Systematic analysis and nomenclature of mammalian F-box proteins (original) (raw)
Much of the targeted protein ubiquitylation that occurs in eukaryotes is performed by cullin-based E3 ubiquitin ligases, which form a superfamily of modular E3s. The best understood cullin-based E3 is the SCF ubiquitin ligase (Feldman et al. 1997; Skowyra et al. 1997), which is composed of a modular E3 core containing CUL1 and RBX1 (also called ROC1), and a substrate specificity module composed of SKP1 and a member of the F-box family of proteins (Cardozo and Pagano 2004). The CUL1/RBX1 complex functions as a scaffold to assemble the E2 ubiquitin conjugating enzyme with the substrate specificity module (Zheng et al. 2002). CUL1 interacts with RBX1 through its C terminus and with SKP1 through its N terminus. The interaction of F-box proteins with SKP1 occurs through the F-box motif, an ∼40-amino acid motif first identified in budding yeast Cdc4p and human cyclin F, the latter giving the name to the entire family (Bai et al. 1996). F-box proteins contain additional protein interaction domains that bind ubiquitylation targets. The overall architecture of SCF complexes is conserved in the superfamily of SCF-like ubiquitin ligases that use cullin proteins as a scaffold. All cullins characterized to date (CUL1-5) are known to interact with RBX1 or RBX2 but use distinct specificity modules, which generally display structural and functional similarities with the SKP1/F-box protein module. For example, CUL2 and CUL5 are known to interact with the SKP1-like protein elongin C, which, in turn, interacts with F-box protein-like specificity factors called BC/SOCS-box proteins (Deshaies 1999; Guardavaccaro and Pagano 2003). In addition, CUL3 interacts with the BTB/POZ family of proteins, which appear to merge the functions of SKP1 and the F-box protein into a single polypeptide (Furukawa et al. 2003; Geyer et al. 2003; Pintard et al. 2003; Xu et al. 2003), with the BTB domain displaying structural relationships with SKP1 (Schulman et al. 2000; Xu et al. 2003). Cul4 forms a complex wherein DDB1/DDB2 and CSA proteins appear to function as substrate specificity modules (Groisman et al. 2003). Thus, the current expectation is that all cullin-containing ligases will share the modular nature of the original SCF family of ligases.
A major strategy employed by the SCF is the use of extended protein families as specificity factors. In 1999, we reported the identification of 47 F-box proteins in mammals (Cenciarelli et al. 1999; Winston et al. 1999). These proteins fell into three major classes, depending on the types of substrate interaction domains identified in addition to the F-box motif. The two largest classes of interaction domains are WD40 repeats (Smith et al. 1999) and leucine-rich repeats (LRRs) (Kobe and Kajava 2001). A third generic class of F-box proteins contained various other types of protein interaction domains or no recognizable domains. These classes of F-box proteins were designated FBWs, FBLs, and FBXs, respectively, followed by a numerical identifier (Cenciarelli et al. 1999; Winston et al. 1999). Paralogous genes in the same species used the same number followed by a letter (a, b,...) representing the individual genes in the paralogous group. The Human Genome Organization (HUGO) Gene Nomenclature Committee adopted a related four-letter gene nomenclature: FBXW, FBXL, and FBXO, respectively, where “O” in FBXO refers to “other” domains. Since this initial work, subsequent efforts, particularly cDNA and genomic sequencing projects, have facilitated the further identification of F-box protein-coding genes. However, the inconsistent use of nomenclature standards has greatly limited the utility of the sequence database. This inconsistency is due in part to the rapid pace of research in this area that has precluded coordination of gene names. A survey of F-box proteins in GenBank revealed several issues: (1) several different F-box protein coding genes have been given the same gene name; (2) multiple individual F-box genes have been given several different names; (3) the nomenclature used for clearly orthologous mouse and human genes is inconsistent; (4) several genes present in GenBank encode F-box proteins but are not annotated as such; (5) mRNA sequence revisions and refinement of algorithms for detection of F-box motifs have led to the removal of some genes from the F-box category; and (6) improvements in structural domain identification suggest that genes previously designated in the FBXO subclass may be more appropriately placed in the FBXL or FBXW subclasses. The need for clear communication in this field necessitates a unified nomenclature for F-box proteins.
