Plastid-Expressed Betaine Aldehyde Dehydrogenase Gene in Carrot Cultured Cells, Roots, and Leaves Confers Enhanced Salt Tolerance (original) (raw)
Abstract
Salinity is one of the major factors that limits geographical distribution of plants and adversely affects crop productivity and quality. We report here high-level expression of betaine aldehyde dehydrogenase (BADH) in cultured cells, roots, and leaves of carrot (Daucus carota) via plastid genetic engineering. Homoplasmic transgenic plants exhibiting high levels of salt tolerance were regenerated from bombarded cell cultures via somatic embryogenesis. Transformation efficiency of carrot somatic embryos was very high, with one transgenic event per approximately seven bombarded plates under optimal conditions. In vitro transgenic carrot cells transformed with the badh transgene were visually green in color when compared to untransformed carrot cells, and this offered a visual selection for transgenic lines. BADH enzyme activity was enhanced 8-fold in transgenic carrot cell cultures, grew 7-fold more, and accumulated 50- to 54-fold more betaine (93–101 _μ_mol g−1 dry weight of _β_-Ala betaine and Gly betaine) than untransformed cells grown in liquid medium containing 100 mm NaCl. Transgenic carrot plants expressing BADH grew in the presence of high concentrations of NaCl (up to 400 mm), the highest level of salt tolerance reported so far among genetically modified crop plants. BADH expression was 74.8% in non-green edible parts (carrots) containing chromoplasts, and 53% in proplastids of cultured cells when compared to chloroplasts (100%) in leaves. Demonstration of plastid transformation via somatic embryogenesis utilizing non-green tissues as recipients of foreign DNA for the first time overcomes two of the major obstacles in extending this technology to important crop plants.
Salt stress is a major abiotic stress in plant agriculture. The problem of soil salinity has been compounded by irrigation and excessive use of fertilizers. About 20% of the world's irrigated lands are affected by salinity (Zhu, 2001). Currently, high salinity limits crop production in 30% of the irrigated land in the United States and 20 million hectares globally. High salinity causes ion imbalance, toxic levels of cytoplasmic sodium, and drought stress (Ward et al., 2003). Plants utilize a number of protective mechanisms to maintain normal cellular metabolism and prevent damage to cellular components (Wood et al., 1996). One of the metabolic adaptations to salt stress is the accumulation of osmoprotectants. Gly betaine and _β_-Ala betaine are quaternary ammonium compounds that accumulate in many plant species in response to salt stress (Hanson et al., 1991; Hanson and Gage, 1991; Rhodes and Hanson, 1993; Rathinasabapathi et al., 2001). Gly betaine protects the cell from salt stress by maintaining an osmotic balance with the environment (Robinson and Jones, 1986) and by stabilizing the quaternary structure of complex proteins (Papageorgiou and Murata, 1995). This substance occurs naturally in some crops, like sugar beet and cotton, as well as in many highly salt- or drought-tolerant wild plants, including halophytes (Rhodes and Hanson, 1993; Nishimura et al., 2001). However, many stress-susceptible crops do not contain significant amounts of Gly betaine or other osmoprotectants. It was proposed that genetic engineering of osmotolerance in plants could be achieved by producing betaine in nonaccumulators (McCue and Hanson, 1990). This has been demonstrated in several reports where transgenic plants accumulating Gly betaine exhibit moderate levels of tolerance to salt stress (Nakamura et al., 1997; Guo et al., 2000; Holmström et al., 2000; Kishitani et al., 2000; Jia et al., 2002).
The metabolic pathway for Gly betaine synthesis in higher plants involves two enzymes, i.e. choline monooxygenase (CMO) and betaine aldehyde dehydrogenase (BADH), which are compartmentalized within the chloroplast (Rathinasabapathi et al., 1997; Nuccio et al., 1998). _β_-Ala betaine is produced after methylation of _β_-Ala that is derived from 3-aminopropionaldehyde in a reaction catalyzed by the BADH enzyme (Rathinasabapathi et al., 2001). Overexpression of betaine by manipulation of badh via chloroplast genetic engineering may prove to be an important strategy in order to confer salt tolerance on desired crops. Expressing a transgene in the chloroplast allows for high-level transgene expression (Daniell et al., 2002a), multigene engineering in a single transformation event (DeCosa et al., 2001; Daniell and Dhingra, 2002; Ruiz et al., 2003), transgene containment via maternal inheritance (Daniell, 2002), lack of gene silencing (DeCosa et al., 2001; Lee et al., 2003), position effect due to site-specific transgene integration (Daniell et al., 2002b), and pleiotropic effects due to subcellular compartmentalization of transgene products (Daniell et al., 2001a; Lee et al., 2003).
Carrot (Daucus carota) is one of the most important vegetable crops used worldwide for human and animal consumption, as it is an excellent source of sugars, vitamins A and C, and fiber in the diet. It is classified as a salt-sensitive plant and there is a 7% growth reduction for every 10 mm increment in salinity above 20 mm salt. Salt stress results in reduced leaf gas exchange and a reduction in apparent photosynthetic capacity in cultivated carrot crops (Gibberd et al., 2002). Therefore, carrot is a good candidate for manipulation of the Gly betaine biosynthetic pathway for increased salt tolerance. Being a biennial plant, carrot completes its life cycle in 2 years, producing an edible fleshy taproot in the first year and flowers in the second year after passing through a cold season (Yan and Hunt, 1999). It has recently been demonstrated that the chloroplast genome shows strict maternal inheritance in cultivated carrot crops (Vivek et al., 1999). Thus, carrot is an ideal crop for genetic engineering because it is doubly protected against transgene flow via pollen or seed dispersal.
We report here successful engineering of the carrot chloroplast genome to overexpress the badh gene that results in enhanced tolerance to salt stress. The chloroplast transgenic line was able to survive in 400 mm NaCl, the levels at which halophytes survive salt stress. Interestingly, BADH-expressing carrot cells appeared green in contrast to nontransgenic yellow cells. In order to achieve chloroplast transformation in carrot and to overexpress the badh gene, we employed appropriate regulatory sequences for both the selectable marker and the gene of interest, which facilitate expression in non-green plastids. The lack of expression of transgenes in non-green plastids has been one of the major obstacles in extending chloroplast transformation to other plant species (Bogorad, 2000; Daniell et al., 2002b). The other major obstacle is the inability to generate chloroplast transgenic plants via somatic embryogenesis and achieve homoplasmy, without the benefit of subsequent rounds of regeneration offered by organogenesis. Both of these obstacles have been successfully overcome in this study.
