Arrangement of Subunits in 20 S Particles Consisting of NSF, SNAPs, and SNARE Complexes (original) (raw)
. Author manuscript; available in PMC: 2017 Jul 4.
Summary
The structure of 20 S particles, consisting of NSF, SNAPs, and SNARE complexes, was analyzed by electron microscopy and fluorescence resonance energy transfer. Structural changes associated with the binding of α-SNAP and NSF to SNARE complexes define the contribution of each component to the 20 S particle structure. The synaptic SNARE complex forms a 2.5 × 15 nm rod. α-SNAP binds laterally to the rod, increasing its width but not its length. NSF binds to one end of the SNAP/SNARE complex; the resulting 20 S particles measure 22 nm in length and vary in width from 6 nm at their narrowest point to 13.5 nm at their widest. The transmembrane domains of VAMP and syntaxin emerge together at the NSF-distal end of 20 S particles, adjacent to the amino terminus of α-SNAP.
Introduction
Despite the constant flow of proteins and lipids into, out of, and through intracellular compartments, each type of organelle maintains a distinct functional and morphological identity. To preserve the organization of cellular compartments, cargo-laden transport vesicles act as intercompartmental shuttles that selectively dock to and fuse with their intended membrane targets. A family of membrane proteins, referred to as SNAREs, forms a core machinery involved in these transport reactions (Söllner et al., 1993a; Rothman, 1994; Südhof, 1995; Hanson et al., 1997b). SNAREs are distributed to distinct intracellular compartments with t-SNAREs mainly present on target membranes and v-SNAREs predominately on vesicle membranes. The v-SNARE VAMP-2/synaptobrevin II (Baumert et al., 1989; Elferink et al., 1989; hereafter referred to as VAMP) and the t-SNAREs syntaxin (Bennett et al., 1992; isoforms 1A and 1B) as well as SNAP-25 (Oyler et al., 1989) are the prototypic members of the SNARE family whose complex and function were identified first at the neuronal synapse (Söllner et al., 1993a).
In the case of heterotypic membrane fusion, a v-SNARE binds to its cognate t-SNARE, forming a SNAREpin, which pairs a transport vesicle to its target membrane for fusion. The specificity of v-/t-SNARE association and their assembly into stable complexes may thereby dictate, at least at this stage of the process, which set of membranes can fuse. Reconstitution experiments indicate that SNAREpins are the minimal fusion machinery (Weber et al., 1998).
v- and t-SNAREs interact via their cytosolic domains that form a parallel four-stranded helical bundle (Poirier et al., 1998b). SNARE complexes found at neuronal synapses are stable up to 90°C and resist denaturation by SDS (Hayashi et al., 1994). On electron micrographs, neuronal v-/t-SNARE complexes appear as rod-like structures with a parallel v- and t-SNARE alignment (Hanson et al., 1997a).
To disassemble otherwise stable SNARE complexes, a general machinery consisting of the _N_-ethylmaleimide-sensitive fusion protein (NSF; Block et al., 1988; Wilson et al., 1989) and soluble NSF attachment proteins (SNAPs; Clary and Rothman, 1990; Whiteheart et al., 1993) acts on v-/t- SNARE complexes at many different intracellular locations (Söllner et al., 1993b; Søgaard et al., 1994; Nagahama et al., 1996; Subramaniam et al., 1996, 1997; Hay et al., 1997; Paek et al., 1997; Nichols et al., 1997; Ungermann et al., 1998). NSF and SNAPs bind to SNARE complexes in a sequential manner, giving rise to 20 S particles, named for their sedimentation coefficient of 20 Svedberg units. 20 S particles are stable under conditions that preclude ATP hydrolysis by NSF. NSF is a hexameric ATPase; each subunit contains three domains, an amino-terminal N domain and two ATP-binding domains, termed D1 and D2 (Tagaya et al., 1993; Whiteheart et al., 1994; Fleming et al., 1998). 20 S particle disassembly is catalyzed by NSF’s ATPase activity, which releases SNAPs and free v- and t-SNAREs (Söllner et al., 1993b; Hayashi et al., 1995). Free v- and t-SNAREs can thereby be recycled for another round of vesicular transport.
In this study, we analyzed the structure of 20 S particles as well as intermediate complexes involved in 20 S particle assembly to assess the contribution of NSF, SNAPs, and (synaptic) SNARE complexes to the 20 S particle structure. To localize distinct domains of NSF, α-SNAP, and SNAREs within 20 S particles we isolated 20 S particles containing epitope-tagged recombinant proteins and examined these particles complexed with antibodies by electron microscopy. This technique allowed us to locate the D2 domain of NSF, the amino terminus of α-SNAP, and the membrane-spanning region of VAMP within 20 S particles. In addition, we assembled SNARE complexes that contained green and blue fluorescent protein moieties in lieu of the trans-membrane domains of VAMP and syntaxin. Fluorescence resonance energy transfer (FRET) studies (Fairclough and Cantor, 1978; Stryer, 1978; Heim and Tsien, 1996) in the presence of α-SNAP and NSF, under conditions of 20 S particle assembly, suggest that the membrane anchors of SNAREs lie in close proximity, irrespective of whether they are incorporated in SNARE complexes (Hanson et al., 1997a; Lin and Scheller, 1997; Poirier et al., 1998b) or in 20 S particles. Our results therefore define the structural layout of 20 S particles.
Results
Purification of Neuronal SNARE Complexes and 20 S Particles
To analyze the structure of SNARE complexes by electron microscopy, synaptic v/t-SNARE complexes were purified from a brain detergent extract based on their α-SNAP binding property. SNARE complexes were isolated by affinity chromatography on GST-α-SNAP, released from the immobilized fusion protein by incubation with _n_-octylglucoside and high salt, sedimented through glycerol gradients, and processed for electron microscopy (see Experimental Procedures).
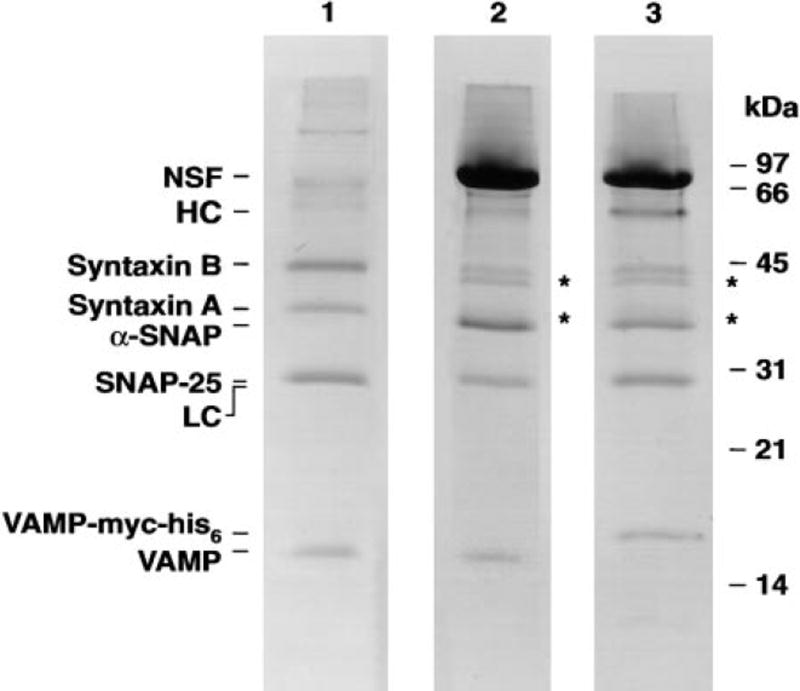
The purification of SNAP/SNARE complexes and 20 S particles for electron microscopy relied on similar preparative procedures. SNARE complexes were bound to GST-α-SNAP as above; in the case of 20 S particles, NSF was added under conditions precluding ATP hydrolysis. Both SNAP/SNARE complexes and 20 S particles were released from the beads by thrombin cleavage of GST-α-SNAP (such that the GST moiety remained on the beads) and purified further by velocity centrifugation. The polypeptide compositions of a typical SNARE complex (Figure 1, lane 1) and a 20 S particle preparation (Figure 1, lane 2) were analyzed by SDS-PAGE.
Figure 1. Ternary SNARE Complex and 20 S Particle Preparations.
