Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs (original) (raw)
Abstract
MicroRNAs (miRNAs) are believed to play important roles in developmental and other cellular processes by hybridizing to complementary target mRNA transcripts. This results in either cleavage of the hybridized transcript or negative regulation of translation. Little is known about the regulation or pattern of miRNA expression. The predicted presence of numerous miRNA sequences in higher eukaryotes makes it highly likely that the expression levels of individual miRNA molecules themselves should play an important role in regulating multiple cellular processes. Therefore, determining the pattern of global miRNA expression levels in mammals and other higher eukaryotes is essential to help understand both the mechanism of miRNA transcriptional regulation as well as to help identify miRNA regulated gene expression. Here, we describe a novel method to detect global processed miRNA expression levels in higher eukaryotes, including human, mouse and rats, by using a high-density oligonucleotide array. Array results have been validated by subsequent confirmation of mir expression using northern-blot analysis. Major differences in mir expression have been detected in samples from diverse sources, suggesting highly regulated mir expression, and specific gene regulatory functions for individual miRNA transcripts. For example, five different miRNAs were found to be preferentially expressed in human kidney compared with other human tissues. Comparative analysis of surrounding genomic sequences of the kidney-specific miRNA clusters revealed the occurrence of specific transcription factor binding sites located in conserved phylogenetic foot prints, suggesting that these may be involved in regulating mir expression in kidney.
INTRODUCTION
MicroRNAs (miRNAs) are ∼22 nt non-coding RNAs that can play important roles in cell function and development by targeting the mRNA sequences of protein-coding transcripts, resulting in either mRNA cleavage or repression of productive translation (1–4). Originally, miRNAs were discovered in the nematode Caenorhabditis elegans through genetic screens for mutants that lacked the ability to control the timing of specific cell fate switches during development (5,6). Several hundred miRNAs from C.elegans, plants, Drosophila melanogaster and mammals have since been identified through computational and cloning approaches (7–20).
There are currently estimated to be ∼200–255 miRNAs (8) present in the human genome. In C.elegans, there are >1000 molecules per cell, with some exceeding 50 000 copies per cell (9). The experimental validation of miRNA target mRNA remains a challenge. Many miRNAs are limited in their expression to certain stages in development or to certain tissues and cell types (4). Recently, it has been reported that human miRNA genes are frequently located in fragile sites and genomics regions involved in cancer (21). Therefore, the ability to determine global miRNA expression in mammalian cells will prove valuable in helping to understand the putative roles played by miRNAs in cell function. The first use of an array-based technology to profile microRNA expression was documented by Krichevsky et al. (22). Since then, a number of miRNA array papers have been published (23–28). In a recent review, Esquela-Kerscher and Slack (29) compared these different array based detection approaches. Our method has some unique features compared with those already published papers (22,23,26), and we discuss these differences in Results and Discussion.
In our study, we describe a robust array based technique, which allows us to identify the expression of 254 miRNAs in mammalian cells. To do this, oligonucleotides (5′-Amino-Modifier C6) corresponding to human and mouse mature sense miRNA sequences were designed to hybridize to biotin end-labeled antisense miRNA targets. We have used this approach to compare miRNA expression in six different human organs, and our array results indicate the presence of differentially expressed groups of miRNA in different human tissues. Wherever possible, we have compared our array data with these published results and these were found to be in high concordance. The array described here offers more comprehensive coverage and higher-throughput than northern blot (NB) approaches, and represents a valuable tool to better understand the recently identified class of gene regulating RNA molecules.