RESULTS AND DISCUSSION
Construction of Carrot Plastid Transformation Vectors
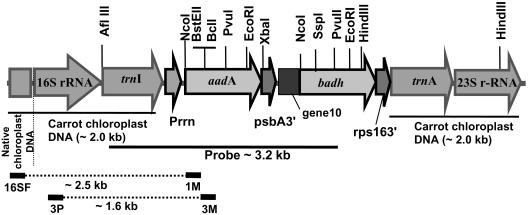
Carrot chloroplast transformation vector targets the expression cassette to the 16S/_trn_I- _trn_A/23S region of the chloroplast genome for integration via homologous recombination. The site of integration is similar to the universal chloroplast transformation vector (pLD CtV) reported earlier from our laboratory (Daniell et al., 1998; Guda et al., 2000). For the construction of carrot-specific chloroplast transformation vector, flanking region was amplified from carrot genomic DNA. In the absence of chloroplast genome sequence information for carrot, primers were designed based on the sequence information available for tobacco (Nicotiana tabacum). PCR amplification of the flanking region from carrot resulted in an approximately 4.0-kb DNA fragment that is about twice the size of the flanking region used in the pLD CtV vector. The size of the flanking sequence was increased in order to enhance the efficiency of homologous recombination. Carrot-specific chloroplast transformation vector (pDD-_Dc_-_aad_A/badh; Fig. 1) harbors the _aad_A gene regulated by the 5′ ribosome-binding site region or the Shine-Dalgarno sequence (GGAGG)/_psb_A 3′ untranslated region (UTR) and the badh gene regulated by the 5′ ribosome-binding site region of the bacteriophage T7 gene 10 leader in order to facilitate expression in green as well as non-green tissues (Guda et al., 2000; Staub et al., 2000; Dhingra et al., 2004)/_rps_16 3′UTR. Transcription of the expression cassette in the carrot chloroplast transformation vector is driven by the full-length 16S rRNA promoter (Shinozaki et al., 1986). The full-length promoter comprises binding sites for both the plastid-encoded and nuclear-encoded RNA polymerase, thereby facilitating transcription in green or non-green tissues. All the 5′ and 3′ regulatory elements were PCR amplified from the tobacco genomic DNA, except for the T7 gene 10 5′UTR, which was PCR amplified from the pET 11 vector (New England Biolabs, Beverly, MA). Details of primers are provided in “Materials and Methods.”
Figure 1.
Physical map of the carrot chloroplast transformation vector pDD-_Dc_-_aad_A/badh. PCR primer landing sites and the probe used for Southern analysis are shown.
Transformation of Carrot Plastid Genomes and Plant Regeneration
Yellow fine-cell suspension cultures induced from stem segments of carrot were bombarded with carrot chloroplast transformation vector pDD_-Dc-aad_A_/badh,_ as described (Daniell, 1997; Kumar and Daniell, 2004; Daniell et al., 2004a). Using the carrot chloroplast transformation vector, several independent transgenic cell lines were recovered using different sets of parameters for particle bombardment (Table I), within 2 to 3 months, from bombarded calli selected on solid medium containing 150 mg L−1 spectinomycin. Later, the transgenic calli were transferred to 350 mg L−1 spectinomycin for a month and subsequently multiplied using 500 mg L−1 spectinomycin. In order to further multiply the transgenic cell cultures, they were either subcultured on solid medium after every 2 to 3 weeks or rapidly multiplied in liquid medium (MSB and 0.1 mg L−1 2,4-D), and maintained at 130 rpm under diffuse light (50 lux) after every week. Transgenic carrot plants produced from somatic embryos on basal MSB medium (containing 500 mg L−1 spectinomycin) were transferred to soil in pots for the development of mature taproots and further molecular characterization.
Table I.
Carrot embryogenic callus was bombarded with the vector pDD-Dc-aadA/badh and coated on 0.6-μm gold particles using indicated parameters
| No. of Platesa | Rupture Disc | Distanceb | Independent Events ± sec | Events per Plate | Efficiency |
|---|---|---|---|---|---|
| %d | |||||
| 30 | 650 psi | 6 cm | 0 ± 0 | 0 | 0 |
| 24 | 9 cm | 1 ± 0 | 1/24 | 4.2 | |
| 26 | 12 cm | 0 ± 0 | 0 | 0 | |
| 36 | 900 psi | 6 cm | 0 ± 0 | 0 | 0 |
| 34 | 9 cm | 1 ± 0 | 1/34 | 2.9 | |
| 36 | 12 cm | 0 ± 0 | 0 | 0 | |
| 38 | 1,100 psi | 6 cm | 0 ± 0 | 0 | 0 |
| 30 | 9 cm | 3 ± 0.71 | 1/10 | 6.7 | |
| 30 | 12 cm | 4 ± 0.96 | 1/7.5 | 13.3 |
Optimization of Plastid Transformation
Plastid transformation efficiency is very high in tobacco (approximately 15 events per bombarded leaf; Fernandez-San Millan et al., 2003), but has been relatively inefficient in other crops, including other solanaceous species (Sidorov et al., 1999; Ruf et al., 2001). Therefore, a plastid transformation protocol was optimized with different bombardment conditions to achieve reproducibility using carrot cell cultures. In order to optimize gene delivery, the carrot-specific chloroplast transformation vector pDD-_Dc_-_aad_A/badh was bombarded using rupture discs of different psi at varying distances between rupture discs and the target tissues. Maximum transformation efficiency (13.3%) was observed with carrot cell cultures bombarded at a pressure of 1,100 psi and a distance of 12 cm. Considerably lower efficiencies were obtained at other parameters used for particle bombardment (Table I).
Visible Selection of Transgenic Carrot Cells
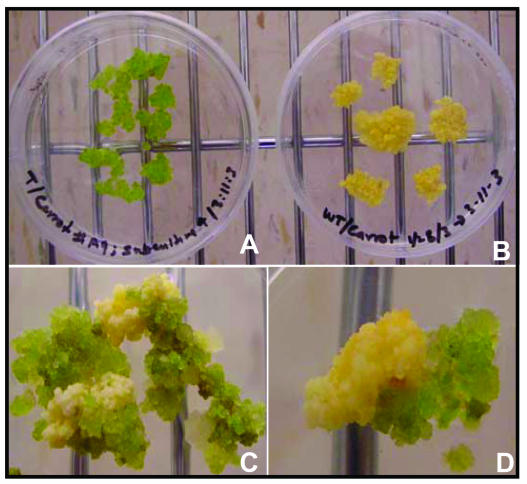
During in vitro cell culture studies of transgenic and nontransgenic carrot, it was interesting to note that chloroplast transgenic carrot cells could be distinguished on the basis of color. Transgenic calli derived from cultured cells expressing the badh transgene were always green in color, whereas nontransgenic cells were yellow in color (see Fig. 2, A and B). To test whether transgenic bright green cells were truly transgenic, heteroplasmic (partially transformed plastids) carrot cell cultures were placed on a growth medium without selection and were allowed to segregate; green and yellow cells visually segregated within 3 to 4 weeks (Fig. 2, C and D). Further, transgene integration in green carrot cells was confirmed by PCR, using a 16SF and _aph_A6-rev primer pair.
Figure 2.
Visual selection of green transgenic calli versus yellow nontransgenic carrot calli. A, Transformed. B, Untransformed. C and D, Heteroplasmic transgenic calli in the absence of a selection agent.
It has been shown that, in the presence of Gly betaine, light-dependent repair of the PSII complex is accelerated and favored over its photoinduced damage (Aro et al., 1993; Alia et al., 1999). Also, Rubisco has been shown to be protected in the presence of Gly betaine (Nomura et al., 1998; Sakamoto and Murata, 2002). Thus, the observed greening of BADH-expressing carrot cells may be a consequence of increased Gly betaine accumulation in the transformed cells, which prevents the photosynthesis apparatus from degradation. Visible distinction based on green color phenotype may be employed in future strategies for maintaining the transgenic status of transformed cell lines after the removal of stably integrated (Fischer et al., 1996; Iamtham and Day, 2000) or transiently cointegrated (Klaus et al., 2004) antibiotic-selectable markers.
