Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation (original) (raw)
Abstract
HIV-1 Vif (viral infectivity factor) protein overcomes the antiviral activity of the DNA deaminase APOBEC3G by targeting it for proteasomal degradation. We report here that Vif targets APOBEC3G for degradation by forming an SCF-like E3 ubiquitin ligase containing Cullin 5 and Elongins B and C (Cul5-EloB-EloC) through a novel SOCS (suppressor of cytokine signaling)-box that binds EloC. Vif binding to EloC is negatively regulated by serine phosphorylation in the BC-box motif of the SOCS-box. Vif ubiquitination is promoted by Cul5 in vitro and in vivo, and requires an intact SOCS-box. Thus, autoubiquitination of Vif occurs within the assembled Vif-Cul5 complex, analogous to F-box proteins that are autoubiquitinated within their SCF (Skp1-Cullin-F-box) complex. These findings suggest mechanisms that regulate the assembly and activity of Cul5 E3 complexes through phosphorylation or autoubiquitination of the SOCS-box protein, and identify interactions between Vif and host cell proteins that may be therapeutic targets.
Keywords: Cullin, SOCS-box, BC-box, HIV, Vif, APOBEC3G
The Vif (viral infectivity factor) protein encoded by HIV-1 and most other lentiviruses is a ∼23 kDa cytoplasmic protein that is required during virus replication to overcome the anti-viral activity of APOBEC3G (for review, see Rose et al. 2004). APOBEC3G, a member of the APOBEC family of cytidine deaminases, is a cellular DNA editing enzyme that deaminates cytidine to uracil in newly synthesized minus-strand viral DNA, inducing G-to-A hypermutation in the viral genome and triggering DNA repair pathways that lead to degradation of viral transcripts (Harris et al. 2003; Mangeat et al. 2003; Mariani et al. 2003; Zhang et al. 2003). Vif is required for replication in “nonpermissive” cells, including primary T cells, macrophages, and certain T-cell lines, but is dispensable for replication in “permissive” cell lines, such as 293T cells (Gabuzda et al. 1992; Rose et al. 2004). APOBEC3G expression is restricted to nonpermissive cells, whereas its expression in permissive cells confers a nonpermissive phenotype (Sheehy et al. 2002). Vif binds directly to APOBEC3G and targets it for degradation via the ubiquitin-proteasome pathway, thereby preventing its incorporation into virions and protecting the viral genome from mutation (Conticello et al. 2003; Marin et al. 2003; Sheehy et al. 2003; Stopak et al. 2003; Yu et al. 2003; Mehle et al. 2004).
Ubiquitination is a post-translational modification that controls the activity, localization, and proteasomal degradation of many cellular proteins (for review, see Ulrich 2002). The E1 ubiquitin activating enzyme transfers ubiquitin to an E2 ubiquitin conjugating enzyme, which together with an E3 ubiquitin ligase transfers ubiquitin to the target protein. Two major families of E3s have been identified: HECT E3s and RING E3s. HECT E3s are defined by their homology to the E6-associated protein C terminus. RING E3s contain an intrinsic zinc-binding RING domain or a tightly associated RING-finger protein that binds the E2. Here, we investigated mechanisms by which Vif targets APOBEC3G for degradation by the ubiquitin-proteasome pathway.
Results and Discussion
A novel SOCS (suppressor of cytokine signaling)-box motif in Vif directs formation of a Cul5-EloBC E3 ubiquitin ligase
We identified Vif-associated cellular proteins that mediate APOBEC3G ubiquitination and degradation by coimmunoprecipitation and mass spectrometry. Several proteins involved in ubiquitination were identified, including Cullin 5 (Cul5), Elongin B (EloB), and Elongin C (EloC) (Supplementary Fig. 1A), which are core subunits of a Skp1-Cullin-F-box (SCF)-like E3 complex (Ulrich 2002). These results confirm those of Yu et al. (2003), who reported the identification of a Vif-Cul5-EloBC-Rbx1 complex.
