Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia (original) (raw)
Abstract
Knockout studies have shown that the transcription factor Nrf1 is essential for embryonic development. Nrf1 has been implicated to play a role in mediating activation of oxidative stress response genes through the antioxidant response element (ARE). Because of embryonic lethality in knockout mice, analysis of this function in the adult knockout mouse was not possible. We report here that mice with somatic inactivation of nrf1 in the liver developed hepatic cancer. Before cancer development, mutant livers exhibited steatosis, apoptosis, necrosis, inflammation, and fibrosis. In addition, hepatocytes lacking Nrf1 showed oxidative stress, and gene expression analysis showed decreased expression of various ARE-containing genes, and up-regulation of CYP4A genes. These results suggest that reactive oxygen species generated from CYP4A-mediated fatty acid oxidation work synergistically with diminished expression of ARE-responsive genes to cause oxidative stress in mutant hepatocytes. Thus, Nrf1 has a protective function against oxidative stress and, potentially, a function in lipid homeostasis in the liver. Because the phenotype is similar to nonalcoholic steatohepatitis, these animals may prove useful as a model for investigating molecular mechanisms of nonalcoholic steatohepatitis and liver cancer.
Keywords: hepatocellular carcinoma, oxidative stress, knockout mouse
Transcription of many cytoprotective genes and phase-2 xenobiotic metabolizing genes is regulated through cis-active sequences known as antioxidant response elements (ARE) (1, 2). Regulation of ARE function is mediated by various basic leucine zipper (bZIP) transcription factors including members of the “cap n collar” (CNC)-bZIP and small-Maf family of proteins. Nrf1 and Nrf2 are CNC-bZIP proteins, and they function as obligate heterodimers by complexing with small-Maf and other bZIP proteins (3). An important role for Nrf2 in xenobiotic metabolism and oxidative stress response had been identified through knockout studies in mice (4–10). These and other studies indicate that Nrf2 is an important activator of AREs.
In contrast, the function of Nrf1 is not fully understood. Mice deficient in Nrf1 function die during development (11). Analysis of nrf1 and nrf1::nrf2 mutant cells suggests that Nrf1 is also involved in the oxidative stress response (12, 13). However, the importance of Nrf1 in this function in an intact animal is not certain because early lethality precludes analysis of Nrf1-deficient animals beyond embryonic development. Chimeric mice generated with Nrf1-deficient embryonic stem cells showed widespread apoptosis in fetal livers at late gestation, demonstrating a cell autonomous role of Nrf1 in the survival of hepatocytes (14). This finding suggests that Nrf1 is required for normal function of hepatocytes. Based on these findings, we hypothesize that Nrf1 is critical to the oxidative stress response in the adult liver, and that it plays an important role in oxidative stress-induced liver disease. To bypass embryonic lethality, we used a Cre-lox system to investigate the function of Nrf1 in adult liver. We report here that the disruption of Nrf1 function in hepatocytes results in oxidative stress, steatohepatitis, liver damage, and spontaneous development liver cancer. This study provides genetic evidence identifying a critical role for Nrf1 attenuating oxidative stress and neoplastic growth in livers of adult mice.
Materials and Methods
Nrf1 Gene Targeting Using the Cre-loxP Strategy. Genomic clones containing the nrf1 gene have been described (11). We generated a targeting construct containing a 5′ loxP site inserted into the nrf1 intron-3, and a phosphoglycerate kinase 1 (PGK-1) promoter-driven neomycin resistance gene cassette flanked by a 3′ loxP site downstream of the terminal exon. An internal ribosome entry site (IRES)-GFP was also incorporated 3′ of the Neo cassette. A PGK-1 diphtheria toxin cassette was included for negative selection. Targeting in JM-1 ES cells has been described (11). Southern blotting of _Nsi_I-digested DNA using a 5′ nrf1 external probe was done. Wild-type and targeted alleles yield a 14-kb and a 16-kb fragment, respectively. Approximately 10 positive clones were identified from 100 clones screened. Chimeric mice were generated as described (11). F1 offspring containing the floxed nrf1 targeted allele were determined by Southern blot analysis and/or PCR amplification. Hepatocyte-specific deletion of the floxed nrf1 allele was accomplished through breeding with the Alb-Cre transgenic mice (The Jackson Laboratory).
Biochemical Analysis and Measurements of Cellular Redox Status. Alanine aminotransferase levels were measured by using Infinity alanine aminotransferase (ALT) reagent (Thermo DMA, Louisville, CO). Liver triglyceride was determined by an enzymatic method (Stanbio Laboratories, Boerne, TX). Lipid peroxidation levels were quantified by measuring thiobarbituric acid-reactive substance (TBARS). Livers from adult mice (3–4 months old) were homogenized in 20 mM phosphate buffer (pH 7.4) containing 5 mM butylated hydroxytoluene, and mixed at a 1:2 ratio with a solution containing 15% trichloroacetic acid, 0.37% thiobarbituric acid, 0.2 M HCl and 0.03% butylated hydroxytoluene, and boiled for 15 min. TBARS was measured spectrophotometrically at 535 nm. Primary hepatocytes from adult mice (2–5 months old) were isolated by a collagenase perfusion protocol (15). Hepatocytes were cultured on collagen-coated dishes in DMEM/F12 with 5% calf serum to remove nonparenchymal cells before analysis. Fluorescent measurements of reactive oxygen species (ROS) were done as described (12).
