Minireview: Autophagy in Pancreatic β-Cells and Its Implication in Diabetes (original) (raw)
Abstract
Autophagy is a conserved system for the degradation of cytoplasmic proteins and organelles. During insulin resistance, in which insulin secretion is enhanced and β-cell mass is increased owing to changes in the expression and function of various proteins in pancreatic β-cells, autophagic activity appears to also be enhanced to adapt to the dynamic changes occurring in β-cells. Indeed, defective autophagy in β-cells recapitulates several features that are observed in islets during the development of type 2 diabetes mellitus. In addition, the dyregulation of autophagic activity appears to occur in the β-cells of type 2 diabetic model mice and type 2 diabetes mellitus patients. These lines of evidence suggest that autophagic failure may be implicated in the pathophysiology of type 2 diabetes mellitus. In this review, we summarized the recent findings regarding how autophagy in β-cells is regulated and how dysfunction of the autophagic machinery may lead to the dysfunction of β-cells.
Autophagy is an evolutionarily conserved catabolic process involving the degradation of cellular components by the lysosomal machinery. At least 3 kinds of autophagy have been defined to date: macroautophagy (1), chaperone-mediated autophagy (2), and microautophagy (3). In macroautophagy, a small part of the cytoplasm is sequestered by a membrane sac, which eventually form a double membrane structure, the autophagosome. The autophagosome fuses with lysosomes to form the autolysosome, in which the sequestered cytoplasm is degraded by lysosomal enzymes. In chaperon mediated autophagy, cytoplasmic substrates are recognized by a chaperon protein, Heat shock cognate (Hsc)70, and directly translocate to lysosome for degradation. In microautophagy, lysosomal membrane directly engulfs small parts of the cytoplasm. Thus, the formation of autophagosome is not required for chaperon-mediated autophagy and microautophagy. Among these, macroautophagy has been most extensively studied (4–6). In this review, we focused on the role of macroautophagy in pancreatic β-cells, and hereafter, we used the term “autophagy” to indicate macroautophagy.
A low level of autophagy is constitutively active in cells to remove misfolded proteins and damaged and senescent organelles (7). In various kinds of cells, such as hepatocytes (8), neurons (9, 10), and cardiac muscle cells (11), autophagy has been shown to be essential for proper cell function. In this regard, autophagy is a fundamental process for cellular homeostasis. On the other hand, enhancement of this process is important for the active reallocation of nutrients from unnecessary processes to more pivotal processes required for survival. Typically, during the shortage of nutrients, cells activate autophagy to supply nutrients from endogenous energy sources to processes required for cell survival. In addition, several stress response signals are known to activate autophagy and play an important role in the homeostasis of cell function under stressful conditions (5).
Insulin resistance, which is the state of an increased demand of insulin secretion, and pancreatic β-cell dysfunction are among the major defects observed in type 2 diabetes mellitus. In the natural history of type 2 diabetes, insulin resistance caused by obesity and/or reduced physical activity appears earlier than the onset of disease. At this stage, pancreatic β-cells adapt to the insulin resistance by increasing glucose responsive insulin secretion from individual islets and probably β-cell mass to prevent a rise in blood glucose (12, 13). After this stage, the gradual failure of β-cells to adaptation occurs and plays an important role in the progression to impaired glucose tolerance and eventually to diabetes (14, 15).
Pancreatic β-cells are specialized cells dedicated to insulin secretion in response to glucose and other various secretagogues. The main mechanism of glucose-stimulated insulin secretion is through an increase in cytosolic ATP levels. Pancreatic β-cells catabolize glucose to increase ATP to levels much higher than that required for cell survival, to induce the secretion of insulin. In addition, owing to unknown reasons, β-cells synthesize more than enough insulin for secretion under normal conditions (16). Indeed, the cytosol of normal β-cells is largely occupied by numerous insulin granules even after the stimulation of insulin secretion. Under these conditions, β-cells appear to be vulnerable to various forms of cellular stress, such as oxidative stress caused by damaged mitochondria and Endoplasmic reticulum (ER) stress caused by misfolded proteins. Insulin resistance enhances the accumulation of these various types of stresses in β-cells (14). Given that autophagy protects cells against oxidative stress and ER stress (5), failure of the autophagic system in β-cells is a potential factor that aggravates β-cell function, particularly under insulin-resistant conditions, and thus could be a cause of hyperglycemia. On the other hand, in certain conditions, activation of autophagy appears to enhance cell death in several cell types (17, 18), as well as in β-cells (19). The aim of this review is therefore to summarize the recent findings regarding the role of autophagy in β-cells in the pathophysiology of diabetes.
Molecular Mechanisms of the Induction and Regulation of Autophagy
Autophagy is a tightly regulated cellular process. To date, more than 30 autophagy-related (ATG) proteins have been discovered. Most of these proteins are conserved from yeast to mammals (4, 20). These proteins work together to accomplish the elaborate process composed of vesicle nucleation (formation of the isolation membrane), vesicle elongation, formation of the double-membrane vesicle, fusion with the lysosome, and degradation of the vesicle content. The detailed process is beyond the scope of this review, and for more information, the reader is advised to refer to previous reviews (4, 5, 20). However, in this review, we briefly summarize the molecular mechanism of autophagy that is essential for understanding its role in β-cells.
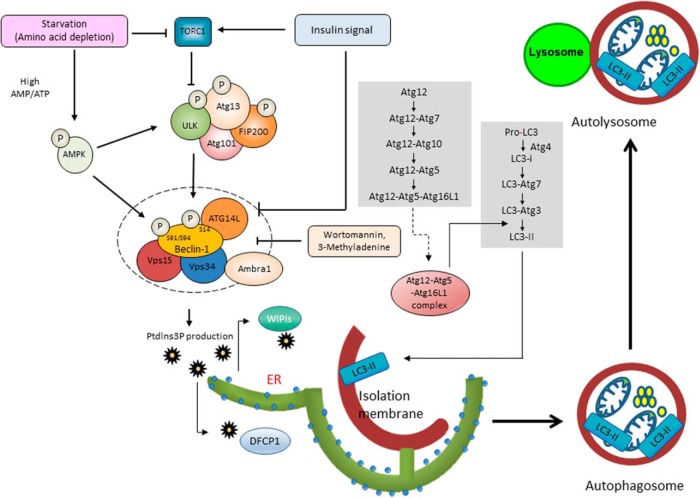
Accumulating evidence suggest that the complex consisting of Uncoordinated (Unc)-51-like kinase 1 (ULK1) and Vacuolar Protein Sorting (Vps34) are key regulators for the initiation and progression of autophagy. ULK1 is the mammalian ortholog of Atg1 and is a serine-threonine kinase. The ULK1 complex is composed of ULK1, Atg13, Atg101, and focal adhesion kinase family interacting protein of 200 kDa (FIP200) (21). Under normal conditions, mammalian target of rapamycin (mTOR) complex (mTORC)1 interacts with the ULK1 complex and inactivates it. Signal of insulin or other growth factors activates class I phosphatidylinositol 3-kinase (PI[3]K) proteins, which in turn signal via the Akt pathway to activate mTOR. Thus, these signaling pathways inactivate ULK1. In the presence of autophagy inducers, such as starvation, rapamycin, and the absence of insulin or growth factors, mTORC1 dissociates from the complex. This dissociation results in the enhancement of ULK1 kinase activity and phosphorylation of Atg13 and FIP200 (Figure 1) (22–24).