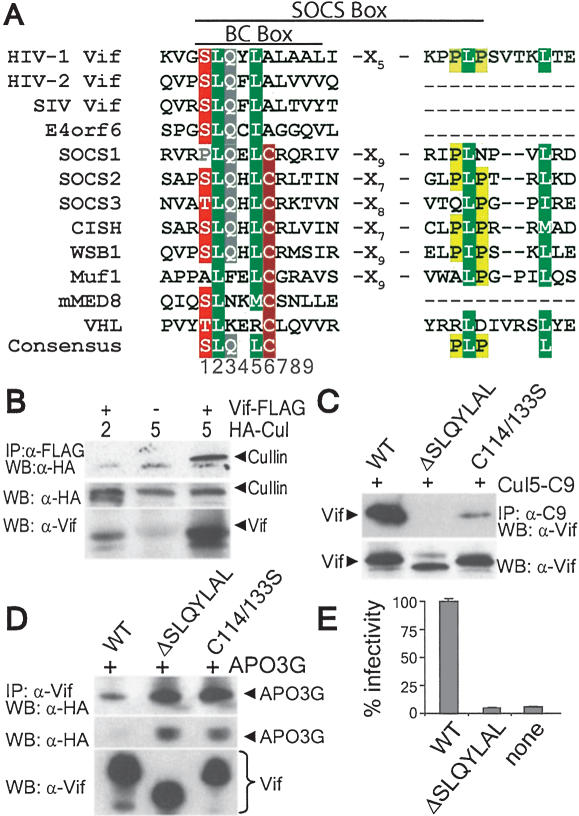
The Vif-Cul5-EloBC complex is analogous to other cullin E3s. The prototype SCF complex consists of Cul1, the RING protein Rbx1, the adaptor subunit Skp1, and an F-box protein that mediates substrate binding and specificity. For Cul5 (or Cul2), the complex contains Rbx1 and the EloB-EloC heterodimer (EloBC), which interacts with a BC-box-containing protein (Kamura et al. 1998; Zhang et al. 1999; Querido et al. 2001). The BC-box is an ∼10-amino acid degenerate sequence motif [(A/P/S/T)L-X3-C-X3-(A/I/L/V)] that mediates direct binding of BC-box proteins to EloC (Fig. 1A; Kile et al. 2002). The BC-box-containing protein mediates substrate binding and specificity, playing a role similar to that of F-box proteins. A well-studied BC-box protein is the von Hippel-Lindau (VHL) tumor suppressor, which interacts with the Cul2-EloBC complex to mediate proteasomal degradation of hypoxia inducible factor-1α (HIF-1α) (Kile et al. 2002). The crystal structure of the VHL-EloBC ternary complex shows that EloC utilizes two distinct interfaces to bind EloB and VHL, while VHL and EloB do not appear to interact (Stebbins et al. 1999). These findings suggest that Vif promotes ubiquitination of APOBEC3G by forming a Vif-Cul5-EloBC E3 ubiquitin ligase.
Figure 1.
The SOCS-box motif of Vif is required for association with the Cul5-EloBC complex and Vif-mediated degradation of APOBEC3G. (A) Sequence alignment of the BC-box and SOCS-box motifs from viral and cellular proteins that bind EloC. Conserved residues with similarity are shaded. (Ad2/5 E4orf6) Human adenovirus 2 or 5 E4orf6 protein; (SOCS) suppressor of cytokine signaling; (CISH) cytokine-inducible SH2 containing protein; (WSB1) WD repeat and SOCS-box containing protein 1; (mMED8) mammalian mediator subunit 8; (VHL) von Hippel-Lindau protein. (B) HA-Cul2 or HA-Cul5 was expressed in 293T cells in the presence or absence of Vif-Flag. (Top) Vif-associated proteins were detected by coimmunoprecipitation followed by Western blot. A nonspecific band detected below Cul5 is present in all three lanes. (Middle and bottom) Vif-Flag and Cul5 or Cul2 expression was confirmed by Western blot. (C) Cul5-C9 was coexpressed in 293T cells with wild-type (WT) or mutant Vif-Flag followed by anti-C9 immunoprecipitation. (Top) Coprecipitating Vif was detected by Western blot. (Bottom) Expression of equivalent levels of Vif was determined by anti-Vif Western blot. (D) APOBEC3G-HA and wild-type (WT) or mutant Vif-Flag were expressed in 293T cells. Vif was immunoprecipitated with anti-Flag antibody. (Top) Coprecipitating APOBEC3G was detected by anti-HA Western blot. APOBEC3G (middle) and Vif (bottom) were detected in cell lysates by Western blot. (E) Virus was produced from 293/APOBEC3G cells following transfection with a _vif-_deleted proviral plasmid and wild-type (WT) or mutant pCDNA1.Vif. Virus was normalized and infectivity was measured using the reporter cell line Cf2-luc.
EloBC can form an E3 ubiquitin ligase complex with Cul2 or Cul5 (Kile et al. 2002; Ulrich 2002). To determine whether Vif interacts selectively with Cul5, coimmunoprecipitations were performed with Vif and Cul2 or Cul5 (Fig. 1B). Cul5, but not Cul2, was coimmunoprecipitated by Vif. Interaction between Vif and EloBC was verified by coimmunoprecipitation with Vif (Supplementary Fig. 1B,C). These data suggest that Vif associates with Cul5 and EloBC to form a cullin E3 ubiquitin ligase complex.
