A futile battle? Protein quality control and the stress of aging (original) (raw)
. Author manuscript; available in PMC: 2019 Jan 22.
Abstract
There exists a phenomenon in aging research whereby early life stress can have positive impacts on longevity. The mechanisms underlying these observations suggest a robust, long-lasting induction of cellular defense mechanisms. These include the various unfolded protein responses of the ER, cytosol, and mitochondria. Indeed, ectopic induction of these pathways, in the absence of stress, is sufficient to increase lifespan in organisms as diverse as yeast, worms, and flies. Here, we provide an overview of the protein quality control mechanisms that operate in the cytosol, mitochondria, and ER, and discuss how they impact cellular health and viability during stress and aging.
Introduction
Stress is the unavoidable pressure and tension experienced by all living organisms throughout life. Stress can be experienced on both a macroscopic scale (e.g. heat stress applied to an entire organism), and a microscopic scale (e.g. a buildup of unfolded proteins in the cell). Because the cells of an organism encounter constant and varied stresses throughout life, several mechanistic stress responses and quality control mechanisms have evolved to deal with a variety of stresses. These include the cytosolic heat shock response (HSR), the mitochondrial unfolded protein response (UPRMT), the endoplasmic reticulum unfolded protein response (UPRER), the ubiquitin-proteasome system, and autophagy. Each of these molecular pathways exists both as a quality control mechanism to preserve cellular homeostasis, and as a response to directly and indirectly damaging stresses, such as exposure to heat or ultraviolet light. Many, if not all, of these stress response pathways are critical to cellular health and organismal survival, and their disruption or inactivation results in decreased lifespan and increased risk of degenerative diseases.
During the aging process, many essential cellular processes begin to break down, and the cellular stress responses are no exception. For example, the HSR has been shown to breakdown at later age, reducing the cellular capacity to respond to heat stress (Morley and Morimoto, 2004). Without a functional HSR, heat-stress can cause irreparable damage to the cell due to an accumulation of damaged and misfolded proteins, which can result in proteotoxicity. A similar age-associated decline occurs in other quality control and stress response pathways, including UPRER, UPRMT, autophagy, and the ubiquitin proteasome system (Cuervo, 2008; Diot et al., 2015; Ekstrand et al., 2007; Taylor and Dillin, 2013; Vilchez et al., 2012). This raises a conundrum analogous to the classical chicken and egg problem: does aging result in a breakdown of protein quality control systems or does the decline of the systems lead to aging? Perhaps the continuous accumulation of damaged proteins and organelles during aging results in increased stress and pressure on these systems. As cellular damage overwhelms the capacity of the quality control mechanisms, they in turn break down as their core components themselves require fully functional organelles and proteins. Indeed, ectopic activation of many quality control mechanisms extends lifespan in many model organisms, suggesting that increased capacity to deal with stress is critical at advanced age.
Here, we attempt to tackle the circular argument of stress response and aging by providing a comprehensive summary of critical protein quality control mechanisms that are responsible for maintaining homeostasis within the cytosol, mitochondria, and ER. As cells have unique and relatively isolated organelles, compartmentalized quality control mechanisms are integral in maintaining protein homeostasis (or proteostasis), especially in the presence of stress and other cellular insults. However, there is increasing evidence that organelles and their quality control mechanisms have significant overlap and are interdependent. Thus, in addition to providing a detailed description of each individual stress response, we also comment on communication between these pathways. Moreover, we speculate how these processes may break down during aging and diseased states due to increased stress and pressure on these systems.
The cytosolic stress response
Many proteins – whether transiently or permanently – reside in the cytoplasm. The cytoplasm hosts a collection of chaperones designated to promote their proper folding and function. Protein misfolding and aggregation can be a serious threat to cellular homeostasis, and perturbations in cellular proteostasis are seen during aging and the pathogenesis of diseases, such as neurodegeneration and cancer (Balch et al., 2008; Dai and Sampson, 2016). To maintain a proper proteostatic landscape, the cell has evolved several cytoplasmic stress response pathways, including the cytoplasmic heat shock response (HSR), autophagy, and the ubiquitin-proteasome system, aimed at preventing and resolving protein damage, misfolding, and aggregation. These pathways complement other compartment specific stress responses, notably the unfolded protein response of the endoplasmic reticulum (UPRER) and unfolded protein response of the mitochondria (UPRmt), and together coordinate proteostasis and cell survival over the lifespan of the organism. In this section, we focus on the HSR and its role in maintaining cytosolic quality control, and briefly cover the roles of autophagy, asymmetric segregation of protein aggregates, and the ubiquitin-proteasome system in relation to HSR, UPRER, and UPRMT (for an in-depth review on general autophagy or the ubiquitin-proteasome system, refer to (Murrow and Debnath, 2013; Tsakiri and Trougakos, 2015).
HSF1 and the heat shock response
In 1962, a unique cytological phenomenon was observed in the polytene chromosomes of the salivary cells of the fruitfly, Drosophila busckii, when the flies were exposed to heat or heavy metals (Ritossa, 1962). Under these stressful conditions, massive regions of the genome appeared to expand relative to others. These expanded, or “puffed,” regions turned out to be regions of active transcription of genes, such as HSP70, required for protection against heat stress (Velazquez and Lindquist, 1984). Now commonly referred to as the HSR, these genes comprise a stress response pathway that is highly conserved from bacteria to humans and found in virtually all organisms examined to date. HSR activation involves upregulation of genes, including chaperones, that promote protein folding and subsequent survival under thermal stress (Tissières et al., 1974).
The transcription factors responsible for eliciting the HSR are known collectively as the heat shock transcription factors, or HSFs. To date, six HSFs have been identified in vertebrates (HSF1-4, X and Y), and up to 50 in plants (Scharf et al., 2012). Invertebrates, on the other hand – for example yeast, worms, and flies – have a single HSF, HSF-1 (heretofore, mammalian HSF1 and invertebrate HSF-1 will be referred to collectively as HSF1). The different HSF proteins in mammals appear to have unique and sometimes tissue-specific functions. HSF1 and 2 are the most broadly expressed and are responsible for activation and modulation of the HSR, respectively. HSF4 is neuronally expressed and plays a role in lens development (Fujimoto et al., 2004), while the function of the remaining paralogs, HSF3, X, and Y, are not as well understood. The invertebrate HSF1, as in vertebrates, serves as the main activator of the HSR.
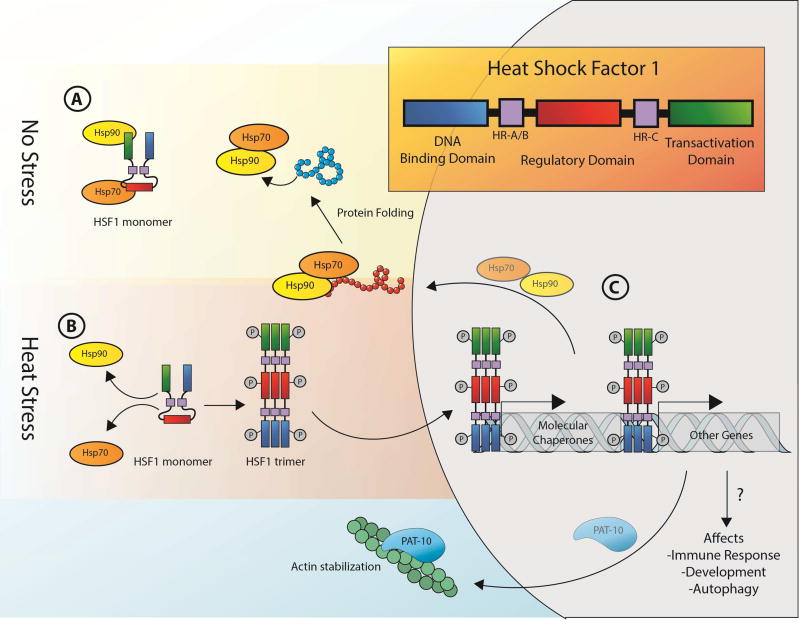
In the absence of stress, HSF1 exists as an inert monomer and cycles between the cytoplasm and the nucleus, bound by the heat shock protein (HSP) chaperones, HSP90, HSP70, and the HSP70 co-chaperone, HSP40 (Fig. 1A). Upon exogenous cellular insult, HSF1 trimerizes and gains DNA binding activity (Fig. 1B). HSF1 has a conserved oligomerization domain, HR-A/B, that is responsible for promoting homo- and heterotypic intermolecular interactions both with other HSF1 molecules, and with HSF2 monomers, resulting in trimerization and subsequent activation (Alastalo et al., 2003; Sarge et al., 1993). Constitutive trimerization is prevented, at least partially, by a repressive domain, HR-C, that interacts with HR-A/B and prevents trimerization. In the nucleus, HSF1 trimers localize to distinct puncta, generally referred to as stress bodies or granules, which are sites of active transcription and correspond to loci of chaperone genes (Jolly et al., 1999; Morton and Lamitina, 2013; Sarge et al., 1993) (Fig. 1C). Activated HSF1 trimers recognize conserved pentameric sequence motifs (nGAAn), termed heat shock elements (HSEs), located near the promoter regions of all heat-induced genes and are necessary and sufficient for HSF1-dependent upregulation (Amin et al., 1988). Recent evidence also supports a model in which HSF1 forms heterotrimers with HSF2. These heterotrimers are important for modulating HSR activity and have also been implicated in development and differentiation (Östling et al., 2007; Sandqvist et al., 2009).
Fig. 1. Role of HSF1 in the heat shock response.
In normal, steady-state conditions, HSF1 exists as an inactive monomer bound by HSP chaperones, including Hsp70 and Hsp90 (A). Under conditions of stress, HSF1 is released from Hsp70 and Hsp90, allowing it to trimerize, become phosphorylated, and gain DNA binding activity (B). In the nucleus, HSF1 trimers localize to conserved pentameric sequence motifs (nGAAn), called HSEs, to promote the expression of HSR genes that are necessary for protein repair and cell survival under heat stress (C).
The regulation of HSF1 activity is both multi-faceted and complex, with its modulation ranging from intrinsic repressive domains, discussed above, to protein-protein and even protein-RNA interactions (for a comprehensive analysis of HSF1 regulation, refer to (Gomez-Pastor et al., 2017)). Under steady state conditions, HSF1 is bound by cytoplasmic chaperones, HSP90 and HSP70/40, keeping it in a repressed, monomeric state. Upon cellular insults that result in increased misfolded or unfolded proteins, the chaperones are titrated away, resulting in the alleviation of repression and subsequent trimerization of HSF1 (Fig. 1). Consistent with this hypothesis, genetic or biochemical perturbations of chaperone levels result in an increased activation of the HSR (Guisbert et al., 2013). It is well accepted that HSP70/40 is a negative regulator of HSF1; indeed, overexpression negatively affects HSF1 transcriptional activity. HSP90 has been reported to have a similar negative regulatory role (Zou et al., 1998). However, purified HSP90 does not affect HSF1 trimerization or DNA binding activity in vitro, suggesting that regulation of HSF1 by HSP90 may be complex (Hentze et al., 2016). It is possible that HSP90 functions as part of a regulated negative feedback mechanism, and that a critical threshold of HSP90 in combination with other downstream targets of HSF1 is required to repress HSF1. In addition to HSP70/40 and 90 impacting HSF1 activation, the cytoplasmic chaperonin complex (TriC/CCT) has also been shown to bind and repress HSF1-mediated upregulation of chaperone genes. Consistent with this, a small molecule inhibitor of TRiC/CCT, HSF1A, which prevents the interaction between HSF1 and the TRiC/CCT complex, releases HSF1 inhibition and upregulates HSF1-dependent gene expression (Neef et al., 2014).
Physiological Roles of Heat Shock Factor
HSF1 is the master transcriptional regulator of stress response genes, including chaperones, ubiquitin, and translational regulators. Perhaps most well-studied is the robust activation of heat-shock proteins (HSPs), which include molecular chaperones essential for protein refolding, such as HSP70, HSP90, DNAJ, and HSP60, and components of the ubiquitin proteasome system to remove damaged proteins (reviewed in (Labbadia and Morimoto, 2015; Li et al., 2017). Since its initial discovery, HSF1 has now been implicated in many other forms of stress response outside of the HSR, including autophagy, actin, and innate immunity as described below. It is clear that HSF1 promotes general cytosolic quality control and protects cellular homeostasis against protein damage, misfolding, and aggregation through multiple pathways. Due to its multi-faceted roles in numerous cellular defense mechanisms, overexpression of HSF1 can delay aging. Indeed overexpression of HSF1 in C. elegans significantly extends lifespan (Baird et al., 2014; Hsu et al., 2013). Conversely, downregulation of HSF1 shortens lifespan and results in premature aging phenotypes (Garigan et al., 2002). Moreover, depletion of downstream targets of HSF1, such as HSR chaperones, Hsp70s and Hsp90s, similarly results in decreased lifespan, suggesting that the chaperone proteostasis network is important for the longevity conferred by HSF1 (Morley and Morimoto, 2004). Interestingly, HSR genes are poorly induced in aged animals (Fargnoli et al., 1990; Fawcett et al., 1994) and several lines of evidence suggest that the HSR declines as a function of age (Gagliano et al., 2007; Kayani et al., 2008; Sherman and Goldberg, 2001). However, the exact connection between HSR capacity and aging is complex. HSF1 activity is diminished in old cells, and recovery after heat-shock is largely attenuated (Heydari et al., 2000; Kern et al., 2010), but whether the expression of HSF1 declines as a function of age depends on the organism and tissue type,. Furthermore, ectopic expression of HSF1 during development promotes lifespan extension, whereas overexpression of HSF1 later on in adulthood has less of an impact (Volovik et al., 2012). It is possible that presence of HSF1 alone is not sufficient to impact lifespan during advanced aging due to the lack of key regulatory elements to promote HSF1 signaling. In-depth analysis of the expression profile of HSF1 target genes in wildtype and HSF1 overexpressing animals during late adulthood would provide intriguing insight to these unanswered questions.
