Antigen Presentation Profiling Reveals Recognition of Lymphoma Immunoglobulin Neoantigens (original) (raw)
. Author manuscript; available in PMC: 2018 Feb 12.
Published in final edited form as: Nature. 2017 Mar 22;543(7647):723–727. doi: 10.1038/nature21433
Abstract
Cancer somatic mutations can generate neoantigens that distinguish malignant from normal cells 1–7. However, the personalized identification and validation of neoantigens remains a major challenge. Here we discover neoantigens in human mantle cell lymphomas using an integrated genomic and proteomic strategy that interrogates tumor antigen peptides presented by major histocompatibility complex (MHC) class I and class II molecules. We applied this approach to systematically characterize MHC ligands from 17 patients. Remarkably, all discovered neoantigenic peptides were exclusively derived from the lymphoma immunoglobulin (Ig) heavy or light chain variable regions. Although we identified MHC presentation of private polymorphic germline alleles, no mutated peptides were recovered from non-Ig somatically mutated genes. Somatic mutations within the immunoglobulin variable region were almost exclusively presented by MHC-II. We isolated circulating CD4 T-cells specific for immunoglobulin-derived neoantigens and found these cells could mediate killing of autologous lymphoma cells. These results demonstrate that an integrative approach combining MHC isolation, peptide identification and exome sequencing is an effective platform to uncover tumor neoantigens. Application of this strategy to human lymphoma implicates immunoglobulin neoantigens as targets for lymphoma immunotherapy.
Main Text
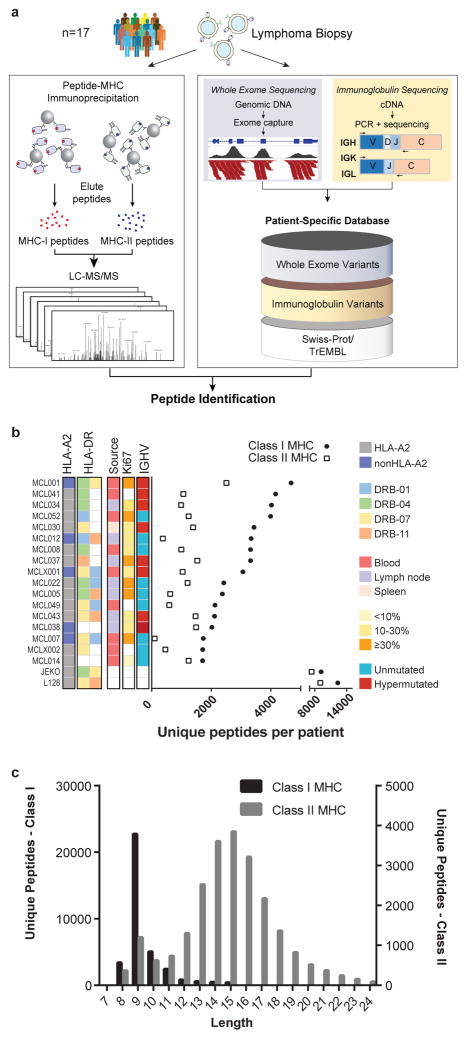
We sought to profile MHC antigen repertoires of primary human lymphomas, with the intent of discovering cancer neoantigens. Typically, “reverse immunology” neoantigen identification strategies have relied first on the isolation of cognate T-cells to then identify the candidate antigens. By contrast, direct proteomic analysis of cancer major histocompatibility complex (MHC) ligands 8–14 by liquid chromatography and tandem mass spectrometry (LC-MS/MS) enables discovery of tumor antigens, including neoantigens, directly from cancer cells. We profiled lymphoma MHC-I and MHC-II ligands from seventeen patients with untreated mantle cell lymphoma (MCL) and additionally from two MCL cell lines (Fig. 1a). We focused on MCL, a subtype of B-cell non-Hodgkin lymphoma with characteristically high expression of both class I and class II MHC molecules, because of the availability of large numbers of these tumor cells that had been collected as part of an ongoing clinical trial of immunotransplantation (NCT00490529). To define candidate somatic neoantigens, we used our previously described approach for whole exome sequencing of DNA from highly pure tumor cells and matched germline, and additionally directly sequenced the expressed lymphoma immunoglobulin heavy and light chain variable regions 15,16.
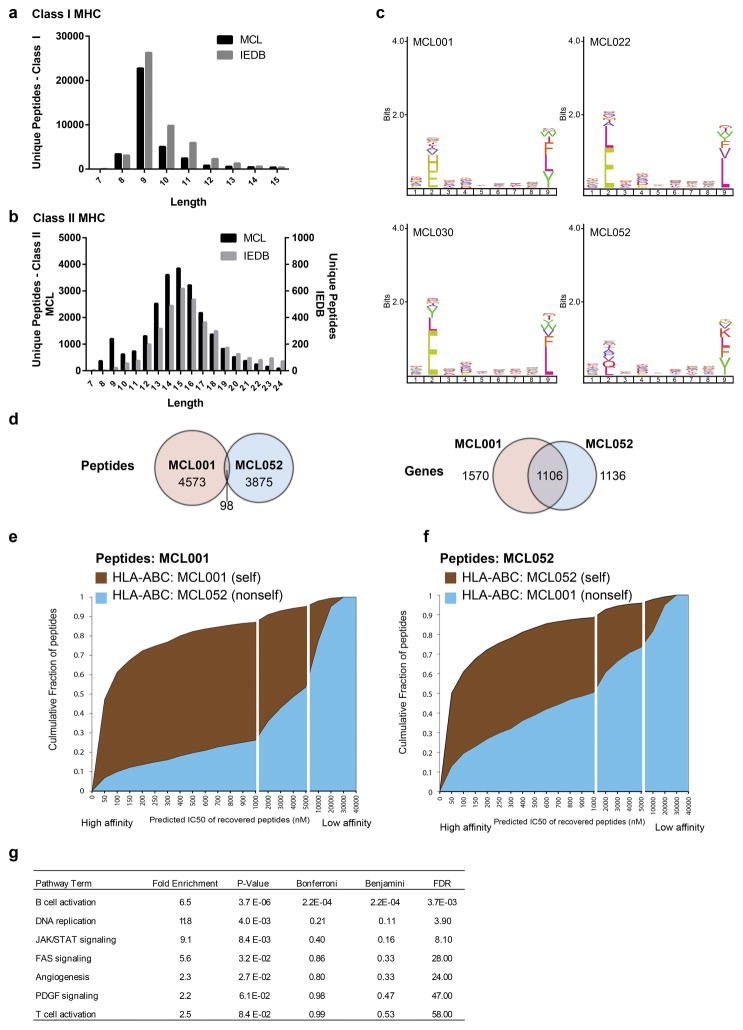
Fig. 1. Integrative genomic and proteomic approach for tumor antigen discovery.
(a) Whole exome and targeted immunoglobulin sequencing of lymphoma tumor specimens and germline DNA was performed for 17 patients. Sequencing data were integrated with a human proteome database to create patient-specific catalogues incorporating somatically mutated proteins, lymphoma-specific immunoglobulins, and germline variants. MHC-ligands were directly immunoprecipitated using both anti-HLA-A,B,C and anti-HLA-DR antibodies. Peptides were then acid-eluted, profiled by LC-MS/MS and identified with reference to patient-specific catalogues. The number of unique peptides per case (b) and the length distribution of identified MHC ligands (c) are depicted.
Peptides bound to MHC-I and MHC-II were purified in parallel via immunoprecipitation with a pan-MHC-I antibody and an antibody specific for HLA-DR, a class II MHC molecule, and analyzed by LC-MS/MS. This strategy identified over 24,000 unique MHC-I associated peptides and over 12,500 unique MHC-II associated peptides (Fig. 1b). Both MHC-I and MHC-II peptide repertoires demonstrated length distributions consistent with those expected for each class (Fig. 1c, Extended Data Fig. 1a–b). Furthermore, MHC-I peptides showed the expected reduced amino acid complexity at anchor residue positions (Extended Data Fig. 1c) and agreed with a widely used binding affinity model (Extended Data Fig. 1d–f).
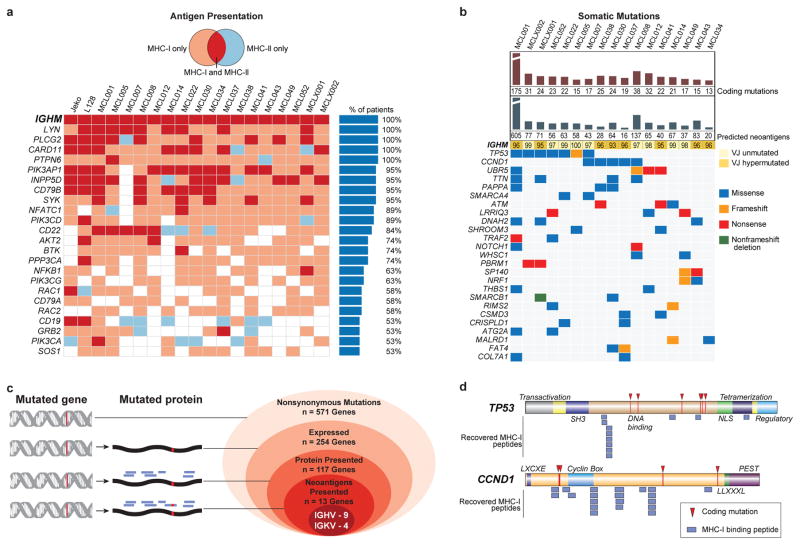
Through whole proteome analysis of two MCL cell lines, we found MHC-I and MHC-II presentation was significantly biased toward abundant proteins (Extended Data Fig. 2). In contrast, we found mutated proteins tended to be significantly less abundant than average. We found a high degree of overlap among genes presented by MHC across patients (Extended Data Fig. 3a–b). However, the specific peptides we recovered were generally private to each individual, with the exception of patients who shared MHC-I and /or MHC-II alleles (Extended Data Fig. 3c–f), further confirming MHC as the source of the recovered peptides. Among the recurrently presented genes were members of the B-cell receptor (BCR) signaling pathway including IGHM, CD79B, BTK, LYN, SYK, MS4A1 (CD20) and CD22, which showed significant enrichment (Fig 2a, Extended Data Fig. 3g).
Fig. 2. Characterization of lymphoma-specific MHC-I and MHC-II epitopes and somatic mutations.