The Vif proteins encoded by HIV-1 and the related lentiviruses HIV-2 and SIVagm contain a highly conserved region with homology to the SOCS-box motif (Fig. 1A). The SOCS-box is a conserved domain found in proteins that bind to EloBC (Fig. 1A; Kile et al. 2002). The SOCS-box consists of ∼25 to 40 amino acids and contains an N-terminal BC-box sequence and a C-terminal P/L-rich region. The BC-box contains a leucine at position 2 and cysteine at position 6 that are important for EloC binding (Kamura et al. 1998, 2001; Ohh et al. 1999; Brower et al. 2002), but the function of the downstream P/L-rich region is unclear (Kamura et al. 1998, 2001; Brower et al. 2002). The SOCS-box-like motif of Vif is similar to that of other viral and cellular proteins, containing a BC-box-like sequence, the invariable leucine at position 2, and a C-terminal PPLP motif (Fig. 1A). The BC-box-like region of HIV-1 Vif is the most highly conserved sequence among all lentivirus Vif proteins (Rose et al. 2004) and is required for viral replication (Goncalves et al. 1994). A BC-box-like motif was recently identified in the adenovirus E4orf6 protein, which also interacts with Cul5-EloBC, but whether the BC-box mediates EloC binding is unknown (Querido et al. 2001). In contrast to cellular BC-box proteins, the viral BC-box motifs of Vif and E4orf6 contain an alanine at position 6 instead of cysteine.
We performed experiments to assess whether the Vif BC-box-like motif is required for binding to the Cul5 complex using the Vif mutants ΔSLQYLAL, which deletes the BC-box motif from residues 142-154, and C114/133S, which mutates two conserved cysteine residues. These mutations were previously shown to abolish Vif-mediated enhancement of HIV infectivity in nonpermissive cells (Goncalves et al. 1994; Sheehy et al. 2003). Cul5 precipitated wild-type Vif, whereas Cul5 association was significantly decreased for Vif C114/133S and abolished for Vif ΔSLQYLAL (Fig. 1C). Thus, the BC-box is required for Cul5 association. The cysteine residues are N-terminal to the SOCS-box and do not share significant homology to other cullin-binding proteins, and thus are unlikely to be directly involved in binding. Rather, the cysteines may be important for protein conformation or other as yet unknown functions. As demonstrated previously, wild-type Vif decreased steady-state levels of APOBEC3G, whereas equivalent levels of the Vif mutants failed to reduce APOBEC3G levels (Fig. 1D, middle). Both mutants retained the ability to bind APOBEC3G (Fig. 1D), demonstrating that Vif associates independently with APOBEC3G and Cul5. Vif ΔSLQYLAL was severely impaired for the production of infectious HIV-1 in the presence of ectopically expressed APOBEC3G, with infectivity indistinguishable from that of Vif-deficient virus (Fig. 1E). These results suggest that association of Vif with Cul5 is required to counteract APOBEC3G and produce infectious virus.
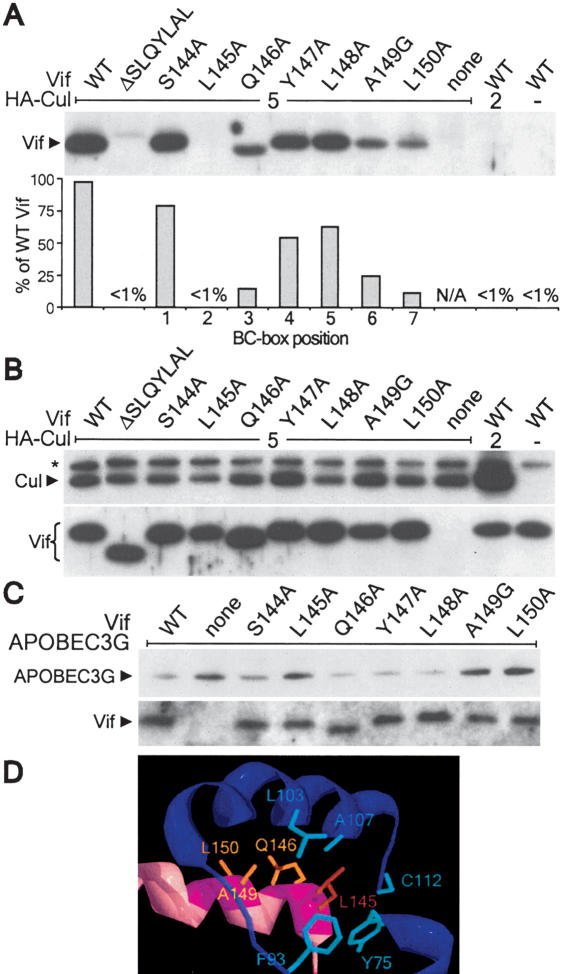
The preceding experiments demonstrate that the Vif BC-box-like motif is required to induce APOBEC3G degradation, bind the Cul5 complex, and enhance viral infectivity. The Vif BC-box-like motif contains several signature hydrophobic residues (i.e., L145, L148, and L163) that are shared with the BC-boxes of cellular proteins, including VHL, EloA, and SOCS-1 (Stebbins et al. 1999; Kile et al. 2002). To determine if Vif contains a functional BC-box, we assessed the ability of point mutants in the BC-box-like motif to interact with the Cul5 complex. In agreement with the preceding experiments (Fig. 1B,C), wild-type Vif was coprecipitated in the presence of Cul5, but not in its absence or with Cul2, and the BC-box mutant Vif ΔSLQYLAL failed to interact with Cul5 (Fig. 2A). Equivalent expression of cullin and Vif proteins was confirmed by Western blot (Fig. 2B). Mutation of the invariant L145 at position 2 in the BC-box completely disrupted binding to the Cul5 complex. Vif Q146A, A149G, and L150A exhibited an ∼75% to 85% decrease in association with Cul5 when compared with wild type. However, S144A, Y147A, and L148A were capable of coprecipitating Cul5, albeit at lower levels than wild-type Vif. Thus, Vif mutants with low or undetectable binding to the Cul5-EloBC complex (L145A, A149G, and L150A) failed to reduce APOBEC3G levels (Fig. 2C). Q146A showed decreased binding to the Cul5 complex but wild-type ability to decrease APOBEC3G levels. While glutamine is the most common residue