MATERIALS AND METHODS
miRNA array design
5′ Amine modified C6 oligonucleotides were resuspended in 1× Micro Spotting Plus buffer (ArrayIt, Sunnyvale, CA) at 20 μM concentration. Each oligonucleotide probe is printed four times on CodeLink-activated slides (GE health/Amersham Biosciences, Piscataway, NJ) by a Pixsys7000 pin-based dispensing system (Genomics Solutions, Irvine, CA) in 2 × 2 pin and 40 × 8 spot configuration of each sub-array, with a spot diameter of 120 μm. The printed slides are further processed according to the manufacturer's recommendations. The array also contains several 23 bp U6 and Drosophila tRNA oligonucleotides specifically designed as labeling and hybridization controls (positive) while 23 bp random oligonucleotides are designed as negative controls. To a limited extent, we have investigated the sensitivity, specificity and isothermal properties of the miRNA probes that we printed on the chip. We studied the secondary structure of the probes as well as the possibility of binding to other closely related probes on the array. Computational searches were also done for global cross hybridization. Even after testing standard parameters for microarray probe designing, it is not possible to design an alternate probe to a given processed miRNA due to the limited sequence length. Therefore, we have decided to include all the available miRNA sequences on the chip.
Sample (RNA) preparation for miRNA array
For target preparation, we obtained total RNA from six normal adult human tissues (prostate, lung, heart, kidney, spleen and skeletal muscle) from Ambion (Austin, TX).
From these samples, 20 μg of total RNA and 20 μl of random hexamer (75 ng/μl), from Invitrogen (Carlsbad, CA) were mixed at a final volume of 60 μl and incubated at 70°C for 10 min and then cooled to room temperature (RT). To this mixture, we performed a reverse transcription reaction to obtain first-strand cDNA products according to the modified SuperScript™ II Reverse Transcriptase Kit from Invitrogen, as follows: to the RNA/primer mixture was added 24 μl of 5× first-strand buffer, 12 μl of 0.1 M DTT, 6 μl of 10 mM dNTP mixture, 3 μl of SuperRase·In and 15 μl of SuperScript II RNaseH− reverse transcriptase (200 U/μl) and incubated for 10 min at RT, 1 h at 37°C and another 1 h at 42°C in a water bath. After incubation, 40 μl of 1 N NaOH was added to the first-strand cDNA reaction mixture and further incubated at 65°C for another 30 min to denature the RNA/DNA hybrids. Finally, 40 μl of 1 N HCl was added to neutralize the reaction mixture. cDNA was purified according to Qiaquick Nucleotide Removal Kit protocol (Qiagen, Valencia, CA). We used Enzo® BioArray Terminal Labeling Kit with Biotin-ddUTP (Enzo Life sciences, Farmingdale, NY) to label the 3′ termini of the first stand cDNA. An aliquot of 40 μl of 5× reaction buffer, 20 μl of 10× CoCl2, 2 μl of 100× Biotin-ddUTP and 4 μl of 50× terminal deoxynucleotide transferase were added to the first-strand cDNA at a final volume of 200 μl and incubated at 37°C for 1 h. The reaction was terminated by adding 4 μl of 0.5 M EDTA. The target is now ready to be hybridized onto miRNA arrays.
Target hybridization and detection
Hybridization buffer consists of 100 mM 2-(_N_-morpholino)ethanesulfonicacid (MES), 1 M [Na+], 20 mM EDTA, 0.01% Tween-20, 0.1 mg/ml Herring sperm DNA and 0.5 mg/ml acetylated BSA. Target hybridization was done at 45°C for 16 h, and slides were washed four times (6 min each) in buffer A (6× SSPE and 0.01% Tween-20) at RT, and then twice with buffer B (100 mM MES, 0.1 M [Na+] and 0.01% Tween-20) for 8 min at 45°C. Slides were then incubated for staining with Streptavidin solution mixture (100 mM MES, 1 M [Na+], 0.05% Tween-20, 2 mg/ml BSA and 10 μg/ml R-Phycoerythrin streptavidin) from Invitrogen at RT for 10 min followed by four washes with buffer A (6 min each) at 30°C.
