Divergence of Spatial Gene Expression Profiles Following Species-Specific Gene Duplications in Human and Mouse (original) (raw)
Abstract
To examine the process by which duplicated genes diverge in function, we studied how the gene expression profiles of orthologous gene sets in human and mouse are affected by the presence of additional recent species-specific paralogs. Gene expression profiles were compared across 16 homologous tissues in human and mouse using microarray data from the Gene Expression Atlas for 1575 sets of orthologs including 250 with species-specific paralogs. We find that orthologs that have undergone recent duplication are less likely to have strongly correlated expression profiles than those that remain in a one-to-one relationship between human and mouse. There is a general trend for paralogous genes to become more specialized in their expression patterns, with decreased breadth and increased specificity of expression as gene family size increases. Despite this trend, detailed examination of some particular gene families where species-specific duplications have occurred indicated several examples of apparent neofunctionalization of duplicated genes, but only one case of subfunctionalization. Often, the expression of both copies of a duplicated gene appears to have changed relative to the ancestral state. Our results suggest that gene expression profiles are surprisingly labile and that expression in a particular tissue may be gained or lost repeatedly during the evolution of even small gene families. We conclude that gene duplication is a major driving force behind the emergence of divergent gene expression patterns.
Gene duplication gives rise to a state of genetic redundancy. The newly formed gene pair enters a period of reduced evolutionary pressure, during which entirely novel functional patterns can emerge. This classic theory of neofunctionalization was proposed by Susumo Ohno more than 30 years ago (Ohno 1970), who famously stated that “natural selection merely modified while redundancy created.” Neofunctionalization can be achieved through changes in amino acid sequence (e.g., leading to the development of novel enzymatic activity), or through changes in the gene's expression pattern (e.g., resulting in expression of a gene in a tissue where the ancestral gene was not previously expressed). The very act of gene duplication can lead to spontaneous neofunctionalization, through loss of a silencer caused by incomplete duplication of the regulatory region of a gene, fortuitous gain of an exogenous promoter or enhancer, or when the parental locus was previously under balancing selection (Lynch et al. 2001; Katju and Lynch 2003).
Given the large numbers of duplicated genes present in most eukaryotic genomes, it seems doubtful that selection for novel functions alone could account for every case of gene duplication. Instead, alternative models were proposed that postulated subdivision of the functions of an ancestral gene. The first such model—the “gene sharing” or “adaptive conflict” model—was initially described for the example of the crystallin family (Piatigorsky and Wistow 1991) and later defined more formally by Hughes (1994, 1999). Under this model, the two daughter genes are preserved in the genome because at least one of them has been subject to positive selection for mutations that were previously disallowed owing to pleiotropic constraints, but which now enable it to perform a subset of the ancestral functions more effectively. This model was originally formulated using the example of multifunctional proteins (e.g., a metabolic enzyme that is also a lens crystallin) whose functions become partitioned among more specialized daughter proteins, but the concept can also be applied to changes in the regulatory regions of genes (Hughes 1994). However, Hughes proposed that only a minority of duplicated genes evolved distinct functions by this mechanism, and that in the majority of cases where duplicated genes are retained, they are subject to purifying selection against dominant deleterious mutations (Hughes 1999).
More recent models—the subfunctionalization and the duplication-degeneration-complementation (DDC) models of Lynch and Force—propose that two daughter genes can accumulate degenerative changes resulting in division of the ancestral function, and hence a requirement to retain both daughter copies in the genome (Force et al. 1999; Lynch and Force 2000; Lynch 2004). These models can describe the situation in which two daughter genes accumulate degenerative changes in their promoter regions, resulting in division of the ancestral expression pattern, or it can be applied to protein region subfunctions encoded by domains in the same protein, or different proteins encoded by splice variants. In either case, the two daughter copies are described as being subfunctionalized. The DDC model is attractive because it suggests a mechanism through which both daughter copies can be preserved in the genome simply through the action of degenerative (not adaptive) mutations. Lynch and Force emphasize the role of subfunctionalization as a preservative mechanism in the early stages after gene duplication, which does not rule out the possibility that at a later stage the daughter genes could gain novel functions that were not present in the ancestral gene (i.e., neofunctionalization). An alternative, quantitative, form of subfunctionalization can also occur through degenerative promoter mutations that reduce the level of expression of the daughter genes to the point where both of them are needed to supply enough protein product, even without any change in the gene's function or tissue specificity of expression.
In complex organisms subfunctionalization can perhaps most readily be detected if it causes changes in the expression profiles of genes. In the long run, this type of subfunctionalization should lead to a decrease in expression breadth and the development of tissue-specific genes, as has been demonstrated in some previous studies on individual genes where subfunctionalization has occurred (Force et al. 1999; Prince and Pickett 2002). Over evolutionary time, duplicated copies of genes will accumulate amino acid substitutions at the same time as they are diverging in expression pattern. Furthermore, new tissue and cell types can be created during the course of evolution while others may disappear. The molecular basis of phenotypic differences between species are beginning to be understood through genome sequence information, and there is a growing awareness that gene duplications are responsible for some of these differences (Emes et al. 2003).
A more global study of the relationship between gene duplication and gene expression changes was recently carried out by Makova and Li (2003), who used microarray data to study the divergence in transcription profiles of large numbers of paralogous genes within the human genome. They showed that the correlation in the spatial expression profiles (across different tissues) of paralogs decreases with increasing age of the paralogs. They also showed that divergence in expression profiles occurred quite rapidly, consistent with a subfunctionalization model of duplicate gene preservation. However, because they only considered expression data from humans, Makova and Li were not able to put the levels of transcription profile change seen in duplicated genes (paralogs) into the context of the changes in transcription profile that occur during evolution even without gene duplication, for example, in comparisons of orthologous genes between species.
Here, we have used microarray data from the Gene Expression Atlas (GEA; Su et al. 2002) to study divergence of transcription profiles of genes between human and mouse, compared across homologous tissues in the two species. We focus on loci where recent species-specific gene duplications have occurred within human or mouse, creating pairs of young paralogs whose transcription profiles can be compared with that of the single-copy ortholog in the other species. We show that the presence of a species-specific gene duplication accelerates the rate of expression divergence between human and mouse, and also that these recent duplicates are subject to reduced constraints on their protein sequences.
Finally, we observe that expression domains become progressively narrower as the number of paralogs increases. This finding is consistent with the subfunctionalization model. However, detailed examination of several gene families in which recent duplications have occurred suggests that in most cases multiple changes in the spatial expression profile have occurred, and neofunctionalization is suggested by the appearance of expression in a new tissue.
