Nuclear speckles: molecular organization, biological function and role in disease (original) (raw)
Abstract
The nucleoplasm is not homogenous; it consists of many types of nuclear bodies, also known as nuclear domains or nuclear subcompartments. These self-organizing structures gather machinery involved in various nuclear activities. Nuclear speckles (NSs) or splicing speckles, also called interchromatin granule clusters, were discovered as sites for splicing factor storage and modification. Further studies on transcription and mRNA maturation and export revealed a more general role for splicing speckles in RNA metabolism. Here, we discuss the functional implications of the localization of numerous proteins crucial for epigenetic regulation, chromatin organization, DNA repair and RNA modification to nuclear speckles. We highlight recent advances suggesting that NSs facilitate integrated regulation of gene expression. In addition, we consider the influence of abundant regulatory and signaling proteins, i.e. protein kinases and proteins involved in protein ubiquitination, phosphoinositide signaling and nucleoskeletal organization, on pre-mRNA synthesis and maturation. While many of these regulatory proteins act within NSs, direct evidence for mRNA metabolism events occurring in NSs is still lacking. NSs contribute to numerous human diseases, including cancers and viral infections. In addition, recent data have demonstrated close relationships between these structures and the development of neurological disorders.
INTRODUCTION
Less than 1.5% of the human genome consists of protein-coding sequences and the number of protein-coding genes is similar across most higher eukaryotes. Notably, differences in developmental programs arise from numerous gene expression regulatory mechanisms, which allow different cell types to respond adequately to specific environmental conditions. These mechanisms depend on the flexibility and dynamics of molecular interactions, which can be promoted or prevented by spatial organization in the nucleus (1–3). The enhancement of necessary interactions and reduction of undesired interactions are facilitated by reversible separation of specific molecules within a spatially restricted area. Indeed, numerous macromolecules from interchromatin regions of the nucleoplasm, predominantly proteins and RNAs, are gathered within nuclear subcompartments (or nuclear bodies), e.g. nucleoli, NSs, paraspeckles, Cajal (coiled) bodies, gemini of Cajal bodies (gems) and promyelocytic leukemia (PML) bodies. An increasing number of additional nuclear domains have been described, including nuclear stress bodies, histone locus bodies, polycomb bodies, DNA damage foci, cleavage bodies, matrix-associated deacetylase bodies and clastosomes (1–6). As a consequence, in addition to the exchange of molecules between the nucleus and cytoplasm, the tightly controlled distribution and movement of factors within the nucleus is an important level of regulation in many nuclear pathways, including RNA maturation.
Because alternative pre-mRNA splicing greatly increases transcriptome diversity in higher eukaryotes, nuclear bodies involved in splicing regulation are key gene expression regulators. These bodies include NSs, which are also known as splicing speckles, B snurposomes, splicing factor compartments, SC-35 domains and interchromatin granule clusters. The first observations of stained NSs using light microscopy were made by Santiago Ramon y Cajal in 1910. Electron microscopy (EM) observations and RNA identification in NSs were made by Hewson Swift in 1959. Two years later, J. Swanson Beck used the term ‘speckles’ for the first time to describe these bodies (4). The earliest identification of splicing factors and small nuclear ribonucleoproteins (snRNPs) in NSs uncovered connections between NSs and splicing (7–9). NSs were thought to play a role predominantly in regulating the availability of splicing factors at transcription sites because alteration of their function or composition led to changes in alternative pre-mRNA splicing. However, as research on NSs progressed, additional NS functions have been revealed and will be discussed in this review.
More recent studies have demonstrated that proteins involved in chromosome localization, chromatin modification, transcription, splicing, 3′ end processing, mRNA modification, mRNA coating with proteins and messenger ribonucleoprotein (mRNP) export are assembled in NSs, supporting the hypothesis that NSs act as a hub to coordinate all of the nuclear gene expression regulation steps. Importantly, all of these steps are coupled with RNA polymerase II transcription, which occurs within perichromatin fibrils in close proximity to NSs (10). Despite many studies aimed at functionally characterizing NS proteins, the precise role of NSs requires further clarification. This need for additional studies also applies to extensively explored processes, such as splicing, because in addition to the conventional view that NSs function in the assembly, modification, temporary storage and recycling of splicing factors, several reports have shown splicing activity within NSs (11,12). Moreover, the majority of NS proteins can also be found at other nuclear locations and their specific roles in NSs, interacting partners and post-translational modifications need to be elucidated.
In this review, we describe the involvement of NS proteins in various nuclear gene expression regulation pathways. We also review recent insights into the role of regulatory proteins, which are enriched in NSs; these proteins include protein kinases, cytoskeletal elements, factors involved in ubiquitination, SUMOylation and phosphoinositide (PI) signaling. Finally, we discuss the connection between NSs and human disease, with an emphasis on neurological disorders and the defects in RNA synthesis and metabolism that contribute to these disorders (13).
MOLECULAR ORGANIZATION
General characteristics
The human interphase nucleus contains 20–50 NSs measuring up to several micrometers in diameter. EM has revealed that a single NS is composed of spots (interchromatin granules) measuring 20–25 nm in diameter connected by fine fibrils to form a cluster (14). Their size or irregular shape can alter dynamically, may vary between different cell types and depend on numerous factors, including cellular ATP levels, the phosphorylation status of various proteins, transcription of stress-activated genes, SWI/SNF chromatin remodeling and RNA polymerase II transcription and splicing (15,16). Inhibition of RNA polymerase II transcription or splicing leads to accumulation of proteins in enlarged NSs, whose normal size can be restored upon elimination of the block (17).
Forces that maintain NSs
The biophysical properties of NSs and the nucleoplasm do not differ substantially. NSs are slightly denser than the surrounding nucleoplasm, and the protein concentrations (ranging from 115 to 162 mg/ml) are similar in both compartments (18) or are slightly higher in NSs (19). Although NSs are highly dynamic structures and their components are constantly in flux (20), NSs remain clearly separated from the nucleoplasm and when isolated from the nuclei of mouse liver cells by Spector and colleagues, they remained stable and resistant to subsequent steps in the purification procedure (21,22).
It is widely accepted that NS assembly and maintenance depend on interactions among NS components. In addition to structured protein domains, very flexible low-complexity regions (LCRs) play an important role in protein–protein and protein–RNA interactions and are over-represented in NS proteins (Supplementary Table S1). LCRs can change the properties of a protein permanently or transiently after post-translational modification or upon protein partner binding. Thus, proteins containing LCRs are regulated and ensure that cellular processes can be adjusted. Depletion or mutation of LCRs alters protein–protein interactions, protein–RNA interactions, protein functions and localization to NSs (23).
Multiple LCRs, e.g. serine/arginine-rich (SR) motifs and folded domains coexisting within individual NS proteins enable interactions with multiple proteins at the same time. Transient and frequent low-specificity interactions can explain why NS proteins move 100-fold more slowly than GFP in the nucleoplasm (20). Thus, separation of proteins in NSs allows escape from unfavorable interactions and ensures the integrity of NSs despite the lack of a lipid boundary.
Another source of the spatial distinction of NSs is cellular crowding; high macromolecule concentrations favor phase separation over the homogeneity of solutions with low macromolecule concentrations (24). Accordingly, liquid droplets with clear boundaries are spontaneously generated in aqueous solutions in vivo and in vitro when concentrated proteins with LCRs are incubated in them. Such phase separation can be affected by temperature, pH, ionic strength and LCR modification (24,25).
Post-translational protein modifications (phosphorylation, methylation and conjugation with PIs (26) or SUMO-1 (27)) within LCRs have been demonstrated to not only regulate protein–protein interactions and recruitment to NSs (28–30) but also to increase structural order (31). Disorder-promoting stretches of repetitive amino acids, such as five or more consecutive histidine residues (32,33), constitute NS-targeting signals. Thus, the level of disorder appears to be the main force regulating NS composition.
Nuclear speckles and the cell cycle
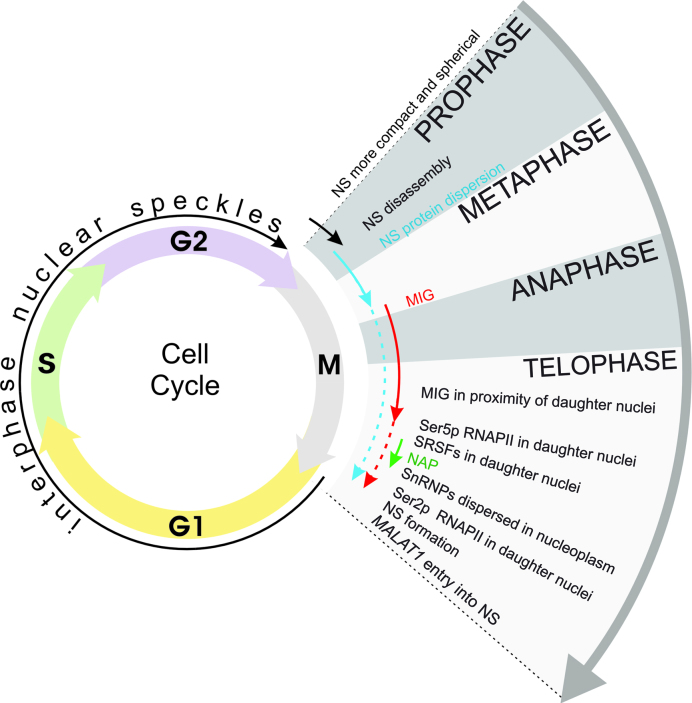
NSs are very stable during interphase. However, the self-organizing properties of their components must be specifically suppressed during cell division. Disruption of the nuclear envelope following mitosis initiation leads to NS disassembly (Figure 1) and cytoplasmic dispersion of NS proteins (34). In addition to diffuse patterns, NS proteins in the cytoplasm assemble into an increasing number of mitotic interchromatin granules (MIGs), which are observed from metaphase to telophase (34–36). Because pre-mRNA splicing factors were found to be in an active state immediately after entry into the nucleus (36), it has been postulated that MIGs, being structurally similar to NSs (35), are required for the modification, assembly and delivery of pre-mRNA processing complexes to transcription sites in daughter nuclei.
Figure 1.
Schematic representation of the dynamics of NS components during the cell cycle. The diverse forms of NS protein assemblies are indicated with arrows: black represents enlarged NSs (possibly caused by transcriptional inhibition), blue represents a diffuse pattern, red represents MIGs and green represents nucleolar organizing region-associated patches (NAP). Importantly, different sets of proteins exhibit diverse patterns of cell cycle-regulated localization and diverse timing of their entry into and their presence in assemblies of NS proteins. Different assemblies of NS proteins can coexist and those in the minority are indicated with dotted lines. Note that NSs and MIG, in contrast to NAP, contain poly(A)+ RNA.
After the reconstitution of the nuclear envelope, most pre-mRNA splicing factors gradually relocalize from MIGs to the nucleus within 10 min, but some of them (e.g. SRSF2) can stay in MIGs until G1 (36). During telophase, before formation of NSs, serine/arginine-rich splicing factors (SRSFs) in daughter nuclei gather transiently (for 15–20 min) in the proximity of active nucleolar organizing regions (NORs), in so-called NOR-associated patches (NAPs). SRSFs colocalize with CLK1 kinase (37), which is responsible for SRSFs leaving NSs (29). The assembly of SRSFs into NAPs and MIGs confirms their strong self-organizing properties, but SRSFs alone are not sufficient to form NSs. Hence, additional factors are required to nucleate NSs.
Because the establishment of transcription in telophase precedes NS formation (38), it has been hypothesized that the recruitment of splicing and processing factors to nascent transcripts triggers the spatial enrichment of NS proteins, followed by NS nucleation in the vicinity of active transcription sites. Accordingly, the ability of nascent pre-mRNA to trigger NS nucleation de novo has been shown in vivo (39).
The gathering of NS proteins can be explained by the self-organization model, which is based on protein–protein and protein–RNA interactions. However, induction of NS disintegration during prophase requires additional unknown factors. Cyclins seem to be potential NS cell cycle regulators because, to date, cyclin L1 is the only immobile protein in interphase NSs (40). Overall, the mechanisms that orchestrate cell cycle-dependent construction and deconstruction of NSs are still enigmatic and require further investigation.
Protein composition
Similar to other membrane-less bodies with liquid-like properties, NSs are characterized by the dynamic exchange of components within the nucleoplasm (24,25) and they share some proteins with other nuclear bodies. For instance, spliceosomal snRNPs are assembled in Cajal bodies before their relocation to NSs (2) and regulate the maturation of 3′ ends of histone transcripts within histone locus bodies (U2 snRNP) (41). NSs contain the paraspeckle proteins PSF (42) and PSP2, and both bodies often localize in close proximity (43). Similarly, Pat1b is found in both NSs and PML nuclear bodies (44), which can merge (45). However, the functional consequences of the close relationships between distinct nuclear bodies remain to be revealed.