Second staining was carried out with antibody solutions (100 mM MES, 1 M [Na+], 0.05% Tween-20, 2 mg/ml BSA, 0.1 mg/ml goat IgG and 5 μg/ml biotin anti-streptavidin) at RT for 10 min followed by washing with buffer A (twice) for 4 min. Third staining was performed with Streptavidin solution mixture at RT for 10 min and slides were washed four times (6 min each) with wash buffer A at 30°C. Finally, slides were washed one time, 5 min each at RT with 0.2× SSC and followed by a similar wash with 0.1× SSC to remove any salt remnant and binding particles to the slides.
Statistical analysis
Axon 4000B scanner and the GenePix Pro 4.0 software (Axon Instruments Union city, CA) were used to scan images. The median intensities of each feature and of the corresponding background were measured. The median intensity of the background was subtracted from the median intensity of the feature. Outliers detected by the ESD procedure (30) were also removed at this stage. The resulting signal intensity values were normalized to per-chip median values. These signal intensity values were then used to obtain geometric means and standard errors for each miRNA. Each miRNA signal was transformed to log base 2 and 1-sample _t_-test was conducted. If the signal was significantly (P ≤ 0.05) high or low compared with the chip median then a Present (P) or Absent (A) call is assigned. A Marginal (M) call is given otherwise. Our method of miRNA array data analysis has some unique features compared with those published previously. For example, our analysis method has built-in automatic outlier detection algorithms, called the ESD procedure (30), and stringent expression call algorithms based on statistics.
Data clustering (_K_-means)
Expression profiles of miRNAs across different human organs are clustered using the _K_-means clustering algorithm (31). In this algorithm, observations are clustered as belonging to one of k groups. Initially, group centroids are randomly assigned. Group membership is then determined by calculating the centroid of each group and assigning each observation to the group with the closest centroid. The algorithm alternates between calculating the centroids based on the current group memberships, and reassigning observations to groups based on the new centroids. The above iterations stop when a predetermined convergence limit is met.
RESULTS AND DISCUSSION
Sensitivity and the specificity of the miRNA array
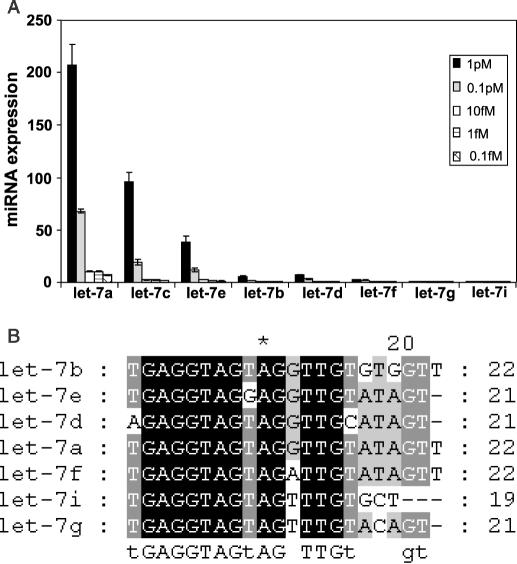
The specificity and the sensitivity of this array based approach were initially determined using a small array designed to detect _Let_-7 miRNA expression (Figure 1A). This array consists of seven different _Let_-7 probes to each of the closely related _Let_-7 miRNAs (Figure 1A). This was used to detect a spiked sample of a Let-7a mir transcript. Compared with the Let-7a sequence, _Let_-7c and _Let_-7e contain one mismatch each, _Let_-7b and _Let_-7d contain two mismatches, while _Let_-7g and _Let_-7i contain multiple mismatches. At both 1 and 0.1 pM, Let-7a signals are clearly distinguishable from Let-7c and Let-7e (Figure 1A). At 1 pM Let-7a signal is more than 2.2- and 5.4-fold higher than that of Let-7c and Let-7e, respectively. These differences are statistically highly significant (P < 0.0001). Similarly, at 0.1 pM Let-7a signal is more than 3.6- and 5.7-fold higher than that of Let-7c and Let-7e, respectively (P < 0.0001). Hence, we have demonstrated here the ability of our arrays to distinguish the expression of miRNAs with one or more mismatches from that of the perfect match with high specificity.