HSF1 dysregulation is not unique to aging given its role in human diseases, such as metabolic syndrome, cancer, and neurodegeneration. For example, downregulating HSF1 can result in neurodegeneration due to accumulation of toxic misfolded species (Garigan et al., 2002; Morley et al., 2002; Wang et al., 2009), whereas activating HSF1 can prevent PolyQ-mediated neurodegeneration in mouse, worm, and fly models (Fujikake et al., 2008; Fujimoto et al., 2005; Neef et al., 2010). The protective effects of HSF1 hyperactivation is thought to be mediated primarily through upregulation of HSPs, as overexpression of HSPs or pharmacological activation of HSPs is sufficient to suppress proteinopathic neurodegeneration (Bonini, 2002; Fujikake et al., 2008). However, HSF1 can also have protective effects in neurons that are not experiencing protein misfolding stress (Verma et al., 2014), suggesting a regulation of lifespan independent of the presence of misfolded proteins. Indeed, HSF1 is essential for lifespan extension in several lifespan extension paradigms, including inhibition of insulin/IGF-1 and mTOR signaling pathways. Specifically, HSF1 knockdown – or knockdown of downstream targets of HSF1 – eliminate the lifespan extension caused by daf-2 (insulin/IGF-1 receptor) and rsks-1 (S6K) mutations in C. elegans (Hsu et al., 2003; Seo et al., 2013). Interestingly, overexpression of HSF-1 in neurons alone is sufficient to extend lifespan in C. elegans. Known as the non-autonomous HSR, activation of HSR in neurons – either in the presence or absence of stress – results in an organism-wide activation of HSF1 (Prahlad et al., 2008). Specifically, neuronal overexpression of HSF1 results in generation of a transcellular signal dependent on serotonergic neurons (Tatum et al., 2015), which activates both DAF-16 (FOXO transcription factor) and HSF-1 in the intestine (Douglas et al., 2015).
The above discussion underlines HSF1’s broad roles in proteome stability and lifespan, and may be taken to suggest that activation of HSF1 may be therapeutically beneficial. Studies suggest, however, that hyperactivation of HSF1 may not be an ideal therapeutic approach due to its proteostatic roles, and potentially also HSP-independent roles. Although it is not classified as an oncogene, several cancers nevertheless rely on HSF1 activity for growth – a state referred to as non-oncogenic addiction (for a thorough review, refer to (Dai and Sampson, 2016)). For example, genetic deletion of HSF1 in mice attenuates chemically and genetically-induced lymphomagenesis (Dai et al., 2007; Min et al., 2007). Moreover, HSF1 knockdown by RNAi is sufficient to decrease the growth and invasion of hepatocellular carcinomas (Chen et al., 2013; Fang et al., 2012) and melanomas (Fujimoto et al., 2012; Nakamura et al., 2014). Conversely, overexpression of HSF1 promotes growth and metastasis of melanoma cells (Scott et al., 2011; Toma-Jonik et al., 2015). While the exact mechanism underlying the role of HSF1 in cancer progression is still under investigation, it is widely recognized that the HSPs are critical for protecting the cancer proteome. HSF1-regulated chaperones play an important role in promoting the stability of oncoproteins, such as BCR-ABL (common in patients with leukemia) and mutant TP53 (oncogenic P53), and HSF1 suppression can result in depletion of oncoproteins and a decrease in cancer cell survival (Dai et al., 2012; Li et al., 2014; Miyajima et al., 2013; Workman and van Montfort, 2014). Additionally, HSF1 mediates a transcriptional program distinct from the HSR to promote tumor malignancy that is associated with decreased survival in patients with breast cancer (Mendillo et al., 2012). Due to the growing number of studies implicating HSF1 in cancer progression, antagonizing HSF1 signaling is quickly becoming an attractive therapeutic intervention. However, since HSF1 signaling is critical for maintaining the cellular proteome, particularly under conditions of thermal stress, broad inhibition of HSF1 may have some detrimental effects. Thus, defining the role of HSF1 in cancer and elucidating how this role may be harnessed or targeted therapeutically will require further work. Would HSF1 inhibition result in loss of protein homeostasis and premature cell death in non-cancer cells? Would HSF1 inhibition alone be sufficient to kill cancer cells, and would they eventually become resistant to HSF1 inhibition? These and many other questions must first be addressed before moving forward with HSF1-targeting drugs as therapies for human disease.
Non-canonical roles of HSF1
Aside from the clear role in HSR modulation, HSF1 affects aging via modulation of additional processes such as autophagy, the immune response, and maintenance of the actin cytoskeleton. Intriguingly, some studies suggest that chaperones are not responsible for HSF1-mediated thermotolerance and longevity. In C. elegans, neither hypomorphic mutations of HSF1 nor perturbations of chaperones had an impact on thermotolerance (Kourtis et al., 2012; McColl et al., 2010). Moreover, overexpression of HSF1 harboring a C-terminal truncation showed dramatic increase in lifespan, despite being incapable of upregulating canonical heat-shock proteins and chaperones involved in HSR, suggesting that HSF1 acts via an alternative pathway in this case (Baird et al., 2014). Proteomic analysis comparing transgenic animals overexpressing the truncated version of HSF1 and the full length protein identified genes involved in cytoskeletal organization. Further analysis suggested a role for HSF1 in mediating actin cytoskeletal maintenance through the troponin-like calcium-binding protein, PAT-10. Specifically, HSF1 promotes PAT-10 expression, which improves actin cytoskeletal integrity, thermotolerance, cellular trafficking, and ultimately extends lifespan independent of chaperone induction through canonical HSR (Baird et al., 2014). Understanding this and other non-canonical functions of HSF1 may have significant therapeutic implications. As discussed above, HSF1 has oncogenic properties that may counteract its beneficial effects. However, targeting of downstream effectors of HSF1 signaling, independent of chaperones, may provide alternative avenues for therapeutic intervention.
HSF1 and autophagy
HSF1 may act both alongside and as a regulator of autophagy. Under heat stress, proteins are denatured, translation is altered, and cellular homeostasis as a whole is disrupted. Ramping up the HSR as described above upregulates expression of chaperones and other molecular players important in remodeling the protein architecture of the cell by promoting protein refolding and removal of irreparable, damaged proteins. However, damaged proteins can also aggregate, or localize to organelles and compartments inaccessible to chaperones. Moreover, organelles themselves may be functionally impaired under heat stress. Autophagy acts as the primary means for a cell to clear these larger debris that are inaccessible to more focal proteolytic systems (Klionsky and Codogno, 2013). Therefore, HSR and autophagy represent functionally distinct, yet complementary, systems of protein quality control with the similar goal of preserving protein homeostasis under stress. Indeed, a recent study has identified that activation of autophagy is essential for the lifespan extension of C. elegans under early exposure to heat shock or through HSF1 overexpression (Kumsta et al., 2017). Similarly, autophagy is upregulated under conditions of heat stress in many in vitro and in vivo mammalian studies (Chao et al., 2014; Han et al., 2013b; Nivon et al., 2009; Oberley et al., 2008). These studies highlight the importance of the interplay between HSR and autophagy in stress resistance and lifespan regulation. However, how HSF1 modulates autophagy and the cross-talk between HSR and autophagy is poorly understood.
Direct mechanistic insight on the role of HSF1 in autophagy is lacking, and molecular models are currently controversial at best. A limited number of studies have implicated HSF1 in direct regulation of autophagy. In mammalian cell lines, knockdown or deletion of HSF1 results in increased basal autophagy (Dokladny et al., 2013), while constitutive activation of HSF1 results in decreased autophagy (Zhao et al., 2009). HSF1 may prevent induction of autophagy by promoting the expression of HSP70, whose overexpression similarly inhibits autophagy. It is also possible that HSF1 can preferentially activate HSR over autophagy (Dokladny et al., 2013). In contrast, overexpression of HSF-1 in C. elegans increases autophagosome formation in muscle, neurons, and intestine. Several genes involved in autophagy are also necessary for HSF-1-mediated lifespan extension (Kumsta et al., 2017). These apparently contradictory results may reflect a difference between acute and chronic stress responses. It is possible that in an acute stress response, HSF1 induces HSR and preferentially promotes damage repair by protein remodeling. However, under chronic stress when damage has exceeded repair capacity or when HSR has failed, autophagy becomes the primary means by which cells promote proteostasis. Indeed, in human peripheral blood mononuclear cells, cells obtained from older patients had compromised ability to upregulate HSP70 and an increase in markers for autophagy (Lanphere et al., 2013). While there is evidence that both HSR and autophagy decline with age (Del Roso et al., 2003; Hsu et al., 2013; Kurapati et al., 2000; Rubinsztein et al., 2011), it would be interesting to investigate whether autophagy can partially compensate for defects in HSR under specific stress conditions, such as advanced aging.
HSF1 and the immune response
High temperature and fever is a principle response to pathogenic infection in metazoans: Homeotherms are capable of increasing their own body temperatures, whereas poikilotherms migrate to warmer temperatures to increase body temperatures during infection. Febrile temperatures can benefit the organism l by promoting expression of HSPs in an HSF1-dependent manner (Hasday et al., 2014; Ostberg et al., 2002). Many of the chaperones induced by heat-stress are also expressed in inflammatory diseases (van Eden et al., 2005), making it clear that the HSR and immunity are tightly linked. For example, during pathogenic infection in C. elegans, expression of HSP-16 and HSP-90 are essential for survival. Moreover, overexpression of HSF1 increases resistance to Pseudomonas aeruginosa infection (Singh and Aballay, 2006). The role of HSF1 in immunity is not limited to HSPs: HSF1 can also induce chemokine CXCL8 (IL-8) activation in models of acute lung inflammation to promote recruitment of neutrophils under conditions of hyperthermia (Singh et al., 2008; Tulapurkar et al., 2012). Other inflammatory chemokines that recruit natural killer cells, T cells, and monocytes (CXCL9, CXCL10, CXCL11, and CXCL12) have also been shown to contain putative HSF1-binding sites (Hasday et al., 2014). These data support the hypothesis that HSF1 can promote immunity under specific stress conditions, such as heat-shock.
In contrast to those studies arguing a positive role for HSF1 in immunity, a study in mouse models showed that HSF1 can be a negative regulator of the inflammatory genes, TNF-α and IL-1β (Cahill et al., 1996). In addition, other studies report that HSP70 can act as a negative regulator of inflammatory responses during systemic infection by preventing the activation of pro-inflammatory cytokines (Aneja et al., 2006; Singleton and Wischmeyer, 2006) and by down-regulating the production of TNF-α and Toll-like receptors in an HSF1-dependent manner (Ferat-Osorio et al., 2014). While the interactions between HSF1 signaling and the immune response are complex, it makes sense that the HSPs involved in proteostasis would be crucial in the highly stressed environment of pathogenic infection. Further studies, with particular focus on immune responses under acute and chronic stress, may help to resolve some of the apparent contradictions in the effects of HSF1 on immunity. For example, it is possible that under acute stress, HSF1 first promotes activation of the inflammatory response. However, as downstream targets of HSF1 accumulate, they themselves can act in a negative feedback loop to dampen the immune response, reminiscent of the negative feedback loop mediated by HSP70 in regulation of HSF1 (Abravaya et al., 1992).
Like the age-dependent decline in many quality control and stress response pathways, advanced age is also associated with a decline in immune function, a process known as immunosenescence (Pawelec and Solana, 1997). Aging is also accompanied by a decline in the capacity to recruit macrophages and other immune cells upon infection (Aprahamian et al., 2008; Ashcroft et al., 1998). It would be interesting to determine whether the perturbations in regulation of the immune response are due to defects in HSF1 signaling and HSR at advanced age. For example, would overexpression of HSF1 or ectopic activation of HSR promote a viable immune response at old age, similar to its effects on lifespan?
Asymmetric segregation of protein aggregates during cell division
As cells age, damaged proteins will begin to accumulate, and will eventually surpass the capacity of existing quality control mechanisms. This is exacerbated even further as quality control mechanisms begin to collapse during advance age. In actively dividing cells, damaged cellular components that cannot be repaired or eliminated within the cell can be partitioned off into one cell, leaving the other cell free of this “cellular debris.” For example, in budding yeast, damaged protein aggregates are preferentially retained in the original mother cell, producing a new daughter cell that is clear of damaged proteins (Aguilaniu et al., 2003; Liu et al., 2010). While the exact mechanism is controversial, it is clear that protein aggregates are preferentially retained in the mother cell. While a comprehensive overview of the field is beyond the scope of this review (refer to (Higuchi-Sanabria et al., 2014; Hill et al., 2017) for more detailed reviews), one model proposes that this process involves two complementary mechanisms: an actin-dependent physical retention of protein aggregates in the mother cell (Hill et al., 2016, 2017; Liu et al., 2010), and clearance of protein aggregates in the daughter cell (Hill et al., 2014). Several chaperones are involved in recognition and clearance of protein aggregates, some of which are direct targets of HSF1, including Hsp70.
Asymmetric segregation and damage retention are conserved in higher eukaryotes. For example, damaged proteins are sequestered in what are called “aggresomes,” which are asymmetrically apportioned in mammalian cells in a vimentin and tubulin-dependent manner (Ogrodnik et al., 2014; Rujano et al., 2006). In mammalian systems, cells destined to have longer longevity are rejuvenated, whereas those with shorter chronological lifespan, such as differentiating progeny of intestinal stem cells, retain damaged proteins (Bufalino et al., 2013). Beyond cell division, there exist evidence that post-mitotic cells also apportion damaged proteins into isolated environments to preserve cellular function. In C. elegans, misfolded proteins and organelles can be exuded out of the cell in membrane-bound vesicles termed “exophers,” which decrease proteotoxicity and promote health and function in neurons (Melentijevic et al., 2017). Analogous processes may promote cellular health in mammalian cells. Indeed there exist some evidence that aggregated poly-Q proteins can transfer between neurons, although the direct physiological consequence of these transfers is unclear (Costanzo et al., 2013).
