Streamlined Regulation and Gene Loss as Adaptive Mechanisms in Prochlorococcus for Optimized Nitrogen Utilization in Oligotrophic Environments (original) (raw)
Abstract
Prochlorococcus is one of the dominant cyanobacteria and a key primary producer in oligotrophic intertropical oceans. Here we present an overview of the pathways of nitrogen assimilation in Prochlorococcus, which have been significantly modified in these microorganisms for adaptation to the natural limitations of their habitats, leading to the appearance of different ecotypes lacking key enzymes, such as nitrate reductase, nitrite reductase, or urease, and to the simplification of the metabolic regulation systems. The only nitrogen source utilizable by all studied isolates is ammonia, which is incorporated into glutamate by glutamine synthetase. However, this enzyme shows unusual regulatory features, although its structural and kinetic features are unchanged. Similarly, urease activities remain fairly constant under different conditions. The signal transduction protein PII is apparently not phosphorylated in Prochlorococcus, despite its conserved amino acid sequence. The genes amt1 and ntcA (coding for an ammonium transporter and a global nitrogen regulator, respectively) show noncorrelated expression in Prochlorococcus under nitrogen stress; furthermore, high rates of organic nitrogen uptake have been observed. All of these unusual features could provide a physiological basis for the predominance of Prochlorococcus over Synechococcus in oligotrophic oceans.
INTRODUCTION
Cyanobacteria are one of the oldest groups of organisms on Earth (69). They have colonized almost every available niche for the last 3.5 billion years, demonstrating an outstanding ability for adaptation to extremely different habitats. Thus, it was somehow not surprising to discover abundant cyanobacteria (27) (namely, Synechococcus [78] and Prochlorococcus [6]) thriving in environments with a very poor nutrient supply, such as the vast intertropical gyres of the oceans (4, 28, 83), which were previously considered to be almost empty of living cells. However, their ecological importance was truly unexpected, since it is currently accepted that about half of the global primary production occurs in the oceans (81) and that marine cyanobacteria contribute two-thirds of it (16, 35). Since the discovery of Prochlorococcus in 1988 (6), its major importance in the ecology of the oceans has become evident, leading to a large number of studies (59) and to the sequencing of the genomes of five representative ecotypes, MED4, MIT9313, MIT9312, and NATL2A (http://www.jgi.doe.gov/JGI_microbial/html/index.html) and SS120 (http://www.sb-roscoff.fr/Phyto/ProSS120/), making Prochlorococcus one of the most-studied microorganisms from a genomic point of view (8, 24, 65).
The striking ecological success of Prochlorococcus has been the subject of many studies, focused on some of the most intriguing abilities of this organism, e.g., to live at depths of up to >200 m. This means that Prochlorococcus has to cope with the natural gradients of different parameters occurring along the water column, including light irradiance, which decreases almost 4 orders of magnitude from the ocean surface to the end of the euphotic zone. These conditions have induced the occurrence of a number of peculiarities and a remarkable diversity in the photosynthetic apparatus of Prochlorococcus (for recent reviews, see references 58 and 73), enabling these microorganisms to efficiently harvest very low levels of light energy. These are some of the keys to the ubiquity and abundance of Prochlorococcus in the oceans (57). However, there also exist other less obvious, but not less important, gradients that could provoke similar adaptive modifications in metabolic pathways, including gradients of temperature, pressure, UV light penetration, or nutrient availability. Nitrogen is one of the key environmental factors in the ocean (29, 83), and nitrogen gradients have been proposed as one of the main forces driving the evolution of Prochlorococcus (36, 47, 73). This review focuses on the adaptive features of the nitrogen assimilatory pathway of Prochlorococcus, with special emphasis on a comparative analysis with other cyanobacteria (in particular, the coexistent marine cyanobacterium Synechococcus) (67, 78).
ADAPTATION TOWARD THE ASSIMILATION OF SELECTED NITROGEN SOURCES AND SIMPLIFICATION OF THE REGULATORY NETWORKS
Cyanobacteria can use a wide variety of nitrogen sources available in nature, including molecular nitrogen, nitrate, nitrite, ammonium, urea, and some amino acids, such as arginine or glutamine (12). The oxidation state of these molecules has direct consequences for the energy required for their assimilation, as the most oxidized forms of nitrogen (molecular nitrogen or nitrate) are rather expensive to utilize (Table 1). The enormous amount of ATP necessary for the breakdown of molecular nitrogen (16 ATPs per N2) explains why nitrogenase is far from ubiquitous in nature and, particularly, its absence in Prochlorococcus. Also, a comparison between the full reduction of nitrate to glutamate versus ammonium to glutamate (Table 1) shows that one ATP is necessary in both cases but that nitrate reduction requires fivefold more electrons than ammonium reduction (i.e., 10 electrons for nitrate reduction to glutamate versus 2 electrons for ammonium reduction to glutamate), making nitrate reduction much more expensive in bioenergetic terms. We can make a simple calculation, based on the estimations proposed by Losada et al. (37). Let us assume that all reducing power from cells is consumed by carbon and nitrogen reduction. If we consider that the C/N ratio in nutrient-replete Prochlorococcus cells is ca. 5 to 10 (2, 21) and that reduction of the carbon atom from CO2 to (CH2O) requires four electrons (37) while the reduction of the nitrogen atom from NO3− to glutamate requires 10 electrons (37), then a hypothetical Prochlorococcus strain utilizing nitrate would expend 20 to 33% of the total reducing power in nitrogen assimilation. If we then consider that Prochlorococcus switches to utilizing only ammonium as a nitrogen source, the reducing power expense in ammonium would be decreased by a factor of 5, i.e., to 4 to 6.6%, liberating the remaining (18 to 26.4%) for other physiological needs. This rough estimation shows the importance of preferential utilization of ammonium when it is available in the environment. Furthermore, if we take into account that nitrate is most abundant at greater depths, where there is a strong energy limitation due to low light penetration, it is clear that avoiding the assimilation of nitrate could be essential to optimize energy utilization in order to fulfill all metabolic requirements.
TABLE 1.
Bioenergetics of nitrogen source assimilation
| Enzyme(s) | Gene(s) | Step | No. of electrons | No. of ATP molecules |
|---|---|---|---|---|
| Nitrogenase | nif cluster | N2 → NH4+ | 8 | 16 |
| Nitrate reductase | narB | NO3− → NO2− | 2 | 0 |
| Nitrite reductase | nirA | NO2− → NH3+ | 6 | 0 |
| Glutamine synthetase/ glutamate synthase | glnA/gltB | NH3+ → Glu | 2 | 1 |
Consequently, ammonium is the preferred nitrogen source for most cyanobacteria. Furthermore, the regulatory mechanisms of nitrogen assimilation in these microorganisms are usually built upon the basis of detecting the presence or absence of ammonium in the environment. This complex regulatory network includes a transcriptional factor, the global nitrogen regulator, NtcA (22), and the signal transduction protein PII (72), which coordinates the metabolism of nitrogen and carbon in the cell.