Figure 1.
miRNA array specificity. The array was probed with a _Let_-7a transcript and detection was performed as described in Materials and Methods. Signal intensity values of the different _Let_-7 mirs are plotted in (A). Sequences of Let-7 mirs are seen in (B).
Validation of miRNA array data with NBs
A total of 254 oligonucleotides (5′-Amino-Modifier C6) corresponding to human and mouse mature sense miRNA sequences designed to hybridize onto biotin end-labeled antisense miRNA targets, at a final concentration of 20 μM are spotted (four chip replicates) on a three-dimensional (3D) CodeLink slides (GE health/Amersham Biosciences) and processed according to the manufacturer's recommendations (see Materials and Methods). An aliquot of 20 μg of total RNA from six different human tissues was taken as targets for each array. As described in Materials and Methods, target labeling is based on first-strand cDNA 3′-ddUTP biotin end-labeling with terminal dideoxy transferase enzyme.
Recently, Liu et al. (23) published an array-based technique to measure miRNA expression in human cell lines. Our protocols are significantly different from the other miRNA array papers published recently, including Liu et al., in terms of target labeling, array hybridization, slide processing as well as statistical data analysis procedure. For example, target labeling (biotin-ddUTP), hybridization and processing are different from published methods. During the slide processing, double staining with R-Phycoerythrin Streptavidin increased the overall signal levels by several folds and therefore, we were able to precisely discriminate raw signal intensity values from the background noise. As discussed above in Materials and Methods, our statistical analysis method of miRNA array data also has unique features, such as outlier detection and expression calls, compared with the recently published methods.
In order to validate our miRNA array data, we have compared our miRNA expression data with the corresponding NB data from Sempere et al. (32). Sempere et al. quantified NB bands using ImageQuant software and calculated a ratio of signal intensity in the band to the signal in the background. In order to compare our results with theirs, a Present call (P) was given if the ratio is >2.0, an Absent call (A), if the ratio is <1.1 and a Marginal call (M) otherwise. After combining their NB and our miRNA array data, we have calculated the sensitivity and the specificity of both experimental results for the two common tissues, human heart and skeletal muscle. Sensitivity is defined as the probability that the miRNA array yields a positive result, given that the NB data are positive. On the other hand, specificity is defined as the probability that the miRNA array yields a negative result, given that the NB data is negative.
Tables 1 and 2 show the comparison between the two tissues, human heart and skeletal muscle, respectively. Fisher's exact test is used to determine if there are non-random associations between categorical variables from the miRNA array and the NB data (e.g. A, M or P calls). For both human heart and skeletal muscle, the association between miRNA array and NB is found to be extremely significant (P < 0.0001). According to Table 1, 25 miRNAs are called to be Present in human heart by NB and 21 of them are also called to be Present by miRNA array. Similarly, 47 miRNAs are called to be Absent in human heart by NB and 33 of them are also called to be Absent by miRNA array. In contrast, 4 miRNAs called to be Present in human heart by NB are called to be Absent by miRNA array. A total of 11 miRNAs called to be Absent in human heart by NB are called to be Present by miRNA array. These results show 84% sensitivity and 75% specificity. Similar analysis on human skeletal muscle (Table 2) shows 89% sensitivity and 83% specificity. This result indicates a high concordance between our miRNA array and the NB data.