RESULTS
Linking Gene Expression Data to Orthologs and Paralogs in Human and Mouse
Gene expression data from human and mouse were obtained from the Gene Expression Atlas (GEA) of Su et al. (2002). These data comprise 101 human (microchip U95A) and 89 mouse (microchip U74A) Affymetrix experiments. From these, we used data from 16 homologous tissues that had been studied in both human and mouse (see Methods). Annotations of the human and mouse genomes were obtained from the Ensembl database using the EnsMart tool (Hubbard et al. 2002). We were able to confidently assign 5261 out of the total of 24,848 human genes (21%), and 4522 of 24,950 mouse genes (18%) to Affymetrix probes, after omitting probes that mapped to multiple genomic locations and other possible artifacts as described in Huminiecki et al. (2003).
A list of putative human/mouse orthologs was also downloaded from Ensembl. We retained only orthologs that were listed in a simple one-to-one relationship between human and mouse. A pair of orthologs was regarded as linked to expression data if both the human and mouse gene were mapped to Affymetrix probes. Of the 13,341 pairs of orthologs in the list, 1833 (14%) were mapped to probes in both human and mouse. Among these, 1575 (94%) were expressed in at least one of the 16 homologous tissues we used for human/mouse comparisons. The minimal signal accepted for expression was an Affymetrix average difference (AD) value of 200 (see Methods). Paralogs within each genome were identified using TRIBE (Enright et al. 2002). Both genes could be linked to Affymetrix probes for 2697 (4%) of the 67,666 paralog pairs in human, and for 2680 (4%) of the 73,843 paralog pairs in mouse.
Orthologs Are More Conserved in Amino Acid Sequence Than Paralogs of Similar Evolutionary Age
Mean and median values of synonymous (_K_s) and nonsynonymous (_K_a) sequence divergence were calculated for 13,341 orthologs between human and mouse, and for paralogs within each species, using the method of Yang and Nielsen (2000; Supplemental Table S1). The median values for orthologs were _K_s = 0.61 and _K_a = 0.08, in good agreement with the medians reported in the mouse genome sequencing paper (0.602 and 0.071, respectively; Waterston et al. 2002) which used the same method and a similar number of orthologs, but our median _K_s is significantly higher than the value of 0.46 reported in the earlier study by Makalowski and Boguski (1998), which used Ina's method (Ina 1995) and 1138 orthologs.
To compare the _K_a/_K_s ratio between orthologs and paralogs, we only looked at gene pairs whose _K_s values were in the interval 0.51-0.71 (i.e., within a narrow range around the median value for orthologs) because comparisons of _K_a/_K_s ratios are only meaningful if the _K_s values are similar (Nembaware et al. 2002). This filtering meant that only 29% of orthologs and 3% of paralog pairs were considered, but the sample sizes remained large (Supplemental Table S1). Among these, there is a very substantial difference in _K_a/_K_s, with mean ratios of 0.71 and 0.88 seen in paralogs of mouse and human, respectively, compared with only 0.17 in orthologs. This fourfold to fivefold increase in the total cumulative amount of nonsynonymous substitutions calculated for relatively old paralogs indicates even more dramatic relaxation in selective constraints in the period immediately following the duplications event, which took place approximately at the same time as the mouse/human speciation date. Similar conclusions were reached by Lynch and Conery (2000), Kondrashov et al. (2002), and Seoighe et al. (2003), using different approaches and data sets.
Identification of Ortholog Pairs With Species-Specific Duplication in Human or Mouse
To investigate whether the relaxation of constraint on protein sequence evolution seen in paralogs is correlated with a tendency to change their spatial patterns of gene expression, we focused on paralogs that have been formed recently by species-specific gene duplication in either human or mouse. The Ensembl list of human/mouse orthologs was merged with human and mouse paralog data sets to identify lineage-specific duplications. Lineage-specific duplications were defined as those where the synonymous substitution distances between paralogs was lower than 0.70 and, in any case, lower than that between the corresponding human/mouse orthologs. We were satisfied that the quality of ortholog assignment was equally high regardless of whether a human or mouse duplication occurred. The average _K_s between orthologs was 0.68 ± 0.42 for the set with human-specific duplications and 0.60 ± 0.45 for the set with mouse-specific duplications, values that are actually lower than the 0.71 ± 0.43 calculated for the set of 1325 one-to-one orthologs.
For this analysis, we considered only ortholog sets for which gene expression data were available from GEA. Expression information was available for 1575 human/mouse ortholog sets including 250 with species-specific paralogs. Among these, there were 1325 simple one-to-one human/mouse orthologs (those without any duplication in the human or mouse lineage), 163 sets with recent duplications in human and 139 with recent duplications in mouse. For most of these sets, GEA expression data were only available for one of the species-specific duplicates, as well as for the ortholog in the other species. Several sets included multiple species-specific duplications: there were 192 sets with two lineage-specific paralogs, 44 with three, 27 with four, 8 with five, 11 with six, and 20 with more than six. In 52 ortholog sets there were both human and mouse lineage-specific duplications.
Orthologs With Lineage-Specific Gene Duplications Tend Not to Have Highly Correlated Expression Profiles
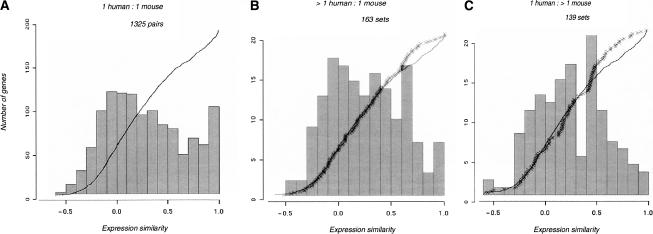
One-to-one orthologous gene pairs are generally expected to be correlated in their expression patterns across different tissues. For example, a human liver-specific gene is expected to have a mouse ortholog that also has liver-specific expression. Figure 1A shows the histogram of expression correlation coefficients (R) for the set of 1325 human/mouse one-to-one orthologs. The distribution of _R_-values appears bimodal with one peak showing orthologs with highly correlated expression profiles (close to the value of R = 1), and a second peak centered on R = 0. The latter group of orthologs presumably includes those that are primarily expressed in tissues other than the 16 that we were able to compare between human and mouse. A similarly bimodal plot of ortholog expression profiles was obtained in the original analysis of GEA data by Su et al. (2002) with 799 ortholog pairs.
Figure 1.