To develop a comprehensive nomenclature for mammalian F-box proteins, we have systematically analyzed F-box proteins in the human and mouse genomes and have organized these genes in a manner that largely conforms to previous nomenclature standards, as explained below. This nomenclature has now been adopted and implemented by the HUGO Gene Nomenclature Committee. Several factors were considered in devising the most appropriate nomenclature for the future. First, genes whose symbols were approved by the nomenclature committee prior to the discovery of these genes as F-box proteins will remain as the approved symbol. Second, the previous nomenclature used letters (a, b,...) to indicate what appeared to be paralogous genes (e.g., FBXL3a and FBXL3b). However, because it is now appreciated that many F-box proteins exist as multiple splicing variants, the use of such a designation scheme has been avoided, necessitating the complete renaming of a small number of F-box proteins. Finally, mouse and human orthologs have been given the same symbols to facilitate comparative studies in the future. A detailed description of how the nomenclature changes have affected individual F-box genes is provided in the Supplemental Material.
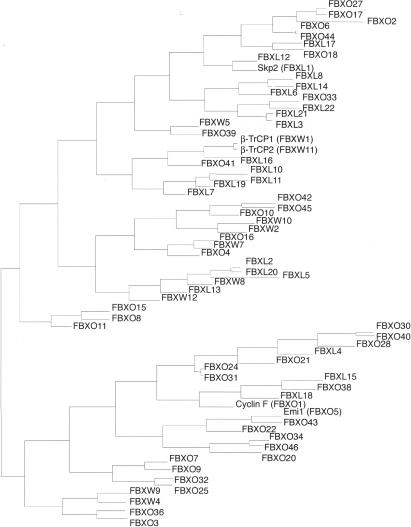
Our analysis led to the identification of 68 human and 74 mouse genes encoding recognizable F-box motifs, as detected by Hidden Markov Models (Table 1; Fig. 1) (Bateman et al. 2004; Letunic et al. 2004). A phylogenetic representation of human F-box motifs is shown in Figure 2. The phylogeny of F-box domain sequences only, which gives the cleanest available view of the evolutionary signature of the family, shows two major groups of F-box proteins (an evolutionary divergence). Different protein interaction domains are scattered throughout the two groups indicating that similar domain swapping mechanisms acted on both, but ruling out that all FBXW subfamily members diverged from a single FBXW ancestor, for example.
Table 1.
Mammalian F-box proteins
| F-box protein | Revised HUGO gene symbol | Aliases | Human Entrez gene ID | Human location | Mouse accession | % identity | Mouse location | Fly ortholog | Worm ortholog | Other domains (c) |
|---|---|---|---|---|---|---|---|---|---|---|
| FBXW1 (β-TRCP1) | BTRC | Fwd1, FBXW1A | 8945 | 10q24.32 | NM 009771 | 99 | 19C3 | slmb (CG3412) | _lin_-23 (K10B2.1) | |
| FBXW2 | FBXW2 | Fwd2, MD6 | 26190 | 9q34 | NM 013890 | 98 | 2B | |||
| FBXW4 (Dactylin) | SHFM3 | FBXW4 | 6468 | 10q24 | NM 13907 | 92 | 19C3 | |||
| FBXW5 | FBXW5 | 54461 | 9q34.3 | NM 013908 | 88 | 2A3 | CG9144 | |||
| FBXW7 | FBXW7 | FBXW6, Cdc4, Sel-10, Fbx30 | 55294 | 4q31.3 | NM 080428 | 97 | 3E3.3 | ago (CG15010) | _sel_-10 (F55B12.3) | transmembrane domain in β-isoform |