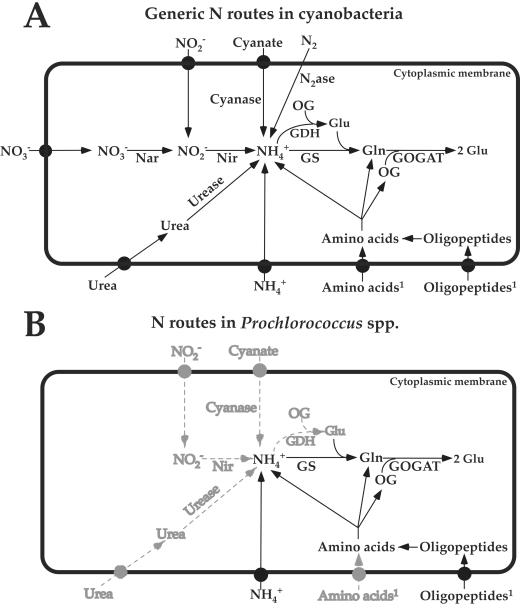
Although some cyanobacteria are able to fix molecular nitrogen, the central nitrogen assimilatory pathway, which is common for most of these microorganisms, is composed of the nitrate reductase (Nar), nitrite reductase (Nir), and glutamine synthetase/glutamate synthase cycle (GS/GOGAT) (Fig. 1A).
FIG. 1.
Comparison between the generic nitrogen assimilation routes in cyanobacteria (A) and the modifications observed in different isolates of Prochlorococcus (B) (12). Black circles represent specific transporters. Gray indicates nitrogen sources or pathways not utilized by all Prochlorococcus strains. Superscript 1 indicates that amino acid uptake by Prochlorococcus has been shown in the field (85), but otherwise utilization of amino acids, oligopeptides, and cyanate is unknown, although it is strongly suggested by genomic analysis.
Some unicellular marine cyanobacteria (assigned to the genera Synechococcus [45] and Synechocystis [84]) perform nitrogen fixation in the oceans. However, no Prochlorococcus isolate has been shown to utilize molecular nitrogen to date, in good agreement with the lack of nif genes (involved in nitrogen fixation) in the studied Prochlorococcus genomes (8, 65). This suggests that the evolutionary constraints that affect the genome size (70) and the low light energy available at depth in the oceans (14) prevented Prochlorococcus from utilizing this ubiquitous (4), but very expensive, nitrogen source.
The first unusual and probably most surprising traits of nitrogen assimilation in all studied strains of Prochlorococcus are their inability to utilize nitrate (9, 36, 47, 56, 63) and the fact that only some low-light-adapted isolates grow on nitrite (47), while most coexistent Synechococcus strains can assimilate both nitrogen sources (7, 13, 47, 67). Physiological studies have shown the lack of nitrate reductase in both high- and low-irradiance-adapted Prochlorococcus strains (36). Furthermore, in nonaxenic Prochlorococcus cultures transferred to media containing nitrate as the sole nitrogen source, it has been observed that no reduced nitrate (either nitrite or ammonium) was transferred from the heterotrophic contaminant bacteria to Prochlorococcus (36). Genomic analysis of strains MED4 (equivalent to PCC 9511 [63]), MIT9313, and SS120 confirmed the absence of several genes required for nitrate assimilation (8, 47, 65). This is an apparent ecological contradiction, as nitrate is considered to be the main nitrogen source at depth in the oceans (4), where Prochlorococcus is abundant. In the lower parts of the euphotic zone, however, the available light is very limited, and amino-acid-like molecules could provide less expensive reduced nitrogen forms to Prochlorococcus (20). Recent observations showing the higher rate of organic nitrogen compound uptake by Prochlorococcus than by Synechococcus (85) fit nicely with this hypothesis. On the other hand, the evolutionary pressure probably induced a fine equilibrium between nutrient and energy requirements. Since the genome of Prochlorococcus is subjected to a process of compaction with deletion of nonessential genes (24, 70), the lack of nitrate utilization could provide selective advantages in ocean niches where there are more convenient nitrogen sources available.
Oceanographic studies in the field could, however, widen this picture: since its discovery, it was noted that maximal abundances of Prochlorococcus occurred slightly above the nitracline (6, 50, 75). Besides, addition of nitrate stimulated Prochlorococcus cell cycling in the Mediterranean Sea (75), and nitrogen enrichment provoked an increase in Prochlorococcus abundance in the North Atlantic (18). Moreover, there are preliminary studies suggesting the possible occurrence of some Prochlorococcus isolates utilizing nitrate (47, 82). Therefore, the existence of specific Prochlorococcus ecotypes that are capable of nitrate assimilation seems rather probable. This would be consistent with the currently accepted model of Prochlorococcus as a recent organism in evolutionary terms, derived from _Synechococcus_-like ancestors adapted to iron-depleted areas of the oceans (31, 73) in a progressive process of genome reduction (24, 70), including the described losses of phycoerythrin (23, 24, 62) and nitrate/nitrite reductases (8, 36, 47, 65) in specific strains. Whatever the case, Prochlorococcus remains to date the only described genus of cyanobacteria in which all of the studied strains are unable to grow on nitrate as the sole nitrogen source (36, 47).
The utilization of organic nitrogen compounds by Prochlorococcus has been hypothesized (36) to explain the lack of nitrate assimilation at depths where nitrate is abundant. Only very recently, however, flow cytometry uptake studies in the field with radiolabeled methionine provided the proof of amino acid uptake by Prochlorococcus (85). Nevertheless, although this observation is consistent with the presence of putative amino acid transporters in the genomes of these microorganisms (8, 65), preliminary results obtained with Prochlorococcus in laboratory cultures suggest that differential amino acid utilization could also occur between different ecotypes (O. Rangel, J. Diez, and J. M. García-Fernández, unpublished results). For example, strain PCC 9511 does not utilize arginine, glutamine, or glutamate for growth, and addition of glycine, proline, glutamine, arginine, tryptophan, or methionine has no beneficial effect on cell growth (63). Interestingly, the rate uptake of organic nitrogen compounds in the field by Prochlorococcus is 10-fold higher than that by marine Synechococcus (85), possibly explaining the dominance of Prochlorococcus in oligotrophic oceans. That study also showed that one-third of the amino acid pool was consumed by Prochlorococcus, which could obtain as much as 10% of its total nitrogen requirements solely from dissolved amino acids (85). Little is known about the actual molecular structure of the total organic matter found at depth in the oceans, but recent methods allowed the demonstration of nonselective preservation of organic matter in sinking organic particles at depth (20). For example, in water samples from the equatorial Pacific and the Arabian Sea, where high Prochlorococcus abundances have been reported (57), the fraction attributed to amino acid-like molecules varied from 11 to 15% of the total organic matter (20). These results further support the hypothesis of organic nitrogen molecules as important nitrogen sources at depth in oligotrophic oceans and provide a possible explanation for the occurrence of low-irradiance Prochlorococcus ecotypes that are unable to utilize either nitrate or nitrite, such as strains SS120 and MIT 9211 (36, 47).
