A target selection of somatic hypermutations is regulated similarly between T and B cells upon activation-induced cytidine deaminase expression (original) (raw)
Abstract
Activation-induced cytidine deaminase (AID) is essential for somatic hypermutations (SHM) and class switch recombination. Overexpression of AID in non-B cells can induce SHM in artificial constructs inserted in various loci in the genome. AID overexpression was thus proposed to introduce mutations in a wide variety of genes with little specificity. We previously showed that AID transgenic mice developed T cell lymphomas in which the variable region β genes of the T cell receptor and c-myc were mutated as frequently as SHM in activated B cells. To understand the target specificity of SHM in AID-expressing T lymphomas, we sequenced six oncogenes (c-myc, pim1, p53, atm, _tgfbr_-2, and _k_-ras) and two genes (cd4 and cd5) that are actively transcribed in T lymphomas. SHM was found only in c-myc, pim1, cd4, and cd5, which share the E47 binding motif in the enhancer/promoter. The rest that are not mutated in B cells were not mutated in AID-induced T lymphomas either, although they are transcribed in T and B cells. Comparison of several features of SHM, including selection of targets and mutation distribution, suggests that the regulatory mechanism of SHM is similar between T and B cells. SHM base specificities in the CD4 and CD5 genes were biased to AT, indicating that the preference of target bases of the mutations generated by overexpression of AID is not always GC bases but variable between target genes.
Keywords: CD4 and CD5 genes, E47 binding motif, T lymphoma
Generation of high-affinity antibodies is critical to effective immune responses against antigen. Germinal center B cells that proliferate extensively after antigen stimulation accumulate point mutations by the mechanism called somatic hypermutation (SHM) in the variable-region gene of Ig. B cells expressing high-affinity antibodies are selected among mutated B cells by limited amounts of antigen. SHM introduces point mutations at an extraordinarily high frequency of 10–3 to 10–4 per base pair per generation. This rate of mutation is ≈106-fold higher than the usual background rate of general mutations (1, 2).
SHM was originally considered to take place specifically in the Ig variable-region genes but not in the Ig constant-region genes. Subsequently, several genes other than Ig genes have been shown to be targets of SHM in activated B cells and human B cell lymphomas. Such target genes include _MYC, Ig_α, _PAX5, BCL_-6, and PIM1 (3–5). Extensive transgenic (Tg) experiments revealed that transcription of target loci is an absolute requirement for SHM (6, 7), and the region that accumulates mutation is limited to the 3′ proximity (≈2 kb) of the promoter (8, 9). Furthermore, Tg studies have also shown that there are no clear primary sequence requirements for the target of SHM (10, 11). Mutated bases are biased to a small motif called RGYW (12, 13). However, such preference is rather weak, and it is difficult to explain all mutations introduced. Thus, there are three major questions about the selection of mutation targets in SHM: (i) how specific target genes are selected, (ii) why transcription is required, and (iii) how regions 3′ proximal to the promoter are specifically mutated.
Activation-induced cytidine deaminase (AID) has been shown to be essential for SHM, gene conversion, and class switch recombination, which are the three genetic alterations induced by antigen stimulation of B lymphocytes (14–18). The mechanism to introduce SHM by AID has been extensively debated. One hypothesis explains that AID deaminates cytidine in an unknown precursor mRNA and converts it to a novel mRNA encoding an endonuclease (RNA editing hypothesis). AID has two separate domains, each separately required for SHM and class switch recombination (19, 20). AID thus appears to associate with a SHM-specific cofactor that edits an mRNA precursor to generate SHM-specific endonuclease. The alternative model postulates that AID deaminates cytidine in DNA and generates a U/G mismatch (DNA deamination hypothesis). Such mismatches may be recognized by uracil DNA glycosylase or Msh2/6 mismatch repair enzymes. This model assumes that aberrant bases are incorporated by the mismatch repair mechanism to correct U/G mismatches generated by AID. Mutations can be also introduced by coupling with replication of U/G mismatches. In both models, the critical question that has to be explained is how AID can distinguish specific targets among thousands of genes that are actively transcribed in activated B cells.
Ectopic expression of AID can induce SHM on Ig genes and artificial constructs of GFP in hybridoma and non-B cells, respectively (21–23). Studies on AID Tg animals revealed that T cell lymphoma is induced in all individual mice and that T cell receptor (TCR) genes in T lymphoma cells accumulate massive mutations in the region 3′ proximal to the promoter (24). This observation raises interesting questions. (i) How can AID cause T cell tumors with such a high frequency? (ii) How can the TCR gene be selected as a target of SHM? Because AID is involved in DNA cleavage (25–27), tumorigenesis induced by AID may be explained by accumulation of mutations on oncogenes or by chromosome translocation to activate oncogenes. In these T cell lymphomas, extensive mutations accumulated in _c_-myc, but no gross chromosomal translocations were observed (24).
To study the regulatory mechanism of SHM in T lymphoma cells induced in AID Tg animals, we compared mutation target specificities in T lymphomas and B cells. Among equally transcribed genes, the same genes accumulated mutations in B cells and T lymphomas. T cell-specific genes cd4 and cd5 were also mutated in T cell lymphomas. All mutated genes share the E47 binding motif somewhere in the genes and their flanks. Analyses of mutations in these genes revealed that the distribution of mutations is similar, but the base specificity of mutations is variable between the target genes in AID-overexpressing cells. From these results, we propose that the target selection of SHM is regulated similarly between T and B cells when AID is expressed.
