Amino-Terminal Region of the Human Immunodeficiency Virus Type 1 Nucleocapsid Is Required for Human APOBEC3G Packaging (original) (raw)
Abstract
APOBEC3G exerts its antiviral activity by targeting to retroviral particles and inducing viral DNA hypermutations in the absence of Vif. However, the mechanism by which APOBEC3G is packaged into virions remains unclear. We now report that viral genomic RNA enhances but is not essential for human APOBEC3G packaging into human immunodeficiency virus type 1 (HIV-1) virions. Packaging of APOBEC3G was also detected in HIV-1 Gag virus-like particles (VLP) that lacked all the viral genomic RNA packaging signals. Human APOBEC3G could be packaged efficiently into a divergent subtype HIV-1, as well as simian immunodeficiency virus, strain mac, and murine leukemia virus Gag VLP. Cosedimentation of human APOBEC3G and intracellular Gag complexes was detected by equilibrium density and velocity sucrose gradient analysis. Interaction between human APOBEC3G and HIV-1 Gag was also detected by coimmunoprecipitation experiments. This interaction did not require p6, p1, or the C-terminal region of NCp7. However, the N-terminal region, especially the first 11 amino acids, of HIV-1 NCp7 was critical for HIV-1 Gag and APOBEC3G interaction and virion packaging. The linker region flanked by the two active sites of human APOBEC3G was also important for efficient packaging into HIV-1 Gag VLP. Association of human APOBEC3G with RNA-containing intracellular complexes was observed. These results suggest that the N-terminal region of HIV-1 NC, which is critical for binding to RNA and mediating Gag-Gag oligomerization, plays an important role in APOBEC3G binding and virion packaging.
The Vif protein, which modulates viral infectivity (15, 21-23, 29, 38, 45, 59, 62, 78-80, 82, 84, 88) and pathogenicity (13, 28, 29, 33, 34, 52), is present in nearly all lentiviruses, excluding equine infectious anemia virus. Recently, it has been shown that APOBEC3G has antiviral activity and that its activity is suppressed by Vif (73). APOBEC3G belongs to a family of proteins that have cytidine deaminase activity (35, 73, 89), and its cellular function is still unknown (35, 73, 89). Vif mutant, but not wild-type, human immunodeficiency virus type 1 (HIV-1) produced in the presence of APOBEC3G undergoes hypermutations in newly synthesized viral DNA (30, 44, 53, 55, 94), presumably due to C-to-U modification during minus-strand viral DNA synthesis (30, 44, 53, 55, 94).
HIV-1 Vif can be coimmunoprecipitated with human APOBEC3G from virus-infected or -transfected cells, suggesting that Vif and APOBEC3G may form a complex (39, 56, 74, 81, 93). However, whether Vif directly interacts with APOBEC3G is still unclear (57). Amino acids 54 to 124 of human APOBEC3G have been shown to mediate interaction with HIV-1 Vif (9). Recent observations indicate that a single amino acid variation (at position 128) between human and monkey APOBEC3G determines the species-specific sensitivity to Vif of primate immunodeficiency viruses (4, 54, 71). However, whether this position is involved in APOBEC3G interaction with Vif (4, 54, 71, 91) or whether it is involved in direct interaction between these two molecules is still controversial (4, 54, 71, 91). Expression of HIV-1 Vif reduced the half-life of human APOBEC3G (9, 56, 57, 74, 81, 93), resulting in a reduced steady-state level of APOBEC3G (9, 39, 55-57, 74, 81, 93). The stability of human APOBEC3G in the presence of Vif was significantly increased when proteasome inhibitors were applied (9, 56, 57, 74, 81, 93), indicating that Vif induced-degradation of human APOBEC3G requires proteasomes. HIV-1 Vif also induces polyubiquitination of human APOBEC3G (9, 56, 57, 74, 93), which is a prerequisite for proteasome-mediated protein degradation.
HIV-1 Vif interacts with cellular proteins Cul5, Elongin B, Elongin C, and Rbx1 to form an E3 ubiquitin ligase complex (93), similar to the Elongin C-Cul2-SOCS box (ECS). Cullin-based E3 ligases display striking similarities, which are typified by SCF complexes (12). In SCF and ECS complexes, Skp1 and Elongin C bridge the interaction between cullin and the substrate recognition protein (F box and SOCS box proteins, respectively). These adaptors bind substrates through a distinct protein-protein interaction domain (e.g., WD40 for the F box protein Cdc4 and the β-domain for VHL). Cullin-containing E3 ubiquitin ligases represent one of the largest families of ubiquitin-protein E3 ligases, ubiquitinating a broad range of proteins involved in cell cycle regulation, signal transduction, transcription, and other cellular processes (12). The highly conserved SLQXLA motif in Vif has some similarities with SOCS box sequences and is required for efficient interaction between HIV-1 Vif and the Cul5-Elongin B-Elongin C complex (56, 93). It is also required for Vif-induced degradation of human APOBEC3G (56, 74, 93). The ability of Vif to suppress the antiviral activity of APOBEC3G is specifically dependent on Cul5-Elongin B-Elongin C function. When Cul5 complex function was inhibited, HIV-1 Vif induced polyubiquitination and degradation of human APOBEC3G was blocked (93).
The ability of HIV-1 Vif to degrade human APOBEC3G (9, 56, 57, 74, 81, 93) and to exclude APOBEC3G from released virions is compromised in the presence of a proteasome inhibitor (51, 93), suggesting that degradation of human APOBEC3G by the proteasome, rather than competition for binding by HIV-1 Vif during virus assembly, is the main mechanism by which APOBEC3G is excluded from released virions. When proteasome activity was compromised, more molecules of Vif than of APOBEC3G were packaged into the released virions (51). However, the net effect of increased packaging of Vif and APOBEC3G in the presence of MG132 was reduced virus infectivity (51, 93). Treatment with MG132 in the absence of APOBEC3G under the same conditions had only a minor effect on virus infectivity (93). Therefore, a low concentration of MG132 selectively blocks virus infectivity in the presence of APOBEC3G. The mere presence of Vif in virions is not sufficient to overcome the APOBEC3G activity (51, 93).
It is conceivable that APOBEC3G exerts its antiviral activity by interacting with certain viral components and that targeting to retroviral particles induces viral DNA hypermutations in the absence of Vif. The mechanism of APOBEC3G packaging remains elusive. In this study, we report that, although the efficiency of virion packaging of human APOBEC3G could be influenced by the level of viral genomic RNA, a major determinant for APOBEC3G packaging is the Gag molecule. Virus-like particles (VLP) containing Gag molecules of diverse HIV-1 subtypes, simian immunodeficiency virus, strain mac (SIVmac), and murine leukemia virus (MuLV) could efficiently package human APOBEC3G. Interaction between human APOBEC3G and HIV-1 Gag was detected by cosedimentation experiments and coimmunoprecipitation analysis. The N-terminal region of NCp7, especially the first 11 amino acids, played an important role in mediating this interaction. The first linker region in APOBEC3G was also required for efficient interaction between Gag and APOBEC3G and virion packaging.