Figure 1. Signaling pathway regulating the initiation of autophagy and the formation of autophagosomes.
Macroautophagy consists of distinct stages: phagophore formation, vesicle elongation and completion, and fusion of the double-membrane autophagosome with the lysosome to form an autolysosome. The ULK1 complex and Vps34 complex are key regulators for the initiation of autophagy. The ULK1 complex is composed of ULK1, Atg13, Atg101, and FIP200. Under normal conditions, mTORC1 interacts with the ULK1 complex and inactivates it. In the presence of autophagy inducers, mTORC1 dissociates from the complex. This results in increased ULK1 kinase activity and the phosphorylation of Atg13 and FIP200. The Vps34 complex is composed of Vps34, Beclin-1, Vps15, Atg14L, and Ambra1. Phosphatidylinositol 3-phosphate, the product of ULK1, recruits the double FYVE domain-containing protein 1 (DFCP1) and WD-repeated protein interacting with phosphoinositides (WIPI) family proteins. Wortmannin and 3-methyladenine inhibit ULK1. ULK1 phosphorylates Beclin-1 at Ser14 and enhances PI(3)K activity of the Vps34 complex. This phosphorylation is essential for the full induction of autophagy. In addition, AMP-activated protein kinase (AMPK) phosphorylates Beclin-1 and activates Vps34 lipid kinase. On the other hand, Akt phosphorylates Beclin-1 and suppresses its kinase activity. Vesicle elongation and the formation of double-membrane vesicles are mediated by 2 ubiquitin-like conjugation systems involving ATG proteins. The first system is the ATG12-ATG5 conjugation reaction. The ATG12-ATG5 conjugates form a multimeric complex with ATG16L1 through ATG7 and ATG10. The second system is the conjugation reaction of LC3, the mammalian homologue of ATG8. LC3-I is generated from pro-LC3 by the Atg4 protease. Then, through ATG7, ATG3, and the ATG12-ATG5-ATG16L complex, LC3 conjugates to the phospholipid phosphatidylethanolamine to form LC3-II. During the formation of the autophagosome from the preautophagosome, ATG12-ATG5 complexes are released from the membranes, and LC3-II localizes on them.
Another key regulator of autophagy is Vps34, a class III PI(3)K. The Vps34 complex is composed of Vps34, Beclin-1, Vps15, Atg14L, and activating molecule in beclin-1-regulated autophagy (Ambra1). Phosphatidylinositol 3-phosphate, the product of Vps34 recruits double FYVE domain-containing protein 1 (DFCP1) and WD-repeated protein interacting with phosphoinositides family proteins (WIPIs) that are essential for double-membrane vesicle nucleation (25, 26). Indeed, wortmannin and 3-methyladenine, which strongly inhibit Vps34, inhibit autophagy. After typical autophagy induction, activated ULK1 phosphorylates Ambra1 and recruits the Vps34 complex to the site of autophagosome formation (27). In addition, phosphorylated Ambra1 stabilizes ULK1 kinase activity by its Lys-63-linked ubiquitination (28). In the Vps34 complex, Beclin-1 regulates Vps34 activity through its phosphorylation states and binding partners. Indeed, ULK1 phosphorylates Beclin-1 at Ser14 and enhances the PI(3)K activity of the Vps34 complex (29). This phosphorylation is essential for the full induction of autophagy. In addition, AMP-activated protein kinase, a target of metformin, phosphorylates Beclin-1 at Ser91 and Ser94 and activates Vps34 lipid kinase (30). On the other hand, Akt phosphorylates Beclin-1 at Ser234 and Ser295 and suppresses its kinase activity (31). Regarding binding partners of Beclin-1, Atg14L, UV irradiation resistance-associated gene, BAX-interacting factor-1 (Bif-1), and Ambra1 interact with Beclin-1 and positively regulate Vps34 activity. On the other hand, rubicon and antiapoptotic family members such as B cell lymphoma 2 (BCL-2), B cell lymphoma-extralarge (Bcl-xL), and myeloid cell leukemia-1 (MCL-1) negatively regulate Vps34 activity (Figure 1) (5).
Vesicle elongation and formation of double-membrane vesicles are mediated by 2 ubiquitin-like conjugation systems of ATG proteins. The first system is the ATG12-ATG5 conjugation reaction. The ATG12-ATG5 conjugates form a multimeric complex with ATG16L1 through ATG7, an E1-like ubiquitin-activating enzyme, and ATG10, an E2-like ubiquitin-activating enzyme. The second system is the conjugation reaction of microtubule-associated protein 1A/1B-light chain 3 (LC3), a mammalian homologue of ATG8. LC3-I is generated from pro-LC3 by the Atg4 protease. Then, through ATG7, ATG3 (an E2-like enzyme), and the ATG12-ATG5-ATG16L complex that possesses E3-like activity, LC3 conjugates to the phospholipid phosphatidylethanolamine to form LC3-II. During the formation of the autophagosome from the preautophagosome, ATG12-ATG5 complexes are released from the membranes, and LC3-II becomes localized on them. Usually, LC3-I is distributed throughout the cytosol and can only localize to the autophagosome membrane when it is lipidated to form LC3-II (Figure 1) (32).
The proteins and organelles destined for degradation are engulfed by the autophagosome. Some such proteins are selectively bound with adaptor proteins such as p62, which is also known as sequestosome/SQSTM1 (33), neighbor of BRCA1 gene 1 (NBR-1) (34), and optineurin (35), which binds with LC3-II. Then, the autophagosome membrane closes and then fuses with the lysosome to form the autolysosome. Fusion of autophagosomes with lysosomes is regulated by soluble NSF attachment protein receptors (SNARE) proteins, such as syntaxin 17, synaptosomal-associated protein (SNAP)-29, and vesicle-associated membrane protein (VAMP) 8. Intriguingly, syntaxin 17 is only localized to the outer membrane of closed autophagosomes, not the unclosed membrane; thus, only closed membranes can fuse with lysosomes (36). Then, the contents of the autolysosome are digested by lysosomal enzymes.