Confirmation of Transgene Integration into Carrot Plastid Genomes
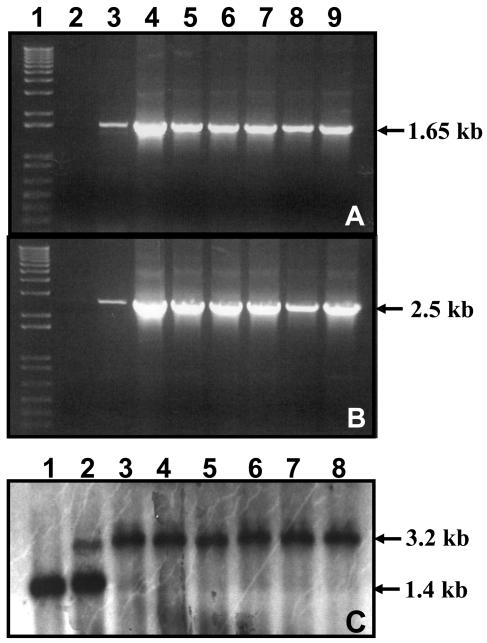
The carrot chloroplast vector pDD_-Dc_-aad_A/badh_ integrates the _aad_A and badh genes into the 16S-23S spacer region of the plastid genome by homologous recombination. Transgene integration into carrot plastid genomes was confirmed by PCR (Fig. 3A) using internal primers 3P (which lands on the 16S gene) and 3M (which lands on the _aad_A gene), producing a 1.6-kb PCR product. This eliminates mutants that may arise due to a mutation in the chloroplast 16S rRNA gene. In order to distinguish between nuclear and chloroplast transgenic cell lines (Fig. 3B), the 16SF primer was landed on the native chloroplast genome, 200 bp upstream of the integration site, and 1M primer was landed on the _aad_A gene; this generated a 2.5-kb PCR product, confirming site-specific integration of the transgene cassette.
Figure 3.
Confirmation of transgene integration into the carrot plastid genome by PCR and Southern-blot analysis. A, PCR product (1.65 kb) from internal primers 3P (land on flanking sequence) and 3M (land on the aad_A gene). B, PCR product (2.5 kb) from an external primer 16SF (land on the native chloroplast genome) and an internal primer 1M (land on the aad_A gene). Lane 1, 1-kb DNA ladder; lane 2, untransformed; lanes 3 to 9, transgenic carrot cell lines. All primer landing sites are shown in Figure 1. C, Southern-blot analysis of untransformed and transformed carrot with the vector pDD_-Dc-aad_A/badh. Carrot genomic DNA (5 _μ_g/lane) digested with _Afl_III and Pvu_II was hybridized with the 3.2-kb radioactive-labeled P32 DNA probe containing a 1.4-kb flanking sequence or a 1.8-kb aad_A/badh sequence (see Fig. 1 for details). Lane 1, Untransformed plant; lanes 2 and 3, heteroplasmic transgenic plants; lanes 4 to 8, homoplasmic transgenic plants derived after repetitive subculture in liquid medium under spectinomycin selection.
Southern-blot analysis was performed using total genomic DNA isolated from untransformed and transformed carrot plants generated from different transgenic cell lines. Total genomic DNA was digested with _Afl_III and _Pvu_II restriction enzymes (Fig. 3C). In order to investigate homoplasmy or heteroplasmy, total genomic DNA from carrot plants, digested with _Afl_III and _Pvu_II, was hybridized with a 3.2-kb radiolabeled DNA fragment isolated from the chloroplast transformation vector pDD-Dc_-aad_A/badh, by digesting it with _Afl_III and _Pvu_II; this fragment includes the 1.4-kb _trn_I flanking sequence and the 1.8-kb transgene sequences of the chloroplast transformation vector. Transgenic plants regenerated after two subcultures in selective liquid medium (350 mg L−1 spectinomycin) showed heteroplasmy, as is evident by the presence of both 1.4-kb wild-type and 3.2-kb transformed chloroplast genomes (Fig. 3C, lane 2). Plants that were regenerated from cell lines after 8 to 10 subcultures in liquid medium supplemented with a high concentration of antibiotic (500 mg L−1 spectinomycin) exhibited almost complete homoplasmy, as only the 3.2-kb DNA fragment (lanes 4–8), representing transformed chloroplast genomes was observed. A very faint signal corresponding to the wild-type fragment (Fig. 3C, lanes 2–3) was observed in cell lines that have not gone through repetitive stringent selection; subsequent rounds of selection eliminated this wild-type fragment (lanes 4–8). Observation of slight heteroplasmy in T0 transgenic lines and conversion to complete homoplasmy in T1 transgenic lines, upon germination of seeds under stringent selection, is of common occurrence in chloroplast transgenic lines (Guda et al., 2000; Daniell et al., 2004b).
BADH Enzyme Activity in Carrot Cells, Roots, and Shoots
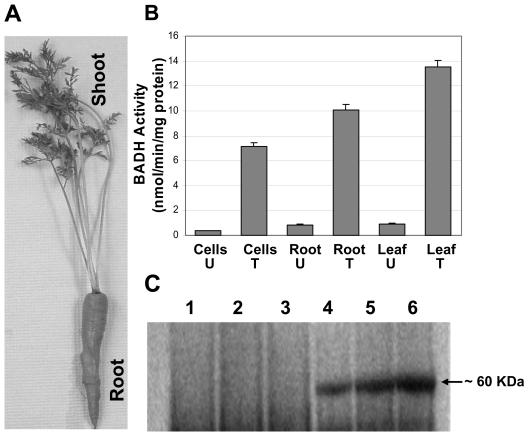
BADH enzyme activity was assayed in crude extracts from untransformed and transformed carrot cell cultures, taproots (carrot), and leaves as described (Daniell et al., 2001b). By assessing BADH enzyme activity in cells and different parts of carrot plants, expression of the badh transgene was characterized. In the presence of betaine aldehyde, BADH enzyme reduces NAD+ to NADH and the rate of this reaction was measured by an increase in A_340 due to the reduction of NAD+. Crude extracts from chloroplast transgenic tissues (cells, taproots, and leaves) showed elevated levels of BADH activity compared to untransformed tissues of carrot (Fig. 4B). High-BADH activity was observed in leaves, taproots of carrot plants, and transgenic cells in suspension culture, confirming that full-length 16S-promoter P_rrn and gene 10 5′UTR are highly suitable for expressing transgenes in different tissues. Because these regulatory elements are anticipated to function uniformly in all tissues, we presume that the observed difference in BADH activity might be due to variation in the plastid genome copy numbers. It is known that plastid genome copy numbers vary significantly in different tissues, with only 5% observed in roots compared to leaves (Sasaki et al., 1990). However, the high BADH enzyme activity observed in carrot taproot (74.8% of leaves) may be due to the large number of chromoplasts present; this was quite evident by their orange color (Fig. 4A).
Figure 4.
BADH enzyme activity and BADH expression in control and pDD_-Dc-aad_A_/badh_ lines. A, A pDD_-Dc-aad_A_/badh_ transgenic line shown with taproot and shoot. B, BADH activity in untransformed (U) and transformed (T) cell suspension, root, and leaf. Note: Mean and errors bars are the average of three replicates. C, Western blot using polyclonal anti-BADH serum. Antigenic peptides were detected using horseradish peroxidase-linked secondary antibody. Lanes 1 to 3, Untransformed cell culture, root, and leaf; lanes 4 to 6, transformed cell culture, root, and leaf.
BADH Protein Expression in Carrot Cells, Roots, and Shoots
To further confirm the results of BADH activity in cells, taproots, and leaves, western-blot analysis was performed using crude extracts of transformed and untransformed carrot tissues. Protein transferred to nitrocellulose membranes was hybridized with polyclonal anti-BADH serum raised in rabbits against native BADH (kindly provided by Dr. Elisa Soto; Figueroa-Soto et al., 1999), and antigenic peptides were detected using horseradish peroxidase-linked secondary antibodies. No badh expression was detected in untransformed carrot tissues (cells, taproots, and leaves; Fig. 4C, lanes 1–3). However, in chloroplast transgenic samples (Fig. 4C), higher expression was observed in leaves (lane 6) and taproots (lane 5) compared to carrot cell suspension cultures (lane 4). BADH protein accumulation in carrot root and leaf tissues was in agreement with the BADH enzyme activity observed in transgenic roots and shoots.