Ternary SNARE complexes (lane 1), 20 S particles (lane 2), and 20 S particles containing VAMP-myc-his6 complexed with anti-myc antibodies (lane 3) were subjected to high Tris-urea-SDS PAGE as described in Söllner et al. (1993a) and stained with Coomassie Brilliant Blue. The abbreviations HC and LC refer to the heavy and light chains of anti-myc antibodies, respectively. Preparations shown in lanes 2 and 3 treated with thrombin yielded both full-length and N-terminally clipped syntaxins (marked by *). Thrombin digestion led to a partial proteolytic cleavage of 9 N-terminal amino acids of syntaxin 1A and 1B (as detemined by N-terminal sequencing). These shortened syntaxins behaved identically to their full-length counterparts in SNARE complex and 20 S particle formation as well as in NSF-mediated SNARE complex disassembly (data not shown).
To localize individual domains of 20 S particle constituents, epitope tags were added to the components of interest (NSF, α-SNAP, and VAMP). For example, to determine the position of the membrane-spanning region of VAMP within 20 S particles, the endogenous VAMP in 20 S particles was replaced with recombinant VAMP-myc-his6, which contains a myc epitope in lieu of the membrane-spanning region (see Experimental Procedures). Anti-myc antibodies were bound to 20 S particles with the myc-tagged constituent (Figure 1, lane 3) and the antibody-decorated particles imaged.
Ternary SNARE Complexes Form Rod-like Structures
Electron micrographs of synaptic v-/t-SNARE complexes contained numerous rod-like structures with a globular mass at one end (data not shown). Image analysis demonstrated that SNARE complexes form rods, 2.5 nm in width and 15 nm in length, including a globular mass with a diameter of ~5 nm at one end (Figure 2B). The size of the globular mass in SNARE complexes is compatible with the presence of a detergent micelle of Triton-X-100 with a molecular weight of ~90 kDa attached to the SNARE membrane anchors (Furth et al., 1984).
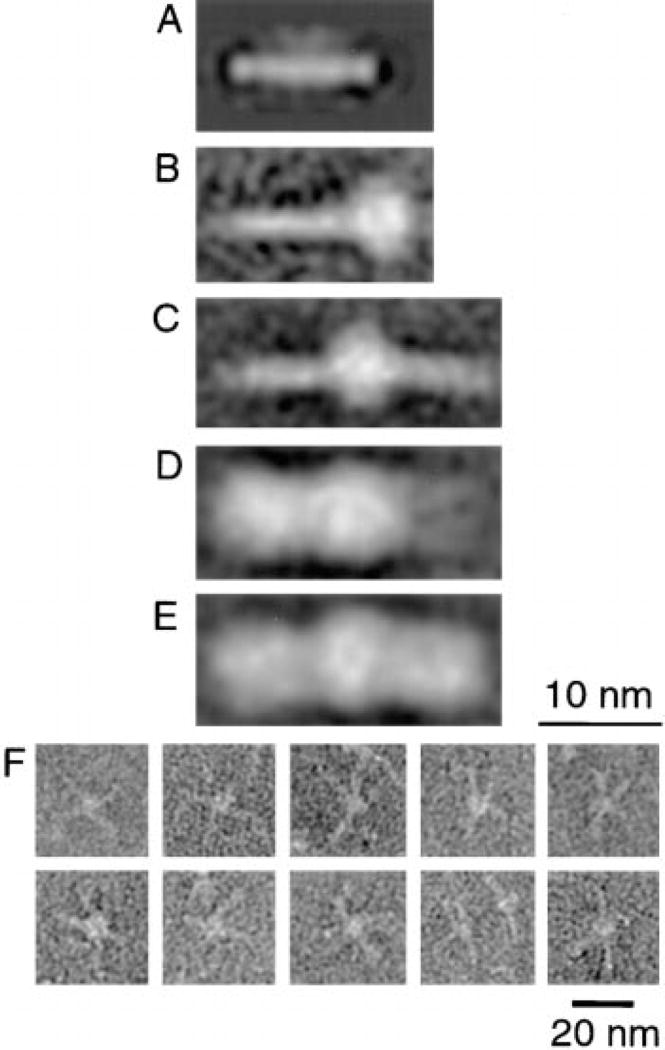
Figure 2. Structure of Ternary SNARE and SNAP/SNARE Complexes.
Protein complexes were purified and analyzed by electron microscopy as described in Experimental Procedures. (A) Average of 343 ternary SNARE complexes assembled from bacterially expressed SNAREs lacking membrane-spanning domains. (B and C) Averages of 207 monomeric (B) and 95 dimeric (C) SNARE complexes isolated from a bovine brain detergent extract. (D and E) Averages of 162 monomeric (D) and 223 dimeric (E) SNAP/SNARE complexes. (F) Examples of oligomeric, nonaveraged SNARE complexes that appear as central plug and spoke structures.
Individual SNARE complex rods showed a tendency to aggregate, forming dimeric (Figure 2C) and higher order oligomeric structures (see gallery, Figure 2F). Linear dimers were selected for image analysis and appear as elongated rods, 23 nm length, with a ~5 nm globular mass in their center (Figure 2C). Dimers, however, did not show a strict prevalence for a linear arrangement. Higher order oligomers (trimers, tetramers, etc.) of SNARE complexes appear as “plug and spoke” structures (Figure 2F) with the “spokes” corresponding to individual SNARE complex rods, emanating from a central “plug” (Figure 2F). This plug is morphologically similar to the globular mass attached to individual and dimeric SNARE complex rods although it occasionally appeared increased in size, perhaps as more SNARE hydrophobic domains aggregate and share a detergent micelle.
Neither the globular mass nor the formation of oligomers was observed with SNARE complexes containing recombinant proteins lacking the SNARE membrane spans (see Figure 2A; Hanson et al., 1997a), which otherwise were similar in size and dimension to those containing full-length SNAREs. The presence of the globular mass at only one end of the SNARE complex rod (with full-length SNAREs) suggests that all SNARE membrane anchors lie at one end of the rod and share the same detergent micelle.
SNAPs Bind to the Lateral Side of SNARE Complex Rods
The addition of α-SNAP to the SNARE complex rod tripled its width (to 7–8 nm) without altering its length (Figure 2D). α-SNAP thus binds laterally to the SNARE complex rod. Linear dimers of SNAP/SNARE complexes were equivalent in length to SNARE complex dimers (Figure 2E), as expected for lateral α-SNAP binding. SNAP/SNARE complexes seem to oligomerize via the hydrophobic SNARE domains, as is the case with ternary SNARE complexes alone.
20 S Particles Have a Sparkplug Shape
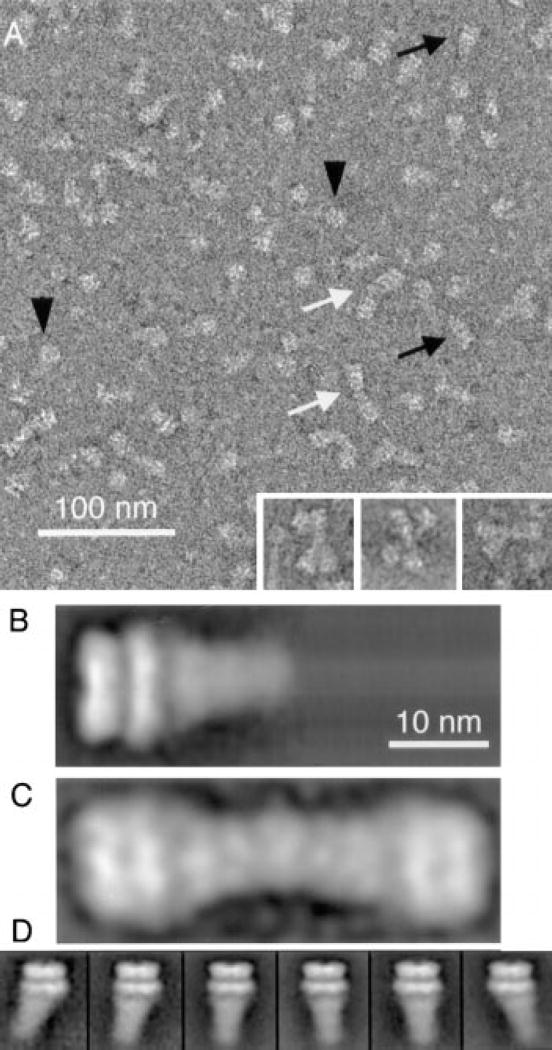
Electron micrographs of 20 S particles reveal numerous asymmetric particles with a typical “sparkplug shape” (Figure 3A, black arrows). These particles possess two distinct structural domains, a stacked double ring structure at one end and a cone-shaped tail that diminishes in width as it projects away from the stacked ring structure. The cone-shaped tail and the double-ring structure are strikingly similar in size and shape to the SNAP/SNARE complex and the side view of NSF (Figure 4A), respectively. Owing to these structural similarities, we conclude that the sparkplug-shaped assemblies are 20 S particles. The micrographs also contain ring-shaped molecules (Figure 3A, black arrowheads), which likely represent top views of NSF (Figure 4B; see also Hanson et al., 1997a; Fleming et al., 1998).