Histology and Immunohistochemistry. Livers were fixed in 10% neutral buffered formalin and imbedded in paraffin. Oil red O staining of neutral lipids was done on frozen sections. For immunohistochemical staining, paraffin sections are dewaxed, and rehydrated through a series of alcohols. Antigen retrieval was done by heating in 10 mM citrate buffer (pH 6.5). Slides were incubated with rabbit polyclonal activated caspase-3 antibody (Cell Signaling Technology, Beverly, MA) or mouse monoclonal proliferating cell nuclear antigen (PCNA) antibody (Santa Cruz Biotechnology), followed by incubation with biotinylated secondary antibody and avidin-peroxidase (Vector Laboratories). Visualization was achieved with 3,3′-diaminobenzidine and counterstaininig with hematoxylin. Oxidative DNA damage was detected by mouse monoclonal 8-oxoG antibody (Trevigen, Gaithersburg, MD), followed by staining with biotinylated secondary antibody and rhodamine avidin-DCS (Vector Laboratories).
Immunoblot and RT-PCR. Immunoblot analysis was done as described (13). Antibodies used include actin (Sigma), Gclc and Gclm (gifts from T. Kavanaugh, University of Washington, Seattle, WA), and HO-1, SOD1, and SOD2 (Stressgen Biotechnologies, Victoria, Canada). Synthesis of cDNAs from total RNA and RT-PCR was done as described (13). Primers for PCR reactions are shown in Table 1.
Table 1. PCR primers.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| 18S | TCGGCGTCCCCCAACTTCTTA | GGTAGTAGCGACGGGCGGTGT |
| Nrf1 | GACAAGATCATCAACCTGCCTGTAG | GCTCACTTCCTCCGGTCCTTTG |
| GSTA1 | AGAATGGAGTGCATCAGGTGGTGGCTC | GGCAGGCAAGTAACGGTTTTTGGT |
| GSTA3 | GAGATCGACGGGATGAAACTGGTG | GCGCTTTCAGGAGAGGGAAGTTGT |
| GSTA4 | CCTTGGTTGAAATCGATGGGATGA | GCAGCAGAGGGAAGTCGGACACAGTA |
| GSTM1 | AGCACCCTGGCCTTCTGCACT | TTCGCAGAAACGGGCTGTGAG |
| GSTM2 | TACACCATGGGGGACGCTCCT | TGGCCAACTGTATGCGGGTGT |
| GSTM3 | TGATTAGGCCCCTGCCATGCT | TTGGGTCTGGGCACCAATGAA |
| GSTM4 | CGGGGGTTCTGGGAACAGTTG | GCTGGCAGGCAAGACCATCAA |
| GSTM6 | TCGGAGTTCCTGGGGAAGCAG | CCTCAAAGCGGGCCATGAAGT |
| GSTP | TTTGGGGGCTTTATGGGAAAAACCA | ACATAGGCAGAGAGCAGGGGGAAG |
| GSTTL | GCCCTGTCGCGCCATTTATATCTT | TTGCTCACCAAGGAAAACAGGGAA |
| PPAR-α | AGGGCCTCCCTCCTACGCTTG | GGGTGGCAGGAAGGGAACAGA |
| Acox1 | GCACCTTCGAGGGGGAGAACA | GCGCGAACAAGGTCGACAGAA |
| Ehhadh | GCCGCTAGCCCAGAGCACATT | CAGCAGCAGCAGCAACAGCAG |
| Catalase | GGCACACTTTGACAGAGAGCGGAT | AGTTTTTGATGCCCTGGTCGGTCT |
| Acaa | GGCTGAGCGGTTTGGCATTTC | TGGGGAGGCCCAGTTCTTCAG |
| Cyp4A10 | TTCAGAGCCTCCTGGGGGATG | GGAGCAGTGTCAGGGCCACAA |
| Cyp4A14 | ATGCCTGCCAGATTGCTCACG | GGGTGGGTGGCCAGAGCATAG |
Statistics. Data are expressed as mean ± SD. Differences in means between groups were analyzed by t test and considered significant at P < 0.05.
Results
Nrf1 Liver Knockout Mice Are Viable. A targeting vector was constructed in which exon-4 of nrf1 was flanked with loxP sequences (Fig. 1a). Exon-4 contains the bZIP domain of Nrf1 (11). Thus, its elimination functionally cripples the nrf1 gene. Targeted clones identified by Southern blotting were injected into blastocyst (Fig. 1_b_). Germ-line transmission was achieved, and F1 animals were obtained. Heterozygous F1 mice bearing the modified (“floxed”) nrf1 allele (nrf1flox/+) were intercrossed to existing nrf1+/– mice to generate mice that bear one floxed nrf1 allele and one _nrf1_-null allele (nrf1flox/–) (Fig. 1_c_). Because nrf1flox/– mice have only one functional allele, only a single recombination event will be required per cell. Thus, expression should be all or none on a single cell basis. Interbreeding of nrf1+/– and nrf1flox/+ resulted in nrf1flox/– offspring that followed the predicted Mendelian frequency, and they were phenotypically indistinguishable from nrf1+/– and wild-type mice (data not shown). Thus, the presence of loxP sequences in the modified nrf1 allele did not seem to interfere with gene function.
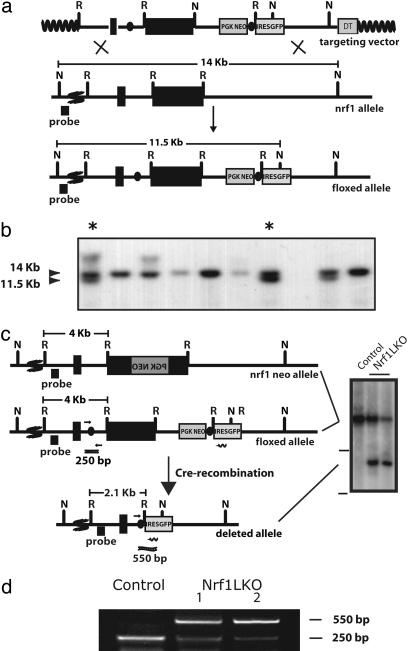
Fig. 1.