Genes highly similar in expression are underrepresented in ortholog sets with recent human- or mouse-specific gene duplications. Histograms of expression correlation coefficients (R) across 16 tissues are shown for (A) 1325 one-to-one orthologs between human and mouse; (B) 163 ortholog sets with one mouse sequence and more than one human co-ortholog; (C) 135 ortholog sets with one human sequence and more than one mouse co-ortholog. Overlaid on each histogram are cumulative lines showing the proportion of ortholog sets having expression similarity lower than a particular value of R, for the one-to-one set (solid line), and for the sets with species-specific duplications (stars).
In contrast, there is no peak at the high end of correlation values when analogous histograms are drawn for ortholog sets that also have lineage-specific duplications in either human or mouse (Fig. 1B,C). In other words, the presence of a species-specific paralog increases the likelihood that two species' expression profiles differ. This result is not an artifact of the choice of bin sizes chosen for the histograms, as shown by the cumulative curves in Figure 1.
A nonparametric randomization procedure was used to estimate the statistical significance of the underrepresentation of highly correlated pairs in orthologs with lineage-specific paralogs. Specifically, we estimated the probability that _R_-value distributions that are skewed similarly to those in Figure 1, B and C, could be obtained by chance alone. Random samples of K ortholog pairs were chosen from the total set of expressed orthologs, where K was 163 and 139 for human and mouse, respectively, and the sampling was repeated 10,000 times. For each random sample, the number of ortholog pairs in four overlapping intervals (R > 0.9, R > 0.8, R > 0.7, and R > 0.6) on the right-hand side of the distribution was counted. The proportion of samples that produced equal or lower numbers of orthologs in a given _R_-interval than observed in the real data defined the _p_-value of the observation. All intervals with R > 0.7 were significant at the α < 0.01 level in both human and mouse, except for one that was significant at α < 0.05 (Table 1).
Table 1.
Statistical Test of the Significance of Underrepresentation of Highly Correlated Gene Pairs in Ortholog Sets With Species-Specific Duplications
| R > 0.6 | R > 0.7 | R > 0.8 | R > 0.9 | |
|---|---|---|---|---|
| Human duplication | 91% | 58% | 52% | 60% |
| p = 0.37 | p = 0.0042 | p = 0.0043 | p = 0.038 | |
| Mouse duplication | 61% | 48% | 36% | 24% |
| p = 0.0111 | p = 0.0027 | p = 0.0015 | p = 0.0018 |
Orthologs With Multiple Duplications Show Even Weaker Correlations of Expression Profiles
From Table 1, we chose a value of R > 0.8 as a threshold for defining “similar” expression profiles and compared the proportions of genes with similar expression among groups of orthologs with different amounts of lineage-specific gene duplication. In the set of 1325 one-to-one orthologs, 181 pairs (13.7%) had similar expression profiles by this criterion. The proportion of pairs having similar profiles was lower in all categories of orthologs with lineage-specific duplications. Among 148 ortholog sets with exactly one human- or mouse-specific duplication, only 7.4% (11 pairs) had similar expression. This was further reduced in ortholog sets with multiple human or mouse duplications (4.9%; 5 pairs out of 102), and in sets with independent lineage-specific duplications in both human and mouse (1.9%; 1 pair out of 52). The reduction in each of these three groups relative to the group of one-to-one orthologs is statistically significant in χ2 tests (calculated as two-by-two contingency tables in pairwise comparisons; P < 0.05 for each of the three tests). Thus, orthologs with multiple lineage-specific duplications or that duplicated independently in both human and mouse are even less likely to be highly correlated in expression than those where only one duplication event occurred. This result is suggestive of either subfunctionalization or neofunctionalization after each gene duplication.
Tissue-Specific Genes Are More Likely to Belong to Large Gene Families
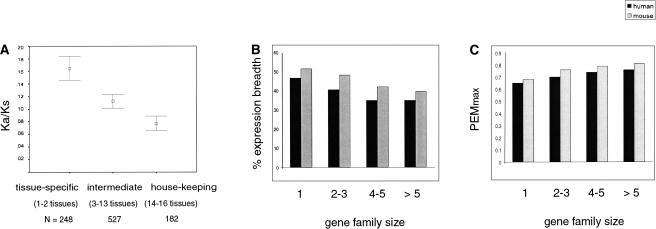
It was previously reported that tissue-specific genes evolve faster than ubiquitously expressed genes, based on analyses of expressed sequence tag data (Duret and Mouchiroud 2000) and a microarray data set of 1581 genes (Zhang and Li 2004). We confirmed this observation using the larger GEA data set (Fig. 2A). Average _K_a/_K_s ratios were calculated for three subsets of human/mouse orthologs, defined on the basis of the expression breadth of both the human and the mouse gene in the 16 tissues analyzed, and using expression in one to two tissues as a working definition of a “tissue-specific” gene and 14-16 tissues as “housekeeping.”
Figure 2.
Tissue-specific genes evolve faster and are more likely to belong to large gene families. (A) Average _K_a/_K_s ratios with 95% confidence intervals for human/mouse orthologs with tissue-specific, intermediate, and housekeeping gene expression profiles. Kruskal-Wallis rank test, p < 0.001. (B) Average breadth of expression, measured as the mean percentage of the 16 tissues in which a gene was expressed, for human and mouse orphan genes and genes in families of growing size. (C) Average PEMMAX values for human and mouse orphan genes and genes in families of growing size.
If subfunctionalization of gene expression were a major driving force behind duplicate gene retention, then there should be a trend toward the development of tissue-specific expression following many cycles of gene duplication and growth of the family size. To test this hypothesis, two separate measures of tissue-specificity were used: percentage breadth of expression (Fig. 2B) and PEMMAX (Fig. 2C). Percentage breadth of expression was defined as the percentage of the 16 tissues studied in which a given gene was expressed above the threshold level. PEMMAX is the maximal value of the Preferential Expression Measure (PEM; Huminiecki et al. 2003) for a gene, which measures the extent to which the gene's transcription profile is concentrated into one tissue. PEM is log10(S/A), where S is the Affymetrix signal for a given gene in a specific tissue, and A is the arithmetic mean signal for the gene across all tissues. PEMMAX is the maximal value of PEM among all tissue scores for a gene. The more tissue-specific a gene's expression is, the higher its PEMMAX score. It can be seen that, in both human and mouse, larger gene families tend to have decreased expression breadth and increased tissue specificity (Fig. 2B,C), consistent with subfunctionalization. This observation differs from Zhang and Li's conclusion that there is little difference in the average family size of housekeeping and tissue-specific genes in mammals. The difference between these results may hinge on the fact that Zhang and Li (2004) grouped genes into only two classes of family size (single-copy and those with two or more copies), whereas we see differences in expression breadth and specificity between smaller and larger multigene families (Fig. 2B,C).