MATERIALS AND METHODS
Plasmid construction.
pNL4-3 was obtained from the AIDS Research Reagents Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (catalog no. 114). pNC2/2 has been described previously (25). pEF-HIV-myc expressing HIV-1 Gag with a Myc tag and pEF-MuLV-Myc expressing MuLV Gag with a Myc tag were gifts from Jeremy Luban (27). pΔPolΔEnv was constructed by inserting a 2.1-kb fragment digested with SalI and BamHI from HXB2ΔBgl (15) into the same restriction sites in HXB2ΔPol (47). Based on the Gag-Pol amino acid sequence of 97CNGX-6F (64), a new gene, termed GagPol INS, was synthesized. By removing the inhibitory sequences (INS) from the original gene, GagPol INS was highly expressed in a mammalian expression system independent of HIV-1 Rev. pGPCINS was constructed by cloning GagPol INS into VR1012 with XbaI and BamHI sites. The same method was used to clone pGagCINS into VR1012. The VR1012 vector was generously provided by Vical Inc. (San Diego, Calif.). pGagINS was constructed as previously described (66). pNC11S and pNC34S were made from pGagINS by generating stop codons after codons for K11 and K34 of the nucleocapsid (NC) protein, respectively. pP2S was made from the same plasmid by introducing a stop codon before the sequence encoding the NC protein. In order to clone the coding sequence for the leucine zipper domain (LZ) of the yeast transcription factor GCN4 into pNCS, a NotI site was introduced between p2 and p7 by the mutagenesis method. After the LZ coding sequence was amplified by primers 5′-ATAAGAATGCGGCCGCCCTCCAACGTATGAAGCAGCTC-3′ and 5′-CGGAATTCTTACTCACCCACAAGCTTTTTC-3′, it was inserted into pNCS by NotI and EcoRI sites. Human APOBEC3G-hemagglutinin (HA) was amplified by reverse transcription-PCT (RT-PCR) using mRNA from H9 cells with the forward primer 5′-CTCGAGACCATGAAGCCTCACTT-3′ and reverse primer 5′-GAATTCTCACGCGTAATCTGGGACGTCGTAAGGGTAGTTTTCCTGATTCTGGAG-3′, containing XhoI and EcoRI sites, respectively. APOBEC3G mutants were constructed by PCR using wild-type APOBEC3G-HA as the template. The following primers were used: Apo-3GΔN (Δ2-46), forward primer 5′-CTCGAGACCATGCCCCCTTTGGACGCAAAG-3′ and reverse primer 5′-GAATTCTCACGCGTAATCTGGGACGTCGTAAGGGTAGTTTTCCTGATTCTGGAG-3′; Apo-3GΔA1 (Δ65-100), forward primer 5′-CAGGTGTATTCCGAACTTAAGTACACAAGGGATATGGCCACG-3′ and reverse primer 5′-CGTGGCCATATCCCTTGTGTACTTAAGTTCGGAATACACCTG-3′; Apo-3GΔL1 (Δ108-161), forward primer 5′-GATATGGCCACGTTCCTGAGCAAGTTCGTGTACAGCCA-3′ and reverse primer 5′-TGGCTGTACACGAACTTGCTCAGGAACGTGGCCATATC-3′; Apo-3GΔPA (Δ162-256), forward primer 5′-ACGAATTTCAGCACTGTTGGCATGCAGAGCTGTGCTTCCT-3′ and reverse primer 5′-AGGAAGCACAGCTCTGCATGCCAACAGTGCTGAAATTCGT-3′; Apo-3GΔA2 (Δ255-292), forward primer 5′-ACATAAACACGGTTTTCTTGAACAGGAAATGGCTAAATTCATT-3′ and reverse primer 5′-AATGAATTTAGCCATTTCCTGTTCAAGAAAACCGTGTTTATGT-3′; Apo-3GΔC (Δ301-384), forward primer 5′-CTCGAGACCATGAAGCCTCACTT-3′ and reverse primer 5′-GAATTCTCACGCGTAATCTGGGACGTCGTAAGGGTATGAAATGAATTTAGCCATT-3′.
Antibodies and cell lines.
The following antibodies were used for this study: anti-p24 monoclonal antibody (MAb) (AIDS Research Reagents Program, Division of AIDS, NIAID, NIH; catalog no. 1513) (7), anti-c-Myc MAb (Sigma;catalog no. M5546), anti-HA MAb (Covance; catalog no. MMS-101R-10000). 293 cells (AIDS Research Reagents Program, Division of AIDS, NIAID, NIH; catalog no. 3522) (6) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum and penicillin-streptomycin (D-10 medium) and passaged upon confluence.
Transfection and virus purification.
DNA transfection was carried out with Lipofectamine 2000 (Invitrogen) as described by the manufacturer. To obtain 293/Apo-3G cells, the pABOBEC3G-HA plasmid was transfected into 293 cells and selected with 1 mg of G418 (Invitrogen)/ml for 2 weeks. Expression of APOBEC3G was detected by immunoblotting with the anti-HA MAb. Virion-associated viral proteins were prepared from cell culture supernatants and separated from cellular debris by centrifugation at 3,000 rpm for 30 min in a Sorvall RT 6000B centrifuge and filtration through a 0.2-μm-pore-size membrane. Virus particles were concentrated by centrifugation through a 30% sucrose cushion at 100,000 × g for 2 h in a Sorvall Ultra80 ultracentrifuge.
Immunoblot analysis.
Cells were collected 48 h after transfection. Cell and viral lysates were prepared as previously described (16). Cells (105) were lysed in 1× loading buffer (0.08 M Tris [pH 6.8], 2.0% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1 M dithiothreitol, 0.2% bromophenol blue). The samples were boiled for 10 min, and proteins were separated by SDS-polyacrylamide gel electrophoresis. For virion lysates, cell culture supernatants were collected 72 h after transfection by removal of cellular debris through centrifugation at 3,000 rpm for 10 min in a Sorvall RT 6000B and filtration through a 0.2-μm-pore-size membrane. Virus particles were concentrated by centrifugation through a 30% sucrose cushion at 100,000 × g for 2 h in a Sorvall Ultra80 ultracentrifuge. Proteins were transferred onto two separate nitrocellulose membranes by passive diffusion for 16 h, producing identical mirror image blots. Membranes were probed with various primary antibodies against proteins of interest. Secondary antibodies were alkaline phosphatase-conjugated anti-human and anti-mouse antibodies (Jackson Immunoresearch, Inc.), and staining was carried out with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium solutions prepared from chemicals obtained from Sigma. The staining of HA- tagged APOBEC3G in virion samples was performed with the ECL Plus kit (Amersham; RPN2132) using a mouse anti-HA MAb as the primary antibody and horseradish peroxidase-conjugated anti-mouse immunoglobulin G as the secondary antibody.
Immunoprecipitation.
For APOBEC3G-HA immunoprecipitation, transfected 293 cells were harvested and washed twice with cold phosphate-buffered saline (PBS) and then lysed with PBS containing 0.5% Triton X-100 and a protease inhibitor cocktail (Roche, Basel, Switzerland) at 4°C for 1 h. Cell lysates were clarified by centrifugation at 10,000 × g for 30 min at 4°C. Anti-HA-agarose (Roche) was mixed with the precleared cell lysates and incubated at 4°C for 3 h. The reaction mixture was then washed three times with cold PBS and eluted with 0.1 M glycine-HCl buffer, pH 2.0. The eluted materials were subsequently analyzed by immunoblotting.
Sucrose gradient analysis.