The number of proteins found in NSs using proteomic approaches (21,22) has greatly added to the numerous proteins found in NSs based on microscopic studies. As listed in Table 1 and Supplementary Table S1, NS proteins are involved in multiple nuclear gene expression regulation steps, such as epigenetic regulation, transcription activator and repressor functions, transcription elongation and termination, splicing, 3′ end processing, mRNA modification and mRNA packing and export (Figure 2). Proteins involved in gene expression regulation will be described in the ‘Biological Function’ section. The localization, interactions and degradation of these proteins is regulated by NS regulatory proteins, which include protein kinases and proteins involved in PI signaling, cytoskeletal organization and ubiquitination and are described in more detail below.
Table 1. Serine/arginine-rich splicing factors localized in NSs.
| Protein encoding gene (with synonyms) | RNA- binding domains | Protein- binding domains or LCR | Recognized sequences | Interacting proteins | Molecular function | Selected references |
|---|---|---|---|---|---|---|
| SRSF1 (ASF; SF2; SRp30a; SFRS1) | RRM; RRMH | RS;LCR | RGAAGAAC; AGGAC[A/G][G/A]AGC; GAAGAA | CCDC55; CCNL1; CCNL2; CDK11B; CIR1;NFYA; NXF1; PSIP1; RSRC1; SAFB;SFRS12; SNRNP70; SRPK1; SRPK2; SRSF1; SRSF2; TRAF5; U2AF1; ZRSR2 | alternative splicing; splicing enhancer; mRNA nuclear export | (47) |
| SRSF2 (SC35; SRp30b; SFRS2) | RRM | RS;LCR | [C/G][C/G]NG; AGGAGAU; GUUCGAGUA; UGCNG[C/U] and more | CCDC55; CCNL1; CCNL2; CIR1; KAT5; SCAF11; SNRNP70; SRSF1; U2AF1; ZRSR2 | splicing: formation of the earliest ATP-dependent splicing complex | (9) |
| SRSF3 (SRp20; SFRS3) | RRM | RS;LCR | [A/U]C[A/U][A/U]C; CUC[U/G]UC[C/U]; CA-rich | CPSF6; NXF1; PCBP2; RBMY1A1; SFRS12; YTHDC1 | promotion of exon-inclusion during alternative splicing; mRNA nuclear export | (131) |
| SRSF4 (SRp75; SFRS4) | RRM; RRMH | RS;LCR | GAAGGA; GA-rich | PNN; SNRNP70; SNRPA1; SRRM1; SRRM2; SRSF5; TRA2B | alternative splicing | (135) |
| SRSF5 (SRp40; HRS; SFRS5) | RRM; RRMH | RS;LCR | GAGCAGUC GGCUCAC[A/C/U]G[G/C] | PHF5A; SNRNP70; SNRPA1; SRRM1; SRRM2; SRSF4 | pre-mRNA splicing | (22) |
| SRSF6 (SRp55; B52; SFRS6) | RRM; RRMH | RS;LCR | U[C/G]CG[U/G] [A/C]UCAACCAGGCGAC | DYRK1A; LUC7L2; SFRS12 | pre-mRNA splicing; modulates the selection of alternative splice sites | (46) |
| SRSF7 (SFRS7; 9G8) | RRM | RS;LCR; ZnF_C2HC | UCAACA; ACGAGAGA[C/U] GGACGACGAG | CCNL1; CCNL2; CDC2L1; CDC2L2; CPSF6; LUC7L2; NXF1; RBBP6; SDCB1; SFRS12; SRPK1; SRPK2 | pre-mRNA splicing; mRNA nuclear export | (136) |
| SRSF10 (SRp38; SRrp40;TASR1; SFRS13A) | RRM | RS;LCR | AAAGACAAA; [A/T/G]GA[A/G][A/G][A/G] | FUS; SNRNP70; TRA2B; YTHDC1 | pre-mRNA splicing; promotes exon skipping during alternative splicing | (131) |
| SRSF11 (SRp54; p54; SFRS11) | RRM | RS;LCR | AAGAAG | ARL6IP4;ETS1; GATC; HYI; PUF60; RBM39; SDCBP; SFRS12; TANK; TERF2 | pre-mRNA splicing | (122) |
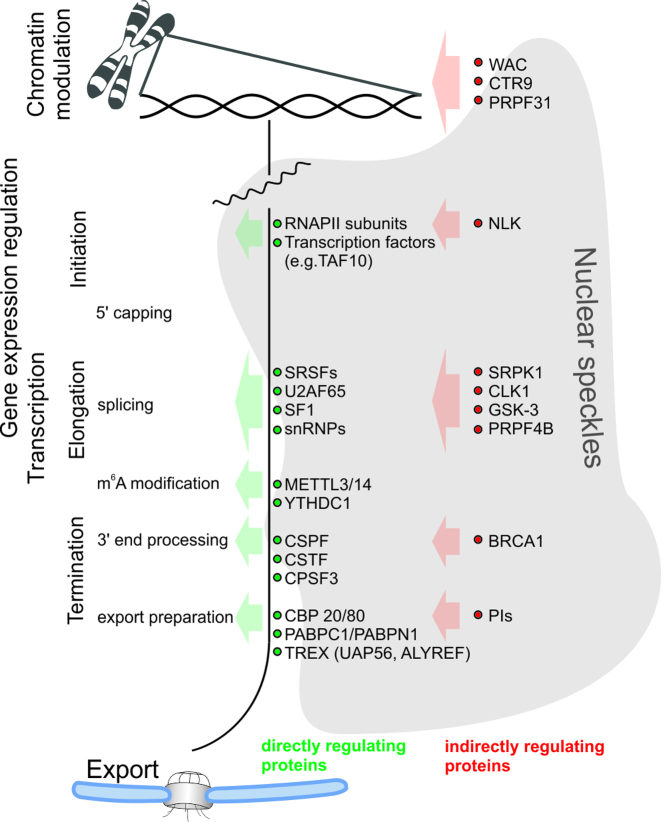
Figure 2.
RNA processing, from the site of transcription to nuclear export, is regulated by multiple proteins localized within NSs. The RNA pathway in the nucleus starts with transcription initiation and multiple proteins, many of which are found in NSs, are responsible for the processing of primary transcripts, including splicing, m6A modification, 3′ end processing and export. In the figure, examples of proteins with known functions in transcription and RNA maturation are presented (green dots). Additionally, a large group of other NS-associated proteins is indirectly involved in the precise regulation of RNA processing and their examples are shown (red dots).
Protein kinases
Post-translational protein modifications that can alter protein properties are among the most important NS functions. Based on their protein composition, NSs seem to be nuclear hubs for protein phosphorylation, methylation, acetylation, ubiquitination and SUMOylation. As more than 30% of human proteins can be phosphorylated and kinase-encoding genes constitute ∼2.4% of protein-coding human genes, protein phosphorylation is a fundamental regulatory mechanism.
In total, 31 protein kinases have been found in NSs (Supplementary Table S1) and the substrates of many NS protein kinases have been identified (46). Because many NS proteins participate in various gene expression steps that occur in different cellular compartments, the control of shuttling of these proteins has emerged as an important instrument of gene expression regulation. Reversible protein phosphorylation plays an important role in proper cellular localization of NS proteins, including SRSFs (47). SRSF1 is targeted to the nucleus (and further to NSs) upon SRPK1-driven phosphorylation (48). The resultant hypophosphorylated RS domain of SRSF1 prevents SNRNP70 binding and the subsequent formation of the spliceosomal E complex (49). In contrast, SRSF1 hyperphosphorylation by CLK1 promotes spliceosome assembly and releases SRSF1 from NSs (50). SRSFs are dephosphorylated during splicing and then serve as adaptors for mRNA export (51) or translation initiation (52). In summary, SRSF phosphorylation/dephosphorylation serves as a critical mechanism affecting both splicing and protein assembly in NSs (30). Accordingly, enhanced activity or overexpression of many NS kinases induces NS disassembly, indicating a role for kinases in NS maintenance.
Numerous kinases can phosphorylate multiple sites within a single NS protein and conversely, individual NS protein kinases can phosphorylate multiple substrates. In addition to splicing- or transcription-specific kinases (e.g. NLK), the action of many NS kinases is not restricted to a single-specific process. Thus, nuclear processes are mutually regulated. NS kinases also ensure communication with the extranuclear and extracellular environment by acting as modules of cellular signaling cascades. Therefore, NS kinases are mediators that integrate and modulate various signals via nuclear gene expression regulation events to adjust cellular homeostasis.
PI signaling
Derivatives of phosphoinositol act as another major group of signaling molecules that are relevant to NS function. In the cell nucleus, PIs are involved in regulation of gene expression, including chromatin modification, pre-mRNA maturation and mRNP export (26). Numerous NS proteins are affected by PIs directly (upon PI binding) or indirectly by the activity of PI-dependent regulatory proteins (e.g. protein kinases or ubiquitin ligases).
Growing evidence has indicated that PIs are localized in NSs (53). Moreover, PI-modifying enzymes, which add or remove phosphate from PIs (kinases and phosphatases) or hydrolyze them (phospholipases C), interact with each other in NSs (54), suggesting that some PI derivatives are produced in NSs. To date, more than twenty proteins implicated in PI signaling have been found in NSs. Among them, speckle-targeted PIP5K1a-regulated poly(A) polymerase has important roles in PI-dependent pre-mRNA 3′ end maturation and in regulation of the expression of specific mRNAs. The complexity of PI signaling may result from regulation of individual proteins by various PIs. For instance, binding of PI(4,5)P2 or phosphatidic acid to nuclear receptor steroidogenic factor 1 (NR5A1) modifies NR5A1-dependent gene expression (27,55).
PIs may have a substantial impact on NS function due to the multifunctionality of their downstream signaling proteins, which include several prominent NS protein kinases. One of them is AKT1, which enhances SRPK2 kinase activity toward SRSF2 and ACIN1. Another substrate of AKT1 is ALY/REF. PIs target ALY/REF to NSs independently of AKT1-driven phosphorylation but can also affect ALY/REF-regulated mRNA nuclear export in cooperation with AKT1 (26).
Moreover, several NS proteins, e.g. PARD6A/TIP-40 and PDLIM7/ENIGMA, contain PIP2-binding PDZ domains, which mediate protein–protein interactions. Disturbances in the PIP2 level or in PIP2-PDZ domain interactions abolish syntenin-2 and ZO-2 guanylate kinase targeting to NSs (56,57). Importantly, impaired ZO-2 (56) or syntenin-2 expression (57) promotes the formation of more dispersed and smaller NSs.
Taken together, these facts reinforce the view that NSs are nuclear hubs for PI production and signaling, which are important for protein targeting to NSs and NS assembly, as well as for the regulation of transcription, transcript maturation, splicing and export (26,27,55,58). However, our understanding of the most basic mechanisms of PI generation and its wide impact on nuclear functions is at an early stage and requires further elucidation.
Nucleoskeletal organization
In proteomic studies (21,22), different types of structural proteins were identified in NSs, but our knowledge at that time was not sufficient to unambiguously assign lamins, myosins and tubulin to NSs. However, later studies revealed that some of these cytoskeletal proteins are indeed NS components. Cytoskeletal rearrangements are regulated by NS proteins involved in PI signaling (PIP5K1A, INPPL1, PDLIM7/ENIGMA and profilin-1) and calcium signaling (L-plastin, PTK2B and EPB41). This suggests that the functional associations between PIs, calcium and cytoskeletal proteins in the cytoplasm are similar in NSs.
Proteins implicated in cytoskeletal organization contribute to nuclear assembly (59) but also regulate transcription (60). Disruption of actin delivery to RNA polymerase II (RNAPII) profoundly impairs transcription elongation (61). In addition to its role in transcriptional elongation, myosin Ic takes part in the formation of the first phosphodiester bond during transcription initiation (62). Furthermore, actin and myosins are cofactors for several transcription and chromatin remodeling complexes (e.g. PCAF, INO80 and BAF, numerous components of which have been found in NSs) and RNA-binding proteins.
Nuclear actin exists in monomers (G-actin) and short polymers (filaments, F-actin). Actin polymerization regulation is important for nuclear transport and transcription because NS reorganization upon RNAPII inhibition and RNAPII recruitment to activated promoters depend on actin polymerization (63). However, despite the fact that modifiers of actin polymerization localize in NSs, primarily monomeric actin was shown to be concentrated in NSs (64). Therefore, deeper insight into the role of actin in NSs is required and whether NSs play a role in actin polymerization needs to be determined.
Ubiquitination and SUMOylation
A substantial portion of NS proteins undergo covalent attachment of ubiquitin or ubiquitin-like proteins, e.g. SUMO1, ISG15 and UBL5. Three sequential ubiquitination steps are performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and E3 ubiquitin ligases. NSs contain more than 20 E3 ligases or constituents of E3 ligase multisubunit complexes. Although active proteasomes have a role in protein degradation in NSs and affect NS assembly (65), only a specific type of polyubiquitination (attachment of four or more ubiquitin molecules) targets a protein for proteasomal degradation. Other forms of polyubiquitination and monoubiquitination adjust the functions (activity, cellular localization and protein–protein interactions) of the conjugated protein.