Mitochondrial quality control and the UPRMT
While cytosolic quality control mechanisms, such as those mediated by HSF1, can have indirect beneficial effects on organelles, such as mitochondria (see below), these distinct environments have dedicated mechanisms to preserve their quality and function. The mitochondrion is an essential, membrane-bound organelle that serves as the primary source of energy production in the cell. Beyond its well-known function in metabolism, it also serves as a key regulator of other important processes, such as calcium signaling and apoptosis. As the central hub of energy production and housing a plethora of redox reactions, mitochondria are under constant oxidative stress. Accumulation of oxidative damage during cellular life is thus inevitable. Indeed, mitochondrial damage and loss of organelle function are hallmarks of advanced aging, implicated in age-related diseases, such as neurodegeneration and muscle myopathies (Letessier et al., 2007; Ramadasan-Nair et al., 2014; Stauch et al., 2014). Not surprisingly, the cell has evolved numerous molecular pathways aimed at preserving the quality and function of this significant organelle. These protective mechanisms range from protein quality control (including UPRMT and mitochondria-associated degradation), mitochondrial fusion and fission, mitophagy, and mitochondrial biogenesis. As all of these mechanisms rely on nuclear-encoded proteins, it is imperative for the mitochondria to be in continuous communication with the nucleus. This cross-talk is achieved via mitochondrial-nuclear signaling. In this section we discuss the UPRMT and other quality control mechanisms, which preserve the mitochondria both basally and under conditions of stress.
The mitochondrial unfolded protein response
Mitochondrial fitness and function is inarguably significant to cell viability, and, as mentioned above, its dysfunction is linked to aging and diseased states. However, counterintuitively, a growing number of studies have shown that a decline in or perturbations of mitochondrial function can lead to lifespan extension (Dillin et al., 2002; Liu et al., 2005; Owusu-Ansah et al., 2013; Yee et al., 2014). The reason for these apparently contradictory observations may have to do with the fact that impairment of mitochondrial fitness and function triggers the UPRMT, which results in beneficial effects on organismal health and lifespan (Durieux et al., 2011). In this section, we describe this response.
The UPRMT was originally identified in mammalian cells in which loss of the mitochondrial genome led to upregulation of mitochondrial chaperones, and was so named due to presumed effects on protein folding within mitochondria (Martinus et al., 1996). In the ensuing years, it has been shown that this stress response pathway can be activated upon mitochondrial insult and that it promotes nuclear transcription of genes involved in protein folding, as well as ROS defense, metabolism, and the innate immune response (Nargund et al., 2015; Schulz and Haynes, 2015). The UPRMT is activated under a variety of conditions including loss of mitochondrial proteostasis, mitonuclear imbalance, impaired mitochondrial physiology (for example, decreased electron transport chain function, perturbed redox environment, and so on), and oxidative damage, many of which are characteristic of advanced age (Dillin et al., 2002; Owusu-Ansah et al., 2013; Yoneda et al., 2004).
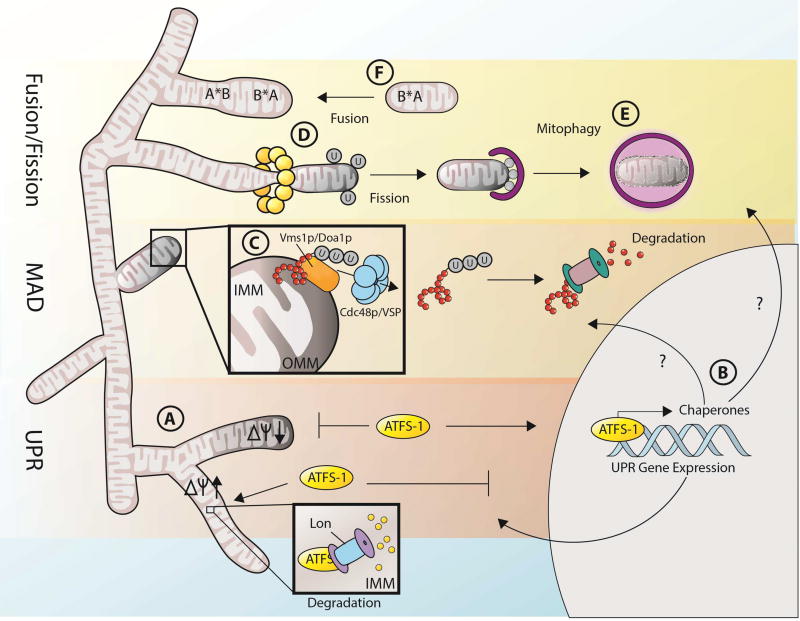
While mitochondria have their own genome, it encodes only a few genes, and they do not include stress-response genes. Therefore, just as stress within the ER or cytosol has to be efficiently communicated to the nucleus to activate a broad program of gene expression for recovery, mitochondrial stress must also be communicated to the nucleus. This process has been studied most extensively in C. elegans, where UPRMT is regulated at least in part by a transcription factor, ATFS-1, which can shuttle between mitochondria and nucleus (Fig. 2A). ATFS-1 contains both a nuclear localization sequence and mitochondrial signal sequence. Under basal conditions, ATFS-1 is imported into mitochondria where it is degraded by the Lon protease. When mitochondrial import is compromised, for example under conditions of mitochondrial stress, ATFS-1 import is reduced, resulting in accumulation and nuclear translocation of ATFS-1, and transcriptional activation of stress response genes (Nargund et al., 2012). ATFS-1 transcriptional activity is further regulated by the ubiquitin-like protein, UBL-5, and the homeobox protein, DVE-1. During conditions of mitochondrial stress, UBL-5 and DVE-1 dimerize, and together with ATFS-1 promote global transcription of genes required to regain mitochondrial homeostasis, including the mitochondrial chaperones, hsp-6 and hsp-60 (Benedetti et al., 2006; Durieux et al., 2011; Haynes et al., 2007; Houtkooper et al., 2013).
Fig. 2. Mitochondrial quality control is maintained by a series of protective mechanisms.
The mitochondria has several dedicated quality control mechanisms, which repair and preserve the organelle both in the presence and absence of stress. UPRMT is regulated by ATFS-1, which is imported into the mitochondria and subsequently degraded by Lon protease under steady-state conditions (A). When the mitochondria is damaged and membrane potential is insufficient to drive import of ATFS-1 into the matrix, ATFS-1 shuttles to the nucleus to act as a transcription factor to promote expression of mitochondrial chaperones and other genes important in repairing damaged mitochondrial proteins and other organelle components (B). Damaged proteins in the mitochondria can also be selectively removed through mitochondria-associated degradation (C). Here, damaged proteins are presented to the outer surface of the mitochondria, ubiquitinated by substrate-specific E3 ubiquitin ligases, then pulled out of the mitochondria by the concerted efforts of Vms1p/Doa1p and Cdc48p. Once outside of the mitochondria, damaged proteins can be degraded by the cytosolic proteasome. On a larger scale, regions of damaged mitochondria can be teased apart from the rest of the mitochondria via fission (D). This damaged mitochondrial unit is subsequently targeted for degradation by the lysosome in a process known as mitophagy (E). Less critical damage, such as minor mutations in mtDNA or damage to specific mitochondrial proteins, can be complemented by fusion to mitochondria harboring fully functional mitochondrial components (F). Here, minor damage is demarcated with an asterisk (*). A mitochondrion harboring a damaged protein B*, but a fully functional protein A, can complement and itself be complemented via fusion to another mitochondrion harboring a fully functional protein B, but a damaged protein A*.
Recent studies have also shown that mitochondrial stress results in widespread changes to chromatin structure, which can be maintained throughout organismal lifespan. Upon exposure to stress, the histone methyltransferase, MET-2, interacts with the nuclear co-factor, LIN-65, to promote H3K9-dimethylation, which promotes recruitment of DVE-1 to specific gene loci, ultimately activating transcription of UPRMT target genes (Tian et al., 2016). It is possible that these epigenetic changes can be carried into adulthood, and that they extend lifespan in organisms experiencing stress during early life. Indeed, an independent study has shown that overexpression of two histone lysine demethylases, PFH8 and JMJD3, is sufficient to activate UPRMT in the absence of stress and extend lifespan, providing further evidence that epigenetic changes under mitochondrial stress can have long-lasting effects throughout lifespan (Merkwirth et al., 2016).
The UPRMT signaling network has not yet been fully characterized in mammals, though it has been proposed that the CHOP-C/EBPβ heterodimer acts to transcriptionally activate mitochondrial chaperones and proteases (Aldridge et al., 2007; Horibe and Hoogenraad, 2007; Zhao et al., 2002). Unfolded proteins in the mitochondrial matrix stimulate the phosphorylation of JNK2, resulting in transcriptional activation of CHOP and activation of UPRMT (Aldridge et al., 2007; Zhao et al., 2002). In addition, the mammalian transcription factor, ATF5, which is induced in response to stressed conditions, such as defects in mtDNA replication (Tyynismaa et al., 2010), has been shown to mediate mammalian UPRMT (Fiorese et al., 2016). Specifically, shRNA knockdown of ATF5 in HEK293T cells suppressed induction of UPRMT target genes, including HSP70, LONP1, and HD-5, when cells were treated with paraquat, a drug that induces ROS production in the mitochondria. Moreover, loss of mitochondrial membrane potential was sufficient to induce ATF5-mediated upregulation of these genes, suggesting that its molecular mechanism was similar to that of ATFS-1. Perhaps most interesting is that ATF5 can rescue defects in UPRMT and promote mitochondrial function in C. elegans lacking ATFS-1. Taken together these data support a role for ATF5 in mediating UPRMT in mammalian cells similar to ATFS-1 function in C. elegans.
The activation of UPRMT is accompanied by a broad transcriptional response with pleiotropic effects on organismal health. UPRMT genes affect NAD+, TOR, ROS, and insulin signaling pathways, all of which have been implicated in lifespan regulation (Gomes et al., 2013; Haynes et al., 2007; Jensen and Jasper, 2014; Zarse et al., 2012). Furthermore, a seminal study in human cells using pharmacological inhibitors of mitochondrial matrix chaperones HSP90/TRAP1 and mitochondrial LON protease to activate UPRMT has identified downstream targets of UPRMT, including factors involved in pre-RNA processing, translation, and protein folding (Münch and Harper, 2016). While an in-depth review of these individual pathways is beyond the scope of this review, in combination these studies provide clear evidence that perturbations in mitochondrial function, such as ETC dysfunction, impact longevity (Dillin et al., 2002).
Interestingly, activation of UPRMT under stress exposure is not limited to the confines of a single cell. Rather, when one cell experiences mitochondrial stress, it can send a signal to distal tissues, promoting further UPRMT in a non-autonomous manner. In C. elegans, ETC disruption or expression of toxic PolyQ aggregates in neurons results in massive induction of UPRMT in the intestine, measured by induction of hsp-6, a mitochondrial chaperone downstream of UPRMT (Berendzen et al., 2016; Durieux et al., 2011). As neurons do not innervate the intestine in C. elegans, the authors of these studies have hypothesized that a specific chemokine (or “mitokine”) must be responsible for this non-autonomous signaling from neurons to intestines. Consistent with this idea, the neuronal transmission of UPRMT to the intestine requires the release of dense core vesicles and the neurotransmitter serotonin. In addition, UPRMT components are required in peripheral tissues to respond to neuronal cues as RNAi knockdown of dve-1, atfs-1, ubl-5, and clpp-1 suppressed UPRMT activation in peripheral tissues under polyQ40-induced mitochondrial stress in neurons (Berendzen et al., 2016). Beyond this, an elaborate neuron-specific CRISPR screen has identified six neuropeptides required for UPRMT across tissues. Only one of these six neuropeptides, FLP-2, was sufficient to induce non-autonomous UPRMT, such that its deletion blocks non-autonomous UPRMT signaling, while its expression solely in neurons is sufficient to induce UPRMT in the intestine (Shao et al., 2016). Evidence of non-autonomous UPRMT signaling is also present in mammals. Human patients with mitochondrial myopathies secrete elevated levels of FGF-21 from muscles, which can promote ketogenesis in the liver and general lipid metabolism, both direct physiological consequences of UPRMT activation (Suomalainen et al., 2011). Similarly, mice with mitochondrial impairment in muscle cells due to defects in autophagy also show increased expression of Fgf21. These mice have a significant decline in fat mass and improved insulin sensitivity (Kim et al., 2013).
While many studies have shown that activation of UPRMT promotes mitochondrial quality and lifespan, other studies suggest that prolonged exposure to a hyperactive stress response pathway can be deleterious. For example, a recent study in C. elegans has shown a role for UPRMT in preserving mutant mitochondrial DNA (mtDNA) harboring a 3.1-kb deletion of four OXPHOS subunits. In these strains, hyperactivation of UPRMT, by expression of a constitutively active ATFS-1, results in enhanced propagation of mutant mtDNA and subsequent loss of efficiency in respiration (Lin et al., 2016). Similarly, there is evidence that UPRMT can promote cancer cell survival. Using superoxide dismutase 2 (SOD2) as a marker for UPRMT, Kenny et al. found that aggressive and metastatic primary breast cancers have elevated UPRMT activity and hence propose a model for UPRMT as a means to maintain mitochondrial fitness and integrity throughout metastasis (Kenny et al., 2017). Furthermore, key UPRMT components, such as ATF5, have been shown to be essential for the progression of glioblastomas (Arias et al., 2012). These data point to UPRMT as a potential candidate for therapeutic intervention against cancer, but it remains unclear whether the repression of UPRMT would be detrimental to human health. A better understanding of the mechanistic pathways involved in both the positive and negative consequences of UPRMT is essential before it can be considered as a target for any human disease.
Mitochondrial housekeeping: maintaining mitochondrial quality control
As described above, the cell can mount the UPRMT to upregulate chaperones to respond to unfolded protein stress and repair protein damage. Housing the electron transport chain and its many redox reactions, the mitochondrion is subject to oxidative damage through production and accumulation of ROS, which can damage proteins, lipids, and DNA (Tal et al., 2009). In addition to UPRMT, damaged mitochondrial proteins can also be removed via mitochondria-associated degradation (MAD), which involves the physical removal of damaged proteins and their degradation through the ubiquitin-proteasome quality control pathway (Tanaka et al., 2010) (Fig. 2B). Briefly, damaged or unnecessary proteins are retrotranslocated to the outer mitochondrial membrane, where they are targeted for MAD by ubiquitination through multiple E3 ubiquitin ligases. These ubiquitinated proteins recruit p97 (Cdc48p in yeast), which extracts the ubiquitinated proteins from the mitochondria and presents them to the proteasome for degradation, reminiscent of the process of ER-associated degradation (ERAD) described below (Chatenay-Lapointe and Shadel, 2010; Xu et al., 2011). Although the exact mechanism of MAD is not fully understood, it is known that in yeast, Vms1p (homologue of mammalian valosin-containing protein, VCP) and/or Doa1p recruit Cdc48p to the mitochondria through specific cofactors, some of which include Ufd2p (Heo et al., 2010; Wu et al., 2016).
