRAP), and readily coalesce with one another upon contact. The phase diagram and biophysical properties of the droplets vary depending upon different mRNA that is present. Additionally, we see that binding RNA in vivo is critical for the normal morphology of Whi3 droplets in cells and we speculate mRNA plays a role in preventing the maturation of droplets into more static gels or fibers. We hypothesize that the differential effect of RNAs on Whi3 droplet properties underlies the capacity of Whi3 to form assemblies with distinct functions in a common cytoplasm.
Results
Whi3 phase separates to form liquid-like droplets in vivo and in vitro
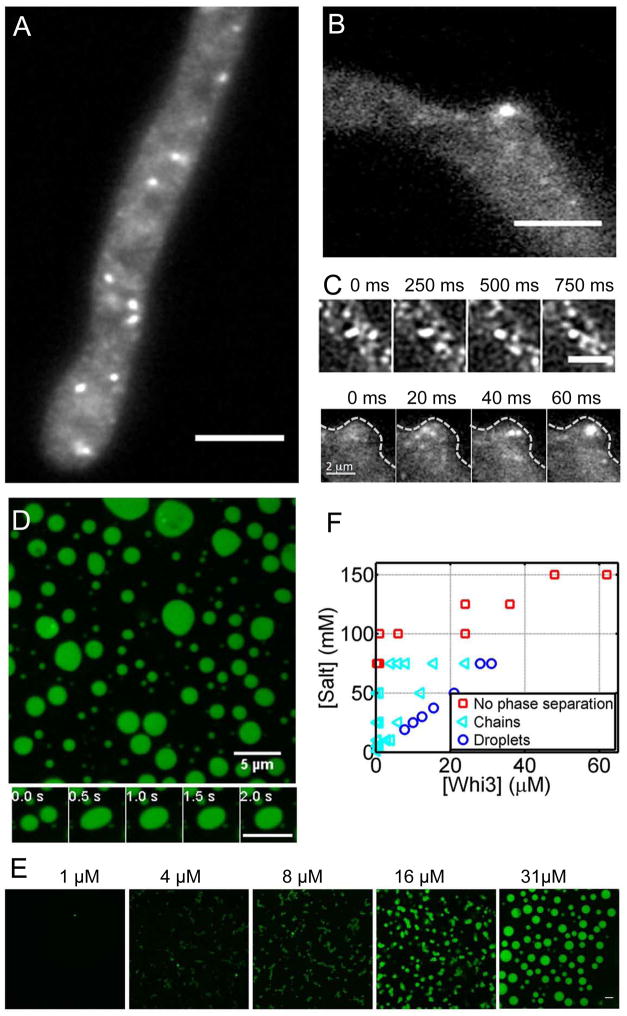
In our previous work using conventional wide-field imaging, we saw that Whi3 protein forms highly heterogeneous structures in the cytoplasm of Ashbya (Lee et al., 2013b). To examine their properties in more detail and to capture dynamics of the assemblies, we used oblique angle total internal reflection fluorescence microscope (TIRF) to visualize these assemblies in live Ashbya cells. Incline TIRF is ideal to minimize photobleaching while filming the assemblies at fast frame rates (Tokunaga et al., 2008). Although this method only captures a subset of the assemblies, and their small size is comparable to the diffraction limit, Whi3 appears to form dynamic droplet-like assemblies in the cytoplasm, which can be readily seen to fuse, as would be predicted if they are liquid droplets (Movie 1, Fig. 1A,C). We see Whi3 droplets in cytosol where nuclei reside and also enriched in foci at nascent branches where polarity is being established, as would be predicted from previous functional analysis linking Whi3 to nuclear division and cell polarity (Lee et al., 2013a; Lee et al., 2015), (Movie 2, Fig. 1 B,C). This raises the question as to how functionally distinct Whi3 assemblies arise and remain discrete in a common cytoplasm if they are behaving as liquid-like compartments which have a propensity to fuse.
Figure 1. Whi3 phase separates in vivo and in vitro.
(A) Whi3 forms liquid-like assemblies in Ashbya cells. Scale bar is 5 μm. (B) Whi3 assemblies at a branch site. Scale bar is 5 μm. (C) Fusion events in cells. Top panel shows a fusion event in cytosol. Scale bar is 2 μm. Bottom panel shows a fusion event at a branch site indicated by dashed line. (D) Recombinant Whi3 (28 μM) forms liquid droplets at 75mM salt. Bottom panel highlights a fusion event. Scale bar is 5 μm. (E) Whi3 forms distinct small round assemblies that are attached to each other at low protein concentration, and forms round droplets at high protein concentration. Images were taken after overnight incubation at room temperature. Scale bar is 5 μm. (F) Whi3 phase diagram with varying salt concentrations. Also see Fig S1 and Movie 1, 2, and 3.
To determine mechanisms for how discrete assemblies may emerge in the same cytoplasm, we established a simplified reconstitution of the assemblies in vitro with purified Whi3 protein and known target mRNAs. We first asked if purified Whi3 can phase separate on its own, in the absence of mRNA. Indeed, at relatively low salt (75 mM) and relatively high protein concentrations (28 μM), recombinantly-expressed, full length Whi3 phase separates to condensed liquid-like droplets, as indicated by the spherical shape and the ability to coalesce (Movie 3, Fig. 1D, visualized with 10% of protein labeled with GFP). We could see similar phase separation behavior with 100% untagged protein indicating that demixing is not a consequence of the GFP tag (Fig. S1). At lower protein concentrations (1–16 μM) and 75 mM salt, distinct small, round assemblies that are attached to each other form after overnight incubations (Fig. 1E). The size of each spherical unit decreases as Whi3 concentration decreases. Thus, at lower protein concentration, the condensed phase appears to nucleate and grow to a certain size, beyond which droplets cannot coalesce when they collide. Notably, however, at physiological salt concentration (150 mM) we see no evidence of phase separation of Whi3 even at very high concentrations of pure protein (Fig. 1F). Thus, Whi3 protein is capable of phase separating on its own, however under physiological salt and protein conditions there must be other factors promoting condensation.