Ubiquitin-dependent regulation is an important mechanism for splicing control and ubiquitination facilitates protein–protein interactions required for spliceosome assembly (66). Little is known regarding the fate of proteins that are ubiquitinated in NSs because, to date, deubiquitinating enzymes have not been identified in NSs. This lack of deubiquitinating enzymes could suggest that after ubiquitination, proteins escape from NSs to be deubiquitinated or undergo proteasomal degradation in NSs. Interestingly, several NS protein complexes contain subunits responsible for interactions with ubiquitin-specific proteases.
In contrast, SUMOylation of NS proteins is better understood. Attachment of SUMO-1 is a typical signal to target proteins to NSs. The presence of deSUMOylating enzymes in NSs suggests that SUMO-1-dependent protein targeting to NSs may be reversible. SUMOylation is involved in the regulation of many other NS-associated processes. For example, CBX4 is the main regulator of the relocation of chromatin to the proximity of NSs upon transcriptional activation (67) and SUMOylation of TCERG1 inhibits its activity in transcription elongation (68).
Together, the presence of numerous components of the ubiquitination/SUMOylation machineries in NSs merits defining NSs as nuclear ubiquitination and SUMOylation centers. However, most of the ubiquitination/SUMOylation mechanisms in NSs have yet to be uncovered, e.g. how SUMOylation and ubiquitination can cooperatively modulate the functions of individual proteins.
RNAs in nuclear speckles
Numerous studies have revealed the presence of RNAs, including poly(A)+ RNAs (69,70) and noncoding RNAs, in NSs. A significant portion of total nuclear poly(A)+ RNA is localized within NSs (71). However, there is no substantial immobile fraction of poly(A)+ transcripts (72,73), as these transcripts are freely exchanged between NSs and the nucleoplasm.
The enrichment of some transcripts in NSs mainly depends on the presence of introns (12) but intronless transcripts also localize to NSs (74); however, they need to contain either additional motifs (e.g. sequences promoting the alternative export or ALREX, pathway) or specific mRNA structures to direct trafficking of intronless mRNAs through NSs.
Nuclear export seems to be the most prominent pathway regulating mRNAs residing in NSs because depletion of transcription export (TREX) complex components leads to enhanced association of mRNAs with NSs (12). Moreover, redirecting mRNAs to the nuclear export pathway is energy-dependent; therefore, this process significantly differs from passive intranuclear diffusion (75).
Unlike the transcription-independent exchange of poly(A)+ RNA between NSs and the nucleoplasm (76), mRNA nuclear export depends on active transcription (70), complete polyadenylation and splicing (77). Because most exons are spliced cotranscriptionally at the transcription site (78,79), NSs seem to be the location of the transcript maturation that is needed for nuclear export (80). Nevertheless, slow splicing or splicing defects lead to enrichment of transcripts in NSs because they inhibit transcript export (80,81). Such ‘difficult’ posttranscriptional splicing might occur in NSs and several reports have suggested that splicing can occur within NSs (11,12). However, this possibility requires further investigation.
Noncoding RNAs present in NSs include spliceosomal small nuclear RNAs (snRNAs), 7SK RNA, whose downregulation leads to mislocalization of NS components (82) and the long noncoding RNA metastasis-associated lung carcinoma transcript 1 (MALAT1), which will be described in more detail in the following section.
Role of MALAT1 RNA
Long noncoding RNA MALAT1 is also known as nuclear-enriched abundant transcript 2 (NEAT2) (83,84). The 8.7 kb human MALAT1 gene locus is on chromosome 11, ∼60 kb downstream of the NEAT1 gene, which gives rise to another long noncoding RNA that is a well-established structural component of paraspeckles. The primary transcript of the MALAT1 gene is cleaved at its 3′ end by RNase P to generate the ∼7.5-kb long MALAT1 RNA, which is stabilized by a highly conserved triple helical structure instead of a poly(A) tail and is retained in the cell nucleus. The remaining short 61 nt tRNA-like mascRNA is localized exclusively to the cytoplasm (85).
Multiple SRSFs associate with MALAT1, including SRSF1, SRSF2 and SRSF3, which directly bind to their recognition sites at the 5′ end of MALAT1. In addition, MALAT1 binds other proteins and its NS localization depends on RNPS1, SRm160 and IBP160 (86–88). Of the nuclear RNAs, MALAT1 interacts directly with U1 snRNA but is unlikely to interact similarly with other noncoding nuclear RNAs. Similarly, MALAT1 does not form direct RNA–RNA contacts with nascent pre-mRNAs; rather, it binds indirectly via protein mediators (89). MALAT1 also interacts with chromatin at actively spliced genes, close to polyadenylation sites, in a transcription-dependent manner, similar to patterns of RNAPII occupancy (89). It also recruits machinery for gene activation and the movement of active chromatin to NSs (see below).
MALAT1 influences alternative splicing by regulating the phosphorylation and resulting nuclear distribution of splicing factors. In addition, MALAT1 downregulation seems to increase the cytoplasmic pool of poly(A)+ RNAs (90). However, in vivo analyses of mice depleted of MALAT1 revealed that MALAT1 is not essential for mouse development, gene expression regulation, alternative splicing, splicing factor phosphorylation or the organization of NSs (91,92). This difference between human cellular and mouse models is surprising as MALAT1 shows strong sequence conservation across mammalian species and is a highly abundant transcript.
BIOLOGICAL FUNCTION
Kinases, cytoskeletal proteins and enzymes of ubiquitin and PI metabolism are prominent nuclear gene expression regulators with a known role in transcription and splicing regulation (Figure 3). However, they represent only a small fraction of the NS proteome; >50% of the NS proteins listed in Supplementary Table S1 are involved in transcription or splicing regulation. Over-representation of splicing proteins in NSs has been apparent from early studies, but the data from recent years have revealed that the largest group of NS proteins is involved in transcription and includes transcription factors and chromatin remodeling factors.
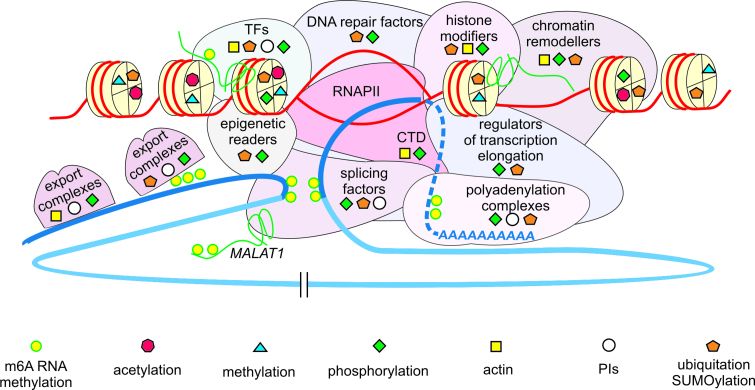
Figure 3.
The role of proteins localized to NSs in pre-mRNA synthesis, maturation and DNA template regulation; impact of NS regulatory proteins on depicted protein complexes is indicated with specific symbols explained in the figure. DNA template is shown in red and primary transcript in blue, exon (dark blue) intron (light blue). TFs: transcription factors; RNAPII: RNA Polymerase II; PIs: phosphoinositides; CTD: C-terminal domain of RNAPII.
NS formation and function are tightly associated with active transcription and RNAPII integrates transcript synthesis with the DNA template and DNA-regulating proteins on one hand and RNA maturation and export on the other (Figure 3). Notably, mRNA 3′ end processing and polyadenylation, mRNA m6A methylation, mRNA coating with proteins, execution of mRNA nuclear export and chromatin regulation are also directly coupled with splicing. Accordingly, more than 30 NS proteins play important roles in both transcription and splicing. These processes are connected not only functionally but also by physical protein–protein interactions. In the following subsection, we discuss the crucial role of RNAPII transcription and splicing in integration of the biological function of NSs.
RNAPII transcription and splicing
NSs are formed in the direct neighborhood of RNAPII transcription sites and contain several RNAPII subunits. Despite the numerous transcription factors residing in NSs (Supplementary Table S1), other proteins that directly regulate RNAPII during transcription initiation are under-represented. In contrast, components of protein complexes that regulate transcription elongation are present. These include subunits of the super elongation complex and the general transcription elongation factor SIII (elongin), which increases the catalytic rate of RNAPII transcription by suppressing transient pausing. NSs also contain the multifunctional epigenetic regulator TRIM28, whose phosphorylation stimulates the transition to transcription elongation, while the non-phosphorylated form causes RNAPII pausing (93).
Direct coupling of multiple mRNA maturation steps with the transcription process occurs through recruitment of suitable factors by the C-terminal domain (CTD) of Rpb1, the largest RNAPII subunit (94). Protein recruitment to the CTD is governed by phosphorylation events on the conserved heptapeptide Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, which is repeated 52-times in the human CTD (95). Ser5 phosphorylation is connected to synthesis of the 5′ end of nascent pre-mRNA, while specific phosphorylation of Ser2 is responsible for transcription elongation and engagement of the pre-mRNA splicing and processing machineries (96).
Among the CTD regulators, components of the Ser2-phosphorylating p-TEFb complex, i.e. CDK9 (as well as CDK12/13) and cyclins have been found in NSs. The CDK9-interacting proteins HEXIM1 and BRD4, which are negative and positive regulators of p-TEFb, respectively, are also localized to NSs (97,98). In contrast, kinases implicated in Ser5 phosphorylation are absent from NSs, supporting the role of NSs in processes coupled to transcription elongation.
Transcription requires the formation of a short hybrid of nascent pre-mRNA and an unwound DNA template, the so-called R loop, which is known to increase DNA instability and affect DNA integrity (99). R loop removal is promoted by mRNA packing with a protein coat (100), a high RNAPII elongation rate, efficient transcription termination or mRNA 3′ end processing. However, the NS-associated mechanisms preventing the harmful effects of R loops on genome stability involve not only tight temporal integration between co-transcriptional steps of pre-mRNA maturation but also numerous DNA repair proteins localized in NSs (Supplementary Table S1).
The spatial organization of chromatin within the cell nucleus serves as an important level of gene expression regulation that is linked to NSs and RNAPII transcription. Genes on different chromosomes can be spatially clustered due to random movement of chromosomes within the nucleoplasm and specific binding of decondensed stretches of chromatin by transcriptional machinery, which is differentially arranged in various cell types. Spatial association occurs within actively transcribed genes and co-regulated genes gather around shared NSs (38,101). This association of chromatin domains with NSs is correlated with gene expression upregulation (102,103). Notably, NS dispersion leads to a decrease in chromatin domains in shared neighborhoods (104), supporting the importance of NSs in the spatial organization of transcriptionally active chromatin.
Most human pre-mRNAs contain introns, which are excised during the splicing process. Splicing is a key step in pre-mRNA maturation and is typically coupled with transcription elongation (105). The excision of introns in eukaryotes is catalyzed and regulated by five spliceosomal snRNPs and more than 100 associated proteins (106), most of which are known NS proteins. The resultant mature transcript can be alternatively spliced in specific cells or environmental conditions due to exon skipping or extending or by intron retention.
Notably, splicing depends on transcription elongation and vice versa: active splicing is required for Ser2 phosphorylation (107) of the CTD. Ser2p- and p-TEFb-induced transcription elongation is positively correlated with splicing efficiency (108). The spatial proximity of NSs and active RNAPII transcription sites allows convenient access to splicing factors because splicing factors are continuously added to and released from RNAPII as RNAPII moves along a gene. In addition to SRSFs, which interact with the CTD in a phosphorylation-dependent manner (109), several other splicing factors have been shown to directly interact with the CTD. Among them, U2AF65 (complexed with PRP19C) is required for CTD-dependent splicing activation (110).
Splicing is thought to affect the rate of nascent RNA synthesis by promoting RNAPII pausing (111). Conversely, RNAPII slowing promotes RNAPII accumulation in the intronic regions flanking the alternative exons, which allows recruitment of activators or repressors of alternative splicing. These can promote exon inclusion or exon skipping, respectively. Pre-mRNA splicing and negative regulation of transcription elongation are coupled by NS-localized proteins: e.g. the splicing factor/transcriptional repressor SF1 interacts with the CTD-binding regulators of transcription elongation (112). Additionally, BRM (the ATPase subunit of the SWI/SNF chromatin remodeler) (113) and ZNF326 (114) have been proposed to modulate connections between transcription pausing and alternative splicing.