Materials and Methods
T Cell Lymphomas in AID Tg Mice. As previously shown, all individuals of AID Tg mice died from T cell lymphomas by 90 weeks (24). Lymphomas 1, 2, 3, and 4 were reported in ref. 24. Lymphomas 1, 2, 3, 4, and 5 were developed in the B1 Tg line, and lymphoma 6 was developed in the B2 Tg line. Flow cytometric analysis classified lymphomas 1, 2, and 3 and lymphomas 4 and 5 into peripheral T cell lymphoma and thymoma, respectively (24). Lymphoma 6 was not available for immunophenotyping.
Analyzed Sequence. The regions of gene sequences analyzed are listed in Table 1. The genomic sequences the regions were aligned to are as follows, with ensembl gene codes in parentheses: c-myc (ENSMUSG00000022346), pim1 (ENSMUSG00000024014), p53 (ENSMUSG00000059552), atm (ENSMUSG00000034218), tgfbr-2 (ENSMUSG00000032440), k-ras (ENSMUSG00000030265), cd4 (ENSMUSG00000023274), and cd5 (ENSMUSG00000024669). The analyzed regions of those genes encompassed the transcriptional starting points and ≈1 kb from them.
Table 1. Mutations in the analyzed genes in lymphoma of AID Tg mice.
| Gene (region, GC composition) | Cell type | Mutations, n | Total bases, n | Mutated clones, n (total) | Mutation frequency | RGYW mutation, n (total) | GC mutations, % |
|---|---|---|---|---|---|---|---|
| c-myc (exon 1 and intron 1, 54.6% GC) | Lymphoma 1* | 24 | 18,183 | 19 (19) | 13.2 | 19 (24)† | 95.6 |
| Lymphoma 2 | 68 | 22,968 | 24 (24) | 29.6 | 24 (68)† | 83.5 | |
| Lymphoma 3 | 19 | 18,183 | 14 (19) | 10.5 | 9 (19)† | 90.1 | |
| Lymphoma 4 | 9 | 18,183 | 11 (19) | 4.95 | 6 (9)† | 88.7 | |
| Lymphoma 5 | 16 | 16,269 | 13 (17) | 9.83 | 9 (16)† | 82.1 | |
| WT | 0 | 20,097 | 0 (21) | <0.50 | — | — | |
| c-myc (exon 2, 61.1% GC) | Lymphoma 1 | 4 | 13,752 | 11 (18) | 2.91 | 4 (4)† | 100 |
| Lymphoma 2 | 5 | 15,280 | 16 (20) | 3.27 | 1 (5) | 50.8 | |
| Lymphoma 3 | 11 | 15,280 | 13 (20) | 7.20 | 2 (11) | 68.6 | |
| Lymphoma 4 | 9 | 11,460 | 14 (15) | 7.85 | 3 (9) | 68.6 | |
| Lymphoma 5 | 8 | 14,516 | 13 (19) | 5.51 | 3 (8) | 60.2 | |
| WT | 1 | 15,280 | 1 (20) | 0.65 | 0 (1) | 0 | |
| pim1 (exons 1-4, 66.6% GC) | Lymphoma 1 | 2 | 10,800 | 12 (14) | 1.85 | 1 (2) | 100 |
| Lymphoma 2 | 3 | 13,680 | 3 (19) | 2.19 | 1 (3) | 33 | |
| Lymphoma 4 | 3 | 15,120 | 12 (21) | 1.98 | 2 (3) | 25 | |
| WT | 0 | 14,400 | 0 (20) | <0.70 | — | — | |
| atm (exon 1 and intron 1) | Lymphomas 1, 2, and 4 | 0 | 5,330 | 0 (10) | <1.88 | — | — |
| p53 (exon 1 and intron 1) | Lymphomas 1, 2, and 4 | 0 | 10,920 | 0 (10) | <0.92 | — | — |
| tgfbr2 (exon 1 and intron 1) | Lymphomas 1, 2, and 4 | 0 | 6,660 | 0 (10) | <1.50 | — | — |
| k-ras (exon 1 and intron 1) | Lymphomas 1, 2, and 4 | 0 | 4,620 | 0 (20) | <2.16 | — | — |
Transcriptional Levels of Oncogenes. Transcriptional levels of those genes are according to the DNA chip data. RNA was extracted from the thymus of AID Tg and WT mice as described in ref. 24. Mouse genome-wide mRNA screening was done by using a Murine Genome 430A probe array (GeneChip, Affymetrix, Santa Clara, CA) containing ≈22,000 probe sets. Target cRNA preparations from total RNA, hybridization to the GeneChip, staining, and scanning were carried out according to the manufacturer's instructions. The level of gene expression was determined as the average difference by using genechip analysis suite, version 4.0 (Affymetrix) as described in ref. 28.