Targeting of the nrf1 locus. (a Top) Map of the targeting construct containing loxP elements (dark ovals) flanking the terminal exon of nrf1 and the positive (PGKNEO) selection cassette. The targeting construct also contains an internal ribosome entry site (IRES)-EGFP cassette and a negative (DT) selection cassette. (Middle) The terminal portion of the wild-type nrf1 gene, and the 5′ external probe used to detect targeted clones. (Bottom) The targeted allele. Sites for restriction enzymes _Eco_RI (R) and _Nsi_I (N) are shown. The predicted sizes of _Nsi_I fragments of wild-type (14-kb) and targeted allele (11.5-kb) are shown. (b) Southern blot analysis of ES clones digested with _Nsi_I and probed with the 5′ external probe. Asterisks indicate targeted clones. (c Top) The nrf1-neo knockout allele described previously (11). (Middle) The floxed nrf1 allele. (Bottom) The Cre-mediated recombined allele. Restriction enzyme sites for _Eco_RI (R), and _Nsi_I (N) are shown. Primers used to detect the nonrecombined floxed nrf1 allele (250-bp product) and the recombined allele (500-bp) are indicated by short straight arrows and wiggly arrow, respectively. (Right) Southern blot analysis of liver DNA obtained from control and Nrf1LKO mice. The top band represents the 4kb _Eco_RI fragment of both the nrf1-neo and floxed nrf1 alleles detected by the indicated probe. The lower band represents the 2.1kb _Eco_RI fragment of the recombined allele. (d) Analysis of recombined and nonrecombined floxed nrf1 allele in liver DNA. The top band represents the 550-bp product amplified from the recombined floxed nrf1 allele. The bottom band represents the 250-bp product amplified from the nonrecombined floxed nrf1 allele.
Alb-Cre transgenic mice (16) were crossed with nrf1flox/– mice to generate liver-specific Nrf1 knockouts. Henceforth AlbCre;nrf1flox/– mice will be referred to as Nrf1LKO mice for convenience. Southern analysis using an intron-3 probe detected a 4.0-kb _Eco_RI fragment derived from the floxed allele and the mutant allele containing the Neo cassette in nrf1flox/– animals (Fig. 1_c_). Liver DNA from Nrf1LKO animals showed an additional 2.1-kb _Eco_RI fragment derived from the deleted allele (Fig. 1_c_). Liver DNA analyzed for Cre-mediated deletion of the floxed allele by PCR using primers flanking the loxP sequences revealed the expected 250-bp product from the intact floxed locus and a 550-bp product representing the deleted allele (Fig. 1_d_). Thus, efficient recombination of the nrf1-floxed allele was detected in livers of Nrf1LKO animals. RT-PCR and real-time RT-PCR analysis were used to determine the effectiveness of Cre-mediated gene deletion on abolishing Nrf1 gene expression. Nrf1 mRNA levels in Nrf1LKO livers were markedly decreased and ranged around 10% of controls (Fig. 4 and data not shown). Although an internal ribosome entry site (IRES)-GFP was incorporated for tracking recombined cells, GFP expression was too weak to be of usefulness for our analysis (data not shown). Mice genotyped as Nrf1LKO mice were recovered at the expected frequency (data not shown). No increase in mortality or morbidity was observed early on in Nrf1LKO mice compared with littermate controls (nrf1flox/– and AlbCre;nrf1+/–).
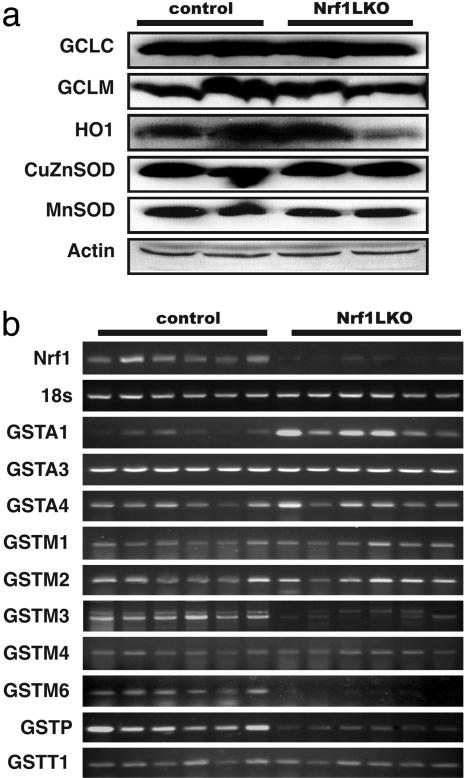
Fig. 4.
Expression of ARE-regulated genes in Nrf1LKO livers. (a) Western blot analysis of control and Nrf1LKO livers. Equal loading of protein in each lane was confirmed by actin. (b) RT-PCR analysis of mRNA encoding various ARE-dependent genes and lipid metabolism. 18s levels were used as control. Figures are representative of six control and six Nrf1LKO liver samples.