Ammonium is the only nitrogen source so far reported to be utilizable by all Prochlorococcus isolates (36, 47, 63). The central position of this nitrogen source in the N assimilatory pathways in cyanobacteria (Fig. 1) explains the pivotal role of glutamine synthetase (GS) in the regulation of nitrogen assimilation in these photosynthetic organisms. Accordingly, the gene glnA, encoding GS, is found in all available genomes (8, 10, 65). Interestingly, the amino acid sequence, isoelectric point, molecular size, and kinetic parameters of GS from Prochlorococcus have been shown to be very similar to those of the enzymes from other cyanobacteria, either freshwater or marine strains (10), indicating very slight modifications of the properties of this enzyme during evolution. Regulation of GS by oxidative modification in Prochlorococcus has also been shown to occur (17), in a process that induces the inactivation and subsequent degradation of the enzyme. Catalase and peroxidase, but not superoxide dismutase, effectively protected GS against inactivation, suggesting the mediation of hydrogen peroxide. This regulatory mechanism is common to other enteric bacteria (32), cyanobacteria (40, 41), green algae (25), and higher plants (53). It has been proposed to be involved in the response to oxidative stress produced by specific conditions in the metabolism (53).
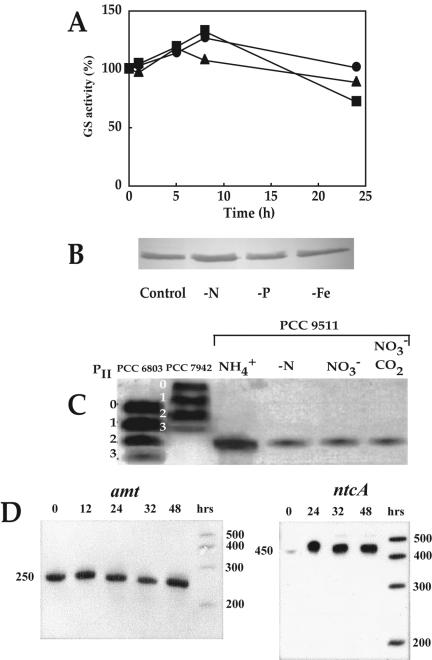
However, when the physiological regulation of this enzyme in the axenic strain Prochlorococcus sp. strain PCC 9511 was studied in detail (9, 10), very unusual regulatory features were found (Fig. 2A and B): GS was not upregulated under conditions of nitrogen starvation (9, 10) and was not inactivated in darkness (9), although both are fairly standard responses observed in other cyanobacteria (11, 12). Preliminary results on the expression of glnA in the same strain confirm these observations (S. El Alaoui, A. López-Lozano, J. Diez, and J. M. García-Fernández, unpublished data). These differences cannot be explained on the basis of structural changes of the enzyme (10) but may rely on modifications of the regulatory networks of nitrogen assimilation. The advantages of having a simplified regulatory system of regulation for a key enzyme like GS might be rooted in the natural environment where Prochlorococcus is most abundant: large oligotrophic intertropical areas of the oceans, without abrupt changes in the kind or concentration of nitrogen sources at a given depth. Under these conditions, the maintenance of a sophisticated and costly regulatory machinery to finely tune the assimilation of ammonium could simply represent an unjustified energetic expense for an organism like Prochlorococcus. In this view, the evolution could have favored an inexpensive, streamlined regulatory network, leading to the observed lack of response of GS to changes in key parameters (such as lack of light and/or nitrogen) whose detection is crucial to other cyanobacteria inhabiting more changing environments. This hypothesis is also supported by the similarly simple behavior of other nitrogen-related regulatory proteins (PII [56] or NtcA [33]) (Fig. 2C and D), enzymes (urease [55]), or transporters (the ammonium transporter encoded by amt1 [33]) (Fig. 2D).
FIG. 2.
Regulatory responses of nitrogen metabolism in Prochlorococcus. (A) Time course of glutamine synthetase activities in Prochlorococcus strain PCC 9511. Squares, control (illuminated culture growing on ammonium); circles, culture subjected to darkness; triangles, cultures subjected to nitrogen starvation. Data are taken from reference 9. (B) Western blotting of Prochlorococcus strain PCC 9511 extracts from cells subjected for 312 h to standard conditions (control) or to starvation in nitrogen, phosphorus, or iron, using antibodies against GS from Synechocystis strain PCC 6803. Reprinted from reference 10 with permission. (C) Western blotting of Prochlorococcus strain PCC 9511 extracts subjected to different N and C conditions, using PII antibodies against PII from Synechococcus strain PCC 7942. The levels of phosphorylation (groups designated 0 to 3) are indicated on the left, using cyanobacterial strains showing this kind of modification. Reprinted from reference 56 with permission. (D) Expression of the ammonium transporter amt1 and the transcriptional regulator ntcA in Prochlorococcus strain PCC 9511 cultures subjected to nitrogen starvation, as determined by RNase protection assay. The times after transfer to no nitrogen medium are indicated. Reprinted from reference 33 with permission.
The ammonium transporter amt1 is highly expressed in Prochlorococcus strain PCC 9511 cultures either growing on ammonium or subjected to nitrogen limitation, and only severe nitrogen starvation leads to a slight decrease in its expression (33) (Fig. 2D). This is another most unusual trait in the regulation of nitrogen assimilation in Prochlorococcus, in sharp contrast with that in other cyanobacteria (46) and other organisms (33). It points to a permanently high capacity for ammonium uptake, thus reinforcing the importance of this nitrogen source for this microorganism. It is noteworthy that the expression of amt1 is not under the control of NtcA (33), in contrast to the situation described for Synechococcus sp. strain PCC 7942 (76).
In much the same way as nitrite, urea is utilized by some (55, 63) but not all Prochlorococcus isolates (e.g., SS120 lacks the ure cluster [8]), indicating the relevance of this nitrogen source in some marine environments, where its concentration varies between 0.1 and 1 μM and it is the dominant component among the dissolved organic nitrogen compounds (1). The measured urease activities were fairly constant irrespective of the nitrogen source supplied to Prochlorococcus strain PCC 9511. This apparent lack of regulation, similar to that observed for GS, led to hypothesize that NtcA was not involved in the control of urease biosynthesis in Prochlorococcus, in spite of the presence of putative NtcA binding sites located next to the ure genes (encoding the urease structural and accessory molecules) (55). These genes are organized in two overlapping, opposite clusters in Prochlorococcus strain PCC 9511 (55) and Synechococcus strain WH 7805 (7), in contrast to the case for most freshwater cyanobacteria, where these genes are scattered throughout the genome (55). Additional differences were observed in the quaternary structure (two heterotrimers) and in the molecular mass of the enzyme (168 kDa), which is the lowest among the described ureases (55). The physiological significance of these observations remains to be further investigated.