Epigenetic mechanisms of gene expression regulation serve as another important link between transcription and splicing. Nucleosomes are barriers for RNAPII during transcription and increase the frequency and duration of RNAPII pauses. Thus, histone modifications that enhance DNA wrapping around nucleosomes induce exon inclusion (115) and exon skipping is triggered by histone modifications that promote relaxation of the chromatin structure (116). Histone modifications also guide the recruitment of splicing factors to chromatin (117) and conversely, splicing factors recruit histone-modifying enzymes in a pre-mRNA-dependent manner (118). Similarly, histone acetyltransferases and methyltransferases interact with U1/2 snRNPs (119). Consequently, splicing inhibition or splicing factor depletion leads to changes in histone modification patterns (111).
These close relationships between splicing and epigenetic mechanisms of gene expression regulation may underlie the robust enrichment of proteins involved in epigenetic regulation in NSs. Notably, histones, histone acetyltransferases, methyltransferases, deacetylases and HP1 protein, which are fundamental players in epigenetic regulation of gene expression, were identified in NSs (Supplementary Table SI). However, the potential function of HP1 in NSs and the role of NSs in histone modification has not been directly studied, despite the fact that particular core catalytic subunits of histone modifiers can localize to non-overlapping NSs, suggesting that specific multisubunit complexes regulate a distinct set of genes (120). Because multiple protein complexes involved in histone modification or histone variant deposition and protein partners of HP1 (including SRSF1/3 (117)) are found in NSs, future research on epigenetics in the context of NS localization may uncover novel mechanisms of gene expression regulation.
In short, transcription and splicing are largely implicated in the regulation of chromatin structure and function. The dependence of splicing on epigenetic regulation is not the only connection that occurs between splicing factors and DNA regulatory proteins because multiple examples of DNA repair and DNA stability regulation by splicing factors has been published recently (reviewed in (121)). Because many DNA-regulating proteins have been found in NSs (122), the role of splicing factors and NSs in DNA regulation is an attractive target for future research.
mRNP maturation and export
Proteins localized in NSs not only participate in numerous aspects of mRNA synthesis and mRNP maturation and are essential for harmonization of these nuclear processes but also influence cytoplasmic mRNA events.
The final step of mRNA synthesis in almost all eukaryotes is the addition of a poly(A) tail at the 3′ end. The most prominent factors regulating and catalyzing endonucleolytic cleavage and the subsequent non-templated poly(A) tail addition are localized in NSs (Supplementary Table S1) and their activity depends on their interaction with the CTD (94). Up to 70% of human mRNA transcripts are thought to be subjected to alternative polyadenylation (123). NS-associated processes, including CTD phosphorylation, epigenetic regulation, N6-methylation of adenosine (m6A) and splicing, are implicated in site selection in alternative polyadenylation (124,125). Relationships between splicing and 3′ end processing have been widely studied and the studies have shown that 3′ end processing is promoted by the intron upstream of the polyadenylation signal, whereas the splicing efficiency of the terminal intron is reduced by mutations at the polyadenylation signal (126). This reciprocal coupling is thought to be realized via interactions of splicing factors (e.g. U2AF65) or spliceosome components (U1A of U1 snRNPs) with nuclear poly(A)-polymerase (127) or cleavage and polyadenylation specificity factor 160 (CPSF160) (128), respectively, among other mechanisms.
m6A mRNA modification has a strong impact on orchestration of mRNP maturation. NSs contain the critical m6A-specific methyltransferases METTL14 and METTL3 that associate with the regulatory Wilms' tumor 1-associating protein (WTAP), whose protein partners are NS proteins. The m6A modification is enzymatically removed by the demethylase ‘eraser’ enzymes fat mass and obesity-associated protein (FTO) (129) and ALKBH5 (130), which are also localized in NSs. The presence of all crucial nuclear elements of the m6A modification system in NSs suggests a prominent role for NSs in m6A regulation. Moreover, the known nuclear readers of m6A YTHDC1 and hnRNPb1a are also localized in NSs. YTHDC1 promotes SRSF3 NS localization and RNA-binding affinity but represses those of SRSF10, thus promoting exon inclusion and inhibiting exon skipping (131). m6A modifies structure (base pairing) and interactions of RNAs with proteins and thus affects numerous aspects of RNA metabolism, as shown for regulation of MALAT1 binding by hnRNP C (132).
All the aforementioned steps of mRNA synthesis and maturation are closely associated and leave signatures on the mRNA (e.g. deposited proteins, poly(A) tail length or modified nucleotides) that regulate its subsequent nuclear export, subcellular localization, stability and translation. More than 1000 different proteins can be recruited to human mRNAs to form mRNP particles. Among them, the core components of the exon–junction complex (EJC) are recruited to the spliceosome during the early steps of splicing (133). The EJC is a key regulator of mRNP stability (134) and attracts multiple EJC peripheral factors, a substantial portion of which are NS proteins (135,136) that specifically enhance subsequent gene expression steps, such as adjacent splicing events, mRNA nuclear export or translation.
The TREX complex plays an essential role in coordinating transcription and splicing with mRNA export (137,138) and TREX can be recruited to mRNA through the EJC in a splicing-dependent manner (139). TREX serves as a platform for binding of the NXF1/NXT1 heterodimer (140), which is a general export receptor and is responsible for translocation of mRNPs through the nuclear pore. In addition to the association of exporting complexes with the EJC, NSs contain additional proteins that interact with NXF1/NXT1 in proximity to the 5′ and 3′ ends of transcripts, e.g. the cap-binding complex (141) and CPSF (142), respectively. Furthermore, several other adaptors localized in NSs, namely, CIP29, LUZP4, UIF, Dbp5 and the SRSFs, have been shown to mediate or modify NXF1/NXT1 recruitment. This observation indicates that multiple NXF1/NXT1 complexes are capable of binding various sites within individual mRNPs.
mRNP export is well known to be enhanced by splicing (12,143). UAP56, which is found in the EJC (133), is a splicing factor essential for spliceosome assembly and for the interaction of U2 snRNP with the branch point during splicing (144). Moreover, TREX components have been shown to interact directly with Ser2-phosphorylated CTD (145). TREX recruitment to Ser2-phosphorylated CTD is mediated by the PRP19 complex (146), which can be considered an important link integrating mRNP export with transcription elongation and splicing. Importantly, NS proteins involved in splicing regulation, i.e. SRSF3/7 (147), the PRP19 complex and U2AF2 (148), are also required for export of intronless mRNAs.
The above mentioned mutual relationship between mRNP maturation and export machineries can be considered a part of the mRNA quality control system. This system impacts nuclear retention of transcripts containing splicing-defective introns that form partial spliceosomes (81) and allows the export of only fully processed mRNPs. However, the protein composition of mRNPs controlled by NSs may also influence cytoplasmic RNA degradation, for example, through the nonsense-mediated mRNA decay pathway, which is regulated by proteins involved in the EJC (149). Thus, NSs regulate mRNP formation and subsequent export efficiency, influencing subcellular targeting of mRNPs and translation.
ROLE OF NUCLEAR SPECKLES IN THE PATHOGENESIS OF HUMAN DISEASES
Multiple diseases are associated with altered NSs or aberrant function of proteins found in these structures. Some rare disorders are caused directly by mutations in genes encoding proteins and noncoding RNAs found in NSs (150–152). Among these diseases are retinitis pigmentosa 13, 18, 33, 60, which is caused by mutation in PRPF8, PRPF3, SNRNP200 and PRPF6 genes; mandibulofacial dysostosis, Guion-Almeida type, which is caused by mutation in the EFTUD2 gene; and thrombocytopenia-absent radius syndrome, which is caused by mutation in the RBM8A gene. However, most diseases associated with NS malfunction, including cancers and viral diseases, are indirectly linked to these structures.
Carcinogenesis is strongly associated with alternative splicing aberrations that are caused by changed expression of proteins involved in alternative splicing that often result in NSs morphology disturbances. Overexpression of NS-forming proteins, including SR proteins, is observed in many types of cancer (153) and abnormally high levels of SRSF1 have been shown to be sufficient to induce tumorigenesis (154). Similar effects have been described for several other splicing and polyadenylation-associated proteins (155,156). In cancers, the altered polyadenylation pattern is explained, in part, by the tendency to shorten transcripts to reduce their ability to undergo regulation, e.g. by miRNAs (157). Moreover, single nucleotide mutations in pre-mRNA splicing factors may be associated with a gain-of-function mechanism (158).
Viruses use multiple host proteins during their life cycle, which includes viral RNA splicing and export (159). Both SRSFs and hnRNP proteins have been shown to control HIV RNA splicing and that of other viruses, thereby strongly influencing the translation of viral proteins (160–162). Consequently, viral infection triggers changes in the cellular levels and localization of splicing factors, e.g. their exclusion from NSs (163,164). Proteins participating in host mRNA 3′ end formation are also involved in the viral reproductive cycle (165). Interestingly, even viruses that replicate in the cytoplasm may affect NS function (166).
Emerging evidence supports the involvement of NSs in the pathogenesis of neurological diseases. NS proteins are involved in neuronal cell differentiation, as well as in the control of neuron-specific splicing and contribute to the abnormalities observed in neurological diseases, which is discussed below.
Neurological diseases
Neurodegeneration that occurs during the progression of neurological disorders is accompanied by disruption of nuclear bodies, including NSs. The involvement of NSs in neurodegeneration often expands into general splicing aberrations and dysfunction of NS proteins. In some neurological diseases, neurodegeneration is attributed to protein and/or RNA aggregates that are formed within the cell nucleus.
Alzheimer’s disease (AD) is associated with β-amyloid, which is formed from an amyloid precursor protein that is regulated by U1 snRNP (167). One of the proteolytic products of this protein is the Cγ fragment, whose phosphorylated form localizes to NSs and is thought to take part in pre-mRNA splicing (168). Neurodegeneration is also linked to NSs by the GSK-3 protein, which accumulates in the cytoplasm of neuronal cells in AD and other tauopathies (169). A lower GSK-3 level in the nucleus of neurons results in aberrant splicing of exon 10 in the τ transcript and enrichment of NSs with SRSF2 (170).
Aberrations in alternative splicing control that may be attributed to proteins found in NSs are also observed in other diseases. In Parkinson’s disease (PD), alternative splicing events, including exon skipping in the leucine-rich pentatricopeptide repeat-containing RNA transcript that binds hnRNP A1-associated poly(A) mRNAs and is regulated by hnRNP A1, may contribute to neurodegeneration (171). Aberrant alternative splicing of transcripts involved in PD pathogenesis has been extensively investigated and includes missplicing of genes directly involved in disease development, as well as downstream effects resulting in the inclusion of alternative exons that alter interactions with miRNAs and lnRNAs (172).
In frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS), which are closely related diseases, a general disruption of nuclear structure that may affect NSs has been observed. The TDP-43 protein has been shown to serve as a cellular scaffold for multiple nuclear bodies, including Cajal bodies, PML bodies, NSs and gems (173). In some FTD patients, TDP-43 formed intracellular inclusions that mostly localized within the cytoplasm, decreasing its level in the nucleus (174). Similar inclusions were also observed in ALS and other neurodegenerative diseases (175). Aberrations in TDP-43 may therefore lead to significant disruption of nuclear structure and affect the function of nuclear compartments. Decreases in TDP-43 were also shown to impair splicing (176,177), triggering the inclusion of cryptic exons (178). Many splicing events of disease-associated transcripts involved in neurodegeneration were disturbed upon depletion of TDP-43. Interestingly, TDP-43 was also shown to bind and regulate MALAT1 levels and depletion of TDP-43 led to decreased expression of this NS-specific lnRNA (177). In addition to TDP-43, ALS is associated with fused in sarcoma (FUS) protein aggregation (179). C-terminal mutations in FUS that disrupt the nuclear localization signal result in cytoplasmic accumulation of the protein (180). Physiologically, FUS is found in NSs and decreased levels of FUS and mutations in its sequence may alter the function of these structures.
In spinal muscular atrophy, in which the SMN1 gene is inactive, a disturbance in nuclear structure has also been observed (181,182). SMN1 is involved in protein transport to NSs and along with close interactions between various nuclear compartments, a lack of the SMN1 protein may lead to disturbance of NSs, especially because the SMN complex is also involved in spliceosome assembly (183).
For several neuropsychiatric diseases, known associations with NSs may explain the mechanisms of disease progression. Known point mutations in, and copy number variants of, genes encoding EJC proteins that are found in NSs are linked to intellectual disabilities, autism and schizophrenia (184). Moreover, mutations in the 5′ UTR of EIF4A3, which is associated with the EJC, are implicated in Richieri-Costa-Pereira syndrome, which causes learning deficits, among other symptoms (185). The EJC has been shown to play an important role in neurodevelopment, which highlights its potential role in neuropsychiatric disorders (186). In schizophrenia patients, aberrant alternative splicing, including splicing of transcripts such as the glutamate transporter and microcephalin, which are important for neuronal function, was observed (187,188). Aberrant splicing was also found in autism and often involved microexons, which are atypical exons only 3–27 nt in length. Interestingly, microexon splicing events often occur in neuronal cells and the functions of proteins found in NSs may also be altered by such microexon-dependent mechanisms, as has been shown for SR100/SRRM4 (189,190). Thus, the current knowledge of the scale of microexon involvement in diseases may be just the tip of the iceberg.