at position 3 in cellular BC-box motifs, a high degree of sequence variability is observed at this site (Kile et al. 2002). Only one human SOCS-box protein, ASB-2, contains an alanine at position 3 [PLAHLCR]. This protein is capable of binding EloBC (Kamura et al. 1998), suggesting that an alanine at position 3 does not prevent EloC binding and raising the possibility that Vif Q146A binds EloC in the Cul5 complex with a reduced affinity that is sufficient to mediate APOBEC3G degradation, but not coprecipitation by Cul5. These findings suggest that specific amino acids in the BC-box required for binding to the Cul5 complex are also required to induce APOBEC3G degradation.
Figure 2.
Identification of critical residues in the Vif BC-box motif required for association with the Cul5-EloBC complex. (A) 293T cells were transfected with the indicated plasmids. Cullin complexes were immunoprecipitated with anti-HA and subjected to anti-Vif Western blot. Coprecipitating Vif was quantitated, normalized for expression (see B), and expressed as a percent of wild-type (WT) Vif binding. (B) Expression of similar levels of Cul5, Cul2, and Vif in transfected 293T cells used in A was confirmed by Western blot. Results for A and B are representative of three independent experiments. (C) 293T cells were transfected with pAPOBEC3G:HA and pCDNA1.Vif as indicated. APOBEC3G and Vif were detected by Western blot. (D) Homology model of the Vif:EloC interface based on the VHL-EloBC crystal structure (Stebbins et al. 1999). Secondary structural elements of EloC and Vif are colored in blue and pink, respectively. EloC amino acids that compose the hydrophobic binding pocket are highlighted in light blue and labeled. Vif amino acids predicted to be involved in EloC binding are highlighted in magenta. Vif L145 (orange) is predicted to project into the EloC hydrophobic binding pocket. Vif Q146, A149, and L150 are shown with yellow side chains.
The crystal structure of the VHL-EloBC complex reveals a binding interface between EloC and the BC-box of VHL dominated by hydrophobic interactions (Stebbins et al. 1999). The BC-box motif forms a helix and is seated on a concave binding surface presented by EloC. The most extensive contacts are made by leucine at position 2 in the BC box (VHL L158), which projects into the central binding pocket of EloC, and by arginine and cysteine at positions 5 and 6, respectively (R161 and C162). In addition, the side chains of residues at positions 3, 9, and 10 (K159, V165, and V166) as well as L169 outside the BC-box and L178 in the downstream P/L region contribute to EloC binding.
Homology modeling of the Vif BC-box using the VHL structure suggests that Vif residues required for Cul5 association cluster at the Vif-EloC interface (Fig. 2D). Residues L145, Q146, A149, and L150 are modeled on the face of the BC-box helix proximal to EloC. L145 at position 2 projects into the hydrophobic binding pocket of EloC, similar to VHL L158. Mutation of these residues disrupted or significantly impaired the association of Vif with Cul5 (Fig. 2A). Similarly, analogous mutations in the BC-box motifs of VHL, SOCS-1, Muf1, and mMED8 disrupted EloC binding (Kamura et al. 1998, 2001; Ohh et al. 1999; Brower et al. 2002). In particular, mutation of leucine at position 2 in the BC-box disrupts function for all characterized BC-box motifs, including that of Vif (Fig. 2). Mutation at A149 in Vif (position 6) also disrupts association with Cul5. This position is important in cellular BC-box proteins as well, even though cellular BC-boxes contain a conserved cysteine at this site in contrast to the conserved alanine in viral BC-boxes. Mutation of Y147 in Vif and the corresponding residue in VHL E160 (Ohh et al. 1999) did not significantly affect association with Cul5. Our results also highlight several differences between the sequence requirements of cellular BC-box proteins and Vif. Whereas Vif L150 at position 7 is important for Vif binding to EloC, mutation of the corresponding residue in VHL does not affect EloC binding (Ohh et al. 1999). Another difference is that L148 at position 5 of Vif is not essential for EloC binding, whereas the corresponding residue in VHL makes hydrophobic contacts to EloC (Stebbins et al. 1999). Thus, although functional studies and homology modeling suggest that the Vif BC-box motif shares a similar structural organization with that of VHL, it also has distinct sequence requirements.