Preparation of Genomic DNA. Genomic DNA from T cell lymphomas in AID Tg mice and lymph nodes in WT mice (C57BL/6J, Shimizu Experimental Material, Kyoto) was extracted according to standard methods. The c-myc, Pim1, p53, ATM, TGFβR-2, K-ras, CD4, and CD5 genes were amplified by genomic PCR with Pyrobest polymerase (Takara Shuzo, Kyoto). The oligonucleotide sequences used for genomic PCR were as follows: c-_myc_-1 (for the c-myc exon 1 fragment), 5′-GCGTTTTTTTCTGACTCGCTGTAG-3′ (forward) and 5′-GCGGGGGTCAGGCTAAATTTTACT-3′ (reverse); _c-myc_-2 (for the c-myc exon 2 fragment), 5′-CAGACAGCCACGACATGCCC-3′ (forward) and 5′-GGTGGTGGGCGGTGTCTCCTCAT-3′ (reverse); _c-myc_-3 (for the c-myc exon 3 fragment), 5′-TAGGAGGTGCTTGGGAATG-3′ (forward) and 5′-TCCAGCTCCTCCTCGAGTTA-3′ (reverse); _pim1_-1 (for the pim1 exons 1–4 fragment), 5′-GCAACGCCACCCGCAGCTCGAG-3′ (forward) and 5′-CCAGCACCTGCCAGAAGAAT-3′ (reverse); _pim1_-2 (for the pim1 exon 6 fragment), 5′-GCCCGAGCTATTGAAGTCTG-3′ (forward) and 5′-CCCAGGCAGAGTTTGAGAAG-3′ (reverse); p53, 5′-GTTCATTGGGACCATCCTGGCTGT-3′ (forward) and 5′-CGGAATGCGTTAAGCAAGGGAAT-3′ (reverse); atm, 5′-GTGGCTATGTTTTGAAGCCTG-3′ (forward) and 5′-GATCTTTGCTCGCTTCAGTCAACC-3′ (reverse); _tgfbr_-2, 5′-CTGCTGCATATCGTCCTGTG-3′ (forward) and 5′-TAGTAGCTGCTGAGATC-3′ (reverse); k-ras, 5′-AAAGTTTTTGATAATCTTGT-3′ (forward) and 5′-CTGCCGTCCTTTACAAGCGC-3′ (reverse); _cd4_-1 (for cd4 exon 1 and the 5′ region of intron 1), 5′-CCCACTGGAATCATGAGCTT-3′ (forward) and 5′-GAAAGGTCTGAGGGTGTGGA-3′ (reverse); _cd4_-2 (for the cd4 3′ region of intron 1), 5′-TTCAGAAAGGGCAGATTGCT-3′ (forward) and 5′-TTTCGTGGAGGTTGGAAGAC-3′ (reverse); _cd4_-3 (for the cd4 3′ region of intron 9 and exon 10), 5′-GGCATTCGACCCTTACAAAA-3′ (forward) and 5′-TTCTGTCAGCTCAGGAAGCA-3′ (reverse); _cd5_-1 (for the cd5 exon 1 and 5′ region of intron 1), 5′-CCCTTTCCACCCCTGTTTAT-3′ (forward) and 5′-GCAGGCATGGTGTACCTCTT-3′ (reverse); _cd5_-2 (for the cd5 3′ region of intron 1), 5′-GAGGAGCTCCGTCAACTGTC-3′ (forward) and 5′-CAGGACTCTGCTTGGAGACC-3′ (reverse); and _cd5_-3 (for the cd5 3′ region of intron 10 and exon 11), 5′-ACACACACACACACCCCATC-3′ (forward) and 5′-GGTTGCTCGTAGGCTCTGTC-3′ (reverse). The PCR conditions for amplification of all of the genes analyzed are available upon request.
Cloning Procedure and Sequencing Analysis. After 35 cycles of PCR reaction, PCR products were gel-purified and cloned into the pCR4-TOPO blunt vector (Invitrogen) as recommended by the manufacturer. The DNA sequence of each clone was determined by using T3 and T7 primers. Nucleotide differences due to the allelic polymorphism were disregarded in the assessment of the mutation frequency. Mutations were counted by the number of events reported by the presumed genealogic trees showing the relationship between the mutations. The presumed genealogic trees were drawn by assuming the order of the mutation development from the number of clones with common mutations (24).
Computer Analysis for Secondary Structures. Folding of the single-stranded DNA was analyzed by using the mfold DNA folding program (29). The temperature for folding conditions was 37°C, and the concentrations of Na+ and Mg2+ were 150 and 0.5 mM, respectively.
Motif Screening in Enhancer/Promoter by Computer. Motif screening was done by using transfac software (30). The group of matrices we used was the vertebrates group, and a cutoff selection for the matrix group was used to minimize the sum of both error rates. The National Center for Biotechnology Information accession numbers of the sequences subjected to enhancer analysis were as follows: tcrb, AE000665; c-myc, M12345; pim1, M13945; p53, AF287146; tgfbr-2, AF118264; and k-ras, MMU49448. The enhancer/promoter region of mouse atm has not yet been reported.
Results and Discussion
Comparison of Regulatory Mechanisms for SHM in B Cells and in the T Lymphoma of AID Tg Mice. Previously, we found that extensive mutations accumulate in the TCR variable region β genes but little, if any, in the TCR constant region β gene of AID-induced T lymphomas. The clustered distribution of mutations within 1–2 kb from the promoter of the TCR gene (24) is similar to that of SHM in the Ig gene in B cells. We also showed that mutations were clustered in exon 1 and intron 1 of _c_-myc as reported in B cells (Fig. 1). We therefore concluded that mutations found in AID-induced T lymphomas are indistinguishable from SHM (24). The target bases in the TCRβ gene and _c_-myc were highly biased to GC bases, as shown in mutations in Ig genes and GFP constructs in hybridomas and fibroblasts, respectively, which overexpress AID (21–24, 31).