Cyclin mRNA promotes Whi3 phase separation in physiological conditions
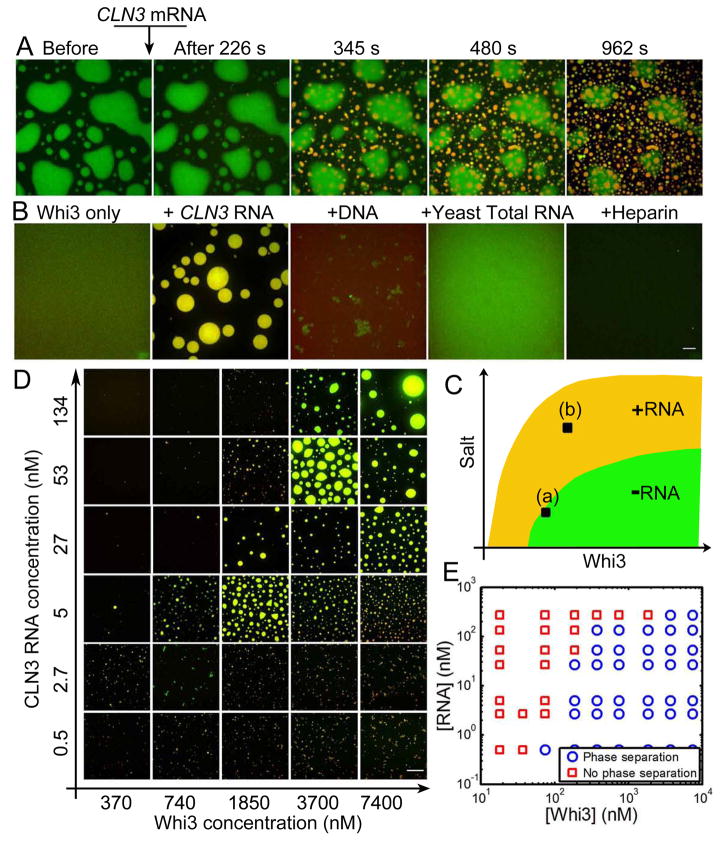
Given that the cellular function of Whi3 involves regulating mRNA, we hypothesized that mRNA may promote phase separation under physiological salt conditions. The first indication that mRNA can promote droplets is clear when an endogenous Whi3-interacting mRNA, CLN3, is added to Whi3-protein droplets (Fig. 2A). Before adding RNA, the bulk concentration outside the protein droplet is the saturation concentration in these conditions, corresponding point “a” in Fig. 2C (Elbaum-Garfinkle et al., 2015; Jones, 2002). Shortly after adding CLN3 mRNA, new droplets consisting of Whi3 and RNA start to form and enlarge (Movie 4, Fig. 2A). Meanwhile, old protein-only droplets begin shrinking and eventually dissolve completely as Whi3 leaves the protein-only droplets and goes into the new protein-RNA droplets. Notably, the new droplets do not fuse with the pre-formed Whi3-only droplets. This shows that at the saturation concentration, where Whi3 alone can no longer phase separate, CLN3 RNA can drive phase separation to form new droplets. To confirm this finding, we started with a low protein and high salt concentration (point ‘b’ in Fig. 2C), in which Whi3 alone does not condense into droplets, and then added CLN3 RNA. We found that droplets consisting of Whi3 and RNA were indeed readily formed (Fig. 2B). In contrast, adding the same amount of DNA, yeast total RNA or heparin did not promote Whi3 droplets (Fig. 2B). This indicates that CLN3 mRNA can shift the phase boundary to lower protein concentration and high salt concentration, enabling Whi3 phase separation in physiological conditions (Fig. 2C).
Figure 2. CLN3 RNA promotes Whi3 phase separation in physiological conditions.
(A) Addition of 53 nM CLN3 mRNA (cy3-labled, red) to phase-separated Whi3 (10%GFP labeled, green) at 60 mM salt. (B) 3.7 μM Whi3 at 150 mM salt is not phase separated, adding 53 nM CLN3 RNA promotes liquid-liquid demixing, resulting in condensed droplets consist with both RNA (red) and protein (green). Adding DNA, total yeast RNA or Heparin that are normalized to 53 nM CLN3 RNA by charge does not promote droplet formation. Some aggregates are formed with DNA. Scale bar is 10 μm. (C) Schematics showing RNA shifts Whi3 phase boundary. (D) Images of droplets formed at various CLN3 mRNA and Whi3 concentration at 150 mM salt. Images were taken overnight after mixing Whi3 and CLN3 mRNA. Scale bar is 20 μm. (E) Phase diagram of Whi3 and CLN3 mRNA at 150 mM salt. See also Fig S2, S3, movie 4.
To assess how RNA may promote phase separation, we next asked how CLN3 mRNA impacts the Whi3 phase behavior at physiological salt conditions. Remarkably, CLN3 RNA can promote detectable formation of droplets at Whi3 concentrations as low as 200nM (Fig. 2 D, E). This is in the same order of magnitude of soluble Whi3 in Ashbya cytosol that we have measured by fluorescence correlation spectroscopy (57±14 nM, Fig. S2, method in SI). For a given Whi3 concentration, as RNA concentration increases, the apparent volume of condensed phase (roughly indicated by the total droplet area in each image) increases up to a point, but surprisingly then decreases, and eventually no droplets can be observed with light microscopy. The RNA concentration at which the largest apparent droplet volume is observed is higher for a higher protein concentration indicating that the RNA to protein molar ratio is likely a critical parameter. At ratios that are either too low or too high, phase separation is not favored. The molar ratio of CLN3 RNA to Whi3 for obtaining the largest apparent droplet volume is estimated to be ~0.02 (Fig. S3). These data show that a native mRNA binding partner of Whi3 is sufficient to drive phase separation in physiological salt and protein concentration.
RNA binding via the RRM is required for Whi3 phase separation
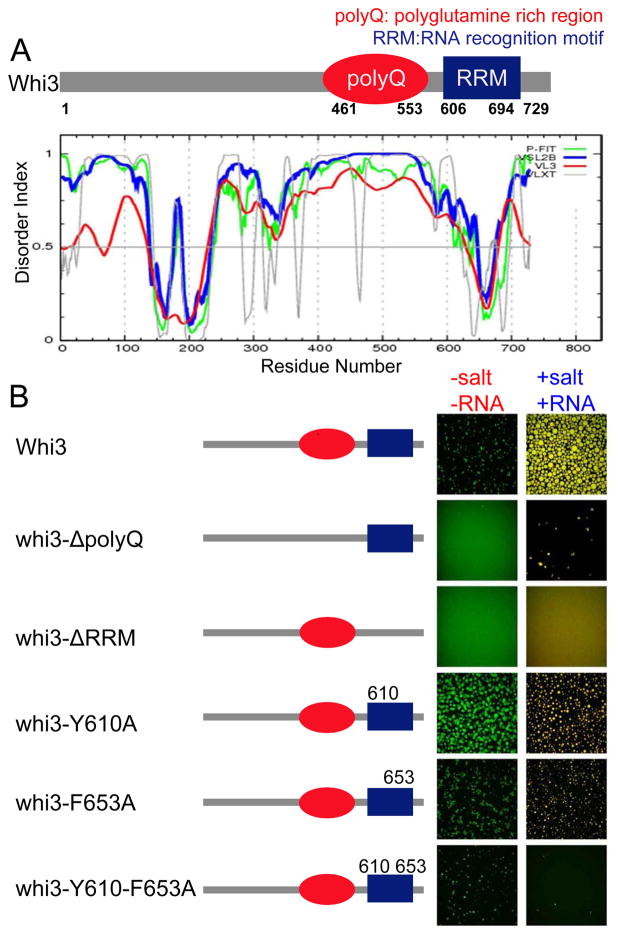
The data thus far show that Whi3 phase separation can be controlled by both salt and mRNA concentration. In addition to its RNA recognition motif (RRM), there are other features of Whi3, including a long polyQ tract, that are functionally relevant in vivo for promoting regionalized cytoplasm and making heterogeneous cytoplasmic assemblies (Lee et al., 2015; Lee et al., 2013b). The polyQ domain in Whi3 is predicted to be disordered, and its disruption imparts loss of function phenotypes in vivo (Fig 3A) (Lee et al., 2013a). To dissect the contributions of the polyQ domain and RNA-binding in Whi3 phase-separation behavior, we made a series of constructs including truncation of the polyQ and RRM domain, and point mutations in the RRM domain which are predicted to be important for transcript binding (Fig. 3B). For analysis of all mutants, we start with high salt concentration where there is no phase separation of the proteins. We then asked if the mutants can phase separate upon either lowering salt concentration without adding mRNA, or upon adding CLN3 mRNA without lowering salt concentration.