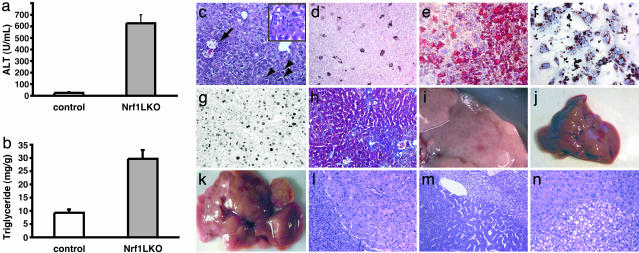
Nrf1LKO Livers Develop Steatohepatitis and Spontaneous Hepatic Cancer. Analysis of Nrf1LKO embryos failed to show any apparent abnormalities (data not shown). This finding is consistent with published results showing that maximal Cre expression driven by the albumin promoter was not detected until after birth (data not shown). At 4 weeks, serum alanine-transaminase levels were elevated in Nrf1LKO mice (Fig. 2_a_). Histological analysis showed apoptotic cells, necrosis, and infiltration of inflammatory cells (Fig. 2 c and d). Vacuolated cells that stained for oil red O were also seen (Fig. 2_e_), and biochemical analysis revealed a 3-fold elevation in triglyceride levels (Fig. 2_b_). Lipid accumulation was similarly seen in cultures of primary hepatocytes derived from Nrf1LKO mice, suggesting that this defect is a direct consequence of Nrf1 inactivation (Fig. 2_f_). Active hepatic proliferation was indicated by mitotic figures and increased proliferating cell nuclear antigen (PCNA)-positive cells (Fig. 2_g_ and data not shown). Pericentral and pericellular fibrosis was seen in older animals (Fig. 2_h_). Abnormal hepatocytes with enlarged nuclei or abnormal mitotic figures were seen in some Nrf1LKO livers (data not shown). These abnormal appearing cells were frequently present in livers of older animals containing frank neoplasm, suggesting that they were probably precancerous in nature. As early as 4 months, foci of neoplastic growth resulting in visible nodules in livers were evident in some Nrf1LKO mice (n = 2/9) (Fig. 2_i_). At 10–12 months, enlarged livers with multiple tumors were seen (compare Fig. 2 j and k). Both hepatocellular adenomas and carcinomas were detected (Fig. 2 l_–_n). PCR of DNA isolated from nodules revealed prominent recombination of the conditional nrf1 allele, indicating that tumors were deficient in Nrf1 (data not shown). The incidence of tumors was 100% (n = 12/12) by 12 months, and occurred equally in both males (n = 6) and females (n = 6). No tumors were observed in control littermates (nrf1flox/– and AlbCre;nrf1+/–) mice as far out as 15 months (n = 8), indicating that these abnormalities were unrelated to adverse effects of Cre, or the loxP sequence. These findings indicate that loss of Nrf1 results in steatohepatitis and spontaneous liver cancer.
Fig. 2.
Steatohepatitis and liver cancer in Nrf1LKO mice. (a) Serum concentration of alanine aminotransferase (ALT) in control and Nrf1LKO mice at 4 weeks of age. Bar graph shows mean ± SD of 5–7 animals per group. *, P < 0.05. (b) Liver triglyceride levels in control and Nrf1LKO mice at 6–8 weeks of age. Bar graph shows mean ± SD of 6–8 animals per group. *, P < 0.05. (c) A representative Nrf1LKO liver at 4–8 weeks of age stained with hematoxylin/eosin showing apoptosis (chevrons), necrosis (black arrow), inflammatory infiltrate (Inset), and vacuolated cells. (d) Immunohistochemical staining for active caspase-3 in Nrf1LKO liver showing multiple apoptotic hepatocytes. (e) Frozen section of Nrf1LKO liver stained with oil red O showing increased number of lipid droplets. (f) Cultured primary Nrf1LKO hepatocytes stained with oil red O showing increased number of lipid droplets. (g) Immunohistochemical detection of proliferating cell nuclear antigen (PCNA) showing increased number of actively dividing hepatocytes in Nrf1LKO liver. (h) Masson's trichrome staining showing fibrosis in a 6-month-old Nrf1LKO animal. (i) Gross appearance of a liver from a 4-month-old Nrf1LKO mouse showing multiple small nodules. (j) Representative liver from a control mouse at 12 months of age. (k) Liver from an Nrf1LKO mouse at 12 months of age showing multiple large, vascularized nodules. (l) Hematoxylin/eosin-stained sections showing hepatocellular adenomas with distinctive borders between tumors and parenchyma. (m) Hematoxylin/eosin stained sections showing HCC with trabecullar features. (n) Section showing HCC-containing clear cells.
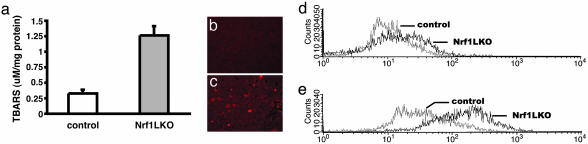
Hepatocytes Lacking Nrf1 Show Elevated ROS and Oxidative Damage. We sought to determine whether oxidative stress was present before development of tumors. Marked elevation in TBARS was detected (Fig. 3_a_). Immunohistochemical staining revealed nuclear accumulation of 8-oxoG in hepatocytes consistent with oxidative DNA damage (compare Fig. 3 b and c). Direct measurement of intracellular ROS indicated that oxidative stress was also intrinsic to hepatocytes. Primary hepatocytes isolated from Nrf1LKO mice showed a 2-fold elevation in fluorescent dichlorofluorescein (DCF) levels compared with controls (Fig. 3_d_). Treatment with _tert_-butylhydroperoxide (tBHP) to induce oxidative stress resulted in marked increase in fluorescence in Nrf1LKO hepatocytes compared with control (Fig. 3_e_). Similar results were obtained by using dihydrorhodamine to detect ROS (data not shown). Thus, loss of Nrf1 results in oxidative stress in hepatocytes, and oxidative injury is present in Nrf1LKO livers before tumor formation.