| FBXW8 | FBXW8 | Fbx29, FBXO29, Fbw6 | 26259 | 12q24.23 | NM_172721 | 71 | 5F | |||
| FBXW9 | FBXW9 | 84261 | 19p13.2 | BC043658.1 | 64 | 9 | T01E8.4 | |||
| FBXW10 | FBXW10 | C17orf1A | 10517 | 17p12 | XM 126264.2 | 62 | 11B2 | |||
| FBXW11 (β-TRCP2) | FBXW11 | Hos, FBXW1B, BTRC2, Fbx1b | 23291 | 5q35.1 | NM 134015 | 99 | 11A4 | slmb (CG3412) | _lin_-23 (K10B2.1) | |
| FBXW12 | FBXW12 | FBXO35 | 285231 | 3p21.31 | ||||||
| Fbxw13 | Fbxw13 | NM 177598 | 9F2 | |||||||
| Fbxw14 | Fbxw14 | Fbx12, Fbxo12 | NM 015793 | 9F2 | ||||||
| Fbxw15 | Fbxw15 | AK087669 | 9F2 | |||||||
| Fbxw16 | Fbxw16 | AK078661 | 9F2 | |||||||
| Fbxw17 | Fbxw17 | AAH40428 | 13A5 | |||||||
| Fbxw18 | Fbxw18 | XM 356193 | 9F2 | |||||||
| Fbwx19 | Fbxw19 | AK087808 | 9F2 | |||||||
| FBXL1 (SKP2) | SKP2 | FBXL1 | 6502 | 5p13 | NM 013787.1 | 86 | 15A2 | CG9772 | ||
| FBXL2 | FBXL2 | Fbl3 | 25827 | 3p22.3 | NM 178624.2 | 95 | 9F3 | CG9003 | C02F5.7 | |
| FBXL3 | FBXL3 | Fbl3a, FBLX3A | 26224 | 13q22 | AF 176521.1 | 93 | 14E2.3 | |||
| FBXL4 | FBXL4 | Fbl5 | 26235 | 6q16.1 | NM 172988.1 | 93 | 4A3 | CG1839 | ||
| FBXL5 | FBXL5 | Fbl4, Fir4 | 26234 | 4p15.33 | AK085100 | 88 | 5B3 | |||
| FBXL6 | FBXL6 | 26233 | 8q24.3 | NM 013909 | 75 | 15 | ||||
| FBXL7 | FBXL7 | Fbl6 | 23194 | 5p15.1 | AK129227.1 | 93 | 15B1 | CG4221 | ||
| FBXL8 | FBXL8 | 55336 | 16q22.1 | NM 015821 | 61 | 8D3 | ||||
| FBXL10 | FBXL10 | 84678 | 12q24.31 | AK129479.1 | 84 | 4A5 | DG11033 | PHD, ZF, Jmjc | ||
| FBXL11 | FBXL11 | Lilina, Fbl7 | 22992 | 11q13.1 | BC057051 | 90 | 19A | CG11033 | PHD, ZF, Jmjc | |
| FBXL12 | FBXL12 | 54850 | 19p13.2 | AF176525.1 | 93 | 9A3 | ||||
| FBXL13 | FBXL13 | 222235 | 7q22.1 | NM 177076.2 | 63 | 5A3 | ||||
| FBXL14 | FBXL14 | 144699 | 12p13.33 | AK084506.1 | 100 | 6F1 | ppa (CG9952) | |||
| FBXL15 | FBXL15 | FBXO37 | 79176 | 10q24.32 | NM 133694.1 | 87 | 19C3 | CG8873 | ||
| FBXL16 | FBXL16 | C16orf22 | 146330 | 16p13.3 | XM 128530.4 | 81 | 17A3.3 | CG32085 | ||
| FBXL17 | FBXL17 | Fbx13, FBXO13 | 64839 | 5q21.3 | XM 128716.2 | 93 | 17E1.1 | |||
| FBXL18 | FBXL18 | 80028 | 7p22.2 | Bl853840 | 89 | 5G2 | ||||
| FBXL19 | FBXL19 | 54620 | 16p11.2 | NM 172748.2 | 95 | 7F3 | PHD, ZF | |||
| FBXL20 | FBXL20 | Fbl2 | 84961 | 17q21.2 | XM 126674.3 | 99 | 11D | CG9003 | CO2F5.7 | |
| FBXL21 | FBXL21 | FBXL3B, Fbl3B | 26223 | 5q31 | AK035290 | 81 | 13B1 | |||
| FBXL22 | FBXL22 | 400380 | 15q22.1 | NM 175206 | 80 | 9C | ||||
| FBXO1 (Cyclin F) | CCNF | FBX1, FBXO1 | 899 | 16p13.3 | NM 007634.2 | 75 | 17A3.3 | cyclin box | ||
| FBXO2 | FBXO2 | Nfb42, Fbs1, Fbg1, Ocp1 | 26232 | 1p35.21 | BC027053.1 | 87 | 4E2.0 | FBA | ||
| FBXO3 | FBXO3 | FBA | 26273 | 11p13 | AK004544.2 | 92 | 2E2.0 | ApaG-like domain (SCOP) | ||
| FBXO4 | FBXO4 | 26272 | 5p12 | NM 134099.1 | 83 | 15A1 | ||||