Figure 3. Field View and Structure of 20 S Particles.
(A) The micrograph shows monomeric 20 S particles (black arrows), dimeric 20 S particles (white arrows), and top views of NSF (black arrowheads). The insets contain examples of trimeric and tetrameric 20 S particles. (B) Average of 574 monomeric 20 S particles (4th individual class in [D]). (C) Average of 133 dimeric 20 S particles. (D) Gallery of six classes of averages that indicate the variability of 20 S particles in the electron micrographs. The individual classes contain averages of 451, 209, 871, 574, 400, and 730 particles (from left to right). 20 S particles were aligned with respect to the double ring structure and analyzed by PCA using the entire particle. The width of each individual image in the gallery corresponds to 27 nm.
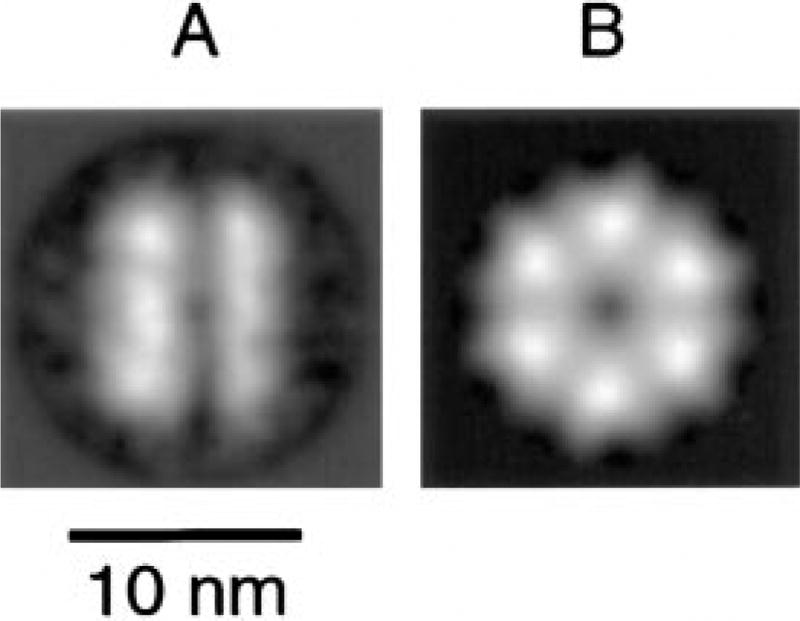
Figure 4. Structure of NSF in Its ATP-Bound State.
(A–B) NSF was imaged in buffer containing 2 mM EDTA and 0.5 mM ATP. (A) Average of 184 NSF side views. (B) Average of 431 NSF top views, illustrating a 6-fold rotational symmetry.
The sparkplug-shaped particles were susceptible to ATP hydrolysis, providing biochemical evidence that they represent 20 S particles. Upon incubation with Mg-ATP to promote their disassembly, these particles disappeared almost completely from electron micrographs; top view projections of NSF became the predominant visible structure (data not shown). NSF forms a double ring structure with a diameter of 12 nm, a height of 9 nm, and a 6-fold rotational symmetry (Figure 4). NSF was imaged under the same nucleotide conditions as 20 S particles.
The length of averaged 20 S particles is 22 nm (Figure 3B), including a 13 nm long tail whose width varies from 6 nm at its tip to 9.5 nm at its edge in contact with NSF. The tail of 20 S particles appears to be shaped like a cone, while SNAP/SNARE complexes resemble a cylinder (Figure 2D). The SNAP/SNARE complex may therefore undergo a conformational change when bound to NSF, leading to the observed change in shape. The individual rings of 20 S particles vary in height, with the inner ring 4 nm and the outer ring 5 nm in height, identical to the height of the rings in free NSF (Figure 4A). In 20 S particles, the outer ring diameter is 12 nm while the inner ring diameter is 13.5 nm. The latter diameter is greater than either ring diameter in free NSF (Figure 4A). This increase in the inner ring diameter is consistent with a structural change in NSF due to its association with SNAPs and SNARE complexes (see Discussion). However, the similarity of the stacked ring arrangement of NSF both in the absence and presence of binding partners indicates that a single NSF hexamer resides within a 20 S particle.
Like SNARE and SNAP/SNARE complexes, 20 S particles associate into dimeric, trimeric, and higher order oligomeric structures (see inset, Figure 3A). Linear 20 S particle dimers have an overall length of 40 nm and represent two 20 Sparticles associated at their tips, with a 4 nm overlap (Figure 3C). 20 S particle oligomerization occurred not only on EM grids but also in solution when the detergent concentration was reduced below the critical micellar concentration. 20 S particles sedimented much more rapidly than expected in velocity centrifugation experiments when glycerol gradients contained only 0.01% (w/v) Triton-X-100 or CHAPS (data not shown). The more rapid sedimentation of the particles was not due to protein denaturation since large aggregates sedimenting into the pellet were not observed.
The image analysis of 20 S particles revealed subclasses that may reflect some variability in the 20 S particle structure (Figure 3D). These subclasses differ mainly in the orientation of the cone-shaped region relative to the double ring. This indicates either a limited flexible link between NSF and SNAP/SNAREs or an inherent asymmetry that might be masked by variations in the way 20 S particles adsorb to EM grids.
NSF and the Membrane-Spanning Region of VAMP Are Located at Opposite Ends of 20 S Particles
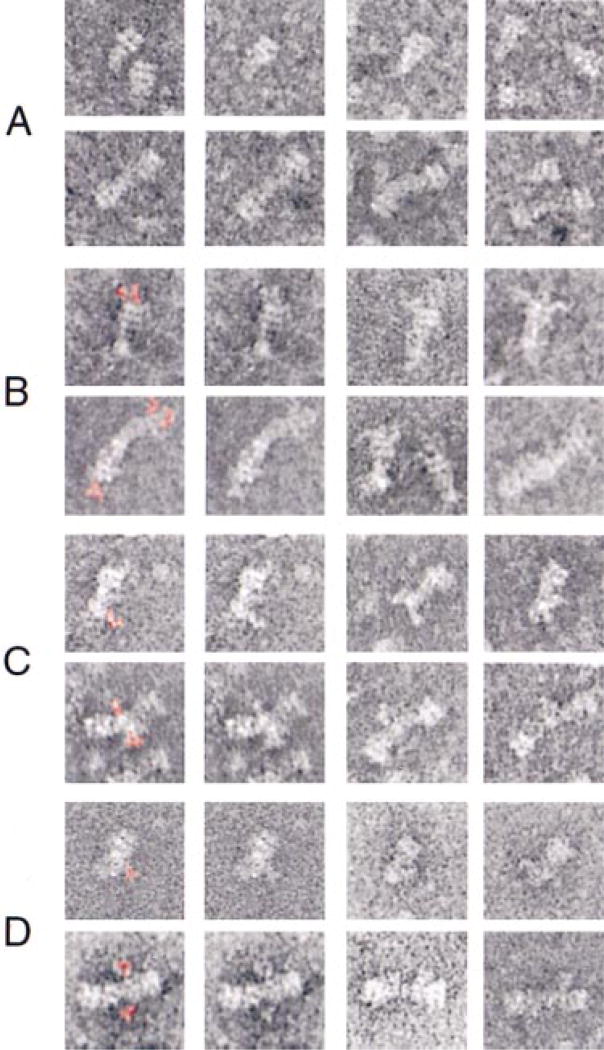
To locate protein domains within 20 S particles, individual proteins were epitope-tagged (with a myc epitope), incorporated into 20 S particles, decorated with antibodies, and imaged. Figure 5A shows examples of monomers and dimers of 20 S particles without bound antibodies. To aid in the recognition of the antibodies, they were colored red in the first column of Figure 5B–5D. For comparison, duplicate images in the second column of Figure 5B–5D were left uncolored. In the case of 20 S particles with carboxy-terminal myc-tagged NSF, the antibodies selectively label the stacked ring structure of NSF, with no additional mass observed in the cone-shaped region of 20 S particles (Figure 5B). In all cases examined, the antibodies bind to the outer of the two stacked rings with no binding observed to the inner ring. This indicates that the outer ring consists of the carboxy-terminal D2 domains of NSF and that D1 (and possibly N) domains form the inner ring of 20 S particles.