Salt Tolerance and BADH Activity in Cell Suspension Cultures of Carrot
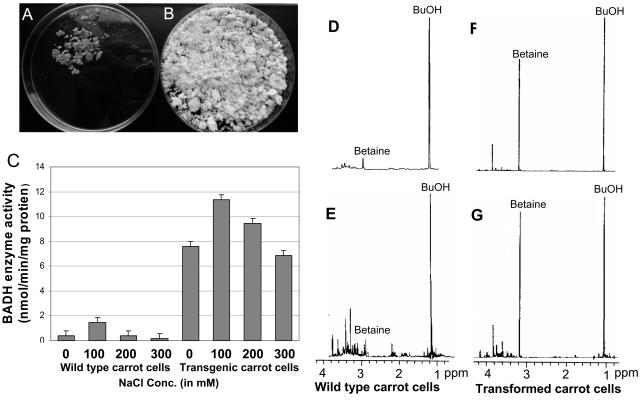
To test whether salt stress affected BADH enzyme activity in chloroplast transgenic cell lines, experiments were performed under different salt concentrations (0–300 mm NaCl). It was observed that transformed cells were able to survive and proliferate at high concentrations of NaCl in the liquid medium when compared to untransformed cells (Fig. 5, A and B). In two replicates, both transgenic and wild-type carrot cultures produced about an average of 11.82 ± 0.18 g of cells (1,475%) in the absence of NaCl while, in the presence of 100 mm NaCl, 8.75 ± 0.13 g (1,096%) and 1.29 ± 0.14 g (161%) of chloroplast transgenic and wild-type cells were produced, respectively, from 0.8 g (control as 100%) of initially inoculated cell culture. Further, BADH enzyme activity was enhanced 8.05-fold in transgenic carrot cell cultures in the presence of 100 mm NaCl when compared to untransformed cells (Fig. 5C). This shows that the full-length P_rrn_ promoter and gene 10 5′UTR facilitate efficient transcription and translation in all tissues, regardless of the developmental stage and despite low copy number of plastid genomes in non-green cells or roots.
Figure 5.
Effect of different salt concentrations on growth of untransformed and transformed cell lines with pDD-Dc-aad_A/badh_ and study of betaine with 1H-NMR spectra. Untransformed (A) and transformed (B) cell cultures grown on 100 mm NaCl. C, Stimulation of BADH activity in the presence of salt. Untransformed and transformed carrot cells in suspension cultures were placed on a shaker at 130 rpm for 2 weeks in liquid medium containing 0, 100, 200, and 300 mm NaCl. 1H-NMR spectra (500 MHz) of extracts from untransformed (D and E) and transformed (F and G) carrot cell suspension cultures grown in the presence of 100 mm NaCl alone (D and F) or in combination with 4 mm choline (E and G). Purified samples were dissolved in D2O and _t_-butanol (an internal standard). Integration of the singlet versus _t_-butanol was used for quantification of betaine. A dominant singlet of betaine is detected at 3.20 ppm.
Betaine Accumulation in Carrot Cells
Because transformed carrot cells expressed BADH (confirmed by western blot) and also showed BADH enzyme activity, it is logical to evaluate accumulation of betaine in these cells. Therefore, betaine concentration was measured by 1H-NMR (Robinson and Jones, 1986). The level of betaine observed was 26.5 _μ_mol g−1 dry weight (DW) in the transgenic carrot cell culture. It was enhanced up to 3-fold (93.1 _μ_mol g−1 DW) when transgenic cell suspension cultures were supplemented with 100 mm NaCl. However, in the presence or absence of choline as well as salt, no significant level of betaine was recorded in the untransformed control carrot cell cultures (Table II). Transformed carrot cells grown in 100 mm NaCl accumulated 50- to 54-fold more betaine than untransformed cells (in the presence or absence of choline), when determined on the basis of DW.
Table II.
Quantification of betaine using 1H-NMR spectra (500 MHz) in the transgenic and nontransgenic carrot cell cultures grown in liquid medium supplemented with choline (0, 4 mm) and NaCl (0, 100 mm) after 2 weeks
| Samples | NaCl | Choline | Betaine |
|---|---|---|---|
| mm | mm | μmol g−1 DW | |
| Untransformed | 0 | 0 | ND |
| 100 | 0 | 1.67 ± 0.87 | |
| 0 | 4 | ND | |
| 100 | 4 | 1.86 ± 1.05 | |
| Transformed | 0 | 0 | 26.5 ± 3.26 |
| 100 | 0 | 93.1 ± 3.52 | |
| 0 | 4 | 29.2 ± 2.18 | |
| 100 | 4 | 101.2 ± 6.32 |
Members of the family Chenopodiaceae can accumulate high levels (>100 _μ_mol g−1 DW) of betaine in leaves when salinized (Weretilnyk et al., 1989). While genetic engineering has allowed engineered plants to produce betaine, there are considerable differences in levels of betaine, on a fresh-weight (FW) basis, among nuclear transgenic plants (0.05–5 _μ_mol g−1 FW; Sakamoto and Murata, 2000) and natural accumulators under stress conditions (4–40 _μ_mol g−1 FW; Rhodes and Hanson, 1993). Recently, Nishimura et al. (2001) reported 167 _μ_mol g−1 DW betaine in sea blite that was collected from a saline area of China that belongs to Chenopodiaceae and is known as a strong halophytic plant. Using 1H-NMR spectroscopy, we have observed about 93 _μ_mol g−1 DW betaine in transgenic tissues when cell cultures were grown in liquid medium containing 100 mm NaCl for 2 weeks. While this level of accumulation is adequate to confer salt tolerance (up to 300 mm NaCl), higher betaine accumulation may occur in the transgenic leaf or root tissues, as BADH activity was much higher in transgenic plants when compared to carrot cell cultures (Fig. 4B), and transgenic plants were able to grow in the presence of 400 mm NaCl (Fig. 6).
Figure 6.
Effect of salt (100–500 mm NaCl) on untransformed (U) and transgenic (T) lines grown at different concentrations of NaCl. Plants were irrigated with water containing different concentrations of NaCl on alternate days for up to 4 weeks.
Previous studies demonstrated that choline-fed transgenic plants synthesized more betaine because endogenous choline supply limits betaine synthesis in transgenic tobacco, Arabidopsis, and Brassica plants (Nuccio et al., 1998; Huang et al., 2000). In order to test the role of endogenous CMO, carrot cell cultures were grown in the presence or absence of choline. We observed a slight enhancement of betaine accumulation in the transgenic carrot cell suspension cultures that were supplemented with 4 mm choline along with 100 mm salt (Table II). Lack of significant increase in betaine in the presence of choline may be due to limitation of CMO or uptake of choline by carrot cells.
BADH is not substrate specific, as had been reported previously. It plays several roles in plants during salt stress (Trossat et al., 1997) and helps in the accumulation of osmolytes like Gly betaine and _β_-Ala betaine (Hanson et al., 1991; Rathinasabapathi et al., 2001). Gly betaine is produced in plants by a two-step oxidation of choline (Nuccio et al., 1998), while _β_-Ala betaine is produced after methylation of _β_-Ala, converted from 3-aminopropionaldehyde by BADH enzyme (Rathinasabapathi et al., 2001). In the salinized plants, quaternary ammonium compounds _β_-Ala betaine [(CH3)3-N+-CH2-CH2-COO−] has shown better osmoprotective properties than Gly betaine [(CH3)3-N+-CH2-COO−]; Hanson et al., 1991; Hanson and Gage, 1991; Rathinasabapathi et al., 2001]. Since 1H-NMR spectra detect both quaternary ammonium compounds as betaine, levels of betaine reported here do not distinguish between _β_-Ala betaine and Gly betaine.