| FBXO5 (EMI1) | FBXO5 | FBXO31 | 26271 | 6q25 | BC053434.1 | 70 | 10A1 | Rca1 (CG10800) | IBR domain | |
| FBXO6 | FBXO6 | Fbs2, Fbg2, Fbx6b | 26270 | 1p36.23 | NM 015797 | 75 | 4E2.0 | C14B1.3 | FBA domain | |
| FBXO7 | FBXO7 | Fbx | 25793 | 22q12-q13 | NM 153195.1 | 70 | 10C1 | UBL-domain (SCOP) | ||
| FBXO8 | FBXO8 | Fbs | 26269 | 4q34.1 | NM 015791.2 | 90 | 8B1.3 | Sec7 | ||
| FBXO9 | FBXO9 | Ny-ren-57 | 26268 | 6p12.3-p11.2 | AK077607.1 | 89 | 9E1.0 | CG5961 | TPR, HNHc (SCOP) | |
| FBXO10 | FBXO10 | 26267 | 9p13.1 | XM_194139.2 | 84 | 4B1 | CG9461 | K04A8.6 | CASH | |
| FBXO11 | FBXO11 | 80204 | 2p21 | XM_110248.4 | 98 | 17E5.0 | CG9461 | K04A8.6 | CASH | |
| FBXO15 | FBXO15 | 201456 | 18q22.3 | AF176530 | 60 | 18E4.0 | ||||
| FBXO16 | FBXO16 | 157574 | 8p21.1 | NM 015795.1 | 81 | 14D1 | ||||
| FBXO17 | FBXO17 | Fbg4, FBXO26 | 115290 | 19q13.2 | AF176532/NM 015796 | 80 | 7A3 | FBA | ||
| FBXO18 | FBXO18 | Fbh1 | 84893 | 10p15.1 | NM 015792 | 87 | 2A1 | Helicase | ||
| FBXO20 | LMO7 | FBXO20 | 4008 | 13q21.33 | AK129231 | 68 | 14E2.2 | CH, PDZ, Lim | ||
| FBXO21 | FBXO21 | 23014 | 12q24.23 | AB093270 | 91 | 5F | ||||
| FBXO22 | FBXO22 | 26263 | 15q23 | NP 028049 | 96 | 9B | ||||
| FBXO24 | FBXO24 | 26261 | 7q22 | XM 132440 | 84 | 5G2 | RCC1-fold (SCOP) | |||
| FBXO25 | FBXO25 | 26260 | 8p23.3 | NM 025785 | 84 | 8A1.1 | CG11658 | DY3.6 | ||
| FBXO27 | FBXO27 | Fbg5 | 126433 | 19q13.2 | AK053292 | 79 | 7A3 | FBA | ||
| FBXO28 | FBXO28 | 23219 | 1q42.12 | NM 175127 | 89 | 1H5 | CG3428 | |||
| FBXO30 | FBXO30 | 84085 | 6q24 | XM 125493 | 87 | 10A1 | ||||
| FBXO31 | FBXO31 | Fbx14, FBXO14 | 79791 | 16 | AU066822/NM 133765.2 | 95 | 8.00E+01 | |||
| FBXO32 | FBXO32 | Mafbx, Atrogin-1 | 114907 | 8q24.13 | NM_026346 | 96 | 15D1 | CG11658 | DY3.6 | |
| FBXO33 | FBXO33 | 254170 | 14q13.3 | XM_127032 | 90 | 12C1 | CG4911 | RNI-like (SCOP) | ||
| FBXO34 | FBXO34 | 55030 | 14q22.2 | NM 030236.1 | 71 | 14B | ||||
| FBXO36 | FBXO36 | 130888 | 2q37.1 | NM 025386 | 78 | 1C5 | ||||
| FBXO38 | FBXO38 | MOKA | 81545 | 5q33.1 | AK 031347 | 86 | 18E2.0 | RNI-like (SCOP) | ||
| FBXO39 | FBXO39 | 162517 | 17p13.2 | XM 282966 | 78 | 11B4 | CG2010 | RNI-like (SCOP) | ||
| FBXO40 | FBXO40 | 51725 | 3q21.1 | XM 156082 | 80 | 16A1 | TDL (SCOP) | |||
| FBXO41 | FBXO41 | 150726 | 2p13.2 | AK129466 | 80 | 6C3 | ||||
| FBXO42 | FBXO42 | 54455 | 1p36.23-p36.11 | AK028867 | 89 | 4D3 | CG6758 | Kelch repeats | ||
| FBXO43 | FBXO43 | 286151 | 8q22.3 | NM_175281 | 70 | 15B3.1 | Rca1 (CG10800) | IBR domain | ||
| FBXO44 | FBXO44 | Fbx30, FBG3, FBXO6a | 93611 | 1p36.21 | NM 173401 | 90 | 4E2.0 | C14B1.3 | FBA domain | |
| FBXO45 | FBXO45 | 20093 | 3q29 | BC026799 | 99 | 16A1 | SPRY | |||
| FBXO46 | FBXO46 | FBXO34L | 23403 | 9q13.3 | NM 175530 | 80 | 7A2 |
Figure 1.