Role of nuclear speckles in simple repeat expansion diseases
Simple sequence repeats occur within numerous human genes and expansion of these sequences beyond a critical number of copies in specific genes causes incurable genetic diseases. In these disorders, pathogenic effects can be triggered by the toxic RNA transcribed from an expanded allele, and nuclear foci formed by mutant RNAs have been found in multiple neurological diseases belonging to this class. The reasons for nuclear retention of mutant RNA are not clear; however, RNA foci localization within or in proximity to NSs suggests that proteins present in these structures may contribute to foci formation.
Expanded CUG repeats present in the 3′ UTR of the DMPK transcript in myotonic dystrophy type 1 (DM1) form RNA foci that localize on the periphery of NSs (191). CUG RNA foci are arrested at the border of NSs due to RNA structure and aberrant interactions with MBNL1 proteins. MBNL1 silencing released mutant RNA foci from the periphery of NSs, proving the involvement of MBNL1 in nuclear retention of mutant transcripts (192). On the other hand, tautomycin-mediated disintegration of NSs did not influence CUG RNA foci (193), showing their structural independence of these nuclear structures. Interestingly, DNA loci of normal and mutant DMPK genes also localized to the periphery of NSs, demonstrating that RNA foci are formed close to the site of mutant RNA transcription. RNA foci in DM1 contain spliced mRNA (194), which suggests that proper splicing of this transcript does not require NS entry and that another mechanism must be responsible for the arrest.
In polyglutamine diseases in which expanded CAG repeats are present in the ORFs of various functionally unrelated genes, mutant RNA is either temporarily or is partially captured within NSs (193,195,196). The presence of mutations did not trigger significant changes in the number or size of NSs in fibroblasts, but disintegration of NSs induced by tautomycin resulted in disassembly of RNA foci (193). In Huntington’s disease (HD), which is one of the polyglutamine disorders, aberrant alternative splicing events were observed, including aberrant splicing of the τ transcript (197). Moreover, aberrant splicing of transcripts formed from the mutant allele in HD was observed and attributed to misregulation of SRSF6-controlled splicing (Figure 4B) (198). Similar to CAG repeats, foci formed by expanded CGG repeats present in the 5′ UTR of FMR1 in FXTAS localize within NSs (199). The CGG foci also disassembled when cells were treated with tautomycin; however, the mechanism proposed implied an interaction between tautomycin and the mutant transcript, preventing foci formation.
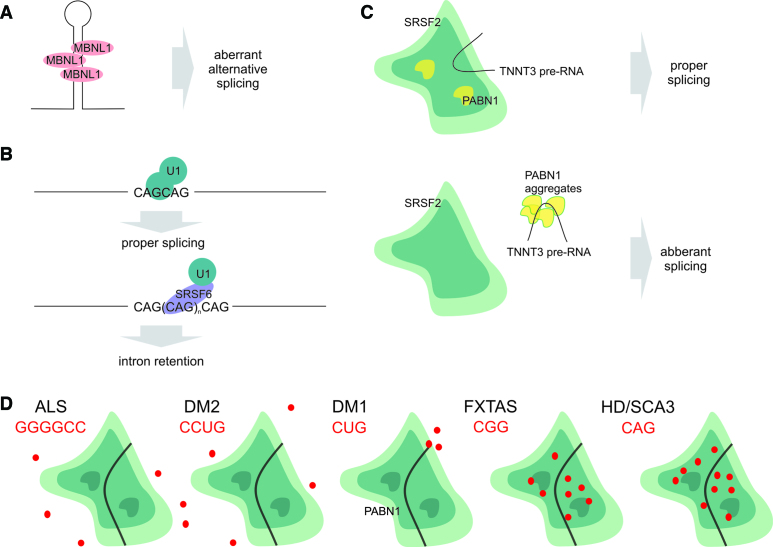
Figure 4.
(A) MBNL1 binds to expanded CUG RNA, resulting in aberrant alternative splicing in myotonic dystrophy type 1. (B) HTT mRNA splicing is disturbed by SRSF6 in a CAG length-dependent manner. (C) PABN1 aggregates bind pre-mRNA, which hinders splicing factor binding and results in aberrant splicing. (D) RNA foci harboring transcripts containing expanded simple repeats have different localization patterns in relation to NSs. Schematic representations of RNA foci localization are shown.
In cellular models of myotonic dystrophy type 2 (DM2), in which expanded CCUG repeats are present in the intronic sequence of the ZNF9 gene, there is no connection between NSs and RNA foci (192). Similar to repeats in DM2, RNAs with expanded GGGGCC repeats localized in the intronic sequences of C9orf72 RNA, which are found in a fraction of ALS patients, localize mostly outside NSs (200). These observations can be explained by both types of repeat-containing RNAs found in RNA foci being excised introns, unlike mutant mRNAs in triplet repeat expansion diseases in which RNA foci are associated with NSs.
As mutant RNAs interact abnormally with various proteins (201), transcripts with expanded repeats may trigger protein dislocation. This dislocation can have a dual effect: the sequestered proteins cannot perform their functions in their normal location as well as they can perform their aberrant function in their new location, but loss of protein function is more common, as shown for the MBNL1 protein in DM1 (Figure 4A). Similar mechanism was recently shown for oculopharyngeal muscular dystrophy (OPMD), which is caused by a GCG expansion within the PABPN1 gene; the mutant PABPN1 protein delocalized transcripts from NSs, resulting in decreased activity of splicing factors residing in NSs toward these transcripts (Figure 4C) (202). Interestingly, in OPMD, a mutant protein is involved in NS aberrations, as the polyadenylation function of the mutant protein is altered (203).
An investigation of the subnuclear localization of foci formed by RNAs containing various expanded repeats showed that their localization differs; transcripts containing expanded CAG and CGG repeats localize within NSs, those containing CUG repeats localize on the periphery of the NSs and those containing GGGGCC and CCUG repeats localize outside of NSs (Figure 4D). A single mechanism that triggers the disturbances in nuclear export of all mutant RNAs is thus unlikely.
SUMMARY AND FUTURE PERSPECTIVES
NSs were initially thought to be sites for storage and modification of splicing factors but have been now recognized as nuclear bodies that facilitate integrated regulation of gene expression. In this review, we summarized the current knowledge regarding the molecular composition and structural organization of these nuclear bodies. We discussed their properties and maintaining forces, fate during the cell cycle, canonical and newly postulated physiological functions and role in human disease. In characterizing these nuclear structures, our insights frequently expanded beyond the borders of NSs themselves into their nuclear environment. In this section, we put studies on NSs in a still broader context of high resolution mapping of the entire nuclear structure and function in the dimensions of space and time, which is the scope of such megaprojects as the 4D Nucleome Project (https://www.4dnucleome.org/) and the Human Protein Atlas (204). One of the goals of these initiatives is to gain deeper insight into the structure and function of nuclear compartments and nuclear bodies, including NSs. The 4D Nucleome Project has great potential for developing new methods capable of determining the molecular composition of NSs and methods for probing protein–protein and RNA–protein interactions within these structures as well as interactions between components of NSs and chromatin.
Also of great importance are new labeling and imaging methods to visualize NS constituents at high resolution. Such methods include CRISPR/Cas9-based labeling systems (205), super-resolution microscopic approaches (206) and cryo_-_EM (cryo-EM) (207) with cryo-electron tomography (cryo-ET) (208) for three-dimensional (3D) reconstruction of biological structures. The latter methods are based on EM imaging of frozen samples and do not require fixation or crystallization and thus, cryo-EM/ET allows native structures to be preserved and avoids artifacts. These methods enabled determination of the structure of the 21 MDa supraspliceosome (four spliceosomes gathered on pre-mRNA, ∼50 nm) (209) and the 100 MDa polyribosome complex consisting of 20 ribosomes bound to mRNA (about one order of magnitude smaller than NSs) (210). Recent progress in cryo-EM instrumentation (electron detectors) and image processing methods allowed the structure of native ribosomes (211) and spliceosomes (212) to be resolved at near-atomic-resolution.
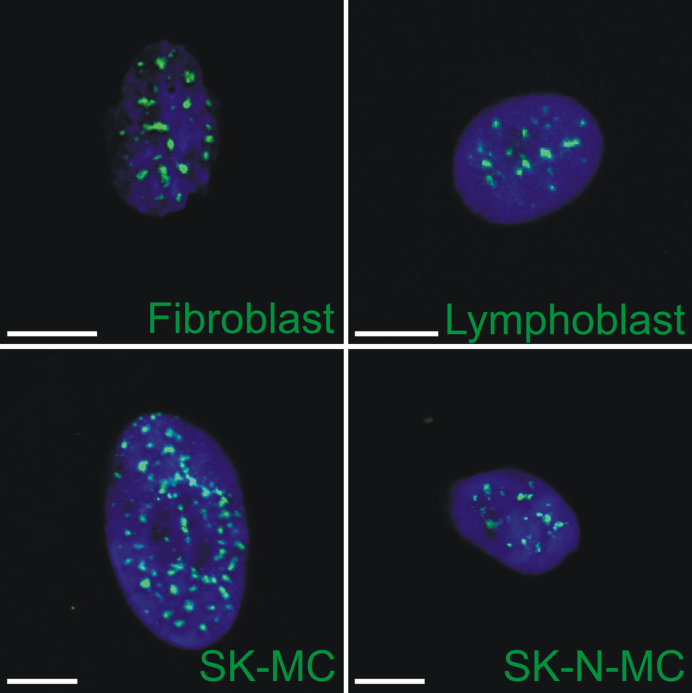
Structural studies of highly dynamic assemblies such as NSs remain challenging because protein composition and the shape and size of distinct NSs differ within individual cells and between various analyzed human cell types (Figure 5). The presence of multiple LCRs and the flexibility of NS proteins adds additional difficulty. However, new developments in cryo-EM/ET allow analysis of difficult biological specimens. Single particle analysis on one hand and averaging, which is obtained by merging corresponding structures, on the other hand, increase structure resolution. Additionally, image classification methods aid in managing sample heterogeneity (213). Furthermore, and most importantly for structures as large as NSs (0.8–1.8 μm), cellular cryo-ET enables visualization of particles in the native state inside cells. Analysis of thick biological samples without biochemical purification is possible due to progress in non-disruptive preparation of thin sections of biological material (214). In the near future, progress in cryo-ET is expected to allow further development of visual proteomics, i.e. 3D reconstruction of the detailed localization, conformation and interactions of proteins in a complex environment based on fitting of known structures from crystallography and nuclear magnetic resonance or solving structures de novo. Recently, the 26S proteasomes in intact neurons were visualized using this technology (215). Therefore, it is reasonable to expect that application of these technologies will bring new insights into the molecular organization of NSs and clarify their role in cell physiology and the molecular basis of diseases.
Figure 5.
NSs differ considerably among various human cell lines. Representative microscopy images of cells labeled for SRSF2 (constitutive protein of NSs) are presented. Due to the different expression levels of SRSF2 in different cell lines, the anti-SRSF2 antibody was used at different dilutions: 1:500 in fibroblasts, 1:500 in SK-MCs, 1:200 in lymphoblasts and 1:500 in SK-N-MCs. Bars = 10 μm (for the fibroblast, SK-MC and SK-N-MC) and 5 μm (for the lymphoblast).
Supplementary Material
Supplementary Data
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre [2012/06/A/NZ1/00094 to W.J.K., 2015/19/B/NZ2/02453 to W.J.K., 2015/17/N/NZ3/03629 to M.O.U.]; L’Oreal Poland Women in Science Fellowship (to M.O.U.); Polish Ministry of Science and Higher Education (KNOW program for years 2014–2019). Funding for open access charge: National Science Centre—Poland [2015/19/B/NZ2/02453].
Conflict of interest statement. None declared.