While BADH activity increased approximately 8-fold in transformed carrot cells compared to untransformed cells, when grown in the presence of 100 mm NaCl, betaine accumulation increased 55-fold. Under similar physiological conditions, transformed cells grew approximately 7-fold more than untransformed cells when grown in the presence of 100 mm NaCl. Even though accumulation of betaine is quite high, the osmoprotection mechanism in combination with other mechanisms (such as antiport) may yield plants with even higher levels of salt tolerance.
Effect of Salt Stress on Carrot Plants
Chloroplast transgenic carrot plants and wild-type plants were subjected to increasing degrees of salt stress, ranging from 100 to 500 mm NaCl. Chloroplast transgenic plants expressing the badh transgene thrive well up to 400 mm NaCl (Fig. 6), whereas untransformed plants exhibited severe growth retardation at 200 mm NaCl. The understanding of metabolic fluxes in plant cells and the ability to synthesize compatible solutes have opened up the possibility of genetically modifying plants to confer stress tolerance. Improved salinity tolerance has been achieved by overexpressing a vacuolar Na+/H= antiport, up to 200 mm NaCl (Zhang and Blumwald, 2001), accumulation of Gly betaine by expression of BADH alone, up to 120 mm NaCl (Jia et al., 2002), or coexpression of BADH and choline dehydrogenase, up to 200 mm NaCl (Holmström et al., 2000). In this study, we report that expression of BADH alone in transgenic plants via the chloroplast genome was adequate to confer higher levels of salinity tolerance (up to 400 mm NaCl). This appears to be the highest level of salt tolerance reported in the literature so far; however, it should be pointed out that the origin of badh genes and the transformed plant species is different in the aforementioned examples.
CONCLUSIONS
There are at least 15 prior reports where attempts have been made to manipulate the Gly betaine biosynthesis pathway via nuclear genetic engineering in order to enhance salt tolerance (Flowers, 2004). This study demonstrates that overexpression of the badh gene via engineering of the carrot chloroplast genome results in significant enhancement of salt tolerance. Carrot chloroplast transgenic lines are able to grow well at 400 mm NaCl, a concentration at which only halophytes are able to thrive. In contrast, the untransformed wild-type line exhibits severe growth retardation even at 200 mm NaCl (Fig. 6). This appears to be the highest level of salt tolerance reported so far when compared to 13 other plant species where enhancement of salt tolerance has been reported (Flowers, 2004).
To our knowledge, this is the first report expressing a useful trait via chloroplast genetic engineering in a non-tobacco crop. So far, only the tobacco chloroplast genome has been engineered to confer herbicide resistance (Daniell et al., 1998), insect resistance (McBride et al., 1995; Kota et al., 1999; DeCosa et al., 2001; Reddy et al., 2002), disease resistance (DeGray et al., 2001), drought tolerance (Lee et al., 2003), or phytoremediation of toxic metals (Ruiz et al., 2003).
There are several reasons that have impeded the extension of chloroplast transformation technology to other plant species. Chloroplast transgenic lines are routinely obtained in tobacco via organogenesis. The chloroplast transformation vectors utilize homologous flanking regions for recombination and insertion of foreign genes. Transformation of Arabidopsis, potato (Solanum tuberosum), and tomato (Lycopersicon esculentum) chloroplast genomes was achieved via organogenesis by bombardment of green leaf tissues, but the efficiency was much lower than tobacco (Sikdar et al., 1998; Sidorov et al., 1999; Ruf et al., 2001). In Arabidopsis, one transgenic line per 40 or 151 bombarded plates was obtained; in potato, one chloroplast transgenic line per 35 bombarded plates was obtained; and in tomato, one transgenic line per 20 bombarded plates was obtained. In contrast, 15 tobacco chloroplast transgenic lines were obtained per bombarded plate (Fernandez-San Millan et al., 2003). In the case of Lesquerella, transgenic shoots had to be grafted onto Brassica napus rootstock to reconstruct transgenic plants (Skarjinskaia et al., 2003). In oilseed rape, direct Southern-blot analysis of transgenic chloroplast genomes was not presented (Hou et al., 2003). The vectors employed for chloroplast transformation of potato, tomato, and Lesquerella contained the flanking sequences from tobacco or Arabidopsis. This may be one of the reasons for lower transformation efficiency. When petunia flanking sequences were used for chloroplast transformation of tobacco, the transformation efficiency decreased drastically (DeGray et al., 2001). In contrast, efficient transformation of carrot chloroplast genomes was achieved (one event per approximately seven bombarded plates) using species-specific chloroplast vectors containing 100% homologous flanking sequences.
The use of non-green explants has often been cited as one of the major obstacles that has limited the chloroplast transformation to solanaceous crops (Bogorad, 2000). In carrot plastid transformation, the expression cassette for the detoxification of antibiotic is functional in non-green cells due to the full-length P_rrn_ promoter used in the cassette that has binding sites for both the nuclear-encoded and plastid-encoded RNA polymerase (Daniell et al., 2002a; Devine and Daniell, 2004). Transformation of carrot plastid genomes is the very first example of successful, stable plastid transformation using non-green explants via somatic embryogenesis. The optimal site of transgene integration may be an additional prerequisite for efficient plastid transformation (Dhingra et al., 2004). In addition, use of long 100% homologous flanking sequences (4 kb) should have facilitated efficient recombination. It has been erroneously claimed earlier that rice plastid transformation was achieved via somatic embryogenesis, but no data were provided to support stable transgene integration into the plastid genome or homoplasmy by Southern-blot analysis (Khan and Maliga, 1999). Therefore, development of protocols that facilitate chloroplast transformation via somatic embryogenesis is a major breakthrough in this field. Because most of the crop species are regenerated via somatic embryogenesis, methods developed here should help in transforming the plastid genomes of other crop plants.
Another significant observation in this study is the high level of transgene expression observed in proplastids of cultured carrot cells. Earlier, 100-fold less green fluorescent protein accumulation in amyloplasts of potato tubers compared to leaves was reported (Sidorov et al., 1999). In sharp contrast, in proplastids and in chromoplasts, 53.1% and 74.8% BADH activity, respectively, was observed when compared to leaf chloroplasts (100%). Such high levels of transgene expression were achieved using appropriate heterologous regulatory sequences in the expression cassette. Both the selectable marker and the gene of interest (aad_A and badh) are transcribed by the plastid P_rrn promoter; this 16S rRNA promoter drives the entire rRNA operon in the native chloroplast and contains binding sites for both the nuclear-encoded and plastid-encoded RNA polymerases (for a recent detailed review and discussion, see Daniell et al., 2002a). Therefore, this promoter is capable of functioning in both proplastids and chloroplasts (green and non-green, in light and dark). The badh gene is further regulated by the T7 gene 10 5′UTR capable of efficient translation in the dark in proplastids present in non-green tissues. It should be noted that the heterologous T7 gene 10 5′UTR that regulates translation of the badh gene was indeed promoterless and the transgene expression could be further enhanced by adding a suitable promoter, which could further enhance salt tolerance. To our knowledge, this is also the first report of stable transgene expression in proplastids.