293/Apo-3G-HA cells were transfected with HXB2 DNA. Forty-eight hours after transfection cells were collected and lysed with 1% Triton X-100 lysis buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1% Triton X-100 plus a protease inhibitor cocktail; Roche). Cell lysates were subjected to 5 to 40% sucrose (in PBS) gradient ultracentrifugation at 260,800 × g for 4 h. Twenty-five fractions were collected for Western blotting analysis by using anti-HA for detection of APOBEC3G-HA and anti-P24 for detection of the HIV-1 Gag protein. For RNase treatment, the cell lysate was divided into two aliquots, one to which RNase A was added to a final concentration of 100 μg/ml and one to which an RNasin inhibitor was added to a final concentration of 1 U/μl; both aliquots were incubated at RT for 30 min and then subjected to 5 to 40% sucrose (in PBS) gradient ultracentrifugation at 260,800 × g for 4 h. Equilibrium density gradient analysis was performed as previously described (47).
RESULTS
Virion packaging of human APOBEC3G was reduced in HIV-1 genomic RNA packaging mutant viruses.
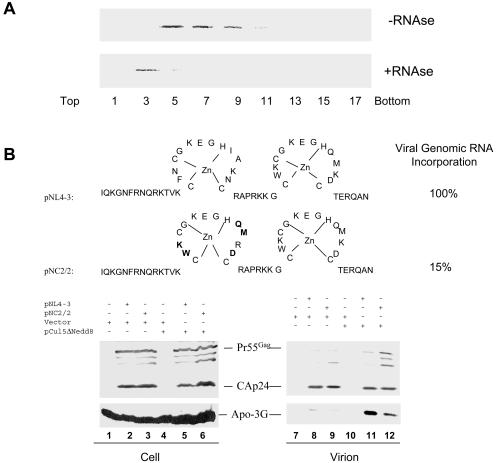
APOBEC3G is potentially an RNA-binding protein (35), or, as is the case for APOBEC1, it may associate with other RNA-binding proteins (35, 89). Consistent with this idea, we observed that human APOBEC3G was associated with high-molecular-weight complexes in 293/Apo-3G cell lysates when analyzed through a sucrose velocity gradient (Fig. 1A, top panel, fractions 5, 7, and 9). In the presence of RNase, human APOBEC3G was largely detected in fraction 3 of the sucrose velocity gradient (Fig. 1A, bottom panel). These results suggest that human APOBEC3G is associated with a cellular complex(es) that contains RNA.
FIG. 1.
Influence of HIV-1 NC on APOBEC3G packaging. (A) Sedimentation of human APOBEC3G in sucrose gradients. 293/Apo-3G cells were lysed, and cell debris was removed by centrifugation at 10,000 × g for 30 min. Supernatants were treated with RNAse A or RNasin and centrifuged at 260,800 × g for 4 h. Different fractions were analyzed by immunoblotting with a MAb against the HA tag for the detection of APOBEC3G-HA. (B) Packaging of human APOBEC3G into wild-type and NC mutant HIV-1 virions. pNL4-3 and pNL2/2 were transfected with control vector or pCul5ΔNedd8 into 293/Apo-3G cells as indicated. Cell lysates and virion pellets were prepared and analyzed by immunoblotting with a MAb against p24 for the detection of viral Gag proteins and a MAb against the HA tag for the detection of APOBEC3G-HA.
To address the question of whether viral genomic RNA plays a role in human APOBEC3G packaging into HIV-1 virions, we compared the levels of virion-associated APOBEC3G in wild-type pNL4-3 and the viral genomic RNA packaging mutant pNC2/2 (25) in the presence of functional Vif or nonfunctional Vif. pNC2/2 (Fig. 1B) contains five amino acid substitutions in the N-terminal zinc finger of the HIV-1 NC region and packages only approximately 15% of the viral genomic RNA (25). Consistent with previous reports (9, 39, 55-57, 74, 81, 93), in the presence of functional Vif, little APOBEC3G was detected in either the pNL4-3 or pNC2/2 virions (Fig. 1B, lanes 8 and 9). When Vif function was blocked by the mutant Cul5 (pCul5ΔNedd8) as previously described (93), a significant level of APOBEC3G was detected in the pNL4-3 virions (Fig. 1B, lane 11); however, a reduced level of APOBEC3G was detected in the pNC2/2 mutant virions (Fig. 1B, lane 12) compared to the level detected in the pNL4-3 virions (lane 11). The reduced packaging of APOBEC3G into pNC2/2 mutant virions could be related to reduced viral genomic RNA packaging or to amino acid substitutions in NC that affected the APOBEC3G interaction.
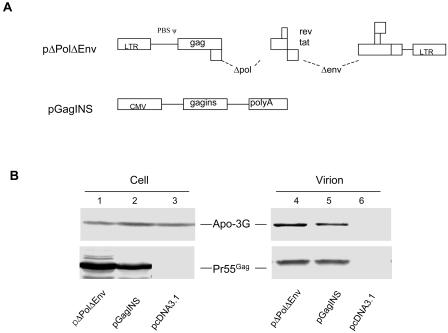
To address more directly whether viral genomic RNA plays a role in virion packaging of APOBEC3G, we next compared the levels of virion-associated APOBEC3G in pΔPolΔEnv virions and in VLP containing only Gag molecules, pGagINS. The pΔPolΔEnv construct contains all the 5′ RNA sequences upstream of the gag coding sequence, including TAR, polyA, PBS, DIS, SD, and the packaging signal (Fig. 2A). pΔPolΔEnv does not express Pol, Env, Vif, Vpr, Vpu, or Nef. The Gag amino acid sequence encoded by pGagINS is identical to that of the Gag encoded by pΔPolΔEnv. However, pGagINS lacks all the 5′ viral RNA sequences, including the viral RNA packaging sequences upstream of the Gag coding region (Fig. 2A). Both constructs lacked the vif coding sequence and should produce VLP consisting only of Gag proteins of viral origin. The levels of expression of APOBEC3G in all the transfected cells were comparable (Fig. 2B, lanes 1 to 3). Gag VLP were produced from pΔPolΔEnv- and pGagINS-transfected 293/Apo-3G cells (Fig. 2B, lanes 4 and 5). VLP-associated APOBEC3G was reduced in Gag VLP from pGagINS-transfected cells (Fig. 2B, lane 5) compared to that in Gag VLP produced from pΔPolΔEnv-transfected cells (Fig. 2B, lane 4). As expected, no APOBEC3G was detected in the supernatant of control vector-transfected cells, which did not generate VLP (Fig. 2B, lane 6). Collectively, these results suggest that viral genomic RNA enhanced packaging and/or virion retention of human APOBEC3G in HIV-1 particles. However, VLP containing only the Gag molecules could still efficiently package human APOBEC3G, suggesting that the Gag molecule contains a major determinant for the packaging of APOBEC3G.
FIG. 2.
Packaging of human APOBEC3G into Gag VLP. (A) Diagrams of pΔPolΔEnv and pGagINS vectors. pΔPolΔEnv contains all the 5′ HIV-1 sequences upstream from the gag coding region that are important for RNA packaging. All the 5′ HIV-1 sequences upstream from the gag coding region were missing in pGagINS. (B) Packaging of human APOBEC3G into pΔPolΔEnv and pGagINS Gag VLP. pΔPolΔEnv and pGagINS were transfected into 293/Apo-3G cells. Cell lysates and virion pellets were prepared and analyzed by immunoblotting with a MAb against p24 for the detection of Pr55Gag and a MAb against the HA tag for the detection of APOBEC3G-HA. LTR, long terminal repeat; CMV, cytomegalovirus promoter.
Gag proteins from a divergent HIV-1 subtype as well as SIVmac and MuLV could package human APOBEC3G into VLP.