(a) B-cell receptor pathway components presented by >50% of patients in the context of MHC-I (pink), MHC-II (blue), or both (red). (b) Nonsynonymous somatic mutations, predicted neoantigens, and the degree of somatic hypermutation in IGVH or IGJH are depicted, along with genes recurrently mutated in the cohort by exome sequencing. (c) Antigen presentation of mutated genes across the cohort. Nested ovals depict increasing evidence levels for candidate neoantigens, starting with nonsynonymous mutations (outermost oval) to direct evidence of neoantigen presentation (innermost oval). (d) TP53- or _CCND1-_derived MHC-I peptides and observed somatic mutations are depicted in relation to the corresponding protein domains.
There were 13–175 nonsynonymous somatic mutations per patient exome, including genes known to recurrently mutated in MCL such as Ig variable regions, TP53, CCND1, ATM, UBR5, SMARCA4, NOTCH1, FAT4, TRAF2, and WHSC1 17,18 (Fig 2b). We also identified novel genes as recurrently mutated in MCL including PBRM1 and SMARCB1, with the latter gene previously found to be mutated in a single MCL patient 19. Most nonsynonymous mutations occurred in genes not characteristically expressed in MCL (Fig 2c)20. Among expressed genes bearing coding mutations, we found 46% with at least one peptide presented by either class I or class II MHC, suggesting active processing of these mutated proteins. However, with the exception of peptides derived from immunoglobulin genes, the MHC presented peptides were derived from the non-mutated regions of these genes, as exemplified by the two most frequently mutated non-immunoglobulin genes, TP53 and CCND1 (Fig 2d).
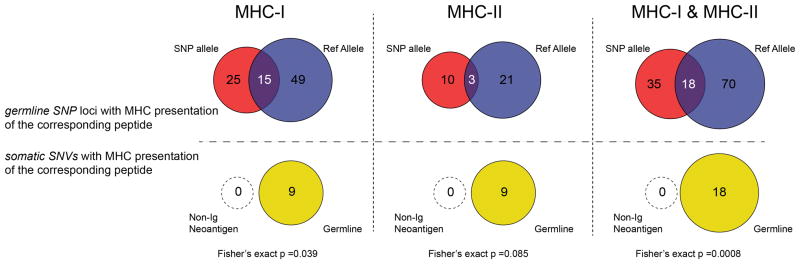
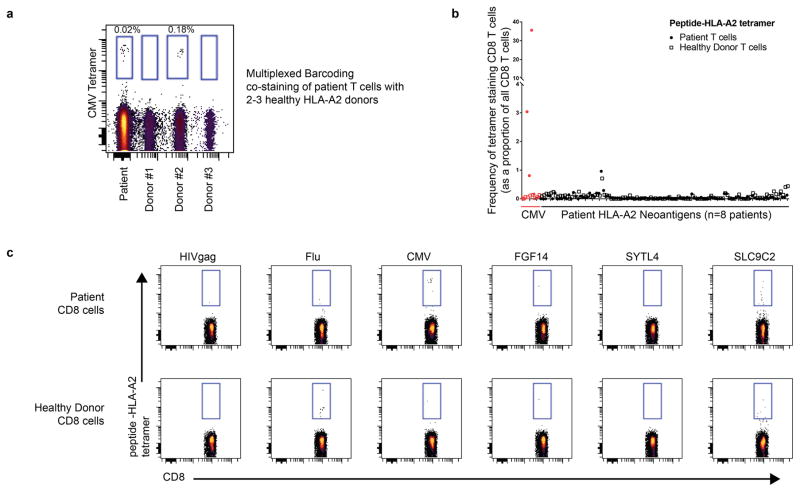
We recovered neoantigen peptides from 13 genes, all of which were derived from immunoglobulin variable regions. To test whether the lack of non-immunoglobulin neoantigens was due to technical limitations in recovering private peptide variants, we assessed the recovery of peptides encoded by heterozygous germline single nucleotide polymorphisms (SNPs) for each patient. This analysis revealed significantly greater presentation of germline versus somatic allelic variants across the genome (p<0.001, Extended Data Fig. 4). To determine whether our approach was insensitive in detecting clinically significant neoantigens, we additionally assayed 8 patients’ CD8 T-cell responses against computationally predicted HLA-A2 restricted neoantigens with peptide-MHC tetramers (Extended Data Fig. 5). No immune responses were detected against any of the 108 putative neoantigens tested.
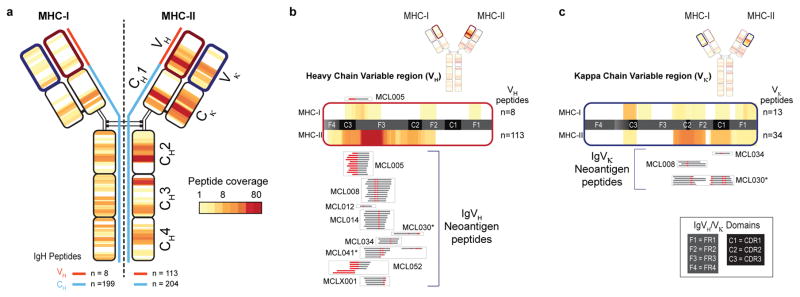
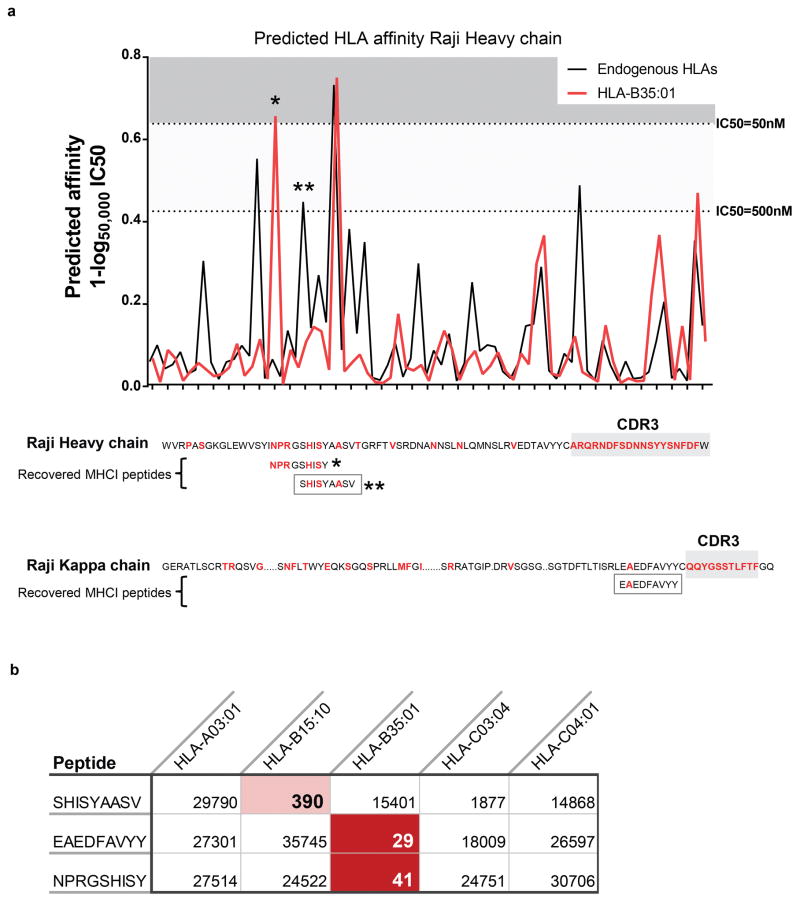
The immunoglobulin heavy chain was presented by both MHC-I and MHC-II in all seventeen patients (Fig. 2b). MHC-I and MHC-II displayed strikingly distinct antigen presentation patterns from this protein. Peptides presented by MHC-I mapped to the Ig constant regions (199 peptides, Supplemental Note 1, Supplemental Table 1), but there were few peptides recovered from the Ig variable regions (8 peptides) (Fig. 3a). In contrast to MHC-I, antigen presentation of immunoglobulin by MHC-II was clustered within the variable region. (Fig. 3a). Specifically, ~93% of MHC-II bound peptides (105/113) spanned the junction of complementary determining region 3 (CDR3) and framework region 3 (Fig. 3a). Nearly half of the MHC-II presented peptides from this hotspot were neoantigens created by somatic hypermutation or VDJ gene rearrangement (52/105 peptides, 49.5%) (Fig. 3b, Supplemental Table 2). In contrast, only a single non-germline peptide was recovered from MHC-I. Peptides derived from light chain variable regions showed similar profiles: 14 neoantigen peptides formed via somatic hypermutation were presented by MHC-II, but none by MHC-I (Fig. 3c). Using the Raji lymphoma cell line, we tested whether our approach did not detect MHC-I restricted immunoglobulin neoantigens as a result of technical obstacles. We ectopically expressed an MHC-I allele with high predicted affinity for Raji immunoglobulin neoantigens. Antigen presentation profiling of the resultant Raji cells revealed 3 immunoglobulin neoantigens, thus confirming our ability to recover MHC-I immunoglobulin neoantigens (Extended Data Fig. 6).
Fig. 3. MHC-I and MHC-II presentation of lymphoma immunoglobulins.
(a) Heat-map reflects the frequency and distribution of MHC-I (left) and MHC-II (right) presentation of IgM-derived peptides across the cohort. Panels (b) and (c) depict expanded views of antigen presentation from variable regions of Ig heavy and kappa light chains, respectively. Neoantigen peptides created by either somatic hypermutation or VDJ rearrangement are aligned to expanded heat-maps, and grouped by patient (boxes). Red, somatically mutated positions within recovered peptides creating neoantigens. Asterisks, patients/peptides selected for subsequent functional immunological studies.