Statistical tests confirmed that each of the three size subclasses of genes in families (Fig. 2B,C) is narrower in its expression domain than singleton genes. The distribution of PEMMAX values was transformed to a normal distribution using a classic Box-Cox transformation with the λ parameter of 0.1, according to the formula f(x) = (_x_λ - 1)/λ, where x is the PEMMAX value, and the Welch one-sided two-sample _t_-test was applied. The difference between the singleton genes and the families is highly significant in all cases. Pairwise comparisons show singletons to have less-specialized expression than genes in families of two to three, four to five, and more than five members with P = 3e - 06, 1e - 09, and 6e - 14, respectively, for human, and P = 8e - 12, 7e - 08, and 1e - 08 for mouse. For percentage breadth of expression values, the distribution was very different from normal, with most genes being far toward either the tissue-specific or the housekeeping end of the distribution. Therefore, the nonparametric Fligner-Killeen test for homogeneity of means was applied (http://www.r-project.org/; “The R Reference Index”). _P_-values were <0.001 in both human and mouse. Additionally, pairwise Wilcoxon tests were applied following a transformation according to the formula f(x) = 1/(50.01 - x), which was used to center the distribution of percentage breadth of expression around zero. The following _P_-values were obtained in order of increasing family size: 0.002, 0.001, and 7e - 10 for human, and 0.024, 0.007, and 1.05e - 06 for mouse.
Examples of Expression Divergence in Sets of Orthologs With Lineage-Specific Duplications
To examine how gene expression changes following species-specific gene duplications, we studied four gene families in detail. In particular, we looked at cases in which GEA expression data were available for two or more paralogs in one species as well as the ortholog in the other species. We used literature and EST data, in addition to GEA, to gain further information about these genes and their expression patterns.
Seven In Absentia Homolog 1
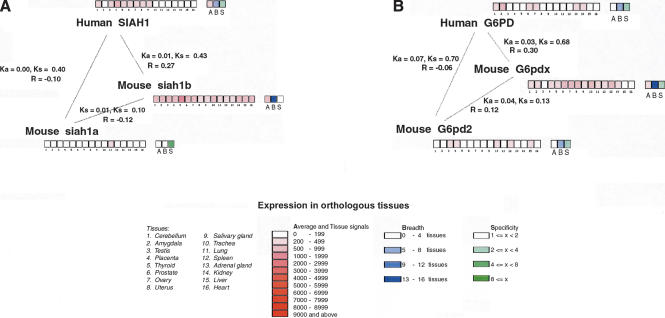
SIAH1 is a broadly expressed human ortholog of the Drosophila Seven in absentia gene involved in the development of the R7 photoreceptor (Hu et al. 1997). SIAH1 has undergone a duplication in the rodent lineage giving rise to two murine orthologs: Siah1a and Siah1b (Della et al. 1993). Siah1b is broadly expressed like its human ortholog, but Siah1a has a dramatically narrowed expression pattern, being expressed only in the lung. Figure 3A shows the relationships among the spatial expression profiles for the three genes in this ortholog set. The profile for Siah1b is positively correlated with the human ortholog, whereas Siah1a has a negative correlation coefficient. Lung expression is seen in both of the mouse co-orthologs but not in the human gene, thus it is not clear whether the ancestral expression profile included lung (tissue #11 in Fig. 3A). For other tissues, the data in Figure 3A allow parsimony arguments to be made in favor of both subfunctionalization and neofunctionalization of gene expression patterns. For example, the gain of amygdala (tissue #2) expression by mouse Siah1b is inferred to be neofunctionalization, whereas the loss of expression of Siah1a in many other tissues (#1, #3-#9) could be viewed as subfunctionalization. However, this is not a classic case of subfunctionalization in which ancestral functions have been divided up among daughters, because Siah1a has not retained any ancestral expression sites (at least, not among the 16 tissues considered here).
Figure 3.
Expression triangles for (A) Seven in absentia homolog genes SIAH1, Siah1a, and Siah2b; and (B) glucose-6-phosphate dehydrogenase genes G6PD, G6pdx, and G6pd2. Values of _K_a, _K_s, and the correlation coefficient (R) for expression similarity are shown for each pairwise comparison. For each gene, the boxes numbered 1-16 show expression intensities (Affymetrix AD) in 16 tissues as named; boxes A, B, and S show the average intensity (A) across all 16 tissues, the breadth (B; the number of tissues in which the gene is expressed), and the specificity (S; the ratio of maximum expression among all tissue scores to average intensity). Tissue signals (ADs) and average intensity are coded in red, breadth in blue, and specificity in green.
Glucose-6-Phosphate Dehydrogenase
G6PD is also a gene with broad expression in human tissues. It is an X-linked gene, whose main function is in the pentose phosphate pathway (Martini et al. 1986). Mouse has two co-orthologs of this gene: the X-linked G6pdx and the autosomal G6pd2, which lacks introns and was formed by retroposition (Zollo et al. 1993; Hendriksen et al. 1997). Like the SIAH1 example, the profile correlation coefficient of the human gene with one of the mouse co-orthologs is much higher than with the other, and one of the _R_-values is negative (Fig. 3B).
G6pd2 was reported to be transcribed in testis in spermatogenic cells where the X-linked gene is not expressed (Hendriksen et al. 1997). Indeed, this is confirmed in the GEA data set, where G6pd2 shows expression in testis (tissue #3 in Fig. 3B), but G6pdx and G6PD do not. Hence, expression in testis (as well as in liver; tissue #15) could be regarded as a neofunctionalization of G6pd2 following its retroposition. Remarkably, none of the five tissues in which mouse G6pd2 is expressed is the same as any of the six tissues in which human G6PD is expressed. Viewed from the human perspective, the expression pattern of G6pd2 could be described as complete neofunctionalization. However, mouse G6pdx shows a very broad expression pattern, covering all six tissues where the human gene is expressed, as well as three of the five where G6pd2 is expressed, and four tissues that are unique to G6pdx. Hence, the two mouse co-orthologs show extensive neofunctionalization (gaining expression in two tissues for G6pd2 and four tissues for G6pdx), but there is no evidence of subfunctionalization of the human gene's expression pattern (it has all been retained by G6pdx).