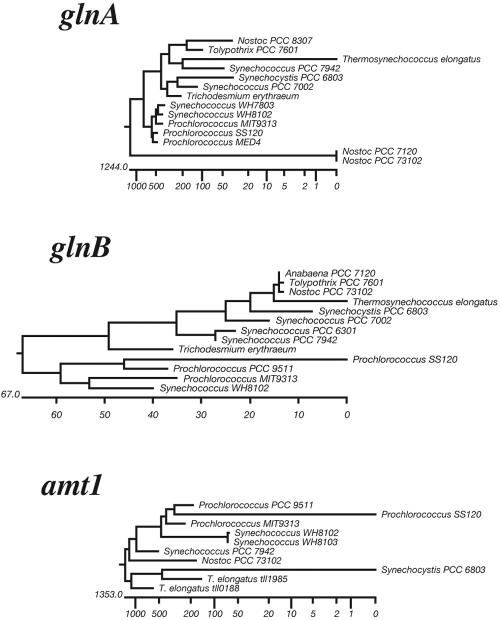
The signal transduction protein PII of Prochlorococcus strain PCC 9511 is encoded by a gene (glnB) that is very similar to its orthologs in cyanobacteria, with a putative NtcA binding site in its promoter regions (56). Moreover, the deduced amino acid sequence shows typical cyanobacterial signatures (56). The Prochlorococcus PII is therefore of the cyanobacterial type, but, interestingly, it forms a separate subclade with other oceanic strains within the glnB cyanobacterial radiation (Fig. 3). In cyanobacteria, the PII protein, whose main function is to coordinate nitrogen and carbon metabolism, is posttranslationally modified by phosphorylation in response to the nitrogen and carbon status of the cells (72). In Prochlorococcus, however, the PII protein is not phosphorylated under any of the tested conditions (56) (Fig. 2C). In agreement with this observation, the pphA gene, encoding the PP2C-like phosphatase that dephosphorylates PII in Synechocystis strain PCC 6803 (26), is absent in the genome of Prochlorococcus strain MED4 (65). PII could act, nevertheless, as a sensor of the intracellular concentration of 2-oxoglutarate. As proposed for Synechocystis strain PCC 6803 (56), both molecules could indeed form a complex involved in the direct or indirect inhibition of a high-affinity inorganic carbon transport system, in a manner independent of the PII phosphorylation state. Since the genome of MED4 lacks orthologs of the genes involved in Ci acquisition in other cyanobacteria, it has been proposed that the CynABD transport system might be utilized for the transport of both bicarbonate and cyanate in these Prochlorococcus strains (56), but this remains to be experimentally demonstrated.
FIG. 3.
Phylogeny of nitrogen metabolism-related genes in marine cyanobacteria. Phylogenetic trees obtained by Jotun-Hein alignment (MegAlign 4.0 software, Lasergene package) of the available amino acid sequences from glutamine synthetase (encoded by glnA), the signal transducer PII (encoded by glnB), and the high-affinity ammonium transporter (encoded by amt1) from marine and freshwater cyanobacteria are shown. Note that Prochlorococcus strains MED4 and PCC 9511 correspond to the same genotype, with the only difference being that PCC 9511 is axenic.
The role of the global regulator NtcA (22) in Prochlorococcus is rather uncertain (33). In several cyanobacteria, the expression of the genes encoding nitrate reductase, nitrite reductase, glutamine synthetase, and the transporters of nitrate, nitrite, and ammonium, as well as that of ntcA itself, is downregulated in the presence of ammonium and induced under conditions of nitrogen stress (22, 34, 38, 39). However, in Prochlorococcus strain PCC 9511, nitrogen limitation induces the expression of ntcA but not that of amt1 (33) (Fig. 2D). Moreover, the expression of GS is not induced under conditions of nitrogen starvation in the same strain (9, 10). In Prochlorococcus strain PCC 9511, the upstream region of ntcA, in contrast to that of amt1, contains a typical −10 box (33), usually found near the NtcA binding site. Although these results confirm that amt1 might not be under NtcA control in this strain, further work is required to convincingly establish a correlation between the lack of a −10 box and the occurrence of an unusual regulatory behavior for a given gene.
The reviewed literature on nitrogen assimilation in Prochlorococcus clearly suggests a simplification of the regulatory networks. This is reflected in different observations, such as the fairly constant concentration and activity of GS (9, 10), the constitutive expression of the ammonium transporter (33), the lack of phosphorylation of the PII protein (56), and the apparent lack of nitrogen control by NtcA (33). This is in good agreement with the small number of response regulators and histidine kinases observed in the Prochlorococcus genomes (8, 42, 65, 68). Since this is also observed in marine Synechococcus (54, 68), it is tempting to speculate that microorganisms require far less complicated systems of detection and transduction of environmental signals and metabolic control when they live in marine environments than when they live in other habitats, such as rivers, lakes, or soils. The rather constant conditions observed in the oligotrophic gyres of intertropical oceans could have provoked a progressive loss of these kinds of genes, in order to produce streamlined regulatory mechanisms for better adaptation. In addition, this could contribute to the reported compactness of the Prochlorococcus genomes (70).
The traditional dilemma of physiological acclimation versus evolutionary adaptation is illuminated on the basis of the current knowledge on the large degree of genetic diversity in several representative isolates of Prochlorococcus. Different studies on the photosynthetic apparatus showed that any Prochlorococcus isolate seems to be able to grow under a large range of irradiances (48, 60), thus supporting the importance of physiological acclimation. However, there exist largely divergent light-harvesting adaptations within the genus Prochlorococcus (since, for example, there are eight pcb genes in the SS120 strain [8, 14] with specific expression patterns [3, 15], versus only one in MED4, and three psbA genes in MIT 9313 versus one in MED4 or SS120 [8, 24]). On the other hand, adaptation with gene loss seems to be the rule in the field of nitrogen assimilation. Indeed, depending on the ecotype (49) considered, the utilizable sources are rather restricted, going from oxidized sources such as nitrite in some low-light-adapted strains (47) to reduced forms such as urea (55), ammonium (9, 36), or amino acids (63, 85). Physiological acclimation is therefore more advantageous in some biological aspects, while in others evolutionary adaptation is more beneficial. Hence, it seems that evolution drove the genomes of Prochlorococcus strains in different, sometimes opposite, directions, such as gene loss, gene multiplication, and extensive genome reorganizations (8, 65). The outcome of all of these processes is the current pool of Prochlorococcus genotypes, where biological selection induced an extremely efficient, yet remarkably simplified, combination of maximum economy by removing genes that were not strictly essential (thus saving the energy required for their replication, transcription, and translation) and considerable expense by multiplying those coding for proteins of paramount importance under conditions of strong limitations (3, 14, 15).