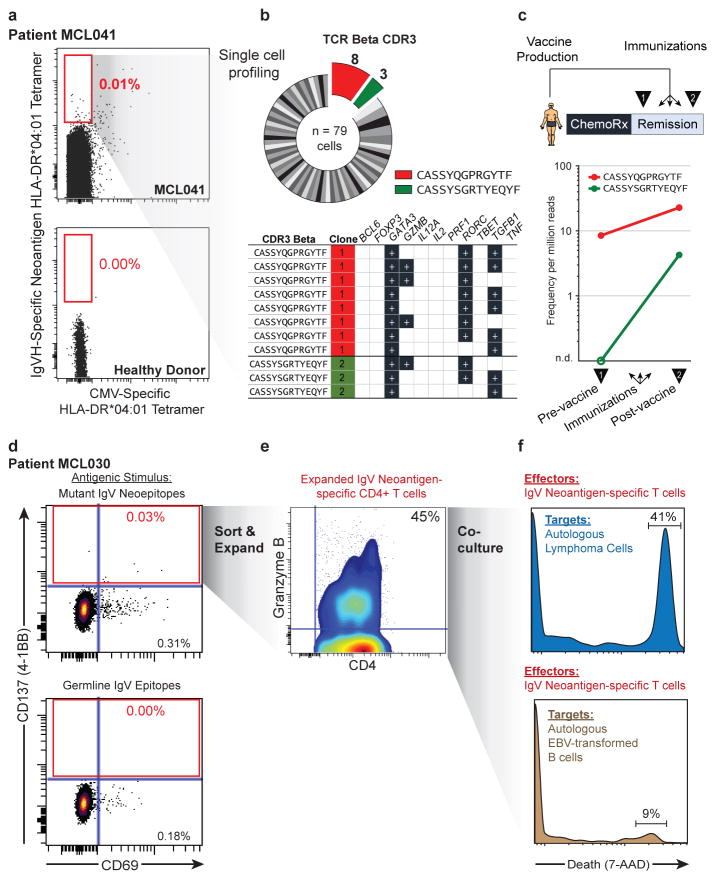
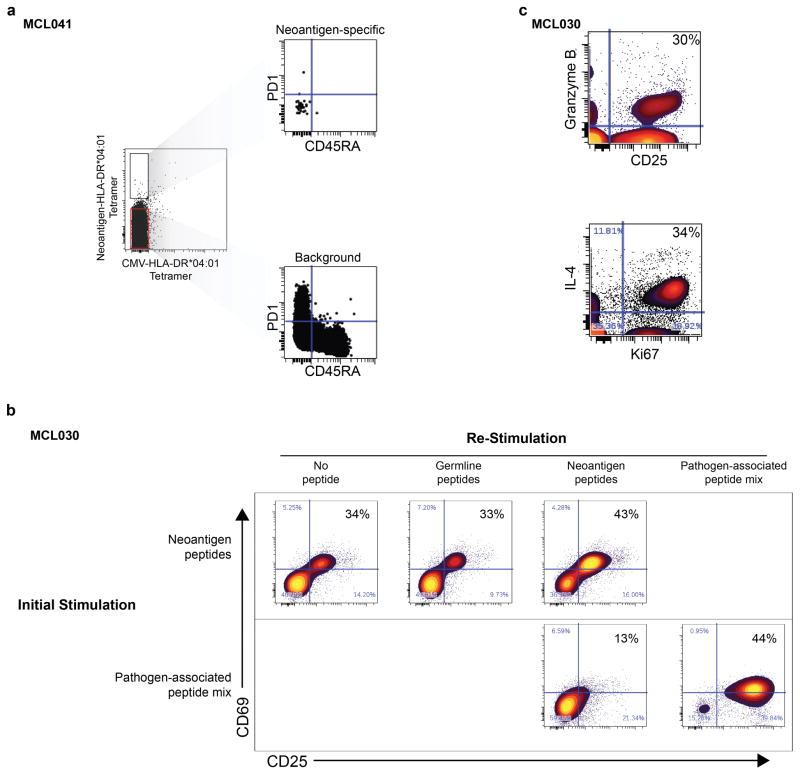
To validate these neoantigens as true HLA-DR ligands, we synthesized neoantigen peptides from three patients with an HLA-DR*0401 allele and confirmed their affinity for HLA-DR by exchange onto recombinant HLA-DR*0401 molecules for five of the six tested peptides (Extended Data Fig. 7). We next determined whether these patients harbored T-cells that recognize their autologous immunoglobulin neoantigens. Neoantigen peptide-HLA-DR*0401 tetramers revealed corresponding neoantigen-specific peripheral blood CD4 T-cells in one of three patients tested (Fig. 4a). These CD4+ T-cells appeared to be memory T-cells based on absence of CD45RA expression, but also lacked PD-1 expression, which has been associated with neoantigen-specific T-cells in melanoma patients (Extended Data Fig. 8a)21. Tetramer-binding T-cells were sorted as single cells for sequencing of T-cell receptor (TCR) alpha and beta chains, and single cell gene expression phenotyping 22. Of 79 cells from which TCR sequences were obtained, we found 8 derived from a single dominant T-cell clone, and three from a second clonotype (Fig. 4b). Neither clone was seen among 192 analyzed cells that did not bind the neoantigen tetramer. All cells expressed GATA3 and RORC, consistent with a Th2/Th17-skewed phenotype, and a subset also expressed granzyme B (Fig. 4b). We next profiled this patient’s neoantigen-specific T-cells following autologous tumor vaccination (Fig. 4c, top). TCR repertoire sequencing of peripheral blood T-cells collected before and after the immunizations revealed that both neoantigen-specific T-cell clones were induced by vaccination (Fig. 4c, bottom).
Fig. 4. Detection and cytololytic activity of neoantigen-specific CD4 T-cells.
(a) Peripheral blood CD4 T-cells from patient MCL041 (top) or a healthy donor (bottom) were stained with neoantigen-specific or CMV-specific HLA-DR tetramers. (b) Neoantigen-specific T-cells were sorted and subjected to single-cell TCRβ sequencing (top) and RNA expression profiling (bottom), with recurrent neoantigen-specific TCRβ clones highlighted. (c) Pre- and post- immunization peripheral blood frequencies of corresponding neoantigen-specific clones from TCRβ repertoire sequencing. (d) CD4 T-cells from a second patient were stimulated with either Ig neoantigen peptides (top) or corresponding unmutated germline counterparts (bottom). (e) Sorted neoantigen-specific cells were expanded and immunophenotyped for granzyme B expression. (f) Cytotoxic activity of neoantigen-specific T-cells against either autologous lymphoma cells (top) or EBV-transformed B-cells (bottom).
We next explored the therapeutic potential of patient-derived neoantigen-specific T-cells. In a second patient, we recovered neoantigen-specific CD4 T-cells, identified by CD137 expression, after stimulation with autologous lymphoma immunoglobulin neoepitopes (Fig. 4d). We expanded these isolated T-cells ex vivo and found they produced both IL-4 and granzyme in response to lymphoma neo-epitopes, suggesting potential cytolytic function as previously described3,23 (Extended Data Fig. 8b–c). Remarkably, we found these CD4 neoantigen-specific T-cells could mediate cell killing of autologous lymphoma cells (Fig. 4f). Autologous EBV-transformed normal B-cells that lacked the lymphoma neoantigen were unaffected, further demonstrating these T-cells’ antigen specificity (Fig. 4f). Similarly, pathogen-specific T-cells that were purified and expanded in parallel were unable to kill lymphoma cells (Extended Data Fig. 9).
The unexpected bias against immunoglobulin variable region presentation by MHC-I has important biologic and therapeutic implications. Immunoediting represents one possible explanation for the observed bias24,25. Although we found the lymphoma immunoglobulin sequences included numerous variable region peptides with predicted high affinity for self-MHC-I, none were measured with our direct profiling (Extended Data Fig. 10). It remains unclear how primary lymphoma cells could avoid presenting high affinity immunoglobulin variable region peptides on MHC-I, while peptides derived from the constant region are prominently presented (Supplemental Note 2 and Supplemental Table 3).
Whereas the lack of MHC-I immunoglobulin variable region presentation may represent a method of immune evasion, the contrasting frequent presentation of variable region peptides by MHC-II suggests that immunoglobulin neoantigens recognition by CD4 does not inhibit lymphoma development. Murine models have suggested that such presentation may serve to promote lymphomagenesis via cognate CD4-expressing helper T-cells 26. While immunoglobulin neoantigen-specific T-cells displayed a Th2-like phenotype similar to these murine models, they could nevertheless be engaged and expanded and demonstrated anti-tumor functions.
These findings also inform upon the previous experience with idiotype therapy. Idiotype vaccines appeared to provoke predominantly CD4 and/or humoral responses, consistent with our finding of selective MHC-II presentation of immunoglobulin antigens and CD4 immunoglobulin neoantigen specific T-cells 27,28. Clinical vaccine therapy trials targeting lymphoma immunoglobulin variable regions showed early promise, but three independent phase III trials of idiotype vaccine found either modest or no overall clinical benefit 29–31. Nevertheless, the subgroup of patients who generated immune responses were noted to derive substantial benefit30. Our results suggest that MHC presentation of variable region epitopes is not equivalent across all individual lymphomas, raising the possibility that the patients who benefited from idiotype vaccination were those with active presentation of their immunoglobulin neoepitopes.
The number of somatic mutations, and therefore potential neoantigens, has been posited as a predictor of immune checkpoint response in malignancies with high mutational burden, such as lung cancer and melanoma4,32,33. Mass spectrometry-based neoantigen discovery has successfully identified MHC class I neoantigens from specimens with large mutation burdens, including cancer cell lines 13 and melanoma11,12. By incorporating MHC class II ligands into our analysis, we uncover a different category of lymphoma-specific neoantigens. We find that, aside from immunoglobulin, mutated antigens are not prominently presented as MHC epitopes in mantle cell lymphoma, similar to other hematologic malignancies with low to moderate mutation burden 8,9,14. It remains unclear whether the lack of recovered MHC neoantigens from primary human tumors reflects their true absence or a limitation of our method in detecting rare epitopes. Nevertheless, our findings suggest that the epitopes uncovered through direct, proteomic antigen profiling represent actionable antigen targets for T-cell therapy. Here, we leverage our ability to recover precise CD4 neoantigens to discover and augment the cytotoxic potential of the neoantigen-specific CD4+ T-cells. Our results suggest that expansion of endogenous immunoglobulin neoantigen-specific CD4+ T-cells may represent a viable lymphoma treatment strategy. We demonstrate that by integrating genomic and proteomic analysis, antigen presentation profiling effectively identifies clinically relevant tumor antigens, and reveals class II MHC restricted immunoglobulin neoantigens as an effective target for lymphoma immunotherapy.
Methods
Tumor Specimens
All specimens were obtained with informed consent in accordance with the Declaration of Helsinki and this study was approved by Stanford University’s Administrative Panels on Human Subjects in Medical Research. Samples were collected from patients with untreated mantle cell lymphoma by either excisional lymph node biopsy, splenectomy, or peripheral blood leukapheresis and cryopreserved.
Vaccine trial
Patient MCL041 was treated as part of an ongoing clinical trial of tumor vaccination (NCT00490529). Vaccine was prepared by ex vivo culture of circulating autologous lymphoma cells in AIM-V media and human AB-negative sera containing 3μg/ml PF-3512676 (CpG molecule) for 72 hours. Cells were then irradiated with 200Gy and cryopreserved. The patients were treated with induction chemotherapy and those with a complete remission then received a series of three weekly autologous tumor vaccinations. Leukapheresis was used to collect peripheral blood mononuclear cells before and after the series of vaccinations.
Cell Lines
JEKO and L128 mantle cell lymphoma lines were originally obtained from the Lymphoma Research Foundation / Mantle Cell Lymphoma Consortium ATCC cell line resource, tested for mycoplasma, and authenticated by STR profiling. Cell lines were grown per ATCC recommendations.