Cysteine-Rich Secretory Proteins
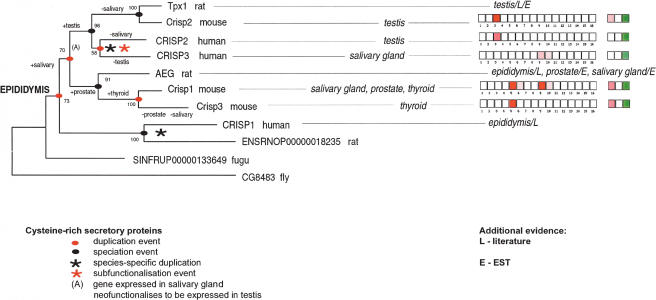
There are three CRISP genes in both human and mouse, but their nomenclature is misleading because none of the genes are simple one-to-one orthologs between human and mouse. Figure 4 illustrates the complex evolutionary history of the CRISP family, including sequences from rat, Fugu, and Drosophila. A single ancestral gene at the base of the vertebrate lineage was most likely subject to two rounds of gene duplication before the human/rodent split, a human-specific duplication (resulting in CRISP2 and CRISP3), a mouse-specific duplication (resulting in Crisp1 and Crisp3), differential gene losses, and multiple shifts in expression patterns.
Figure 4.
Phylogenetic relationships and expression profiles in the cysteine-rich secretory protein (CRISP) family. The phylogenetic tree was constructed by the minimum evolution method after CLUSTALW sequence alignment of protein sequences. Bootstrap values are shown. Human CRISP1, CRISP2, CRISP3, and mouse Crisp1, Crisp2, Crisp3, were compared with three rat genes: testis-specific protein 1, Tpx1 (Maeda et al. 1998; O'Bryan et al. 1998); epididymal glycoprotein, AEG (Charest et al. 1988); and an Ensembl novel gene prediction, as well as outgroups from Fugu rubripes and Drosophila melanogaster (used to root the tree). Where GEA data were not available (CRISP1 and all rat genes), expression evidence from the literature (L) or EST databases (E) was used. The numbering and color scheme for gene expression data is the same as in Figure 3.
Northern blots indicated that human CRISP1 is epididymis-specific, CRISP2 is expressed in testis, and the most prominent expression site for CRISP3 is in salivary gland (Kratzschmar et al. 1996). Using information from the literature and EST expression patterns as well as data from GEA, and interpreting expression patterns in the context of the phylogenetic tree in Figure 4, we suggest that the ancestral vertebrate CRISP gene was primarily expressed in the epididymis. This is agreement with what is reported to be the classic biological role of CRISP: facilitating plasma membrane recognition and fusion between sperm and egg (Brooks et al. 1986; Evans 2002). This role is known to be carried out by CRISP1 in human and AEG in rat.
Testicular expression, as seen in the ortholog trio of human CRISP2, mouse Crisp2, and rat Tpx1, appears to be a neofunctionalization following duplication (node A in Fig. 4) of a gene expressed in the salivary gland shortly before the human/rodent split. These testis-specific genes play a role in sperm maturation and are responsible for interactions between spermatogenic precursors and Sertoli cells (Maeda et al. 1998). The precise function of CRISP proteins in thyroid and prostate remains to be established, but it is interesting that there are several homologous cysteine-rich peptides in snake venoms, which are products of modified salivary glands (Chang et al. 1997; Yamazaki et al. 2002, 2003). The human-specific CRISP2/CRISP3 duplication is a putative subfunctionalization event. The ancestral human gene was expressed in both testis and salivary gland. Following duplication, CRISP2 and CRISP3 subfunctionalized to become expressed specifically in testis and salivary gland, respectively. The CRISP phylogenetic tree also shows several lineage-specific gene losses. If all duplicate genes were preserved following duplications, there should be four CRISP proteins in human and mouse. Gene losses are inferred, for example, in the case of the mouse ortholog of human CRISP1, and human ortholog of the murine Crisp1/Crisp3 pair.
The overall impression from Figure 4 is that a surprisingly high number of changes in gene expression patterns have occurred during the evolution of the CRISP family. Using a parsimony approach and assuming ancestral expression in epididymis alone, we infer that there have been at least nine changes in expression site during the evolution of this family in vertebrates (Fig. 4), including several reversals of state. Alternative assumptions about the ancestral expression profile require even more changes and so are less parsimonious.
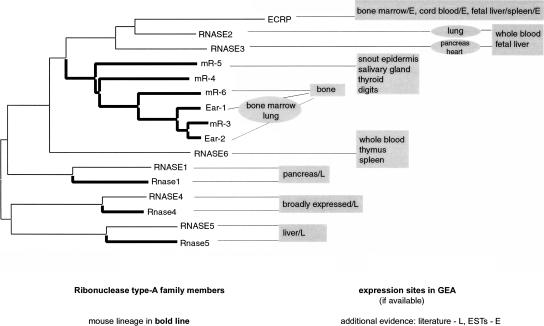
Eosinophil-Associated Ribonucleases
These are the classic example of multiple species-specific duplications, and have been the subject of intense investigation in the field of molecular evolution. In human, the eosinophil-expressed subfamily expanded through two lineage-specific duplications to give rise to RNASE2 (eosinophil-derived neurotoxin), RNASE3 (eosinophil cationic protein; Hamann et al. 1990), and ECRP (GenBank X55989). Multiple mouse-specific duplications produced at least six genes (Ear-1, Ear-2, and mR-3 through mR-6; Batten et al. 1997). Zhang and co-workers have previously shown that the eosinophil-associated subfamily has been subject to very strong positive selection (Zhang et al. 1998, 2000; Zhang and Rosenberg 2002). To investigate whether expression pattern shifts are likely to occur in a family whose evolution is well documented to be driven by positive selection at the protein sequence level, we examined the family of eosinophil-associated ribonucleases using expression data from GEA, ESTs, and literature (Fig. 5). We found that this family contrasts sharply with the CRISPs because of few expression profile changes. Almost all the expression profiles were consistent with expression in single adult cell-type eosinophils (immune cells infiltrating other tissues). However, we also found expression in immune and blood progenitor tissues such as bone marrow, CD34+ cells from cord blood, and fetal liver and spleen. mR-5 was a sole exception, not being expressed in any of the immune tissues.
Figure 5.
Type-A ribonucleases. Amino acid _p_-distance neighbor-joining tree with sites of expression in GEA for the eosinophil-associated subfamily. Seven human genes and nine mouse genes were compared. Mouse lineages are drawn as bold lines. Amino acid sequences were aligned using CLUSTALW, and the tree was drawn using Mega v. 2.1.