DIVERSITY AND PHYLOGENY OF NITROGEN ASSIMILATION PATHWAYS IN THE GENOMES OF PROCHLOROCOCCUS STRAINS
Comparative analysis of two Prochlorococcus genomes provided interesting insights into the photosynthetic apparatus of these cyanobacteria (24). The current availability of three Prochlorococcus genomes (8, 65), including two from low-irradiance-adapted ecotypes (MIT9313 and SS120) and an additional genome from the closely related marine Synechococcus clade (54), now permits a detailed analysis of the nitrogen assimilation strategies utilized by Prochlorococcus at different water depths, which correspond to different conditions of nitrogen supply. Table 2 summarizes some of the most interesting features from this comparison; Fig. 1 outlines the comparison between the generic N routes in cyanobacteria and those observed in Prochlorococcus. The central entry point for nitrogen assimilation in Prochlorococcus is ammonium, since no nitrogenase-related or nitrate reductase genes exist in Prochlorococcus, thus eliminating the possibility of assimilation of the two most abundant nitrogen sources in nature. Moreover, only one of the low-light-adapted ecotypes (MIT9313) possesses the gene nirA, encoding nitrite reductase. The genes encoding the ammonium transporter and the glutamine synthetase/glutamate synthase are found in all three genomes. The three strains also share the presence of genes encoding oligopeptide transporters, and amino acid transporters are found in the low-light-adapted strains (SS120 and MIT9313) but not in the high-light-adapted strain (MED4) (8, 65). Utilization of amino acids (either derived from oligopeptides or directly imported as single molecules into the cell) is therefore a common feature in Prochlorococcus. Given that amino acids are molecules containing reduced nitrogen and carbon skeletons, their utilization could provide an important advantage (51, 52) under conditions of oligotrophy and very low energy input from light at depth.
TABLE 2.
Presence of nitrogen metabolism-related genes in the genomes of Prochlorococcus strains MED4, MIT9313, and SS120 and Synechococcus strain WH8102
| Gene(s) | Protein(s) | Presencea in strain: | |||
|---|---|---|---|---|---|
| MED4 | MIT9313 | SS120 | WH8102 | ||
| nif cluster | Nitrogenase and cofactors | − | − | − | − |
| narB | Nitrate reductase | − | − | − | + |
| nirA | Nitrite reductase | − | + | − | + |
| glnA | Glutamine synthetase | + | + | + | + |
| gdhA | Glutamate dehydrogenase | − | + | − | − |
| glsF | Fd-glutamate synthase | + | + | + | + |
| ure cluster | Urease and cofactors | + | + | − | + |
| amt1 | Ammonium transporter | + | + | + | + |
| Amino acid transporters | − | + | + | + | |
| Oligopeptide transporters | + | + | + | + | |
| cynABD | Cyanate transporter | + | − | − | + |
| cynS | Cyanate lyase | + | − | − | + |
A remarkable outcome from comparative genome analysis is the presence in Synechococcus strain WH8102 and Prochlorococcus strain MED4 (but not in SS120 or MIT9313) of genes coding for cyanate ABC transporters (cynABD) and cyanate lyase (cynS) (8, 54, 56, 65). Furthermore, cyanate utilization has been reported for Synechococcus strain WH8102 (54). Initial results strongly suggest that cultures of Prochlorococcus strain MED4, in contrast to those of SS120, can grow by utilizing cyanate as the sole nitrogen source (Rangel et al., unpublished data). This points to the utilization of a potentially important nitrogen source, since cyanate is a degradation product of urea (19), which in turn is an excretion product of different organisms (77), reaching concentrations of 0.1 to 1 μM in the ocean (1, 4, 43).
The presence of the gene gdhA, encoding glutamate dehydrogenase, in strain MIT9313 but not in MED4, SS120, or even Synechococcus strain WH8102 is intriguing. This enzyme catalyzes an alternate route for ammonium incorporation directly into oxoglutarate, to produce glutamate (Fig. 1). Nevertheless, due to the higher Michaelis constant for ammonium of this enzyme with respect to that of glutamine synthetase, the main route for ammonium assimilation in cyanobacteria is that composed by GS/GOGAT (11, 12, 44). Preliminary investigations on Prochlorococcus showed that this activity is detected at relatively high levels in crude extracts from the MIT9313 strain and not in those from MED4 or SS120 (Rangel et al., unpublished data). It has been proposed that glutamate dehydrogenase confers selective advantages to Synechocystis strain PCC 6803 under nonexponential growth conditions (5). Whether this is also the case in Prochlorococcus strain MIT9313 remains to be studied.
Phylogenetic studies on different key genes from the nitrogen assimilation pathway, such as those shown in Fig. 3 for glnA, glnB, and amt1 (similar trees have been constructed for other genes from this pathway [not shown]), are in good agreement with the current model of the evolution of Prochlorococcus within the cyanobacterial radiation: they show a rather recent speciation, leading to the appearance of a cluster corresponding to marine cyanobacteria, in which Prochlorococcus is closely related to marine Synechococcus isolates (30, 64, 74, 80). This suggests that the genes involved in nitrogen assimilation in Prochlorococcus have not been the result of lateral gene transfer (71), as has been observed for other important genes (8, 24, 65), but have evolved from their counterparts in ancestral cyanobacteria. There is, however, at least one exception to this rule: the proteobacterial-like nitrite transporter found in the genome of MIT9313 (65). Therefore one can speculate that the genome of a common ancestor of Prochlorococcus and marine Synechococcus strains contained a set of genes involved in nitrogen assimilation that was sufficient to allow fine metabolic tuning in order to optimize nitrogen assimilation in oligotrophic environments. This tuning would involve either modification of the regulatory networks or removal of unessential genes, without a major import of foreign components, leading to the appearance of specific genotypes for the different niches inhabited by Prochlorococcus and marine Synechococcus.
CONCLUDING REMARKS
The discovery of Prochlorococcus has led to a profound reconsideration of established models in marine ecology, from the composition of the marine phytoplankton to its contribution to the global primary production (59). While initial observations explained its ecological success mainly on the basis of photosynthetic adaptations to efficiently colonize most of the photic zone of the oceans (3, 14, 48, 58, 60, 61), it is now becoming clear that other major metabolic changes contribute significantly to such success, as evidenced in the present review on the nitrogen assimilation pathways in Prochlorococcus. Further work is required to understand the importance of these changes for Prochlorococcus fitness in oligotrophic regions of the oceans and their specific relationship with the niche differentiation described for the different Prochlorococcus ecotypes (49, 79, 80). (For further information online, see the genome databases available at http://www.jgi.doe.gov/JGI_microbial/html/ for Prochlorococcus strains MED4, MIT9313, MIT9312, and NATL2A and Synechococcus strain WH8102 and at http://www.sb-roscoff.fr/Phyto/ for Prochlorococcus strain SS120 and Synechococcus strain WH7803. [Note that sequencing and/or annotation of the genomes from Prochlorococcus strains MIT9312 and NATL2A and Synechococcus strain WH7803 is in progress, and consequently their genome databases are still not available at the corresponding websites.] Websites for research projects concerning Prochlorococcus are http://www.sb-roscoff.fr/PROMOLEC/ [the European project “Prochlorococcus Molecular Ecology”] and http://arep.med.harvard.edu/DOEGTL/ [the U.S. project “Microbial Ecology, Proteogenomics and Computational Optima”].)