Assays for Monitoring Autophagy
Based on the morphology of the components and molecular mechanism of autophagy, several assays have been developed to monitor autophagy. For the detailed interpretation of the assays, the reader is advised to refer to its guidelines (37). Here, we briefly introduce the frequently used assays to monitor autophagy in β-cells.
When autophagy is activated, the number of autophagosome increases in cytosol. This accumulation of autophagosome can be directly observed by electron microscopy. In addition, when autophagy is activated, LC3 is lipidated and localized in autophagosome membrane, thus monitoring of Green fluorescent protein (GFP)-LC3 dots in cells and quantification of LC3-II by Western blotting are helpful to estimate the state of autophagy. However, accumulation of autophagosome can be observed, not only when autophagy is activated but also when autophagosome turnover is reduced. Thus, the evaluation of autophagic flux is more important to evaluate autophagic state in cells. One of the frequently used method to evaluate autophagic flux is to evaluate the further accumulation of LC3-II after the addition of lysosomal inhibitors. Accumulation of p62 frequently reflects relative decrease of degradation of autophagy; however, recent data revealed that the expression level of p62 in starved cells is determined not only by autophagic degradation, but also by transcriptional up-regulation, suggesting that the expression level of p62 does not always inversely correlate with autophagic activity (38). More reliable approach to evaluate autophagic state is to measure the degradation rate of long lived protein that is degraded by autophagy.
Autophagy in β-Cells Under Diabetic Conditions
Any structure in the cytosol can be a substrate of autophagy. In β-cells, various types of cellular structures, including insulin granules, mitochondria, and endoplasmic reticulum membranes, are observed in autophagic vacuoles (39, 40), although the formation of autophagosomes is rarely detected under steady-state conditions. In most other organs, such as liver and muscle (41), overnight starvation increases autophagic vacuole formation; however, in β-cells, there is no prominent increase in autophagic vacuoles after starvation (39). This is similar to the lack of autophagic up-regulation in neuronal tissues under starvation conditions. However, a previous study reported that starvation increases LC3-positive dots that represent the formation of autophagosomes in β-cells (19). Taken together, starvation-induced autophagy is present also in β-cells but may be less prominent than in other tissues.
Mice in an insulin-resistant state induced by a high-fat diet have more autophagosomes in their β-cells (39). Increased numbers of autophagosomes are also observed in several diabetic model rodents, such as ob/ob mice (42), db/db mice (39), Akita mice (43), and Zucker diabetic fatty rats (44). Although more autophagosomes may be caused by either enhanced autophagic flux or the inhibition of autophagic degradation, in ob/ob mice (42) and Akita mice (43), enhanced autophagic flux was reported. However, the accumulation of p62/SQSTM1, which is an adaptor protein interacting with LC3-II, is also observed in ob/ob mice (42) and in db/db mice (45). These results suggest that autophagic flux may not be activated sufficiently to deal with the increased demand of proteolysis in these type 2 diabetic model mice.
Regarding human type 2 diabetes mellitus, a study using islets from organ donors showed that β-cells in type 2 diabetic patients have extensive accumulation of autophagic vacuoles (46). Other studies using autopsy samples showed increased numbers of p62-positive β-cells in patients with type 2 diabetes (45, 47). Although further assessment of autophagic flux in β-cells is required, these data suggest that autophagic dysfunction occurs in the β-cells of patients with type 2 diabetes.
Consequences of Autophagic Dysfunction in β-Cells
The β-cell-specific Atg7 knockout (Atg7f/f:RIP-Cre) mouse is a model that is useful for assessing the consequences of autophagic dysfunction in β-cells. Two groups independently reported the phenotype of these mutant mice (39, 40). Decreased LC3-II levels and marked accumulation of p62 observed in the β-cells of these mice showed the efficient inhibition of autophagy in these mice. β-cells of Atg7f/f:RIP-Cre mice developed progressive degeneration characterized by cellular hypertrophy and the accumulation of polyubiquitinated proteins in β-cells. These features are similarly observed in the liver, central nervous system, and heart of each tissue-specific Atg5 or Atg7 knockout mice (8–11). Intriguingly, this phenotype was also observed in human patients with steatohepatitis (48) and neurodegenerative diseases such as Parkinson's disease and Alzheimer's disease (49). In β-cells of Zucker diabetic fatty rats, age-dependent polyubiquitinated protein aggregates are formed and this aggregate formation is accelerated by hyperglycemia-induced oxidative stress (50). However, evident accumulation of polyubiquitinated protein aggregates was not found in β-cells of autopsy samples from patients with type 2 diabetes mellitus (H. Mizukami and H.W. unpublished data).
Atg7f/f:RIP-Cre mice showed increased blood glucose levels and reduced glucose tolerance with reduced glucose responsive insulin secretion. Experiments using isolated islets showed reduced glucose responsive insulin secretion with preserved KCl-induced insulin secretion. In addition, a reduction in glucose-stimulated ATP production and Ca2+ influx in β-cells was also observed (39, 40). Furthermore, accumulation of distended mitochondria with distorted cristae was observed in the β-cells of Atg7f/f:RIP-Cre mice. These results suggest that mitochondrial dysfunction may be a cause of reduced glucose responsive insulin secretion in β-cells of Atg7f/f:RIP-Cre mice.
In insulin-resistant states, β-cells increase their number to secrete more insulin. This is one of the most important mechanisms of β-cells to compensate for insulin resistance. In fact, a 12-week high-fat diet increased β-cell mass by approximately 2-fold in wild-type mice (39). Although β-cell mass of Atg7f/f:RIP-Cre mice was comparable with that of wild-type mice on a normal diet, the increase in β-cell mass upon a high-fat diet was severely inhibited. The islets of Atg7f/f:RIP-Cre mice fed a high-fat diet are characterized by a reduced number of Ki67-positive replicating cells and more caspase-3-positive apoptotic cells (39). These results showed that autophagy in β-cells affects cell proliferation and apoptosis, particularly in insulin-resistant states (51, 52).