DISCUSSION
After a gene duplication, the daughter sequences tend to undergo a period of accelerated protein sequence change, as evidenced by the markedly increased _K_a/_K_s ratio in paralogs as compared with orthologs of similar ages (Supplemental Table S1). In addition, paralogs tend to diverge in their gene expression patterns, which we documented in several ways using transcription profiles from 16 homologous tissues from the GEA data set (Su et al. 2002). One persistent concern with microarray experiments is that cross-hybridization between closely related transcripts and probes may affect the results. However, we do not think that cross-hybridization is a serious problem in our analysis, because the primary data come from Affymetrix oligonucleotide arrays as opposed to cDNA arrays, and we excluded oligonucleotide probes that did not map to a unique gene in human or mouse. Also, our detailed examination of the GEA data showed that, although a small number of probes that were flagged by Affymetrix as being susceptible to cross-hybridization did, indeed, show evidence of cross-hybridization, the great majority of the data was free of this artifact and there was no correlation between the sequence similarity of a pair of genes and the correlation of their expression profiles (see Methods).
We found that among orthologs, a relatively small number of genes show strongly conserved expression profiles between human and mouse. For the one-to-one ortholog class (those lacking recent species-specific duplications in either human or mouse), only 14% of pairs show a strong correlation coefficient of R ≥ 0.8 across the 16 tissues compared. However, this is significantly more conservation than in the class of orthologs that have recent gene duplications. Because the ortholog pairs are all the same age (corresponding to the date of the human/mouse speciation), the increased divergence in expression profiles in ortholog sets having recent gene duplications can be directly attributed to the presence of those duplications. Moreover, the divergence in expression profiles becomes more pronounced in gene families with increasing numbers of duplications.
We propose that in many cases both genes of the duplicated pair have changed their expression pattern in comparison to the ancestral state. The overall trend is toward expression subfunctionalization and the development of tissue-specific expression in large gene families (Fig. 2B,C). Despite this trend, examining the expression profiles of three gene families in detail uncovered several examples of apparent neofunctionalization, and only one case of putative subfunctionalization (the CRISP2/CRISP3 duplication). It should be noted, however, that possible neofunctionalizations are relatively easy to detect (i.e., the appearance of gene expression by one paralog in a tissue where neither the other paralog, nor the ortholog in the other species, is expressed—although this does not demonstrate that the change was fixed by positive selection). In contrast, inferring expression subfunctionalization requires that the tissues of expression of a gene in one species become more-or-less perfectly divided up among multiple recent paralogs in the other species, which is a much more complicated set of conditions. Moreover, partitioning-out of the tissues of expression is only one of several ways by which two daughter genes could become subfunctionalized (Lynch 2004). We should also bear in mind that the present-day expression profiles may not be the same as those shortly after gene duplication. We feel that at present there are insufficient data to attempt to quantify the relative rates of neo- versus subfunctionalization, and we note that these processes are not mutually exclusive. However, our observations do suggest that the concepts of subfunctionalization and neofunctionalization may be too simple a vocabulary to describe what has actually happened at many loci after duplication, and that it may be possible for two paralogs to simultaneously become subfunctionalized as regards expression in one group of tissues, and neofunctionalized in other tissues.
Microarray data have further potential to enhance our understanding of the patterns of subfunctionalization and neofunctionalization of duplicated genes. Furthermore, neofunctionalization does not necessarily have to be secondary to a gene duplication event. The existence of many one-to-one orthologs that are poorly correlated in expression between human and mouse suggests that expression diversification also occurs independently of duplication. This may be a neutral evolutionary phenomenon, or it could reflect genuine physiological differences between human and mouse, such as in the biology of reproduction or olfaction. Further investigation is also warranted into the evolution and interdependency of arbitrary expression domains (tissue categories) investigated in microarray studies, such as somatic versus reproductive tissues, or tissues derived from the same embryonic layer. Finally, from a cell biology perspective, tissue expression is simply a cumulative function of heterogeneous cellular transcriptomes. The expression states of a tissue may therefore vary, depending on the proportions and interactions of the constituent cell types. On the other hand, organs and tissues with very different biological roles might share a surprisingly high proportion of cellular elements, such as vascular cells (endothelial cells, vascular smooth muscles), infiltrating white cells, or cells of the connective tissue.
METHODS
Sequence Data
The Ensembl-confirmed gene protein and nucleotide set were extracted using the EnsMart tool (Hubbard et al. 2002). There were 24,848 human genes, and 24,950 mouse genes. In case of multiple transcripts being mapped to one gene, only the transcript resulting in the longest protein product was accepted. A list of human/mouse putative orthologs was downloaded from the Ensembl database. Only reciprocally unique pairs were retained resulting in 13,341 pairs of high-quality orthologs. Human and mouse paralogs were determined within the Ensembl data set using the TRIBE algorithm described by Enright et al. (2002).
Proteins coded for by ortholog and paralog gene pairs were aligned using CLUSTALW with default parameters. Protein-coding nucleotide sequences were then aligned using protein alignments as a guide with the program TRANALIGN from the Emboss suite (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/Apps/tranalign.html). Pairwise _K_a and _K_s distances were calculated using the method of Yang and Nielsen implemented in the program Yn00 from the PAML suite (version 3.13).
Expression Data
Human gene and mouse expression data were derived from the Gene Expression Atlas (GEA) of Su et al. (2002), downloaded from http://expression.gnf.org. This data set comprises 101 human U95A and 89 mouse U74A Affymetrix microchip experiments. Many of the measurements were performed in duplicate or triplicate on the identical or the same tissue-type sample. These replicates show very good correlation (Huminiecki et al. 2003) with Pearson _R_-squared values of 0.94 ± 0.04 for the same RNA sample (N = 45 pairwise comparisons) and of 0.87 ± 0.06 (N = 17 pairwise comparisons) for repeat hybridizations of different RNA preparations from the same tissue type. We calculated arithmetic means of the average difference (AD) values and used these in further analyses.
For orthology comparisons, 16 tissue categories shared by the mouse and human GEA data sets were used (namely, adrenal gland, amygdala, cerebellum, heart, kidney, liver, lung, ovary, placenta, prostate, salivary gland, spleen, testis, thyroid, trachea, and uterus).
An average difference (AD) value of 200 was used as a threshold for a gene to be regarded as “on” in a given tissue. Several methods of data transformation were experimented with. For example, two-state (on/off) and four-state (off/weak/strong/very strong) expression scales were investigated. However, these data transformations resulted in a lower overall Pearson _R_-value for the total set of human/mouse orthologs and were not used in further analysis. Pearson R was chosen as a preferred pairwise distance measure of expression correlation because it is sensitive to shape (not absolute values) of expression profiles. As such, it corresponds well to the biologist's intuitive understanding of what coexpressed or coregulated genes are. This is particularly true for tissue-specific genes, which are most interesting from the biological point of view, being widely used as markers in tissue staining or clinical diagnosis, as drug targets, or homing molecules for drug delivery systems.