Acknowledgments
J.M.G.-F. and J.D. are thankful to Sabah El Alaoui, Guadalupe Gómez-Baena, Antonio López-Lozano, Lourdes Humanes, Fermín Toribio, and Oriol Rangel; without their effort, this work would not have been possible. The availability of the axenic Prochlorococcus strain PCC 9511, kindly provided by R. Rippka (Unité des Cyanobactéries, Institut Pasteur, Paris, France), has been an invaluable tool to us. Other Prochlorococcus strains were generously provided by F. Partensky (Station Biologique de Roscoff, Roscoff, France) (strains MED4, TAK9803-2, GP2, SS120, NATL1A, and NATL2A) and S. W. Chisholm (Massachusetts Institute of Technology, Cambridge, Mass.) (strain MIT9313). The kind collaboration of Carlos Massó de Ariza and the Instituto Español de Oceanografía allowed us to obtain seawater for culturing Prochlorococcus. We thank F. Partensky and L. R. Moore (University of Maine, Bangor) and D. J. Scanlan (University of Warwick, Warwick, United Kingdom) for critical reading of the manuscript and helpful discussions.
Our research on nitrogen assimilation in Prochlorococcus has been funded by the European Union MASTIII program (PROMOLEC, MAS3-CT97-0128), the Spanish Ministerio de Ciencia y Tecnología (BMC2003-09218-CO2-01, cofunded by the FEDER program from the European Union), the University of Córdoba (Programa Propio de Investigación), and the Junta de Andalucía (III Plan Andaluz de Investigación). J.M.G.-F. received a postdoctoral fellowship from the University of Córdoba and a “Ramon y Cajal” grant from the Spanish Ministerio de Ciencia y Tecnología.
REFERENCES
- 1.Antia, N. J., P. J. Harrison, and L. Oliveira. 1991. The role of dissolved organic nitrogen in phytoplankton nutrition, cell biology and ecology. Phycologia 30**:**1-89. [Google Scholar]
- 2.Bertilsson, S., O. Berglund, D. M. Karl, and S. W. Chisholm. 2003. Elemental composition of marine Prochlorococcus and Synechococcus: implications for the ecological stoichiometry of the sea. Limnol. Oceanogr. 48**:**1721-1731. [Google Scholar]
- 3.Bibby, T. S., I. Mary, J. Nield, F. Partensky, and J. Barber. 2003. Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424**:**1051-1054. [DOI] [PubMed] [Google Scholar]
- 4.Capone, D. G. 2000. The marine microbial nitrogen cycle, p. 455-493. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, Inc., New York, N.Y.
- 5.Chávez, S., J. M. Lucena, J. C. Reyes, F. J. Florencio, and P. Candau. 1999. The presence of glutamate dehydrogenase is a selective advantage for the cyanobacterium Synechocystis sp. strain PCC 6803 under nonexponential growth conditions. J. Bacteriol. 181**:**808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chisholm, S. W., R. J. Olson, E. R. Zettler, R. Goericke, J. B. Waterbury, and N. A. Welschmeyer. 1988. A novel free living prochlorophyte abundant in the oceanic euphotic zone. Nature 334**:**340-343. [Google Scholar]
- 7.Collier, J. L., B. Brahamsha, and B. Palenik. 1999. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC-3.5.1.5) to utilize urea as a nitrogen source—molecular genetic and biochemical analysis of the enzyme. Microbiology 145**:**447-459. [DOI] [PubMed] [Google Scholar]
- 8.Dufresne, A., M. Salanoubat, F. Partensky, F. Artiguenave, I. M. Axmann, V. Barbe, S. Duprat, M. Y. Galperin, E. V. Koonin, F. Le Gall, K. S. Makarova, M. Ostrowski, S. Oztas, C. Robert, I. B. Rogozin, D. J. Scanlan, N. Tandeau de Marsac, J. Weissenbach, P. Wincker, Y. I. Wolf, and W. R. Hess. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 100**:**10020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Alaoui, S., J. Diez, L. Humanes, F. Toribio, F. Partensky, and J. M. García-Fernández. 2001. In vivo regulation of glutamine synthetase activity in the marine chlorophyll _b_-containing cyanobacterium Prochlorococcus sp. strain PCC 9511 (Oxyphotobacteria). Appl. Environ. Microbiol. 67**:**2202-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Alaoui, S., J. Diez, F. Toribio, G. Gómez-Baena, A. Dufresne, and J. M. García-Fernández. 2003. Glutamine synthetase from the marine cyanobacteria Prochlorococcus spp.: characterization, phylogeny and response to nutrient limitation. Environ. Microbiol. 5**:**412-423. [DOI] [PubMed] [Google Scholar]
- 11.Florencio, F. J., and J. C. Reyes. 2002. Regulation of ammonium assimilation in cyanobacteria, p. 93-113. In C. H. Foyer and G. Noctor (ed.), Photosynthetic nitrogen assimilation and associated carbon metabolism, vol. 12. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 12.Flores, E., and A. Herrero. 1994. Assimilatory nitrogen metabolism and its regulation, p. 487-517. In D. A. Bryant (ed.), The molecular biology of cyanobacteria, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 13.Fuller, N. J., D. Marie, F. Partensky, D. Vaulot, A. F. Post, and D. J. Scanlan. 2003. Clade-specific 16S rDNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69**:**2430-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garczarek, L., W. Hess, J. Holtzendorff, G. van der Staay, and F. Partensky. 2000. Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote. Proc. Natl. Acad. Sci. USA 97**:**4098-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garczarek, L., G. W. van der Staay, W. R. Hess, F. Le Gall, and F. Partensky. 2001. Expression and phylogeny of the multiple antenna genes of the low-light-adapted strain Prochlorococcus marinus SS120 (Oxyphotobacteria). Plant Mol. Biol. 46**:**683-693. [DOI] [PubMed] [Google Scholar]
- 16.Goericke, R., and N. A. Welschmeyer. 1993. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep-Sea Res. I 40**:**2283-2294. [Google Scholar]
- 17.Gómez-Baena, G., J. Diez, J. M. García-Fernández, S. El Alaoui, and L. Humanes. 2001. Regulation of glutamine synthetase by metal-catalyzed oxidative modification in the marine oxyphotobacterium Prochlorococcus. BBA Gen. Subjects 1568**:**237-244. [DOI] [PubMed] [Google Scholar]
- 18.Graziano, L. M., R. J. Geider, W. K. W. Li, and M. Olaizola. 1996. Nitrogen limitation of North Atlantic phytoplankton—analysis of physiological condition in nutrient enrichment experiments. Aquat. Microb. Ecol. 11**:**53-64. [Google Scholar]
- 19.Hargel, P., J. J. T. Gerding, W. Fieggen, and H. Bloemendal. 1971. Cyanate formation in solutions of urea. I. Calculation of cyanate concentrations at different temperature and pH. Biochim. Biophys. Acta 243**:**366-373. [DOI] [PubMed] [Google Scholar]
- 20.Hedges, J. I., J. A. Baldcock, Y. Gélinas, C. Lee, M. Peterson, and S. G. Wakeham. 2001. Evidence for non-selective preservation of organic matter in sinking marine particles. Nature 409**:**801-804. [DOI] [PubMed] [Google Scholar]
- 21.Heldal, M., D. J. Scanlan, S. Norland, F. Thingstad, and N. H. Mann. 2003. Elemental composition of single cells of various strains of marine Prochlorococcus and Synechococcus using X-ray microanalysis. limnol. Oceanogr. 48**:**1732-1743. [Google Scholar]
- 22.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183**:**411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess, W. R., F. Partensky, G. W. M. Van der Staay, J. M. García-Fernández, T. Börner, and D. Vaulot. 1996. Coexistence of phycoerythrin and a chlorophyll a/b antenna in a marine prokaryote. Proc. Natl. Acad. Sci. USA 93**:**11126-11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hess, W. R., G. Rocap, C. S. Ting, F. Larimer, S. Stilwagen, J. Lamerdin, and S. W. Chisholm. 2001. The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosynth. Res. 70**:**53-71. [DOI] [PubMed] [Google Scholar]
- 25.Humanes, L., J. M. García-Fernández, A. López-Ruiz, and J. Diez. 1995. Glutamine synthetase from the green alga Monoraphidium braunii is regulated by oxidative modification. Plant Sci. 110**:**269-277. [Google Scholar]
- 26.Irmler, A., and K. Forchhammer. 2001. A PP2C-type phosphate dephosphorylates the PII signaling protein in the cyanobacterium Synechocystis PCC 6803. Proc. Natl. Acad. Sci. USA 98**:**12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, P. W., and J. M. Sieburth. 1979. Chroococcoid cyanobacteria in the sea: a ubiquitous and diverse phototrophic biomass. Limnol. Oceanogr. 24**:**928-935. [Google Scholar]
- 28.Karl, D. M. 2002. Nutrient dynamics in the deep blue sea. Trends Microbiol. 10**:**410-418. [DOI] [PubMed] [Google Scholar]
- 29.Kirchman, D. L. 2000. Microbial ecology of the oceans. Willey-Liss, New York, N.Y.