As a dedicated mechanism to preserving the mitochondria, modulation of MAD is likely to have repercussions in aging and disease pathology. For example, mutations in critical components of MAD, such as Cdc48p and VCP result in decreased membrane potential and cell death, both of which are hallmarks of aging and disease (Bartolome et al., 2013; Braun et al., 2006). Moreover, Vms1p overexpression and ectopic activation of MAD can counteract mitochondrial dysfunction and cell death in yeast expressing a variant of ubiquitin B associated with Alzheimer’s disease (Braun et al., 2015). Several lines of evidence suggest that the efficiency of the ubiquitin proteasome system declines as a function of age and in age-related diseases, and that its ectopic activation can alleviate proteotoxic stress and have beneficial effects on lifespan (Ekstrand et al., 2007; Kujoth et al., 2005; Vilchez et al., 2012). These changes are likely to have direct effects on MAD. It remains unclear whether other critical components of MAD also become dysfunctional with age, and whether they may be targeted to correct age-related deficiencies in protein homeostasis and mitochondrial deficiencies.
Beyond protein repair, mitochondria can also undergo dramatic alterations in structure and function via fusion and fission of the compartment, which reorganize the organelle under conditions of stress. The first discovery that alteration of fusion and fission dynamics can alter lifespan came from a study in two fungi models, which showed that a push towards mitochondrial fusion can extend lifespan by perturbing apoptosis (Scheckhuber et al., 2007). Since then, a number of studies have linked the beneficial effects of both fusion and fission to lifespan and disease (Chaudhari and Kipreos, 2017; Higuchi-Sanabria et al., 2016a; Rana et al., 2017; Weir et al., 2017). While a comprehensive analysis of fusion and fission is beyond the scope of this review, below we highlight the impact of mitochondrial dynamics on some quality control pathways, including complementation, mitophagy, and asymmetric cell division (refer to the following reviews for comprehensive analysis of mitochondrial fusion and fission (Trotta and Chipuk, 2017; Westermann, 2010, 2012).
Mitochondrial fusion can promote complementation between damaged mitochondria, while mitochondrial fission can compartmentalize damage to a single area of the organelle (Fig. 2C–F). The mitochondrial genome encodes subunits of ATP synthase, electron transport chain, and tRNAs/rRNAs, and these genes can collect mutations over time. However, mitochondria containing mutant DNA can fuse with healthy mitochondria within a single cell to complement the mutated genes (Nakada et al., 2001; Yoneda et al., 1994). Complementation is not unique to DNA, and mitochondrial fusion can promote cross-complementation of lipids and proteins to mitigate damage (Huang and Frohman, 2012; Westermann, 2002).
Damage beyond repair: poorest quality mitochondria are eliminated from the cell
When mitochondrial damage exceeds the repair capacity of the cell, it can have deleterious effects. To prevent cellular damage, dysfunctional mitochondria are subject to autophagy (also known as mitophagy) (Fig. 2E). Current models suggest that mitochondrial fission can segregate regions of mitochondria harboring excessive damage and partition it from an otherwise healthy organelle for mitophagy. Mitophagy can preferentially eliminate damaged mitochondria through the activity of PINK1 and Parkin. Similar to the ATFS-1 mechanism described above, PINK1 is imported into healthy mitochondria via the translocases of the outer and inner membrane (TOM/TIM), where it is subsequently cleaved by mitochondrial proteases and ultimately degraded by the cytosolic proteasome or directly in the matrix (Lazarou et al., 2012; Yamano and Youle, 2013). In damaged, depolarized mitochondria, PINK1 import is perturbed, leading to its accumulation on the outer surface of the mitochondria, where it phosphorylates ubiquitin to activate the E3 ubiquitin ligase Parkin (Koyano et al., 2014). Parkin then ubiquitylates the surface of the mitochondrion, marking the it for autophagic elimination (Narendra et al., 2008). Because mitophagy eliminates only the lowest quality (i.e. lowest membrane potential, accumulation of damaged proteins and lipids, etc.) mitochondria, this process is intimately tied to mitochondrial fission, and defects in fission prevent mitophagy (Twig et al., 2008).
Similar to other mechanisms of mitochondrial quality control, mitochondrial fusion, fission and mitophagy are also linked to aging and lifespan regulation, and their efficiency declines as a function of age (Diot et al., 2015; García-Prat et al., 2016). This loss of efficiency may underlie some age-related diseases, such as cancer, neurodegeneration, and metabolic syndrome (Kitada et al., 1998; Valente et al., 2004). A significant decline in mitophagy activity is found in brain tissues of aged mice, which results in compromised stress resistance and increased mitochondrial dysfunction (Sun et al., 2015). Moreover, a suppression of mitophagy decreases lifespan in Drosophila and C. elegans (Palikaras et al., 2015; Park et al., 2006), whereas simultaneous loss of mitochondrial fusion and fission decreases lifespan in S. cerevisiae due to defects in mitophagy (Bernhardt et al., 2015). In contrast, pharmacological activation of mitophagy by treatment with urolithin A (Ryu et al., 2016) or by tomatidine (Fang et al., 2017) prolongs lifespan and healthspan in C. elegans. Urolithin A also increases muscle function in rodents. Defects in mitophagy are also observed in neurodegenerative disorders, such as Parkinson’s disease, Alzheimer’s disease, and Huntington’s disease. Both PINK1 and Parkin mutations are associated with hereditary Parkinson’s disease (Kitada et al., 1998; Valente et al., 2004) as loss of PINK1 or Parkin function results in degeneration of dopaminergic neurons due to mitochondrial dysfunction (Greene et al., 2003; Park et al., 2006).
The link between mitophagy and cancer is complex as both defects and augmentation of autophagy have been identified in specific cancers. For example, Parkin, a key player in mitophagy, is a potential tumor suppressor that is commonly deleted in multiple cancers, and its deficiency promotes the progression and aggressiveness of breast, colon, and liver cancers (Cesari et al., 2003; Letessier et al., 2007; Poulogiannis et al., 2010; Veeriah et al., 2010). More specifically, knockout of Parkin in mice increases spontaneous liver tumor formation (Zhang et al., 2011). PINK1 has also been linked to the survival and prognosis of adrenocortical tumors (Fragoso et al., 2012; de Reyniès et al., 2009). By contrast, an increase in mitophagy in some solid tumors promotes oxygen homeostasis, lower ROS generation, and ultimately the survival of cancer cells in hypoxic conditions (Sowter et al., 2001; Zhang et al., 2008). In addition, ectopic activation of mitophagy regulatory genes leads to proliferation and tumor growth in nude mice.
Leaving the worst behind: asymmetric segregation of mitochondria in dividing cells
For dividing cells, such as yeast, quality control can be accomplished by asymmetric cell division – that is, by partitioning damaged, dysfunctional mitochondria to one cell to ensure the survival of the other. In S. cerevisiae, mitochondria undergo asymmetric inheritance based on their functional state, such that mitochondria inherited by the daughter (the newly forming cell) is highly functional with higher membrane potential and redox state (Higuchi et al., 2013; Higuchi-Sanabria et al., 2014; McFaline-Figueroa et al., 2011). This asymmetric segregation is, in part, accomplished by the actin cytoskeleton. During cell division, mitochondria traffic along dynamic actin cables, which constantly undergo a retrograde flux in the bud to mother direction (i.e. from the polar end of the newly budding daughter cell to the opposite polar end of the original cell). Mitochondria being inherited by the daughter cell must overcome this retrograde flow. Therefore, damaged, dysfunctional mitochondria are incapable of overcoming the retrograde flow of actin cables cannot be inherited by the daughter cell and are ultimately sequestered in the mother cell (Higuchi et al., 2013). Recent evidence also suggests that mitochondrial fusion and fission contribute to asymmetric cell division by modulating the shape and size of the “mitochondrial unit” that is transported between cells (Böckler et al., 2017). Asymmetric mitochondrial division is not unique to budding yeast: human mammary stem-like cells also asymmetrically apportion their mitochondria, in such a way that the daughter cell destined to differentiate inherits aged mitochondria, while the cell maintaining stemness inherits the younger mitochondrial pool (Katajisto et al., 2015). It is not known whether these pathways decline with age. There is evidence that short-lived mutant cells exhibit perturbations in asymmetric segregation of mitochondria due to collapse of the cytoskeletal networks involved in maintaining this segregation, whereas the opposite is true for long-lived mutants (Higuchi et al., 2013; Higuchi-Sanabria et al., 2016b). Moreover, daughter cells of older mothers have a shorter lifespan than those from younger mothers, suggesting the capacity of the mother cell to produce healthy offspring declines with age (McFaline-Figueroa et al., 2011). It would be interesting to understand whether the process of asymmetric cell division as a whole declines with age.
These studies bring to question whether non-dividing, post-mitotic cells can benefit from similar mechanistic pathways, or whether they rely solely on other mitochondrial quality control pathways. Indeed, mitochondria are found to accumulate at the immunological synapse that forms between T-cells and antigen-presenting cells during T-cell activation (Quintana et al., 2007) and at neurological synapses during neuronal transmission (Guo et al., 2005; Stowers et al., 2002; Verstreken et al., 2005), but it is yet unknown whether these populations of mitochondria exhibit differences in quality compared to other populations. However, in both dividing and non-diving cells, mitochondrial fusion and fission have been tightly linked to mitophagy, as described above.
Maintenance of the endoplasmic reticulum and UPRER
Similar to the mitochondria, the ER is a membrane-bound organelle with a distinct environment and has its own dedicated stress response pathways. As the site for the synthesis and folding for the vast majority of transmembrane and secreted proteins, the endoplasmic reticulum (ER) has evolved mechanisms for sensing and communicating protein misfolding stress. The best understood of these is the unfolded protein response (UPRER), in which transmembrane proteins sense the accumulation of misfolded proteins leading to signaling events that activate transcription of genes to mitigate the stress, such as chaperones or redox factors. Terminally misfolded proteins in the ER are extracted by ATP-driven motors and polyubiquitylated to promote their degradation by the proteasome by a process called ER-associated degradation (ERAD). When damage has exceeded repair, parts of the ER can be targeted for large-scale degradation through ERphagy (also known as reticulophagy). Here we focus on recent findings and emerging themes in ER quality control, touching upon their impacts on aging and disease pathology with particular attention to UPRER, ERAD, ERphagy, and ER diffusion barriers.
The unfolded protein response of the endoplasmic reticulum
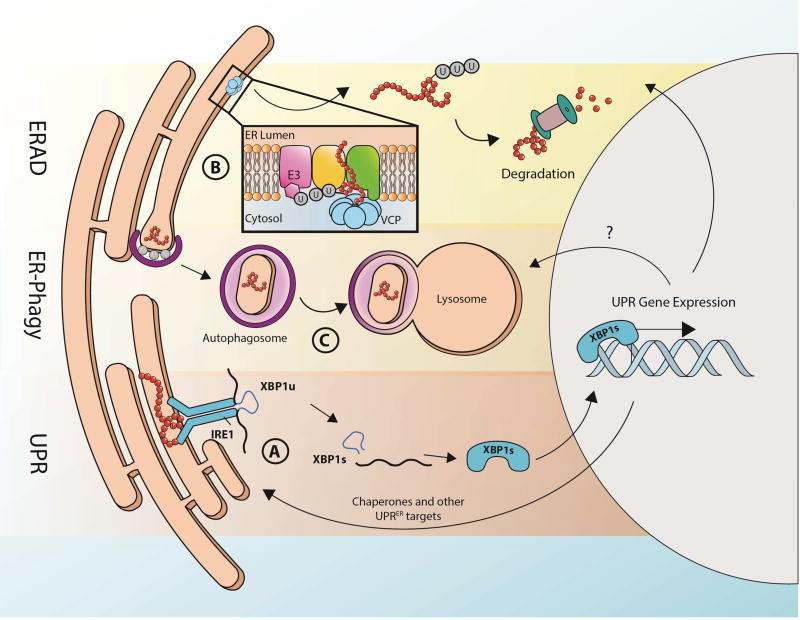
In metazoans, the UPRER consists of three distinct signaling pathways that ultimately coordinate gene expression at the level of transcription and translation to mitigate protein folding stress in the ER. Each arm of the UPRER consists of a luminal facing sensor, which induces a signal to the nucleus upon detection of protein misfolding (Fig. 3A). IRE1 is the most ancient of the sensors (conserved throughout eukaryota). It exists as a monomer until its luminal domain directly interacts with unfolded proteins to trigger homodimerization, followed by autophosphorylation by its cytosolic kinase domain. A third domain in IRE1, with RNase activity, executes an unconventional splicing reaction, removing an intron from the mRNA encoding the transcription factor, XBP1. The spliced mRNA is then translated into XBP1s, which subsequently translocates to the nucleus to activate transcription of chaperones, ERAD factors, and enzymes in the lipid biosynthetic pathway (reviewed by (Frakes and Dillin, 2017; Walter and Ron, 2011)). Similarly, another transmembrane UPR component, PERK also undergoes homodimerization and phosphorylation in response to unfolded proteins in the lumen. PERK then phosphorylates translation initiation factor eIF2α, leading to global downregulation of translation by limiting eIF2. At the same time, specific translation of activating transcription factor 4 (ATF4) occurs, which similarly induces a transcriptional program to cope with ER stress (reviewed in (Hetz, 2012)). A third arm of the UPR involves ATF6, an ER transmembrane protein that is activated by unknown mechanisms. Once activated, it translocates to the Golgi, where it is cleaved to create an N-terminal fragment that functions as a transcription factor (Haze et al., 1999; Schindler and Schekman, 2009; Ye et al., 2000).
Fig. 3. Multi-faceted quality control of the endoplasmic reticulum.