Figure 5. Localization of 20 S Particle Constituents by Immunoelectron Microscopy.
(A) Series of monomeric (top row) and dimeric 20 S particles (bottom row) in the absence of bound antibody (same magnification as Figure 3A). (B–D) 20 S particles with different myc-tagged constituents complexed with antibodies. The first two columns show duplicate images with antibodies colored red in the first column and left uncolored in the second. The final two columns provide additional examples of antibody-decorated particles. The antibodies were directed against carboxy-terminal myc-tagged NSF (B), carboxy-terminal myc-tagged cytosolic VAMP (C), and amino-terminal myc-tagged α-SNAP (D). Note the presence of one to several additional mass(es) attached to the outer ring of 20 S particles (B), the tip of the tail region of 20 S particles (C), and the region adjacent to the tips of 20 S particles (D).
To localize the membrane anchor region of VAMP within 20 S particles, the endogenous VAMP was exchanged for a recombinant counterpart, VAMP-myc-his6, with a myc epitope in lieu of the membrane span. Anti-myc antibodies bound to the tip of individual 20 S particles or at the junction between 20 S particle dimers (Figure 5C). 20 S particles lacking the membrane anchor of VAMP still oligomerize at their tips, suggesting that the hydrophobic regions of t-SNAREs are sufficient for oligomerization and reside at the tips of 20 S particles.
In order to locate the amino terminus of α-SNAP within 20 S particles, we isolated 20 S particles containing myc-α-SNAP decorated with anti-myc antibodies (Figure 5D). The amino terminus of α-SNAP is located near the tip of 20 S particles, in close proximity to the SNARE membrane anchors.
The Membrane Anchors of SNAREs Emerge from the Same End of 20 S Particles
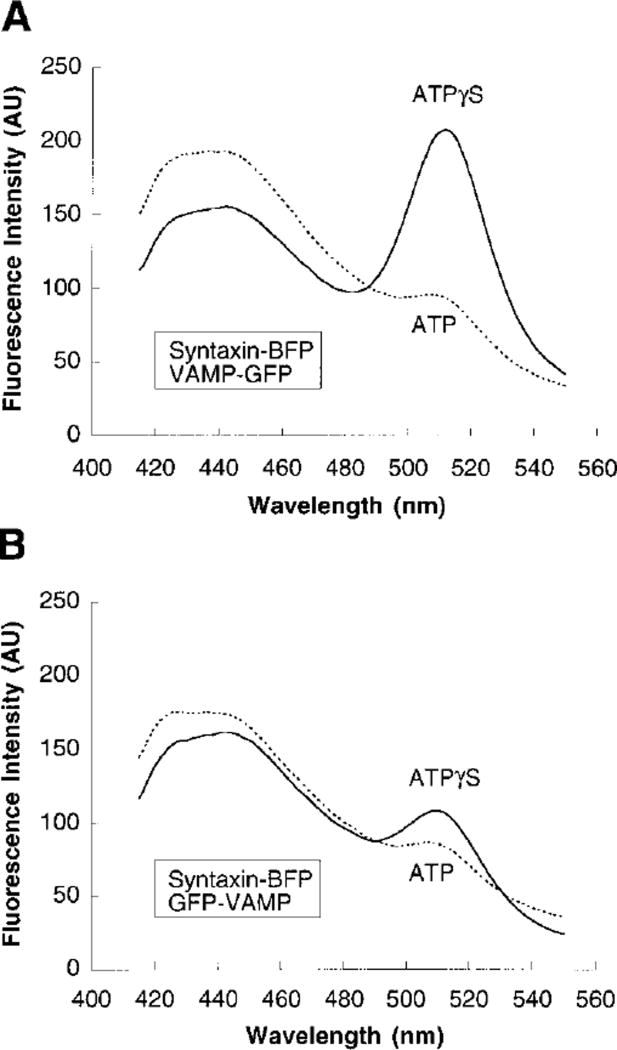
Our data indicate that the membrane anchors of the v-SNARE and t-SNAREs emerge together at the NSF-distal end of 20 S particles. To provide further evidence for such an arrangement, we constructed chimerae containing the cytosolic domains of VAMP or syntaxin fused either to blue or green fluorescent protein (BFP/GFP; see Experimental Procedures). We formed ternary SNARE complexes with these fusion proteins and monitored fluorescence resonance energy transfer between BFP and GFP under conditions of 20 S particle assembly and disassembly. FRET is a highly distance-dependent process; the efficiency of energy transfer from the donor BFP (λexc. 389 nm, λemis. 446 nm) to the acceptor GFP (λexc. 489 nm, λemis. 508 nm) decreases with the sixth power of the distance between the fluorophores (Fairclough and Cantor, 1978; Stryer, 1978; Heim and Tsien, 1996). If the carboxyl termini of VAMP and syntaxin lie in close proximity in 20 S particles, FRET between the donor (syntaxin-BFP) and the acceptor (GFP-VAMP or VAMP-GFP) fluorophore should be most prominent when the fluorophores are fused to the carboxyl termini of both the v- and t-SNARE.
SNAREs were present in approximately equimolar amounts in both sets of ternary complexes (data not shown). α-SNAP and Mg-ATPγS were added to both sets of ternary complexes, followed by addition of NSF. None of the additions had an effect on the emission spectra of the ternary complexes shown in Figures 6A and 6B (solid lines). However, the sample containing a C-terminal fluorophore fused to VAMP and to syntaxin (Figure 6A, solid line) exhibited a larger FRET signal than the sample with an N-terminal fluorophore fused to VAMP and a C-terminal fluorophore fused to syntaxin (Figure 6B, solid line). This indicates that VAMP and syntaxin align with their membrane anchors in close proximity, irrespective of whether they are incorporated in ternary SNARE complexes (Hanson et al., 1997a; Lin and Scheller, 1997; Poirier et al., 1998b) or in 20 S particles.
Figure 6. Fluorescence Resonance Energy Transfer between GFP-and BFP-Tagged SNAREs Assembled in 20 S Particles.
(A and B) The emission spectra of ternary SNARE complexes containing SNAP-25, syntaxin-BFP, and either VAMP-GFP (A) or GFP-VAMP (B) in the presence of α-SNAP, NSF, and either Mg-ATPγS (solid lines) or Mg-ATP (dashed lines). The emission spectra reported in A and B were recorded 30 min after addition of NSF (see Experimental Procedures). Both sets of measurements were confirmed in 11 (A) and 3 (B) independent experiments.
AU, arbitrary units.
To verify that the FRET observed was due to SNARE complex formation and not due to a SNARE-independent association of GFP and BFP, duplicate samples of both ternary SNARE complexes were incubated with α-SNAP and Mg-ATP. No change in the emission spectra was detected as compared to the solid lines in Figures 6A and 6B. However, when NSF was added, SNARE complexes were disassembled and FRET disappeared in both sets of samples (dashed lines in Figures 6A and 6B). This demonstrates that the FRET signal was dependent on SNARE complex formation.
The efficiency of energy transfer was calculated for both sets of samples (for C-terminal fluorophores and fluorophores at opposite termini) by comparing the fluorescence intensity of the BFP donor peak under conditions of 20 S particle assembly (Mg-ATPγS) and disassembly (Mg-ATP) (Fairclough and Cantor, 1978). The efficiency of energy transfer between syntaxin-BFP and VAMP-GFP was 21.6% ± 2.6% (n = 11), whereas the efficiency of energy transfer between syntaxin-BFP and GFP-VAMP was 6.0% ± 1.9 (n = 3). These results are consistent with a largely parallel arrangement of VAMP and syntaxin in 20 S particles.