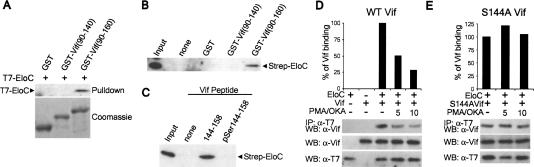
The sequences in Vif required for interaction with the Cul5 complex were further investigated by assessing the ability of recombinant GST-Vif fusion proteins to interact with EloC. We showed that purified GST-Vif(90-160), which contains the BC-box but not the downstream P/L-rich sequence, interacts with EloC expressed in 293T cells (Fig. 3A). However, neither GST nor GST-Vif(90-140), which lacks the BC-box motif, bound detectable amounts of EloC. Using purified recombinant Strep-EloC, we then showed that GST-Vif(90-160) specifically coprecipitated Strep-EloC (Fig. 3B), implying that Vif and EloC interact directly and do not require additional components of the Cul5 complex. These data further suggest that the BC-box motif of Vif, but not the P/L-rich region, is required for its interaction with EloC and the Cul5 complex.
Figure 3.
Direct binding of Vif to EloC is negatively regulated by phosphorylation. (A) Lysates were prepared from 293T cells expressing T7-EloC and incubated with recombinant GST, GST-Vif(90-140), or GST-Vif(90-160). GST proteins were recovered with glutathione-Sepharose. T7-EloC was detected by Western blot, and GST proteins were detected by staining with Coomassie Blue. (B) Recombinant Strep-EloC was incubated with GST proteins, recovered with glutathione-Sepharose, and detected by Western blot. (C) Strep-EloC was incubated with biotinylated Vif peptides (amino acids 144-158 or pSer144-158) bound to streptavidin-Sepharose. Bound Strep-EloC was detected by Western blot. (D,E) pCDNA3.T7-EloC and pCDNA1.Vif (D) or S144AVif (E) were transfected into 293T cells. Cells were stimulated with 0.2 μM PMA and 0.5 μM okadaic acid for 5 or 10 min prior to harvesting where indicated. (Top) T7-EloC was immunoprecipitated and coprecipitating Vif was detected by Western blot and quantitated. Binding was normalized to the amount of Vif present without stimulation. (Middle and bottom) Equivalent levels of expression were confirmed by Western blot of cell lysates. Results are representative of three independent experiments.
Phosphorylation of the Vif SOCS-box negatively regulates EloC binding
Vif is phosphorylated on several serine and threonine residues, which plays an important role in regulating HIV-1 replication and infectivity (Yang et al. 1996; Yang and Gabuzda 1998). However, the specific functional consequences of Vif phosphorylation are unclear. Unexpectedly, mutation of the conserved phosphorylation sites T96, S144, and T188 did not decrease the ability of Vif to induce APOBEC3G degradation (Mehle et al. 2004). S144 in the Vif BC-box is a major phosphorylation site. The S144A mutation in Vif prevents phosphorylation at this site and significantly impairs but does not abolish Vif function and viral replication (Yang et al. 1996; Yang and Gabuzda 1998). Given that the BC-box is tightly associated with EloC via hydrophobic interactions (Fig. 2D; Stebbins et al. 1999), we hypothesized that a phosphorylated BC-box might not be accommodated at the Vif-EloC interface.