Mapping Expression Data to Genes
Affymetrix tag/UniGene cluster mapping was extracted from the Affymetrix probe consensus sequence file, which lists UniGene clusters used to design microarray probes (http://www.affymetrix.com/analysis/downloadcenter.affx). In some cases, two or more Affymetrix tags were targeted against the same UniGene cluster. In these cases, only the tag with strongest average expression across all libraries from the human Gene Expression Atlas data set was retained. UniGene clusters were mapped to the human genome (NCBI build 31) using the LocusLink and Ensembl databases. Firstly, UniGene to LocusLink mapping was extracted from the UniGene release file Hs.data (human build U150). Secondly, LocusLink/Ensembl gene id mapping was extracted from the Ensembl database (release 14.31.1) using the EnsMart tool (http://www.ensembl.org/EnsMart/). LocusLink clusters mapping to multiple UniGene clusters or multiple Ensembl genes were discarded to ensure that the resulting mapping was unique and nonredundant. A similar procedure was followed for the mouse genome (NCBI build 30).
Cross-Hybridization in Microarray Data
The primary data in this study are derived from the Affymetrix oligonucleotide array experiments of Su et al. (2002). In these arrays, 16 oligonucleotide probes are used for each gene, and probes likely to cause cross-hybridization are omitted from the chip during the design stage. In addition, Affymetrix chips have a very stringent built-in internal subtraction mechanism in which every antisense probe is coupled with a single mismatch probe (Wodicka et al. 1997). Target abundance is estimated by the difference between the hybridization signals from the perfect match probe and a single mismatch probe. Because the probes are 25 nt in length, we can make the approximation that even 1 nt difference in 25 (96% identity) is sufficient to safeguard differential hybridization for a single probe, although this does depend on the location of the mismatching base (mismatches in the middle of the target/probe duplex are most effective). When all Affymetrix tags were compared against each other using BLAST, the average percentage identity between probes for pairs of paralogs with _K_s < 0.2 was only 93%, and 88% for _K_s between 0.2 and 1. Furthermore, we detected no correlation between the measured _R_-values (expression profile correlation coefficients) for pairs of paralogs and their percentage sequence identity, number of matching, or number of mismatching base pairs.
Affymetrix probes with name suffixes _f_at (“sequence family”) and _s_at (“similarity constraint”) are thought to be more prone to cross-hybridization than others (see Affymetrix manual, Data Analysis Fundamentals, Appendix B). We refer to these probes collectively as suboptimal probes. To investigate whether inclusion of data from these probes was affecting our results, we performed a separate analysis of ortholog and paralog pairs that mapped to suboptimal probes. Out of 1575 human/mouse orthologs, only in two cases did both the human and the mouse gene map to suboptimal probes. In addition, there were 155 cases in which one of the orthologs linked to a suboptimal probe, but the average R for this subset was not different from that calculated for the total set (0.296 and 0.283, respectively; P = 0.686). In the analysis of ortholog sets with duplication, there was only one mouse set in which both the human and the mouse gene were mapped to suboptimal probes. Additionally, there were 24 and 20 sets, in groups with human-specific and mouse-specific duplications, respectively, in which one of the orthologs linked to a suboptimal probe. However, in both of these subsets there was no significant difference in the average R compared with the remaining orthologs (_t_-test; P = 0.388 and 0.196, respectively). In other words, pairs in which one gene mapped to a suboptimal probe behaved just like pairs mapped to probes with full cross-hybridization control.
For paralogs within human, there were 17 pairs of paralogs in which both genes were mapped to suboptimal probes, and 227 pairs in which one of the genes was mapped to a suboptimal probe. The average pairwise expression similarity (Pearson R) between these paralogs was _R_avg = 0.348 for two suboptimal probes, _R_avg = 0.145 for one suboptimal probe, as compared with _R_avg = 0.121 for paralog pairs in which neither gene mapped to a suboptimal probe. Similarly for mouse, _R_avg was 0.383 for 33 paralog pairs with two suboptimal probes, and 0.108 for 297 pairs with one suboptimal probe, as compared with 0.079 for pairs with no suboptimal probes. The average R for pairs mapping to one suboptimal probe was not significantly different from those without suboptimal probes (_t_-test, P = 0.27 and 0.09 for human and mouse, respectively). In summary, although there is a tendency toward cross-hybridization in pairs of human and mouse paralogs in which both genes were linked to suboptimal probes, the number of such pairs was very limited.
Acknowledgments
This study was supported by Science Foundation Ireland. We thank Greg Singer and Andrew Lloyd for many helpful discussions and lively exchange of ideas, and are grateful for helpful comments from anonymous referees.
Footnotes
[Supplemental material is available online at www.genome.org.\]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2705204.
References
- Batten, D., Dyer, K.D., Domachowske, J.B., and Rosenberg, H.F. 1997. Molecular cloning of four novel murine ribonuclease genes: Unusual expansion within the ribonuclease A gene family. Nucleic Acids Res. 25**:** 4235-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, D.E., Means, A.R., Wright, E.J., Singh, S.P., and Tiver, K.K. 1986. Molecular cloning of the cDNA for two major androgen-dependent secretory proteins of 18.5 kilodaltons synthesized by the rat epididymis. J. Biol. Chem. 261**:** 4956-4961. [PubMed] [Google Scholar]
- Chang, T.Y., Mao, S.H., and Guo, Y.W. 1997. Cloning and expression of a cysteine-rich venom protein from Trimeresurus mucrosquamatus (Taiwan habu). Toxicon 35**:** 879-888. [DOI] [PubMed] [Google Scholar]
- Charest, N.J., Joseph, D.R., Wilson, E.M., and French, F.S. 1988. Molecular cloning of complementary deoxyribonucleic acid for an androgen-regulated epididymal protein: Sequence homology with metalloproteins. Mol. Endocrinol. 2**:** 999-1004. [DOI] [PubMed] [Google Scholar]
- Della, N.G., Senior, P.V., and Bowtell, D.D. 1993. Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development 117**:** 1333-1343. [DOI] [PubMed] [Google Scholar]
- Duret, L. and Mouchiroud, D. 2000. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17**:** 68-74. [DOI] [PubMed] [Google Scholar]
- Emes, R.D., Goodstadt, L., Winter, E.E., and Ponting, C.P. 2003. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum. Mol. Genet. 12**:** 701-709. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., Van Dongen, S., and Ouzounis, C.A. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30**:** 1575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J.P. 2002. The molecular basis of sperm-oocyte membrane interactions during mammalian fertilization. Hum. Reprod. Update 8**:** 297-311. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151**:** 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, K.J., Ten, R.M., Loegering, D.A., Jenkins, R.B., Heise, M.T., Schad, C.R., Pease, L.R., Gleich, G.J., and Barker, R.L. 1990. Structure and chromosome localization of the human eosinophil-derived neurotoxin and eosinophil cationic protein genes: Evidence for intronless coding sequences in the ribonuclease gene superfamily. Genomics 7**:** 535-546. [DOI] [PubMed] [Google Scholar]
- Hendriksen, P.J., Hoogerbrugge, J.W., Baarends, W.M., de Boer, P., Vreeburg, J.T., Vos, E.A., van der Lende, T., and Grootegoed, J.A. 1997. Testis-specific expression of a functional retroposon encoding glucose-6-phosphate dehydrogenase in the mouse. Genomics 41**:** 350-359. [DOI] [PubMed] [Google Scholar]
- Hu, G., Chung, Y.L., Glover, T., Valentine, V., Look, A.T., and Fearon, E.R. 1997. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46**:** 103-111. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30**:** 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L. 1994. The evolution of functionally novel proteins after gene duplication. Proc. R Soc. Lond. B Biol. Sci. 256**:** 119-124. [DOI] [PubMed] [Google Scholar]
- ____. 1999. Adaptive evolution of genes and genomes. Oxford University Press, New York.