Three different pathways are suggested to mediate salinity tolerance in plants, which include maintenance of ion and osmotic homeostasis, regulation of cell division and growth, and detoxification of toxic byproducts and cellular repair (Zhu, 2002). The protective properties of betaine are provided by supporting the osmotic homeostasis in a plant. Even though it has been suggested that salt stress is a multigenic trait, there are experimental data to prove otherwise (Kasuga et al., 1999; Saijo et al., 2000; Zhang and Blumwald, 2001; Zhang et al., 2001; Mukhopadhyay et al., 2004). In order to provide broad-range salinity tolerance to plants, a better strategy may be to engineer genes that confer other mechanisms involved in signal transduction of salt stress, in addition to osmoprotection. Therefore, engineering plants for salt tolerance either by large accumulation of betaine via the chloroplast genome (50- to 54-fold higher than the untransformed control) or in combination with an antiport mechanism (Zhang and Blumwald, 2001) should be an attractive option for future strategies. Furthermore, effective engineering strategies leading to greater salinity tolerance may also be devised using the information available from comparative and functional genomic studies of model organisms (Cushman and Bohnert, 2000) in conjunction with the chloroplast transformation strategy reported in this study.
MATERIALS AND METHODS
Construction of Carrot Plastid Transformation Vectors
DNA fragments representing a carrot flanking sequence were amplified from carrot genomic DNA that was isolated from the leaves using DNeasy Plant Mini kit (Qiagen, Valencia, CA), following the manufacturer's protocol. The flanking sequence fragment was amplified with the primers designed based on a tobacco (Nicotiana tabacum) chloroplast genome sequence information using Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA). The forward primer, ADLF, and the reverse primer, ADLR, amplified a 4.0-kb DNA fragment representing the 16S/_trn_I-_trn_A/23S region of the carrot chloroplast genome. The PCR-amplified DNA fragment was treated with T4 polynucleotide kinase (Promega, Madison, WI), cloned into Pvu_II-digested pBluescript II KS (Stratagene, La Jolla, CA), and dephosphorylated with shrimp alkaline phosphatase (Promega). The kinase and dephosphorylation reactions were performed as per the manufacturer's instructions. The chloroplast promoters and regulatory sequences were amplified using PCR based on the information available for the tobacco chloroplast genome (accession no. NC_001879). The primers used were as follows: ADLF (5′-CACTCTGCTGGGCCGACACTGACAC-3′); ADLR (5′-CACTAGCCGACCTTGACCCCTGTT-3′); P_rrn (Forward, 5′-ATCGATGAGCCTGATTATCCTAAG-3′; Reverse, 5′-CAGCAGGTAGACAAAGCGGATTC-3′); P_psb_A (Forward, 5′-GATATCGTCGACGTAGAGAAGTCCG-3′; Reverse, 5′-CATATGAAAATCTTGGTTTATTTAA-3′); T_psb_A (Forward, 5′-TCTAGAGCGATCCTGGCCTAG-3′; Reverse, 5′-GAGCTCGCAGCCCAAACAAATAC-3′); T_rps_16 (Forward, 5′-ACTAGTCCTAATCAACCGAAATTC-3′; Reverse, 5′-GAGCTCGAACACGGAATTCAATGGAAGC-3′); T7 gene 10 (Forward, 5′-GGTAACCCCGGGAGACCACAACGGTTTCCCTCTAGAAATAATTTTGTTTA-3′; Reverse, 5′-CATATGTATATCTCCTTCTTAAAGTTA-3′); 3P (5′-AAAACCCGTCCTCAGTTCGGATTGC-3′); 1M (5′-CGCGCTTAGCTGGATAACGCCACGGAA-3′); and 16SF (5′-CAGCAGCCGCGGTAATACAGAGGA-3′). The carrot-specific chloroplast transformation vector pDD-_Dc_-_aad_A-badh (Fig. 1) was constructed by inserting a blunt-ended fragment representing the _aad_A-badh expression cassette into the _Pvu_II site of the carrot chloroplast DNA flanking sequences. All general bacterial and DNA manipulations were performed as per standard molecular biology protocols.
Transformation and Regeneration Protocol for Carrot
Sterile carrot plants (Daucus carota L. cv Half long) were raised in plant tissue culture tubes containing MSB, Murashige and Skoog salts (Murashige and Skoog, 1962), B5 vitamins (Gamborg et al., 1968), 2% Suc, and 0.8% agar in the medium. The stems were cut into 0.5-mm segments and were placed on MSB solid medium supplemented with 3 mg L−1 2,4-diphenoxyacetic acid (2,4-D) and 1 mg L−1 kinetin for induction of callus. After bombardment with pDD-_Dc_-_aad_A-badh, embryogenic callus was incubated for 2 d in the dark and selected on MSB (3 mg L−1 2,4-D and 1 mg L−1 kinetin) containing different concentrations of spectinomycin (150, 250, 350, and 450 mg L−1). Cultures were incubated in a 16-h day/8-h night cycle at 50 to 100 lux light intensity and 26°C ± 2°C temperature. Transgenic cultures were multiplied using both solid and liquid medium supplemented with a selection agent. Transgenic plants produced on MSB medium containing 0.2 mg L−1 kinetin were transferred to soil in pots.
Optimization of Transformation Parameters in Carrot
For optimization of gene delivery, an embryogenic cell culture of carrot was placed on Whatman Number 1 filter paper (Whatman, Clifton, NJ), supported by MSB medium (3 mg L−1 2,4-D and 1 mg L−1 kinetin). Gene delivery was optimized using a pDD-_Dc_-_aad_A/badh vector coated on 0.6-_μ_m gold particles using different rupture discs (Bio-Rad Laboratories, Hercules, CA) and at different distances between rupture discs and target tissues. Bombarded cell cultures were incubated in the dark for 2 d and transferred to a selection medium containing 150 mg L−1 spectinomycin. Transgenic calli obtained at different bombardment parameters were tested for site-specific transgene integration into the plastid genome by PCR.
BADH Enzyme Activity and Imunoblot Analysis in Carrot
Protein extraction and BADH activity assays were done as described earlier (Daniell et al., 2001b). One gram of carot tissues was homogenized in 2 mL homogenization buffer containing 50 mm HEPES-KOH, pH 8.0, 1 mm EDTA, 20 mm sodium metabisulfite, 10 mm sodium borate, 5 mm ascorbic acid, and 5 mm dithiothreitol. Crude extract was centrifuged at 10,000_g_ at 4°C for 10 min and the supernatant was desalted using Sephadex G-25 columns (Amersham-Pharmacia Biotech, Uppsala). Reduction of NAD+ by BADH was measured spectrophotometrically at 340 nm after 1- and 10-min intervals in 1 mL assay buffer (50 mm HEPES-KOH, pH 8.0; 1 mm EDTA, 5 mm dithiothreitol, 1 mm NAD+) at 25°C, supplemented with 1 mm betaine aldehyde) to start the reaction.
For immunoblot analysis, total soluble protein was isolated using 2× Laemmli buffer from 100-mg carrot tissues. The mixture was boiled for 5 min and centrifuged for 5 min at 10,000_g_. Supernatant containing 50 _μ_g total soluble protein (quantified with Bradford assay) was loaded on a 10% SDS-PAGE gel and transferred to nitrocellulose membrane (Bio-Rad). The membrane was hybridized with polyclonal anti-BADH serum raised in rabbits against BADH (provided by Dr. Elisa Soto). Hybridizing peptides were detected using horseradish peroxidase-linked secondary antibody, with Lumi-Phos WB chemiluminescent reagent (Pierce Chemical, Rockford, Illinois).
Salt Tolerance in Cell Suspension Cultures of Carrot
To assess the effect of salt stress on chloroplast transgenic cell suspension cultures of carrot, cells were grown in liquid MSB media (0.1 mg L−1 2,4-D) supplemented with 0 to 300 mm NaCl. Cultures were maintained at 130 rpm under diffuse light at 28°C ± 2°C for 2 weeks. Cells were harvested on a filter disc in a filtration apparatus and their relative weight was recorded.