Discussion
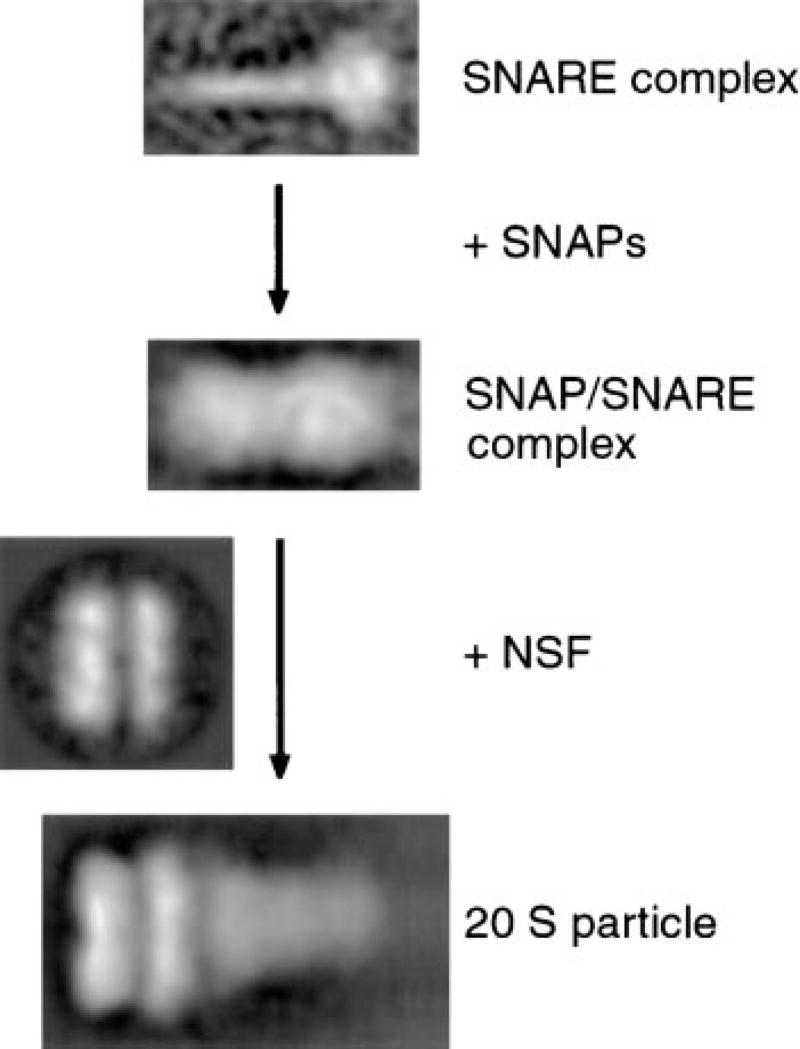
In this study, we illustrate the assembly of 20 S particles on a structural level (Figure 7). The stepwise addition of α-SNAP and NSF to SNARE complexes allowed us to define the contribution of each of these three constituents to the 20 S particle structure. Most results were obtained with endogenous SNARE complexes containing full-length proteins. This excludes potential structural alterations, which might occur with the use of recombinant, bacterially expressed SNAREs lacking the membrane-spanning domains or posttranslational modifications such as the palmitoylation of SNAP-25 (Hess et al., 1992).
Figure 7. Structural Assembly Pathway of 20 S Particles.
Stepwise assembly of 20 S particles, illustrating the structural contributions of NSF, α-SNAP, and SNARE complexes to 20 S particles. α-SNAP binds laterally to the SNARE complex rod in a sheath-like manner. NSF binds as a double ring to the SNAP/SNARE complex at the end opposite to the SNARE membrane anchors.
SNARE complexes appear as rod-like structures. Protein domains involved in this structure include the carboxy-terminal domain of syntaxin, the majority of the cytosolic domain of VAMP, and the amino- and carboxy-terminal domains of SNAP-25 (Hayashi et al., 1994; Pevsner et al., 1994; Fasshauer et al., 1998; Poirier et al., 1998a, 1998b). The membrane anchors of SNAREs emerge together at one end of the rod and appear as a globular domain in the presence of detergent (Figure 2B). This result is consistent with the FRET data (Figure 6; Lin and Scheller, 1997) as well as previous electron microscopy (Hanson et al., 1997a) and spin labeling electron paramegnetic resonance spectroscopy studies (Poirier et al., 1998b) of recombinant SNARE complexes. The SNARE membrane anchors seem to be prone to aggregation, causing SNARE complex oligomerization, in particular with detergent concentrations below the critical micellar concentration. The amino terminus of syntaxin was visible in SNARE complex field views as a slender hook-like appendage to the SNARE complex rod (data not shown; Hanson et al., 1997a); however, this domain was lost during the image averaging procedure (Figure 2A) owing to its flexible linkage to the rest of the SNARE complex.
The 60–70 residue helical juxtamembrane domain of syntaxin that interacts with the other synaptic SNAREs also binds α-SNAP (Hanson et al., 1995; Hayashi et al., 1995; McMahon and Südhof, 1995). These interactions may explain the lateral association of SNAP with the SNARE complex rod. Both the N-terminal 63 and the C-terminal 37 residues of α-SNAP are required for SNARE complex binding, indicating that these regions are either indispensable for the correct folding of α-SNAP or are part of its SNARE-interacting surface that may include one or more α-helical domains of SNAP. Regardless of the exact nature of SNARE-SNAP interaction it appears that the SNARE complex rod (given the likely assumption that α-SNAP binding does not alter the conformation of the rod) is surrounded by α-SNAP along its entire length in a sheath-like manner. This spatial arrangement may create an environment required for the dissociation of the extraordinarily stable synaptic SNARE complex. α-SNAP may in fact interact with SNAREs along the entire length of the rod in order to translate the energy generated by NSF’s ATPase activity into conformational changes in one or several SNAREs, leading to the dissociation of the entire complex.
The NSF double ring binds to the SNAP/SNARE complex at the end opposite to the SNARE membrane anchors. The cone-shaped tail of the 20 S particle has a length of 13 nm in contrast to the 15 nm length of SNARE and SNAP/SNARE complexes. This leaves the possibility that the SNARE complex rod partially inserts into the inner ring of NSF, consistent with the presence of a stain-filled pore or cavity in free NSF (Figure 4B). However, the major portion of NSF is stacked above the SNAP/SNARE complex. Although gross rearrangements of the SNAP/ SNARE complex do not seem to occur upon NSF binding (as judged by morphology and by FRET studies), subtle changes are clearly detectable. The cylindrical shape of the SNAP/SNARE complex, with a constant width of 7–8 nm, changes to a cone-like shape in 20 S particles; its width ranges from 9.5 nm adjacent to the NSF rings to 6 nm at its tip. The diameter of one of NSF rings increases from 12 nm to 13.5 nm upon 20 S particle assembly. This change occurs in the inner NSF ring at the interface with the SNAP/SNARE complex.
Our localization of NSF’s carboxyl terminus to the outer ring indicates that the inner ring likely contains the N or D1 domains of NSF, which are involved in SNAP and SNARE interactions (Nagiec et al., 1995), while the outer ring consists of D2 hexamerization domains (Lenzen et al., 1998; Yu et al., 1998). Furthermore, platinum replicas of uncomplexed NSF under conditions permitting 20 S particle assembly demonstrate that the N domains are arranged radially as six small globules surrounding the central core of two stacked rings (Hanson et al., 1997a). It is therefore possible that a movement of the six NSF N-terminal domains upon SNAP/SNARE binding increases the diameter of the inner NSF ring and/or the mass of the SNAP/SNARE complex adjacent to the inner NSF ring. Unfortunately, all attempts to decorate 20 S particles with antibodies directed against the N domains of NSF were unsuccessful.