Exome sequencing
Tumor whole exome sequencing was performed for seventeen patients and the JEKO and L128 mantle cell lymphoma cell lines as described previously 15,16. Tumor DNA was collected from either peripheral blood or lymph node biopsy. Tumor cells were purified by either flow cytometry or by magnetic bead purification for CD19 expression, or in some cases were not purified if the biopsy sample had at least >90% tumor purity. Germline DNA was obtained either from peripheral blood collected at time of complete remission or else by fluorescence activated cell sorting of CD3 positive T-cells from the initial tumor specimen. DNA and RNA were extracted from fresh single cell suspensions using the Qiagen Allprep kit (Qiagen). Library preparation and exome capture were performed as previously described16. Next generation sequencing (NGS) libraries were prepared using KAPA Libarary Preparation kits (KAPA Biosystems). Briefly, 100ng-1.5μg of DNA per sample was sheared by sonication using a Covaris S-series instrument (Covaris). Sheared DNA was end-repaired according to the manufacturer’s protocol and ligated with Illumina sequencing adaptors overnight at 16°C. Adaptor-ligated product was enriched using 8–10 cycles of PCR with Phusion High-Fidelity PCR Master Mix (New England Biolabs), and size-selected using Agencourt Ampure XP beads (Beckman Coulter). Purified libraries were evaluated for size distribution using a BioAnalyzer High-Sensitivity DNA kit (Agilent), and quantified using spectrophotometry and Qubit dsDNA HS assay (Invitrogen). NGS libraries were pooled and enriched for coding sequences using the SeqCap EZ Exome v3.0 capture reagent (NimbleGen). Hybrid captures were performed according to the manufacturer’s protocol, with all 47°C steps conducted on a thermal cycler to ensure maximal temperature stability. Pooled libraries were amplified postenrichment using 6–8 cycles of PCR. Illumina sequencing services were performed by the Stanford Center for Genomics and Personalized Medicine, with 101bp paired-end reads on a HiSeq 2000 instrument (Illumina) loading 1 capture pool per lane. Exome sequencing achieved a median depth of coverage over this region 94X in purified tumor B-cells and 71X in germline samples.
Variant calling and annotation
Raw sequences were demultiplexed and adapter sequences trimmed. Reads were aligned to UCSC hg19 reference genome with the BWA mem aligner 34 and duplicate reads were removed. SNV and indel variants were called by VarScan2 35 using reads with mapping quality > 10, minimum base quality >20, minimum variant frequency 0.1, minimum of 3 variant reads, strand bias <90%, minimum normal coverage ≥6 reads, and minimum tumor coverage ≥8 reads. High confidence somatic SNVs were then realigned around indels by Varscan2 and filtered using the false positive filter script fpffilter.pl 36. Variants were annotated and coding changes were predicted using Annovar 37. Germline SNPs were called using VarScan2 with the above parameters. For cell lines, SNPs were assigned as germline if they were present in the NCBI dbSNP137 database.
Neoantigen prediction
Coding variants were annotated to provide the corresponding mutated protein sequences using Annovar. Patient HLA alleles were extracted from sequencing of autologous normal cells using Phlat 38. Neoantigen predictions were performed with NetMHC 3.4 39 or else when the HLA allele was not included in the NetMHC model then NetMHCpan2.8 40 was utilized. Unless otherwise specified, peptides with predicted IC50<500nM were considered neoantigens.
Targeted immunoglobulin variable region sequencing
Sequencing of immunoglobulin heavy and light chains was performed using modification of the BIOMED-2 protocol to reliably sequence the full length variable region 41. Tumor RNA was used to create cDNA using Superscript Reverse Transcriptase (ThermoFisher Scientific) according to standard protocol using random hexamers. PCR was performed with Kapa HiFi HotStart ReadyMix (Kapa Biosystems) with an annealing temperature of 68–70 °C using a mix of forward V-leader family primers and a reverse primer in the constant region (Supplemental Table 4). PCR products were purified by agarose gel electrophoresis and extraction. PCR products were sequenced by Sanger sequencing either directly or after subcloning into the pCR-Blunt vector (ThermoFisher Scientific). VH usage and germline homology was determined using IMGT®, the international ImMunoGeneTics information system® http://www.imgt.org 42.
Generation of Custom Reference Proteome Databases
Two protein sequence databases were constructed. Both were built upon the human proteome UniprotKB database including Swiss-Prot and TrEMBL databases (version May 2015) with common contaminant protein sequences included (e.g. staphylococcus protein A). The first was a somatic personalized database which included predicted mutated peptides inferred from somatic SNVs and indels from exome sequencing, as well as the lymphoma-specific IGHV and light chain variable sequences as inferred from immunoglobulin V(D)J sequencing. The second database consisting of the same human proteome UniProt database but with added peptides derived from patient-specific coding, heterozygous single nucleotide polymorphisms (SNPs) and cell line specific SNPs. Personalized entries consisted of the mutated amino acid(s) with flanking 21 amino acids both N-terminal and C-terminal to the mutated residue(s). Reversed “decoy” protein sequences were appended to both of these databases for “target-decoy” error estimation 43.
Purification of HLA-peptides from cells
MHC-class I and class II peptidomes were extracted from 17 MCL-patients’ tumors and from two cell lines (JEKO, L128). The MHC class I or MHC class II molecules were isolated and the associated peptides extracted as previously described 44,45 with some modifications. Prior to lysis all cells were washed twice with PBS. For all patient samples, 1x108 cells were lysed per MHC preparation, while for the cell lines 1x108 and 1x109 cells were used to evaluate the extent to which limited input material may reduce the sensitivity ligand detection. In brief, cells were lysed for 20 min on ice in 20 mM Tris-HCl (pH8), 150 mM NaCl, 1 % (w/v) CHAPS, 0.2 mM PMSF, 1x Halt™ Protease and Phosphatase Inhibitor Cocktail (ThermoFisher Scientific) supplemented with complete protease inhibitor cocktail (Roche). The lysate was subjected to centrifugation (2x30 min, 13,200 rpm at 4°C) and the resulting supernatant was precleared for 30 min using rProtein A Sepharose fast-flow beads (GE Healthcare). For all MHC-1 immunoprecipitations the precleared lysate was incubated with the pan HLA-A-, B-, and C- antibody W6/32 (purified in-house from hybridoma kindly provided by Betsy Mellins and with immunoglobulin produced by Genentech 46) coupled to rProtein A Sepharose fast-flow beads for 5h at 4°. For MHC-II captures the precleared lysate was incubated with the HLA-DR specific antibody L243 (produced and purified by Genentech from our hybridoma) 47 coupled to rProtein A Sepharose fast-flow beads for 5h at 4°C. Following immunoprecipitation, the beads were washed with TBS (pH 7.4) and peptides were eluted from the purified MHC-molecules using 10% acetic acid. Eluted peptides were further purified with a 10 kDa MWCO size filter, followed by a concentration step using vacuum centrifugation, desalted on C18 based STAGE tips 48. Peptides were lyophilized and stored at −80°C until LC-MS/MS analysis.
Mass Spectrometry Analysis of HLA Associated Peptides
For MS analysis, isolated peptides were reconstituted in 0.1 % formic acid and analyzed on an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) in three replicate injections of 25% of the total specimen amount. Samples were separated by capillary reverse phase chromatography on a 18 cm reversed phase column (100 μm inner diameter, packed in-house with ReproSil-Pur C18-AQ 3.0 m resin (Dr. Maisch GmbH)) over a total run time of 160 min using a two-step linear gradient with 4–25 % buffer B (0.2% (v/v) formic acid, 5% DMSO, and 94.8% (v/v) acetonitrile) for 120 min followed by 25–40 % buffer B for 30 min using the Eksigent ekspert nanoLC-425 system (Sciex, Framingham, USA). Three injections were made per sample to utilize multiple fragmentation modes (HCD (higher energy collisional dissociation) or CID (collision induced dissociation). The third injection was performed with CID including singly-charged species. Acquisition was executed in data dependent mode with the full MS scans acquired in the Orbitrap mass analyzer with a resolution of 60000 and m/z scan range 340–1600. The top ten most intense ions were then selected for sequencing and fragmented in the Orbitrap mass analyzer at a resolution of 15,000 (FWHM). Data-dependent scans were acquired from precursors with masses ranging from 700–1800 Da for all MHC class 1 samples and from 700–2750 Da for MHC class II samples. Precursor ions were fragmented with a normalized collision energy of 35% and an activation time of 5 ms for CID and 30ms for HCD. Repeat count was set to 2 and fragmented m/z values were dynamically excluded from further selection for a period of 30 seconds. The minimal signal threshold was set to 500 counts.
Computational Identification of HLA Peptides From Mass Spectra
All tandem mass spectra were queried against personalized somatic and personalized germline proteome databases described above using both SEQUEST 49 and PEAKS DB search engines (PEAKS Studio 7, Bioinformatics Solutions Inc.)50. Spectra were also interpreted by de novo sequencing (PEAKS Studio 7, Bioinformatics Solutions Inc.) to improve high-confidence peptide identification. The msConvert program (v3.0.45) was used to generate peak lists from the RAW data files, and spectra were then assigned to peptides using the SEQUEST (v28.12) algorithm. RAW data files were imported into PEAKS Studio 7 prior to PEAKS DB and PEAKS de novo searching and subjected to default data refinement (deisotoping, charge deconvolution, peak centroiding). For all searches, the parent mass error tolerance was set to 20 ppm and the fragment mass error tolerance to 0.02 Da. For SEQUEST and PEAKS DB, enzyme specificity was set to none and oxidation of methionines and deamidation (N,Q), cysteinylation, and phosphorylation (S, T, Y) were considered as variable modifications.
High-confidence peptide identifications were selected at a 1% false discovery rate with a version of the Percolator algorithm, which we modified for proteogenomic immunopeptide analysis51. Percolator implements a linear-SVM to score peptide identifications based on multiple peptide attributes. We modified the Percolator method to incorporate peptide attributes that influence the confidence in immunopeptide identifications. These include whether the de novo sequence matches database assignments; the magnitude of scores assigned by all three search algorithms; the source of the identified peptide (Swiss-Prot, TrEMBL, or our own sequencing); and the presence of one or more post-translational modifications in the identified peptide. Unlike conventional proteome analysis, false discovery rates were not evaluated at the level of assembled proteins, as this would unnecessarily penalize proteins identified by small numbers of peptide.