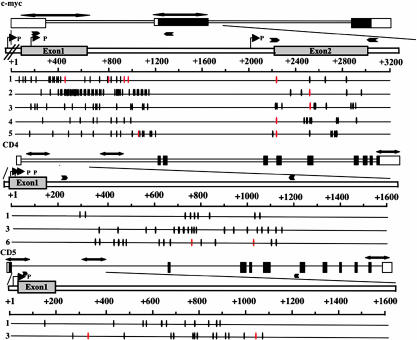
Fig. 1.
Distribution of mutations in c-myc, cd4, and cd5 in lymphoma cells. The mutations described in Tables 1 and 5 are plotted on respective genomic sequences. Genomic loci are shown with untranslated and translated sequences (open and filled boxes, respectively). The bold double-headed arrows above the genes show the regions sequenced. The numbers at left and above the thin horizontal lines indicate the lymphoma numbers and the position from the transcriptional starting sites, respectively. Red vertical lines indicate mutations common to more than half of the total clones analyzed from the same individual mice. Black vertical lines indicate sporadic mutations, and arrowheads labeled with a P indicate transcriptional start sites. Primers used for amplification are shown by arrowheads above genomic loci. Data for c-myc in lymphoma 1 are taken from ref. 24.
To examine the specificity of mutation target genes in T cell lymphomas, we chose c-myc and pim1 among the oncogenes that are mutated frequently in B cells because the c-myc and pim1 are expressed in T cells (24, 32). BCL6, RhoH/_TTF, Ig_α, _Ig_β, and PAX5, which are also known to have SHM in B cells (5, 33), are not expressed at significant levels in T cells. As controls, the p53, ATM, TGFβR-2 and K-ras oncogenes are selected because they are expressed in T cells but not mutated within 1–2 kb from the promoters in B cells (34, 35). In the six oncogenes analyzed, only c-myc and pim1 had SHM in all T cell lymphomas analyzed (Table 1). The other oncogenes, p53, atm, tgfbr-2, and k-ras, which have only sporadic mutations in cancers, showed no mutations among 10 or 20 clones isolated from three independent lymphomas that accumulated mutations in the TCRβ gene. It is clear that SHM target genes are not random but highly selective even in non-B cells with overexpressed AID. It is important to note that none of genes that are not mutated in B cells or B lymphomas did not accumulate SHM in the T lymphomas.
Exon 1 and intron 1 of _c_-myc, which were reported to accumulate SHM (24), showed high mutation frequencies (4.95 × 10–4 to 29.6 × 10–4 mutations per base pair) in all individual lymphomas (Fig. 1 and Table 1). Almost all mutations were different among clones, and their targets were biased to GC bases and the RGYW motif, as reported in ref. 24. Some of mutations were reported to alter _c_-myc transcription by releasing a block on transcriptional elongation (36, 37). The mutation frequencies in exon 2 of c-myc and pim1 were less than those in exon 1 and intron 1 of c-myc. However, these mutations are common to more than half of the clones sequenced, suggesting that these mutations may have selective advantages to generate lymphomas (Fig. 1). The mutation frequency of pim1 appears to be lower than that of the B lymphomas reported in ref. 5 because we considered that mutations were common in clones as a single event, and Pasqualucci et al. determined mutations by sequencing genomic PCR product without cloning.
Some of these mutations resulted in amino acid substitutions with potential functional consequences. In particular, _c_-myc missense mutations located in the transactivation domain (exon 2) coincide with a mutational hot spot in translocated MYC alleles in human Burkitt's lymphoma and mouse plasmacytoma (38, 39). Notably, one of the changes (Leu-56 in lymphomas 3 and 5) was located in proximity to the evolutionally conserved and functionally critical phosphorylation sites Thr-58 and Ser-62 (exon 2). The mutations flanking these residues abolish phosphorylation of the c-myc transactivation domain, resulting in defective regulation of p107 retinoblastoma 1 protein stabilization and increased transforming activity (40). It is likely that some of these _c_-myc mutations are involved in lymphomagenesis of T cell lymphomas in AID Tg mice. The lower mutation frequency might be partially due to the selection for the clones with functional missense mutations for tumor evolution and against the additional mutations.
The above features of SHM in AID-induced T lymphomas, including selection of the target genes, the locations of mutation clusters within the mutated genes, targeted base bias, and possible functional consequences by mutations, are indistinguishable from SHM in human B cell lymphoma (5). The results suggest that the regulatory machinery for SHM may not be B cell-specific but shared by T cells.
Transcriptional Regulation of SHM Target Genes. To investigate the mechanism of target selection for SHM in AID Tg mice, we compared the transcription levels of the six genes tested because expression of AID and active transcription of the target loci were required for SHM (41). The DNA chip data of the thymic mRNA from three AID Tg mice showed that all of the six oncogenes were transcribed at substantial levels (n = 1,171–2,556 transcripts) compared with the transcriptional levels of GAPDH (n = 18,567 transcripts) and the median (n = 1,758 transcripts) of 22,692 transcripts. Each of the six genes was transcribed to a similar extent between AID Tg and WT mice (Table 2).