- 30.Laloui, W., K. A. Palinska, R. Rippka, F. Partensky, N. Tandeau De Marsac, M. Herdman, and I. Iteman. 2002. Genotyping of axenic and non-axenic isolates of the genus Prochlorococcus and the OMF-‘_Synechococcus_’ clade by size, sequence analysis or RFLP of the internal transcribed spacer of the ribosomal operon. Microbiology 148**:**453-465. [DOI] [PubMed] [Google Scholar]
- 31.La Roche, J., G. W. van der Staay, F. Partensky, A. Ducret, R. Aebersold, R. Li, S. S. Golden, R. G. Hiller, P. M. Wrench, A. W. Larkum, and B. R. Green. 1996. Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc. Natl. Acad. Sci. USA 93**:**15244-15248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine, R. L., C. N. Oliver, R. M. Fulks, and E. R. Stadtman. 1981. Turnover of bacterial glutamine synthetase: oxidative inactivation precedes proteolysis. Proc. Natl. Acad. Sci. USA 78**:**2120-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindell, D., D. Erdner, D. Marie, O. Prasil, M. Koblizek, F. Le Gall, R. Rippka, F. Partensky, D. J. Scanlan, and A. F. Post. 2002. Nitrogen stress response of Prochlorococcus strain PCC 9511 (Oxyphotobacteria) involves contrasting regulation of ntcA and amt1. J. Phycol. 38**:**1113-1124. [Google Scholar]
- 34.Lindell, D., E. Padan, and A. F. Post. 1998. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J. Bacteriol. 180**:**1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, H. B., H. A. Nolla, and L. Campbell. 1997. Prochlorococcus growth rate and contribution to primary production in the Equatorial and Subtropical North Pacific ocean. Aquat. Microb. Ecol. 12**:**39-47. [Google Scholar]
- 36.López-Lozano, A., J. Diez, S. El Alaoui, C. Moreno-Vivián, and J. M. García-Fernández. 2002. Nitrate is reduced by heterotrophic bacteria but not transferred to Prochlorococcus in non axenic cultures. FEMS Microbiol. Ecol. 41**:**151-160. [DOI] [PubMed] [Google Scholar]
- 37.Losada, M., M. A. Vargas, M. A. de la Rosa, and F. J. Florencio. 1999. Los elementos y moléculas de la vida, vol. II, p. 1023-1058. Rueda, Madrid, Spain.
- 38.Luque, I., E. Flores, and A. Herrero. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 13**:**2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marqués, S., F. J. Florencio, and P. Candau. 1992. Purification and characterization of the ferredoxin-glutamate synthase from the unicellular cyanobacterium Synechococcus sp. PCC 6301. Eur. J. Biochem. 206**:**69-77. [DOI] [PubMed] [Google Scholar]
- 40.Martin, G., and P. Böger. 1997. Two ways of hydrogen peroxide formation in the oxidative inactivation of cyanobacterial glutamine synthetase. Z. Naturforsch. 52c**:**812-816. [Google Scholar]
- 41.Martin, G., W. Haehnel, and P. Böger. 1997. Oxidative inactivation of glutamine synthetase from the cyanobacterium Anabaena variabilis. J. Bacteriol. 179**:**730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mary, I., and D. Vaulot. 2003. Two-component systems in Prochlorococcus MED4: genomic analysis and differential expression under stress. FEMS Microbiol. Lett. 226**:**135-144. [DOI] [PubMed] [Google Scholar]
- 43.McArthy, M., T. Pratum, J. Hedges, and R. Benner. 1997. Chemical composition of dissolved organic nitrogen in the ocean. Nature 390**:**150-154. [Google Scholar]
- 44.Meeks, J. C., C. P. Wolk, J. Thomas, W. Lockau, P. W. Shaffer, S. M. Austin, W. S. Chien, and A. Galonsky. 1977. The pathways of assimilation of 13NH4+ by the cyanobacterium, Anabaena cylindrica. J. Biol. Chem. 252**:**7894-7900. [PubMed] [Google Scholar]
- 45.Mitsui, A., S. Kumazawa, A. Takahashi, H. Ikemoto, S. Cao, and T. Arai. 1986. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature 323**:**720-722. [Google Scholar]
- 46.Montesinos, M. L., A. M. Muro-Pastor, A. Herrero, and E. Flores. 1998. Ammonium/methylammonium permeases of a cyanobacterium—identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J. Biol. Chem. 273**:**31463-31470. [DOI] [PubMed] [Google Scholar]
- 47.Moore, L., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47**:**989-996. [Google Scholar]
- 48.Moore, L. R., R. Goericke, and S. W. Chisholm. 1995. Comparative physiology of Synechococcus and _Prochlorococcus_—influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 116**:**259-275. [Google Scholar]
- 49.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393**:**464-467. [DOI] [PubMed] [Google Scholar]
- 50.Olson, R. J., E. R. Zettler, M. A. Altabet, J. A. Dusenberry, and S. W. Chisholm. 1990. Spatial and temporal distributions of prochlorophyte picoplankton in the North Atlantic Ocean. Deep-Sea Res. I 37**:**1033-1051. [Google Scholar]
- 51.Paerl, H. W. 1991. Ecophysiological and trophic implications of light-stimulated amino acid utilization in marine picoplankton. Appl. Environ. Microbiol. 57**:**473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paerl, H. W. 2000. Marine plankton, p. 121-148. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 53.Palatnik, J. F., N. Carrillo, and E. M. Valle. 1999. The role of photosynthetic electron transport in the oxidative degradation of chloroplastic glutamine synthetase. Plant Physiol. 121**:**471-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palenik, B., B. Brahamsha, F. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. B. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424**:**1037-1042. [DOI] [PubMed] [Google Scholar]
- 55.Palinska, K. A., T. Jahns, R. Rippka, and N. Tandeau de Marsac. 2000. Prochlorococcus marinus strain PCC 9511, a picoplanktonic cyanobacterium, synthesizes the smallest urease. Microbiology 146**:**3099-3107. [DOI] [PubMed] [Google Scholar]
- 56.Palinska, K. A., W. Laloui, S. Bedu, S. Loiseaux-De Goer, A. M. Castets, R. Rippka, and N. Tandeau De Marsac. 2002. The signal transducer PII and bicarbonate acquisition in Prochlorococcus marinus PCC 9511, a marine cyanobacterium naturally deficient in nitrate and nitrite assimilation. Microbiology 148**:**2405-2412. [DOI] [PubMed] [Google Scholar]