Fig. 3.
Oxidative stress in Nrf1LKO livers. (a) Liver TBARS levels in control and Nrf1LKO mice. Values are expressed as mean ± SD for six control (open bar) and five Nrf1LKO (filled bar); P < 0.05. (b) Immunohistochemical staining for 8-oxoG showing no reactivity in normal liver. (c) Immunohistochemical detection of 8-oxoG-positive cells in Nrf1LKO liver. Shown are flow cytometric determination of dichlorofluorescein (DCF) fluorescence reporter DCF dye in untreated hepatocytes (d) and in hepatocytes treated with 100 μM _tert_-butylhydroperoxide (tBHP) (e). The x axis shows a 4-decade log scale representing fluorescent emission, and the y axis represents relative number of cells. Genotypes are indicated. Figures are representative overlays.
Expression of ARE-Dependent Genes Is Altered in Nrf1LKO Mice. Nrf1 has been shown to regulate expression of both the catalytic and regulatory subunits of glutamyl-cysteine ligase (Gclc and Gclm). Thus, we wished to determine whether oxidative stress resulted from altered regulation of these genes. However, no difference was detected by RT-PCR and immunoblot analysis (Fig. 4_a_ and data not shown). Similarly, RT-PCR and immunoblot did not detect altered expressions of heme-oxygenase-1 (HO-1) and other major cytoprotective genes including MnSOD and CuZn-SOD (Fig. 4_a_ and data not shown). Thus, oxidative stress does not seem to correlate with decreased expression of these genes. We therefore extended our analysis to determine whether expression of other known ARE-dependent genes was altered in Nrf1LKO livers. RT-PCR analysis showed no significant change in GSTA3, GSTA4, GSTM1, GSTM2, GSTM4, and GSTT1 (Fig. 4_b_). In contrast, transcripts for GSTM3, GSTM6, and GSTP2 were clearly reduced in Nrf1LKO samples. Interestingly, GSTA1 and metallothionein-1 and -2 were elevated in Nrf1LKO livers, indicating that other counteractive mechanisms against oxidative stress were induced (Fig. 4_b_ and data not shown). These results indicate that a subset of ARE-containing genes was affected in Nrf1LKO livers.
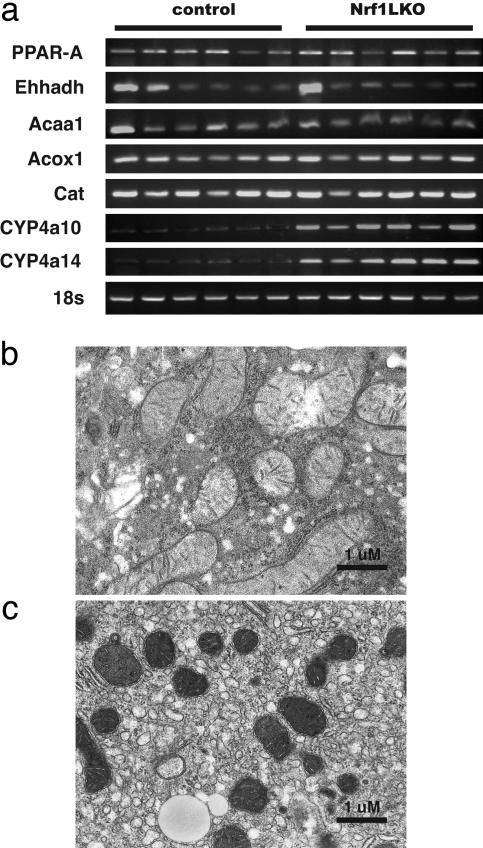
Microsomal Fatty Acid Oxidation Is Induced in Nrf1LKO Livers. Fatty acid metabolism by peroxisomal and microsomal pathways generates a significant source of ROS (17). The fatty liver phenotype in Nrf1LKO mice prompted us to examine whether these pathways were induced. RT-PCR failed to reveal increased peroxisome proliferator-activated receptor α (PPAR-α) expression in Nrf1LKO (Fig. 5_a_). In addition, PPAR-α did not seem to be transcriptionally activated because expression of its target genes, including acyl-CoA oxidase (Acox1), enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase (Ehhadh), and acetyl-CoA acyltransferase 1 (Acaa1) was not altered (Fig. 5_a_). Thus, β-oxidation pathway does not seem to be induced in Nrf1LKO livers. These findings are also consistent with electron microscopic analysis indicating that peroxisomes were not increased in Nrf1LKO livers (Fig. 5_b_). In contrast, Nrf1LKO livers showed proliferation of smooth endoplasmic reticulum (Fig. 5_c_). RT-PCR showed a significant induction of cytochrome P450 4a10 and 4a14 (CYP4a10 and CYP4a14) (Fig. 5_a_). These results suggest that ω-oxidation of fatty acids by means of microsomal CYP4A enzymes participates in the generation of oxidative stress in Nrf1LKO livers.
Fig. 5.
Microsome proliferation and induction of ω-fatty acid oxidation in Nrf1LKO livers. (a) RT-PCR analysis of mRNA encoding various enzymes associated with β- and ω-fatty acid oxidation. 18s levels were used as control. Representative electron micrograph of control (b) and Nrf1LKO (c) livers at 2 months. Note lipid droplets, smaller and darker mitochondria, and proliferation of smooth ER in the Nrf1LKO liver.