Figure 3. RNA binding through RRM domain is critical for Whi3 phase separation.
(A) Schematics of Whi3 protein with a polyQ domain and an RNA recognition motif. Whi3 is predicted to be disordered in the polyQ region. (B) Schematics illustrate the mutated Whi3 constructs. In the no-RNA experiment (left column of images), salt concentration was lowered from 150mM to 75mM by adding buffer with no salt, final protein concentration was 5μM of Whi3 and for fragments and mutants the highest protein concentration each construct could be purified was used: 25 μM of whi3ΔpolyQ, 39μM of whi3-Y610A, 23 μM of whi3-F653A, 11 μM whi3-Y610-F653A, 23 μM Whi3ΔRRM. In the CLN3 mRNA experiment (right column of images), 7.4 μM protein at 150mM salt was used for all mutants and 53 nM CLN3 mRNA was added. All images were taken after 4 hours of either adding no-salt buffer or mRNA.
Indeed, when the polyQ domain is deleted, Whi3 cannot phase separate by simply lowering the salt concentration (Fig. 3B) indicating a role for polyQ-driven assembly. However, upon addition of CLN3 mRNA, ΔpolyQ can form droplets suggesting that the polyQ domain is not strictly essential in mRNA-mediated assembly. Interestingly, deletion of the RRM domain and remaining C-terminus abrogates droplet formation with or without mRNA. To further probe the role of the RRM domain in droplet assembly, we introduced point mutations (Y610A or F653A) predicted to be directly involved in RNA binding (Maris et al., 2005; Nash et al., 2001). Individual single point mutations showed minimal effect on droplet formation with or without mRNA. However, the RRM double mutant demonstrates a marked reduction in mRNA-mediated assembly. Thus, direct binding of the RRM domain with CLN3 mRNA is necessary for mRNA-mediated assembly of droplets. Purified individual RRM domain, polyQ domain and RRM-polyQ domains are also not able to form droplets, neither by lowering salt nor adding CLN3 mRNA. Taken together we find that 1) both polyQ and RRM domains are necessary for droplet formation, however neither domain is sufficient at low salt and 2) RRM binding to CLN3 mRNA is required for mRNA-mediated assembly of Whi3 droplets.
RNA influences droplet physical properties
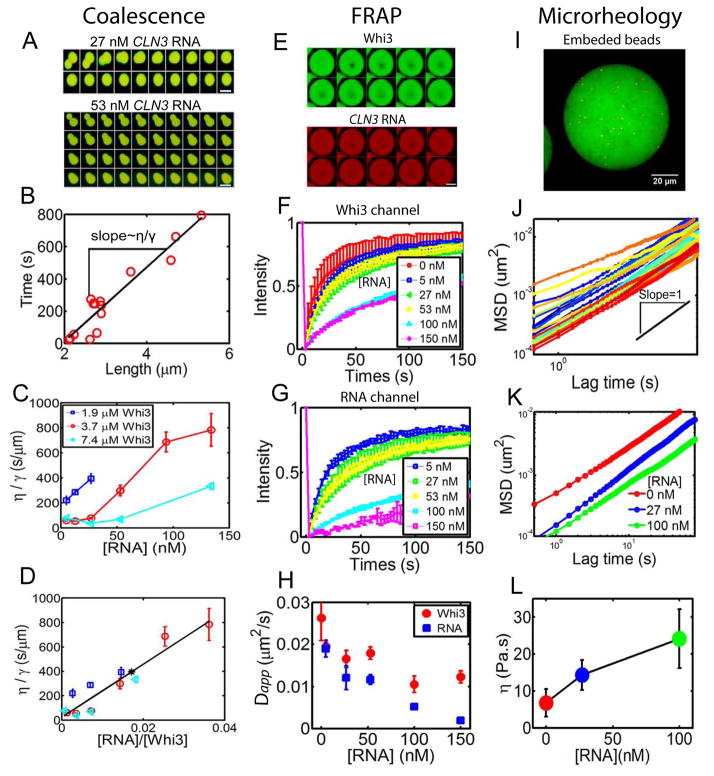
Given the potent role of mRNA in promoting Whi3-phase separation, we next asked if mRNA also impacts the biophysical properties of droplets. We first noticed that at high RNA concentration droplets coalesce more slowly, suggesting that mRNA indeed strongly impacts droplet properties (Fig. 4 A, Fig. S4). Coalescence is driven by surface tension γ and resisted by droplet viscosity η; the rate of coalescence can therefore be used to estimate the ratio of the two (η/γ) (Brangwynne et al., 2011; Elbaum-Garfinkle et al., 2015; Jones, 2002). Plotting the fusion relaxation time against the characteristic fusion length yields a linear relation, with the slope providing an estimate of η/γ (Fig. 4B). For a given RNA concentration, we find that η/γ decreases with increasing protein concentration (Fig. 4C), while for a given protein concentration, η/γ increases with increasing RNA concentration. Interestingly, the monotonic increase of η/γ with RNA concentration contrasts with the phase behavior observed in Fig. 2, where the droplet volume fraction first increases with RNA concentration and then decreases. However, after plotting η/γ against the ratio of RNA to protein concentration, all data points roughly collapse (Fig. 4 D). This indicates that, as with the phase separation behavior (Fig. 2), the molar ratio of RNA to protein is a key parameter, which in this case affects the biophysical properties of the droplets.
Figure 4. CLN3 RNA influence Whi3 droplet physical properties.