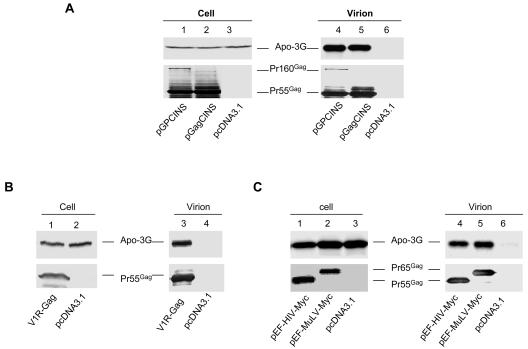
We next examined whether Gag from a divergent HIV-1 subtype and SIVmac alone could package human APOBEC3G. The SIVmac Gag expression vector, V1R-SIVgag, has been previously described (19). The modified SIVmac gag coding sequence is flanked by the cytomegalovirus promoter and BGHpA signal sequences. The HIV-1 B/C recombinant CRF-08 gag sequence was modified to remove INS sequences and cloned into VR1012 to generate pGagCINS. The gag sequence of CRF-08 is largely subtype C with a small amount of subtype B sequence in the p24 region (64). Both V1R-SIVgag and pGagCINS lack the viral RNA packaging signal upstream of the gag coding sequences. VLP were produced from pGagCINS (Fig. 3A, lane 5)- and V1R-SIVgag (Fig. 3B, lane 3)-transfected 293/Apo-3G cells, as detected by the anti-p24 antibody or SIV+ monkey serum, respectively. We were able to detect packaging of human APOBEC3G into Gag VLP produced from pGagCINS-transfected 293/Apo-3G cells (Fig. 3A, lane 5). In contrast, no APOBEC3G was detected in the supernatant of control vector-transfected 293/Apo-3G cells, which did not generate VLP (Fig. 3A, lane 6). Packaging of human APOBEC3G into SIVmac Gag VLP produced from V1R-SIVgag-transfected cells (Fig. 3B, lane 3) but not from control vector (Fig. 3B, lane 4)-transfected 293/Apo-3G cells was also detected.
FIG. 3.
Packaging of human APOBEC3G into divergent HIV-1, SIVmac, and MuLV Gag VLP. Various constructs were transfected into 293/Apo-3G cells. Cell lysates and virion pellets were prepared and analyzed by immunoblotting with a MAb against the HA tag for the detection of APOBEC3G-HA and a MAb against p24 for the detection of Pr55Gag and Pr160Gag-Pol (A), with SIVmac-infected monkey sera for the detection of SIVmac Pr55Gag (B), or with a MAb against the c-Myc tag for the detection of HIV-1 Pr55Gag-Myc and MuLV Pr65Gag-Myc (C).
Packaging of human APOBEC3G into MuLV particles has been reported (30, 53, 55). However, it is not clear whether MuLV Gag alone can package human APOBEC3G. Therefore, we examined the efficiency of human APOBEC3G packaging into HIV-1 Gag and MuLV Gag VLP. HIV-1 Gag and MuLV Gag were both tagged with the c-Myc epitope (27) so that the amounts of released HIV-1 Gag and MuLV particles could be compared. Efficient production of HIV-1 Gag or MuLV Gag VLP from transfected 293/Apo-3G cells was detected (Fig. 3C). Compared to HIV-1 Gag (Fig. 3C, lane 4), MuLV Gag was also able to efficiently package human APOBEC3G (Fig. 3C, lane 5). Thus, human APOBEC3G was able to be packaged into VLP formed by divergent retroviral Gag molecules that have little primary sequence homology.
VLP containing Gag and Gag-Pol proteins did not enhance human APOBEC3G packaging.
A previous study has reported an enhanced interaction between the Gag domain in Gag-Pol precursor molecules and HIV-1 Vif, compared to that between Gag and HIV-1 Vif (32). Therefore, we next examined whether VLP containing both Gag and Gag-Pol precursor molecules might enhance human APOBEC3G packaging. The construct pGPCINS contained gag and pol sequences modified to remove INS, but the amino acid sequences in Gag and Pol that it encoded were identical to those in the parental CRF08 Gag and Pol, with the exception of the protease active site Asp being changed to Ala. The frameshifting signal and surrounding 506 nucleotides were not altered, so that the expression of Gag-Pol would not be affected. The Pr160Gag-Pol precursor was expressed from pGPCINS and incorporated into released VLP (Fig. 3A, lane 4). The level of APOBEC3G in VLP containing both Gag and Gag-Pol precursors (Fig. 3A, lane 4) was not significantly higher than that in VLP containing Gag alone (Fig. 3A, lane 5). These results suggest that, at least in the Gag VLP system, the presence of Gag-Pol molecules did not significantly influence APOBEC3G packaging.
APOBEC3G cosediments with detergent-resistant intracellular Gag complexes.
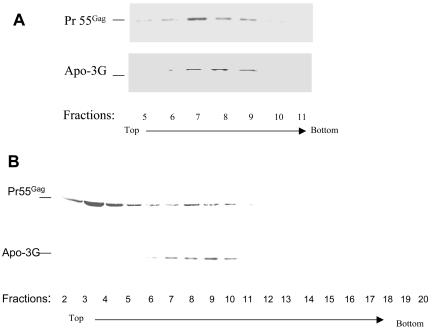
We (46, 47) and others (50, 76) have previously shown that HIV-1 Gag forms detergent-resistant cytoplasmic complexes. Incorporation of APOBEC3G into Gag VLP suggested that APOBEC3G and Gag colocalize at the site of virus assembly. To examine whether Gag and APOBEC3G indeed colocalize intracellularly, cell lysates from HXB2-transfected 293/Apo-3G cells were prepared as previously described (47) and centrifuged through a 20 to 60% sucrose equilibrium density gradient. Fractions were collected, pelleted by ultracentrifugation, and analyzed by immunoblotting with anti-p24 to detect Pr55Gag and anti-HA to detect APOBEC3G-HA. Detergent-resistant Gag complexes were detected in fractions 7 to 9 (from top to bottom of the gradient) with a density of 1.10 to 1.13 g/ml (Fig. 4A), as previously described (46). Cosedimentation of APOBEC3G with Pr55Gag was detected in fractions 7 to 9 in the same gradient (Fig. 4A). Cofractionation of APOBEC3G and HIV-1 Gag was also detected by velocity gradient analysis. Cell lysates were prepared from HXB2-transfected 293/Apo-3G cells as previously described (47), loaded onto a 5 to 40% sucrose gradient, and centrifuged at 260,800 × g for 4 h. Fractions were collected and analyzed by immunoblotting with anti-p24 to detect Pr55Gag and anti-HA to detect APOBEC3G-HA. Peaks of HIV-1 Gag were detected at fractions 8 to 10 and 2 to 4. Cosedimentation of APOBEC3G with Pr55Gag was detected in fractions 8 to 10 (Fig. 4B). However, APOBEC3G did not colocalize with all HIV-1 Gag, as indicated by the relative lack of APOBEC3G in fractions 2 to 4 (Fig. 4B).
FIG. 4.