Table 1. Comparison between miRNA array data and NB data (32) for human heart.
| Northern blot | Array | ||
|---|---|---|---|
| A | M | P | |
| A | 33 | 3 | 11 |
| M | 10 | 8 | 17 |
| P | 4 | 0 | 21 |
Table 2. Comparison between miRNA array data and NB data (32) for human skeletal muscle.
| Northern blot | Array | ||
|---|---|---|---|
| A | M | P | |
| A | 33 | 9 | 7 |
| M | 14 | 5 | 19 |
| P | 2 | 1 | 17 |
At this early developmental stage it is hard to make any conclusions on which method is better. Such a conclusion must surely be based on direct head-to-head comparisons, and any other method will be speculative, therefore, likely misleading. Also it is important to take into account not only the processed mir expression but also the possible pre- and pri-mir expression in cells. Designing pre- and pri-mir probes along side with a processed mir on the array may answer this question.
Organ-specific and enriched miRNAs in six human organs (heart, kidney, spleen, skeletal muscle, lung and prostate)
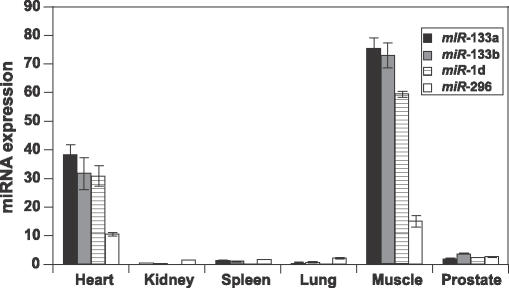
Array miRNA expression profiling has identified differentially expressed miRNAs in six different human organs. Two chip replicates and quadruple spots for each miRNA provided adequate signal intensity information for data comparison across organs. We applied unsupervised _K_-means clustering technique to characterize array data and identified several organ-specific and organ-enriched expression clusters. Figure 2 shows a human heart and skeletal muscle specific cluster that contains _miR_-133a, _miR_-133b, _miR_-1d and _miR_-296. _miR_-133a and _miR_-1d are located on chromosome 18, 3312 bp apart from each other, and may originate from a common transcript. Copies of these miRNAs (_miR_-133a-2 and _miR_-1d) reside in chromosome 20 and the distance between them is 10 618 bp. _miR_-296 also resides in chromosome 20, but its location is not close to _miR_-133a-2 and _miR_-1d. Array data also confirmed that _miR_-133b and _miR_-206d are co-regulated and highly expressed in skeletal muscles and not in any other tissues tested. Both of these miRNAs are located in chromosome 6 and the distance between them is 4576 bp. Again it is possible that these miRNAs may have a common transcriptional regulation. Tissue-specific miRNA expression has been discussed previously by several other groups (13,14,22,32). _MiR_-1 was previously shown by northern-blot analysis to be strongly expressed in human adult heart but not in brain, liver, kidney, lung or colon (13).
Figure 2.
Human heart and skeletal muscle enriched mir cluster. Each bar represents the expression of an average signal intensity value of a given mir. Expression of _miR_-133a, _miR_-133b and _miR_-1d are higher in skeletal muscle compared with that in heart. There is no or very little expression of these mirs in other tested organs.
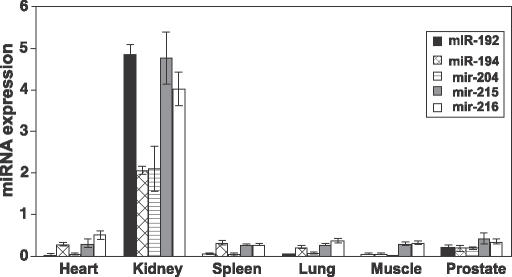
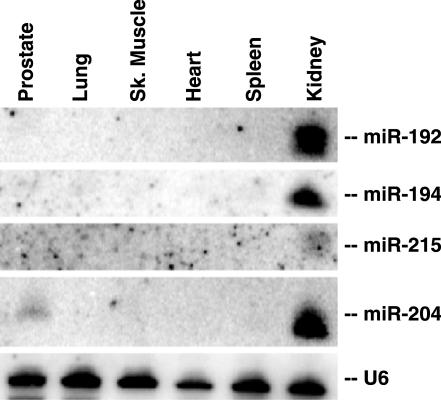
Another interesting group of miRNAs obtained by _K_-means clustering is a highly kidney-specific miRNA cluster (Figure 3), representing five miRNAs. These are _miR_-192, _miR_-194, _miR_-204, _miR_-215 and _miR_-216. _miR_-194-1 and _miR_-215 are located in chromosome 1 and the distance between them is only 195 bp. _miR_-192 is located in chromosome 11 and 109 bp upstream of this is _miR_-194-2. It is likely that _miR_-192 and _miR_-194-2 are also regulated as a common transcriptional unit. We have also confirmed the expression of four of these miRNAs by northern-blot analysis (Figure 4) and the data were found to be in good agreement with the array, with enriched expression in kidney. We could not detect the signal of _miR_-216 by northern-blot analysis.