Discussion
To determine whether Nrf1 plays an important role in the oxidative stress response in the adult liver, conditional knockout of the nrf1 gene was done. Because Cre-recombinase is not expressed at a substantial level until after birth in Alb-Cre transgenics, the massive cell death and degeneration observed in livers of chimeric embryos derived from _nrf1_–/– ES cells was not seen here. Whether Nrf1's role during fetal liver development is different from its role in the mature hepatocytes cannot be determined here. However, the Alb-Cre transgenic line used in this study allowed us to examine adult mouse that is deficient in Nrf1 function in the liver. Strikingly, hepatic deficiency of Nrf1 in adult animals leads to liver cancer. Before cancer formation, livers showed oxidative stress that was associated with varying degrees of inflammation, hepatic cell death, and fibrosis. We propose a mechanism whereby loss of Nrf1 results in the sensitization of hepatocytes to oxidative stress-induced cellular toxicity and damage due to impaired expression of ARE-regulated genes, and we speculate that several interrelated processes resulting from oxidative stress, including steatosis, inflammation, cell death, and proliferation promote tumorigenesis in Nrf1LKO mice.
An important finding here is that Nrf1-deficient hepatocytes showed increased susceptibility to oxidative stress and damage. Loss of Nrf1 is associated with a significant increase in intracellular ROS levels that was associated with elevated lipid and DNA oxidation. Treatment with _tert_-butylhydroperoxide (tBHP) raised ROS levels further and caused increased cell death in cultures of primary hepatocytes derived from Nrf1LKO livers (Fig. 3_e_ and unpublished data). Thus, mutant hepatocytes also showed a severe impairment in handling additional oxidant stress. Gene expression analysis revealed that expression of several ARE-regulated genes, such as GSTP2, GSTM3, and GSTM6, was lower in Nrf1LKO livers. Although GST enzymes are critical in the detoxification of xenobiotics, they are also regarded as a major cellular defense against oxidative stress through their roles in the inactivation of endogenous electrophiles derived from various metabolic processes including the metabolism of fatty acids, phospholipids, and DNA (18). Thus, impairment in GST gene expression is likely to contribute to oxidative stress in Nrf1LKO livers. Moreover, diminished GSTs may also contribute to apoptosis in Nrf1LKO livers because these enzymes also modulate signaling pathways through interaction with cellular kinases (19, 20). For example, GSTP1 and GSTM1 have been shown to bind Jun N-terminal kinase (JNK) and apoptosis signaling kinase (ASK), respectively, suggesting that GSTs are involved in protecting cells from apoptotic signals.
It is interesting that expression of genes that were previously implicated as Nrf1 targets, such as the catalytic and regulatory subunits of glutamyl-cysteine ligase (Gclc and Gclm) that are involved in glutathione biosynthesis was not affected here. Although the basis of this discrepancy is not known, our analysis suggests that only a subset of ARE-dependent genes depends on Nrf1 in adult hepatocytes. These findings raise the possibility that there may be a differential selectivity between Nrf1 and other “cap n collar” (CNC)-bZIP proteins in driving expression through the ARE. Thus, Nrf1's function in fibroblasts may be different compared with its function in hepatocytes, and this function may also vary during different stages of development. In addition, it is possible that Nrf2 may functionally compensate for the loss of Nrf1, because several of the ARE-regulated genes examined here are also known targets of Nrf2 (21–23). In addition, we cannot exclude the possibility that other ARE-containing target genes involved in the oxidative stress response are also affected by loss of Nrf1 in the liver. It is possible that impaired expression of these other ARE-dependent genes, in combination with the reduction in GSTs, results in oxidative stress in Nrf1LKO livers.
In addition, deficiency of Nrf1 function results in the development of fatty liver. Thus the phenotype of Nrf1LKO mice also incorporates features of nonalcoholic steatohepatitis (NASH) in humans. NASH is a chronic condition that encompasses varying degrees of hepatic injury, necrosis, and inflammation (24, 25). NASH may progress to hepatocellular carcinoma (HCC), and it is recognized as another risk factor for liver cancer (26, 27). However, a precise understanding of the pathogenesis of NASH is still lacking. One prevailing theory suggests a two-hit mechanism (24). The first hit is accumulation of hepatic fat, and the second hit is thought to result from several interrelated processes, including peripheral insulin resistance, oxidative stress, and inflammation (28). The basis for steatosis in Nrf1LKO is not known. Nrf1 may directly regulate genes involved in maintaining lipid homeostasis in the liver. Thus, fat accumulation in Nrf1LKO livers may be caused by increased fatty acid synthesis or uptake from the circulation. Alternatively, steatosis may be due to failure in up-regulation of fatty acid oxidation genes, or it could be secondary to oxidative stress-induced impairment of mitochondrial β-oxidation leading to accumulation of fatty acids (29–31). Regardless of the mechanism, the fatty phenotype is also likely to contribute to oxidative stress. First, steatosis predisposes the liver to oxidative stress as a result of increased ROS production from oxidative metabolism of fatty acid (17, 31). Second, it has been shown that steatosis increases the vulnerability of hepatocytes to injury initiated by oxidative stress (32, 33). Third, accumulated fat can also cause direct toxicity to cells and induce apoptosis in hepatocytes (34). These effects in turn lead to cellular death, inflammation, and induction of cytokines such as TNF-α, thereby perpetuating the pathogenic process by causing mitochondrial damage and ROS production (24).