Table 2. Transcriptional level of the genes analyzed in the thymus of AID Tg and WT mice.
| Transcriptional level | ||
|---|---|---|
| Gene | Tg | WT |
| c-myc | 2,347 | 1,147 |
| pim1 | 1,171 | 1,018 |
| p53 | 2,038 | 1,122 |
| atm | 1,272 | 722 |
| tgfbr-2 | 2,566 | 2,693 |
| k-ras | 2,059 | 2,214 |
| tcrbv8.2 | 4,367 | 5,388 |
| cd4 | 2,918 | 4,094 |
| cd5 | 1,919 | 2,646 |
| gapdh | 18,567 | 14,879 |
| Median of 22,692 transcripts | 1,758 | 1,479 |
The enhancer of the Ig gene is critical for SHM (42, 43), and several transcription regulatory elements have also been proposed to regulate the efficiency of SHM (44, 45). We thus searched common regulatory elements among the genes that accumulated SHM in T cell lymphomas of AID Tg mice and have known enhancer/promoter sequences. The murine enhancer/promoter sequences of the TCRβ, c-myc, Pim1, p53, TGFβR-2 and K-ras genes were subjected to a search of known transcription elements by using transfac software (30). Among the identified transcription elements, we picked up those shared by at least two SHM-positive genes but none by SHM-negative genes. We found six transcriptional factors whose binding motifs were found only in the SHM target genes (Table 3). Freac-7, Gfi-1, and HFH3 have not been extensively investigated, and their expression in lymphocyte is not reported, whereas GATA3 and c-rel are expressed ubiquitously. We focused on E47 because E47 is known to have not only a critical role in B cell development (46) but also enhancing activity of SHM in Igκ transgenes (45).
Table 3. Enhancer elements specific for the genes that have mutations.
| Freac-7 | E47 | GATA3 | HFH3 | Gfi-1 | c-rel | |
|---|---|---|---|---|---|---|
| Tcrb | + | + | + | + | - | - |
| c-myc | + | + | + | - | + | + |
| pim1 | + | -† | - | + | + | + |
| p53 | - | - | - | - | - | - |
| tgfbr2 | - | - | - | - | - | - |
| k-ras | - | - | - | - | - | - |
E47 Binding Motif in SHM Target Genes. So far, nine genes (_Ig_κ, IgH, MB-1(_Ig_α), B29(_Ig_β), PAX5, BCL6, c-MYC, PIM1, and RhoH/TTF) are known to have SHM in B cells (3–5, 33). All of these genes have the E47 binding motif in their enhancer/promoter regions except for RhoH/TTF and PIM1. The enhancer/promoter of RhoH/TTF has not been analyzed. Although the known enhancer/promoter sequence of the Pim1 gene does not contain the E47 binding motif, the enhancer/promoter activity for pim1 has not been extensively investigated as compared with c-myc and k-ras (47). The E47 binding motif exists in intron 2 of pim1, like Bcl6, which has the E47 binding motif in the intronic regulatory region (45). Igκ, IgH, MB-1(Igα), B29(Igβ), and PAX5 are shown to be regulated by E47 (48). Moreover, as shown many B lineage-specific genes regulated by E47 are known to be the target of SHM in Table 4 (3–5, 33, 45, 48).
Table 4.
E47 target genes in B lymphocyte and T lymphocyte (24, 45, 48, 59)
| SHM-positive* | Not tested | |
|---|---|---|
| B lymphocyte-specific | Igκ, IgH, Pax5, Bcl6*, mb-1 (Igα), B29(Igβ) | VpreB, EBF, λ5, AID |
| T lymphoyte-specific | TCRβ | TCRα, TCRγ, TCRδ, CD4, CD5, preTα |
Because T lineage-specific genes regulated by E47 include CD4, CD5, pre-Tα, TCRαβ, and TCRγδ (48), we examined SHM in the CD4 and CD5 genes, which are expressed in the T cell lymphoma of AID Tg mice. We found massive mutations in CD4 and CD5 genes in T cell lymphomas (lymphomas 1, 3, and 6 in cd4 and lymphomas 1 and 3 in cd5). Mutations in the CD4 and CD5 genes did not show clear bias to the RGYW motif (Table 5). CD4 and CD5 genes accumulated SHM with frequencies on the order of 10–3 per base pair. Mutations were clustered in the region within ≈1–2 kb from the promoter but little in the region 3 kb away from the promoter (Fig. 1 and Table 6, which is published as supporting information on the PNAS web site). The CD4 and CD5 genes continued to be targets of SHM after tumorigenesis because most of the mutations differed between the DNA clones sequenced. The fact that the CD4 and CD5 genes are the targets of SHM in AID-expressing T cell lymphomas is consistent with the hypothesis that E47 regulates the target specificity of SHM. It is interesting that many genes that accumulate SHM in B cells or AID-expressing non-B cells are regulated by E47 or contain the E47 element in the enhancer/promoter region. In addition, the E47 protein is particularly abundant in the centroblasts of germinal center B cells (49). Moreover, Michael et al. (45) reported that the E47 binding motif enhanced SHM without transcriptional changes in transgenes. Taken together, these data suggest that E47 might have a role in the targeting process of SHM. It is unlikely that E47 directs AID to the target locus because the protein complex associated with the E47 binding motif in B cells does not contain AID (45). The E47 protein may play a role in SHM targeting by chromatin remodeling. E47 associates with coactivators such as histone acetyltransferases and collaboratively regulates B cell development (50). In fact, hyperacetylation of histones in the Ig variable region gene locus increased the frequency of SHM in a B cell line (51).