We therefore investigated the possibility that binding between Vif and EloC is regulated by phosphorylation at S144 in the Vif BC-box. Binding reactions were performed with recombinant EloC and phosphorylated or nonphosphorylated peptides corresponding to the Vif BC-box (amino acids 144-158). The nonphosphorylated BC-box peptide was capable of precipitating EloC (Fig. 3C). However, the phosphorylated peptide failed to interact with EloC (Fig. 3C). We then performed experiments to determine if phosphorylation of Vif affects EloC binding in cells. We previously showed that stimulation of 293T cells with PMA and okadaic acid (OKA) results in phosphorylation of Vif at S144, in addition to T155 and T188 (Yang et al. 1996). EloC and wild-type or S144A Vif were coexpressed in 293T cells treated with PMA/OKA for 0, 5, or 10 min to induce Vif phosphorylation, followed by cell lysis and immunoprecipitation of EloC (Fig. 3D). Stimulation for 5 or 10 min decreased coprecipitation of Vif by EloC by ∼50% and 75%, respectively, compared with unstimulated cells (Fig. 3D). In contrast, similar levels of S144A were precipitated by EloC in the presence or absence of PMA/OKA treatment (Fig. 3E). These findings suggest that phosphorylation at S144 negatively regulates binding of the Vif BC-box to EloC. Phosphorylation of the cellular SOCS-box proteins SOCS1 and SOCS3 disrupts their association with EloC, but neither of these proteins are known to be phosphorylated in the BC-box motif (Chen et al. 2002; Haan et al. 2003). Whereas nonphosphorylated SOCS3 can bind EloC, phosphorylated SOCS3 is selectively recognized by p120 RasGAP, Nck, and Crk-L, activating downstream signaling pathways (Cacalano et al. 2001; Sitko et al. 2004). These findings raise the possibility that phosphorylation of Vif, in addition to negatively regulating the association with EloC, may also regulate another as yet unknown function that is important for HIV replication. For example, by analogy with SOCS3, phosphorylated Vif might be selectively recognized by an as yet unknown cellular protein. Alternatively, Vif phosphorylation may alter its subcellular localization, as is the case for many phosphoproteins (Blenis and Resh 1993), leading to APOBEC3G sequestration. Such alternative functions of Vif could interfere with the antiviral activity of APOBEC3G without inducing its degradation. However, we cannot exclude the possibility that the S144A mutant contributes significantly to APOBEC3G degradation in a manner that only becomes obvious after multiple rounds of infection.
Vif is autoubiquitinated within the Cul5 complex
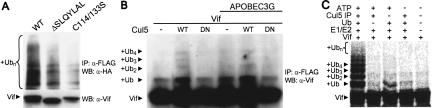
In addition to phosphorylation, Vif is post-translationally modified by ubiquitin (Mehle et al. 2004). The SOCS-box proteins SOCS3 and CIS are also ubiquitinated and rapidly degraded (Verdier et al. 1998; Zhang et al. 1999), and the F-box proteins Grr1 and Cdc4 are autoubiquitinated within their SCF complex (Zhou and Howley 1998; Galan and Peter 1999). Although Yu et al. showed that Cul5 is the E3 for APOBEC3G (Yu et al. 2003), it is unclear whether Vif is also ubiquitinated by Cul5. Therefore, we performed experiments to determine if Cul5 can ubiquitinate Vif. Vif mutants ΔSLQYLAL and C114/133S are defective for binding to the Cul5 complex (Fig. 1C). Wild-type and mutant Vif were expressed at high levels to permit detection of polyubiquitinated forms in the absence of APOBEC3G. The ubiquitination of these mutants was reduced when compared with wild type (Fig. 4A), suggesting that association with the Cul5 complex is important for Vif ubiquitination. Furthermore, overexpression of Cul5 enhanced both mono- and polyubiquitination of Vif, whereas overexpression of the dominant negative Cul5ΔNedd8 did not (Fig. 4B). Overexpression of Cul5ΔNedd8 also prevented the increase in Vif ubiquitination associated with APOBEC3G coexpression. The small amounts of ubiquitinated Vif observed in the presence of Cul5ΔNedd8 may result from the activity of endogenous wild-type Cul5 or another E3. Immunopurified Cul5 was capable of directing the in vitro ubiquitination of Vif (Fig. 4C). The ubiquitination of Vif was specific to Cul5, as omitting ubiquitin, ATP, or Cul5 prevented ubiquitination. These results suggest that the target, APOBEC3G, and the specificity-determining factor, Vif, are ubiquitinated by the same E3, Cul5-EloBC. This is similar to the F-box proteins Grr1 and Cdc4, which can be ubiquitinated and degraded by their own SCF complex (Zhou and Howley 1998; Galan and Peter 1999). Thus, the degradation of SOCS-box proteins by their cullin complexes may permit dynamic substrate recognition by initiating a rapid exchange of the recognition module within the E3 complex.
Figure 4.
Cul5 is an E3 ubiquitin ligase for Vif. (A) Wild-type (WT) and mutant Vif-Flag were coexpressed with HA-ubiquitin in 293T cells. (Top) Ubiquitinated Vif was detected by anti-Flag immunoprecipitation followed by anti-HA Western blot. (Bottom) Vif was detected by Western blot. (B) Vif was expressed in 293T cells in the presence of HA-Cul5, HA-Cul5ΔNedd8 (DN), and APOBEC3G-HA as indicated. Ubiquitinated Vif was detected by anti-Flag immunoprecipitation and anti-Vif Western blot. Ubiquitinated species are indicated. (C) Cul5 was immunoprecipitated from HeLa cells and used as a source of E3 ligase for in vitro ubiquitination reactions with 35S-labeled Vif prepared by in vitro translation, E1, E2 (UbcH5a), ubiquitin, and ATP as indicated. Reaction products were separated by SDS-PAGE and detected by PhosphorImaging. Vif and ubiquitinated species are identified.