Our data also demonstrate that microsomal pathway for fatty acid oxidation is induced in Nrf1LKO livers, and excess production of ROS from this pathway is likely to synergize with decreased GST enzymes (and other ARE-genes) in promoting oxidative stress. Ultrastructural analysis showed proliferation of smooth endoplasmic reticulum associated with up-regulation of CYP4a10 and 4A14 genes. CYP4A enzymes have been shown to play a role in diet-induced models of steatohepatitis (35). CYP4A enzymes are also up-regulated in obesity, and they generate ROS, and cause lipid peroxidation (36). Lipid peroxides undergo autocatalytic chain reaction and can cause membrane damage (37). Lipid peroxides also generate stable aldehydes that can recruit inflammatory cells, and stimulate fibrosis, and these end products have potent alkylating activity that can cause DNA damage (38, 39). Combined with the vigorous cell proliferation in Nrf1LKO livers, the DNA damage is likely to be fixed, causing rapid acquisition of cancer-causing mutations.
Inflammation is likely to be elicited by hepatic damage and necrosis in Nrf1LKO livers. However, it is possible that Nrf1 may repress inflammation through regulating genes involved in the metabolism of proinflammatory molecules. Inflammation is an important source of ROS, and it has been implicated as a causative factor in a variety of cancers, including HCC (40, 41). Inflammatory cells secrete various mediators and ROS that can induce cell damage, apoptosis and proliferation. Thus, inflammation is an important cause of oxidative stress, and it is likely to contribute to the pathology in Nrf1LKO livers. Finally, we cannot rule out that Nrf1 may function as a tumor suppressor in hepatocytes, and it is also possible that Nrf1 may accelerate disease progression by regulating yet to be identified pathways. Further experiments are required to sort out these possibilities.
In summary, here we provide a link between Nrf1 and cancer. The changes observed in Nrf1LKO livers characterized by the initial presence of cell death, proliferation, and inflammation, followed by the appearance of dysplastic cells and ultimately cancer, closely mimic the salient features of NASH. Thus, these animals may prove useful as a model to study the mechanisms in the development and progression of NASH and liver cancer.
Acknowledgments
We thank Sandra Huling and the San Francisco Veterans Affairs Medical Center Microscopy and Advanced Imaging Core Facility for the electron microscopy work. This work was supported by National Institutes of Health Grants CA91907, DK60465, and 5PO DK26743.
Author contributions: Z.X., L.C., L.L., T.S.B.Y., C.L., and J.Y.C. performed research; Z.X., T.S.B.Y., and J.Y.C. analyzed data; J.Y.C. designed research; and J.Y.C. wrote the paper.
Abbreviations: ARE, antioxidant response element; bZIP, basic leucine zipper; TBARS, thiobarbituric acid-reactive substance; ROS, reactive oxygen species; PPAR-α, peroxisome proliferator-activated receptor α; NASH, nonalcoholic steatohepatitis; HCC, hepatocellular carcinoma.
References
- 1.Hayes, J. D., Ellis, E. M., Neal, G. E., Harrison, D. J. & Manson, M. M. (1999) Biochem. Soc. Symp. 64**,** 141–168. [PubMed] [Google Scholar]
- 2.Nguyen, T., Sherratt, P. J. & Pickett, C. B. (2003) Annu. Rev. Pharmacol. Toxicol. 43**,** 233–260. [DOI] [PubMed] [Google Scholar]
- 3.Motohashi, H., O'Connor, T., Katsuoka, F., Engel, J. D. & Yamamoto, M. (2002) Gene 294**,** 1–12. [DOI] [PubMed] [Google Scholar]
- 4.Chan, K., Lu, R., Chang, J. C. & Kan, Y. W. (1996) Proc. Natl. Acad. Sci. USA 93**,** 13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, K. & Kan, Y. W. (1999) Proc. Natl. Acad. Sci. USA 96**,** 12731–12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, J. Y. & Kwong, M. (2000) Biochim. Biophys. Acta 1517**,** 19–26. [DOI] [PubMed] [Google Scholar]
- 7.Enomoto, A., Itoh, K., Nagayoshi, E., Haruta, J., Kimura, T., O'Connor, T., Harada, T. & Yamamoto, M. (2001) Toxicol. Sci. 59**,** 169–177. [DOI] [PubMed] [Google Scholar]
- 8.Itoh, K., Chiba, T., Takahashi, S., Ishii, T., Igarashi, K., Katoh, Y., Oyake, T., Hayashi, N., Satoh, K., Hatayama, I., et al. (1997) Biochem. Biophys. Res. Commun. 236**,** 313–322. [DOI] [PubMed] [Google Scholar]
- 9.Ramos-Gomez, M., Kwak, M. K., Dolan, P. M., Itoh, K., Yamamoto, M., Talalay, P. & Kensler, T. W. (2001) Proc. Natl. Acad. Sci. USA 98**,** 3410–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, H. Y., Jedlicka, A. E., Reddy, S. P., Kensler, T. W., Yamamoto, M., Zhang, L. Y. & Kleeberger, S. R. (2002) Am. J. Respir. Cell Mol. Biol. 26**,** 175–182. [DOI] [PubMed] [Google Scholar]