Table 5. Mutations in cd4 and cd5 in lymphoma of AID Tg mice.
| Gene (region, GC composition) | Cell type | Mutations, n | Total bases, n | Mutated clones, n (total) | Mutation frequency | RGYW mutation, n (total) | GC mutations, % |
|---|---|---|---|---|---|---|---|
| cd4 (5′ side of intron 1, 43.1% GC) | Lymphoma 1 | 10 | 10,920 | 8 (13) | 9.16 | 1 (8) | 38.3 |
| Lymphoma 3 | 20 | 7,560 | 9 (9) | 26.5 | 4 (20) | 31.6 | |
| Lymphoma 6 | 16 | 8,400 | 10 (10) | 19.1 | 2 (16) | 51.1 | |
| cd5 (exon 1 and 5′ side of intron 1, 53.6% GC) | Lymphoma 1 | 11 | 8,400 | 6 (10) | 13.1 | 3 (11) | 39.3 |
| Lymphoma 3 | 14 | 8,400 | 9 (9) | 16.6 | 8 (14)* | 100 |
The cd4 and cd5 mutations are probably not due to genomic instability caused by cancer because (i) the localization of mutations is similar to AID-dependent SHM in the Ig gene; (ii) the mutations of the CD4 and CD5 genes have not been found in T cell lymphoma, such as EL4 (data not shown); (iii) the mutation rates in the CD4 and CD5 genes were much higher than the mutations associated with cancers that carry usually only one or two mutations in the total coding regions (52–54); and (iv) the mutations located in 767C and 1018T of CD4 in lymphoma 6 and 382C and 1028C of CD5 in lymphoma 3 were found in more than half of the clones analyzed (Fig. 1), indicating that these mutations were introduced before lymphomagenesis, because CD4 and CD5 were neither oncogenic nor subjected to selection pressure of tumors.
Base Preference of SHM Is Variable Among Target Genes. The strong GC base bias has been reported to be one of the features of SHM introduced by overexpression of AID (21–23, 31). All individual T lymphomas analyzed showed strong GC base bias mutations in the TCRβ gene and c-myc exon 1 and intron 1. By contrast, the CD4 and CD5 genes showed AT base bias except for the CD5 gene in lymphoma 3. SHM in non-Ig genes of human B cell lymphomas has been postulated to result from deregulation of AID and has shown GC base bias mutations and association with the RGYW/WRCY motif (55). However, primary mediastinal B cell lymphoma, a subtype of human non-Hodgkin's lymphoma, which is believed to arise from thymic medullary B cells, had SHM in the Bcl6 gene at AT bases but not GC bases. Their mutations are not associated with the RGYW/WRCY motif (56).
It is not clear why the CD5 gene had more SHM at AT bases in lymphoma 1 and only GC bases in lymphoma 3. Lymphomas 1 and 3 were categorized in peripheral CD4+CD8– T cell lymphoma; however, peripheral T cell lymphoma is a diverse group of diseases with various clinical manifestations arising from various stages of differentiation (57). Depending on the subtype of T cells from which the tumor derives, different combinations of transcription factors and error-prone polymerases might have affected the target bases of mutations in the CD5 gene. The other enhancer elements, PU.1 and NF-TM3, have been reported to regulate not only the mutation rate but also base specificity, such as biases to RGYW and GC base in the Igκ transgene (44). Disruption of the PU.1 and NF-TM3 binding motifs in the Igκ gene showed SHM with marked reduction of mutations in G, C, and A bases, indicating that the transcriptional factors binding to the 3′ enhancer have effects on base specificity. These studies, together with AT base bias mutations in the CD4 and CD5 genes in T lymphomas generated by overexpression of AID, do not agree with the prediction of the DNA deamination model that the GC base bias mutations are generated by overexpression of AID (58).
Supplementary Material
Supporting Table
Acknowledgments
We thank Drs. R. Shinkura and H. Nagaoka for critically reading the manuscript and T. Nishikawa and Y. Shiraki for preparation of the manuscript. This work was supported by Center of Excellence Grant 12CE2006 from the Ministry of Education, Science, Sports, and Culture of Japan.
Author contributions: A.K., I.-m.O., and T.H. designed research; A.K., I.-m.O., M.M., K.K., N.A.B., T.N., and H.S. performed research; A.K., I.-m.O., T.N., H.S., and T.H. analyzed data; and A.K. and T.H. wrote the paper.
Abbreviations: AID, activation-induced cytidine deaminase; Tg, transgenic; SHM, somatic hypermutation; TCR, T cell receptor.