Domain structures of mammalian F-box proteins. Domains identified by the Hidden Markov Model algorithms of SMART or PFam include F-box motif (F), WD40 repeat (WD), leucine-rich repeat (L), transmembrane domain (T), F-box-associated domain (FBA), between-ring domain (IBR), domain in carbohydrate binding proteins and sugar hydrolases (CASH), kelch repeat (K), calponin homology domain (CH), domain found in cupin metalloenzyme family (Jmjc), domain present in PSD-95, Dlg, and ZO-1 (PDZ), zinc-binding domain found in Lin-11, Isl-1, and Mec-3 (Lim), HNH nuclease family (HNHc), novel eukaryotic zinc-binding domain (CHORD), and tetratrico peptide repeat (TPR). The following domains were found via the Structural Classification of Proteins (SCOP) database, which can be used to predict protein sequences that can adopt known protein folds: ApaG-like, which is structurally similar to bacterial ApaG; Apolipophorin, the apolipophorin-III-like fold; Ubl, the ubiquitin-like fold; TDL, which is Traf-domain like; RNI-like, which may form structure similar to that of leucine-rich repeats in placental RNase inhibitor; and RCC1, which is a possible regulator of chromatin condensation-1 fold.
Figure 2.
Phylogenetic tree depiction of interrelationships between human F-box proteins. The tree is generated from the pairwise ZEGA distances (Abagyan and Batalov 1997) within the set of amino acid sequences comprising the F-box domain only by the neighbor-joining method (Saitou and Nei 1987) as adapted in ICM software (Molsoft LLC; http://www.molsoft.com).
Clear mouse orthologs were identified for all human F-box proteins except FBXW12, with the majority of mouse genes displaying >80% identity with their human counterparts (Table 1). In the mouse, _FBXW12_-related sequences have been dramatically expanded to seven genes (one at chromosome 13A5 [_Fbxw17_] and a cluster of six genes at chromosome 9F2 [_Fbxw13, Fbxw14, Fbxw15, Fbxw16, Fbxw18, Fbxw19_]). Each of these seven mouse genes is equally related to FBXW12, and, therefore, we are unable to unambiguously designate a mouse ortholog of human FBXW12. The mechanism and significance of expansion of this subclass of F-box proteins in the mouse are unknown. Three human proteins with F-box like motifs—Tome-1 (CDCA3), TBL1, and TBLR1 (TBL1XR1)—were not included because the presumptive F-box sequence did not reach the threshold sufficient for this classification.
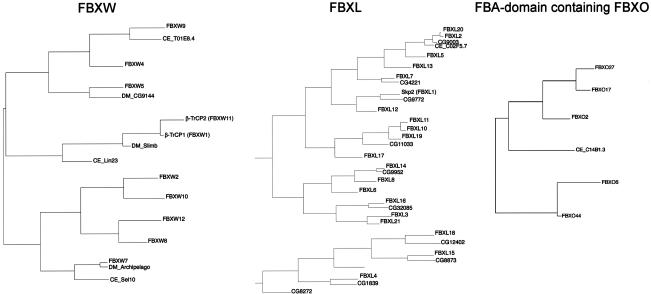
A combination of BLAST analyses and phylogenetic tree construction using putative substrate interaction domains together with the F-box motif revealed possible orthologs of mammalian F-box proteins in Drosophila melanogaster and Caenorhabditis elegans (Table 1; Fig. 3). The inclusion of substrate interaction domains allows confirmation of some relationships with the mammalian proteins (e.g., FBXL12 with SKP2), but also demonstrates, in comparison to the F-box domain only tree, that the phylogenetic spread of each subgroup is as wide as that of the whole family. Interestingly, the D. melanogaster genome contains several possible orthologs of the human FBXL series that are not found in C. elegans (Table 1; Fig. 3). The fact that C. elegans has more than 300 F-box proteins but that only a few display relationships with mammalian genes indicates significant diversification of the F-box proteins in this organism. This expansion is species-specific because the Caenorhabditis briggsae genome is predicted to encode a similar number of F-box proteins as found in human and mouse genomes (Stein et al. 2003). Six genes encoding F-box proteins appear to be conserved in C. elegans, D. melanogaster, and mammals: BTRC (FBXW1), FBXW7, FBXL2, FBXO10, FBXO25, and FBXO45 (Table 1; Fig. 3). Interestingly, in mammals four of these six genes have a paralog: FBXW1 (BTRC, β-TRCP1) for FBXW11 (β_-TRCP2_), FBXL20 for FBXL2, FBXL11 for FBXL10, and FBXO32 for FBXO25, respectively. The FBA-containing subclass of FBXO proteins are