(A) Montages show a fast fusion event with low RNA concentration (27 nM) and a slow fusion event with high RNA concentration (53 nM) for 3.7 μM Whi3 at 150 mM salt. Time interval between images is 50 seconds and scale bar is 5 μm. (B) An example plotting characteristic fusion length when two droplets initially meet against the fusion relaxation time (circles) yields a linear relation, with the slope scales with the ratio of viscosity over surface tension η/γ. (C) Viscosity over surface tension ratio (η/γ) obtained from fusion events plotted against RNA concentration for various Whi3 concentrations. Mean±SEM (D) η/γ scales roughly with RNA to Whi3 molar ratio. Mean±SEM. Black line is a linear fit. Black asterisk on line corresponds to the ratio that gives the largest apparent droplet volume from Fig. S3 that’s based on Fig. 2D. (E) FRAP images show recovery of CLN3 RNA and Whi3. Scale bar is 5 μm. Time interval is 20 seconds. (F) Normalized FRAP curves in Whi3 channel show slower recovery as RNA concentration increases in droplets with 25 μM Whi3 at 60mM salt. Mean±STD. (G) Normalized FRAP curves in RNA channel also show slower recovery as RNA concentration increases. Mean±STD. (H) Decrease in apparent diffusion coefficients with increasing RNA concentration estimated from FRAP data. Mean±STD. (I) An example of fluorescent tracer beads (red) embedded in Whi3 droplets (green). RNA not labeled. (J) An example of MSD for tracer beads in droplets. Black line shows slope=1. (K) Mean MSD with varying RNA concentration in droplets with 25 μM Whi3 at 60mM salt. (L) Viscosity calculated from MSD data. Mean±STD. See also Fig S4.
These fusion data suggest that RNA has the potential to alter the viscoelastic properties of droplets. Since fusion kinetics only give the ratio of η/γ and surface tension is difficult to measure, we set out to probe the properties within droplets directly. We first used FRAP (fluorescence recovery after photobleaching) to monitor molecular dynamics of Whi3 and CLN3 RNA within the droplets (Fig. 4 E). Both Whi3 and CLN3 RNA are exchanging, as indicated by recovery after photobleaching. Consistent with the increased η/γ obtained from fusion dynamics, increased RNA concentration causes both RNA and protein to exhibit slower FRAP recovery (longer characteristic recovery time, τ) (Fig. 4 F,G). In all cases, protein recovers faster than RNA. We estimate the apparent diffusion coefficients from FRAP, using Dapp = _a_2/τ where a is the radius of the photobleached region. Plotting Dapp against RNA concentration shows that RNA indeed leads to slowed dynamics within the droplet (Fig. 4H).
To further examine the physical properties of droplets, we used microrheology. Particle tracking microrheology is a technique that utilizes the motion of tracer beads to determine the viscoelasticity of soft materials. We recently applied this technique to protein droplets to directly measure droplet viscosity (Elbaum-Garfinkle et al., 2015). Fluorescent tracer beads were embedded in the droplets (Fig. 4I) and their motion was traced over time to obtain the mean-squared displacement, < (r⃗_(t) − _r⃗_0)2 > MSD, Fig. 4J). As RNA concentration increases, the MSD of tracer beads shifts down, indicating slower bead motion (Fig. 4K). Fitting the MSD over time to a power law < (_r⃗_(_t_) − _r⃗_0)2 > ~ tα gives the diffusion exponent α ≈ 1, as expected for Brownian motion of beads in a purely viscous droplet. The bead diffusion coefficient Dbead can then be obtained by fitting < (_r⃗_(_t_) − _r⃗_0)2 >= 4_Dbeadt. Knowing tracer bead size R, droplet viscosity can then be determined through the Stokes-Einstein relation η = kBT/6_π_D_beadR_. For Whi3-only droplets, a viscosity of 6 Pa.s is obtained, which is similar to that of honey. Moreover, as RNA concentration increases, droplet viscosity indeed increases (Fig.4 L), which is consistent with both the FRAP and droplet fusion data showing decreased dynamics with increasing CLN3 RNA. Thus, the presence of mRNA not only shifts the phase boundary, but also significantly changes the droplets’ biophysical properties.
Different mRNAs create droplets with different properties
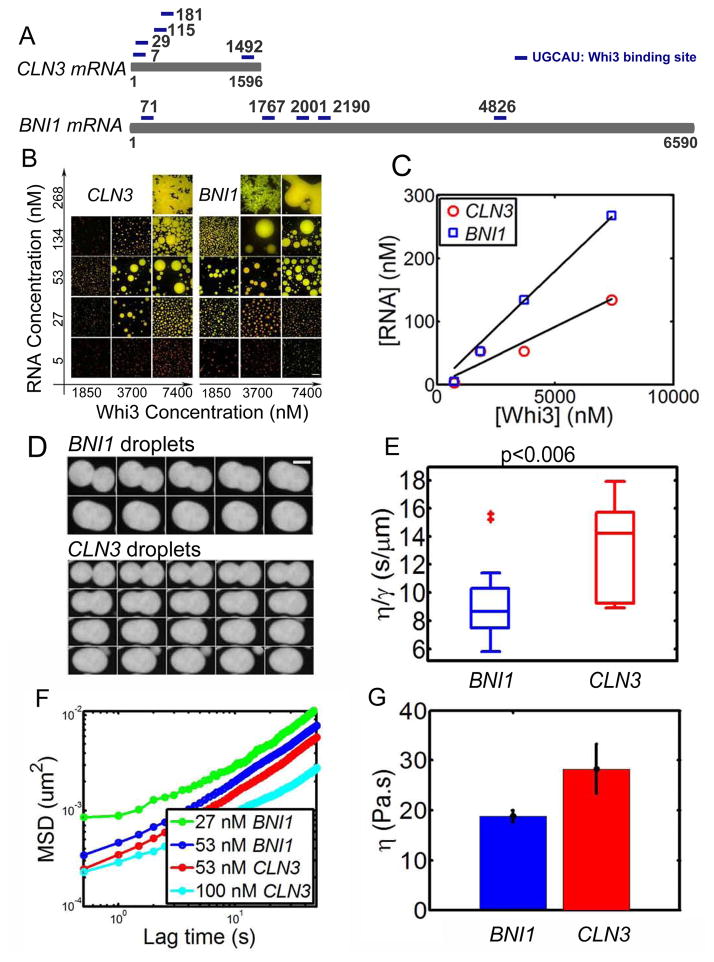
In cells, many distinct liquid compartments can coexist without all coalescing into one giant compartment. In Ashbya, Whi3 forms functionally distinct assemblies such that Whi3 binds to CLN3 mRNA to regulate cell cycle and binds to the formin-encoding _BNI1_mRNA to establish polarity sites (Lee et al., 2015; Lee et al., 2013b). We hypothesized that different target mRNAs could produce Whi3 assemblies that have distinct biophysical properties. This idea is attractive because the mRNAs could be the basis for functional specificity, with subcellular localization patterns arising from distinct biophysical properties dictated by the mRNAs themselves. Having observed that CLN3 mRNA can drive Whi3 phase separation at physiological conditions, and can tune droplet physical properties, we next asked how an alternative Whi3 target, BNI1, impacts droplet behavior. Like CLN3, the BNI1 mRNA has five predicted Whi3-binding sites, but it is four times longer than CLN3 (Fig. 5A). We see that BNI1 mRNA can drive Whi3 phase separation at 150 mM salt in a similar fashion as CLN3 RNA, in that for a given protein concentration Whi3, the apparent droplet volume fraction increases then decreases with increasing RNA concentration (Fig. 5B). Similarly, the RNA concentration at which the largest droplet volume is observed for each protein concentration depends on the molar ratio of RNA to Whi3 (Fig. 5C). However, the optimal RNA to Whi3 molar ratio for BNI1 (~0.04) is larger than that estimated for CLN3 (~0.02). This difference is also clear at high concentrations of mRNA, where the inhibitory concentration of BNI1 for droplet formation is twice that of CLN3 mRNA for a given Whi3 concentration. This reflects a shift of the phase boundary to higher RNA concentration by BNI1 compared to CLN3 mRNA.