Mitophagy in β-Cells
Mitophagy is an important mitochondrial quality control mechanism that eliminates damaged mitochondria (53). Depolarized, dysfunctional mitochondria are selectively recognized by PTEN-induced putative kinase 1 (PINK1) and Parkin, which are responsible for familial Parkinson's disease (54, 55). These molecules are important for a certain type of mitophagy, and are not responsible for all types of mitophagy (53, 56–59). PINK1 is a serine/thereonine kinase that localizes to mitochondria with reduced membrane potential (60, 61). Accumulation of PINK1 on the mitochondrial surface activates the translocation of Parkin, an E3 ubiquitin ligase, from the cytosol to damaged mitochondria (57). Although the kinase activity of PINK1 is essential for the recruitment of Parkin to damaged mitochondria, the mechanism involved has not yet been clarified. After its localization on mitochondria, Parkin mediates the formation of polyubiquitin chains on specific mitochondrial outer membrane proteins. Previous data demonstrated that ubiquitination of the voltage-dependent anion channel-1, a component of the permeability transition pore, is essential for mitophagy (58), although another study showed that mitophagy can occur in the absence of voltage-dependent anion channel-1 (62). Accordingly, it has not been determined whether specific ubiquitinated proteins are required for mitophagy. After the formation of polyubiquitin chains on mitochondrial outer membrane proteins, the ubiquitin-binding adaptor protein p62/SQSTRM1 accumulates on depolarized mitochondria and is considered to facilitate the recruitment of damaged mitochondria to autophagosomes by binding to LC3-II (53, 60). In addition, a recent data showed that optineurin plays an indispensable role as an autophagy receptor in Parkin-mediated mitophagy (35).
The cooccurrence of decreased glucose responsive insulin secretion with the accumulation of distended mitochondria in Atg7f/f:RIP-Cre mice suggests that a dysfunction of mitophagy may be a cause of reduced glucose responsive insulin secretion. In another model of β-cell-specific autophagic dysfunction (Atg7f/f:RIP2-Cre), the administration of an antioxidant ameliorated the impairment in glucose-stimulated insulin secretion without improvement of autophagic flux (63). Increased oxidative stress from accumulated depolarized mitochondria may decrease the function of healthy mitochondria and may aggravate glucose responsive insulin secretion in the autophagy-deficient β-cells.
Recently, Hoshino et al (64) compared β-cell ATP content and insulin secretion in global Parkin-deficient mice and wild-type mice. They first found that p53 deficiency in β-cells protects against the development of diabetes by multiple low doses injection of streptozotocin. In this study, to avoid the antiinflammatory effect by p53 deficiency, they performed bone marrow transplantation before injection of streptozotocin. Then, to investigate its mechanism, they crossed p53-deficient mice and Parkin-deficient mice and found that the protective effect of p53 was not observed when Parkin is deleted. As a control experiment of this study, they investigated β-cell function with multiple low doses injection of streptozotocin after bone marrow transplantation. Although this is unusual condition, Parkin-deficient mice showed lower glucose-responsive increases in ATP levels as well as lower insulin secretion than wild-type mice with same treatment (64). More recently, the phenotype of C-type lectin domain family 16 member A (Clec16a) knockout mice was reported (65). Clec16a is a gene associated with type 1 diabetes, multiple sclerosis, and adrenal dysfunction. It encodes an endosomal protein that interacts with the E3 ubiquitin ligase Nrdp1. Loss of Clec16a induces an increase in the level of Parkin, an Nrdp1 target. Intriguingly, islets from _Clec16a_-deficient mice showed enhanced expression of Parkin; however, they also showed accumulation of abnormal mitochondria with reduced ATP concentration as well as reduced glucose responsive insulin secretion (65). These result support the possibility that the dysfunction of mitophagy in β-cells accompanied with the decreased or increased expression of Parkin could be the cause of β-cell failure in a certain type of diabetes.
Free Fatty Acids (FFAs) and Autophagy
Increased LC3-II levels and autophagosomes in the β-cells of mice on a high-fat diet suggest that autophagy in β-cells is enhanced by insulin resistance (45). Candidate mediators for sensing systemic insulin resistance in β-cells to enhance autophagy are long-chain FFAs. Previous studies showed that palmitate stimulates autophagic flux and enhances the formation of autophagosomes and autolysosomes in β-cells (66–68).
Regarding the mechanism of FFA-induced autophagy, Komiya et al (69) demonstrated that FFAs activate protein kinase RNA-activated (PKR) and that FFA-induced autophagy is partially blocked by a c-Jun-NH2-terminal kinase (JNK) inhibitor, suggesting the involvement of the PKR-JNK pathway in β-cells. Recent data showed that palmitate disrupts inhibitory Signal tranducer and activator of transcription (STAT)3-PKR interactions and induces PKR-dependent eukaryotic Initiation Factor (eIF)2α phosphorylation, which facilitates the induction of autophagy, although this was not tested in β-cells (70). Another study reported that FFA-induced autophagy in β-cells is regulated by Protein kinase C (PKC)-α (71). Taken together, multiple mechanisms may be involved in FFA-induced autophagy in β-cells. On the other hand, long-term exposure to FFAs suppresses autophagy in β-cells, suggesting the involvement of lipotoxicity in the regulation of autophagy (72).
Autophagy, ER Stress, and β-Cell Death
Pancreatic β-cells undergo extensive proinsulin biosynthesis, and therefore face a high protein-folding burden. An increased demand of insulin biosynthesis in insulin-resistant states readily increases the accumulation of misfolded proteins in the ER of β-cells (73). This stimulates the unfolded protein response by releasing Binding immunoglobulin protein (Bip)/glucose-regulated protein 78 from protein kinase R-like endoplasmic reticulum kinase (PERK), inositol-requiring 1 (IRE1), and activating transcription factor 6. Indeed, the enhanced expression of ER stress markers was observed in β-cells in autopsy samples of the patients with type 2 diabetes (74).
An increase of ER stress induces autophagy. PERK was shown to activate the transcription of LC3 and Atg5 through the action of the transcription factors activating transcription factor (ATF) 4 and CCAAT-enhancer-binding protein (C/EBP)-homologous protein (CHOP) in a tumor hypoxia cell model (75). In addition, although the mechanism has not yet been clarified, phosphorylation of eIF2α on Ser51 by PERK plays a major role in the activation of autophagy (76). Furthermore, previous data showed that the autophagy system is activated by the IRE1-JNK pathway in response to ER stress in neuroblastoma cells (77). On the other hand, the IRE1-X-box-binding protein 1 (XBP1) signal inhibits autophagy, probably to decrease the excessive autophagy activated by other ER stress signaling pathways in neuronal cells (78). Thus, multiple systems may be involved in the activation and regulation of autophagy by ER stress. The underlying mechanism of ER stress induced autophagy in β-cells could be different from that in other cells.