REFERENCES
- 1.Dundr M., Misteli T.. Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010; 2:a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao Y., Zhang B., Spector D.. Biogenesis and function of nuclear bodies. Trends Genet. 2011; 27:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr. Opin. Cell Biol. 2012; 24:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spector D.L., Lamond A.I.. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011; 3:a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin U., Lallemand-Breitenbach V., de Thé H.. PML nuclear bodies: regulation, function and therapeutic perspectives. J. Pathol. 2014; 234:289–291. [DOI] [PubMed] [Google Scholar]
- 6.Sleeman J.E., Trinkle-Mulcahy L.. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr. Opin. Cell Biol. 2014; 28:76–83. [DOI] [PubMed] [Google Scholar]
- 7.Spector D.L., Schrier W.H., Busch H.. Immunoelectron microscopic localization of snRNPs. Biol. Cell. 1983; 49:1–10. [DOI] [PubMed] [Google Scholar]
- 8.Perraud M., Gioud M., Monier J.. Intranuclear struc- tures recognized by autoantibodies against ribonucleo- proteins: Study on monkey kidney cells in culture using immunofluorescent techniques and immunoelectron microscopy. Ann. Immunol. 1979; 130:635–647. [Google Scholar]
- 9.Spector D.L., Fu X.D., Maniatis T.. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991; 10:3467–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biggiogera M., Fakan S.. Fine structural specific visualization of RNA on ultrathin sections. J. Histochem. Cytochem. 1998; 46:389–395. [DOI] [PubMed] [Google Scholar]
- 11.Girard C., Will C.L., Peng J., Makarov E.M., Kastner B., Lemm I., Urlaub H., Hartmuth K., Lührmann R.. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012; 3:994. [DOI] [PubMed] [Google Scholar]
- 12.Dias A.P., Dufu K., Lei H., Reed R.. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 2010; 1:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck J.S. Variations in the morphological patterns of ‘autoimmune’ nuclear fluorescence. Lancet. 1961; 1:1203–1205. [DOI] [PubMed] [Google Scholar]
- 14.Monneron A., Bernhard W.. Fine structural organization of the interphase nucleus in some mammalian cells. J. Ultrastruct. Res. 1969; 27:266–288. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q., Kota K.P., Alam S.G., Nickerson J.A., Dickinson R.B., Lele T.P.. Coordinated dynamics of RNA splicing speckles in the nucleus. J. Cell. Physiol. 2016; 231:1269–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007; 128:787–800. [DOI] [PubMed] [Google Scholar]
- 17.Sinclair G.D., Brasch K.. The reversible action of alpha-amanitin on nuclear structure and molecular composition. Exp. Cell Res. 1978; 111:1–14. [DOI] [PubMed] [Google Scholar]
- 18.Pliss A., Peng X., Liu L., Kuzmin A., Wang Y., Qu J., Li Y., Prasad P.N.. Single cell assay for molecular diagnostics and medicine: monitoring Intracellular concentrations of macromolecules by two-photon fluorescence lifetime imaging. Theranostics. 2015; 5:919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pliss A., Kuzmin A.N., Kachynski A. V., Prasad P.N.. Nonlinear optical imaging and raman microspectrometry of the cell nucleus throughout the cell cycle. Biophys. J. 2010; 99:3483–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phair R.D., Misteli T.. High mobility of proteins in the mammalian cell nucleus. Nature. 2000; 404:604–609. [DOI] [PubMed] [Google Scholar]
- 21.Mintz P.J., Patterson S.D., Neuwald A.F., Spahr C.S., Spector D.L.. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999; 18:4308–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitoh N., Spahr C.S., Patterson S.D., Bubulya P., Neuwald A.F., Spector D.L.. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell. 2004; 15:3876–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marzahn M.R., Marada S., Lee J., Nourse A., Kenrick S., Zhao H., Ben-Nissan G., Kolaitis R.-M., Peters J.L., Pounds S. et al. . Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016; 35:1254–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V, King D.S., Banani S.F. et al. . Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012; 483:336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D. et al. . Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015; 57:936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada M., Jang S.-W., Ye K.. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8649–8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W.-Y., Lee W.-C., Hsu N.-C., Huang F., Chung B.-C.. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 2004; 279:38730–38735. [DOI] [PubMed] [Google Scholar]
- 28.Gui J.F., Lane W.S., Fu X.D.. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994; 369:678–682. [DOI] [PubMed] [Google Scholar]
- 29.Colwill K., Pawson T., Andrews B., Prasad J., Manley J.L., Bell J.C., Duncan P.I.. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996; 15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 30.Misteli T., Spector D.L.. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol. Cell. 1996; 7:1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang S., Gapsys V., Kim H.-Y., Bessonov S., Hsiao H.-H., Möhlmann S., Klaukien V., Ficner R., Becker S., Urlaub H. et al. . Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure. 2013; 21:2162–2174. [DOI] [PubMed] [Google Scholar]
- 32.Salichs E., Ledda A., Mularoni L., Albà M.M., de la Luna S.. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009; 5:e1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarez M., Estivill X., de la Luna S.. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 2003; 116:3099–3107. [DOI] [PubMed] [Google Scholar]
- 34.Ferreira J.A., Carmo-Fonseca M., Lamond A.I.. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J. Cell Biol. 1994; 126:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leser G.P., Fakan S., Martin T.E.. Ultrastructural distribution of ribonucleoprotein complexes during mitosis. snRNP antigens are contained in mitotic granule clusters. Eur. J. Cell Biol. 1989; 50:376–389. [PubMed] [Google Scholar]
- 36.Prasanth K. V., Sacco-Bubulya P.A., Prasanth S.G., Spector D.L.. Sequential entry of components of gene expression machinery into daughter nuclei. Mol. Biol. Cell. 2003; 14:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bubulya P.A., Prasanth K. V., Deerinck T.J., Gerlich D., Beaudouin J., Ellisman M.H., Ellenberg J., Spector D.L.. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 2004; 167:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown J.M., Green J., das Neves R.P., Wallace H.A.C.C., Smith A.J.H.H., Hughes J., Gray N., Taylor S., Wood W.G., Higgs D.R. et al. . Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 2008; 182:1083–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shevtsov S.P., Dundr M.. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2010; 13:167–173. [DOI] [PubMed] [Google Scholar]
- 40.Herrmann A., Fleischer K., Czajkowska H., Müller-Newen G., Becker W.. Characterization of cyclin L1 as an immobile component of the splicing factor compartment. FASEB J. 2007; 21:3142–3152. [DOI] [PubMed] [Google Scholar]
- 41.Liu J.-L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J.G.. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006; 172:875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki Y.T.F., Ideue T., Sano M., Mituyama T., Hirose T.. MEN / noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox A.H., Lam Y.W., Leung A.K.L., Lyon C.E., Andersen J., Mann M., Lamond A.I.. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002; 12:13–25. [DOI] [PubMed] [Google Scholar]
- 44.Marnef A., Weil D., Standart N.. RNA-related nuclear functions of human Pat1b, the P-body mRNA decay factor. Mol. Biol. Cell. 2012; 23:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatron-Colliet A., Lalun N., Terryn C., Kurdykowski S., Lorenzato M., Rusciani A., Ploton D., Duca L., Bobichon H.. The elastin peptide (VGVAPG)3 induces the 3D reorganisation of PML-NBs and SC35 speckles architecture, and accelerates proliferation of fibroblasts and melanoma cells. Histochem. Cell Biol. 2015; 143:245–258. [DOI] [PubMed] [Google Scholar]
- 46.Yin X., Jin N., Gu J., Shi J., Zhou J., Gong C.-X., Iqbal K., Grundke-Iqbal I., Liu F.. Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) modulates serine/arginine-rich protein 55 (SRp55)-promoted Tau exon 10 inclusion. J. Biol. Chem. 2012; 287:30497–30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cáceres J.F., Screaton G.R., Krainer A.R.. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998; 12:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koizumi J., Okamoto Y., Onogi H., Mayeda A., Krainer A.R., Hagiwara M.. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs). J. Biol. Chem. 1999; 274:11125–11131. [DOI] [PubMed] [Google Scholar]
- 49.Cho S., Hoang A., Sinha R., Zhong X.-Y., Fu X.-D., Krainer A.R., Ghosh G.. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Misteli T., Cáceres J.F., Clement J.Q., Krainer A.R., Wilkinson M.F., Spector D.L.. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998; 143:297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y., Yario T.A., Steitz J.A.. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9666–9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michlewski G., Sanford J.R., Cáceres J.F.. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell. 2008; 30:179–189. [DOI] [PubMed] [Google Scholar]
- 53.Boronenkov I.V., Loijens J.C., Umeda M., Anderson R.A.. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell. 1998; 9:3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tabellini G., Bortul R., Santi S., Riccio M., Baldini G., Cappellini A., Billi A.M., Berezney R., Ruggeri A., Cocco L. et al. . Diacylglycerol kinase-theta is localized in the speckle domains of the nucleus. Exp. Cell Res. 2003; 287:143–154. [DOI] [PubMed] [Google Scholar]
- 55.Li D., Urs A.N., Allegood J., Leon A., Merrill A.H., Sewer M.B.. Cyclic AMP-stimulated interaction between steroidogenic factor 1 and diacylglycerol kinase theta facilitates induction of CYP17. Mol. Cell. Biol. 2007; 27:6669–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meerschaert K., Tun M.P., Remue E., De Ganck A., Boucherie C., Vanloo B., Degeest G., Vandekerckhove J., Zimmermann P., Bhardwaj N. et al. . The PDZ2 domain of zonula occludens-1 and -2 is a phosphoinositide binding domain. Cell. Mol. Life Sci. 2009; 66:3951–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mortier E., Wuytens G., Leenaerts I., Hannes F., Heung M.Y., Degeest G., David G., Zimmermann P.. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J. 2005; 24:2556–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mellman D.L., Gonzales M.L., Song C., Barlow C.A., Wang P., Kendziorski C., Anderson R.A.. A PtdIns4, 5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008; 451:1013–1017. [DOI] [PubMed] [Google Scholar]
- 59.Krauss S.W., Chen C., Penman S., Heald R.. Nuclear actin and protein 4.1: essential interactions during nuclear assembly in vitro. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:10752–10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hofmann W.A., Stojiljkovic L., Fuchsova B., Vargas G.M., Mavrommatis E., Philimonenko V., Kysela K., Goodrich J.A., Lessard J.L., Hope T.J. et al. . Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 2004; 6:1094–1101. [DOI] [PubMed] [Google Scholar]
- 61.Percipalle P., Fomproix N., Kylberg K., Miralles F., Bjorkroth B., Daneholt B., Visa N.. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:6475–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofmann W.A., Vargas G.M., Ramchandran R., Stojiljkovic L., Goodrich J.A., de Lanerolle P.. Nuclear myosin I is necessary for the formation of the first phosphodiester bond during transcription initiation by RNA polymerase II. J. Cell. Biochem. 2006; 99:1001–1009. [DOI] [PubMed] [Google Scholar]
- 63.Wang I.-F., Chang H.-Y., James Shen C.-K.. Actin-based modeling of a transcriptionally competent nuclear substructure induced by transcription inhibition. Exp. Cell Res. 2006; 312:3796–3807. [DOI] [PubMed] [Google Scholar]
- 64.Belin B.J., Cimini B.A., Blackburn E.H., Mullins R.D.. Visualization of actin filaments and monomers in somatic cell nuclei. Mol. Biol. Cell. 2013; 24:982–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baldin V., Militello M., Thomas Y., Doucet C., Fic W., Boireau S., Jariel-Encontre I., Piechaczyk M., Bertrand E., Tazi J. et al. . A novel role for PA28gamma-proteasome in nuclear speckle organization and SR protein trafficking. Mol. Biol. Cell. 2008; 19:1706–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song E.J., Werner S.L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J.W., Elledge S.J., Kirschner M.W., Rape M.. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010; 24:1434–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang L., Lin C., Liu W., Zhang J., Ohgi K.A., Grinstein J.D., Dorrestein P.C., Rosenfeld M.G.. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011; 147:773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sánchez-Álvarez M., Montes M., Sánchez-Hernández N., Hernández-Munain C., Suñé C.. Differential effects of sumoylation on transcription and alternative splicing by transcription elongation regulator 1 (TCERG1). J. Biol. Chem. 2010; 285:15220–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carter K.C., Taneja K.L., Lawrence J.B.. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 1991; 115:1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shopland L.S., Johnson C.V., Lawrence J.B.. Evidence that all SC-35 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J. Struct. Biol. 2002; 140:131–139. [DOI] [PubMed] [Google Scholar]