Figure 5. BNI1 mRNA drives Whi3 phase separate into droplets with different properties.
(A) Schematics show CLN3 mRNA and BNI1 mRNA, each with five Whi3 binding sites. (B) Whi3 (green) phase diagram with BNI1 mRNA (red) and CLN3 RNA (red), images were taken after 4 hours of mixing Whi3 with RNA at 150 mM salt. Scale bar is 20 μm. (C) RNA concentration at which the largest apparent droplet volume is observed in B for each Whi3 concentration. The optimal RNA/Whi3 molar ratio estimated from linear fit (black line) for CLN3 is ~0.02 (similar to that obtained from overnight droplets for CLN3 in Fig. S3) and BNI1 is ~0.04. (D) Example of fusion images for 50 nM CLN3 and BNI1 RNA with 9 μM Whi3 at150 mM salt shows a faster fusion rate for BNI1. Time interval between images is 10 second. Scale bar is 5 μm. (E) Box plot of viscosity to surface tension ratio (η/γ) for CLN3 and BNI1 droplets obtained from fusion events. (F) Mean MSD from microrheology for CLN3 and BNI1 with 8 μM Whi3 at 150 mM salt, showing faster movement of tracer beads for less RNA both for BNI1 and CLN3 droplets. (G) Viscosity of 53 nM BNI1 and CLN3 droplets obtained from MSD data, showing BNI1 droplets are less viscous. Mean±SEM.
We also found that BNI1 droplets fuse faster than CLN3 droplets (Fig 5 D). Quantifying the fusion process reveals a smaller η/γ for BNI1 droplets (Fig. 5 E, p< 0.006). Microrheology data show that similar to CLN3, BNI1 droplet viscosity increases as BNI1 RNA concentration increases (Fig. 5 F). However, at 50 nM RNA, BNI1 droplet viscosity is 17 Pa·s, which is much smaller that of CLN3 droplets (28 Pa·s) at the same RNA concentration (Fig. 5 G). Thus, different known physiological mRNA targets of Whi3 can produce droplets of substantially different biophysical properties, even with the same predicted valency in the target mRNA. These results implicate mRNA features other than predicted valency are important for specifying the biophysical properties of intracellular droplets.
Whi3 droplets mature and can appear fibrillar
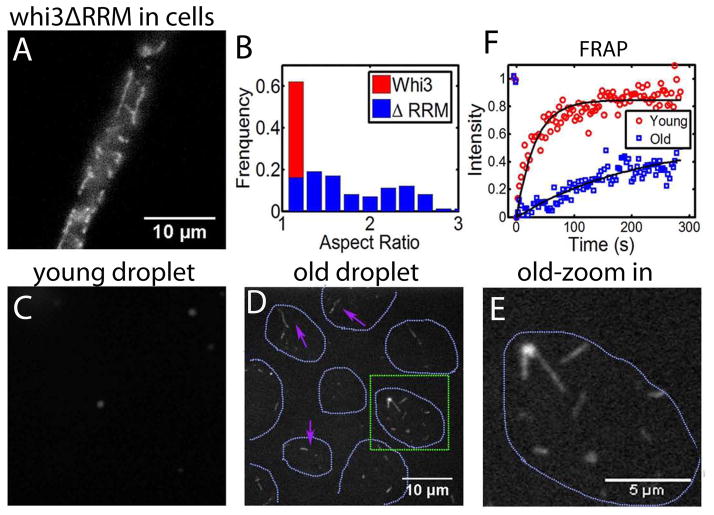
Many polyQ containing proteins are associated with the propensity to form fibrous amyloids (Williams and Paulson, 2008). Recently, the Q-rich ALS protein FUS was shown to transition from a liquid-like compartment to a fibrous aggregate with time, and that disease-associated mutations accelerate this liquid to solid transition (Patel et al., 2015). Despite the extensive polyQ tract in Whi3, for the full-length construct we never observed fibrous structures in vivo or in vitro. Remarkably, however, when Whi3 is no longer able to bind to RNA (Whi3ΔRRM), we observe that in vivo Whi3 assemblies have a markedly different morphology, now forming larger, more filamentous structures (Movie 5, Fig. 6A,B). These elongated assemblies do not generally colocalize with ER or elongated mitochondria (Fig. S5), so we suspect that they reflect a different structural organization than the more round droplet-like structures observed with full length Whi3. These in vivo findings prompted us to look further into whether in vitro droplets of Whi3 may contain the seeds of more stable fibers, as has been seen for other RNA binding proteins with IDP/LCS domains (Han et al., 2012; Kato et al., 2012; Patel et al., 2015). Upon addition of high salt (2M) to freshly prepared Whi3 full-length droplets, all droplets dissolve, consistent with expectations from the equilibrium phase diagram. However, for “old” droplets that have been incubated for >7 hours, we detect elongated, salt-resistant fibers (Fig. 6 C,D,E). Consistent with a change in the molecular organization through time, FRAP indicates a slower recovery for proteins in aged droplets (Fig. 6 F). This suggests that the liquid state represents a metastable precursor to more stable, and potentially toxic fibrous states seen in aged droplets. However, RNA binding to polyQ proteins may prevent or slow the transition to more solid-like assemblies, helping cells avoid these pathological states (Patel et al., 2015; Weber and Brangwynne, 2012).
Figure 6. Maturation of droplets over time.
(A) Whi3 structure after deleting RNA binding domain (Whi3ΔRRM) in Ashbya cells. (B) Aspect ratio of Whi3ΔRRM in comparison with wide type Whi3 in Ashbya cells. (C) Adding high salt (2M) to young droplets (1h) disrupted droplets. Salt concentration increased from 150 mM to 300 mM. Droplets were formed with 8 μM full length Whi3 and 200 nM RNA. (D) Adding high salt (2M) to old droplets (≥7h) disrupted droplets but fibers were left. Magenta arrows point at fibers. Green square highlights the region that is zoomed in and shown in (E). Cyan dotted line indicates where droplets were before adding salt. Droplets were formed with 8 μM full length Whi3 and 200 nM RNA. (F) FRAP shows slower Whi3 recovery in old droplets than in young droplets. See also Fig S5.