Cosedimentation of human APOBEC3G with HIV-1 Gag. (A) Cell lysates from HXB2Vif-transfected 293/Apo-3G cells were prepared and analyzed by 15 to 60% sucrose equilibrium density gradients as previously described (46, 47). Eighteen fractions were collected, and fractions 5 to 10, which represent peak fractions of Gag intracellular complexes, were analyzed by immunoblotting with a MAb against p24 for the detection of Pr55Gag and a MAb against the HA tag for the detection of APOBEC3G-HA. (B) Cell lysates from HXB2Vif-transfected 293/Apo-3G cells were prepared and analyzed by 5 to 40% sucrose velocity gradients as described in Materials and Methods. Twenty fractions were collected, and fractions 2 to 20, which represent several peak fractions of Gag intracellular complexes, were analyzed by immunoblotting with a MAb against p24 for the detection of Pr55Gag and a MAb against the HA tag for the detection of APOBEC3G-HA.
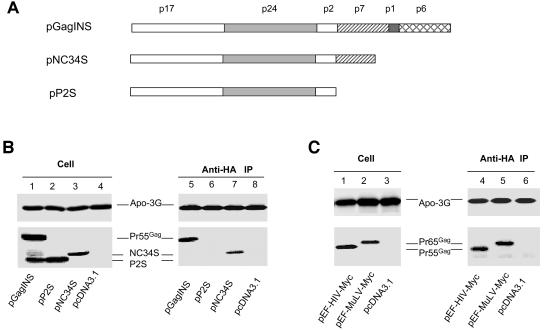
The N-terminal region of HIV-1 NC is important for interaction with APOBEC3G.
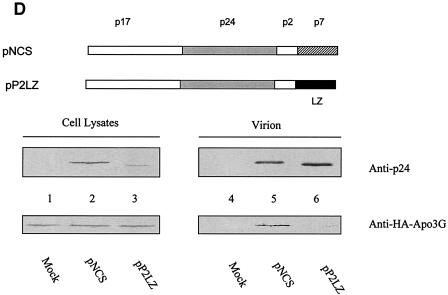
The intracellular interaction between Gag and APOBEC3G was further examined by coimmunoprecipitation experiments. Full-length HIV-1 Gag was expressed in 293/Apo-3G cells, and interaction of HIV-1 Gag with human APOBEC3G-HA was detected by immunoprecipitation with anti-HA followed by immunoblotting with the anti-p24 antibody (Fig. 5B, lane 5). Interaction between HIV-1 Gag and human APOBEC3G apparently did not require p6, p1, or the C-terminal region of NCp7, since an NCp7-truncated Gag molecule (NC34S) could still be coimmunoprecipitated with APOBEC3G-HA (Fig. 5B, lane 7). On the other hand, interaction between a truncated Gag containing only MAp17, CAp24, and p2 (P2S) and APOBEC3G was not detected under the same experimental conditions (Fig. 5B, lane 6), suggesting that the N-terminal region of NCp7 plays an important role in mediating Gag-APOBEC3G interactions. As for HIV-1 Gag, interaction between MuLV Gag and human APOBEC3G was also detected by the similar coimmunoprecipitation experiments (Fig. 5C, lane 5). The construct P2S lacked NC and released VLP inefficiently (data not shown). It has been reported that HIV-1 virus assembly mediated through NC can be replaced by that mediated by protein motifs that promote dimer formation (1, 37, 96). When the NC of HIV-1 Gag was replaced with the LZ domain of the yeast GCN4 (1), virion packaging of APOBEC3G was not detected (Fig. 5D, lane 6), unlike what was found for VLP containing HIV-1 NC (Fig. 5D, lane 5). These data suggest that formation of VLP alone is not sufficient for APOBEC3G packaging.
FIG. 5.
Coimmunoprecipitation of human APOBEC3G and HIV-1 Gag. (A) Diagrams of HIV-1 full-length Gag, NC34S, and P2S Gag constructs. (B) Various Gag constructs were transfected into 293/Apo-3G cells. Cell lysates were prepared and immunoprecipitated with anti-HA beads, and precipitated samples were analyzed by immunoblotting with a MAb against the HA tag for the detection of APOBEC3G-HA and a MAb against p24 for the detection of HIV-1 full-length Gag and truncated NC34S and P2S Gag molecules. (C) pEF-HIV-Myc expressing HIV-1 Gag with a Myc tag and pEF-MuLV-Myc expressing MuLV Gag with a Myc tag were transfected into 293/Apo-3G cells. Cell lysates were prepared and immunoprecipitated with anti-HA beads. Precipitated samples were analyzed by immunoblotting with a MAb against the HA tag for the detection of APOBEC3G-HA or a MAb against the c-Myc tag for the detection of HIV-1 Pr55Gag-Myc and MuLV Pr65Gag-Myc. (D) NCS and P2SLZ constructs were transfected into 293/Apo-3G cells. Cell lysates and virion pellets were prepared and analyzed by immunoblotting with a MAb against the HA tag for the detection of APOBEC3G-HA and a MAb against p24 for the detection of NCS and P2SLZ Gag molecules.
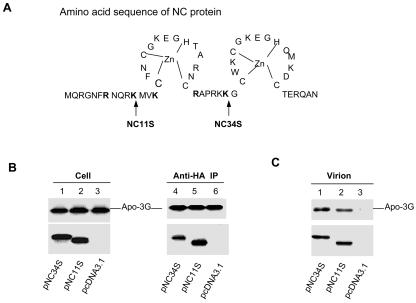
To further map the region in NCp7 that is required for APOBEC3G interaction (Fig. 6A), a stop codon was introduced after the first 11 amino acids (NC11S) of NCp7. As shown in Fig. 6B, C-terminally truncated Gag molecules containing the first 11 amino acids (lane 5) were still able to interact with human APOBEC3G. Deletion of the first zinc finger of NCp7 did not affect the interaction of HIV-1 Gag with APOBEC3G (data not shown), suggesting that the defect in APOBEC3G packaging of mutant NC2/2 (Fig. 1) is unlikely to be due to amino acid substitutions in the first zinc finger of NCp7. Consistent with the ability of NC34S and NC11S to interact with APOBEC3G intracellularly, VLP formed by these mutant Gag molecules also packaged human APOBEC3G (Fig. 6C). Previous work has identified the C-terminal regions of NCp7 and p1 and the N-terminal region of p6 as being important for the interaction between HIV-1 Gag and Vif (32). Since packaging of APOBEC3G was detected with the full-length as well as the NC Gag truncation mutant VLP (Fig. 6C), it appears that the C-terminal region of NCp7, the P1 space peptide, and p6, which were important for interaction with HIV-1 Vif, are not essential for interaction with human APOBEC3G.
FIG. 6.
The N-terminal region of HIV-1 NCp7 is required for interaction with human APOBEC3G and virion packaging. (A) Diagrams of NC34S and NC11S constructs. (B) NC34S and NC11S constructs were transfected into 293/Apo-3G cells. Cell lysates were prepared and immunoprecipitated with anti-HA beads, and precipitated samples were analyzed by immunoblotting with a MAb against the HA tag for the detection of APOBEC3G-HA and a MAb against p24 for the detection of truncated NC34S and P2S Gag molecules. (C) NC34S and NC11S constructs were transfected into 293/Apo-3G cells. Cell lysates and virion pellets were prepared and analyzed by immunoblotting with a MAb against the HA tag for the detection of APOBEC3G-HA and a MAb against p24 for the detection of truncated NC34S and NC11 Gag molecules.
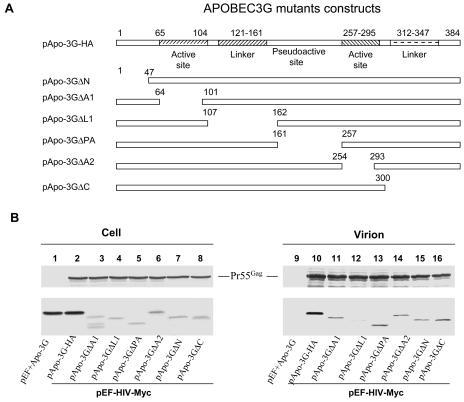
The first linker region of APOBEC3G is required for virion packaging.