contained in the C. elegans genome but are absent in D. melanogaster (Table 1; Fig. 3). Thus, it is possible that much of the core SCF signaling common to metazoans is performed by a relatively small number of highly conserved F-box proteins. To date, conserved degradation pathways have been found for targets of mammalian FBXW7 and β-TRCP1/2 in both C. elegans and Drosophila. c-MYC and cyclin E are targeted by ago/FBXW7 in both Drosophila and mammals (Koepp et al. 2001; Moberg et al. 2001, 2004; Strohmaier et al. 2001; Tetzlaff et al. 2004; Welcker et al. 2004), and Notch is targeted by sel-10/FBXW7 in both mammals and C. elegans (Hubbard et al. 1997; Wu et al. 2001; Tetzlaff et al. 2004; Tsunematsu et al. 2004). Similarly, β-TRCP1/2/slmb has been linked to the β-catenin, IκB, and cell cycle pathways in both Drosophila and mammals (for review, see Maniatis 1999; Guardavaccaro and Pagano 2003).
Figure 3.
Phylogenetic trees for FBXW, FBXL, and FBA-domain-containing subfamilies of F-box proteins, along with orthologous sequences from D. melanogaster and C. elegans. Only the contiguous portions of the sequence corresponding to the F-box domain followed by the indicated protein interaction domain were included and aligned.
Despite the large number of mammalian F-box proteins, in addition to β-TRCP1/2 and FBW7, only one other mammalian F-box protein has been matched to its downstream substrates, namely, SKP2 (Ang and Harper 2004; Cardozo and Pagano 2004). Interestingly, SKP2 is the product of a proto-oncogene, FBW7 is a tumor suppressor (Pagano and Benmaamar 2003; Yamasaki and Pagano 2004), and overexpression of β-TRCP1 can contribute to transformation at least in some epithelial tissues (Kudo et al. 2004). Finally, EMI1/FBXO5, an inhibitor of the mitotic ubiquitin ligase APC/C, is overexpressed in tumor cell lines and certain breast tumors (Hsu et al. 2002; van 't Veer et al. 2002). Other F-box proteins appear to play a role in different diseases. For example, Dactylin/FBW4 is encoded by SHFM3, the split hand-foot malformation syndrome gene 3 (Basel et al. 2003). FBXO3 expression is increased in proliferating synovium of patients with rheumatoid arthritis (Masuda et al. 2002). FBXO32 is up-regulated during muscle atrophy (Bodine et al. 2001; Gomes et al. 2001). Thus, F-box proteins are attractive candidates for drug discovery because they play crucial roles in many important signaling pathways.
Validated protein structure prediction tools revealed inappropriately classified F-box proteins as well the association of new functional or structural domains with the F-box motif (Fig. 1). For example, certain F-box proteins previously placed in the FBXO class (e.g., FBXO13) were found to have LRRs and were reclassified accordingly (Table 1; also see Supplemental Material). FBXO14 was found to have WD40 repeats and was reclassified as FBXW12 (Table 1). Three FBXO members (FBXO33, FBXO38, and FBXO39) may display structural similarity to RNase inhibitor, the prototypical LRR, but these sequences do not reach the threshold required to be fingered as authentic LRRs based on sequence information alone (Fig. 1). Additional protein folds new to the mammalian FBX class include ubiquitin-like folds (FBXO7), TPR-like domain (FBXO9), RCC1 (FBXO24), and Kelch repeats (FBXO42). In addition to the five FBA-containing F-box proteins that bind glycosylated proteins (Cardozo and Pagano 2004), two additional proteins (FBXO10 and FBXO11) contain the CASH domain frequently found in carbohydrate-binding proteins and hydrolases (Fig. 1). Both D. melanogaster and C. elegans contain possible orthologs of FBXO10 and/or FBXO11 (Table 1). Finally, F-box proteins containing a SPRY domain (FBXO45 in mammals) are found in all metazoans. The SPRY domain is of unknown function but is frequently present in ryanodine receptors. Recent studies have linked the C. elegans SPRY domain F-box protein (C26E6.5) with presynaptic differentiation (Liao et al. 2004).