Discussion
Our study was motivated by a desire to understand how the assembly of one protein, Whi3, can spatially pattern transcripts for distinct cell functions-nuclear division and polarity establishment. We found that Whi3 can form phase-separated assemblies in cells and in vitro and that the RNAs it binds to can promote the formation of droplets with different biophysical properties. Using purified Whi3 in vitro, we demonstrated that the known binding partners CLN3 mRNA and BNI1 mRNA can drive Whi3 phase separation at physiological salt and protein conditions. Our results show that instead of being a passive passenger, mRNA plays an active role in driving the phase separation of RNA-binding proteins. These data raise the interesting possibility that mRNA encodes not only “information” in the traditional sense of the genetic code but can also, along with the coassembled proteins, encode biophysical properties in the higher-order assemblies of phase-separated compartments.
How do specific target mRNAs promote the phase separation of Whi3? There is increasing evidence that relatively weak, disordered (IDP/LCS) (Elbaum-Garfinkle et al., 2015; Nott et al., 2015) and/or multivalent (Li et al., 2012) interactions promote intracellular phase transitions. We could see that Whi3 phase separated specifically with known targets that contain multiple predicted binding sites for Whi3 rather than based simply on exposure to negative charge from a bulk pool of non-specific mRNA. Interestingly, however, different phase diagram and droplet physical properties were observed for CLN3 mRNA and BNI1 mRNA. This difference is despite the fact that both RNAs have five Whi3 binding sites and thus theoretically the same valency. Thus, features other than the valency of the mRNAs might be at play for giving rise to the difference in droplet assembly and properties. For example, CLN3 mRNA is almost four times shorter than BNI1 RNA, yielding a much higher density of Whi3 binding sites (~1 per 320 bases) than that of BNI1 RNA (~1 per 1318 bases). In addition, the distribution of Whi3 binding sites is different for the two transcripts with binding sites on CLN3 clustered on the 5′ and that for BNI1 more evenly distributed along the transcript (Fig. 5 A). Moreover, secondary structure of mRNAs can influence function as has been seen with translation efficiency in yeast and other organisms (Kertesz et al., 2010; Pop et al., 2014). We predict that differences in RNA secondary structure and other features such as size, charge distribution of CLN3 and BNI1 mRNAs, and the spacing patterns of protein binding sites are important in dictating protein-RNA interactions that drive phase transitions. It is not yet unclear which of these features are critical or if it is a combination of the above traits and other features of mRNAs that contribute to the process. An exciting future area of work will be dissecting how the different chemical and structural features of mRNAs impact phase transitions.
We also found that the molar ratio of RNA to Whi3 is critical for RNA-driven phase behavior, with high RNA concentration actually inhibiting Whi3 phase separation. This could be because at high RNA concentration, the negatively charged RNA acts like salt to screen charge-based interactions that drive phase separation. We also show that RNA can tune droplet physical properties. As RNA concentration increases, droplets fuse slower, droplet dynamics decreases and viscosity increases. Interestingly, these findings contrast with our recent work showing that a non-specific polyU RNA can decrease the viscosity, effectively “fluidizing” droplets of the disordered P granule protein LAF-1 (Elbaum-Garfinkle et al., 2015). Moreover, for the case of LAF-1, polyU RNA did not significantly shift the phase boundary. Another interesting difference between these experiments is RNA length, the CLN3 and BNI1 RNA used here are 32 and 125 times larger than the polyU50 used in our previous study. Furthermore, other features like secondary structure and charge distribution are expected to be different based on difference in the sequences of RNAs used in the two studies. These differences suggest that different RNAs may facilitate precise control over the location of the phase boundary (Berry, 2015), and can either increase or decrease droplet viscoelasticity depending on the features of the RNA and/or the specific protein components (Elbaum-Garfinkle et al., 2015).
A key unknown for all reconstituted RNA-protein droplets is how well in vitro produced mRNA recapitulates native transcripts, which are coated with a variety of additional proteins. The degree to which these associated proteins may limit the valency of binding sites, alter charge landscape for assembly, and impact the in vivo biophysical properties of liquid-like compartments remains a critical open challenge for the field. In simplified systems, however, it is clear different behavior is observed for different RNAs. It is widely accepted that mRNAs code for proteins and non-coding RNAs play regulatory roles (Rinn and Chang, 2012). Our work in compartment-level organization, along with work in other cellular processes like translation efficiency (Pop et al., 2014) and the formation of nuclear bodies (Shevtsov and Dundr, 2011), support the idea that mRNAs encode not only genetic information but also structural information.
It is increasingly clear that there are different kinds of liquid-like compartments that can coexist either in the same cell or depending on the stress or disease state. For example, recent work has shown evidence that yeast P-bodies are liquid-like while stress granules are more solid-like (Kroschwald et al., 2015). Considering the capacity of the same protein to exist in different states may explain contrasts between the in vivo and in vitro behavior of Whi3. For example, in vitro, the Whi3ΔRRM mutant cannot form droplets either in the presence of RNA or by lowering salt (Fig. 3 B). In vivo, the Whi3ΔRRM does make some assemblies that appear highly elongated and fibrous, possibly with gel-like properties. The difference in phase behavior of Whi3ΔRRM in vivo and in vitro indicates that other factors are likely at play in promoting Whi3 assembly in vivo. One possibility is the interactions between the polyQ domain and other disordered proteins. Recent work shows that promiscuous interactions between IDP/LCS domains can drive the formation of solid-like yeast stress granules under certain stress conditions (Kroschwald et al., 2015). Our finding that in the absence of RNA, Whi3 forms chains at low protein concentration (Fig.1 D) indicates a more stable microstructure formed within the small droplet, preventing further droplet fusion upon collision (Pawar et al., 2012). Therefore, it is conceivable that polyQ-driven droplets can form in vivo due to associating with other factors and these thus have different properties and assembly requirements than the homomeric polyQ-driven droplets formed in vitro. We suspect that the polyQ-driven droplets are likely more representative of gel-like assemblies and could be closer to solid, amyloid states. Indeed, we could detect the presence of fibers under certain conditions in droplets with time indicating the potential to mature to less fluid states (Fig. 6).
These observations are consistent with very recent studies on the ALS-proteins FUS and hnRNPA1, as well as other RNP body proteins, which exist in a liquid-like state that evolves with time into more solid-like and fibrous states (Patel et al., 2015) (Lin, 2015; Molliex, 2015). Taken together with our findings on Whi3, this emerging body of work suggests a shared capacity for liquid-like compartments to change with time, and the need to maintain weak-heterotypic protein-protein and/or protein-RNA interactions to maintain fluidity and functionality, and avoid toxicity. It is then not surprising that a large number of IDP/LCS-containing (or prion-like, aggregation-prone domain-containing) RNA-binding proteins are associated with neurodegenerative diseases (King et al., 2012). We anticipate given the evolutionary distance and different functions of Whi3 compared with FUS, hnRNPA1, and other RNP body proteins, these similar effects likely reflect universal properties of membrane-free organelles.