- 11.Chan, J. Y., Kwong, M., Lu, R., Chang, J., Wang, B., Yen, T. S. & Kan, Y. W. (1998) EMBO J. 17**,** 1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwong, M., Kan, Y. W. & Chan, J. Y. (1999) J. Biol. Chem. 274**,** 37491–37498. [DOI] [PubMed] [Google Scholar]
- 13.Leung, L., Kwong, M., Hou, S., Lee, C. & Chan, J. Y. (2003) J. Biol. Chem. 278**,** 48021–48029. [DOI] [PubMed] [Google Scholar]
- 14.Chen, L., Kwong, M., Lu, R., Ginzinger, D., Lee, C., Leung, L. & Chan, J. Y. (2003) Mol. Cell. Biol. 23**,** 4673–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leffert, H. L., Koch, K. S., Moran, T. & Williams, M. (1979) Methods Enzymol. 58**,** 536–544. [DOI] [PubMed] [Google Scholar]
- 16.Gannon, M., Shiota, C., Postic, C., Wright, C. V. & Magnuson, M. (2000) Genesis 26**,** 139–142. [DOI] [PubMed] [Google Scholar]
- 17.Reddy, J. K. & Hashimoto, T. (2001) Annu. Rev. Nutr. 21**,** 193–230. [DOI] [PubMed] [Google Scholar]
- 18.Hayes, J. D. & Strange, R. C. (2000) Pharmacology 61**,** 154–166. [DOI] [PubMed] [Google Scholar]
- 19.Adler, V., Yin, Z., Fuchs, S. Y., Benezra, M., Rosario, L., Tew, K. D., Pincus, M. R., Sardana, M., Henderson, C. J., Wolf, C. R., Davis, R. J. & Ronai, Z. (1999) EMBO J. 18**,** 1321–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho, S. G., Lee, Y. H., Park, H. S., Ryoo, K., Kang, K. W., Park, J., Eom, S. J., Kim, M. J., Chang, T. S., Choi, S. Y., et al. (2001) J. Biol. Chem. 276**,** 12749–12755. [DOI] [PubMed] [Google Scholar]
- 21.Kwak, M. K., Itoh, K., Yamamoto, M., Sutter, T. R. & Kensler, T. W. (2001) Mol. Med. 7**,** 135–145. [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii, T., Itoh, K., Takahashi, S., Sato, H., Yanagawa, T., Katoh, Y., Bannai, S. & Yamamoto, M. (2000) J. Biol. Chem. 275**,** 16023–16029. [DOI] [PubMed] [Google Scholar]
- 23.Chanas, S. A., Jiang, Q., McMahon, M., McWalter, G. K., McLellan, L. I., Elcombe, C. R., Henderson, C. J., Wolf, C. R., Moffat, G. J., Itoh, K., et al. (2002) Biochem. J. 365**,** 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day, C. & Saksena, S. (2002) J. Gastroenterol. Hepatol. 17**,** Suppl. 3, S377–S384. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, J., Viggiano, T. R., McGill, D. B. & Oh, B. J. (1980) Mayo Clin. Proc. 55**,** 434–438. [PubMed] [Google Scholar]
- 26.Brunt, E. M. (2004) Semin. Liver Dis. 24**,** 3–20. [DOI] [PubMed] [Google Scholar]
- 27.Powell, E. E., Cooksley, W. G., Hanson, R., Searle, J., Halliday, J. W. & Powell, L. W. (1990) Hepatology 11**,** 74–80. [DOI] [PubMed] [Google Scholar]
- 28.Browning, J. D. & Horton, J. D. (2004) J. Clin. Invest. 114**,** 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koteish, A. & Diehl, A. M. (2001) Semin. Liver Dis. 21**,** 89–104. [DOI] [PubMed] [Google Scholar]
- 30.Jaeschke, H., Gores, G. J., Cederbaum, A. I., Hinson, J. A., Pessayre, D. & Lemasters, J. J. (2002) Toxicol. Sci. 65**,** 166–176. [DOI] [PubMed] [Google Scholar]
- 31.Pessayre, D., Mansouri, A. & Fromenty, B. (2002) Am. J. Physiol. 282**,** G193–G199. [DOI] [PubMed] [Google Scholar]
- 32.Letteron, P., Fromenty, B., Terris, B., Degott, C. & Pessayre, D. (1996) J. Hepatol. 24**,** 200–208. [DOI] [PubMed] [Google Scholar]
- 33.Yang, S. Q., Lin, H. Z., Lane, M. D., Clemens, M. & Diehl, A. M. (1997) Proc. Natl. Acad. Sci. USA 94**,** 2557–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaffer, J. E. (2003) Curr. Opin. Lipidol. 14**,** 281–287. [DOI] [PubMed] [Google Scholar]
- 35.Leclercq, I. A., Farrell, G. C., Field, J., Bell, D. R., Gonzalez, F. J. & Robertson, G. R. (2000) J. Clin. Invest. 105**,** 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, G., Leclercq, I. & Farrell, G. C. (2001) Am. J. Physiol. 281**,** G1135–G1139. [DOI] [PubMed] [Google Scholar]
- 37.Girotti, A. W. (1998) J. Lipid Res. 39**,** 1529–1542. [PubMed] [Google Scholar]
- 38.Hussain, S. P., Hofseth, L. J. & Harris, C. C. (2003) Nat. Rev. Cancer 3**,** 276–285. [DOI] [PubMed] [Google Scholar]
- 39.Marnett, L. J. (2000) Carcinogenesis 21**,** 361–370. [DOI] [PubMed] [Google Scholar]
- 40.Pikarsky, E., Porat, R. M., Stein, I., Abramovitch, R., Amit, S., Kasem, S., Gutkovich-Pyest, E., Urieli-Shoval, S., Galun, E. & Ben-Neriah, Y. (2004) Nature 431**,** 461–466. [DOI] [PubMed] [Google Scholar]
- 41.Coussens, L. M. & Werb, Z. (2002) Nature 420**,** 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]