Determination of Betaine Concentration by 1H-NMR
Transgenic and nontransgenic carrot cell cultures were grown in the presence of NaCl (0 and 100 mm) and choline (0 and 4 mm) in liquid medium to measure betaine accumulation. Plant samples were prepared as described previously (Robinson and Jones, 1986; Bessieres et al., 1999). The 1H-NMR spectra (500 MHz; Varian Instruments, Palo Alto, CA) were recorded at 25°C, at 32 pulses, with a pulse repletion time of 5 and a radiofrequency pulse angle of 30°. For betaine determinations with 1H-NMR, purified samples were dried via rotary evaporator and dissolved in 0.6 mL of D2O. _t_-Butanol was added as an internal standard (Robinson and Jones, 1986; Holmström et al., 2000). A dominant singlet (peak) assignable to the authentic betaine methyl groups [R-N+(CH3)3] was detected at 3.20 ppm. Integration of the singlet versus _t_-butanol was used for quantification.
Analysis of Transgenic Plants for Salt Tolerance in Carrot
Transgenic and nontransgenic carrot plants of similar age and height were assayed for salt tolerance after transfer to soil in pots containing 0, 100, 200, 300, 400, and 500 mm NaCl. Plants were maintained in a growth chamber and irrigated daily with saline water containing the above-mentioned levels of salt for 1 month.
Acknowledgments
The authors are grateful to Dr. Elisa Miriam Valenzuela Soto (Centero de Investigacion en Alimentacion y Desarrollo A.C., Mexico) for kindly providing the anti-BADH serum for this study, and to Professor Andrew Hanson (University of Florida, Gainesville) for providing the BADH coding sequence. We are also grateful to Dr. Otto Phanstiel, Associate Professor, Department of Chemistry, University of Central Florida, for his help with the 1H-NMR study. We thank Dr. Bala Rathinasbapathi, Associate Professor, University of Florida, for helpful discussions on salt tolerance.
1
This work was supported, in part, by the National Institutes of Health (grant no. R–01–GM63879) and by the U.S. Department of Agriculture (grant no. 3611–21000–017–00D) to H.D.
References
- Alia, Kondo Y, Sakamoto A, Nonaka H, Hayashi H, Saradhi PP, Chen THH, Murata N (1999) Enhanced tolerance to light stress of transgenic Arabidopsis plants that express the _cod_A gene for a bacterial choline oxidase. Plant Mol Biol 40**:** 279–288 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem-2—inactivation, protein damage and turnover. Biochim Biophys Acta 1143**:** 113–134 [DOI] [PubMed] [Google Scholar]
- Bessieres MA, Gibon Y, Lefeuvre JC, Larher L (1999) A single-step purification for glycine betaine determination in plant extracts by isocratic HPLC. J Agric Food Chem 47**:** 3718–3722 [DOI] [PubMed] [Google Scholar]
- Bogorad L (2000) Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol 18**:** 257–263 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3**:** 117–124 [DOI] [PubMed] [Google Scholar]
- Daniell H (1997) Transformation and foreign gene expression in plants mediated by micoprojectile bombardment. Methods Mol Biol 62**:** 463–489 [DOI] [PubMed] [Google Scholar]
- Daniell H (2002) Molecular strategies for gene containment in transgenic crops. Nat Biotechnol 20**:** 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB (1998) Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol 16**:** 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A (2002) Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol 13**:** 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A, Allison L (2002. a) Chloroplast transformation: from basic molecular biology to biotechnology. Rev Plant Physiol Biochem 1**:** 1–20 [Google Scholar]
- Daniell H, Khan MS, Allison L (2002. b) Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends Plant Sci 7**:** 84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO (2001. a) Expression of cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol 311**:** 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Muthukumar B, Lee SB (2001. b) Marker free transgenic plants: the chloroplast genome without the use of antibiotic selection. Curr Genet 39**:** 109–116 [DOI] [PubMed] [Google Scholar]
- Daniell H, Ruiz ON, Dhingra A (2004. a) Chloroplast genetic engineering to improve agronomic traits. Methods Mol Biol 286**:** 111–137 [DOI] [PubMed] [Google Scholar]
- Daniell H, Watson J, Koya V, Leppla S (2004. b) Expression of Bacillus anthracis protective antigen in transgenic chloroplasts of tobacco, a non-food/feed crop. Vaccine (in press) [DOI] [PMC free article] [PubMed]
- DeCosa B, Moar W, Lee SB, Miller M, Daniell H (2001) Overexpression of the Bt Cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19**:** 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H (2001) Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol 127**:** 852–862 [PMC free article] [PubMed] [Google Scholar]
- Devine AL, Daniell H (2004) Chloroplast genetic engineering for enhanced agronomic traits and expression of proteins for medical/industrial applications. In SG Moller, ed, Plastids, Vol 13. Blackwell Scientific, Oxford, pp 283–323
- Dhingra A, Portis AR Jr, Daniell H (2004) Enhanced translation of a chloroplast expressed _Rbc_S gene restores SSU levels and photosynthesis in nuclear antisense _Rbc_S plants. Proc Natl Acad Sci USA 101**:** 6315–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-San Millan A, Mingo-Castel A, Miller M, Daniell H (2003) A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J 1**:** 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Soto CG, Lopez-Cervantes G, Valenzuela-Soto EM (1999) Immunolocalization of betaine aldehyde dehydrogenase in porcine kidney. Biochem Biophys Res Commun 258**:** 732–736 [DOI] [PubMed] [Google Scholar]
- Fischer N, Stampacchia O, Redding K, Rochaix JD (1996) Selectable marker recycling in the chloroplast. Mol Gen Genet 251**:** 373–380 [DOI] [PubMed] [Google Scholar]
- Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55**:** 307–319 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50**:** 151–158 [DOI] [PubMed] [Google Scholar]
- Gibberd MR, Turner NC, Storey R (2002) Influence of saline irrigation on growth, ion accumulation and partitioning, and leaf gas exchange of carrot (Daucus carota L.). Ann Bot (Lond) 90**:** 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda C, Lee SB, Daniell H (2000) Stable expression of biodegradable protein based polymer in tobacco chloroplasts. Plant Cell Rep 19**:** 257–262 [DOI] [PubMed] [Google Scholar]
- Guo BH, Zhang YM, Li HJ, Du LQL, Yin XC, Shou Y, Chu CC (2000) Transformation of wheat with a gene encoding for the betaine aldehyde dehydrogenase (BADH). Acta Bot Sin 42**:** 279–283 [Google Scholar]
- Hanson AD, Gage DA (1991) Identification and determination by fast atom bombardment mass spectrometry of the compatible solute choline-_O_-sulfate in Limonium species and other halophytes. Aust J Plant Physiol 18**:** 317–327 [Google Scholar]
- Hanson AD, Rathinasabapathi B, Chamberlin B, Gage DA (1991) Comparative physiological evidence that beta-alanine betaine and choline-_O_-sulfate act as compatible osmolytes in halophytic Limonium species. Plant Physiol 97**:** 1199–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmström KO, Somersalo S, Mandal A, Palva TE, Welin B (2000) Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J Exp Bot 51**:** 177–185 [DOI] [PubMed] [Google Scholar]
- Hou BK, Zhou YH, Wan LH, Zhang ZL, Shen GF, Chen ZH, Hu ZM (2003) Chloroplast transformation in oilseed rape. Transgenic Res 12**:** 111–114 [DOI] [PubMed] [Google Scholar]
- Huang J, Hirji R, Adam L, Rozwadowski KL, Hammerlindl JK, Keller WA, Selvaraj G (2000) Genetic engineering of glycinebetaine production toward enhancing stress tolerance in plants: metabolic limitations. Plant Physiol 122**:** 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iamtham S, Day A (2000) Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat Biotechnol 18**:** 1172–1176 [DOI] [PubMed] [Google Scholar]