The use of this systematic nomenclature should facilitate comparative genomics and drug discovery approaches, as well as the communication of experiments designed to elaborate the functional properties of F-box proteins.
Acknowledgments
This work was supported by NIH grant AG11085 (to J.W.H. and S.J.E), and by the Department of Defense (DAMD17-01-1-0135) to J.W.H., and by NIH grants CA76584 and GM57587 to M.P. J.J. was supported by postdoctoral fellowship DAMD17-02-1-0284. S.J.E. is an investigator of the Howard Hughes Medical Institute.
Supplemental material is available at http://www.genesdev.org.
Corresponding authors.
Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1255304.
References
- Abagyan R.A. and Batalov, S.V. 1997. Do aligned sequences share the same fold? J. Mol. Biol. 273**:** 355-368. [DOI] [PubMed] [Google Scholar]
- Ang X.L. and Harper, J.W. 2004. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci. STKE 2004**:** pe31. [DOI] [PubMed] [Google Scholar]
- Bai C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86**:** 263-274. [DOI] [PubMed] [Google Scholar]
- Basel D., DePaepe, A., Kilpatrick, M.W., and Tsipouras, P. 2003. Split hand foot malformation is associated with a reduced level of Dactylin gene expression. Clin. Genet. 64**:** 350-354. [DOI] [PubMed] [Google Scholar]
- Bateman A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32**:** D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine S.C., Latres, E., Baumhueter, S., Lai, V.K., Nunez, L., Clarke, B.A., Poueymirou, W.T., Panaro, F.J., Na, E., Dharmarajan, K., et al. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294**:** 1704-1708. [DOI] [PubMed] [Google Scholar]
- Cardozo T. and Pagano, M. 2004. The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell. Biol. 5**:** 739-753. [DOI] [PubMed] [Google Scholar]
- Cenciarelli C., Chiaur, D.S., Guardavaccaro, D., Parks, W., Vidal, M., and Pagano, M. 1999. Identification of a family of human F-box proteins. Curr. Biol. 9**:** 1177-1179. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15**:** 435-467. [DOI] [PubMed] [Google Scholar]
- Feldman R.M., Correll, C.C., Kaplan, K.B., and Deshaies, R.J. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91**:** 209-219. [DOI] [PubMed] [Google Scholar]
- Furukawa M., He, Y.J., Borchers, C., and Xiong, Y. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell. Biol. 5**:** 1001-1007. [DOI] [PubMed] [Google Scholar]
- Geyer R., Wee, S., Anderson, S., Yates, J., and Wolf, D.A. 2003. BTB/POZ domain proteins are putative substrate adaptors for Cullin 3 ubiquitin ligases. Mol. Cell 12**:** 783-790. [DOI] [PubMed] [Google Scholar]
- Gomes M.D., Lecker, S.H., Jagoe, R.T., Navon, A., and Goldberg, A.L. 2001. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. 98**:** 14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A.F., Tanaka, K., and Nakatani, Y. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113**:** 357-367. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D. and Pagano, M. 2003. Oncogenic aberrations of Cullin-dependent ubiquitin ligases. Oncogene 23**:** 2037-2049. [DOI] [PubMed] [Google Scholar]
- Hsu J.Y., Reimann, J.D., Sorensen, C.S., Lukas, J., and Jackson, P.K. 2002. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nat. Cell Biol. 4**:** 358-366. [DOI] [PubMed] [Google Scholar]
- Hubbard E.J., Wu, G., Kitajewski, J., and Greenwald, I. 1997. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes & Dev. 11**:** 3182-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B. and Kajava, A.V. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11**:** 725-732. [DOI] [PubMed] [Google Scholar]
- Koepp D.M., Schaefer, L.K., Ye, X., Keyomarsi, K., Chu, C., Harper, J.W., and Elledge, S.J. 2001. Phosphorylation-dependent ubiquitination of cycin E by SCFFbw7 ubiquitin ligase. Science 294**:** 173-177. [DOI] [PubMed] [Google Scholar]
- Kudo Y., Guardavaccaro, D., Santamaria, P.G., Koyama-Nasu, R., Latres, E., Bronson, R., Yamasaki, L., and Pagano, M. 2004. Role of F-box protein βTrcp1 in mammary gland development and tumorigenesis. Mol. Cell. Biol. 24**:** 8184-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P. 2004. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32**:** D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao E.H., Hung, W., Abrams, B., and Zhen, M. 2004. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430**:** 345-350. [DOI] [PubMed] [Google Scholar]