Activation of autophagy by increased ER stress plays a protective role in β-cell damage. Indeed, stimulation of autophagy using mTOR inhibitors improved β-cell function in the Akita mouse, which is a diabetic mouse model resulting from increased ER stress (43). In addition, β-cell-specific deletion of tuberous sclerosis complex 2 (TSC2) that induces mTORC1 hyperactivation eventually caused β-cell failure with accumulation of p62/SQSTM1 and an impaired autophagic response (79). Conversely, Atg7f/f:RIP-Cre mice showed a compromised unfolded protein response and increased cell death by ER stressors (42, 45). Accordingly, increased β-cell death caused by autophagic failure especially under insulin-resistant state could be at least in part due to increased vulnerability of β-cells to ER stress. On the other hand, there appears to be a mechanism to compensate for cell death by autophagic failure. Indeed, expression of ERp57/GRP58 is enhanced in islets isolated from Atg7f/f:RIP-Cre mice. ERp57 is a member of the protein disulfide isomerase family that supports the proper folding of glycoproteins. Knock-down of ERp57 in β-cells with autophagic failure enhances apoptotic cell death, suggesting the role of ERp57 in protecting cells from apoptosis induced by autophagic failure (80).
On the other hand, a study reported that reduced β-cell mass in Pancreatic and duodenal homeobox (Pdx)1 hetero knockout mice was associated with increased autophagy in pancreatic islets. Crossing with Beclin-1 hetero knockout mice, which are a model of decreased autophagy, restored β-cell function and preserved β-cell mass in Pdx1 hetero knockout mice (19). Given that Beclin-1 is considered to also have functions in processes other than autophagy, such as in membrane trafficking (81), the reduced expression of Beclin-1 may preserve β-cell function through a mechanism independent of autophagy. However, it is still possible that accelerated autophagy may contribute to β-cell death under particular conditions, such as in Pdx1 hetero knockout mice.
Human Islet Amyloid Polypeptide (IAPP) and Autophagy
The presence of extracellular amyloid plaques is a morphological change that is observed in the islets of patients with type 2 diabetes. Amyloid plaques consist of IAPP, which is specifically expressed in β-cells. In normal β-cells, IAPP is present in insulin-containing granules and are cosecreted with insulin, and an increase in the demand of insulin enhances IAPP secretion. IAPP is a 37-amino acid peptide, and amino acids 20–29 are responsible for its amyloidogenic property. Recent data suggest that intracellular IAPP oligomer formation rather than the extracellular deposition of amyloid plaques is toxic to β-cells (82). Amino acids 25, 28, and 29 of rodent IAPPs are different from human IAPP (substituted to prolines), and thus, rodent IAPPs do not have amyloidogenicity and do not form toxic oligomers (82). Although the mechanism of toxic oligomer formation has not been clarified, the increased proteolysis of IAPP oligomers may prevent its toxicity. Intriguingly, autophagic flux in β-cells is enhanced by the forced expression of human IAPP, but not rat IAPP (83, 84). These data suggest the presence of a mechanism to eliminate the toxicity of human IAPP by the induction of autophagy. In addition, recent data revealed that IAPP is ubiquitinated and digested by autophagy through its interaction with p62 (85). The importance of this system is supported by the fact that a deficiency of ubiquitin carboxyl-terminal esterase L1, which results in dysfunction of the ubiquitin/proteasome system exacerbates human IAPP toxicity in β-cells with increased accumulation of p62 (86). Butler's group found that mice resulting from crossing transgenic mice expressing human IAPP with Atg7f/f:RIP-Cre show decreased glucose tolerance, accumulation of toxic human IAPP oligomers, increased oxidative damage, and decreased β-cell mass (85). Lee's group also reported a similar phenotype in the same model mice. In addition, they demonstrated that trehalose, an autophagy enhancer ameliorates the glucose intolerance of human IAPP transgenic mice (87). Considering that human IAPP transgenic mice express much larger amounts of IAPP than physiological levels, our group crossed human IAPP knock-in mice that carry human IAPP in the endogenous mouse IAPP locus with Atg7f/f:RIP-Cre mice and found that the resultant mice had a modest but similar phenotype to Butler's group and Lee's group (84). These results suggest that autophagy in β-cells is essential for the protection from toxic oligomer formation of human IAPP. Therefore, reduced autophagy in β-cells may elicit more damage in humans than in rodents.
α-synuclein, which is a protein involved in the pathogenesis of Parkinson's disease, also forms oligomers. α-synuclein is a component of the Lewy bodies observed in the neurons of Parkinson's disease patients and was reported to impair autophagy (88). Interestingly, this protein is expressed also in β-cells. A recent study revealed that loss of the Ide gene, which is located in a locus that is associated with the onset of type 2 diabetes, enhanced the β-cell expression of α-synuclein oligomers, because Ide somehow by interacting with α-synuclein reduce the formation of α-synuclein oligomers (89). Indeed, in islets from the patients with type 2 diabetes, Ide expression is decreased and expression of α-synuclein oligomers is increased. In addition, overexpression of α-synuclein in β-cells caused reduced glucose tolerance with decreased insulin secretion with increased autophagic flux in the islets. These results suggest that the increased autophagy may be a protective mechanism against toxicity of α-synuclein oligomers; however, the increased autophagic activity in β-cells is not enough to prevent the toxicity of α-synuclein oligomers in this model. It is interesting to note that α-synuclein knockout mice also showed reduced glucose tolerance with reduced insulin secretion. These data show that the role of α-synuclein in β-cells is more complex than predicted.
β-Cell Inflammation and Autophagy
β-Cell inflammation has now been proposed as a mechanism of the deterioration of β-cell function not only in type 1 diabetes, but also in type 2 diabetes. Previous studies showed increased immune-cell infiltration in the islets of patients with type 2 diabetes (90, 91). In addition, up-regulation of proinflammatory cytokines and chemokines was observed in the islets from patients with type 2 diabetes (92, 93). In this regard, disruption of Atg16L1, an essential molecule for the formation of autophagosomes in macrophages, enhances the lipopolysaccharide-induced production of the inflammatory cytokines IL-1β and IL-18 through activation of the inflammasome in macrophage (94). In addition, depletion of LC3B and Beclin-1 enhances the activation of caspase-1 and the secretion of IL-1β and IL-18 in macrophage (95). Furthermore, reduced autophagy in the hypothalamus by site-specific delivery of a lentiviral small hairpin RNA (shRNA) against Atg7 impairs hypothalamic control of energy balance by activation of the Nuclear factor-kappa B (NFκB) signaling pathway (96). Therefore, a large amount of evidence has accumulated regarding the tight link between autophagy and immunity (97).