Like the mitochondria, the ER also has a collection of unique quality control mechanisms to maintain ER function. There are three branches of UPRER, each consisting of a transmembrane protein with a luminal facing sensor for unfolded proteins, which signals damage to the nucleus through a unique transcription factor. Here, the IRE1-XBP1 arm of UPRER serves as a general representation of UPRER (A). When IRE-1 senses ER stress (e.g. presence of misfolded proteins), it produces homodimers, is phosphorylated, and promotes splicing of XBP1u mRNA localized at the ER. The resulting splice variant of XBP1 is capable of producing functional protein, XBP1s, which can shuttle to the nucleus, acting as a transcription factor to upregulate UPRER genes. Some proteins of downstream UPRER genes, such as chaperones, will shuttle back to the ER to repair damaged components. Damaged proteins in the ER can also be marked by ubiquitination, removed from the ER by the protein VCP, and targeted for degradation by the cytosolic proteasome in a process known as ERAD (B). Larger regions of ER damage can be resolved by sequestering the damage to a specific region of the ER. This damaged unit can then be specifically targeted for degradation upon binding of ER-phagy receptors (C). After this region of the ER buds into a vesicle, it fuses with a lysosome to destroy and recycle its contents.
Similar to the findings in C. elegans, that perturbations to neuronal mitochondria can trigger stress to distal tissues of an organism, the UPRER possesses cell-nonautonomous properties (Taylor and Dillin, 2013). Neuronal UPRER plays a critical role in this process, leading to a hypothesis that neurons are particularly sensitive to stress and efficient at communicating stress to the rest of an organism. For example, the UPRER controls innate immunity in C. elegans dependent on a G-protein coupled receptor, OCTR-1, in sensory neurons that signals upregulation of non-canonical UPR genes beneficial against infection in distal tissues (Sun et al., 2011, 2012). Moreover, ectopic activation of UPRER in neurons by overexpression of XBP1s extends overall lifespan and ER-stress resistance in C. elegans through a still unidentified signaling molecule released in small clear vesicles from neurons to receiving cells (e.g. intestine) (Taylor and Dillin, 2011). Remarkably, these neuronal functions of the UPRER appear to be conserved across metazoans, as XBP1s overexpression in a region within the hypothalamus upregulates metabolic activity in the liver and improves insulin sensitivity in rodents (Williams et al., 2014).
These findings suggest a link between one of the most well conserved stress signaling pathways to the health of an organism. What aspects of multicellularity necessitate the evolution of this phenomenon? Do other, more recently evolved arms of the UPRER also contribute to its cell non-autonomous properties? Additionally, given that the microbiota secretes neurotransmitters and may play a role in brain function, could intestinal signals affect neuronal UPRER to trigger UPR activation in other tissues or the intestine itself? Elucidating this unexpected feature of the UPRER will provide insight into the networks of signals that control homeostasis at the organismal level.
Generally, the ability of an organism to respond to environmental stress decreases with age. It is not surprising, then, that many age-related diseases such as neurodegeneration, metabolic disease, and cancer all share common features of perturbed UPRER. In C. elegans, the ability to activate UPRER in response to stress declines sharply during aging, which has direct implications for lifespan regulation (Taylor and Dillin, 2013). Similarly, mammals also experience a breakdown of ER protein homeostasis as a function of age. Expression of genes involved in ER quality control, such as the chaperones BiP and calnexin decline in aged mouse cerebral cortex and hippocampus, respectively (Naidoo et al., 2008; Nuss et al., 2008). Moreover, aging results in accumulation of oxidative and nitrosative stress, which damages and decreases enzymatic activity of key ER chaperones, including BiP and protein disulfide isomerase (PDI) (Rabek et al., 2003; Yang et al., 2015). The loss of key ER chaperones results in increased accumulation of immature and denatured proteins, and ultimately proteotoxicity and cell death. ER stress and accumulation of damaged proteins are a prominent feature in neurodegenerative disorders (Hu et al., 2000), and upregulation of ER chaperones can actually protect neuronal cells during stress (Ko et al., 2002; Tanaka et al., 2000). Therefore, hyperactivation of specific branches of UPRER can have beneficial effects on lifespan and healthspan: ectopic expression of the IRE1 branch of UPRER by overexpression of the spliced XBP1s results in extended lifespan and stress resistance in C. elegans (Taylor and Dillin, 2013), and PERK-eIF2 α signaling provides neuroprotection by decreasing global translation and protecting the ER from accumulation of damaging misfolded and mutant proteins (Das et al., 2015; Saxena et al., 2009).
Despite these studies providing evidence for a beneficial role in UPRER, chronic or irreversible UPRER can shift from being protective to pro-apoptotic. For example, sustained activation of PERK can promote upregulation of the transcription factor CHOP, which can promote ER-stress induced apoptosis through ATF4 and ATF6 (Cao and Kaufman, 2012). CHOP inhibits the survival protein, BCL-2, and can promote apoptosis by enhancing BAX- and BAK-dependent apoptosis through the mitochondrial caspase cascade (Hetz and Mollereau, 2014; Soo et al., 2009). Beyond apoptosis, over-activation of the PERK arm of UPRER can result in sustained translational repression through eIF2α, which can promote synaptic failure and neuronal death, effects observed in prion-disease mouse models. In this context, decreasing UPRER activity was proposed as a therapeutic intervention. Indeed, reduction of PERK activity using a PERK inhibitor restored protein synthesis in neurons and prevented neurodegeneration and behavior impairment caused by prion-disease in mice (Moreno et al., 2013). In another study, the IRE1-XBP1 arm of UPRER was considered as a therapeutic target for breast cancer. XBP1 is activated in triple negative breast cancer, and its inhibition prevented tumor growth and relapse (Chen et al., 2014). However, it is important to remember that UPRER is an indispensable quality control mechanism, and chronic PERK inhibition can result in pancreatic toxicity and loss of B cell function (Harding et al., 2001; Moreno et al., 2013). Taken together, these studies highlight the complexity of UPRER signaling and show that it can have both positive and negative effects on aging and diseases pathology. Thus an important question is which branches of UPRER, if any, can be reasonably targeted for therapeutic intervention. Perhaps more realistic would be to dissect the downstream targets of UPRER, and target specific chaperones and molecular pathways, rather than inhibiting or activating master transcriptional regulators.
Keeping the factory clean: concerted efforts of ERAD and autophagy in ER quality control
Despite the highly coordinated ER quality control machinery described above, some proteins synthesized in the ER or native to the ER may still become terminally misfolded, unable to be repaired by ER-resident or UPRER-induced chaperones. These damaged proteins need to be degraded. However, given its major role in protein synthesis, the number of proteases in the ER is limited, and thus misfolded proteins must be translocated out of the ER to be degraded by the cytosolic ubiquitin-proteasome system through ERAD (Brodsky, 2012) (Fig. 3B). The machinery of ERAD and its substrate recognition mechanism are complex and under many levels of control (reviewed in (Brodsky, 2012)) and the precise mechanism by which proteins exit the ER during ERAD remains unknown, although a model involving formation of a channel involving multiple molecular players has been proposed (reviewed thoroughly in (Zattas and Hochstrasser, 2015)). Luminal and transmembrane unfolded proteins are recognized by a large transmembrane protein complex centered around the RING ubiquitin ligases, Hrd1p and Doa10p (Ast et al., 2014; Habeck et al., 2015). This complex also includes the transmembrane proteins Hrd3p, Usa1p, and Der1p and the luminal protein Yos9p (Carvalho et al., 2006). A recently solved Cryo-EM structure of the ERAD machinery revealed that Hrd1p dimers form a protein conducting channel in complex with Hrd3p to retro-translocate misfolded proteins after ubiquitylation (Schoebel et al., 2017). Upon poly-ubiquitylation of the unfolded substrate by this complex, the AAA+ ATPase Cdc48 (p97 or VCP in Homo sapiens) drives its extraction from the ER membrane into the cytosol where it is subjected to proteasomal degradation (Xia et al., 2016). Recently, the mechanism by which Cdc48p unfolds and translocates its targets was described using an elegant in vitro system, revealing that Cdc48p pulls polyubiquitylated substrates through its central pore and requires a deubiquitylase for release of substrate (Bodnar and Rapoport, 2017). Study of the mechanism of protein translocation of p97/VCP has also revealed that multisystem proteinopathy-causing mutations in the gene (Johnson et al., 2010; Watts et al., 2004) might increase it unfoldase activity in in vitro assays (Blythe et al., 2017), suggesting a gain-of-function might underlie proteostasis perturbation in multisystem proteinopathy.
Beyond its role in protein quality control in the ER, ERAD also maintains protein quantity control by regulating protein turnover (Kreft, 2015). ERAD is essential for maintaining proper levels of ER-resident proteins, and of proteins that function as part of stoichiometric multi-protein complexes (reviewed in (Printsev et al., 2016)). In both cases, ERAD efficiency depends on tagging of proteins and their targeting to the cytosolic ubiquitin-proteasome system.
Misfolded proteins that are resistant to ERAD can be shunted from ERAD to quality control autophagy via a Vps34/beclin-1 initiation machinery (Houck et al., 2014). Other ER components, such as lipids, which cannot be cleared through ERAD and conventional proteostasis mechanisms, are also under quality control surveillance via autophagy. Under ER stress induced by chronic phospholipid imbalance, the ER and mitochondrial membranes undergo dramatic perturbations in morphology and function. This activates the UPRER, promoting Hsp104p recruitment to the ER, as well as lipid droplet formation at ER aggregates. It is likely that excess lipids and damaged proteins are removed from the organelle into lipid droplets, which can be shuttled to the vacuole for degradation in a selective degradation machinery, known as microlipophagy (Vevea et al., 2015). Lastly, the ER itself can be degraded by selective autophagy via ER-phagy (Fig. 3C). Recent observations suggest that ER-phagy occurs via interactions of ER-bound autophagy receptors with cleaved LC3, LC3-I. LC3-I is conjugated onto phospholipids of the phagophore membrane (also called the isolation membrane, which initiates macroautophagy) by ubiquitylation-like chemistry and becomes LC3B–II. This interaction leads to engulfment of the ER by the tethered phagophore. There are three ER-bound receptors for LC3-II including FAM134B, SEC62 (Khaminets et al., 2015), and RTN3 (Grumati et al., 2017). In yeast, ER stress causes massive ER expansion and formation of “ER whorls,” which are subject to autophagy (Bernales et al., 2006). The ER whorls are degraded through the vacuole in a distinct form of ER-phagy independent of known core autophagy machinery, providing a means for the cell to control organelle size by degrading excess ER membranes (Schuck et al., 2014).
Analogous to MAD and mitophagy, it is expected that when the function of the cytosolic ubiquitin-proteasome system or lyososomes are compromised during aging and diseased states, ERAD/ER-phagy efficiency would also decline. However, the direct impact of aging or disease pathology on ERAD/ER-phagy, and conversely, the impact of ERAD and ER-phagy on aging and disease, are still unclear. Circumstantial evidence links ERAD deficiency with age-related diseases. For example, a study in mice fed alcohol or high fat diets for prolonged periods of time found distinct alterations in expression of ERAD genes due to a significant decline in DNA methylation of promoter regions of major ERAD genes (Han et al., 2013a). Moreover, yeast cells and cultured neuron-like PC12 cells expressing poly-Q huntingtin protein had dramatic dysfunction in ERAD. In these cells, overexpression of major ERAD players, such as Npl4p and Ufd1p, ameliorated polyQ-mediated proteotoxicity (Duennwald and Lindquist, 2008). Finally, reduced expression of ERAD components significantly shorten lifespan in C. elegans. The link between ER-phagy and aging or disease is even less clear. Future work should examine whether ERAD/ER-phagy components decline with age, and whether ectopic activation can rescue loss of proteostasis at advanced age. In addition, it would be important to determine whether selective activation of ERAD/ER-phagy components represents a therapeutic avenue for protein-folding diseases, such as Alzheimer’s and Huntington’s disease.
Diffusion barriers in quality control of the endoplasmic reticulum
Like mitochondria, the ER also segregates asymmetrically on the basis of function and quality during cell division. As an interconnected organelle that generally remains topologically connected between the two dividing cells until cytokinesis, it is surprising that the ER can maintain two functionally distinct environments. Fluorescence loss in photobleaching (FLIP) experiments where fluorescent proteins were targeted to the ER revealed that a single ER reticulum can maintain two functionally distinct regions by formation of a diffusion barrier at the ER membranes. This phenomenon was originally identified in yeast, where a diffusion barrier forms at the ER membranes at the bud neck. The barrier divides the ER into two distinct compartments, such that the daughter cell contains higher quality ER with correctly folded proteins, while the mother cell retains damaged, misfolded proteins and protein deposits within the ER (Chao et al., 2014; Clay et al., 2014; Luedeke et al., 2005; Saarikangas et al., 2017). This functional segregation also extends to the Golgi and plasma membrane (Clay et al., 2014; Shepard et al., 2003; Takizawa et al., 2000). Production and maintenance of the diffusion barrier is dependent on many factors. Briefly, a septin ring is formed and recruits polarity proteins, including Bud1p and Bud5p. These proteins can activate essential polarisome (the machinery required for sequestration of protein aggregates to the mother cell during yeast cell division (Liu et al., 2010)) proteins, such as Pea2p and Bud6p, which promote actin cable assembly, providing the foundation and scaffold for sphingolipid accumulation at the bud neck (Chao et al., 2014). Mutations that disrupt many of the components essential for maintaining the diffusion barrier in the ER result in free diffusion of damaged proteins from the mother into the daughter cell, and ultimately, decreased lifespan of the daughter cell (Clay et al., 2014).
Mammalian cells also show an overlap between sub-cellular localization of septins and regions of restrictive diffusion. For example, septin localization is found at the dendritic spines of neurons where diffusion of ER proteins is relatively restricted (Cui-Wang et al., 2012; Tada et al., 2007). Thus, it is possible that diffusion barriers mediated by septins are also functionally significant in the ER of higher eukaryotes. Similar FLIP studies have found that diffusion barriers exist in the ER membranes of both C. elegans embryos and of mouse neural stem cells (Lee et al., 2016; Moore et al., 2015). In mouse neural stem cells undergoing mitosis, damaged, ubiquitinated proteins and other unfavorable components, such as ectopically expressed progerin, asymmetrically segregate into non-stem progeny to maintain a “pristine” state in the stem cell lineage (Moore et al., 2015). Perhaps most intriguing, the efficiency of the diffusion barrier diminishes with age, and older neural stem cells lose the capacity to partition damaged proteins into a single daughter cell. As the cell ages, critical cellular components, such as the actin cytoskeleton and lipid homeostasis, collapse, and it is possible that the collapse of these units – essential for ER diffusion barrier maintenance – leads to the loss of asymmetric cell division at advanced age. Still to be determined is whether similar diffusion barriers exist in post-mitotic cells to compartmentalize specific regions of the ER, and if they have implications in aging or disease pathology.