- Huminiecki, L., Lloyd, A.T., and Wolfe, K.H. 2003. Congruence of tissue expression profiles from Gene Expression Atlas, SAGEmap and TissueInfo databases. BMC Genomics 4**:** 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ina, Y. 1995. New methods for estimating the numbers of synonymous and nonsynonymous substitutions. J. Mol. Evol. 40**:** 190-226. [DOI] [PubMed] [Google Scholar]
- Katju, V. and Lynch, M. 2003. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165**:** 1793-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F.A., Rogozin, I.B., Wolf, Y.I., and Koonin, E.V. 2002. Selection in the evolution of gene duplications. Genome Biol. 3**:** RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzschmar, J., Haendler, B., Eberspaecher, U., Roosterman, D., Donner, P., and Schleuning, W.D. 1996. The human cysteine-rich secretory protein (CRISP) family. Primary structure and tissue distribution of CRISP-1, CRISP-2 and CRISP-3. Eur. J. Biochem. 236**:** 827-836. [DOI] [PubMed] [Google Scholar]
- Lynch, M. 2004. Gene duplication and evolution. In Evolution: From molecules to ecosystems (eds. A. Moya and E. Font), pp. 33-47. Oxford University Press, Oxford, UK.
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290**:** 1151-1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Force, A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154**:** 459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., O'Hely, M., Walsh, B., and Force, A. 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159**:** 1789-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, T., Sakashita, M., Ohba, Y., and Nakanishi, Y. 1998. Molecular cloning of the rat Tpx-1 responsible for the interaction between spermatogenic and Sertoli cells. Biochem. Biophys. Res. Commun. 248**:** 140-146. [DOI] [PubMed] [Google Scholar]
- Makalowski, W. and Boguski, M.S. 1998. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. 95**:** 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova, K.D. and Li, W.M. 2003. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 13**:** 1638-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini, G., Toniolo, D., Vulliamy, T., Luzzatto, L., Dono, R., Viglietto, G., Paonessa, G., D'Urso, M., and Persico, M.G. 1986. Structural analysis of the X-linked gene encoding human glucose 6-phosphate dehydrogenase. EMBO J. 5**:** 1849-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nembaware, V., Crum, K., Kelso, J., and Seoighe, C. 2002. Impact of the presence of paralogs on sequence divergence in a set of mouse-human orthologs. Genome Res. 12**:** 1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bryan, M.K., Loveland, K.L., Herszfeld, D., McFarlane, J.R., Hearn, M.T., and de Kretser, D.M. 1998. Identification of a rat testis-specific gene encoding a potential rat outer dense fibre protein. Mol. Reprod. Dev. 50**:** 313-322. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene and genome duplication. Springer, Berlin.
- Piatigorsky, J. and Wistow, G. 1991. The recruitment of crystallins: New functions precede gene duplication. Science 252**:** 1078-1079. [DOI] [PubMed] [Google Scholar]
- Prince, V.E. and Pickett, F.B. 2002. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3**:** 827-837. [DOI] [PubMed] [Google Scholar]
- Seoighe, C., Johnston, C.R., and Shields, D.C. 2003. Significantly different patterns of amino acid replacement after gene duplication as compared to after speciation. Mol. Biol. Evol. 20**:** 484-490. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 99**:** 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420**:** 520-562. [DOI] [PubMed] [Google Scholar]
- Wodicka, L., Dong, H., Mittmann, M., Ho, M.H., and Lockhart, D.J. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15**:** 1359-1367. [DOI] [PubMed] [Google Scholar]
- Yamazaki, Y., Koike, H., Sugiyama, Y., Motoyoshi, K., Wada, T., Hishinuma, S., Mita, M., and Morita, T. 2002. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 269**:** 2708-2715. [DOI] [PubMed] [Google Scholar]
- Yamazaki, Y., Hyodo, F., and Morita, T. 2003. Wide distribution of cysteine-rich secretory proteins in snake venoms: Isolation and cloning of novel snake venom cysteine-rich secretory proteins. Arch. Biochem. Biophys. 412**:** 133-141. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Nielsen, R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17**:** 32-43. [DOI] [PubMed] [Google Scholar]
- Zhang, L. and Li, W.H. 2004. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol. Biol. Evol. 21**:** 236-239. [DOI] [PubMed] [Google Scholar]
- Zhang, J. and Rosenberg, H.F. 2002. Complementary advantageous substitutions in the evolution of an antiviral RNase of higher primates. Proc. Natl. Acad. Sci. 99**:** 5486-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Rosenberg, H.F., and Nei, M. 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. 95**:** 3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Dyer, K.D., and Rosenberg, H.F. 2000. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl. Acad. Sci. 97**:** 4701-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollo, M., D'Urso, M., Schlessinger, D., and Chen, E.Y. 1993. Sequence of mouse glucose-6-phosphate dehydrogenase cDNA. DNA Seq. 3**:** 319-322. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://expression.gnf.org; Gene Expression Atlas.
- http://www.affymetrix.com/analysis/downloadcenter.affx; Affymetrix.
- http://www.ensembl.org/EnsMart/; EnsMart and Ensembl.
- http://www.hgmp.mrc.ac.uk/Software/EMBOSS/Apps/tranalign.html; Emboss.
- http://www.r-project.org/; The R Reference Index.