- 71.Huang S., Deerinck T.J., Ellisman M.H., Spector D.L.. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol. 1994; 126:877–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Politz J.C.R., Tuft R.A., Prasanth K.V., Baudendistel N., Fogarty K.E., Lifshitz L.M., Langowski J., Spector D.L., Pederson T.. Rapid, diffusional shuttling of Poly(A) RNA between nuclear speckles and the nucleoplasm. Mol. Biol. Cell. 2006; 17:1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shav-Tal Y., Darzacq X., Shenoy S.M., Fusco D., Janicki S.M., Spector D.L., Singer R.H.. Dynamics of single mRNPs in nuclei of living cells. Science. 2004; 304:1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palazzo A.F., Springer M., Shibata Y., Lee C.-S., Dias A.P., Rapoport T.A.. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007; 5:e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calapez A., Pereira H.M., Calado A., Braga J., Rino J., Carvalho C., Tavanez J.P., Wahle E., Rosa A.C., Carmo-Fonseca M.. The intranuclear mobility of messenger RNA binding proteins is ATP dependent and temperature sensitive. J. Cell Biol. 2002; 159:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Molenaar C., Abdulle A., Gena A., Tanke H.J., Dirks R.W.. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J. Cell Biol. 2004; 165:191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tokunaga K., Shibuya T., Ishihama Y., Tadakuma H., Ide M., Yoshida M., Funatsu T., Ohshima Y., Tani T.. Nucleocytoplasmic transport of fluorescent mRNA in living mammalian cells: nuclear mRNA export is coupled to ongoing gene transcription. Genes Cells. 2006; 11:305–317. [DOI] [PubMed] [Google Scholar]
- 78.Baurén G., Wieslander L.. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994; 76:183–192. [DOI] [PubMed] [Google Scholar]
- 79.Zhang G., Taneja K.L., Singer R.H., Green M.R.. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994; 372:809–812. [DOI] [PubMed] [Google Scholar]
- 80.Ishihama Y., Tadakuma H., Tani T., Funatsu T.. The dynamics of pre-mRNAs and poly(A)+ RNA at speckles in living cells revealed by iFRAP studies. Exp. Cell Res. 2008; 314:748–762. [DOI] [PubMed] [Google Scholar]
- 81.Johnson C., Primorac D., McKinstry M., McNeil J., Rowe D., Lawrence J.B.. Tracking COL1A1 RNA in osteogenesis imperfecta: splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 2000; 150:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prasanth K. V., Camiolo M., Chan G., Tripathi V., Denis L., Nakamura T., Hubner M.R., Spector D.L.. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol. Biol. Cell. 2010; 21:4184–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A.. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007; 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bernard D., Prasanth K. V, Tripathi V., Colasse S., Nakamura T., Xuan Z., Zhang M.Q., Sedel F., Jourdren L., Coulpier F. et al. . A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010; 29:3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilusz J.E., Freier S.M., Spector D.L.. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008; 135:919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miyagawa R., Tano K., Mizuno R., Nakamura Y., Ijiri K., Rakwal R., Shibato J., Masuo Y., Mayeda A., Hirose T. et al. . Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012; 18:738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Änkö M.-L., Müller-McNicoll M., Brandl H., Curk T., Gorup C., Henry I., Ule J., Neugebauer K.M.. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012; 13:R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanford J.R., Wang X., Mort M., Vanduyn N., Cooper D.N., Mooney S.D., Edenberg H.J., Liu Y.. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009; 19:381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Engreitz J.M., Sirokman K., McDonel P., Shishkin A.A., Surka C., Russell P., Grossman S.R., Chow A.Y., Guttman M., Lander E.S.. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014; 159:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. et al. . The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010; 39:925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang B., Arun G., Mao Y.S., Lazar Z., Hung G., Bhattacharjee G., Xiao X., Booth C.J., Wu J., Zhang C. et al. . The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012; 2:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakagawa S., Ip J.Y., Shioi G., Tripathi V., Zong X., Hirose T., Prasanth K. V.. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012; 18:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bunch H., Zheng X., Burkholder A., Dillon S.T., Motola S., Birrane G., Ebmeier C.C., Levine S., Fargo D., Hu G. et al. . TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat. Struct. Mol. Biol. 2014; 21:876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L.. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997; 385:357–361. [DOI] [PubMed] [Google Scholar]
- 95.Buratowski S. The CTD code. Nat. Struct. Biol. 2003; 10:679–680. [DOI] [PubMed] [Google Scholar]
- 96.Gu B., Eick D., Bensaude O.. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 2013; 41:1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dow E.C., Liu H., Rice A.P.. T-loop phosphorylated Cdk9 localizes to nuclear speckle domains which may serve as sites of active P-TEFb function and exchange between the Brd4 and 7SK/HEXIM1 regulatory complexes. J. Cell. Physiol. 2010; 224:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Itzen F., Greifenberg A.K., Bösken C.A., Geyer M.. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014; 42:7577–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012; 149:1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li X., Manley J.L.. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 101.Brown J.M., Leach J., Reittie J.E., Atzberger A., Lee-Prudhoe J., Wood W.G., Higgs D.R., Iborra F.J., Buckle V.J.. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006; 172:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khanna N., Hu Y., Belmont A.S.. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr. Biol. 2014; 24:1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meggendorfer M., Weierich C., Wolff H., Brack-Werner R., Cremer T.. Functional nuclear topography of transcriptionally inducible extra-chromosomal transgene clusters. Chromosome Res. 2010; 18:401–417. [DOI] [PubMed] [Google Scholar]
- 104.Rieder D., Ploner C., Krogsdam A.M., Stocker G., Fischer M., Scheideler M., Dani C., Amri E.-Z., Müller W.G., McNally J.G. et al. . Co-expressed genes prepositioned in spatial neighborhoods stochastically associate with SC35 speckles and RNA polymerase II factories. Cell. Mol. Life Sci. 2014; 71:1741–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beyer A.L., Osheim Y.N.. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988; 2:754–765. [DOI] [PubMed] [Google Scholar]
- 106.Wahl M.C., Will C.L., Lührmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 107.Koga M., Hayashi M., Kaida D.. Splicing inhibition decreases phosphorylation level of Ser2 in Pol II CTD. Nucleic Acids Res. 2015; 43:8258–8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barboric M., Lenasi T., Chen H., Johansen E.B., Guo S., Peterlin B.M.. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7798–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Das R., Yu J., Zhang Z., Gygi M.P., Krainer A.R., Gygi S.P., Reed R.. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007; 26:867–881. [DOI] [PubMed] [Google Scholar]
- 110.David C.J., Boyne A.R., Millhouse S.R., Manley J.L.. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011; 25:972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alexander R.D., Innocente S.A., Barrass J.D., Beggs J.D.. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell. 2010; 40:582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goldstrohm A.C., Albrecht T.R., Suñé C., Bedford M.T., Garcia-Blanco M.A.. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 2001; 21:7617–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Batsché E., Yaniv M., Muchardt C.. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006; 13:22–29. [DOI] [PubMed] [Google Scholar]
- 114.Close P., East P., Dirac-Svejstrup A.B., Hartmann H., Heron M., Maslen S., Chariot A., Söding J., Skehel M., Svejstrup J.Q.. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012; 484:386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saint-André V., Batsché E., Rachez C., Muchardt C.. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat. Struct. Mol. Biol. 2011; 18:337–344. [DOI] [PubMed] [Google Scholar]
- 116.Schor I.E., Rascovan N., Pelisch F., Allo M., Kornblihtt A.R.. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:4325–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Loomis R.J., Naoe Y., Parker J.B., Savic V., Bozovsky M.R., Macfarlan T., Manley J.L., Chakravarti D.. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol. Cell. 2009; 33:450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Khan D.H., Gonzalez C., Cooper C., Sun J.-M., Chen H.Y., Healy S., Xu W., Smith K.T., Workman J.L., Leygue E. et al. . RNA-dependent dynamic histone acetylation regulates MCL1 alternative splicing. Nucleic Acids Res. 2014; 42:1656–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martinez E., Palhan V.B., Tjernberg A., Lymar E.S., Gamper A.M., Kundu T.K., Chait B.T., Roeder R.G.. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 2001; 21:6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee J.-H., Tate C.M., You J.-S., Skalnik D.G.. Identification and characterization of the human set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 2007; 282:13419–13428. [DOI] [PubMed] [Google Scholar]
- 121.Shkreta L., Chabot B.. The RNA splicing response to DNA damage. Biomolecules. 2015; 5:2935–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee J.H., Jeong S.A., Khadka P., Hong J., Chung I.K.. Involvement of SRSF11 in cell cycle-specific recruitment of telomerase to telomeres at nuclear speckles. Nucleic Acids Res. 2015; 43:8435–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kwan T., Benovoy D., Dias C., Gurd S., Provencher C., Beaulieu P., Hudson T.J., Sladek R., Majewski J.. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008; 40:225–231. [DOI] [PubMed] [Google Scholar]
- 124.Ke S., Alemu E.A., Mertens C., Gantman E.C., Fak J.J., Mele A., Haripal B., Zucker-Scharff I., Moore M.J., Park C.Y. et al. . A majority of m 6 A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015; 29:2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lian Z., Karpikov A., Lian J., Mahajan M.C., Hartman S., Gerstein M., Snyder M., Weissman S.M.. A genomic analysis of RNA polymerase II modification and chromatin architecture related to 3′ end RNA polyadenylation. Genome Res. 2008; 18:1224–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rigo F., Martinson H.G.. Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the Poly(A) signal couples to splicing before committing to cleavage. Mol. Cell. Biol. 2008; 28:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vagner S., Vagner C., Mattaj I.W.. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000; 14:403–413. [PMC free article] [PubMed] [Google Scholar]
- 128.Lutz C.S., Murthy K.G., Schek N., O’Connor J.P., Manley J.L., Alwine J.C.. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996; 10:325–337. [DOI] [PubMed] [Google Scholar]
- 129.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y.-G.Y., Yi C., Lindahl T., Pan T., Yang Y.-G.Y. et al. . N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågb⊘ C.B., Shi Y., Wang W.-L., Song S.-H. et al. . ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J.. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014; 10:927–929. [DOI] [PubMed] [Google Scholar]
- 132.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T.. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015; 518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Le Hir H., Izaurralde E., Maquat L.E., Moore M.J.. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000; 19:6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kataoka N., Yong J., Kim V.N., Velazquez F., Perkinson R.A., Wang F., Dreyfuss G.. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 2000; 6:673–682. [DOI] [PubMed] [Google Scholar]
- 135.Zimowska G., Shi J., Munguba G., Jackson M.R., Alpatov R., Simmons M.N., Shi Y., Sugrue S.P.. Pinin/DRS/memA interacts with SRp75, SRm300 and SRrp130 in corneal epithelial cells. Invest. Ophthalmol. Vis. Sci. 2003; 44:4715–4723. [DOI] [PubMed] [Google Scholar]
- 136.Dreumont N., Bourgeois C.F., Lejeune F., Liu Y., Ehrmann I.E., Elliott D.J., Stévenin J.. Human RBMY regulates germline-specific splicing events by modulating the function of the serine/arginine-rich proteins 9G8 and Tra2-{beta}. J. Cell Sci. 2010; 123:40–50. [DOI] [PubMed] [Google Scholar]
- 137.Luo M.L., Zhou Z., Magni K., Christoforides C., Rappsilber J., Mann M., Reed R.. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001; 413:644–647. [DOI] [PubMed] [Google Scholar]
- 138.Strässer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondón A.G., Aguilera A., Struhl K., Reed R. et al. . TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002; 417:304–308. [DOI] [PubMed] [Google Scholar]
- 139.Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R.. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005; 19:1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Viphakone N., Hautbergue G.M., Walsh M., Chang C.-T., Holland A., Folco E.G., Reed R., Wilson S.A.. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 2012; 3:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cheng H., Dufu K., Lee C.-S., Hsu J.L., Dias A., Reed R.. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006; 127:1389–1400. [DOI] [PubMed] [Google Scholar]
- 142.Rougemaille M., Dieppois G., Kisseleva-Romanova E., Gudipati R.K., Lemoine S., Blugeon C., Boulay J., Jensen T.H., Stutz F., Devaux F. et al. . THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008; 135:308–321. [DOI] [PubMed] [Google Scholar]
- 143.Valencia P., Dias A.P., Reed R.. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3386–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Libri D., Graziani N., Saguez C., Boulay J.. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 2001; 15:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Meinel D.M., Burkert-Kautzsch C., Kieser A., O’Duibhir E., Siebert M., Mayer A., Cramer P., Söding J., Holstege F.C.P., Sträßer K.. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA Polymerase II. PLoS Genet. 2013; 9:e1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chanarat S., Seizl M., Strasser K.. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 2011; 25:1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang Y., Gattoni R., Stévenin J., Steitz J.A.. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003; 11:837–843. [DOI] [PubMed] [Google Scholar]