Confirmation of HLA Identifications with Synthetic Peptides and Mass Spectrometry
Synthetic peptides corresponding to the idiotype peptides from MCL-41 (NMLFLQMNSLKTEDT, DDSKNMLFLQMNSLKTEDT, FLQMNSLKTEDT, IKNKIDGETTDYAAPV) were purchased from JPT (JPT Peptide Technologies, Berlin, Germany) with a purity of >70 %, dissolved and resuspended into 0.1 % FA. 250 fmol of each peptide was analyzed on an LTQ-Orbitrap Elite mass spectrometer (ThermoFisher, Bremen, Germany) using the same instrument settings as described above. Synthetic peptides were measured with two replicate LC-MS analyses, using either the CID or HCD fragmentation methods as defined above. Both included singly-charged species.
Quality assessment of HLA ligands
Sequence logos for recovered MHC-I peptides were produced using Icelogo http://iomics.ugent.de/icelogoserver to reveal position weighting at anchor residues52. The size distribution of recovered MHC-I and MHC-II bound peptides was compared to size distributions of known MHC-I and MHC-II ligands from the Immune Epitope Database and Analysis resource (www.iedb.org, January 2016) using filters for host (human) and assays (MHC ligand assays only) 53. To assess compatibility of recovered MHC-I ligands with the netMHCpan 40 prediction algorithm, we tested cross-prediction for two patients with completely private HLA-A, B, and C alleles. Affinities for peptides recovered from one patient were predicted against self or unrelated HLA-A, B, C alleles.
Correlation of MHC presentation with gene expression
To estimate gene expression, we used previous transcriptome profiling of mantle cell lymphoma performed by Kridel et al. 20. The median log2 transformed RPKM from the 15 patients from that study was used for predicted gene expression, and genes with a median log2(RPKM) > 0 were considered to be expressed. Every recovered MHC bound peptide was assigned the log2 RPKM of its corresponding gene.
Total Cell Line Proteome Sample Preparation and high pH Reversed Phase Fractionation for MS-analysis
1x108 cells from each of two MCL cell lines (JEKO and L128) were used for protein extractions. In brief, cells were lysed in 8M urea, 150 mM NaCl, 5 mM DTT, 50 mM Tris pH 8 supplemented with Complete Protease Inhibitor Cocktail tablet (Roche) and 1x Halt™ Protease and Phosphatase Inhibitor Cocktail (ThermoFisher Scientific). Following lysis, the lysate was centrifuged at 13,200 rpm for 15 min, the supernatant was transferred to fresh tubes and a second round of centrifugation was performed. The resulting supernatant was alkylated using 14 mM iodoacetamide for 45 minutes at room temperature in the dark and the reaction was then quenched with DTT. Proteins were methanol-chloroform precipitated and washed twice with acetone before being re-suspended in 300 μl 8M urea, 50 mM Tris pH8. Total protein concentration was determined using the Pierce™ BCA Protein Assay Kit (Pierce, Rockford, IL). The samples were diluted to 1 M urea using 50 mM Tris pH8 prior to digestion with Trypsin/Lys-C Mix (Promega, Madison, WI) at a ratio of 1:25 enzyme: substrate for 16 hours at 37 °C. The reaction was quenched with formic acid and the peptides desalted using a Sep-Pak C18 1 cc Vac Cartridge, 50 mg (Waters, Milford, MA). Digested peptides were resuspended in 130 μl 10 mM ammonium formate, pH 10 followed by high-pHRP fractionation 54,55 using a 65 min + 15 min step-gradient (buffer A (10 mM ammonium formate, pH 10) and buffer B (10 mM ammonium formate, 90 % ACN, 10 % H2O, pH 10) using an Agilent 1200 HPLC (Agilent Technologies). In total 84 fractions were collected, concatenated and combined 55 into a total of 12 fractions and lyophilized by vacuum centrifugation. All fractions were cleaned using a C18 based STAGE Tips 48, lyophilized and stored at -80°C until final LC-MS/MS measurement.
Mass Spectrometry Analysis of Cell Line Proteomes
Each concatenated peptide fraction was resuspended in 20 μl 0.1 % formic acid. In total 10 % of the material was injected and separated on a 18 cm reversed phase column (100 μm inner diameter, packed in-house with ReproSil-Pur C18-AQ 3.0 m resin (Dr. Maisch GmbH) over a total run of 160 min using a two-step linear gradient with 4–25 % buffer B (0.2% (v/v) formic acid, 5% DMSO, and 94.8% (v/v) acetonitrile) for 120 min followed by 25–40 % buffer B for 30 min using the Eksigent ekspert nanoLC-425 system (Sciex, Framingham, U.S.A). The LC system was coupled on-line with an Orbitrap Elite instrument (Thermo Fisher Scientific, Bremen, Germany) via a nano-electrospray source. Full scans were acquired in the Orbitrap mass analyzer with resolution 60,000 at 340–1600 m/z. Unassigned charge states were rejected and the top 20 most intense ions with charge states 2 and up were sequentially isolated for MS/MS analysis using CID fragmentation. A minimal signal of 500 was required, the normalized collision energy was set to 35% and the fragmented peptide masses were collected in the ion-trap. Dynamic exclusion was enabled with a repeat count of 1 with the repeat duration set to 30 seconds.
HLA-B*35 construct and genetic transduction of Raji cells
The cDNA sequence of full-length HLA-B*35:01:01:01:01 was synthesized (IDT) with 25 bp of overlapping vector sequence on either end and cloned into pLenti-CMV-Puro-DEST (w118-1, a gift from Eric Campeau) at the EcoRV sites using Gibson assembly (NEB). A two-step PCR was used to insert an N-terminal FLAG tag downstream of the signal peptide. Lentivirus produced in HEK293FT-cells was used to transduce Raji cells overnight. Transduced cells were selected by treatment with puromycin (1 μg/ml, InvivoGen) for 7 days. Cell culture of Raji cells overexpressing the viral entry receptor DC-SIGN (gift from Dr. Eva Harris, UC Berkeley) with and without HLA*B35:01 overexpression were cultured in RPMI medium supplemented with 5% Fetal Bovine Serum and 1x Penicillin/Streptomycin and L-glutamine.
Computational interpretation of Cell Line Proteomes
Data analysis was performed using ProteomeDiscover v2.1.0.81 (Thermo Fisher Scientific) and SEQUEST-HT with the following settings: The parent mass error tolerance was set to 20 ppm and the fragment mass error tolerance to 0.6 Da. Strict trypsin specificity was required allowing for up to two missed cleavages. Carbamidomethylation of cysteine was set as fixed modification and oxidation of methionines as a variable modification. The minimum required peptide length was set to seven amino acids. All spectra were queried in two independent searches against the patient specific or the SNPs derived databases as earlier described for the ligandome analysis. A false discovery rate of 1% was required at both the peptide level and protein level. For each protein group, summed peptide intensities were used to estimate copies-per-cell abundance using the histone ruler approach 56.
Three subsets of proteins were compared to the complete set of all recovered proteins: 1) Proteins for which at least one corresponding peptide was recovered from the MHC-I immuno-peptidome dataset; 2) Proteins for which at least one corresponding peptide was recovered from the MHC-II immuno-peptidome dataset; 3) Proteins which were predicted to be mutated by exome sequencing and for which the predicted mutated peptide was identified by proteome analysis. The distributions of abundance for each of these subsets was compared to the distribution of all proteins using a two-tailed Kolmogorov-Smirnov test for each cell line, given the unequal variance between groups.
Interpatient similarity for MHC presentation of Peptide and Genes
The list of unique peptides or genes presented by MHC-I and MHC-II for each patient were used to calculate interpatient similarity. Peptides with a post-translation modification were considered distinct from unmodified peptides (e.g. a peptide with a phosphorylated residue was not identical to the same unphosphorylated peptide). Sørensen similarity index was calculated between patients as 1 - (Bray-Curtis dissimilarity) using the R “vegan” package based on peptide or gene similarity. A force-directed graph was generated based upon MHC peptide presentation similarity used in the R “igraph” package. A weighted adjacency matrix was created and plotted with a Fruchterman-Reingold layout. For visualization, a minimum similarity of 0.15 was set as a threshold to produce an edge. HLA allele families for each patient were manually superimposed over the layout to highlight the relationship between peptide similarity and shared HLA alleles.
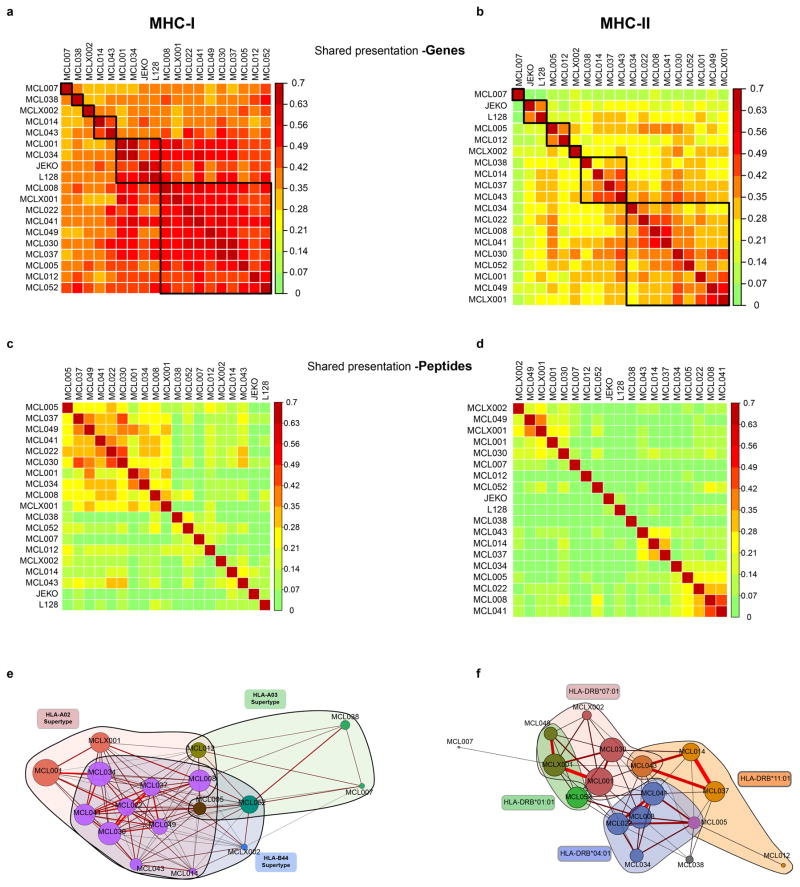
The identified MHC-associated peptide repertoires showed relatively low interpatient similarity (MHC-I: Sorenson similarity 0.01–0.45, median 0.12; MHC-II: Sorenson similarity 0.05–0.55, median 0.17), but similarity was increased between patients with shared or similar HLA alleles. Genes represented in the antigen repertoire were more similar between patients (MHC-I: Sorenson similarity 0.27–0.60, median 0.41; MHC-II: Sorenson similarity 0.04–0.48, median 0.26).