Figure 3.
Kidney enrich mir cluster. This cluster contains five mirs (_miR_-192, _miR_-194, _miR_-204, _miR_-215 and _miR_-216).
Figure 4.
Northern blotting of human tissue RNA (Ambion) for kidney enrich mir cluster. U6 snRNA was used to verify the equal RNA loading.
Clustal W analysis of kidney-specific miRNA cluster revealed that _miR_-192 and _miR_-215 share a high-sequence homology between their precursor mir sequences and contain a 18 bp common sequence (UGACCUAUGAAUUGACAG) in their mature mir sequence region (Figure 5).
Figure 5.
Cluster W alignment of _miR_-192 and _miR_-215. There is an 18 bp conserved DNA sequence (UGACCUAUGAAUUGACAG) common to each of these mirs in their processed mir sequence region.
Statistical analysis shows that 24.3% of all the miRNAs on our microarray give Present calls in all six human organs while 45.1% did not give a Present call in any organs tested. Present calls are showed by 5.5, 3.5, 6.3 and 4.3% of the miRNAs in 5, 4, 3 and 2 tissues, respectively, and 11.0% of the miRNAs on the microarray show Present calls only in one tissue.
Comparative genomic analysis of the upstream sequences of organ-enriched miRNAs
We next analyzed cross-species (human, chimpanzee, mouse, rat and chicken) conserved sequences in a 10 kb region upstream of the kidney-enriched miRNAs, _miR_-192, _miR_-194, _miR_-204, _miR_-215 and _miR_-216. This was completed to search for the presence of CpG islands and known transcription factor binding sites that might drive transcriptional regulation of these kidney enriched miRNAs.
Located ∼2158 bp upstream of the _miR_-192/_miR_-194-2 complex in chromosome 11 is a non-coding region of high cross-species conservation. In this region, we find a high-scoring occurrence of a transcription factor binding site for the proto-oncogene, ets-1 (Figure 6). This transcription factor is present in a cross-species conserved upstream region (1155 bp) of _miR_-204. Numerous published studies have shown that ets-1 gene is essential for the normal development of mammalian kidneys and the maintenance of glomerular integrity (33) and that Ets-1 protein may act as an upstream regulator for the expression of FREAC-4, a winged helix transcriptional factor detected during nephrogenesis (34). Ets-1 Knock-out mouse kidney studies have also demonstrated various glomerular abnormalities, including sclerosis, atrophy and markedly fewer and immature glomeruli (35). These results denote an essential role for Ets-1 in the normal structural development of the kidney.
Figure 6.
Cross-species conservation of proto-oncogene ets-1 binding site located 2158 bp upstream of miR-192/miR-194-2 complex and 1155 bp upstream of miR-204.