- Jia GX, Zhu ZQ, Chang FQ, Li YX (2002) Transformation of tomato with the BADH gene from Atriplex improves salt tolerance. Plant Cell Rep 21**:** 141–146 [Google Scholar]
- Kasuga M, Liu Q, Minura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17**:** 287–291 [DOI] [PubMed] [Google Scholar]
- Khan MS, Maliga P (1999) Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat Biotechnol 17**:** 910–915 [DOI] [PubMed] [Google Scholar]
- Kishitani S, Takanami T, Suzuki M, Oikawa M, Yokoi S, Ishitani M, Alvarez-Nakase AM, Takabe T, Takabe T (2000) Compatibility of glycinebetaine in rice plants: evaluation using transgenic rice plants with a gene for peroxisomal betaine aldehyde dehydrogenase from barley. Plant Cell Environ 23**:** 107–114 [Google Scholar]
- Klaus SMJ, Huang FC, Golds TJ, Koop HU (2004) Generation of marker-free plastid transformants using a transiently cointegrated selection gene. Nat Biotechnol 22**:** 225–229 [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski SF, Gould F, William MJ (1999) Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and _Bt_-resistant insects. Proc Natl Acad Sci USA 96**:** 1840–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Daniell H (2004) Engineering the chloroplast genome for hyper-expression of human therapeutic proteins and vaccine antigens. Methods Mol Biol 267**:** 365–383 [DOI] [PubMed] [Google Scholar]
- Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H (2003) Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breeding 11**:** 1–13 [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogan PS, Stalker DM, Maliga P (1995) Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology 13**:** 362–365 [DOI] [PubMed] [Google Scholar]
- McCue KF, Hanson AD (1990) Drought and salt tolerance—towards understanding and application. Trends Biotechnol 8**:** 358–362 [Google Scholar]
- Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101**:** 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15**:** 473–497 [Google Scholar]
- Nakamura T, Yokota S, Muramoto Y, Tsutsui K, Oguri Y, Fukui K, Takabe T (1997) Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J 11**:** 1115–1120 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Zhang J, Abo M, Okubo A, Yamazaki S (2001) Application of capillary electrophoresis to the simultaneous determination of betaines in plants. Anal Sci 17**:** 103–106 [DOI] [PubMed] [Google Scholar]
- Nomura M, Hibino T, Takabe T, Sugiyama T, Yokota A, Miyake H (1998) Transgenically produced glycinebetaine protects ribulose 1,5-bisphosphate carboxylase/oxygenase from inactivation in Synechococcus sp. PCC7942 under salt stress. Plant Cell Physiol 39**:** 425–432 [Google Scholar]
- Nuccio ML, Russell BL, Nolte KD, Rathinasabapathi B, Gage DA, Hanson AD (1998) The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J 16**:** 487–496 [DOI] [PubMed] [Google Scholar]
- Papageorgiou GC, Murata N (1995) The unusually strong stabilizing effects of glycinebetaine on the structure and function in the oxygen-evolving photosystem II complex. Photosynthetica 44**:** 243–252 [DOI] [PubMed] [Google Scholar]
- Rathinasabapathi B, Burnet M, Russell BL, Gage DA, Liao PC, Nye GJ, Scott P, Golbeck JH, Hanson AD (1997) Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: prosthetic group characterization and cDNA cloning. Proc Natl Acad Sci USA 94**:** 3454–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinasabapathi B, Fouad WM, Sigua CA (2001) _β_-Alanine betaine synthesis in the Plumbaginaceae. Purification and characterization of a trifunctional, _S_-adenosyl-l-methionine-dependent _N_-methyltransferase from Limonium latifolium leaves. Plant Physiol 126**:** 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Leelavathi S, Selvapandiyan A, Raman R, Giovanni F, Shukla V, Bhatnagar RK (2002) Analysis of chloroplast transformed tobacco plants with cry1Ia5 under rice _psb_A transcriptional elements reveals high level expression of Bt toxin without imposing yield penalty and stable inheritance of transplastome. Mol Breed 9**:** 259–269 [Google Scholar]
- Rhodes D, Hanson AD (1993) Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol 44**:** 357–384 [Google Scholar]
- Robinson SP, Jones GP (1986) Accumulation of glycinebetaine in chloroplasts provides osmotic adjustment during salt stress. Aust J Plant Physiol 13**:** 659–668 [Google Scholar]
- Ruf S, Hermann M, Berger I, Carrier H, Bock R (2001) Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol 19**:** 870–875 [DOI] [PubMed] [Google Scholar]
- Ruiz ON, Hussein H, Terry N, Daniell H (2003) Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol 132**:** 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23**:** 319–327 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Murata N (2000) Genetic engineering of glycinebetaine synthesis in plants: current status and implication for enhancement of stress tolerance. J Exp Bot 51**:** 81–88 [PubMed] [Google Scholar]
- Sakamoto A, Murata N (2002) The role of glycine betaine in the protection of plants from stress: clues from transgenic plants. Plant Cell Environ 25**:** 163–171 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Morioka S, Matsuno R (1990) Correlation of plastid DNA copy number with plastid gene expression in various organs in mature pea-plants (Pisum-sativum L). Plant Cell Physiol 31**:** 925–931 [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5**:** 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PTJ, Staub JM, Nehra NS (1999) Technical advance: stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J 19**:** 209–216 [DOI] [PubMed] [Google Scholar]
- Sikdar SR, Serino G, Chaudhuri S, Maliga P (1998) Plastid transformation in Arabidopsis thaliana. Plant Cell Rep 18**:** 20–24 [Google Scholar]
- Skarjinskaia M, Svab Z, Maliga P (2003) Plastid transformation in Lesquerella fendleri, an oilseed Brassicacea. Transgenic Res 12**:** 115–122 [DOI] [PubMed] [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, et al (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol 18**:** 333–338 [DOI] [PubMed] [Google Scholar]
- Trossat C, Rathinasabapathi B, Hanson AD (1997) Transgenically expressed betaine aldehyde dehydrogenase efficiently catalyzes oxidation of dimethylsulfoniopropionaldehyde and w-aldehydes. Plant Physiol 113**:** 1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivek BS, Ngo QA, Simon PW (1999) Evidence for maternal inheritance of the chloroplast genome in cultivated carrot (Daucus carota L. ssp. Sativus). Theor Appl Genet 98**:** 669–672 [Google Scholar]
- Ward JM, Hirschi KD, Sze H (2003) Plants pass the salt. Trends Plant Sci 8**:** 200–201 [DOI] [PubMed] [Google Scholar]
- Weretilnyk EA, Bednarek S, McCue KF, Rhodes D, Hanson AD (1989) Comparative biochemical and immunological studies of the glycine betaine synthesis pathway in diverse families of dicotyledons. Planta 178**:** 342–352 [DOI] [PubMed] [Google Scholar]
- Wood AJ, Saneola H, Rhides D, Joly RJ, Goldbrough PB (1996) Betaine aldehyde dehydrogenase in Sorghum. Plant Physiol 110**:** 1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WK, Hunt LA (1999) Reanalysis of vernalization data of wheat and carrot. Ann Bot (Lond) 84**:** 615–619 [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19**:** 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson J, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98**:** 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6**:** 66–71 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53**:** 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]