Gene set enrichment of MHC-presented peptides
All recovered MHC-I bound peptides from all seventeen patients were included. The total number of unique MHC-presented peptides recovered per gene was used to rank gene presentation. If an identical peptide was recovered from multiple patients, it was treated as an additional unique peptide mapping to that gene. The top 5th percentile of genes ranked in this manner were used for gene set enrichment analyzed by the PANTHER statistical overrepresentation test 57.
Comparison of Germline SNP peptide presentation versus somatically mutated peptide presentation
Germline SNPs for all 17 patients were identified as above. Recovered MHC-I and MHC-II associated peptides were matched with 100% identity against both the SNP variant allele and the corresponding reference allele as listed in hg19. Peptide expression of both alleles was considered true if peptides from both the SNP and corresponding reference allele were recovered in the same patient. The same analysis was performed for non-Ig somatic single nucleotide variants (SNVs). No non-Ig somatically mutated peptides were presented by MHC-I or MHC-II. However, 9 MHC-I peptides and 9 MHC-II peptides from the corresponding germline allele were recovered, which overlapped the predicted mutated amino acid position. A 2x3 Fisher’s exact test was used to compare SNP recovery vs SNV recovery (where the 3 outcomes are recovery of variant peptide, recovery of the reference peptide, and recovery of both reference and variant peptide).
Comparison between Immunoglobulin Variable versus Constant Region Presentation
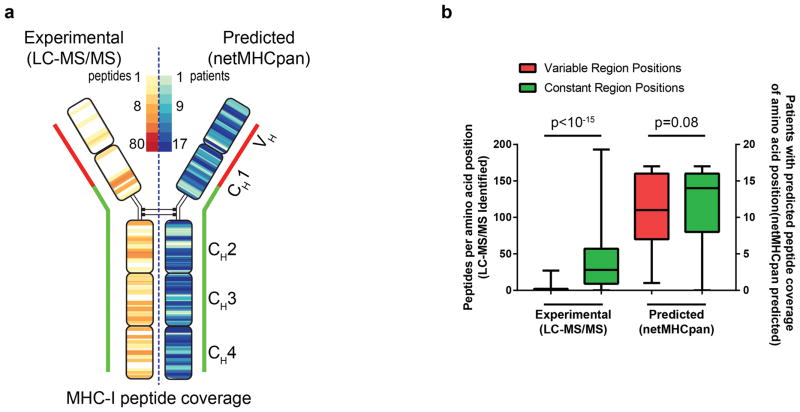
Unique MHC-presented peptides were aligned to the patients’ IGHV (variable region) and IGHM (constant region) sequence. IGHV sequences from all patients were aligned to each other using IMGT nomenclature and gap insertion when required for discrepant lengths. For each position on the immunoglobulin sequence, the number of unique peptides which overlapped that position were summed for all 17 patients and presented as a heatmap. The number of experimentally recovered peptides and the number of predicted peptides were determined for each position along the consensus immunoglobulin molecule. A two-sided Mann-Whitney-Wilcoxon test was used to compare the number of peptides per position for N-terminal (variable region plus N-terminal 50 amino acids of CH1) vs C-terminal (last 350 amino acids in constant regions) positions. This was performed separately for experimentally identified peptides and predicted peptides.
Prediction of immunoglobulin presentation
The full length immunoglobulin heavy chain sequence for each patient was used as input into netMHCpan and predictions were performed for all 8–11mers for each of the patients’ HLA-A, HLA-B, and HLA-C alleles. Two IC50 thresholds were evaluated: 50nM and 500nM. For each Ig position, a patient was counted as presenting that position if at least one peptide spanning that position was predicted to meet the threshold for self-MHC prediction. The number of patients with predicted presentation of that position was then summed, with a maximum value of 17 patients presenting a position.
We evaluated whether the reduced MHC-I presentation of the N-terminal 170aa region of the heavy chain (corresponding to the variable region plus N-terminal portion of the CH1 constant domain) was related to differences in the predicted presentation between this region and the rest of the constant regions. Peptide-MHC binding predictions were performed as above for each patient’s immunoglobulin heavy chain sequence. The total number of predicted peptides for each position was summed across all 17 patients for the N-terminal 170 aa and the C-terminal 400 aa.
HLA-DR tetramer preparation
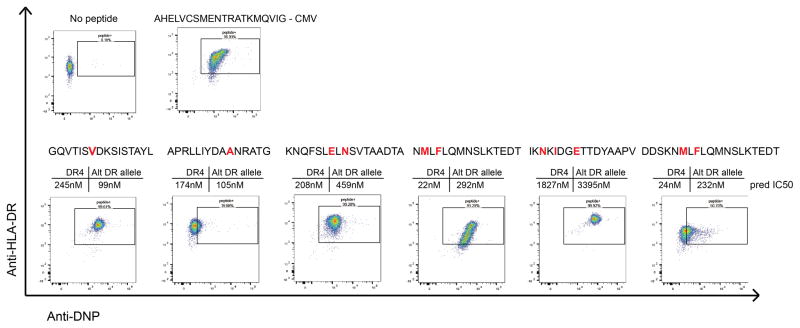
A baculovirus construct encoding Jun/Fos dimerization domain-modified HLA-DRB1*04:01 with tethered CLIP peptide was used to produce DR4 protein in insecT-cells. The product was FPLC purified, biotinylated and peptide exchanged as described by Day and colleagues 58. Candidate peptides predicted to interact with DR4 were assayed for binding by flow cytometry, and those that successfully exchanged into the peptide-binding groove were used in subsequent class II tetramer studies. Peptide-MHC monomers were tetramerized using PE- or APC-linked streptavidin (eBioscience), and residual non-tetramer components were eliminated with biotin agarose (Sigma) and streptavidin agarose (ThermoFisher). The peptides used in this study were MCL008_IgK1 APRLLIYDAANRATG, MCL008_IgH1 KNQFSLELNSVTAADTA, MCL034_IgH1 GQVTISVDKSISTAYL, MCL041_IgH1 NMLFLQMNSLKTEDT, MCL041_IgH2 IKNKIDGETTDYAAPV, and MCL041_IgH3 DDSKNMLFLQMNSLKTEDT. To test binding affinity to HLA-DR, peptides were also synthesized with an N-terminal DNP modification. After exchanging the peptides unto HLA-DR as described above, the biotinylated HLA-DR molecules were bound to streptavidin coated microsphere beads and co-stained with both anti-HLA-DR and anti-DNP antibodies. Beads were then analyzed by flow cytometry. One peptide which was not computationally predicted to bind either of the patient’s HLA-DR alleles (IKNKIDGETTDYAAPV) was found to have high affinity for HLA-DR*0401, highlighting the sensitivity of proteomic methods for MHC ligand discovery.
HLA-DR tetramer staining and single cell sequencing
Total PBMCs or purified CD4 T-cells were treated with 50 nM dasatinib (CST) for 30 minutes at 37°C to inhibit TCR downregulation. Samples were then stained with tetramers (0.5 ug/ml each specificity) and Fc block for one hour at room temperature, with agitation. Antibodies and live/dead staining reagents were added during the last 30 minutes of tetramer staining. Samples were washed and tetramer positive CD4 T-cells (and tetramer negative control cells) were single cell sorted with a FACSAria sorter (BD). The TCR and select phenotypic mRNA were amplified from each cell by nested PCR, the products were pooled and purified, and sequenced using Illumina MiSeq as previously described 22. For single cell sequencing, reads were assembled, debarcoded, and TCR alpha and beta chains were called for each cell that met acceptable confidence thresholds using a customized pipeline 22. For phenotyping analysis, cells with at least 10 reads of a particular mRNA were considered positive.
TCR beta repertoire sequencing
DNA was extracted from peripheral blood mononuclear cells, then TCRβ CDR3 regions were amplified and sequenced using ImmunoSEQ™ (Adaptive Biotechnologies, Seattle, WA) from 400 ng of DNA template59. Bias-controlled V and J gene primers were used to amplify rearranged V(D)J segments for high throughput sequencing at ~20x coverage. After correcting sequencing errors via a clustering algorithm, CDR3 segments were annotated according to the International ImMunoGeneTics collaboration, identifying which V, D, and J genes contributed to each rearrangement 60.
HLA-A2 tetramer staining and preparation
HLA-A02:01 tetramer containing a UV-cleavable peptide 61 were synthesized by the NIH Tetramer Facility. Potential HLA-A2 binding (predicted IC50 <500nM by netMHC 62) neoantigen peptides from eight HLA-A2 patients were synthesized (Sigma-Aldrich or JPT Peptide Technologies) and exchanged and multimerized with streptavidin-PE as previously described 63. Vaccine-primed PBMCs were collected by apheresis. Cells were pre-treated with 50nM dasatinib for 60–180 minutes. To determine background staining, we used T-cells from leukocyte reduction system cones from 2–3 healthy HLA-A2 donors from the Stanford Blood Center. These cells were barcode-labeled with distinct levels of CellTrace Violet dye (ThermoFisher), to enable multiplexing. Donor cells were treated with dasatinib in parallel and then pooled with patienT-cells immediately prior to tetramer staining. Cells were mixed so that the total number of pooled healthy donor cells equaled that of the patienT-cells. Background staining was determined as the highest frequency seen in a single donor. Tetramer staining was otherwise performed as previously described 63. HIV-gag SLYNTVATL peptide was used as a negative control and CMV-pp65 NLVPMVATV and Flu-M1 GILGFVFTL peptides were used as positive controls.
Neoantigen T-cell purification and expansion
Patient leukocytes were collected by leukapheresis approximately 2 weeks after the series of autologous tumor vaccinations. CD4-expressing T-cell were purified by negative magnetic bead selection using a MACS CD4+ T-cell isolation kit (Miltenyi Biotec). CD4 T-cells were co-incubated with autologous PBMCs collected at a later time, at a ratio of 1:1. Cells were cultured in a 1:1 mix of AIM-V media and RPMI1640 (Thermo Fisher) with 10% autologous plasma and 50uM beta-mercaptoethanol. Neoantigen peptides or matching germline peptides were synthesized (ElimBio) and added to a final concentration of 10ug/mL. For each of the 3 MHC-II peptide families sharing a core sequence, a single 29–30mer peptide was constructed with the shared core sequence in the center. As a positive control, cells were stimulated with a mixture of pathogen-associated peptides, CEFT pool (JPT Peptide Technologies). Cells were incubated for 30 hours prior to FACS sorting. Cells were sorted on live, CD4+, CD137+ cells using a FACS Aria sorter (BD Biosciences). A total of 278 neoantigen specific cells were collected and co-cultured with a pooled mix of 50,000 feeder PBMCs from 3 irradiated (40Gy) allogeneic healthy donors in 1:1 AIM-V/RPMI media with 10% pooled human AB sera (Gemini Bio), beta-mercaptoethanol, 30ng/mL anti-CD3 antibody (OKT3, eBiosciences), 1ug/mL anti-CD28 antibody (CD28.2 eBiosciences), and 10 U/mL recombinant human IL2 (Peprotech). Media was supplemented every 1–3 days with media containing fresh IL2 and beta-mercaptoethanol. Every 7 days, cells were co-incubated with fresh irradiated allogeneic feeder cells, anti-CD3 antibody, and anti-CD28 antibody.