It is possible that α-SNAP contributes to these structural changes as well. There are indications that α-SNAP contains at least two NSF binding sites that are distributed to the amino- and carboxy-terminal half of the molecule (Barnard et al., 1997). The occupancy of these binding sites by NSF could cause molecular rearrangements throughout the entire SNAP molecule that may explain the decreased diameter in the SNAP/SNARE complex at the tip of 20 S particles. Another possibility is that the detergent (Triton X-100) images less prominently with 20 S particles than with the smaller SNAP/SNARE and SNARE complexes. To unambiguously resolve the binding interactions between individual domains NSF, SNAPs, and SNAREs will require high resolution data from crystallographic or NMR studies.
SNARE complex formation is not only involved in heterotypic membrane fusion but also plays a role in homotypic membrane fusion (Nichols et al., 1997; Sato and Wickner, 1998; Patel et al., 1998; Rabouille et al., 1998). The hexameric ATPase p97 (Peters et al., 1990) and p47 (Kondo et al., 1997) act on t-/t-SNARE complexes in a manner similar to NSF and SNAP on v-/t-SNARE complexes. p47 and α-SNAP both interact directly with t-SNAREs and compete with each other for a binding site on syntaxin 5 (Rabouille et al., 1998). p97/p47/GST-syntaxin 5 complexes have been visualized by electron microscopy (Rabouille et al., 1998). Similar to the arrangement of 20 S particles, p47 and syntaxin 5 emerge from one ring of the p97 double ring structure (Peters et al., 1992; Rabouille et al., 1998), which resembles NSF in side view. However, p97/p47/GST-syntaxin 5 complexes contain two “legs” (likely consisting of p47 and GST-syntaxin 5) in contrast to the cone-shaped tail of 20 S particles. Remarkably, all structural studies with p97/p47/GST-syntaxin 5 complexes were performed in the presence of Mg-ATP, conditions that favor 20 S particle disassembly. Assuming that NSF and p97 both catalyze SNARE complex disassembly by a similar mechanism involving ATP hydrolysis, the p97/p47/GST-syntaxin 5 images may capture structures following p97-mediated t-SNARE complex dissociation.
NSF and p97 are hexameric ATPases that seem to act by switching the conformational states of their protein substrates. Recent data indicate that NSF has a broader substrate spectrum than first anticipated, as has been demonstrated by its interaction with the carboxy-terminal domain of the GluR2 subunit of AMPA receptors (Nishimune et al., 1998; Osten et al., 1998; Song et al., 1998), which in turn binds to proteins that contain PDZ domains (Dong et al., 1997). It remains to be seen whether NSF and SNAP interact with this and other potential substrates in a similar manner as with SNAREs.
Experimental Procedures
Plasmids
We amplified rat syntaxin 1A, rat VAMP-2, and Aequoria victoria GFP (S65T) and BFP (Y66H/Y145F) cDNAs (Heim and Tsien, 1996) to construct expression vectors for syntaxin-BFP, VAMP-GFP, and GFP-VAMP. The constructs encode the cytoplasmic domains of the SNAREs (residues 1–265 for syntaxin and 1–94 for VAMP) fused at their amino (GFP-VAMP) or carboxyl terminus (VAMP-GFP and syntaxin-BFP) to GFP or BFP (residues 1–238) with an intervening (SGG)3 linker. The syntaxin-BFP and VAMP-GFP PCR products were cloned into the SphI and BglII sites of pQE-70 (Qiagen), while the GFP-VAMP PCR fragment was inserted into the SphI and HindIII sites of pQE-30 (Qiagen).
Expression vectors for syntaxin-HA-his6 or VAMP-myc-his6 were generated by amplifying the cytoplasmic domains of syntaxin (residues 1–265) and VAMP (residues 1–94), and ligating the PCR products into the BamHI and NdeI sites of a pET3A–based vector (Søgaard et al., 1994) modified to encode a carboxy-terminal myc (Evan et al., 1985) or HA (Wilson et al., 1984) tag followed by six histidine residues.
Expression vectors for his6-NSF, his6-NSF-myc, and GST-SNAP-25 have been described elsewhere (Söllner et al., 1993b; Whiteheart et al., 1993; Weber et al., 1998). A plasmid coding for glutathione-S-transferase(GST)-myc-α-SNAP was produced by ligating a double-stranded oligonucleotide encoding the myc epitope into the BamHI site of a GST-α-SNAP expression vector (Nagahama et al., 1996).
Protein Expression
Syntaxin-BFP, GFP-VAMP, and VAMP-GFP were expressed in Escherichia coli strain BL21/DE3 at 25°C, and all other constructs were expressed at 37°C. Bacterial cells were lysed in PBS-D (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.4], and 1 mM dithiothreitol [DTT]) for GST-tagged proteins and in 50 mM Hepes-KOH [pH 7.0], 200 mM KCl, and 2 mM β-mercaptoethanol (β-ME) for his6-tagged proteins in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF) by two passages through a French Press. The lysates were clarified by centrifugation at 105 × g for 1 hr. GST-SNAP-25 was purified by affinity chromatography on glutathione agarose (GSHA) beads (Sigma). GST-SNAP-25 was eluted from the beads in PBS-D, 10 mM glutathione, and 20% (w/v) glycerol, dialyzed into the same buffer lacking glutathione, and stored on ice up to 2 weeks prior to use. His6-tagged proteins were purified by affinity chromatography on Ni2+-NTAagarose beads (Qiagen), eluted with an imidazole gradient in 25 mM Hepes-KOH [pH 7.5], 200 mM KCl, 10% (w/v) glycerol, and 2 mM β-ME, and dialyzed into 25 mM Hepes-KOH [pH 7.5], 100 mM KCl, 10% (w/v) glycerol, and 1 mM DTT.
His6-NSF and his6-NSF-myc were purified as described (White-heart et al., 1994). Protein concentrations were estimated by the method of Bradford using IgG as a standard.
Isolation of SNARE Complexes, SNAP/SNARE Complexes, and 20 S Particles
An E. coli lysate (100 µl) containing approximately 4 mg/ml GST-α-SNAP was incubated with 100 µl GSHA beads for 1 hr at 4°C in 0.5 ml PBS-D containing 0.05% (w/v) Tween-20 (PBS-TD). The beads were washed twice with 1 ml each of PBS-TD, PBS-TD containing 1 M KCl, and buffer A (25 mM Hepes-KOH [pH 7.0], 100 mM KCl, 10% (w/v) glycerol, 1% (w/v) Triton-X-100, and 1 mM DTT), and incubated for 2–16 hr at 4°C with bovine brain extract (50–100 mg protein; Söllner et al., 1993a) in 30 ml buffer A. The beads were washed three times with 1 ml each of buffer A and buffer B (25 mM Hepes-KOH [pH 7.4], 100 mM NaCl, 0.5% (w/v) Triton-X-100, and 1 mM DTT).
SNARE complexes were released from the beads by incubating them four times for 5 min at 25°C in 100 µl 25 mM Hepes-KOH [pH 7.4], 1 M NaCl, 1% (w/v) _n_-octylglucoside, and 1 mM DTT. The eluates were collected and pooled. Under these elution conditions GST-α-SNAP remained on the beads.
SNAP/SNARE complexes were isolated by cleaving GST-α-SNAP with 20 units of thrombin (Sigma) in 100 µl buffer B for 30 min at 25°C. The eluate containing SNAP/SNARE complexes was pooled with three 100 µl washes of the beads in buffer B and PMSF was added to a final concentration of 1 mM.
To assemble 20 S particles, 900 µg his6-NSF-myc was added to the GST-α-SNAP beads in the presence of brain extract; the incubation was continued for 1–2 hr. Whenever NSF was present, all buffers were adjusted to 2 mM EDTA and 0.5 mM ATP. The beads were washed three times each with 1 ml buffer A and B. 20 S particles were released by thrombin cleavage of GST-α-SNAP as above. An additional 80 µg of NSF was added and the sample was incubated on ice for 15 min.
All protein complexes were sedimented in linear 10%–35% (w/v) glycerol gradients in buffer C (25 mM Hepes-KOH [pH 7.0], 100 mM NaCl, 0.05% (w/v) Triton-X-100, and 1 mM DTT) in an SW41 rotor (Beckman) at 4°C and 40,000 RPM for 12–18 hr. Fractions containing the appropriate protein complexes were concentrated by dialysis against buffer C containing 10% (w/v) polyethylene glycol (MW 35,000) prior to electron microscopy.