Regarding the inflammation of islets in type 2 diabetic models, the source of the inflammatory mediators in islets has not been determined. However, β-cells are regarded as candidate cells for the production of inflammatory cytokines in islets as well as in macrophages. Accordingly, autophagic failure in β-cells may enhance the inflammatory reaction and the secretion of inflammatory cytokines. Conversely, recent data showed that overexpression of Atg7 induces the activation of autophagy and NACHT, LRR, and PYD domains-containing protein 3 (NLRP3)-dependent IL-1β secretion, suggesting the harmful effect of autophagic activation in β-cells (98). Thus, further studies are required to clarify the association between autophagy and immunity in β-cells.
Crinophagy in pancreatic β-Cells
“Crinophagy,” the process of direct fusion of secretory vesicles with lysosomes has been known for years as an ATG protein degradation mechanism in endocrine cells (99). Classically, Orci et al described that insulin granules are degraded by this process (100). Given that a more than sufficient number of insulin-containing granules are present in β-cells (16) and that the half-life of insulin-containing granules are estimated as 3–5 days (101), the degradation of insulin is an important process to maintain a constant number of insulin-containing granules in β-cells. Crinophagy seems to play an important role in the homeostasis of insulin-containing granules.
Rab3A is an essential molecule for transporting insulin granules to the cell surface for exocytosis in β-cells. _Rab3_−/− islets show reduced insulin secretion without affecting insulin production (102), suggesting a relatively increased number of insulin-containing granules in β-cells. However, Marsh et al reported that in this mouse, normal number of insulin granules is maintained by marked increase of insulin degradation by crinophagy (103). In addition, in these mice, crinophagy is further activated by treatment with diazoxide, an agent that inhibits insulin secretion (103). These results suggest that crinophagy is a process to maintain a constant number of insulin-containing granules in β-cells. However, the molecular mechanism of crinophagy has not been clarified at all. Considering that crinophagy is the process of direct fusion of lysosomes with hormone-containing granules, the mechanism regulating crinophagy is likely to be completely different from that of macroautophagy. In addition, because there are no established methods to date to specifically activate or inhibit crinophagy, the physiological role of crinophagy remains a matter of speculation.
Concluding Remarks
Insulin resistance triggers an increase in β-cell mass and an enhancement of insulin secretion to preserve normoglycemia. Failure of this compensation is regarded as the fundamental cause of hyperglycemia observed in type 2 diabetes mellitus. In insulin-resistant states caused by a high-fat diet, physiological inactivity, pregnancy, and the exposure of β-cells to harmful factors such as FFAs, glucose, inflammatory cytokines, and IAPP is increased. These factors induce oxidative stress, ER stress, and inflammatory stress and result in the dysfunction of and enhanced death of β-cells.
Most data suggest that enhanced autophagy induced by insulin resistance may be a protective mechanism against the dysfunction of and enhanced death of pancreatic β-cells. However, the precise mechanism of autophagic regulation and its protective mechanism against cell death have not yet been elucidated. Identifying the role of autophagy in cell survival or cell death during the progression to diabetes is an important task for researchers in this field. Furthermore, the development of techniques that allow manipulation of autophagy should have a significant impact on the future development of therapeutic approaches to diabetes.
Acknowledgments
This work was supported by The Ministry of Education, Sports and Culture of Japan Grants 23390244, 26293220, and 26111518 (to H.W.) and 22590996 (to Y.F.), Daiichi-Sankyo Foundation of Life Science (H.W. and Y.F.), and the UBE Foundation (H.W.).
Disclosure Summary: H.W. is a member of advisory panel of Novo Nordisk Pharma, Sanofi, Dainippon Sumitomo Pharma, Mochida Pharmaceutical Co, MSD, Takeda Pharmaceutical Co, Boehringer Ingelheim, Ono Pharmaceutical Co, Novartis Pharmaceuticals, Mitsubishi-Tanabe Pharma, AstraZeneca, Kowa Co Astellas Pharma, Inc, and Pfizer, Inc; has received lecture fees from Novo Nordisk Pharma, Eli Lilly Japan, Sanofi, Dainippon Sumitomo Pharma, Fujifilm, Bayer, Kissei Pharmaceutical Co, Mochida Pharmaceutical Co, MSD, Takeda Pharmaceutical Co, Boehringer Ingelheim, Daiichi Sankyo, Inc, Ono Pharmaceutical Co Novartis Pharmaceuticals, Boehringer Ingelheim, Mitsubishi-Tanabe Pharma, AstraZeneca, Kyowa Hokko Kirin Co, Sanwa Kagaku Kenkyusho Co, Kowa Co Astellas Pharma, Inc, and Pfizer, Inc; and received research funds from Johnson & Johnson, Kyowa Hokko Kirin Co, Kissei Pharmaceutical Co, Bristol-Myers Squibb, Novo Nordisk Pharma, Astellas Pharma, Inc, MSD, Dainippon Sumitomo Pharma., AstraZeneca, Teijin Pharma, Mochida Pharmaceutical Co, Sanofi, Sanwa Kagaku Kenkyusho Co, Boehringer Ingelheim, Pfizer, Inc, Novartis Pharmaceuticals, Ono Pharmaceutical Co, Mitsubishi-Tanabe Pharma, Daiichi Sankyo, Inc, Takeda Pharmaceutical Co, Eli Lilly Japan, and Taisho Toyama Pharmaceutical Co. Y.F. has received lecture fees from Novartis and Eli Lilly and research funding from MSD and Takeda.
Funding Statement
This work was supported by The Ministry of Education, Sports and Culture of Japan Grants 23390244, 26293220, and 26111518 (to H.W.) and 22590996 (to Y.F.), Daiichi-Sankyo Foundation of Life Science (H.W. and Y.F.), and the UBE Foundation (H.W.).
Footnotes
Abbreviations:
Ambra1
activating molecule in beclin-1-regulated autophagy
ATG
autophagy related
Clec16a
C-type lectin domain family 16 member A
ER
endoplasmic reticulum
FFA
free fatty acid
FIP200
focal adhesion kinase family interacting protein of 200 kDa
IAPP
islet amyloid polypeptide
IRE1
inositol-requiring 1
JNK
c-Jun-NH2-terminal kinase
LC3
microtubule-associated protein 1A/1B-light chain 3
mTOR
mammalian target of rapamycin
mTORC
mTOR complex
Pdx1
Pancreatic and duodenal homeobox 1
PERK
dsRNA-activated protein kinase-like ER kinase
PI(3)K
phosphatidylinositol 3-kinase
PINK1
PTEN-induced putative kinase 1
PKR
protein kinase RNA activated
ULK1
Unc-51-like kinase 1
Vps34
Vacuolar Protein Sorting.