Confirmation of neoantigen T-cell specificity
After ~2.5 weeks of expansion, a fraction of the T-cell culture was rested for 5 days in 1:1 AIM-V/RPMI media with 10% pooled human AB sera (Gemini Bio), beta-mercaptoethanol, but lacking feeder cells, anti-CD3, anti-CD28, or rhIL2. The T-cell cultures were labeled with CellTrace Violet (Thermo Fisher), incubated with autologous PBMCs loaded with either a mix of pathogen-derived peptides, a pool of neoantigen peptides, or a pool of matching germline derived peptides at a concentration of 10ug/mL. Cells were co-cultured for 24hrs prior to antibody staining and FACS analysis. Cells undergoing intracellular staining were treated with monensin (Golgi Stop, BD Bioscience) for 6 hours prior to antibody staining.
T-cell mediated lymphoma cytotoxicity
Neoantigen or viral-specific T-cell cultures were produced as above. After 3 weeks of expansion, activated T-cell cultures were mixed with autologous lymphoma cells at an effector to target ratio of 10:1. Autologous lymphoma cells were labeled with CellTrace Violet dye (Thermo Fisher). After 24 hours, cells were stained with 7-AAD to determine cell death. Target lymphoma cells were gated by CellTrace Violet dye labeling.
Glycosylation site prediction
N-linked glycosylation motifs in patient immunoglobulin heavy and light chain variable regions are predicted based on NXS/T motif. Briefly, we computationally searched for peptide sequences containing proline followed by a random amino acid and a serine or threonine. The first random amino acid cannot be proline.
Data Availability
Peptide data has been deposited in the PRIDE Archive at www.ebi.ac.uk/pride/archive under accession numbers PXD004746 and PXD005704.
Extended Data
Extended Data Figure 1. MHC ligand characteristics.
(a) Length distribution of recovered MHC-I peptides plotted alongside the completed Immuno Epitope Database (IEDB) of human HLA ligands. (b) Length distribution of recovered MHC-I peptides plotted with IEDB human HLA ligand dataset. (c) Weblogo from all 9-mer MHC-I ligands for four representative patients. (d) The overlap of MHC-I ligands for two patients with a completely distinct MHC-I profile (i.e. no shared HLA-A, HLA-B, or HLA-C alleles). The overlap of peptides (left) and genes (right) presented by MHC-I for each patient is shown. (e) MHC-I ligands from patient MCL001 were analyzed for their predicted affinity to either the patient’ own HLA alleles (“self”, brown) or the HLA alleles of patient MCL052 (“nonself”, blue) using netMHC. (f) MHC-I ligands from patient MCL052 were analyzed for predicted affinity to either the patient’s own HLA alleles (“self”, brown) or the HLA alleles of patient MCL001 (“nonself”, blue). White breaks indicate a change in scale of x-axis. (g) The number of unique peptides presented by MHC-I across all patients was determined for each gene. The genes which produced the most unique peptides (top 5th percentile) were analyzed by gene set enrichment using the PANTHER pathways database, revealing enrichment of B-cell activation genes.
Extended Data Figure 2. Correlation of protein abundance with MHC-I and MHC-II presentation.
Two mantle cell lymphoma cell lines, JEKO (left) and L128 (right) were digested with trypsin and peptides identified by LC-MS/MS. (a) Violin plots: distribution of protein abundance is plotted as kernel density violin plots with mean value indicated by a bar for all proteins (grey), proteins presented by MHC-I (blue), proteins presented by MHC-II (red), and mutated proteins where a mutated peptide was identified from whole proteome analysis (green). The distribution of protein abundance for each set of proteins (MHC-I presented, MHC-II presented, and mutated proteins) were compared to the distribution for all proteins by Kolmogorov-Smirnov test. For each line: *p<10−3; **p<10−5; ***p<10−9. (b) The abundance of each protein was estimated using the Histone ruler approach (black). Each protein with at least one peptide presented by MHC-I (blue) or MHC-II (red) is marked with an adjacent tick to the right and left (respectively) of the estimated protein abundance line. Proteins for which a mutated peptide was recovered by whole proteomic analysis (not from MHC immunoprecipitation) are indicated by ticks on the left of the figure (green). Noteworthy genes involved in MCL pathogenesis are highlighted. Proteins for which a mutated peptide was recovered by proteomic analysis are indicated by ticks on the left of the figure.
Extended Data Figure 3. Comparison of antigen presentation between patients.
(a, b) Heatmap of Sorenson similarity coefficient between patients for the set of genes presented by MHC-I (a) and MHC-II (b). Patients were clustered by hierarchical clustering. Gene presentation by MHC was true if one or more peptides encoded by the gene were presented by MHC. (c, d) Heatmap of Sorenson similarity index between patients for peptide ligands presented from MHC-I (c) or MHC-II (d). Patients were clustered by hierarchical clustering. (e, f) Two dimensional visualization of the similarity in MHC ligands between patients. Relationship between samples is shown through a Fruchterman–Reingold layout of a force-directed graph of Sorenson similarity (edges) between patients (nodes) for MHC-I (e) or MHC-II (f). For MHC-I, patients with at least one HLA allele belonging to the HLA-A02 (pink), HLA-B44 (blue) or HLA-A03 (green) supertype family are colored accordingly. For HLA-DR, patients are colored by presence of four specific HLA-DR alleles. Edge weight and color are determined by strength of Sorenson similarity (minimum Sorenson similarity 0.15 per edge). Node size is determined by number of MHC ligands for each patient. Nodes are colored by membership to MHC-I supertype family (e) or HLA-DR allele (f).
Extended Data Figure 4. Presentation of heterozygous single nucleotide polymorphisms.
(Top) Germline heterozygous nonsynonymous SNPs were determined for all seventeen patients. MHC-I and MHC-II associated peptides that span each SNP were identified from our LC-MS/MS dataset. The number of SNP loci which produced an MHC-bound peptide bearing the respective SNP is shown in red. The number of SNP loci which produced an MHC-bound peptide bearing the corresponding hg19 reference allele is shown in blue. Overlap (purple) indicates a SNP loci for which both SNP and hg19 reference allele were identified from the same patient. (Bottom) Analogous depiction of recovery of peptides containing a somatic variant (null set; open circle) or the corresponding germline peptide of the variant (yellow). Comparison of recovery for germline SNPs and somatic variants was performed by Fisher’s exact test.
Extended Data Figure 5. Peptide-MHC tetramer T-cell responses against predicted HLA-A2 neoantigens.
HLA-A2 restricted neoantigens were predicted for 8 patients. Peptide-MHC tetramers were synthesized for 108 predicted HLA-A2 neoantigens. Patient peripheral blood cells were pre-treated with dasatinib and then stained with each tetramer. (a) To enable a more accurate estimation of background staining, patienT-cells were mixed and co-stained with fluorescently-barcoded cells from 2 or 3 healthy donors. A representative reaction with multiplexing of 3 healthy donors using cytomegalovirus peptide-MHC tetramer is shown. (b) Frequency of neoantigen specific CD8 T-cells from peripheral blood T-cells is shown (closed circles). The frequency of neoantigen-specific T-cells from healthy donor PBMCs for each neoantigen is also shown (open squares). Staining of a CMV peptide-MHC tetramer for each patient is shown in red. (c) Representative results for 3 viral peptides derived from HIV, influenza A (flu), and CMV are shown along with patient-specific neoantigen staining. The neoantigen SLC9C2 tetramer is shown as an example of a tetramer with high background staining of both patient and healthy donor CD8 cells.
Extended Data Figure 6. MHC-I presentation of lymphoma immunoglobulin neoantigens in the Raji lymphoma cell line.
(a) Top, MHC-I binding affinity predictions were performed for all potential 9-mer peptides from the Raji immunoglobulin heavy chain. The best binding affinity for an endogenous class I MHC allele is shown in black. The predicted binding to an ectopically expressed HLA-B35:01 allele is shown in red. Bottom, The Raji lymphoma cell line was transfected with the HLA-B35:01 allele. MHC-I antigen presentation profiling was performed for both the parental cell line and the B35-transfected line. Recovered peptides were matched against the Raji immunoglobulin heavy and light chain sequences. Positions differing from the germline variable region were altered by either somatic hypermutation or VDJ recombination are shown in red. Three peptides corresponding to Raji immunoglobulin were identified, shown in boxes. All three were only found in the B35-transfected Raji cells. Asterisks indicate peptides that were identified proteomically. (b) The predicted binding IC50 (nM) based on netMHCpan is shown for each recovered peptide for each of the Raji HLA alleles. Red shading indicates a predicted high affinity interaction with the corresponding HLA allele.
Extended Data Figure 7. Experimental determination affinity of HLA-DRB1*04:01 with associated immunoglobulin neoantigens.
Six neoantigen peptides identified from 3 patients were synthesized with an N-terminal DNP modification and tested for binding to recombinant HLA-DR4 molecules. Recombinant, biotinylated HLA-DR4 molecules were produced with a thrombin-cleavable CLIP peptide. Neoantigen peptides were exchanged unto the DR4 molecules. HLA-DR4 molecules were then bound to streptavidin coated microsphere beads and co-stained anti-HLA-DR antibody and anti-DNP antibody. Beads were then washed and analyzed by flow cytometry for dual staining against HLA-DR and DNP-labelled peptide. A known CMV-derived peptide ligand of HLA-DR4 was used as a positive control. Shown above each plot is the predicted affinity of each peptide for both HLA-DR4 and the associated patient’s alternative HLA-DR allele as predicted by netMHCII. Red letters indicate amino acids that differ from the germline variable gene sequence due to somatic hypermutation events.
Extended Data Figure 8. Phenotyping of neoantigen-specific T-cells.