MiRNAs, _miR_-194-1, _miR_-215 and _miR_-216 also contain conserved phylogenetic footprints in their upstream regions. Like _miR_-192 and _miR_-194-2 in chromosome 11, _miR_-194-1 and _miR_-215 are also very close together—separated by 195 bp on chromosome 1. Pockets of high-scoring, non-coding, cross-species conservation lie 226–1123 bp upstream of the start of the _miR_-194-1/_miR_-215 complex. In this region, we see multiple, high-scoring copies of transcription factor binding sites, including Nkx-2.5/Csx, SRY and CAAT boxes. Homeo domain factor, Nkx-2.5/Csx binding site is located in the upstream of 1117 bp of _miR_-194-1/_miR_-215 complex and 3496 bp upstream of _miR_-216 where SRY (sex-determining region Y gene) product binding site is located 600 bp upstream of _miR_-194-1/_miR_-125 complex and 3484 bp upstream of miR-216. We also found a CAAT box binding site located in a cross-species conservation site at 937 and 227 bp upstream of _miR_-194-1/_miR_-215 complex and 3351 bp upstream of _miR_-216. See additional information on cross-species conservation of the above transcription factor sequences in the Supplementary Material.
CONCLUSIONS
The microarray described here is a novel, selective and sensitive approach to monitor miRNA expression in mammalian cells and tissues. We have identified miRNAs that are selectively expressed in kidney, heart and skeletal muscle. By performing comparative sequence analysis, we have proposed the presence of a putative transcriptional regulatory mediator for miRNAs in kidney. It is anticipated that future use of this array-based approach will lead to a greater understanding of recently identified class of gene regulating RNA molecules.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
[Supplementary Material]
REFERENCES
- 1.Ambros V. (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell, 113, 673–676. [DOI] [PubMed] [Google Scholar]
- 2.Bartel B. and Bartel,D.P. (2003) MicroRNAs: at the root of plant development? Plant Physiol., 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palatnik J.F., Allen,E., Wu,X., Schommer,C., Schwab,R., Carrington,J.C. and Weigel,D. (2003) Control of leaf morphogenesis by microRNAs. Nature, 425, 257–263. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 5.Pasquinelli A.E., Reinhart,B.J., Slack,F., Martindale,M.Q., Kuroda,M.I., Maller,B., Hayward,D.C., Ball,E.E., Degnan,B., Muller,P. et al. (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature, 408, 86–89. [DOI] [PubMed] [Google Scholar]
- 6.Lee R.C., Feinbaum,R.L. and Ambros,V. (1993) The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart B.J., Weinstein,E.G., Rhoades,M.W., Bartel,B. and Bartel,D.P. (2002) MicroRNAs in plants. Genes Dev., 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim L.P., Glasner,M.E., Yekta,S., Burge,C.B. and Bartel,D.P. (2003) Vertebrate microRNA genes. Science, 299, 1540. [DOI] [PubMed] [Google Scholar]
- 9.Lim L.P., Lau,N.C., Weinstein,E.G., Abdelhakim,A., Yekta,S., Rhoades,M.W., Burge,C.B. and Bartel,D.P. (2003) The microRNAs of Caenorhabditis elegans. Genes Dev., 17, 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambros V., Lee,R.C., Lavanway,A., Williams,P.T. and Jewell,D. (2003) MicroRNAs and other tiny endogenous RNAs in C.elegans. Curr. Biol., 13, 807–818. [DOI] [PubMed] [Google Scholar]
- 11.Grad Y., Aach,J., Hayes,G.D., Reinhart,B.J., Church,G.M., Ruvkun,G. and Kim,J. (2003) Computational and experimental identification of C.elegans microRNAs. Mol. Cell, 11, 1253–1263. [DOI] [PubMed] [Google Scholar]