References
- 1.Diaz, M. & Storb, U. (2003) DNA Repair 2**,** 623–627. [DOI] [PubMed] [Google Scholar]
- 2.Wagner, S. D. & Neuberger, M. S. (1996) Annu. Rev. Immunol. 14**,** 441–457. [DOI] [PubMed] [Google Scholar]
- 3.Migliazza, A., Martinotti, S., Chen, W., Fusco, C., Ye, B. H., Knowles, D. M., Offit, K., Chaganti, R. S. & Dalla-Favera, R. (1995) Proc. Natl. Acad. Sci. USA 92**,** 12520–12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen, H. M., Peters, A., Baron, B., Zhu, X. & Storb, U. (1998) Science 280**,** 1750–1752. [DOI] [PubMed] [Google Scholar]
- 5.Pasqualucci, L., Neumeister, P., Goossens, T., Nanjangud, G., Chaganti, R. S., Kuppers, R. & Dalla-Favera, R. (2001) Nature 412**,** 341–346. [DOI] [PubMed] [Google Scholar]
- 6.Betz, A. G., Milstein, C., Gonzalez-Fernandez, A., Pannell, R., Larson, T. & Neuberger, M. S. (1994) Cell 77**,** 239–248. [DOI] [PubMed] [Google Scholar]
- 7.Peters, A. & Storb, U. (1996) Immunity 4**,** 57–65. [DOI] [PubMed] [Google Scholar]
- 8.O`Brien, R. L., Brinster, R. L. & Storb, U. (1987) Nature 326**,** 405–409. [DOI] [PubMed] [Google Scholar]
- 9.Hackett, J., Jr., Rogerson, B. J., O'Brien, R. L. & Storb, U. (1990) J. Exp. Med. 172**,** 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azuma, T., Motoyama, N., Fields, L. E. & Loh, D. Y. (1993) Int. Immunol. 5**,** 121–130. [DOI] [PubMed] [Google Scholar]
- 11.Yelamos, J., Klix, N., Goyenechea, B., Lozano, F., Chui, Y. L., Gonzalez Fernandez, A., Pannell, R., Neuberger, M. S. & Milstein, C. (1995) Nature 376**,** 225–229. [DOI] [PubMed] [Google Scholar]
- 12.Golding, G. B., Gearhart, P. J. & Glickman, B. W. (1987) Genetics 115**,** 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogozin, I. B. & Kolchanov, N. A. (1992) Biochim. Biophys. Acta 1171**,** 11–18. [DOI] [PubMed] [Google Scholar]
- 14.Muramatsu, M., Sankaranand, V. S., Anant, S., Sugai, M., Kinoshita, K., Davidson, N. O. & Honjo, T. (1999) J. Biol. Chem. 274**,** 18470–18476. [DOI] [PubMed] [Google Scholar]
- 15.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102**,** 553–563. [DOI] [PubMed] [Google Scholar]
- 16.Revy, P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., et al. (2000) Cell 102**,** 565–575. [DOI] [PubMed] [Google Scholar]
- 17.Harris, R. S., Sale, J. E., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Curr. Biol. 12**,** 435–438. [DOI] [PubMed] [Google Scholar]
- 18.Arakawa, H., Hauschild, J. & Buerstedde, J. M. (2002) Science 295**,** 1301–1306. [DOI] [PubMed] [Google Scholar]
- 19.Ta, V. T., Nagaoka, H., Catalan, N., Durandy, A., Fischer, A., Imai, K., Nonoyama, S., Tashiro, J., Ikegawa, M., Ito, S., et al. (2003) Nat. Immunol. 4**,** 843–848. [DOI] [PubMed] [Google Scholar]
- 20.Shinkura, R., Ito, S., Begum, N. A., Nagaoka, H., Muramatsu, M., Kinoshita, K., Sakakibara, Y., Hijikata, H. & Honjo, T. (2004) Nat. Immunol. 5**,** 707–712. [DOI] [PubMed] [Google Scholar]
- 21.Martin, A., Bardwell, P. D., Woo, C. J., Fan, M., Shulman, M. J. & Scharff, M. D. (2002) Nature 415**,** 802–806. [DOI] [PubMed] [Google Scholar]
- 22.Yoshikawa, K., Okazaki, I. M., Eto, T., Kinoshita, K., Muramatsu, M., Nagaoka, H. & Honjo, T. (2002) Science 296**,** 2033–2036. [DOI] [PubMed] [Google Scholar]
- 23.Wang, C. L., Harper, R. A. & Wabl, M. (2004) Proc. Natl. Acad. Sci. USA 101**,** 7352–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okazaki, I. M., Hiai, H., Kakazu, N., Yamada, S., Muramatsu, M., Kinoshita, K. & Honjo, T. (2003) J. Exp. Med. 197**,** 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaoka, H., Muramatsu, M., Yamamura, N., Kinoshita, K. & Honjo, T. (2002) J. Exp. Med. 195**,** 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Begum, N. A., Kinoshita, K., Muramatsu, M., Nagaoka, H., Shinkura, R. & Honjo, T. (2004) Proc. Natl. Acad. Sci. USA 101**,** 13003–13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen, S., Casellas, R., Reina-San-Martin, B., Chen, H. T., Difilippantonio, M. J., Wilson, P. C., Hanitsch, L., Celeste, A., Muramatsu, M., Pilch, D. R., et al. (2001) Nature 414**,** 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakajima, T., Iikura, M., Okayama, Y., Matsumoto, K., Uchiyama, C., Shirakawa, T., Yang, X., Adra, C. N., Hirai, K. & Saito, H. (2004) J. Allergy Clin. Immunol. 113**,** 528–535. [DOI] [PubMed] [Google Scholar]