(a) Peripheral blood CD4 T-cells were isolated from patient MCL041 and stained with HLA-DR*04:01 tetramers loaded with patient specific neoantigen and a CMV peptide. Right, Gated PD1 and CD45RA expression is shown for neoantigen-specific CD4 T-cells (top) and for non-specific CD4 T-cells (bottom). (b) Vaccine-primed CD4 T-cells from patient MCL030 were stimulated with either a pool of neoantigen peptides or a pool of pathogen-associated (CMV, EBV, influenza, tetanus) peptides and were sorted for CD137 upregulation. The sorted population was expanded ex vivo for 2.5 weeks, then rested for 5 days in the presence of low dose IL-15. Cells were then fluorescently labeled and stimulated for 24 hours with unlabeled autologous PBMCs loaded with either a pool of 3 immunoglobulin neoantigen peptides, a pool of 3 corresponding peptides with somatic alterations reverted back to the variable gene sequence, or a pool of pathogen-associated peptides. Activation of the T-cells was determined by induction of CD25 and CD69 in response to peptide stimulation. (c) Neoantigen specific CD4 T-cells from patient MCL030 were expanded, labeled, and re-stimulated as in panel b. Expression of CD25, Ki67, IL4, and granzyme B is shown for CD4-gated T-cells re-stimulated with neoantigen peptide-loaded PBMC.
Extended Data Figure 9. Lack of cytotoxic activity by pathogen-specific T-cells against autologous lymphoma cells.
(Left) CD4 T-cells were purified from patient MCL030 after autologous tumor vaccination. T-cells were stimulated with autologous PBMCs loaded with either a pool of pathogenic peptides including antigens derived from CMV, EBV, influenza A, and tetanus. After 30 hours, cells were sorted for CD137 expression. (Right) Sorted pathogen antigen-specific cells were expanded using anti-CD3, anti-CD28, IL2, and allogeneic feeders for 3 weeks. The expanded T-cells were co-cultured with fluorescently-labeled autologous lymphoma cells for 24 hours (Top right). Background cell death of the lymphoma cells is also shown (Bottom right). Cytotoxicity of lymphoma cells was determined by 7-AAD uptake of the labeled population.
Extended Data Figure 10. Predicted MHC-I presentation of lymphoma immunoglobulin molecules.
(a) Left, Peptides recovered from MHC-I purification were mapped to the immunoglobulin heavy and light chain. The color heatmap corresponds to the number of peptides recovered at each position by LC-MS/MS. Right, For each patient’s unique lymphoma immunoglobulin sequence and HLA profile, the predicted peptide-HLA affinity was calculated for all possible 8–11mers created from their immunoglobulin using netMHCpan. A heatmap illustrating the number of patients (from a total of 17) with at least 1 peptide predicted to bind self-HLA (IC50 ≤ 500nM) at each position across the immunoglobulin heavy chain is shown. (b) For each position along the immunoglobulin molecule, the number of peptides that were experimentally recovered per position (left y-axis) was determined. Similarly, for each position, the number of patients (of 17 total) with at least one peptide with predicted peptide-MHC affinity IC50 <500nM was determined (right y-axis). Positions from the variable region through the N-terminal 50 amino acids of CH1 (red) were compared to the rest of the constant region (green). p value was calculated using Mann-Whitney test. Error bars show range, black bar shows median.
Supplementary Material
supp_guide
supp_info
Acknowledgments
We are grateful to the patients and their families who participated in this study, as well as Max Diehn, Shoshana Levy and members of their laboratories for critical feedback. Supported by grants: NIH U01 CA194389 (M.M.D., R.L., J.E.E., A.A.A.), NIH PPG CA49605 (R.L.), American Society of Hematology Scholar Award (A.A.A.), V-Foundation (A.A.A.), Damon Runyon Cancer Research Foundation (A.A.A.), J.E.E. is a Damon Runyon-Rachleff Innovation Awardee, W.M. Keck Foundation Medical Research Grant (J.E.E.), Conquer Cancer Foundation Young Investigator Award (M.S.K.); Fellow Award from Leukemia & Lymphoma Society. (M.S.K.); Knut and Alice Wallenberg Foundation Postdoctoral Fellowship (N.O); Stanford Translational Research and Applied Medicine Pilot Grant (M.R.G., H.E.K., and A.A.A); NIH S10 RR02933801. We thank the NIH Tetramer Facility for providing recombinant HLA-A0201 for tetramer experiments.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions: M.S.K., N.O., M.M.D., R.L., J.E.E., and A.A.A. developed the concept, designed the experiments, and analyzed the data. R.L. provided patient material from the associated clinical trial. M.S.K., M.R.G., and A.A.A. performed molecular biology, genomic analyses and neoantigen classification with assistance from C.L.L., H.S., H.E.K., D.S., and B.C. N.O., K.R., M.S.K., and J.E.E. performed proteomic studies and analyses with assistance from B.C., S.R., L.Z., K.S., P.L., C.M., and J.C. M.S.K., L.E.W., O.A.W.H., and D.K.C. performed immunologic experiments and analyses. V.E.H.C., M.M., and M.F. performed TCR repertoire sequencing. M.S.K., N.O., B.C., K.R., A.M.N., C.L.L., D.S., J.E.E., and A.A.A. performed integrative data analyses. M.S.K., N.O., R.L., J.E.E. and A.A.A. wrote the manuscript with contributions from all authors, who commented on the manuscript at all stages.
Conflict of interest: V.E.H.C. and M.M. are employees of Adaptive Biotechnologies. M.F. is a former employee of Adaptive Biotechnologies. Other authors declare no conflict of interest.
References and Notes
- 1.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linnemann C, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21:81–85. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- 3.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajasagi M, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014 doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khodadoust MS, Alizadeh AA. Tumor antigen discovery through translation of the cancer genome. Immunol Res. 2014;58:292–299. doi: 10.1007/s12026-014-8505-4. [DOI] [PubMed] [Google Scholar]
- 7.Kreiter S, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kowalewski DJ, et al. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL) Proc Natl Acad Sci U S A. 2015;112:E166–175. doi: 10.1073/pnas.1416389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berlin C, et al. Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. Leukemia. 2015;29:647–659. doi: 10.1038/leu.2014.233. [DOI] [PubMed] [Google Scholar]
- 10.Dutoit V, et al. Exploiting the glioblastoma peptidome to discover novel tumour-associated antigens for immunotherapy. Brain. 2012;135:1042–1054. doi: 10.1093/brain/aws042. [DOI] [PubMed] [Google Scholar]
- 11.Kalaora S, et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget. 2016;7:5110–5117. doi: 10.18632/oncotarget.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassani-Sternberg M, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun. 2016;7:13404. doi: 10.1038/ncomms13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 14.Walz S, et al. The antigenic landscape of multiple myeloma: mass spectrometry (re)defines targets for T-cell-based immunotherapy. Blood. 2015;126:1203–1213. doi: 10.1182/blood-2015-04-640532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green MR, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green MR, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112:E1116–1125. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beà S. Cyclin D1 transcriptional activation in MCL. Blood. 2014;123:1979–1980. doi: 10.1182/blood-2014-02-553057. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123:2988–2996. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beà S, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kridel R, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119:1963–1971. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- 21.Gros A, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quezada SA, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med. 2010;207:637–650. doi: 10.1084/jem.20091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trojan A, et al. Immunoglobulin framework-derived peptides function as cytotoxic T-cell epitopes commonly expressed in B-cell malignancies. Nat Med. 2000;6:667–672. doi: 10.1038/76243. [DOI] [PubMed] [Google Scholar]
- 25.Strothmeyer AM, et al. Comparative analysis of predicted HLA binding of immunoglobulin idiotype sequences indicates T cell-mediated immunosurveillance in follicular lymphoma. Blood. 2010;116:1734–1736. doi: 10.1182/blood-2010-02-270199. [DOI] [PubMed] [Google Scholar]
- 26.Zangani MM, et al. Lymphomas can develop from B cells chronically helped by idiotype-specific T cells. J Exp Med. 2007;204:1181–1191. doi: 10.1084/jem.20061220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwak LW, et al. Induction of immune responses in patients with B-cell lymphoma against the surface-immunoglobulin idiotype expressed by their tumors. N Engl J Med. 1992;327:1209–1215. doi: 10.1056/NEJM199210223271705. [DOI] [PubMed] [Google Scholar]
- 28.Neelapu SS, et al. Vaccine-induced tumor-specific immunity despite severe B-cell depletion in mantle cell lymphoma. Nat Med. 2005;11:986–991. doi: 10.1038/nm1290. [DOI] [PubMed] [Google Scholar]
- 29.Freedman A, et al. Placebo-controlled phase III trial of patient-specific immunotherapy with mitumprotimut-T and granulocyte-macrophage colony-stimulating factor after rituximab in patients with follicular lymphoma. J Clin Oncol. 2009;27:3036–3043. doi: 10.1200/JCO.2008.19.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy R, et al. Active Idiotypic Vaccination Versus Control Immunotherapy for Follicular Lymphoma. J Clin Oncol. 2014 doi: 10.1200/JCO.2012.43.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuster SJ, et al. Vaccination with patient-specific tumor-derived antigen in first remission improves disease-free survival in follicular lymphoma. J Clin Oncol. 2011;29:2787–2794. doi: 10.1200/JCO.2010.33.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for Germline Variant Calling and Somatic Mutation Detection. Curr Protoc Bioinformatics. 2013;44:15.14.11–17. doi: 10.1002/0471250953.bi1504s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai Y, Ni M, Cooper B, Wei Y, Fury W. Inference of high resolution HLA types using genome-wide RNA or DNA sequencing reads. BMC Genomics. 2014;15:325. doi: 10.1186/1471-2164-15-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundegaard C, et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36:W509–512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen M, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Dongen JJ, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 42.Lefranc MP, et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 44.Hunt DF, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 45.Hunt DF, et al. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 46.Barnstable CJ, et al. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 47.Lampson LA, Levy R. Two populations of Ia-like molecules on a human B cell line. J Immunol. 1980;125:293–299. [PubMed] [Google Scholar]
- 48.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 49.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics. 2012;11:M111.010587. doi: 10.1074/mcp.M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 52.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- 53.Vita R, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiśniewski JR, Hein MY, Cox J, Mann M. A “proteomic ruler“ for protein copy number and concentration estimation without spike-in standards. Mol Cell Proteomics. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Day CL, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112:831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirsch IR, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med. 2015;7:308ra158. doi: 10.1126/scitranslmed.aaa9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodenko B, et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat Protoc. 2006;1:1120–1132. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- 62.Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods. 2009;6:497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supp_guide
supp_info
Data Availability Statement
Peptide data has been deposited in the PRIDE Archive at www.ebi.ac.uk/pride/archive under accession numbers PXD004746 and PXD005704.