In conclusion, we have demonstrated that Vif is a novel SOCS-box protein that recruits APOBEC3G to a Cul5-EloBC E3 for ubiquitination and degradation. Vif functions like an F-box protein, both binding to the Cul5 complex and recognizing specific targets for ubiquitination. Our findings demonstrate that phosphorylation in the BC-box negatively regulates Vif binding to EloC. Thus, phosphorylation can regulate not only E3 target recognition, as is the case for many F-box protein targets (Ulrich 2002), but also assembly of the SCF complex itself (Chen et al. 2002; Haan et al. 2003). The finding that phosphorylation of Vif is important for productive HIV-1 infection but not APOBEC3G degradation suggests that like other SOCS-box proteins (Cacalano et al. 2001; Sitko et al. 2004), Vif may be a multifunctional protein. Vif is ubiquitinated by the same E3 as its target APOBEC3G, similar to F-box proteins that are autoubiquitinated within their own SCF complex (Zhou and Howley 1998; Galan and Peter 1999). Thus, our studies suggest mechanisms that can regulate Cul5 E3 function in response to different physiological conditions through phosphorylation or autoubiquitination of the SOCS-box protein that mediates substrate binding and specificity. Understanding the molecular mechanisms of Vif function will provide new insights into the regulation and assembly of cullin E3 complexes and may also identify new targets for therapeutic intervention.
Materials and methods
Transfections, immunoprecipitations, infections, and in vitro ubiquitination assays followed standard and published protocols, and are detailed in the Materials and Methods section in the Supplemental Material.
Acknowledgments
We thank M. Fay, J. Conaway, M. Malim, K. Strebel, H. Gottlinger, W. Kaelin, and X. Yu for reagents, and H. Wilson for technical assistance. A.M. was supported in part by a NSF Predoctoral Fellowship. This work was supported by NIH grants AI36186 and AI62555 and a grant from AmfAR. J.G. and M.S.M. were supported by PSIDA/MGI/49729/2003. Core facilities were supported by Center for AIDS Research and Cancer Center grants.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1249904.
References
- Blenis J. and Resh, M.D. 1993. Subcellular localization specified by protein acylation and phosphorylation. Curr. Opin. Cell Biol. 5**:** 984-989. [DOI] [PubMed] [Google Scholar]
- Brower C.S., Sato, S., Tomomori-Sato, C., Kamura, T., Pause, A., Stearman, R., Klausner, R.D., Malik, S., Lane, W.S., Sorokina, I. et al. 2002. Mammalian mediator subunit mMED8 is an Elongin BC-interacting protein that can assemble with Cul2 and Rbx1 to reconstitute a ubiquitin ligase. Proc. Natl. Acad. Sci. 99**:** 10353-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacalano N.A., Sanden, D., and Johnston, J.A. 2001. Tyrosine-phosphorylated SOCS-3 inhibits STAT activation but binds to p120 RasGAP and activates Ras. Nat. Cell Biol. 3**:** 460-465. [DOI] [PubMed] [Google Scholar]
- Chen X.P., Losman, J.A., Cowan, S., Donahue, E., Fay, S., Vuong, B.Q., Nawijn, M.C., Capece, D., Cohan, V.L., and Rothman, P. 2002. Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc. Natl. Acad. Sci. 99**:** 2175-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello S.G., Harris, R.S., and Neuberger, M.S. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13**:** 2009-2013. [DOI] [PubMed] [Google Scholar]
- Gabuzda D.H., Lawrence, K., Langhoff, E., Terwilliger, E., Dorfman, T., Haseltine, W.A., and Sodroski, J. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66**:** 6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.-M. and Peter, M. 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. 96**:** 9124-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves J., Jallepalli, P., and Gabuzda, D.H. 1994. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J. Virol. 68**:** 704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan S., Ferguson, P., Sommer, U., Hiremath, M., McVicar, D.W., Heinrich, P.C., Johnston, J.A., and Cacalano, N.A. 2003. Tyrosine phosphorylation disrupts elongin interaction and accelerates SOCS3 degradation. J. Biol. Chem. 278**:** 31972-31979. [DOI] [PubMed] [Google Scholar]
- Harris R.S., Bishop, K.N., Sheehy, A.M., Craig, H.M., Petersen-Mahrt, S.K., Watt, I.N., Neuberger, M.S., and Malim, M.H. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113**:** 803-809. [DOI] [PubMed] [Google Scholar]