Formation of 20 S Particles Decorated with Anti-myc Antibodies
20 S particles containing myc-tagged α-SNAP were prepared by replacing GST-α-SNAP and his6-NSF-myc with GST-myc-α-SNAP and his6-NSF, respectively. To incorporate VAMP-myc-his6 into 20 S particles, GST-α-SNAP beads with bound SNARE complexes were washed with 30 ml of buffer A and resuspended in 50 ml buffer A containing 2 mM MgCl2, 0.5 mM ATP, and 2 mg his6-NSF, as well as 400 µg VAMP-myc-his6. After 2 hr, EDTA was added to a final concentration of 8 mM to favor 20 S particle reassembly and the incubation proceeded overnight. In the presence of excess VAMP-myc-his6, SNARE complexes and 20 S particles reassembled with VAMP-myc-his6 instead of endogenous VAMP (see Figure 1, lane 3). All subsequent buffers contained 2 mM EDTA and 0.5 mM ATP. Immobilized 20 S particles containing either his6-NSF-myc or VAMP-myc-his6 were washed three times with 1 ml buffer B, incubated with 1.2 mg of 9E10 (anti-myc) monoclonal antibody (Evan et al., 1985) for 2 hr, and washed again. 20 S particles with bound antibodies were released from the beads as above with no subsequent NSF addition and sedimented into glycerol gradients. 20 S particles containing myc-α-SNAP were sedimented through glycerol gradients prior to antibody binding to remove myc-α-SNAP that was not incorporated in 20 S particles. These particles were concentrated, incubated with 200 µg of 9E10 antibody, and separated from unbound antibodies by a second round of velocity centrifugation.
Isolation of Recombinant SNARE Complexes and Fluorometric Studies
250 µg of GST-SNAP-25 was bound to 100 µl GSHA beads in 0.5 ml PBS-D. The beads were washed twice with 1 ml each of PBS-D and buffer D (25 mM Hepes-KOH [pH 7.4], 100 mM NaCl, 2% (w/v) glycerol, 0.05% (w/v) Triton-X-100, and 1 mM DTT), incubated for 2 hr with 900 µg Syn-HA-his6 and 1.5 mg VAMP-myc-his6 in a final volume of 10 ml buffer D, and washed again. Ternary SNARE complexes were released from the beads by thrombin cleavage and sedimented for 18 hr in 7%–25% (w/v) glycerol gradients in buffer C lacking detergent as above.
For fluorometric studies, ternary SNARE complexes were formed on 100 µl GSHA beads with 30 µg GST-SNAP-25, 50 µg syntaxin-BFP, and either 50 µg VAMP-GFP or 50 µg GFP-VAMP in 1 ml buffer E (25 mM Tris-HCl [pH 7.5], 125 mM NaCl, 10% (w/v) glycerol, and 1 mM DTT) and released from the beads by thrombin cleavage. Each eluate was divided into duplicate samples (in 600 µl buffer E) and transferred into cuvettes held at 37°C. 300 µg his6-α-SNAP and 2 mM MgCl2 were added to all samples and duplicates received either 1 mM ATP or 1 mM ATPγS. 120 µg NSF was added 5 min later. Emission spectra were monitored before and after all additions with a Perkin Elmer LS 50B fluorimeter using an excitation wavelength of 375 nm and a bandwidth of 15 nm.
FRET efficiency for BFP was calculated from the fluorescence intensities (F) at 440 nm as 1- (FATPγS/FATP). FATPγS represents the fluorescence intensity under 20 S particle assembly (solid lines, Figures 6A and 6B), and FATP the fluorescence intensity under 20 S particle disassembly conditions (dashed lines, Figures 6A and 6B). Therefore, FATPγS reflects the BFP fluorescence intensity with BFP and GFP in a SNARE-mediated complex, while FATP represents the BFP fluorescence intensity with BFP dissociated from GFP.
Electron Microscopy and Image Averaging
Samples (3–5 µl) at a protein concentration of <0.05 mg/ml were applied to electron microscopy grids rendered hydrophilic by a 15–60 sec exposure to glow discharge in a plasma cleaner (Harrick Scientific Corp., New York). The grids were washed once either in distilled water or in 0.5 mM ATP (pH 7.0) and 2 mM EDTA and negatively stained with 2% (w/v) uranyl acetate (pH 4.5) or 2% (w/v) methylamine tungstate (pH 6.5). Electron micrographs were recorded with a Phillips EM 420 at a magnification of 36,000 and an accelerating voltage of 100 kV. The electron micrographs were preselected using a laser diffractometer for evaluating the focus conditions and digitized by means of an Eikonix 1412 camera with a 15 × 15 µm2 pixel size or a LeafScan 45 with a 10 × 10 µm2 pixel size.
Particles were selected by hand and extracted for image processing using programs for single particle averaging and principle component analysis (PCA) contained in the SEMPER and EM system program packages (Hegerl, 1996; Saxton, 1996). Alignment of single particles was performed by means of correlation methods starting with randomly chosen molecules as references. If the preliminary averages were consistent, they were used as references for refinement of the orientational and lateral alignment. PCA was performed with data sets from which the average images had been subtracted (van Heel and Frank, 1981), following the procedures outlined in detail in Frank, 1996. The most significant eigenvectors were used for classification, as judged by their corresponding eigenvalues and their contribution in separating image sets that differ in the orientation, conformation, staining variability, and structural integrity of the molecules. The number of particles classified was 1343 for recombinant SNARE complexes, 510 for endogenous SNARE complexes, 624 for SNAP/SNARE complexes, 296 for NSF side views, 1448 for NSF top views, 3567 for monomeric 20 S particles, and 335 for dimeric 20 S particles.
Selective averages from the most representative and consistent image classes were used to refine the alignment of the whole data sets. These data were reanalyzed by PCA and classification. In particular, 20 S particles were aligned with respect to the double ring structure of the particles, using a reference average masked for the SNAP/SNARE portion of the complex. Images of SNARE and SNAP/SNARE complexes were preselected for smaller and larger particles. PCA and subsequent classification of both types of particles revealed monomeric and dimeric SNARE and SNAP/SNARE complexes that were separated from each other and aligned using particular class averages. The “central-plug and spoke” multimeric SNARE complexes were not processed further. NSF particles, either in top or side view orientation, were selected separately from micrographs and individually processed. The resolution of averages was assessed according to the radial correlation function criterion, by comparing equivalent Fourier components of averaged molecular images calculated from two independent data sets (Saxton and Baumeister, 1982). The values were usually between 1.7 nm for data sets containing ≥300 images and 2.3 nm for smaller data sets. All averaged images (Figures 2A-2E, 3B-3C, 4A-4B, and 7) are presented at the same magnification with the exception of Figure 3D.
Acknowledgments
We thank Wolfgang Baumeister for support, Gero Miesenböck for thoughtful comments and critical reading of the manuscript, and Susanne Volker-Mürkl for technical assistance. This work was supported by the Mathers Charitable Foundation (to J. E. R), a National Institutes of Health grant (to J. E. R.), the Andrew W. Mellon Foundation to the Tri-Institutional M.D.-Ph.D. Program (to T. M. H.), and postdoctoral fellowships of the Medical Research Council of Canada (to F. P.) and the Deutsche Forschungsgemeinschaft (to C. W.).