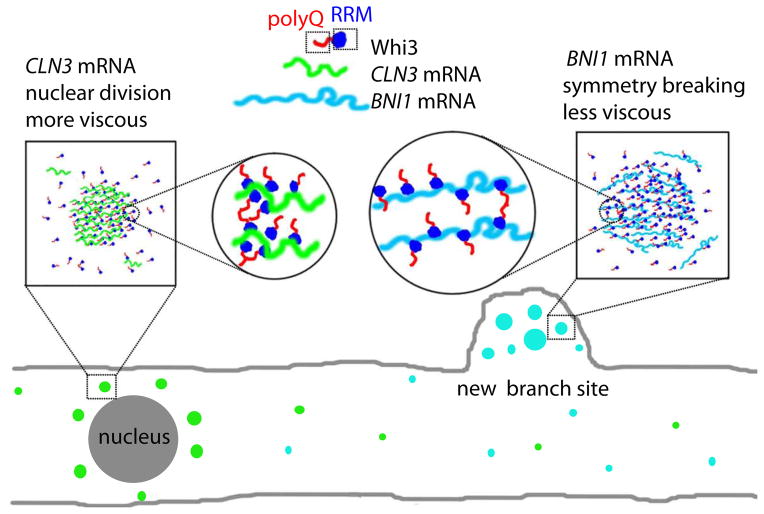
A key result from this minimal, reconstituted system is that RNA can tune both assembly and physical properties of droplets. Which features of the RNA are critical is not yet clear but will be revealed upon further dissection of the phase-properties with different RNA constructs. However, our findings suggest a model for cytoplasmic organization in Ashbya, in which differences in local concentrations of Whi3, CLN3 mRNA and BNI1 mRNA can drive Whi3 into droplets with different physical properties. We speculate that these assemblies of distinct composition are in turn different in how they are subcellularly distributed, particularly in their proximity to nuclei and polarity sites, and in what other components may be recruited (Fig. 7). For example, other RNA interacting partners, such as Puf2 which has disordered regions and associates with Whi3-BNI1 but not Whi3-CLN3 complexes (Lee et al., 2015; Lee et al., 2013b), can be used to further tune droplet assembly, properties and thus functions. This study reveals a key role for mRNA in driving protein phase transitions, and demonstrates that mRNAs can control the biophysical properties of these assemblies.
Figure 7. RNAs control protein phase separation to pattern cytosol in Ashbya.
Model for linking biophysical properties of droplets to differences in cell function. We hypothesize that more viscous Whi3 droplets formed with CLN3 mRNA are adjacent to nuclei for controlling nuclear division timing and less viscous Whi3 droplets formed with BNI1 mRNA are at new branch sites or growth tips to establish polarity sites.
Experimental Procedures
Recombinant protein expression and purification
Full length and fragments of Whi3 were tagged with 6-His and expressed in E. coli using standard procedures. Cells were lysed in lysis buffer (1.5 M NaCl, 20 mM Tris pH 8, 20 mM Imidazole pH8, 1 mM DTT, 1 tablet of Roche protease inhibitor cocktail, 5 μl of Benzonase nuclease). The supernatant of lysates was incubated with Ni-NTA (Qiagen) in gravity columns. The columns were then washed with 10CV lysis buffer without protease inhibitor and Benzonase nuclease. Protein was eluted with 6CV elution buffer(150 mM NaCl, 20 mM Tris pH8.0, 200 mM Imidazole pH8 1 mM DTT). See SI for more details.
In vitro transcription
To obtain DNA template for transcription for CLN3/BNI1 RNA, T7 promotor TAATACGACTCACTATAGGG was cloned to the 5′ of CLN3/BNI1. The plasmids were then digested with restriction enzymes in front of the T7 promotor and after CLN3/BNI1 to obtain DNA template that contained the T7 promotor-CLN3/BNI1. The DNA template was gel extracted (Qiagen) and eluted in RNAse-free water for in vitro transcription. In vitro transcription was done with HiScribe T7 high yield RNA synthesis kit (NEB) following protocols provided in kit. To transcribe labeled RNA for imaging, a trace amount of cy3-UTP was added into the mixture for transcription. Transcribed RNAs were then ethanol-precipitated and resuspended in RNase-free water and stored at −80 °C.
Droplet assembly and imaging
Unless otherwise stated, all proteins used in experiments contain 10:1 unlabeled to GFP-labeled versions. To lower salt concentration, proteins were mixed with no salt buffer (50 mM Tris pH8, 1mM DTT) to obtain desired protein and salt concentrations. For RNA experiments, proteins were diluted with droplet buffer (50 mM Tris pH8, 1mM DTT, 150mM KCl) to desired concentration and then unlabeled RNA or cy3-labeld RNA were added. All mixing was performed at room temperature. The mixing and imaging was done in glass wells (Grace Bio-Labs) that were treated with 30mg/ml BSA (Sigma) for 30min. Screening for droplets was done on a spinning disc confocal microscope (Nikon), with VC Plan Apo 60X/1.4 NA oil objective.
Fusion kinetics
Droplet formation movies were recorded on a spinning disc confocal microscope (Nikon) with VC Plan Apo 60X/1.4 NA oil objective. Fusion events were manually spotted and cropped out in ImageJ. The cropped image series containing fusion events were analyzed in MATLAB with Imaging Processing Toolbox. As we have previously described (Brangwynne et al., 2011), we plot the change in aspect ratio = l/w, where l is the long axis and w is the short axis of fusing droplets, over time. The decay of aspect ratio over time was fitted to an exponential curve to obtain decay time t (Fig. S4). Plotting the decay time against droplet characteristic length ℓ=w(l-w) gives a linear relationship (Fig. 4B), with the slope scales with the ratio of viscosity over surface tension.
FRAP
FRAP were performed on a Nikon A1 laser scanning confocal microscope with a100x Plan Fluor oil objective. Spots of radius a = 1μm were bleached with a 405 laser in the center of droplets and intensity change over time was collected with 488nm laser for GFP labeled Whi3 and 561nm laser for cy3-labeled RNA in Elements. Data in ROIs were exported from Elements and then imported into MATLAB for further processing. After background deduction, the intensity change over time was normalized to pre-bleach level and fitted to an exponential curve F = A(1−e−t/τ) to obtain the apparent recovery time τ The apparent diffusion coefficient was then estimated as Dapp = a2/τ.