By analogy to APOBEC1, the functional domains of APOBEC3G (Fig. 7A) could be separated into an N-terminal domain, a first active site, a first linker region, a pseudoactive site, a second active site, a second linker region, and a C-terminal domain (35). Human APOBEC3G deletion mutant constructs were generated to identify regions important for APOBEC3G virion packaging (Fig. 7A). Mutant APOBEC3G with a deletion of the N-terminal domain (Apo-3GΔN), the first active site (Apo-3GΔA1), the pseudoactive site (Apo-3GΔPA), the second active site (Apo-3GΔA2), or the second linker region plus the C-terminal domain (Apo-3GΔC) could still be packaged into HIV-1 Gag VLP (Fig. 7B). However, the packaging of a mutant APOBEC3G with a deletion of the first linker region (Apo-3GΔL1) was significantly reduced (Fig. 7B, lane 12) compared to that of other mutant APOBEC3G constructs.
FIG. 7.
Packaging of mutant APOBEC3G into HIV-1 Gag VLP. (A) Diagrams of mutant human APOBEC3G constructs. The numbers indicate amino acid positions in human APOBEC3G. Positions of amino acids that were deleted in mutant APOBEC3G constructs are indicated above each construct. (B) Various APOBEC3G constructs were transfected with pEF-HIV-Myc into 293 cells. Cell lysates and viruses were purified and analyzed by immunoblotting 48 h after transfection. Full-length and mutant APOBEC3G-HA was detected by the anti-HA antibody. Viral Gag proteins were detected by an anti-p24 antibody. Comparable amounts of VLP were used, as indicated by immunoblotting with the anti-p24 MAb.
DISCUSSION
APOBEC3G is a major cellular antiviral factor that needs to be neutralized by HIV-1 Vif if successful virus production is to occur (30, 39, 44, 53, 55, 56, 73, 74, 81, 93, 94). Virion-packaged APOBEC3G may negatively influence the reverse transcription complex through its RNA components and/or protein components. It may also induce instability in the newly synthesized viral DNA by causing a C-to-U modification in the minus-strand viral DNA (30, 44, 53, 55, 94). Finally, base substitutions in the viral DNA (30, 44, 53, 55, 94) may also alter translation (e.g., through premature stop codons) and the functions of translated proteins (e.g., through amino acid substitutions). Therefore, interaction between APOBEC3G and one or more viral components and packaging into released virions are critical for the antiviral activity of APOBEC3G.
Results presented here indicate that interaction between human APOBEC3G and HIV-1 Gag either directly or indirectly occurs during virus assembly and is likely to be a major driving force for APOBEC3G virion packaging. HIV-1 Gag VLP lacking all the upstream RNA packaging sequences were still able to package human APOBEC3G, albeit less efficiently than Gag VLP that contained all the RNA packaging sequences. These results suggest that viral genomic RNA can enhance human APOBEC3G virion packaging and/or retention. However, viral genomic RNA was not essential for APOBEC3G packaging.
An interaction between human APOBEC3G and HIV-1 Gag was demonstrated by cosedimentation of these two molecules in sucrose gradients (velocity and equilibrium density) and by coimmunoprecipitation analysis. The N-terminal region of HIV-1 NCp7, especially the first 11 amino acids in NCp7, is critical for mediating this interaction. It is interesting that this region of NCp7 has previously been identified as the I domain of HIV-1 Gag (11). The I domain of retroviral Gag plays important roles in mediating Gag-Gag interaction, particle assembly, and the formation of dense particles (83). It is believed that the I domain, by binding to nucleic acids of either viral genomic or nonspecific cellular RNA, facilitates Gag-Gag dimerization and subsequently Gag oligomerization (83). The role of NC in virus assembly can also be performed by protein interaction motifs that mediate dimer formation (1, 37, 96).
Efficient packaging of human APOBEC3G could be detected in VLP formed by Gag molecules from divergent retroviruses that include HIV-1, SIVmac, and MuLV. The amino acid sequences of these retroviral Gag molecules have a very low degree of sequence homology. Therefore, it is difficult to imagine a direct interaction between human APOBEC3G and linear conserved domains of the various Gag molecules. A common feature of these Gag VLP is the packaging of nonspecific RNA molecules during virus assembly. It is therefore possible that the packaging of human APOBEC3G into diverse retroviral Gag VLP is mediated through a nonspecific interaction of APOBEC3G with RNA. Consistent with this idea, we found that at least a fraction of the intracellular APOBEC3G appeared to be associated with RNA-containing complexes (Fig. 1). In vitro binding of APOBEC3G to RNA has also been reported previously (35). Furthermore, the N-terminal region of HIV-1 NC, especially positively charged amino acids in the first 11 amino acids, has been shown to be important for RNA binding (8, 65, 86, 95). Data presented here indicated that this region is also important for APOBEC3G packaging. When NC of HIV-1 Gag was replaced with the LZ domain of the yeast GCN4, virion packaging of APOBEC3G was abolished, again supporting a role of RNA in mediating HIV-1 Gag and APOBEC3G interaction. However, a possible protein-protein interaction between HIV-1 Gag and human APOBEC3G, either directly or indirectly, cannot be formally excluded at this time.
Although APOBEC3G has been shown to directly bind RNA (35), the regions of APOBEC3G that are important for RNA binding have not yet been mapped. For APOBEC1, two phenylalanine residues in the active site are critical for RNA binding (3, 60). We have determined that the first linker region between the two active sites of human APOBEC3G plays an important role in mediating the virion packaging of APOBEC3G. It is not clear whether this region of APOBEC3G is important for RNA binding. APOBEC1 has been shown to interact with other RNA binding proteins that are critical for apolipoprotein B mRNA binding and APOBEC1-mediated RNA editing (10, 31, 48, 58). Therefore, the first linker region in human APOBEC3G could also bind to other RNA binding proteins to mediate Gag and APOBEC3G interaction through RNA.
Recently, it has been reported that mutations in the first active site of APOBEC3G inhibit cytidine deaminase activity and the induction of viral DNA hypermutation but that the mutant constructs retain substantial antiviral activity, suggesting that cytidine deaminase activity is not the sole determinant of the inhibition of HIV-1 replication (75). It is not clear whether interaction between human APOBEC3G and HIV-1 Gag (particularly the NCp7 domain), either directly or indirectly through RNA, contributes to the antiviral activity of APOBEC3G. HIV-1 NCp7 has been reported to possess chaperone activity (17, 42, 43, 68, 85, 87, 90). The NCp7 domain has also been shown to enhance annealing of the tRNA primer to the primer-binding site of viral genomic RNA (5, 20, 40, 70), to stimulate minus and plus strand viral DNA transfer (2, 14, 26, 41, 63, 67, 92), and to influence the processivity of reverse transcriptase (36, 49, 69). Defects in these steps have been observed in HIV-1 Vif mutant virions produced from nonpermissive cells that contain APOBEC3G (15, 18, 24, 61, 72, 77, 80, 88).