- 57.Partensky, F., J. Blanchot, and D. Vaulot. 1999. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review, p. 457-476. In L. Charpy and A. W. D. Larkum (ed.), Marine cyanobacteria, vol. 19. Bulletin de l'Institut Océanographique, Numéro Spécial, Monaco. [Google Scholar]
- 58.Partensky, F., and L. Garczarek. 2003. The photosynthetic apparatus of chlorophyll _b_- and _d_-containing oxyphotobacteria. In A. W. Larkum, S. Douglas, and J. A. Raven (ed.), Photosynthesis in algae, vol. 13. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 59.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63**:**106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Partensky, F., N. Hoepffner, W. K. W. Li, O. Ulloa, and D. Vaulot. 1993. Photoacclimation of Prochlorococcus sp (Prochlorophyta) strains isolated from the North Atlantic and the Mediterranean sea. Plant Physiol. 101**:**285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Partensky, F., J. Laroche, K. Wyman, and P. G. Falkowski. 1997. The divinyl chlorophyll a/b protein complexes of 2 strains of the oxyphototrophic marine prokaryote _Prochlorococcus_—characterization and response to changes in growth irradiance. Photosynth. Res. 51**:**209-222. [Google Scholar]
- 62.Penno, S., L. Campbell, and W. R. Hess. 2000. Presence of phycoerythrin in two strains of Prochlorococcus (Cyanobacteria) isolated from the subtropical North Pacific ocean. J. Phycol. 36**:**723-729. [DOI] [PubMed] [Google Scholar]
- 63.Rippka, R., T. Coursin, W. Hess, C. Lichtlé, D. J. Scanlan, K. A. Palinska, I. Iteman, F. Partensky, J. Houmard, and M. Herdman. 2000. Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll _a_2/_b_2-containing cyanobacterium (Oxyphotobacteria). Int. J. Syst. Evol. Microbiol. 50**:**1833-1847. [DOI] [PubMed] [Google Scholar]
- 64.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68**:**1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, d. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424**:**1042-1047. [DOI] [PubMed] [Google Scholar]
- 66.Rocap, G., L. R. Moore, and S. W. Chisholm. 1999. Molecular phylogeny of Prochlorococcus ecotypes. Bulletin de l'Institut Océanographique, Numéro spécial, Monaco. 19**:**107-116.
- 67.Scanlan, D. J. 2003. Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microb. Physiol. 47**:**1-64. [DOI] [PubMed] [Google Scholar]
- 68.Scanlan, D. J., and N. J. West. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40**:**1-12. [DOI] [PubMed] [Google Scholar]
- 69.Schopf, J. W. 2000. The fossil record: tracing the roots of the cyanobacterial lineage, p. 13-35. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 70.Strehl, B., J. Holtzendorff, F. Partensky, and W. R. Hess. 1999. A small and compact genome in the marine cyanobacterium Prochlorococcus marinus CCMP 1375: lack of an intron in the gene for tRNA(Leu)UAA and a single copy of the rRNA operon. FEMS Microbiol. Lett. 181**:**261-266. [DOI] [PubMed] [Google Scholar]
- 71.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424**:**1047-1051. [DOI] [PubMed] [Google Scholar]
- 72.Tandeau de Marsac, N., H. M. Lee, M. Hisbergues, A. M. Castets, and S. Bédu. 2001. Control of nitrogen and carbon metabolism in cyanobacteria. J. Appl. Phycol. 13**:**287-292. [Google Scholar]
- 73.Ting, C. S., G. Rocap, J. King, and S. W. Chisholm. 2002. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10**:**134-142. [DOI] [PubMed] [Google Scholar]
- 74.Toledo, G., and B. Palenik. 1997. Synechococcus diversity in the California current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl. Environ. Microbiol. 63**:**4298-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaulot, D., and F. Partensky. 1992. Cell cycle distributions of prochlorophytes in the North Western Mediterranean sea. Deep-Sea Res. I 39**:**727-742. [Google Scholar]
- 76.Vázquez-Bermúdez, M. F., J. Paz-Yepes, A. Herrero, and E. Flores. 2002. The NtcA-activated amt1 gene encodes a permease required for uptake of low concentrations of ammonium in the cyanobacterium Synechococcus sp. PCC 7942. Microbiology 148**:**643-647. [DOI] [PubMed] [Google Scholar]
- 77.Walsh, P. J. 1997. Evolution and regulation of urea synthesis and ureotely in (Batrachoidid) fishes. Annu. Rev. Physiol. 59**:**299-323. [DOI] [PubMed] [Google Scholar]
- 78.Waterbury, J. B., S. W. Watson, R. R. Guillard, and L. E. Brand. 1979. Widespread occurrence of a unicellular, marine planktonic, cyanobacterium. Nature 277**:**293-294. [Google Scholar]
- 79.West, N. J., and D. J. Scanlan. 1999. Niche partitioning of Prochlorococcus populations in a stratified water column in the Eastern North Atlantic ocean. Appl. Environ. Microbiol. 65**:**2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.West, N. J., W. A. Schonhuber, N. J. Fuller, R. I. Amann, R. Rippka, A. F. Post, and D. J. Scanlan. 2001. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147**:**1731-1744. [DOI] [PubMed] [Google Scholar]
- 81.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95**:**6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Williams, E. Z., L. Campbell, and G. DiTullio. 1999. Presented at the ASLO Aquatic Sciences Meeting, 1999, Santa Fe, N.Mex.
- 83.Zehr, J. P., and B. B. Ward. 2002. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68**:**1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zehr, J. P., J. B. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Karl. 2001. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412**:**635-638. [DOI] [PubMed] [Google Scholar]
- 85.Zubkov, M. V., B. M. Fuchs, G. A. Tarran, P. H. Burkill, and R. Amann. 2003. High rate of uptake of organic nitrogen compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Appl. Environ. Microbiol. 69**:**1299-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]