Cross-talk among stress response pathways
The cell is compartmentalized into specific organelles and subregions that are seemingly isolated, each equipped with its own specific stress response pathway to preserve homeostasis during times of stress. However, it is becoming clearer that the fitness, functions, and stress responses of cellular sub-compartments are interconnected and interdependent (Dillin et al., 2014). A steadily increasing number of studies highlights the importance of each organelle upon every other organelle in the cell. For example, proper mitochondrial function is dependent on a functionally acidic vacuole (Hughes and Gottschling, 2012), mito-nuclear balance of proteins encoded in both the mitochondrial and nuclear genome (Houtkooper et al., 2013), and on organelle restructuring through ER-dependent mitochondrial fission (Friedman et al., 2011). Here, we focus on cross communication between the cytosol, ER, and mitochondria and speculate on their significance in aging and disease pathology.
The inseparable duo: mitochondria and ER communication
The mitochondria and ER are two distinct organelles that do not share intra-organelle environments. However, they are highly intertwined and linked via strong contact sites. These sites are tethered by a protein complex consisting of Mmm1p, Mdm10p, Mdm12p, and Mdm34p (ERMES: the ER-mitochondria encounter structure) in yeast (Kornmann et al., 2009), and IP3 receptors and Mfn2 in mammals (de Brito and Scorrano, 2008; Szabadkai et al., 2006). These contact sites, termed mitochondria-ER associated membranes (MAMs) are essential for cellular physiology, as their disruption results in defects in calcium signaling in both yeast and mammals, and defects in energy metabolism, synaptic transmission, ROS generation in mammals (Hayashi et al., 2009; Vance, 2003; Voelker, 2009; Wiedemann et al., 2009). MAMs are essential for both micro- and macro-molecular exchange. For example, Ca2+ cycling occurs between mitochondria and ER, where the sarcoplasmic reticulum acts as a consistent source of Ca+ essential for maintaining mitochondrial Ca2+ levels. Changes in the distance of then mitochondria-ER interface, even on the order of several nanometers can cause dynamic shifts in Ca2+ cycling between the two organelles, as well as Ca2+ efflux into the cytosol (Csordás et al., 2010). Abnormalities in Ca2+ homeostasis within the ER and mitochondria can have major consequences, such as protein folding dysfunction in the ER or mitochondrial permeability transition pore opening (the process which promotes apoptosis by release of pro-apoptotic components from the mitochondria), ultimately leading to excitotoxicity of neurons in ALS (Grosskreutz et al., 2010; Lautenschläger et al., 2013). In addition to Ca2+ cycling, MAMs are a site of lipid exchange, required for the synthesis of phospholipids. For example, mitochondria act as a site for phosphatidylethanolamine synthesis, which is an essential step in the phosphatidylethanolamine N-methyltransferase (PEMT) pathway to synthesize phosphatidylcholine, and requires lipid exchange between the ER and mitochondria (Gaigg et al., 1995; Vance, 2003; Voelker, 2009). Perturbations in these pathways result in phospholipid imbalance, which can have dramatic effects on ER and mitochondrial morphology, fitness, and function (Testerink et al., 2009; Vevea et al., 2015), and ultimately result in the progression of major diseases, including muscular dystrophy (Mitsuhashi et al., 2011) and steatohepatitis (Li et al., 2006).
As organelles with such close proximity and constant communication, it is logical that quality control mechanisms of the ER or mitochondria would affect the quality and function of the other. Indeed, some components of the UPRER and UPRMT are shared: GCN2 activation during mitochondrial stress and PERK activation during ER stress both lead to phosphorylation of eIF2α and to a decrease in global translation (reviewed in (Arnould et al., 2015)). Moreover, GCN2 and PERK can both activate ATF4, which acts as a transcription factor to upregulate genes involved in mitochondrial and ER quality control, including Parkin and CHOP, respectively (Bernales et al., 2012; Bouman et al., 2011). Parkin itself can also directly affect UPRER by enhancing XBP1s signaling. Parkin promotes XBP1s signaling by alleviating the transcriptional, translational, and post-translational repression of XBP1s by p53, which has direct implications in Parkinson’s Disease (Duplan et al., 2013). ERAD and MAD also share critical components: Vms1p and Ufd1p work together with Cdc48p to recruit ubiquitin-marked substrates within the ER and mitochondria for proteasomal degradation (Heo et al., 2010; Wu et al., 2016). Under general stress, Vms1p and Cdc48p promote degradation of nuclear and ER substrates (Baek et al., 2012; Tran et al., 2011), but upon mitochondrial stress, Doa1p and/or Vms1p bind to Cdc48p to mediate clearance of damaged mitochondrial substrates (Wu et al., 2016). Beyond sharing critical components, it would be interesting to determine whether there is direct communication between ERAD and MAD, and whether they have implications in aging and disease.
Although numerous studies have characterized these ER-mitochondria interactions, the direct contributions and implications of these interactions to aging are yet to be determined. It is known, however, that ER-mitochondria interactions are disrupted in Alzheimer’s disease, Parkinson’s disease, ALS, and other neurodegenerative and age-related diseases. Several aggregation-prone proteins, including Aβ, TDP43, and α-synuclein all alter ER-mitochondria associations, often perturbing their critical functions, for example in CA2+ exchange (Area-Gomez et al., 2009; Calì et al., 2012; Tsao et al., 2012). However, there are some inconsistencies in these data, and current research is limited to disease models. Moving forward, it would be interesting to determine what happens to ER-mitochondria contacts during aging. Do they, like other organelles and quality control mechanisms, break down and become dysfunctional with age? What mechanisms are involved in protecting MAMs during stress and aging, and could their ectopic activation have implications for lifespan? These and many other questions concerning ER-mitochondria communication could open up new avenues for therapeutic intervention.
Influencing the environment: communication between mitochondrial and cytosolic stress responses
Given its role as a hub for energy metabolism, it is intuitive that the health and function of the mitochondria directly impacts the health of the entire cell. It turns out, however, that energy generation is not the only way by which mitochondria can affect the function of other organelles; quality control mechanisms originally considered specific to the mitochondria are now known to have direct effects on extra-mitochondrial compartments as well. For example, the MAD machinery that is localized to the outer mitochondrial membrane is exposed to the cytosol and can potentially contribute to cytosolic quality control. Damaged cytosolic proteins and cytosolic protein aggregates are generally cleared by cytosolic chaperones regulated by HSF1. However, protein aggregates can also be associated with mitochondria and take advantage of the MAD machinery. For example, pathogenic variants of ataxin-3, which form cytosolic aggregates and can cause neurodegenerative phenotypes, have been shown to localize to mitochondrial surfaces (Pozzi et al., 2008). Moreover, knockdown of the mitochondrial E3 ubiquitin ligase results in accumulation of the aggregates and subsequent cell death (Sugiura et al., 2011). Protein aggregates also attach to mitochondria in dividing yeast to promote retention of protein aggregates in the mother cell (Zhou et al., 2014). While these aggregates can be cleared by cytosolic chaperones, disaggregases, and metacaspases, such as Hsp104p and Mca1p (Hill et al., 2014; Parsell et al., 1994), they can also be imported into the mitochondria for degradation by mitochondrial proteases (Ruan et al., 2017). More specifically, Hsp104p promotes disaggregation of damaged proteins from mitochondria-associated aggregates, allowing their import into the mitochondria through mitochondrial import proteins, Tom70p and Tom40p. Once inside the mitochondria, these proteins can be cleared by mitochondrial proteases, such as Pim1p. Ruan et al. also showed that unstable, aggregation-prone cytosolic proteins are imported into mitochondria of mammalian RPE1 cells (Ruan et al., 2017). An exciting avenue of research would be to determine whether mitochondria have the capacity to resolve age-associated protein aggregates and whether this mechanistic pathway has implications in lifespan and age-related diseases.
Beyond these studies, direct damage to mitochondria or perturbation of mitochondrial function have been shown to upregulate the cytosolic HSR in an HSF1-dependent manner (Ho et al., 2006; Kim et al., 2016). Specifically, knockdown of the mitochondrial chaperone, hsp-6 (mtHSP70 in mammals), which activates UPRMT, was sufficient to also induce the HSR as measured by induction of the cytosolic chaperone, HSP-16.2. This mitochondrial-to-cytosolic stress response (MCSR) is dependent on the UPRMT and HSR transcription factors, DVE-1 and HSF-1, respectively, which coordinate global changes in fat metabolism. Together, DVE-1 and HSF-1 promote fatty acid synthesis (mainly cardiolipin), while inhibiting ceramide synthesis. This change in lipid architecture is essential for activation of MCSR, and pharmacological manipulation of lipid levels (such as perhexiline treatment to inhibit fatty acid oxidation) is sufficient to induce MCSR. In both C. elegans and human primary fibroblasts, the activation of MCSR via hsp-6/mtHSP70 knockdown or perhexiline treatment decreased sensitivity to polyQ-induced proteoxicity, suggesting this interconnected stress response between the mitochondria and the cytosol has implications in age-related disease (Kim et al., 2016). Could fatty acid metabolism be a target for therapeutic intervention of protein-misfolding diseases by promoting both mitochondrial and cytosolic stress responses? What happens to MCSR activity during aging, and are any components of the MCSR critical for lifespan regulation?
Whether direct communication from the HSR to mitochondria exists is less clear. Intuitively, it makes sense that cytosolic quality control pathways would have direct impact on mitochondrial quality. The ubiquitin proteasome system is certainly essential for MAD (Azzu and Brand, 2010; Maharjan et al., 2014), induction of general autophagy is critical for the efficiency of mitophagy (Ding and Yin, 2012), cytosolic chaperones and disaggregases can aid in the proper folding and refolding of proteins destined for mitochondrial import (Ruan et al., 2017), and HSF1-mediated regulation of the oxidative stress response likely preserves mitochondrial membrane potential and promotes redox control of the organelle (Douglas et al., 2015). What remains unclear is whether HSF1 or other players involved in HSR can directly signal to the mitochondria to activate mitochondrial quality control pathways. A recent study has found that two mitochondrial folding proteins, Hsp78p and Mdj1p, are the direct targets of HSF1 in yeast, but whether this activation has direct impacts on mitochondrial quality and function has not been investigated (Solís et al., 2016). Still to be answered is if activation of mitochondrial quality control under heat stress is simply a secondary effect due to mitochondrial damage caused by heat. Or does the activation of HSR actually drive induction of critical players in mitochondrial regulation? Interestingly, gene ablation or knockdown of HSF1 caused mitochondrial damage due to impaired fission rates in mouse livers. Qiao et al. found that HSF1 deficiency led to a sharp decline in NAD+ levels, and the authors speculated that this decline in NAD+ is responsible for mitochondrial dysfunction and decreased ATP production (Qiao et al., 2017). Thus, it is likely that HSF1 has a much more direct contribution to mitochondrial quality control mechanisms than is currently appreciated.
The environmental influence: cross-talk between the cytoplasmic and ER stress responses
Similarly to mitochondria, the ER also shares some resources with the cytoplasmic HSR, most notably the proteasome. In addition, nine stress response genes are induced by both HSR and UPRER ((Liu and Chang, 2008a)), including the ER molecular chaperone, Kar2/BiP (Kohno et al., 1993), and the COPII cargo receptor, Erv29 (Caldwell et al., 2001). Heat-shock can also directly promote UPRER activation, for instance the splicing of XBP1 (Adachi et al., 2009). Thus, there appears to be significant cross-talk between ER and cytoplasmic quality control. Indeed, overexpression of HSF1 can promote ER proteostasis via ERAD by the induction of cytoplasmic chaperones and the ubiquitin/proteasome system (Han et al., 2007). Moreover, yeast cells deficient in UPRER can be rescued by expression of a constitutively active form of HSF1. Specifically, overexpression of hyperactive HSF1 rescues the survival of IRE1 mutant cells under tunicamycin-induced ER stress. HSR relieved ER stress by promoting translocation of damaged proteins out of the ER, inducing clearance of damaged proteins through ERAD, and promoting protein translocation both into and out of the ER (Hou et al., 2014; Liu and Chang, 2008b). Interestingly, induction of UPRER via heat-shock in mammalian cells can promote activation of typical ER stress markers, such as the splicing of XBP1 and synthesis of ATF4, but traditional transcription factors responsible for UPRER are not essential for heat-induced activation of downstream UPRER genes such as DNAJB9 and HSPA5 (Heldens et al., 2011).
These findings are provocative, and suggest that it would be worth investigating whether HSR and UPRER communicate directly to coordinate stress responses. On a broader scale, despite known interactions of the ER with many general signaling pathways, how changes in the ability of the ER to communicate with other organelles during a stress response may help coordinate cellular processes is not well understood,. For example, the UPRER interacts with the Akt/mTORC signaling axis to affect cell survival during ER stress (Appenzeller-Herzog and Hall, 2012). A major output of active Akt/mTOR signaling is increased cell growth by virtue of globally increased levels of anabolism and protein synthesis, and decreasing autophagy, including reticulophagy (Saxton and Sabatini, 2017). Thus, constitutive activation of the Akt/mTOR pathway could lead to ER stress by overloading its chaperone machinery. Furthermore, given that the ER interacts with multiple organelles around the cell, including late endosomes, it is possible that the ER also physically interacts with lysosomes, the central site of the mTORC1 pathway. Exploring the signaling logic underlying the interactions between UPRER and growth pathways during ER stress will reveal factors that promote survival or stimulate cell death.
Pharmacological modulation of protein quality control during aging
Given the wide involvement of protein quality control pathway in age-related disease, their components are attractive therapeutic targets. One hypothesis is that boosting protein quality control systems may be beneficial for ameliorating age-related diseases such as neurodegeneration or diabetes, where these systems are thought to break down with age. In turn, antagonizing protein quality control systems in human cancers might be viable therapeutic option, considering that protein quality control machinery is often hijacked by cancer cells to facilitate increased proliferation. These seemingly contradictory aims complicate the therapeutic targeting of protein quality control. Here, we briefly highlight some of the most recent pharmacological modulations of protein quality control (refer to (Pilla et al., 2017) and book chapter (Fujikake et al. 2016 Heat Shock Factor, Springer) for a comprehensive review).