References
- 1.Klionsky DJ, Cuervo AM, Dunn WA Jr, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy. 2007;3(5):413–416. [DOI] [PubMed] [Google Scholar]
- 2.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23(2):184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahu R, Kaushik S, Clement CC, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20(1):131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. [DOI] [PubMed] [Google Scholar]
- 5.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim KH, Lee MS. Autophagy–a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10(6):322–337. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. [DOI] [PubMed] [Google Scholar]
- 8.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. [DOI] [PubMed] [Google Scholar]
- 10.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. [DOI] [PubMed] [Google Scholar]
- 11.Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13(5):619–624. [DOI] [PubMed] [Google Scholar]
- 12.Saisho Y, Butler AE, Manesso E, Elashoff D, Rizza RA, Butler PC. β-Cell mass and turnover in humans: effects of obesity and aging. Diabetes Care. 2013;36(1):111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahier J, Guiot Y, Goebbels RM, Sempoux C, Henquin JC. Pancreatic β-cell mass in European subjects with type 2 diabetes. Diabetes Obes Metab. 2008;10 Suppl 4:32–42. [DOI] [PubMed] [Google Scholar]
- 14.Ogihara T, Mirmira RG. An islet in distress: β cell failure in type 2 diabetes. J Diabetes Investig. 2010;1(4):123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prentki M, Nolan CJ. Islet β cell failure in type 2 diabetes. J Clin Invest. 2006;116(7):1802–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rorsman P, Renström E. Insulin granule dynamics in pancreatic β cells. Diabetologia. 2003;46(8):1029–1045. [DOI] [PubMed] [Google Scholar]
- 17.Koike M, Shibata M, Tadakoshi M, et al. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am J Pathol. 2008;172(2):454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohmuraya M, Hirota M, Araki M, et al. Autophagic cell death of pancreatic acinar cells in serine protease inhibitor Kazal type 3-deficient mice. Gastroenterology. 2005;129(2):696–705. [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto K, Hanson PT, Tran H, et al. Autophagy regulates pancreatic β cell death in response to Pdx1 deficiency and nutrient deprivation. J Biol Chem. 2009;284(40):27664–27673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. [DOI] [PubMed] [Google Scholar]
- 21.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5(7):973–979. [DOI] [PubMed] [Google Scholar]
- 22.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polson HE, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6(4):506–522. [DOI] [PubMed] [Google Scholar]
- 27.Di Bartolomeo S, Corazzari M, Nazio F, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191(1):155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nazio F, Strappazzon F, Antonioli M, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15(4):406–416. [DOI] [PubMed] [Google Scholar]
- 29.Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, Kim YC, Fang C, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152(1–2):290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang RC, Wei Y, An Z, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338(6109):956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283(33):22847–22857. [DOI] [PubMed] [Google Scholar]
- 34.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. [DOI] [PubMed] [Google Scholar]
- 35.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci USA. 2014;111(42):E4439–E4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–1269. [DOI] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10(3):431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebato C, Uchida T, Arakawa M, et al. Autophagy is important in islet homeostasis and compensatory increase of β cell mass in response to high-fat diet. Cell Metab. 2008;8(4):325–332. [DOI] [PubMed] [Google Scholar]
- 40.Jung HS, Chung KW, Won Kim J, et al. Loss of autophagy diminishes pancreatic β cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8(4):318–324. [DOI] [PubMed] [Google Scholar]
- 41.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. [DOI] [PubMed] [Google Scholar]
- 42.Quan W, Hur KY, Lim Y, et al. Autophagy deficiency in β cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia. 2012;55(2):392–403. [DOI] [PubMed] [Google Scholar]
- 43.Bachar-Wikstrom E, Wikstrom JD, Ariav Y, et al. Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes. Diabetes. 2013;62(4):1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Zhang L, Meshinchi S, et al. Islet microvasculature in islet hyperplasia and failure in a model of type 2 diabetes. Diabetes. 2006;55(11):2965–2973. [DOI] [PubMed] [Google Scholar]
- 45.Abe H, Uchida T, Hara A, et al. Exendin-4 improves β-cell function in autophagy-deficient β-cells. Endocrinology. 2013;154(12):4512–4524. [DOI] [PubMed] [Google Scholar]
- 46.Masini M, Bugliani M, Lupi R, et al. Autophagy in human type 2 diabetes pancreatic β cells. Diabetologia. 2009;52(6):1083–1086. [DOI] [PubMed] [Google Scholar]
- 47.Mizukami H, Takahashi K, Inaba W, et al. Involvement of oxidative stress-induced DNA damage, endoplasmic reticulum stress, and autophagy deficits in the decline of β-cell mass in Japanese type 2 diabetic patients. Diabetes Care. 2014;37(7):1966–1974. [DOI] [PubMed] [Google Scholar]
- 48.Zatloukal K, Stumptner C, Fuchsbichler A, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160(1):255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakaso K, Yoshimoto Y, Nakano T, et al. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson's disease. Brain Res. 2004;1012(1–2):42–51. [DOI] [PubMed] [Google Scholar]
- 50.Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH. Ubiquitinated-protein aggregates form in pancreatic β-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes. 2007;56(4):930–939. [DOI] [PubMed] [Google Scholar]
- 51.Fujitani Y, Ebato C, Uchida T, Kawamori R, Watada H. β-Cell autophagy: a novel mechanism regulating β-cell function and mass: lessons from β-cell-specific Atg7-deficient mice. Islets. 2009;1(2):151–153. [DOI] [PubMed] [Google Scholar]
- 52.Fujitani Y, Kawamori R, Watada H. The role of autophagy in pancreatic β-cell and diabetes. Autophagy. 2009;5(2):280–282. [DOI] [PubMed] [Google Scholar]
- 53.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304(5674):1158–1160. [DOI] [PubMed] [Google Scholar]
- 55.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. [DOI] [PubMed] [Google Scholar]
- 56.Kawajiri S, Saiki S, Sato S, et al. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584(6):1073–1079. [DOI] [PubMed] [Google Scholar]
- 57.Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8(1):e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geisler S, Holmström KM, Skujat D, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. [DOI] [PubMed] [Google Scholar]