Acknowledgments
We thank R. Garten, T. Sarkis, R. Markham, and Markus Dettenhofer for useful discussions. We are grateful to Jeremy Luban for the plasmids pEF-HIV-myc and pEF-MuLV-Myc and to Vical, Inc., for the VR1012 plasmid. The following reagents were obtained through the AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, NIH: MAb against HIV-1 p24 (Bruce Chesebro and Hardy Chen), pNL4-3 (Malcolm Martin), and 293 cells (Andrew Rice).
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74**:**5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain, B., M. Lapadat-Tapolsky, C. Berlioz, and J. L. Darlix. 1994. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 13**:**973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anant, S., A. J. MacGinnitie, and N. O. Davidson. 1995. apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J. Biol. Chem. 270**:**14762-14767. [PubMed] [Google Scholar]
- 4.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101**:**3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brule, F., R. Marquet, L. Rong, M. A. Wainberg, B. P. Roques, S. F. Le Grice, B. Ehresmann, and C. Ehresmann. 2002. Structural and functional properties of the HIV-1 RNA-tRNA(Lys)3 primer complex annealed by the nucleocapsid protein: comparison with the heat-annealed complex. RNA 8**:**8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71**:**3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66**:**6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74**:**3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13**:**2009-2013. [DOI] [PubMed] [Google Scholar]
- 10.Dance, G. S., M. P. Sowden, L. Cartegni, E. Cooper, A. R. Krainer, and H. C. Smith. 2002. Two proteins essential for apolipoprotein B mRNA editing are expressed from a single gene through alternative splicing. J. Biol. Chem. 277**:**12703-12709. [DOI] [PubMed] [Google Scholar]
- 11.Derdowski, A., L. Ding, and P. Spearman. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78**:**1230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev Biol. 15**:**435-467. [DOI] [PubMed] [Google Scholar]
- 13.Desrosiers, R. C., J. D. Lifson, J. S. Gibbs, S. C. Czajak, A. Y. Howe, L. O. Arthur, and R. P. Johnson. 1998. Identification of highly attenuated mutants of simian immunodeficiency virus. J. Virol. 72**:**1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeStefano, J. J. 1995. Hum. immunodeficiency virus nucleocapsid protein stimulates strand transfer from internal regions of heteropolymeric RNA templates. Arch Virol. 140**:**1775-1789. [DOI] [PubMed] [Google Scholar]
- 15.Dettenhofer, M., S. Cen, B. A. Carlson, L. Kleiman, and X. F. Yu. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 74**:**8938-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dettenhofer, M., and X. F. Yu. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73**:**1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dib-Hajj, F., R. Khan, and D. P. Giedroc. 1993. Retroviral nucleocapsid proteins possess potent nucleic acid strand renaturation activity. Protein Sci. 2**:**231-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dornadula, G., S. Yang, R. J. Pomerantz, and H. Zhang. 2000. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J. Virol. 74**:**2594-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egan, M. A., W. A. Charini, M. J. Kuroda, J. E. Schmitz, P. Racz, K. Tenner-Racz, K. Manson, M. Wyand, M. A. Lifton, C. E. Nickerson, T. Fu, J. W. Shiver, and N. L. Letvin. 2000. Simian immunodeficiency virus (SIV) gag DNA-vaccinated rhesus monkeys develop secondary cytotoxic T-lymphocyte responses and control viral replication after pathogenic SIV infection. J. Virol. 74**:**7485-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, Y. X., S. Campbell, D. Harvin, B. Ehresmann, C. Ehresmann, and A. Rein. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 73**:**4251-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher, A. G., B. Ensoli, L. Ivanoff, M. Chamberlain, S. Petteway, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237**:**888-893. [DOI] [PubMed] [Google Scholar]
- 22.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66**:**6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retroviruses 10**:**607-616. [DOI] [PubMed] [Google Scholar]
- 24.Goncalves, J., Y. Korin, J. Zack, and D. Gabuzda. 1996. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J. Virol. 70**:**8701-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67**:**4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo, J., L. E. Henderson, J. Bess, B. Kane, and J. G. Levin. 1997. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 71**:**5178-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurer, C., A. Cimarelli, and J. Luban. 2002. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J. Virol. 76**:**4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harmache, A., P. Russo, F. Guiguen, C. Vitu, M. Vignoni, M. Bouyac, C. Hieblot, M. Pepin, R. Vigne, and M. Suzan. 1996. Requirement of caprine arthritis encephalitis virus vif gene for in vivo replication. Virology 224**:**246-255. [DOI] [PubMed] [Google Scholar]
- 29.Harmache, A., P. Russo, C. Vitu, F. Guiguen, J. F. Mornex, M. Pepin, R. Vigne, and M. Suzan. 1996. Replication in goats in vivo of caprine arthritis-encephalitis virus deleted in vif or tat genes: possible use of these deletion mutants as live vaccines. AIDS Res. Hum. Retroviruses 12**:**409-411. [DOI] [PubMed] [Google Scholar]
- 30.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113**:**803-809. [DOI] [PubMed] [Google Scholar]
- 31.Henderson, J. O., V. Blanc, and N. O. Davidson. 2001. Isolation, characterization and developmental regulation of the human apobec-1 complementation factor (ACF) gene. Biochim. Biophys. Acta 1522**:**22-30. [DOI] [PubMed] [Google Scholar]
- 32.Huvent, I., S. S. Hong, C. Fournier, B. Gay, J. Tournier, C. Carriere, M. Courcoul, R. Vigne, B. Spire, and P. Boulanger. 1998. Interaction and co-encapsidation of human immunodeficiency virus type 1 Gag and Vif recombinant proteins. J. Gen. Virol. 79**:**1069-1081. [DOI] [PubMed] [Google Scholar]
- 33.Inoshima, Y., M. Kohmoto, Y. Ikeda, H. Yamada, Y. Kawaguchi, K. Tomonaga, T. Miyazawa, C. Kai, T. Umemura, and T. Mikami. 1996. Roles of the auxiliary genes and AP-1 binding site in the long terminal repeat of feline immunodeficiency virus in the early stage of infection in cats. J. Virol. 70**:**8518-8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoshima, Y., T. Miyazawa, and T. Mikami. 1998. The roles of vif and ORF-A genes and AP-1 binding site in in vivo replication of feline immunodeficiency virus. Arch. Virol. 143**:**789-795. [DOI] [PubMed] [Google Scholar]
- 35.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79**:**285-296. [DOI] [PubMed] [Google Scholar]
- 36.Ji, X., G. J. Klarmann, and B. D. Preston. 1996. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry 35**:**132-143. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76**:**11177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kan, N. C., G. Franchini, F. Wong-Staal, G. C. DuBois, W. G. Robey, J. A. Lautenberger, and T. S. Papas. 1986. Identification of HTLV-III/LAV sor gene product and detection of antibodies in human sera. Science 231**:**1553-1555. [DOI] [PubMed] [Google Scholar]
- 39.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77**:**11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan, R., and D. P. Giedroc. 1992. Recombinant human immunodeficiency virus type 1 nucleocapsid (NCp7) protein unwinds tRNA. J. Biol. Chem. 267**:**6689-6695. [PubMed] [Google Scholar]
- 41.Kim, J. K., C. Palaniappan, W. Wu, P. J. Fay, and R. A. Bambara. 1997. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 272**:**16769-16777. [DOI] [PubMed] [Google Scholar]
- 42.Lapadat-Tapolsky, M., H. De Rocquigny, D. Van Gent, B. Roques, R. Plasterk, and J. L. Darlix. 1993. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 21**:**831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapadat-Tapolsky, M., C. Pernelle, C. Borie, and J. L. Darlix. 1995. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 23**:**2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300**:**1112. [DOI] [PubMed] [Google Scholar]