- Kamura T., Sato, S., Haque, D., Liu, L., Kaelin Jr., W.G., Conaway, R.C., and Conaway, J.W. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes & Dev. 12**:** 3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T., Burian, D., Yan, Q., Schmidt, S.L., Lane, W.S., Querido, E., Branton, P.E., Shilatifard, A., Conaway, R.C., and Conaway, J.W. 2001. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276**:** 29748-29753. [DOI] [PubMed] [Google Scholar]
- Kile B.T., Schulman, B.A., Alexander, W.S., Nicola, N.A., Martin, H.M., and Hilton, D.J. 2002. The SOCS box: A tale of destruction and degradation. Trends Biochem. Sci. 27**:** 235-241. [DOI] [PubMed] [Google Scholar]
- Mangeat B., Turelli, P., Caron, G., Friedli, M., Perrin, L., and Trono, D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424**:** 99-103. [DOI] [PubMed] [Google Scholar]
- Mariani R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H., and Landau, N.R. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114**:** 21-31. [DOI] [PubMed] [Google Scholar]
- Marin M., Rose, K.M., Kozak, S.L., and Kabat, D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9**:** 1398-1403. [DOI] [PubMed] [Google Scholar]
- Mehle A., Strack, B., Ancuta, P., Zhang, C., McPike, M., and Gabuzda, D. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279**:** 7792-7798. [DOI] [PubMed] [Google Scholar]
- Ohh M., Takagi, Y., Aso, T., Stebbins, C.E., Pavletich, N.P., Zbar, B., Conaway, R.C., Conaway, J.W., and Kaelin Jr., W.G. 1999. Synthetic peptides define critical contacts between elongin C, elongin B, and the von Hippel-Lindau protein. J. Clin. Invest. 104**:** 1583-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E., Blanchette, P., Yan, Q., Kamura, T., Morrison, M., Boivin, D., Kaelin, W.G., Conaway, R.C., Conaway, J.W., and Branton, P.E. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes & Dev. 15**:** 3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K.M., Marin, M., Kozak, S.L., and Kabat, D. 2004. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10**:** 291-297. [DOI] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis, N.C., Choi, J.D., and Malim, M.H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418**:** 646-650. [DOI] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis, N.C., and Malim, M.H. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9**:** 1404-1407. [DOI] [PubMed] [Google Scholar]
- Sitko J.C., Guevara, C.I., and Cacalano, N.A. 2004. Tyrosine phosphorylated SOCS3 interacts with the Nck and Crk-L adapter proteins and regulates Nck activation. J. Biol. Chem. 279**:** 37662-37669. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E., Kaelin Jr., W.G., and Pavletich, N.P. 1999. Structure of the VHL-ElonginC-ElonginB complex: Implications for VHL tumor suppressor function. Science 284**:** 455-461. [DOI] [PubMed] [Google Scholar]
- Stopak K., de Noronha, C., Yonemoto, W., and Greene, W.C. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 12**:** 591-601. [DOI] [PubMed] [Google Scholar]
- Ulrich H.D. 2002. Natural substrates of the proteasome and their recognition by the ubiquitin system. Curr. Top. Microbiol. Immunol. 268**:** 137-174. [DOI] [PubMed] [Google Scholar]
- Verdier F., Chretien, S., Muller, O., Varlet, P., Yoshimura, A., Gisselbrecht, S., Lacombe, C., and Mayeux, P. 1998. Proteasomes regulate erythropoietin receptor and signal transducer and activator of transcription 5 (STAT5) activation. Possible involvement of the ubiquitinated cis protein. J. Biol. Chem. 273**:** 28185-28190. [DOI] [PubMed] [Google Scholar]
- Yang X. and Gabuzda, D. 1998. Mitogen-activated protein kinase phosphorylates and regulates the HIV-1 Vif protein. J. Biol. Chem. 273**:** 29879-29887. [DOI] [PubMed] [Google Scholar]
- Yang X., Goncalves, J., and Gabuzda, D. 1996. Phosphorylation of Vif and its role in HIV-1 replication. J. Biol. Chem. 271**:** 10121-10129. [DOI] [PubMed] [Google Scholar]
- Yu X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P., and Yu, X.F. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302**:** 1056-1060. [DOI] [PubMed] [Google Scholar]
- Zhang J.-G., Farley, A., Nicholson, S.E., Willson, T.A., Zugaro, L.M., Simpson, R.J., Moritz, R.L., Cary, D., Richardson, R., Hausmann, G. et al. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. 96**:** 2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Yang, B., Pomerantz, R.J., Zhang, C., Arunachalam, S.C., and Gao, L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424**:** 94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. and Howley, P.M. 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell. 2**:** 571-580. [DOI] [PubMed] [Google Scholar]