Microrheology
Microrheology was performed as previously described (Elbaum-Garfinkle et al., 2015). Red microspheres (Invitrogen) were mixed with protein solution (10% GFP labeled) before adding low salt buffer or unlabeled RNA. Bead diffusion was tracked on a spinning disc confocal microscope for 1000 s with 500 ms intervals. MSD data was fit to MSD(t) = 4Dtα + NF where α is the exponent, D is the diffusion coefficient and NF is the noise floor. Microrheology for CLN3 droplets was done in glass wells (Grace Bio-Labs), and for CLN3 RNA and BNI1 RNA comparison was done a device, the design of which will be published elsewhere. All data processing was done in MATLAB (MathWorks).
Cell culture and preparation for microscopy
Ashbya cells were grown in 10 ml Ashbya full media (AFM) under selection of either G418 (200 μg/ml) or clonat (100 μg/ml) in a 125 ml baffled flask shaking at 30°C ~16 h. _whi3Δ_RRM cells were grown at 22°C due to temperature sensitivity and for ~30h to obtain similar biomass to wild-type. The Ashbya culture was then transferred to a 15-ml conical tube (VWR) for centrifugation at 350 rpm for 2 min. AFM was removed and cells were resuspended in 10ml 2x low fluorescence media to reduce the auto-fluorescence of the medium. Cells were then placed on a 2x low fluorescence media gel-pad containing 1% agarose on the top of a slide, covered with a coverslip, sealed with VALAP and imaged.
Oblique angle total internal reflection fluorescence microscopy
Ashbya cells were grown and mounted onto gel pads as described above. TIRF was performed as previously described (Tokunaga et al., 2008). Time-lapse microscopy was performed on a Nikon Ti motorized inverted microscope equipped with a 100X Plan Apo NA 1.5 oil immersion lens and the Perfect Focus System. Whi3-GFP was excited using a 488 nm solid state laser line (Andor). Images were acquired with Hamamatsu ORCA-R2 cooled CCD camera controlled by Nikon elements software. All movies were background subtracted and linearly contrasted in imageJ.
Mitochondria and ER staining
Ashbya cells were grown in 10 ml AFM with proper selection in a 125-ml baffled flask, shaking overnight. Cells were stained with either Mitotracker Red CMXRos or ER-Tracker Red (Life technologies) to a final concentration of 1 mM. After a 30-minute shaking incubation in 30°, cells were centrifuged and washed several times with PBS. Cells were then re-suspended in low fluorescence media before being placed on the gel pad and imaged by epifluorescence microscopy. See SI for more details on microscopy and image processing.
Supplementary Material
1
2
3
4
5
6
Highlights.
- RNA drives phase transition of Whi3, an RNA-binding protein with a long polyQ tract
- RNA alters Whi3 droplet viscosity, dynamics and their propensity to fuse
- Different target mRNAs drive Whi3 to form droplets with distinct properties
- Whi3 droplets mature and appear fibrillar over time
Acknowledgments
We thank Jane Lipson, Charles Barlowe, the HHMI Summer Institute at the Marine Biological laboratory in Woods Hole, MA, members in Heather True lab, the Gladfelter lab and Brangwynne lab for helpful discussion, and Ann M. Lavanway for microscopy support. HZ, EL, PO and ASG were supported by National Institutes of Health (R01-GM081506), CPB, SEG, and NT was supported by Human Frontier Science Program Grant RGP0007/2012, National Institutes of Health New Innovator Awards 1DP2GM105437-01 (to C.P.B.), the Searle Scholars Program (C.P.B.), National Science Foundation CAREER Award 1253035 (to C.P.B.).
Footnotes
Author Contributions
HZ: designed and performed experiments, analyzed data, prepared figures and drafted manuscript, SE: analyzed data and edited manuscript, EL performed experiments, NT performed experiments, PO: prepared constructs, AB: developed protein purification pipeline, CB: designed experiments and edited manuscript, AG: designed and performed experiments, prepared figures and drafted manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson P, Kedersha N. RNA granules. The Journal of Cell Biology. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. P Natl Acad Sci USA. 2015 doi: 10.1073/pnas.1509317112. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. P Natl Acad Sci USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F, Barral Y. A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell. 2013;155:1244–1257. doi: 10.1016/j.cell.2013.10.046. [DOI] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CCH, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. P Natl Acad Sci USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Brangwynne CP. A nuclear F-actin scaffold stabilizes RNP droplets against gravity in large cells. Nature cell biology. 2013;15 doi: 10.1038/ncb2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Hungerbuehler AK, Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. The Journal of cell biology. 2006;172:347–362. doi: 10.1083/jcb.200507003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hubstenberger A, Noble SL, Cameron C, Evans TC. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Developmental cell. 2013;27:161–173. doi: 10.1016/j.devcel.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RAL. Soft condensed matter. Oxford; New York: Oxford University Press; 2002. [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain research. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CF, Brangwynne CP, Gharakhani J, Hyman AA, Julicher F. Spatial organization of the cell cytoplasm by position dependent phase separation. Physical Review Letters. 2013a;111:088101. doi: 10.1103/PhysRevLett.111.088101. [DOI] [PubMed] [Google Scholar]
- Lee C, Occhipinti P, Gladfelter AS. PolyQ-dependent RNA-protein assemblies control symmetry breaking. The Journal of Cell Biology. 2015;208:533–544. doi: 10.1083/jcb.201407105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Zhang H, Baker AE, Occhipinti P, Borsuk ME, Gladfelter AS. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Developmental cell. 2013b;25:572–584. doi: 10.1016/j.devcel.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, David SW, Rosen Michael K, Parker Roy. Formation and Maturation of Phase Separated Liquid Droplets by RNA Binding Proteins. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.08.018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. The FEBS journal. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Molliex AT, Lee J, Coughlin J, Kanagara M, jAP, Kim HJ, Mittag T, Taylor JP. Phase separation by low-complexity domains contributes to stress granule assembly and drives pathological fibrillization. Cell. 2015 doi: 10.1016/j.cell.2015.09.015. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash RS, Volpe T, Futcher B. Isolation and characterization of WHI3, a size-control gene of Saccharomyces cerevisiae. Genetics. 2001;157:1469–1480. doi: 10.1093/genetics/157.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs T, Bazett-Jones D, Pawson T, Forman-Kay J, et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Molecular cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee Hyun O, Jawerth L, Maharana S, Jahnel M, Hein Marco Y, Stoynov S, Mahamid J, Saha S, Franzmann Titus M, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Pawar AB, Caggioni M, Hartel RW, Spicer PT. Arrested coalescence of viscoelastic droplets with internal microstructure. Faraday discussions. 2012;158:341–350. doi: 10.1039/c2fd20029e. [DOI] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Molecular Systems Biology. 2014;10 doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. Journal of cell science. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81 doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nature cell biology. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nature methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS letters. 2015;589:15–22. doi: 10.1016/j.febslet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Weber Stephanie C, Brangwynne Clifford P. Inverse Size Scaling of the Nucleolus by a Concentration-Dependent Phase Transition. Current Biology. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends in neurosciences. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6