Boosting HSF1-mediated chaperone activation to combat misfolded proteins in a variety of age-related disease contexts has been an intense area of interest (Neef et al., 2011). The rationale for this approach stems from the dissection of the chaperone networks induced by HSF1 transactivation which includes a wide range of chaperones such as Hsp40s, Hsp70s, Hsp90s, Hsp110s, TRiC, and small Hsps. Additionally, chaperone expression is thought to decline with age and in neurodegenerative conditions. In line with this hypothesis, inhibitors of HSP90, an HSF1 inhibitor, such as geldanamycin protect metazoans across phyla from neurodegeneration induced by misfolded proteins (Luo et al., 2010). Another drug, riluzole, which is thought to extend lifespan in ALS patients, inhibits degradation of HSF1 in cultured cells (Liu et al., 2011). Several other strategies to increase HSF1 activity and function in disease have been employed but with little success in disease models. Despite these promising findings, HSF1 activation aimed at increasing cells’ protein folding capacity might be detrimental if administered at an organism-wide level by sensitizing cells to oncogenesis (Dai and Sampson, 2016). Future research might emphasize the identification of cell-type specific regulators (e.g. neuronal) of HSF1 or pathways acting in parallel with HSF1 to promote proteostasis in age-related disease without promoting oncogenic HSF1 properties.
Beyond HSF1, therapeutic targeting of the integrated stress response has emerged as another potential area of translational interest. Specifically, inhibitors of the PERK-eukaryotic translation initiation factor 2 (eIF2) axis have garnered recent interest. Inhibition of eIF2α, regulator of eIF2B dimerization and activation, rescues multiple models of neurodegenerative disease from death, potentially by releasing the block on general translation mediated by eIF2α phosphorylation by PERK. Genetic or chemical inhibition of eIF2α phosphorylation has been shown to ameliorate deleterious effects of protein misfolding in ALS, tauopathy, prion disease, and traumatic brain injury (Halliday et al., 2017; Kim et al., 2013; Ma et al., 2013; Moreno et al., 2013; 2012; Sidrauski et al., 2013). Of particular pharmacologic interest is a small molecule, ISRIB, which was initially discovered in a chemical screen for drugs to regulate ATF4 expression (Sidrauski et al., 2015). ISRIB binds to and promotes dimerization of eIF2B, thereby bypassing eIF2α. In mice, ISRIB-mediated inhibition of the ISR conferred increased cognitive recovery from traumatic brain injury (Chou et al., 2017), suggesting that its therapeutic benefit in mammals should be a focus of future studies in other conditions where the ISR is implicated. While these studies suggest an important role for the ISR in multiple age-related disease states, other kinases such as GCN2, protein kinase R, and Heme-regulated eIF2α kinase, may also participate in phosphorylation of eIF2α.
There are several therapeutic avenues under investigation to regulate proteostasis in cancer, where protein quality control machinery is often hijacked to allow for cell proliferation. The majority of these treatments aim to inhibit stress response pathways and the protein clearance machinery that they induce, including inhibition of IRE1/XBP1 signaling axis, as well as decreasing Cdc48p/p97/VCP or proteasome activity (reviewed by (Hetz and Saxena, 2017)). However, a major caveat to these approaches is that, unless specifically targeted to cancerous cells, the treatment could perturb the proteostasis network in other tissues, perhaps making them more vulnerable to perturbations that might underlie other age-related disease. Collectively, these are promising approaches to treat some of the most devastating age-related diseases in humans, but entail major trade-offs as far as deleterious off-target effects.
Similar to targeting HSR and ISR, targeting mitochondrial quality and fitness is a hot topic in therapeutic studies (refer to (Andreux et al., 2013) for a comprehensive review). Perturbations in mitochondrial fitness and dynamics lead to a myriad of diseases, and point to several potential pharmacological targets. For example, muscle degenerative disorders, like Charcot-Marie-Tooth (Alexander et al., 2000; Züchner et al., 2004) are caused by defects in the mitochondrial fusion machinery, which makes mitochondrial dynamics an attractive target for therapeutic intervention. Chemical compounds that either promote fusion (M1 hydrazone (Wang et al., 2012)) or inhibit fission (MDIVI-1 (Cassidy-Stone et al., 2008)) can have beneficial effects on neurodegenerative disease by balancing mitochondrial fusion and fission dynamics towards beneficial fusion (Lackner and Nunnari, 2010; Wang et al., 2012). As described above, shifts towards fusion can also have beneficial effects on lifespan in several model organisms, making it of particular interest to determine whether promoting fusion can be a viable therapeutic option.
In addition to direct modulation of mitochondrial dynamics, activation of the UPRMT is an attractive target for therapeutic intervention as UPRMT can have several positive effects on cellular health and lifespan as described above. For example, promoting UPRMT by nicotinamide riboside, a precursor of NAD(+) biosynthesis, reverses the effects of high-fat, high-sucrose diets on hepatic steatosis, offering a potential therapeutic method to prevent – or even reverse –nonalcoholic fatty liver disease (Gariani et al., 2016). In another study, UPRMT activation is manipulated in cancer cells using an inhibitor of mitochondrial Hsp90 by Gamitrinib. Gamitrinib-induced UPRMT resulted in suppression of NF-kB activation, sensitizing cancer cells to pro-apoptotic anticancer drugs, such as TRAIL (a death receptor agonist) (Siegelin et al., 2011). These studies demonstrate the potential of promoting UPRMT as a viable therapeutic approach. However, as described above, chronic activation of UPRMT can also have detrimental effects, and thus further study is essential to determine whether specific downstream components of UPRMT can be activated to promote beneficial effects of UPRMT without activating detrimental or damaging components.
Concluding remarks
Maintenance of cellular homeostasis involves a complex interplay of many critical stress response pathways, which have implications in lifespan regulation. Each organelle has a collection of dedicated quality control pathways to maintain its function both basally and under various stresses. Yet to be determined is whether bonafide stress response pathways exist in other organelles. There is some limited evidence of a lysosomal stress response mediated by TFEB, which induces genes essential for macroautophagy (Martina et al., 2016; Santaguida and Amon, 2015). In addition, organelle autoregulation, defined as an organelle’s capacity to respond to cellular demands and alter its form and function, has been described in several organelles, including the Golgi (Sasaki and Yoshida, 2015; Taniguchi and Yoshida, 2017). These findings raise the question of whether protein quality control pathways, similar to UPRER or UPRMT exist in these and other organelles, such as the nucleus and peroxisome. Even more interesting would be to determine whether master regulators for quality control pathways in these organelles can be identified, and whether they impact lifespan regulation.
Beyond dedicated, organelle-specific quality control mechanisms, current research has made it clear that communication between these pathways is not only common, but is a key feature of restoring and preserving cellular homeostasis. Thus the notion that proper function of one organelle is critical for the function of another is increasingly better appreciated. For example, the energy produced by the mitochondria is essential for proper protein folding in the ER, while the mitochondria is dependent on proper protein manufacturing in the ER. Moreover, the available pool of cellular machinery essential to respond to stress, is limiting. Thus, different organelle quality control pathways must communicate and cooperate to avoid competitive inhibition and inefficient stress response. This interconnected stress signaling response raises a very interesting question: under conditions of general stress where several cellular compartments are simultaneously damaged, which stress response pathways are preferentially upregulated? For example, when every organelle sustains damage, does the cell preferentially allocate its resources to UPRER, UPRMT, or HSR? Does the cell attempt to repair a specific compartment first? Are some cellular compartments ignored to preserve the quality of “more important” cellular structures? If so, how does the cell communicate the preferential activation of one stress response over the other?
Perhaps even more interesting would be to gain a better understanding of the cross-talk among stress response pathways during the process of aging. As the cell ages, resources are consumed, cellular damage increases, and stress response pathways become less efficient overall. As these compounding effects take place, how is the cross-communication between stress response pathways affected? Is it possible that the breakdown of crucial communication between these pathways is what results in their independent dysfunction? For example, does a young cell have a sequential and highly ordered activation of stress response pathways, prioritizing one over the other to ensure the fastest and most efficient return to cellular homeostasis? Does this structured prioritization breakdown during aging, such that aged cells attempt to activate all stress response pathways at once, resulting in failure to repair damage? Is the cross-communication between stress response pathways essential for healthy lifespan? We believe that moving forward, studies on the role of quality control mechanisms in aging will have to focus on this complex interplay and interconnectedness of quality control pathways rather than isolating them into discrete mechanisms.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- Adachi M, Liu Y, Fujii K, Calderwood SK, Nakai A, Imai K, Shinomura Y. Oxidative stress impairs the heat stress response and delays unfolded protein recovery. PloS One. 2009;4:e7719. doi: 10.1371/journal.pone.0007719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Alastalo T-P, Hellesuo M, Sandqvist A, Hietakangas V, Kallio M, Sistonen L. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J. Cell Sci. 2003;116:3557–3570. doi: 10.1242/jcs.00671. [DOI] [PubMed] [Google Scholar]
- Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PloS One. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat. Rev. Drug Discov. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneja R, Odoms K, Dunsmore K, Shanley TP, Wong HR. Extracellular heat shock protein-70 induces endotoxin tolerance in THP-1 cells. J. Immunol. Baltim. Md 1950. 2006;177:7184–7192. doi: 10.4049/jimmunol.177.10.7184. [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Hall MN. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 2012;22:274–282. doi: 10.1016/j.tcb.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Aprahamian T, Takemura Y, Goukassian D, Walsh K. Ageing is associated with diminished apoptotic cell clearance in vivo. Clin. Exp. Immunol. 2008;152:448–455. doi: 10.1111/j.1365-2249.2008.03658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Area-Gomez E, de Groof AJC, Boldogh I, Bird TD, Gibson GE, Koehler CM, Yu WH, Duff KE, Yaffe MP, Pon LA, et al. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am. J. Pathol. 2009;175:1810–1816. doi: 10.2353/ajpath.2009.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias A, Lamé MW, Santarelli L, Hen R, Greene LA, Angelastro JM. Regulated ATF5 loss-of-function in adult mice blocks formation and causes regression/eradication of gliomas. Oncogene. 2012;31:739–751. doi: 10.1038/onc.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould T, Michel S, Renard P. Mitochondria Retrograde Signaling and the UPR mt: Where Are We in Mammals? Int. J. Mol. Sci. 2015;16:18224–18251. doi: 10.3390/ijms160818224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft GS, Horan MA, Ferguson MW. Aging alters the inflammatory and endothelial cell adhesion molecule profiles during human cutaneous wound healing. Lab. Investig. J. Tech. Methods Pathol. 1998;78:47–58. [PubMed] [Google Scholar]
- Ast T, Aviram N, Chuartzman SG, Schuldiner M. A cytosolic degradation pathway, prERAD, monitors pre-inserted secretory pathway proteins. J. Cell Sci. 2014;127:3017–3023. doi: 10.1242/jcs.144386. [DOI] [PubMed] [Google Scholar]
- Azzu V, Brand MD. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 2010;123:578–585. doi: 10.1242/jcs.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek GH, Cheng H, Kim I, Rao H. The Cdc48 protein and its cofactor Vms1 are involved in Cdc13 protein degradation. J. Biol. Chem. 2012;287:26788–26795. doi: 10.1074/jbc.M112.351825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, Yates JR, Manning G, Dillin A. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. 2014;346:360–363. doi: 10.1126/science.1253168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting Proteostasis for Disease Intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Bartolome F, Wu H-C, Burchell VS, Preza E, Wray S, Mahoney CJ, Fox NC, Calvo A, Canosa A, Moglia C, et al. Pathogenic VCP mutations induce mitochondrial uncoupling and reduced ATP levels. Neuron. 2013;78:57–64. doi: 10.1016/j.neuron.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen KM, Durieux J, Shao L-W, Tian Y, Kim H-E, Wolff S, Liu Y, Dillin A. Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell. 2016;166:1553–1563.e10. doi: 10.1016/j.cell.2016.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Soto MM, McCullagh E. Unfolded protein stress in the endoplasmic reticulum and mitochondria: a role in neurodegeneration. Front. Aging Neurosci. 2012;4:5. doi: 10.3389/fnagi.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt D, Müller M, Reichert AS, Osiewacz HD. Simultaneous impairment of mitochondrial fission and fusion reduces mitophagy and shortens replicative lifespan. Sci. Rep. 2015;5:7885. doi: 10.1038/srep07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe EE, Olson KC, Chau V, Deshaies RJ. Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP•NPLOC4•UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc. Natl. Acad. Sci. U.S.a. 2017;114:E4380–E4388. doi: 10.1073/pnas.1706205114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böckler S, Chelius X, Hock N, Klecker T, Wolter M, Weiss M, Braun RJ, Westermann B. Fusion, fission, and transport control asymmetric inheritance of mitochondria and protein aggregates. J. Cell Biol. 2017 doi: 10.1083/jcb.201611197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar NO, Rapoport TA. Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell. 2017;169:722–735.e9. doi: 10.1016/j.cell.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini NM. Chaperoning brain degeneration. Proc. Natl. Acad. Sci. U. S. A. 2002;99(Suppl. 4):16407–16411. doi: 10.1073/pnas.152330499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman L, Schlierf A, Lutz AK, Shan J, Deinlein A, Kast J, Galehdar Z, Palmisano V, Patenge N, Berg D, et al. Parkin is transcriptionally regulated by ATF4: evidence for an interconnection between mitochondrial stress and ER stress. Cell Death Differ. 2011;18:769–782. doi: 10.1038/cdd.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun RJ, Zischka H, Madeo F, Eisenberg T, Wissing S, Büttner S, Engelhardt SM, Büringer D, Ueffing M. Crucial mitochondrial impairment upon CDC48 mutation in apoptotic yeast. J. Biol. Chem. 2006;281:25757–25767. doi: 10.1074/jbc.M513699200. [DOI] [PubMed] [Google Scholar]