References
- Barnard RJ, Morgan A, Burgoyne RD. Stimulation of NSF ATPase activity by α-SNAP is required for SNARE complex disassembly and exocytosis. J. Cell Biol. 1997;139:875–883. doi: 10.1083/jcb.139.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumert M, Maycox PR, Navone F, De Camilli P, Jahn R. Synaptobrevin: an integral membrane protein of 18,000 daltons present in small synaptic vesicles of rat brain. EMBO J. 1989;8:379–384. doi: 10.1002/j.1460-2075.1989.tb03388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at pre-synaptic active zones. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc. Natl. Acad. Sci. USA. 1988;85:7852–7856. doi: 10.1073/pnas.85.21.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary DO, Rothman JE. Purification of three related peripheral membrane proteins needed for vesicular transport. J. Biol. Chem. 1990;265:10109–10117. [PubMed] [Google Scholar]
- Dong H, O’Brien RJ, Tung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- Elferink LA, Trimble WS, Scheller RH. Two vesicle-associated membrane protein genes are differentially expressed in the rat central nervous system. J. Biol. Chem. 1989;264:11061–11064. [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough RH, Cantor CR. The use of singlet-singlet energy transfer to study macromolecular assemblies. Methods Enzymol. 1978;48:347–379. doi: 10.1016/s0076-6879(78)48019-x. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Eliason WK, Brünger AT, Jahn R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 1998;37:10354–10362. doi: 10.1021/bi980542h. [DOI] [PubMed] [Google Scholar]
- Fleming KG, Hohl TM, Yu RC, Müller SA, Wolpensinger B, Engel A, Engelhardt H, Brünger AT, Söllner TH, Hanson PI. A revised model for the oligomeric state of the N-ethylmaleimide-sensitive fusion protein, NSF. J. Biol. Chem. 1998;273:15675–15681. doi: 10.1074/jbc.273.25.15675. [DOI] [PubMed] [Google Scholar]
- Frank J. Three Dimensional Electron Microscopy of Macromolecular Assemblies. San Diego, CA: Academic Press; 1996. Multivariate statistical analysis and classification of images; pp. 121–182. [Google Scholar]
- Furth AJ, Bolton H, Potter J, Priddle JD. Separating detergent from proteins. Methods Enzymol. 1984;104:318–328. doi: 10.1016/s0076-6879(84)04098-2. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Otto H, Barton N, Jahn R. The N-ethyl-maleimide sensitive fusion protein and α-SNAP induce a conformational change in syntaxin. J. Biol. Chem. 1995;270:16955–16961. doi: 10.1074/jbc.270.28.16955. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997a;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Heuser JE, Jahn R. Neurotransmitter release—four years of SNARE complexes. Curr. Opin. Neurobiol. 1997b;7:310–315. doi: 10.1016/s0959-4388(97)80057-8. [DOI] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Südhof TC, Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Yamasaki S, Nauenburg S, Binz T, Niemann H. Disassembly of the reconstituted synaptic vesicle membrane fusion complex in vitro. EMBO J. 1995;14:2317–2325. doi: 10.1002/j.1460-2075.1995.tb07226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegerl R. The EM program package: a platform for image processing in biological electron microscopy. J. Struct. Biol. 1996;116:30–34. doi: 10.1006/jsbi.1996.0006. [DOI] [PubMed] [Google Scholar]
- Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Hess DT, Slater TM, Wilson MC, Skene JHP. The 25 kDa synaptosomal-associated protein SNAP-25 is the major methionine-rich polypeptide in rapid axonal transport and a major substrate for palmitoylation in adult CNS. J. Neurosci. 1992;12:4634–4641. doi: 10.1523/JNEUROSCI.12-12-04634.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- Lenzen CU, Steinmann D, Whiteheart SW, Weis WI. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- Lin RC, Scheller RH. Structural organization of the synaptic exocytosis core complex. Neuron. 1997;19:1087–1094. doi: 10.1016/s0896-6273(00)80399-2. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Südhof TC. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity α-SNAP binding site. J. Biol. Chem. 1995;270:2213–2217. doi: 10.1074/jbc.270.5.2213. [DOI] [PubMed] [Google Scholar]
- Nagahama M, Orci L, Ravazzola M, Amherdt M, Lacomis L, Tempst P, Rothman JE, Söllner TH. A v-SNARE implicated in intra-Golgi transport. J. Cell Biol. 1996;133:507–516. doi: 10.1083/jcb.133.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec EE, Bernstein A, Whiteheart SW. Each domain of the N-ethylmaleimide-sensitive fusion protein contributes to its transport activity. J. Biol. Chem. 1995;270:29182–29188. doi: 10.1074/jbc.270.49.29182. [DOI] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Nishimune A, Isaac JTR, Molnar E, Noel J, Nash SR, Tagaya M, Collingridge GL, Nakanishi S, Henley JM. NSF binding to GluR2 regulates synaptic transmission. Neuron. 1998;21:87–97. doi: 10.1016/s0896-6273(00)80517-6. [DOI] [PubMed] [Google Scholar]
- Osten P, Srivastava S, Inman GJ, Vilim FS, Khatri L, Lee LM, States BA, Einheber S, Milner TA, Hanson PI, et al. The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and α- and β-SNAPs. Neuron. 1998;21:99–110. doi: 10.1016/s0896-6273(00)80518-8. [DOI] [PubMed] [Google Scholar]
- Oyler GA, Higgins GA, Hart RA, Battenberg E, Billingsley M, Bloom FE, Wilson MC. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J. Cell Biol. 1989;109:3039–3052. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek I, Orci L, Ravazzola M, Erdjument-Bromage H, Amherdt M, Tempst P, Söllner TH, Rothman JE. ERS-24, a mammalian v-SNARE implicated in vesicle traffic between the ER and the Golgi. J. Cell Biol. 1997;137:1017–1028. doi: 10.1083/jcb.137.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SK, Indig FE, Olivieri N, Levine ND, Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Peters JM, Walsh MJ, Franke WW. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. EMBO J. 1990;9:1757–1767. doi: 10.1002/j.1460-2075.1990.tb08300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Harris JR, Lustig A, Müller S, Engel A, Volker S, Franke WW. Ubiquitous soluble Mg2+-ATPase complex. A structural study. J. Mol. Biol. 1992;223:557–571. doi: 10.1016/0022-2836(92)90670-f. [DOI] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Braun JE, Calakos N, Ting AE, Bennett MK, Scheller RH. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- Poirier MA, Hao JC, Malkus PN, Chan C, Moore MF, King DS, Bennett MK. Protease resistance of syntaxin/SNAP-25/VAMP complexes. Implications for assembly and structure. J. Biol. Chem. 1998a;273:11370–11377. doi: 10.1074/jbc.273.18.11370. [DOI] [PubMed] [Google Scholar]
- Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 1998b;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a common component of the NSF-and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Sato K, Wickner W. Functional reconstitution of ypt7p GTPase and a purified vacuole SNARE complex. Science. 1998;281:700–702. doi: 10.1126/science.281.5377.700. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsci. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Saxton WO. Semper: distortion compensation, selective averaging, 3-D reconstruction, and transfer function correction in a highly programmable system. J. Struct. Biol. 1996;116:230–236. doi: 10.1006/jsbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- Søgaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Söllner TH. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993a;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993b;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Song I, Kamboj S, Xia J, Dong H, Liao D, Huganir RL. Interaction of the n-ethylmaleimide-sensitive factor with AMPA receptors. Neuron. 1998;21:393–400. doi: 10.1016/s0896-6273(00)80548-6. [DOI] [PubMed] [Google Scholar]
- Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- Subramaniam VN, Peter F, Philp R, Wong SW, Hong W. GS28, a 28-kilodalton Golgi SNARE that participates in ER-Golgi transport. Science. 1996;252:1161–1163. doi: 10.1126/science.272.5265.1161. [DOI] [PubMed] [Google Scholar]
- Subramaniam VN, Loh E, Hong W. N-ethylmaleimide-sensitive factor (NSF) and α-soluble NSF attachment proteins (SNAP) mediate dissociation of GS28-Syntaxin 5 Golgi SNAP receptor (SNARE) complexes. J. Biol. Chem. 1997;272:25441–25444. doi: 10.1074/jbc.272.41.25441. [DOI] [PubMed] [Google Scholar]
- Südhof TC. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Tagaya M, Wilson DW, Brunner M, Arango N, Rothman JE. Domain structure of an N-ethylmaleimide-sensitive fusion protein involved in vesicular transport. J. Biol. Chem. 1993;268:2662–2666. [PubMed] [Google Scholar]
- Ungermann C, Nichols BJ, Pelham HR, Wickner W. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J. Cell Biol. 1998;140:61–69. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Söllner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Whiteheart SW, Griff IC, Brunner M, Clary DO, Mayer T, Buhrow SA, Rothman JE. SNAP family of NSF attachment proteins includes a brain-specific isoform. Nature. 1993;362:353–355. doi: 10.1038/362353a0. [DOI] [PubMed] [Google Scholar]
- Whiteheart SW, Rossnagel K, Buhrow SA, Brunner M, Jaenicke R, Rothman JE. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Wilson DW, Wilcox CA, Flynn GC, Chen E, Kuang WJ, Henzel WJ, Block MR, Ullrich A, Rothman JE. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 1989;339:355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
- Yu RC, Hanson PI, Jahn R, Brünger AT. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat. Struct. Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]