- 59.Vives-Bauza C, Zhou C, Huang Y, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107(1):378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22(2):320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6(8):1090–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu JJ, Quijano C, Chen E, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY). 2009;1(4):425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoshino A, Ariyoshi M, Okawa Y, et al. Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic β-cell function in diabetes. Proc Natl Acad Sci USA. 2014;111(8):3116–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soleimanpour SA, Gupta A, Bakay M, et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. 2014;157(7):1577–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi SE, Lee SM, Lee YJ, et al. Protective role of autophagy in palmitate-induced INS-1 β-cell death. Endocrinology. 2009;150(1):126–134. [DOI] [PubMed] [Google Scholar]
- 67.Lupi R, Dotta F, Marselli L, et al. Prolonged exposure to free fatty acids has cytostatic and pro-apoptotic effects on human pancreatic islets: evidence that β-cell death is caspase mediated, partially dependent on ceramide pathway, and Bcl-2 regulated. Diabetes. 2002;51(5):1437–1442. [DOI] [PubMed] [Google Scholar]
- 68.Martino L, Masini M, Novelli M, et al. Palmitate activates autophagy in INS-1E β-cells and in isolated rat and human pancreatic islets. PLoS One. 2012;7(5):e36188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komiya K, Uchida T, Ueno T, et al. Free fatty acids stimulate autophagy in pancreatic β-cells via JNK pathway. Biochem Biophys Res Commun. 2010;401(4):561–567. [DOI] [PubMed] [Google Scholar]
- 70.Shen S, Niso-Santano M, Adjemian S, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012;48(5):667–680. [DOI] [PubMed] [Google Scholar]
- 71.Tan SH, Shui G, Zhou J, et al. Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin). J Biol Chem. 2012;287(18):14364–14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Las G, Serada SB, Wikstrom JD, Twig G, Shirihai OS. Fatty acids suppress autophagic turnover in β-cells. J Biol Chem. 2011;286(49):42534–42544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leibowitz G, Kaiser N, Cerasi E. β-Cell failure in type 2 diabetes. J Diabetes Investig. 2011;2(2):82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marchetti P, Bugliani M, Lupi R, et al. The endoplasmic reticulum in pancreatic β cells of type 2 diabetes patients. Diabetologia. 2007;50(12):2486–2494. [DOI] [PubMed] [Google Scholar]
- 75.Rouschop KM, van den Beucken T, Dubois L, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120(1):127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14(2):230–239. [DOI] [PubMed] [Google Scholar]
- 77.Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26(24):9220–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hetz C, Thielen P, Matus S, et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23(19):2294–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bartolomé A, Kimura-Koyanagi M, Asahara S, et al. Pancreatic β-cell failure mediated by mTORC1 hyperactivity and autophagic impairment. Diabetes. 2014;63(9):2996–3008. [DOI] [PubMed] [Google Scholar]
- 80.Yamamoto E, Uchida T, Abe H, et al. Increased expression of ERp57/GRP58 is protective against pancreatic β cell death caused by autophagic failure. Biochem Biophys Res Commun. 2014; 453(1):19–24 [DOI] [PubMed] [Google Scholar]
- 81.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20(6):355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haataja L, Gurlo T, Huang CJ, Butler PC. Islet amyloid in type 2 diabetes, and the toxic oligomer hypothesis. Endocr Rev. 2008;29(3):303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rivera JF, Gurlo T, Daval M, et al. Human-IAPP disrupts the autophagy/lysosomal pathway in pancreatic β-cells: protective role of p62-positive cytoplasmic inclusions. Cell Death Differ. 2011;18(3):415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shigihara N, Fukunaka A, Hara A, et al. Human IAPP-induced pancreatic β cell toxicity and its regulation by autophagy. J Clin Invest. 2014;124(8):3634–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rivera JF, Costes S, Gurlo T, Glabe CG, Butler PC. Autophagy defends pancreatic β cells from human islet amyloid polypeptide-induced toxicity. J Clin Invest. 2014;124(8):3489–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Costes S, Gurlo T, Rivera JF, Butler PC. UCHL1 deficiency exacerbates human islet amyloid polypeptide toxicity in β-cells: evidence of interplay between the ubiquitin/proteasome system and autophagy. Autophagy. 2014;10(6):1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim J, Cheon H, Jeong YT, et al. Amyloidogenic peptide oligomer accumulation in autophagy-deficient β cells induces diabetes. J Clin Invest. 2014;124(8):3311–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winslow AR, Chen CW, Corrochano S, et al. α-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol. 2010;190(6):1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Steneberg P, Bernardo L, Edfalk S, et al. The type 2 diabetes-associated gene ide is required for insulin secretion and suppression of α-synuclein levels in β-cells. Diabetes. 2013;62(6):2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ehses JA, Perren A, Eppler E, et al. Increased number of islet-associated macrophages in type 2 diabetes. Diabetes. 2007;56(9):2356–2370. [DOI] [PubMed] [Google Scholar]
- 91.Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG. Islet-associated macrophages in type 2 diabetes. Diabetologia. 2009;52(8):1686–1688. [DOI] [PubMed] [Google Scholar]
- 92.Gunton JE, Kulkarni RN, Yim S, et al. Loss of ARNT/HIF1β mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122(3):337–349. [DOI] [PubMed] [Google Scholar]
- 93.Bugliani M, Liechti R, Cheon H, et al. Microarray analysis of isolated human islet transcriptome in type 2 diabetes and the role of the ubiquitin-proteasome system in pancreatic β cell dysfunction. Mol Cell Endocrinol. 2013;367(1–2):1–10. [DOI] [PubMed] [Google Scholar]
- 94.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456(7219):264–268. [DOI] [PubMed] [Google Scholar]
- 95.Nakahira K, Haspel JA, Rathinam VA, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meng Q, Cai D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IκB kinase β (IKKβ)/NF-κB pathway. J Biol Chem. 2011;286(37):32324–32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li S, Du L, Zhang L, et al. Cathepsin B contributes to autophagy-related 7 (Atg7)-induced nod-like receptor 3 (NLRP3)-dependent proinflammatory response and aggravates lipotoxicity in rat insulinoma cell line. J Biol Chem. 2013;288(42):30094–30104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Farquhar MG. Secretion and crinophagy in prolactin cells. Adv Exp Med Biol. 1977;80:37–94. [DOI] [PubMed] [Google Scholar]
- 100.Orci L, Ravazzola M, Amherdt M, et al. Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B-cell. J Cell Biol. 1984;98(1):222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Halban PA, Wollheim CB. Intracellular degradation of insulin stores by rat pancreatic islets in vitro. An alternative pathway for homeostasis of pancreatic insulin content. J Biol Chem. 1980;255(13):6003–6006. [PubMed] [Google Scholar]
- 102.Yaekura K, Julyan R, Wicksteed BL, et al. Insulin secretory deficiency and glucose intolerance in Rab3A null mice. J Biol Chem. 2003;278(11):9715–9721. [DOI] [PubMed] [Google Scholar]
- 103.Marsh BJ, Soden C, Alarcón C, et al. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine β-cells. Mol Endocrinol. 2007;21(9):2255–2269. [DOI] [PubMed] [Google Scholar]