- Maniatis T. 1999. A ubiquitin ligase complex essential for the NF-κB, Wnt/Wingless, and Hedgehog signaling pathways. Genes & Dev. 13**:** 505-510. [DOI] [PubMed] [Google Scholar]
- Masuda K., Masuda, R., Neidhart, M., Simmen, B.R., Michel, B.A., Muller-Ladner, U., Gay, R.E., and Gay, S. 2002. Molecular profile of synovial fibroblasts in rheumatoid arthritis depends on the stage of proliferation. Arthritis Res. 4**:** R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg K.H., Bell, D.W., Wahrer, D.C.R., Haber, D.A., and Hariharan, I.K. 2001. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413**:** 311-316. [DOI] [PubMed] [Google Scholar]
- Moberg K.H., Mukherjee, A., Veraksa, A., Artavanis-Tsakonas, S., and Hariharan, I.K. 2004. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr. Biol. 14**:** 965-974. [DOI] [PubMed] [Google Scholar]
- Pagano M. and Benmaamar, R. 2003. When protein destruction runs amok, malignancy is on the loose. Cancer Cell 4**:** 251-256. [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis, J.H., Willems, A., Johnson, J.L., Srayko, M., Kurz, T., Glaser, S., Mains, P.E., Tyers, M., Bowerman, B., et al. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425**:** 311-316. [DOI] [PubMed] [Google Scholar]
- Saitou N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4**:** 406-425. [DOI] [PubMed] [Google Scholar]
- Schulman B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R., Finnin, M.S., Elledge, S.J., Harper, J.W., Pagano, M., and Pavletich, N.P. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408**:** 381-386. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91**:** 209-219. [DOI] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. 1999. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24**:** 181-185. [DOI] [PubMed] [Google Scholar]
- Stein L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 1**:** E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmaier H., Spruck, C.H., Kaiser, P., Won, K.-A., Sangfelt, O., and Reed, S.I. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413**:** 316-322. [DOI] [PubMed] [Google Scholar]
- Tetzlaff M.T., Yu, W., Li, M., Zhang, P., Finegold, M., Mahon, K., Harper, J.W., Schwartz, R.J., and Elledge, S.J. 2004. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. 101**:** 3338-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu R., Nakayama, K., Oike, Y., Nishiyama, M., Ishida, N., Hatakeyama, S., Bessho, Y., Kageyama, R., Suda, T., and Nakayama, K.I. 2004. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279**:** 9417-9423. [DOI] [PubMed] [Google Scholar]
- van 't Veer L.J., Dai, H., van de Vijver, M.J., He, Y.D., Hart, A.A., Mao, M., Peterse, H.L., van der Kooy, K., Marton, M.J., Witteveen, A.T., et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415**:** 530-536. [DOI] [PubMed] [Google Scholar]
- Welcker M., Orian, A., Jin, J., Grim, J.A., Harper, J.W., Eisenman, R.N., and Clurman, B.E. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. 101**:** 9085-9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston J.T., Koepp, D.M., Zhu, C., Elledge, S.J., and Harper, J.W. 1999. A family of mammalian F-box proteins. Curr. Biol. 9**:** 1180-1182. [DOI] [PubMed] [Google Scholar]
- Wu G., Lyapina, S., Das, I., Li, J., Gurney, M., Pauley, A., Chui, I., Deshaies, R.J., and Kitajewski, J. 2001. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21**:** 7403-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.H., Vidal, M., Elledge, S.J., and Harper, J.W. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425**:** 316-321. [DOI] [PubMed] [Google Scholar]
- Yamasaki L. and Pagano, M. 2004. Cell cycle, proteolysis and cancer. Curr. Opin. Cell Biol. (in press). [DOI] [PubMed]
- Zheng N., Schulman, B.A., Song, L., Miller, J.J., Jeffrey, P.D., Wang, P., Chu, C., Koepp, D.M., Elledge, S.J., Pagano, M., et al. 2002. Structure of the Cul1-Rbx1-Skp1-F box Skp2 SCF ubiquitin ligase complex. Nature 416**:** 703-709. [DOI] [PubMed] [Google Scholar]