- 148.Lei H., Zhai B., Yin S., Gygi S., Reed R.. Evidence that a consensus element found in naturally intronless mRNAs promotes mRNA export. Nucleic Acids Res. 2013; 41:2517–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Woodward L.A., Mabin J.W., Gangras P., Singh G.. The exon junction complex: a lifelong guardian of mRNA fate. Wiley Interdiscip. Rev. RNA. 2016; 8:e1411. [DOI] [PubMed] [Google Scholar]
- 150.Morimoto M., Boerkoel C.F.. The role of nuclear bodies in gene expression and disease. Biology (Basel). 2013; 2:976–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Quesada V., Conde L., Villamor N., Ordóñez G.R., Jares P., Bassaganyas L., Ramsay A.J., Beà S., Pinyol M., Martínez-Trillos A. et al. . Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011; 44:47–52. [DOI] [PubMed] [Google Scholar]
- 152.Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M. et al. . Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011; 478:64–69. [DOI] [PubMed] [Google Scholar]
- 153.Anczuków O., Krainer A.R.. Splicing-factor alterations in cancers. RNA. 2016; 22:1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Anczuków O., Rosenberg A.Z., Akerman M., Das S., Zhan L., Karni R., Muthuswamy S.K., Krainer A.R.. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 2012; 19:220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Maimon A., Mogilevsky M., Shilo A., Golan-Gerstl R., Obiedat A., Ben-Hur V., Lebenthal-Loinger I., Stein I., Reich R., Beenstock J. et al. . Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts ras-induced transformation. Cell Rep. 2014; 7:501–513. [DOI] [PubMed] [Google Scholar]
- 156.Golan-Gerstl R., Cohen M., Shilo A., Suh S.S., Bakàcs A., Coppola L., Karni R.. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011; 71:4464–4472. [DOI] [PubMed] [Google Scholar]
- 157.Mayr C., Bartel D.P.. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009; 138:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jenkins J.L., Kielkopf C.L.. Splicing factor mutations in myelodysplasias: insights from spliceosome structures. Trends Genet. 2017; 33:336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mor A., White A., Zhang K., Thompson M., Esparza M., Muñoz-Moreno R., Koide K., Lynch K.W., García-Sastre A., Fontoura B.M.A.. Influenza virus mRNA trafficking through host nuclear speckles. Nat. Microbiol. 2016; 1:16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marchand V., Santerre M., Aigueperse C., Fouillen L., Saliou J.M., Van Dorsselaer A., Sanglier-Cianférani S., Branlant C., Motorin Y.. Identification of protein partners of the human immunodeficiency virus 1 tat/rev exon 3 leads to the discovery of a new HIV-1 splicing regulator, protein hnRNP K. RNA Biol. 2011; 8:325–342. [DOI] [PubMed] [Google Scholar]
- 161.Tazi J., Bakkour N., Marchand V., Ayadi L., Aboufirassi A., Branlant C.. Alternative splicing: regulation of HIV-1 multiplication as a target for therapeutic action. FEBS J. 2010; 277:867–876. [DOI] [PubMed] [Google Scholar]
- 162.Bedard K.M., Daijogo S., Semler B.L.. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007; 26:459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dowling D., Nasr-Esfahani S., Tan C.H., O’Brien K., Howard J.L., Jans D.a, Purcell D.F.J., Stoltzfus C.M., Sonza S.. HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology. 2008; 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bridge E., Xia D.X., Carmo-Fonseca M., Cardinali B., Lamond A.I., Pettersson U.. Dynamic organization of splicing factors in adenovirus-infected cells. J. Virol. 1995; 69:281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nemeroff M.E., Barabino S.M., Li Y., Keller W., Krug R.M.. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell. 1998; 1:991–1000. [DOI] [PubMed] [Google Scholar]
- 166.Rivera-Serrano E.E., Fritch E.J., Scholl E.H., Sherry B.. A cytoplasmic RNA virus alters the function of the cell splicing protein SRSF2. J. Virol. 2017; 91, doi:10.1128/JVI.02488-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bai B., Hales C.M., Chen P.C., Gozal Y., Dammer E.B., Fritz J.J., Wang X., Xia Q., Duong D.M., Street C. et al. . U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:16562–16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Muresan Z., Muresan V.. A phosphorylated, carboxy-terminal fragment of beta-amyloid precursor protein localizes to the splicing factor compartment. Hum. Mol. Genet. 2004; 13:475–488. [DOI] [PubMed] [Google Scholar]
- 169.Ferrer I., Barrachina M., Puig B.. Glycogen synthase kinase-3 is associated with neuronal and glial hyperphosphorylated tau deposits in Alzheimer's disease, Pick's disease, progressive supranuclear palsy and corticobasal degeneration. Acta Neuropathol. 2002; 104:583–591. [DOI] [PubMed] [Google Scholar]
- 170.Hernández F., Pérez M., Lucas J.J., Mata A.M., Bhat R., Avila J.. Glycogen synthase kinase-3 plays a crucial role in tau exon 10 splicing and intranuclear distribution of SC35. Implications for Alzheimer's disease. J. Biol. Chem. 2004; 279:3801–3806. [DOI] [PubMed] [Google Scholar]
- 171.Gaweda-Walerych K., Mohagheghi F., Zekanowski C., Buratti E.. Parkinson's disease-related gene variants influence pre-mRNA splicing processes. Neurobiol. Aging. 2016; 47:127–138. [DOI] [PubMed] [Google Scholar]
- 172.La Cognata V., D’Agata V., Cavalcanti F., Cavallaro S.. Splicing: is there an alternative contribution to Parkinson's disease. Neurogenetics. 2015; 16:245–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang I.F., Reddy N.M., Shen C.K.. Higher order arrangement of the eukaryotic nuclear bodies. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Neumann M., Kwong L.K., Sampathu D.M., Trojanowski J.Q., Lee V.M.-Y.. TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: protein misfolding diseases without amyloidosis. Arch. Neurol. 2007; 64:1388–1394. [DOI] [PubMed] [Google Scholar]
- 175.Wang W., Li L., Lin W.L., Dickson D.W., Petrucelli L., Zhang T., Wang X.. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum. Mol. Genet. 2013; 22:4706–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A.L., Župunski V. et al. . Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011; 14:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.-C., Sun E., Wancewicz E., Mazur C. et al. . Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011; 14:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ling J.P., Pletnikova O., Troncoso J.C., Wong P.C.. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015; 349:650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.-C.C., Liang T.Y., Mazur C. et al. . Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 2012; 15:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vance C., Scotter E.L., Nishimura A.L., Troakes C., Mitchell J.C., Kathe C., Urwin H., Manser C., Miller C.C., Hortobágyi T. et al. . ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum. Mol. Genet. 2013; 22:2676–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Young P.J., Le T.T., thi Man N., Burghes A.H., Morris G.E.. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 2000; 256:365–374. [DOI] [PubMed] [Google Scholar]
- 182.Young P.J., Le T.T., Dunckley M., Nguyen T.M., Burghes A.H., Morris G.E.. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp. Cell Res. 2001; 265:252–261. [DOI] [PubMed] [Google Scholar]
- 183.Woulfe J. Nuclear bodies in neurodegenerative disease. Biochim. Biophys. Acta. 2008; 1783:2195–2206. [DOI] [PubMed] [Google Scholar]
- 184.Nguyen L.S., Kim H.G., Rosenfeld J.A., Shen Y., Gusella J.F., Lacassie Y., Layman L.C., Shaffer L.G., Gécz J.. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum. Mol. Genet. 2013; 22:1816–1825. [DOI] [PubMed] [Google Scholar]
- 185.Favaro F.P., Alvizi L., Zechi-Ceide R.M., Bertola D., Felix T.M., de Souza J., Raskin S., Twigg S.R., Weiner A.M., Armas P. et al. . A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira syndrome, a craniofacial disorder associated with limb defects. Am. J. Hum. Genet. 2014; 94:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.McMahon J.J., Miller E.E., Silver D.L.. The exon junction complex in neural development and neurodevelopmental disease. Int. J. Dev. Neurosci. 2016; 55:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.O’Donovan S.M., Hasselfeld K., Bauer D., Simmons M., Roussos P., Haroutunian V., Meador-Woodruff J.H., McCullumsmith R.E.. Glutamate transporter splice variant expression in an enriched pyramidal cell population in schizophrenia. Transl. Psychiatry. 2015; 5:e579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oldmeadow C., Mossman D., Evans T.-J., Holliday E.G., Tooney P.A., Cairns M.J., Wu J., Carr V., Attia J.R., Scott R.J.. Combined analysis of exon splicing and genome wide polymorphism data predict schizophrenia risk loci. J. Psychiatr. Res. 2014; 52:44–49. [DOI] [PubMed] [Google Scholar]
- 189.Irimia M., Weatheritt R.J., Ellis J.D., Parikshak N.N., Gonatopoulos-Pournatzis T., Babor M., Quesnel-Vallières M., Tapial J., Raj B., O’Hanlon D. et al. . A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014; 159:1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Quesnel-Vallières M., Irimia M., Cordes S.P., Blencowe B.J.. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev. 2015; 29:746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H.. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995; 128:995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Holt I., Mittal S., Furling D., Butler-Browne G.S., Brook J.D., Morris G.E., David Brook J., Morris G.E.. Defective mRNA in myotonic dystrophy accumulates at the periphery of nuclear splicing speckles. Genes Cells. 2007; 12:1035–1048. [DOI] [PubMed] [Google Scholar]
- 193.Urbanek M.O., Jazurek M., Switonski P.M., Figura G., Krzyzosiak W.J.. Nuclear speckles are detention centers for transcripts containing expanded CAG repeats. Biochim. Biophys. Acta. 2016; 1862:1513–1520. [DOI] [PubMed] [Google Scholar]
- 194.Smith K.P., Byron M., Johnson C., Xing Y., Lawrence J.B.. Defining early steps in mRNA transport: mutant mRNA in myotonic dystrophy type I is blocked at entry into SC-35 domains. J. Cell Biol. 2007; 178:951–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Urbanek M.O., Krzyzosiak W.J.. RNA FISH for detecting expanded repeats in human diseases. Methods. 2016; 98:115–123. [DOI] [PubMed] [Google Scholar]
- 196.Jain A., Vale R.D.. RNA phase transitions in repeat expansion disorders. Nature. 2017; 546:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fernández-Nogales M., Cabrera J.R., Santos-Galindo M., Hoozemans J.J.M., Ferrer I., Rozemuller A.J.M., Hernández F., Avila J., Lucas J.J., Fernandez-Nogales M. et al. . Huntington's disease is a four-repeat tauopathy with tau nuclear rods. Nat. Med. 2014; 20:881–885. [DOI] [PubMed] [Google Scholar]
- 198.Gipson T.A., Neueder A., Wexler N.S., Bates G.P., Housman D.. Aberrantly spliced HTT, a new player in Huntington's disease pathogenesis. RNA Biol. 2013; 10:1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sellier C., Rau F., Liu Y., Tassone F., Hukema R.K., Gattoni R., Schneider A., Richard S., Willemsen R., Elliott D.J. et al. . Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010; 29:1248–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C. et al. . Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013; 5:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Galka-Marciniak P., Urbanek M.O., Krzyzosiak W.J.. Triplet repeats in transcripts: structural insights into RNA toxicity. Biol. Chem. 2012; 393:1299–1315. [DOI] [PubMed] [Google Scholar]
- 202.Klein P., Oloko M., Roth F., Montel V., Malerba A., Jarmin S., Gidaro T., Popplewell L., Perie S., Lacau St Guily J. et al. . Nuclear poly(A)-binding protein aggregates misplace a pre-mRNA outside of SC35 speckle causing its abnormal splicing. Nucleic Acids Res. 2016; 44:10929–10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kim Y.J., Noguchi S., Hayashi Y.K., Tsukahara T., Shimizu T., Arahata K.. The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression. Hum. Mol. Genet. 2001; 10:1129–1139. [DOI] [PubMed] [Google Scholar]
- 204.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. et al. . Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010; 28:1248–1250. [DOI] [PubMed] [Google Scholar]
- 205.Zhou Y., Wang P., Tian F., Gao G., Huang L., Wei W., Xie X.S.. Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging. Cell Res. 2017; 27:298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sydor A.M., Czymmek K.J., Puchner E.M., Mennella V.. Super-resolution microscopy: from single molecules to supramolecular assemblies. Trends Cell Biol. 2015; 25:730–748. [DOI] [PubMed] [Google Scholar]
- 207.Murata K., Wolf M.. Cryo-electron microscopy for structural analysis of dynamic biological macromolecules. Biochim. Biophys. Acta. 2017; doi:10.1016/j.bbagen.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 208.Oikonomou C.M., Jensen G.J.. Cellular electron cryotomography: toward structural biology in situ. Annu. Rev. Biochem. 2017; 86:873–896. [DOI] [PubMed] [Google Scholar]
- 209.Azubel M., Wolf S.G., Sperling J., Sperling R.. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol. Cell. 2004; 15:833–839. [DOI] [PubMed] [Google Scholar]
- 210.Myasnikov A.G., Afonina Z.A., Ménétret J.-F., Shirokov V.A., Spirin A.S., Klaholz B.P.. The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat. Commun. 2014; 5:5294. [DOI] [PubMed] [Google Scholar]
- 211.Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P.. Structure of the human 80S ribosome. Nature. 2015; 520:640–645. [DOI] [PubMed] [Google Scholar]
- 212.Zhang X., Yan C., Hang J., Finci L.I., Lei J., Shi Y.. An atomic structure of the human spliceosome. Cell. 2017; 169:918–929. [DOI] [PubMed] [Google Scholar]
- 213.Jonić S. Cryo-electron microscopy analysis of structurally heterogeneous macromolecular complexes. Comput. Struct. Biotechnol. J. 2016; 14:385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Marko M., Hsieh C., Schalek R., Frank J., Mannella C.. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat. Methods. 2007; 4:215–217. [DOI] [PubMed] [Google Scholar]
- 215.Asano S., Fukuda Y., Beck F., Aufderheide A., Förster F., Danev R., Baumeister W.. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science. 2015; 347:439–442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data