- 12.Kim J., Krichevsky,A., Grad,Y., Hayes,G.D., Kosik,K.S., Church,G.M. and Ruvkun,G. (2004) Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl Acad. Sci. USA, 101, 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M., Rauhut,R., Yalcin,A., Meyer,J., Lendeckel,W. and Tuschl,T. (2002) Identification of tissue-specific microRNAs from mouse. Curr. Biol., 12, 735–739. [DOI] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M., Rauhut,R., Meyer,J., Borkhardt,A. and Tuschl,T. (2003) New microRNAs from mouse and human. RNA, 9, 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mourelatos Z., Dostie,J., Paushkin,S., Sharma,A., Charroux,B., Abel,L., Rappsilber,J., Mann,M. and Dreyfuss,G. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev., 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dostie J., Mourelatos,Z., Yang,M., Sharma,A. and Dreyfuss,G. (2003) Numerous microRNPs in neuronal cells containing novel microRNAs. RNA, 9, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 20.Lai E.C., Tomancak,P., Williams,R.W. and Rubin,G.M. (2003) Computational identification of Drosophila microRNA genes. Genome Biol., 4, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin G.A., Sevignani,C., Dumitru,C.D., Hyslop,T., Noch,E., Yendamuri,S., Shimizu,M., Rattan,S., Bullrich,F., Negrini,M. et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA, 101, 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krichevsky A.M., King,K.S., Donahue,C.P., Khrapko,K. and Kosik,K.S. (2003) A microRNA array reveals extensive regulation of microRNAs during brain development. RNA, 9, 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C.G., Calin,G.A., Meloon,B., Gamliel,N., Sevignani,C., Ferracin,M., Dumitru,C.D., Shimizu,M., Zupo,S., Dono,M. et al. (2004) An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl Acad. Sci. USA, 101, 9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calin G.A., Liu,C.G., Sevignani,C., Ferracin,M., Felli,N., Dumitru,C.D., Shimizu,M., Cimmino,A., Zupo,S., Dono,M. et al. (2004) MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA, 101, 11755–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babak T., Zhang,W., Morris,Q., Blencowe,B.J. and Hughes,T.R. (2004) Probing microRNAs with microarrays: tissue specificity and functional inference. RNA, 10, 1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miska E.A., Alvarez-Saavedra,E., Townsend,M., Yoshii,A., Sestan,N., Rakic,P., Constantine-Paton,M. and Horvitz,H.R. (2004) Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol., 5, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomson J.M., Parker,J., Perou,C.M. and Hammond,S.M. (2004) A custom microarray platform for analysis of microRNA gene expression. Nature Methods, 1, 47–53. [DOI] [PubMed] [Google Scholar]
- 28.Nelson P.T., Baldwin,D.A., Scearce,L.M., Oberholtzer,J.C., Tobias,J.W. and Mourelatos,Z. (2004) Microarray-based, high-throughput gene expression profiling of microRNAs. Nature Methods, 2, 155–161. [DOI] [PubMed] [Google Scholar]
- 29.Esquela-Kerscher A. and Slack,F.J. (2004) The age of high-throughput microRNA profiling. Nature Methods, 2, 106–107. [DOI] [PubMed] [Google Scholar]
- 30.Rosner B. (2000) Fundamentals of Biostatistics. Duxbury, NY. [Google Scholar]
- 31.Jain A.K. and Dubes,R.C. (1988) Algorithms for Clustering Data. Prentice Hall, Englewood Cliffs, NJ, 320 p. [Google Scholar]
- 32.Sempere L.F., Freemantle,S., Pitha-Rowe,I., Moss,E., Dmitrovsky,E. and Ambros,V. (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol., 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razzaque M.S., Naito,T. and Taguchi,T. (2001) Proto-oncogene Ets-1 and the kidney. Nephron, 89, 1–4. [DOI] [PubMed] [Google Scholar]
- 34.Cederberg A., Hulander,M., Carlsson,P. and Enerback,S. (1999) The kidney-expressed winged helix transcription factor FREAC-4 is regulated by Ets-1. A possible role in kidney development. J. Biol. Chem., 274, 165–169. [DOI] [PubMed] [Google Scholar]
- 35.Gomez R.A., Sequeira Lopez,M.L., Fernandez,L., Chernavvsky,D.R. and Norwood,V.F. (1999) The maturing kidney: development and susceptibility. Ren. Fail., 21, 283–291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]