- 29.Zuker, M. (2003) Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingender, E., Chen, X., Fricke, E., Geffers, R., Hehl, R., Liebich, I., Krull, M., Matys, V., Michael, H., Ohnhäuser, R., et al. (2001) Nucleic Acids Res. 29**,** 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, A. & Scharff, M. D. (2002) Proc. Natl. Acad. Sci. USA 99**,** 12304–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selten, G., Cuypers, H. T., Boelens, W., Robanus-Maandag, E., Verbeek, J., Domen, J., van Beveren, C. & Berns, A. (1986) Cell 46**,** 603–611. [DOI] [PubMed] [Google Scholar]
- 33.Gordon, M. S., Kanegai, C. M., Doerr, J. R. & Wall, R. (2003) Proc. Natl. Acad. Sci. USA 100**,** 4126–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerbauy, F. R., Colleoni, G. W., Saad, S. T., Regis Silva, M. R., Correa Alves, A., Aguiar, K. C., Albuquerque, D. M., Kobarg, J., Seixas, M. T. & Kerbauy, J. (2004) Leuk. Lymphoma 45**,** 2071–2078. [DOI] [PubMed] [Google Scholar]
- 35.Gronbaek, K., Worm, J., Ralfkiaer, E., Ahrenkiel, V., Hokland, P. & Guldberg, P. (2002) Blood 100**,** 1430–1437. [DOI] [PubMed] [Google Scholar]
- 36.Raffeld, M., Yano, T., Hoang, A. T., Lewis, B., Clark, H. M., Otsuki, T. & Dang, C. V. (1995) Curr. Top. Microbiol. Immunol. 194**,** 265–272. [DOI] [PubMed] [Google Scholar]
- 37.Cesarman, E., Dalla-Favera, R., Bentley, D. & Groudine, M. (1987) Science 238**,** 1272–1275. [DOI] [PubMed] [Google Scholar]
- 38.Gregory, M. A. & Hann, S. R. (2000) Mol. Cell. Biol. 20**,** 2423–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoang, A. T., Lutterbach, B., Lewis, B. C., Yano, T., Chou, T. Y., Barrett, J. F., Raffeld, M., Hann, S. R. & Dang, C. V. (1995) Mol. Cell. Biol. 15**,** 4031–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niklinski, J., Claassen, G., Meyers, C., Gregory, M. A., Allegra, C. J., Kaye, F. J., Hann, S. R. & Zajac-Kaye, M. (2000) Mol. Cell. Biol. 20**,** 5276–5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Besmer, E., Gourzi, P. & Papavasiliou, F. N. (2004) Curr. Opin. Immunol. 16**,** 241–245. [DOI] [PubMed] [Google Scholar]
- 42.Giusti, A. M. & Manser, T. (1993) J. Exp. Med. 177**,** 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klix, N., Jolly, C. J., Davies, S. L., Bruggemann, M., Williams, G. T. & Neuberger, M. S. (1998) Eur. J. Immunol. 28**,** 317–326. [DOI] [PubMed] [Google Scholar]
- 44.Kodama, M., Hayashi, R., Nishizumi, H., Nagawa, F., Takemori, T. & Sakano, H. (2001) Int. Immunol. 13**,** 1415–1422. [DOI] [PubMed] [Google Scholar]
- 45.Michael, N., Shen, H. M., Longerich, S., Kim, N., Longacre, A. & Storb, U. (2003) Immunity 19**,** 235–242. [DOI] [PubMed] [Google Scholar]
- 46.Kee, B. L., Quong, M. W. & Murre, C. (2000) Immunol. Rev. 175**,** 138–149. [PubMed] [Google Scholar]
- 47.Davoodi-Semiromi, A., Laloraya, M., Kumar, G. P., Purohit, S., Jha, R. K. & She, J. X. (2004) J. Biol. Chem. 279**,** 11553–11561. [DOI] [PubMed] [Google Scholar]
- 48.Greenbaum, S. & Zhuang, Y. (2002) Semin. Immunol. 14**,** 405–414. [DOI] [PubMed] [Google Scholar]
- 49.Roberts, V. J., Steenbergen, R. & Murre, C. (1993) Proc. Natl. Acad. Sci. USA 90**,** 7583–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradney, C., Hjelmeland, M., Komatsu, Y., Yoshida, M., Yao, T. P. & Zhuang, Y. (2003) J. Biol. Chem. 278**,** 2370–2376. [DOI] [PubMed] [Google Scholar]
- 51.Woo, C. J., Martin, A. & Scharff, M. D. (2003) Immunity 19**,** 479–489. [DOI] [PubMed] [Google Scholar]
- 52.Albanese, I., Scibetta, A. G., Migliavacca, M., Russo, A., Bazan, V., Tomasino, R. M., Colomba, P., Tagliavia, M. & La Farina, M. (2004) Biochem. Biophys. Res. Commun. 325**,** 784–791. [DOI] [PubMed] [Google Scholar]
- 53.Leroy, K., Haioun, C., Lepage, E., Le Metayer, N., Berger, F., Labouyrie, E., Meignin, V., Petit, B., Bastard, C., Salles, G., et al. (2002) Ann. Oncol. 13**,** 1108–1115. [DOI] [PubMed] [Google Scholar]
- 54.Oliveira, C., Westra, J. L., Arango, D., Ollikainen, M., Domingo, E., Ferreira, A., Velho, S., Niessen, R., Lagerstedt, K., Alhopuro, P., et al. (2004) Hum. Mol. Genet. 13**,** 2303–2311. [DOI] [PubMed] [Google Scholar]
- 55.Pasqualucci, L., Guglielmino, R., Houldsworth, J., Mohr, J., Aoufouchi, S., Polakiewicz, R., Chaganti, R. S. & Dalla-Favera, R. (2004) Blood 104**,** 3318–3325. [DOI] [PubMed] [Google Scholar]
- 56.Malpeli, G., Barbi, S., Moore, P. S., Scardoni, M., Chilosi, M., Scarpa, A. & Menestrina, F. (2004) Haematologica 89**,** 1091–1099. [PubMed] [Google Scholar]
- 57.Dearden, C. E. & Foss, F. M. (2003) Hematol. Oncol. Clin. North Am. 17**,** 1351–1366. [DOI] [PubMed] [Google Scholar]
- 58.Reynaud, C. A., Aoufouchi, S., Faili, A. & Weill, J. C. (2003) Nat. Immunol. 4**,** 631–638. [DOI] [PubMed] [Google Scholar]
- 59.Sayegh, C. E., Quong, M. W., Agata, Y. & Murre, C. (2003) Nat. Immunol. 4**,** 586–593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Table