- 45.Lee, T. H., J. E. Coligan, J. S. Allan, M. F. McLane, J. E. Groopman, and M. Essex. 1986. A new HTLV-III/LAV protein encoded by a gene found in cytopathic retroviruses. Science 231**:**1546-1549. [DOI] [PubMed] [Google Scholar]
- 46.Lee, Y. M., B. Liu, and X. F. Yu. 1999. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J. Virol. 73**:**5654-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, Y. M., and X. F. Yu. 1998. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology 243**:**78-93. [DOI] [PubMed] [Google Scholar]
- 48.Lellek, H., R. Kirsten, I. Diehl, F. Apostel, F. Buck, and J. Greeve. 2000. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 275**:**19848-19856. [DOI] [PubMed] [Google Scholar]
- 49.Lener, D., V. Tanchou, B. P. Roques, S. F. Le Grice, and J. L. Darlix. 1998. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 273**:**33781-33786. [DOI] [PubMed] [Google Scholar]
- 50.Lingappa, J. R., R. L. Hill, M. L. Wong, and R. S. Hegde. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136**:**567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, B., X. Yu, K. Luo, Y. Yu, and X. F. Yu. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 78**:**2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lockridge, K. M., M. Chien, G. A. Dean, K. S. Cole, R. C. Montelaro, P. A. Luciw, and E. E. Sparger. 2000. Protective immunity against feline immunodeficiency virus induced by inoculation with vif-deleted proviral DNA. Virology 273**:**67-79. [DOI] [PubMed] [Google Scholar]
- 53.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424**:**99-103. [DOI] [PubMed] [Google Scholar]
- 54.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279**:**14481-14483. [DOI] [PubMed] [Google Scholar]
- 55.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114**:**21-31. [DOI] [PubMed] [Google Scholar]
- 56.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9**:**1398-1403. [DOI] [PubMed] [Google Scholar]
- 57.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2003. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279**:**7792-7798. [DOI] [PubMed] [Google Scholar]
- 58.Mehta, A., M. T. Kinter, N. E. Sherman, and D. M. Driscoll. 2000. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 20**:**1846-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michaels, F. H., N. Hattori, R. C. Gallo, and G. Franchini. 1993. The human immunodeficiency virus type 1 (HIV-1) vif protein is located in the cytoplasm of infected cells and its effect on viral replication is equivalent in HIV-2. AIDS Res. Hum. Retroviruses 9**:**1025-1030. [DOI] [PubMed] [Google Scholar]
- 60.Navaratnam, N., S. Bhattacharya, T. Fujino, D. Patel, A. L. Jarmuz, and J. Scott. 1995. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell 81**:**187-195. [DOI] [PubMed] [Google Scholar]
- 61.Ohagen, A., and D. Gabuzda. 2000. Role of Vif in stability of the human immunodeficiency virus type 1 core. J. Virol. 74**:**11055-11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park, I. W., K. Myrick, and J. Sodroski. 1994. Effects of vif mutations on cell-free infectivity and replication of simian immunodeficiency virus. J. Acquir. Immune Defic. Syndr. 7**:**1228-1236. [PubMed] [Google Scholar]
- 63.Peliska, J. A., S. Balasubramanian, D. P. Giedroc, and S. J. Benkovic. 1994. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry 33**:**13817-13823. [DOI] [PubMed] [Google Scholar]
- 64.Piyasirisilp, S., F. E. McCutchan, J. K. Carr, E. Sanders-Buell, W. Liu, J. Chen, R. Wagner, H. Wolf, Y. Shao, S. Lai, C. Beyrer, and X. F. Yu. 2000. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J. Virol. 74**:**11286-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poon, D. T., J. Wu, and A. Aldovini. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70**:**6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qiu, J. T., R. Song, M. Dettenhofer, C. Tian, T. August, B. K. Felber, G. N. Pavlakis, and X. F. Yu. 1999. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J. Virol. 73**:**9145-9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raja, A., and J. J. DeStefano. 1999. Kinetic analysis of the effect of HIV nucleocapsid protein (NCp) on internal strand transfer reactions. Biochemistry 38**:**5178-5184. [DOI] [PubMed] [Google Scholar]
- 68.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23**:**297-301. [DOI] [PubMed] [Google Scholar]
- 69.Rodriguez-Rodriguez, L., Z. Tsuchihashi, G. M. Fuentes, R. A. Bambara, and P. J. Fay. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 270**:**15005-15011. [DOI] [PubMed] [Google Scholar]
- 70.Rong, L., C. Liang, M. Hsu, L. Kleiman, P. Petitjean, H. de Rocquigny, B. P. Roques, and M. A. Wainberg. 1998. Roles of the human immunodeficiency virus type 1 nucleocapsid protein in annealing and initiation versus elongation in reverse transcription of viral negative-strand strong-stop DNA. J. Virol. 72**:**9353-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101**:**3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97**:**13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418**:**646-650. [DOI] [PubMed] [Google Scholar]
- 74.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9**:**1404-1407. [DOI] [PubMed] [Google Scholar]
- 75.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278**:**44412-44416. [DOI] [PubMed] [Google Scholar]
- 76.Simon, J. H., E. A. Carpenter, R. A. Fouchier, and M. H. Malim. 1999. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73**:**2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71**:**5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simon, J. H., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70**:**5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sodroski, J., W. C. Goh, C. Rosen, A. Tartar, D. Portetelle, A. Burny, and W. Haseltine. 1986. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science 231**:**1549-1553. [DOI] [PubMed] [Google Scholar]
- 80.Sova, P., and D. J. Volsky. 1993. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with _vif_-negative human immunodeficiency virus type 1. J. Virol. 67**:**6322-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12**:**591-601. [DOI] [PubMed] [Google Scholar]
- 82.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV ′A' (sor) gene product is essential for virus infectivity. Nature 328**:**728-730. [DOI] [PubMed] [Google Scholar]
- 83.Swanstrom, R. A., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 84.Tomonaga, K., J. Norimine, Y. S. Shin, M. Fukasawa, T. Miyazawa, A. Adachi, T. Toyosaki, Y. Kawaguchi, C. Kai, and T. Mikami. 1992. Identification of a feline immunodeficiency virus gene which is essential for cell-free virus infectivity. J. Virol. 66**:**6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsuchihashi, Z., and P. O. Brown. 1994. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 68**:**5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Urbaneja, M. A., B. P. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. R. Casas-Finet. 1999. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 287**:**59-75. [DOI] [PubMed] [Google Scholar]
- 87.Urbaneja, M. A., M. Wu, J. R. Casas-Finet, and R. L. Karpel. 2002. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J. Mol. Biol. 318**:**749-764. [DOI] [PubMed] [Google Scholar]
- 88.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67**:**4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wedekind, J. E., G. S. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19**:**207-216. [DOI] [PubMed] [Google Scholar]
- 90.Williams, M. C., I. Rouzina, J. R. Wenner, R. J. Gorelick, K. Musier-Forsyth, and V. A. Bloomfield. 2001. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl. Acad. Sci. USA 98**:**6121-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101**:**5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.You, J. C., and C. S. McHenry. 1994. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 269**:**31491-31495. [PubMed] [Google Scholar]
- 93.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302**:**1056-1060. [DOI] [PubMed] [Google Scholar]
- 94.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424**:**94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71**:**6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72**:**1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]