- Braun RJ, Sommer C, Leibiger C, Gentier RJG, Dumit VI, Paduch K, Eisenberg T, Habernig L, Trausinger G, Magnes C, et al. Accumulation of Basic Amino Acids at Mitochondria Dictates the Cytotoxicity of Aberrant Ubiquitin. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufalino MR, DeVeale B, Kooy D, van der The asymmetric segregation of damaged proteins is stem cell–type dependent. J Cell Biol. 2013;201:523–530. doi: 10.1083/jcb.201207052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill CM, Waterman WR, Xie Y, Auron PE, Calderwood SK. Transcriptional repression of the prointerleukin 1beta gene by heat shock factor 1. J. Biol. Chem. 1996;271:24874–24879. [PubMed] [Google Scholar]
- Caldwell SR, Hill KJ, Cooper AA. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- Calì T, Ottolini D, Negro A, Brini M. α-Synuclein controls mitochondrial calcium homeostasis by enhancing endoplasmic reticulum-mitochondria interactions. J. Biol. Chem. 2012;287:17914–17929. doi: 10.1074/jbc.M111.302794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao SS, Kaufman RJ. Unfolded protein response. Curr. Biol. CB. 2012;22:R622–626. doi: 10.1016/j.cub.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JT, Wong AKO, Tavassoli S, Young BP, Chruscicki A, Fang NN, Howe LJ, Mayor T, Foster LJ, Loewen CJR. Polarization of the endoplasmic reticulum by ER-septin tethering. Cell. 2014;158:620–632. doi: 10.1016/j.cell.2014.06.033. [DOI] [PubMed] [Google Scholar]
- Chatenay-Lapointe M, Shadel GS. Stressed-out mitochondria get MAD. Cell Metab. 2010;12:559–560. doi: 10.1016/j.cmet.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari SN, Kipreos ET. Increased mitochondrial fusion allows the survival of older animals in diverse C. elegans longevity pathways. Nat. Commun. 2017;8:182. doi: 10.1038/s41467-017-00274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen J, Loo A, Jaeger S, Bagdasarian L, Yu J, Chung F, Korn J, Ruddy D, Guo R, et al. Targeting HSF1 sensitizes cancer cells to HSP90 inhibition. Oncotarget. 2013;4:816–829. doi: 10.18632/oncotarget.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Buvelot Frei S, Snapp EL, Barral Y. A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. eLife. 2014;3:e01883. doi: 10.7554/eLife.01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Abounit S, Marzo L, Danckaert A, Chamoun Z, Roux P, Zurzolo C. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J Cell Sci. 2013;126:3678–3685. doi: 10.1242/jcs.126086. [DOI] [PubMed] [Google Scholar]
- Csordás G, Várnai P, Golenár T, Roy S, Purkins G, Schneider TG, Balla T, Hajnóczky G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging. Trends Genet. TIG. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Sampson SB. HSF1: Guardian of Proteostasis in Cancer. Trends Cell Biol. 2016;26:17–28. doi: 10.1016/j.tcb.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Dai S, Cao J. Proteotoxic stress of cancer: implication of the heat-shock response in oncogenesis. J. Cell. Physiol. 2012;227:2982–2987. doi: 10.1002/jcp.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das I, Krzyzosiak A, Schneider K, Wrabetz L, D’Antonio M, Barry N, Sigurdardottir A, Bertolotti A. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science. 2015;348:239–242. doi: 10.1126/science.aaa4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, Pollera M, Bergamini E. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp. Gerontol. 2003;38:519–527. doi: 10.1016/s0531-5565(03)00002-0. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu A-L, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Dillin A, Gottschling DE, Nyström T. The good and the bad of being connected: the integrons of aging. Curr. Opin. Cell Biol. 2014;0:107–112. doi: 10.1016/j.ceb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W-X, Yin X-M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diot A, Hinks-Roberts A, Lodge T, Liao C, Dombi E, Morten K, Brady S, Fratter C, Carver J, Muir R, et al. A novel quantitative assay of mitophagy: Combining high content fluorescence microscopy and mitochondrial DNA load to quantify mitophagy and identify novel pharmacological tools against pathogenic heteroplasmic mtDNA. Pharmacol. Res. 2015;100:24–35. doi: 10.1016/j.phrs.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Dokladny K, Zuhl MN, Mandell M, Bhattacharya D, Schneider S, Deretic V, Moseley PL. Regulatory coordination between two major intracellular homeostatic systems: heat shock response and autophagy. J. Biol. Chem. 2013;288:14959–14972. doi: 10.1074/jbc.M113.462408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Baird NA, Simic MS, Uhlein S, McCormick MA, Wolff SC, Kennedy BK, Dillin A. Heterotypic Signals from Neural HSF-1 Separate Thermotolerance from Longevity. Cell Rep. 2015;12:1196–1204. doi: 10.1016/j.celrep.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplan E, Giaime E, Viotti J, Sévalle J, Corti O, Brice A, Ariga H, Qi L, Checler F, Alves da Costa C. ER-stress-associated functional link between Parkin and DJ-1 via a transcriptional cascade involving the tumor suppressor p53 and the spliced X-box binding protein XBP-1. J. Cell Sci. 2013;126:2124–2133. doi: 10.1242/jcs.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eden W, van der Zee R, Prakken B. Heat-shock proteins induce T-cell regulation of chronic inflammation. Nat. Rev. Immunol. 2005;5:318–330. doi: 10.1038/nri1593. [DOI] [PubMed] [Google Scholar]
- Ekstrand MI, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, Thams S, Bergstrand A, Hansson FS, Trifunovic A, et al. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Waltz TB, Kassahun H, Lu Q, Kerr JS, Morevati M, Fivenson EM, Wollman BN, Marosi K, Wilson MA, et al. Tomatidine enhances lifespan and healthspan in C. elegans through mitophagy induction via the SKN-1/Nrf2 pathway. Sci. Rep. 2017;7:46208. doi: 10.1038/srep46208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Chang R, Yang L. Heat shock factor 1 promotes invasion and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer. 2012;118:1782–1794. doi: 10.1002/cncr.26482. [DOI] [PubMed] [Google Scholar]
- Fargnoli J, Kunisada T, Fornace AJ, Schneider EL, Holbrook NJ. Decreased expression of heat shock protein 70 mRNA and protein after heat treatment in cells of aged rats. Proc. Natl. Acad. Sci. U. S. A. 1990;87:846–850. doi: 10.1073/pnas.87.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett TW, Sylvester SL, Sarge KD, Morimoto RI, Holbrook NJ. Effects of neurohormonal stress and aging on the activation of mammalian heat shock factor 1. J. Biol. Chem. 1994;269:32272–32278. [PubMed] [Google Scholar]
- Ferat-Osorio E, Sánchez-Anaya A, Gutiérrez-Mendoza M, Boscó-Gárate I, Wong-Baeza I, Pastelin-Palacios R, Pedraza-Alva G, Bonifaz LC, Cortés-Reynosa P, Pérez-Salazar E, et al. Heat shock protein 70 down-regulates the production of toll-like receptor-induced pro-inflammatory cytokines by a heat shock factor-1/constitutive heat shock element-binding factor-dependent mechanism. J. Inflamm. Lond. Engl. 2014;11:19. doi: 10.1186/1476-9255-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW, Haynes CM. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol. CB. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso MCBV, Almeida MQ, Mazzuco TL, Mariani BMP, Brito LP, Gonçalves TC, Alencar GA, Lima L, de O, Faria AM, Bourdeau I, et al. Combined expression of BUB1B, DLGAP5, and PINK1 as predictors of poor outcome in adrenocortical tumors: validation in a Brazilian cohort of adult and pediatric patients. Eur. J. Endocrinol. 2012;166:61–67. doi: 10.1530/EJE-11-0806. [DOI] [PubMed] [Google Scholar]
- Frakes AE, Dillin A. The UPR(ER): Sensor and Coordinator of Organismal Homeostasis. Mol. Cell. 2017;66:761–771. doi: 10.1016/j.molcel.2017.05.031. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T. Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J. Biol. Chem. 2008;283:26188–26197. doi: 10.1074/jbc.M710521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S-I, Kato K, Yonemura S, Inouye S, Nakai A. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 2004;23:4297–4306. doi: 10.1038/sj.emboj.7600435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S, Nakai A. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J. Biol. Chem. 2005;280:34908–34916. doi: 10.1074/jbc.M506288200. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Takaki E, Takii R, Tan K, Prakasam R, Hayashida N, Iemura S, Natsume T, Nakai A. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol. Cell. 2012;48:182–194. doi: 10.1016/j.molcel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- Gagliano N, Grizzi F, Annoni G. Mechanisms of aging and liver functions. Dig. Dis. Basel Switz. 2007;25:118–123. doi: 10.1159/000099475. [DOI] [PubMed] [Google Scholar]
- Gaigg B, Simbeni R, Hrastnik C, Paltauf F, Daum G. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim. Biophys. Acta. 1995;1234:214–220. doi: 10.1016/0005-2736(94)00287-y. [DOI] [PubMed] [Google Scholar]
- García-Prat L, Muñoz-Cánoves P, Martinez-Vicente M. Dysfunctional autophagy is a driver of muscle stem cell functional decline with aging. Autophagy. 2016;12:612–613. doi: 10.1080/15548627.2016.1143211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu A-L, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJY, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017 doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskreutz J, Van Den Bosch L, Keller BU. Calcium dysregulation in amyotrophic lateral sclerosis. Cell Calcium. 2010;47:165–174. doi: 10.1016/j.ceca.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Hölper S, Mari M, Harwardt M-LI, Yan R, Müller S, Reggiori F, Heilemann M, Dikic I. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife. 2017;6 doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisbert E, Czyz DM, Richter K, McMullen PD, Morimoto RI. Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 2013;9:e1003466. doi: 10.1371/journal.pgen.1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Habeck G, Ebner FA, Shimada-Kreft H, Kreft SG. The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J. Cell Biol. 2015;209:621. doi: 10.1083/jcb.20140808804292015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Hu J, Lau MY, Feng M, Petrovic LM, Ji C. Altered methylation and expression of ER-associated degradation factors in long-term alcohol and constitutive ER stress-induced murine hepatic tumors. Front. Genet. 2013a;4:224. doi: 10.3389/fgene.2013.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Xu X, Qin H, Liu A, Fan Z, Kang L, Fu J, Liu J, Ye Q. The molecular mechanism and potential role of heat shock-induced p53 protein accumulation. Mol. Cell. Biochem. 2013b;378:161–169. doi: 10.1007/s11010-013-1607-9. [DOI] [PubMed] [Google Scholar]
- Han S, Liu Y, Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J. Biol. Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/−mice reveals a role for translational control in secretory cell survival. Mol. Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Hasday JD, Thompson C, Singh IS. Fever, immunity, and molecular adaptations. Compr. Physiol. 2014;4:109–148. doi: 10.1002/cphy.c130019. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su T-P. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldens L, Hensen SMM, Onnekink C, van Genesen ST, Dirks RP, Lubsen NH. An atypical unfolded protein response in heat shocked cells. PloS One. 2011;6:e23512. doi: 10.1371/journal.pone.0023512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze N, Le Breton L, Wiesner J, Kempf G, Mayer MP. Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. eLife. 2016;5 doi: 10.7554/eLife.11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J-M, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, et al. A stress-responsive system for mitochondrial protein degradation. Mol. Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Hetz C, Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 2014;15:233–249. doi: 10.1038/nrn3689. [DOI] [PubMed] [Google Scholar]
- Heydari AR, You S, Takahashi R, Gutsmann-Conrad A, Sarge KD, Richardson A. Age-related alterations in the activation of heat shock transcription factor 1 in rat hepatocytes. Exp. Cell Res. 2000;256:83–93. doi: 10.1006/excr.2000.4808. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Vevea JD, Swayne TC, Chojnowski R, Hill V, Boldogh IR, Pon LA. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr. Biol. CB. 2013;23:2417–2422. doi: 10.1016/j.cub.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Pernice WMA, Vevea JD, Alessi Wolken DM, Boldogh IR, Pon LA. Role of asymmetric cell division in lifespan control in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14:1133–1146. doi: 10.1111/1567-1364.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Charalel JK, Viana MP, Garcia EJ, Sing CN, Koenigsberg A, Swayne TC, Vevea JD, Boldogh IR, Rafelski SM, et al. Mitochondrial anchorage and fusion contribute to mitochondrial inheritance and quality control in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2016a;27:776–787. doi: 10.1091/mbc.E15-07-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Vevea J, Charalel J, Sapar M, Pon L. The transcriptional repressor Sum1p counteracts Sir2p in regulation of the actin cytoskeleton, mitochondrial quality control and replicative lifespan in Saccharomyces cerevisiae. Microb. Cell. 2016b;3:79–88. doi: 10.15698/mic2016.02.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SM, Hao X, Liu B, Nyström T. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science. 2014;344:1389–1392. doi: 10.1126/science.1252634. [DOI] [PubMed] [Google Scholar]
- Hill SM, Hao X, Grönvall J, Spikings-Nordby S, Widlund PO, Amen T, Jörhov A, Josefson R, Kaganovich D, Liu B, et al. Asymmetric Inheritance of Aggregated Proteins and Age Reset in Yeast Are Regulated by Vac17-Dependent Vacuolar Functions. Cell Rep. 2016;16:826–838. doi: 10.1016/j.celrep.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SM, Hanzén S, Nyström T. Restricted access: spatial sequestration of damaged proteins during stress and aging. EMBO Rep. 2017:e201643458. doi: 10.15252/embr.201643458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HK, Jia Y, Coe KJ, Gao Q, Doneanu CE, Hu Z, Bammler TK, Beyer RP, Fausto N, Bruschi SA, et al. Cytosolic heat shock proteins and heme oxygenase-1 are preferentially induced in response to specific and localized intramitochondrial damage by tetrafluoroethylcysteine. Biochem. Pharmacol. 2006;72:80–90. doi: 10.1016/j.bcp.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PloS One. 2007;2:e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Tang H, Liu Z, Österlund T, Nielsen J, Petranovic D. Management of the endoplasmic reticulum stress by activation of the heat shock response in yeast. FEMS Yeast Res. 2014;14:481–494. doi: 10.1111/1567-1364.12125. [DOI] [PubMed] [Google Scholar]
- Houck SA, Ren HY, Madden VJ, Bonner JN, Conlin MP, Janovick JA, Conn PM, Cyr DM. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol. Cell. 2014;54:166–179. doi: 10.1016/j.molcel.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A-L, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hsu S-F, Chao C-M, Huang W-T, Lin M-T, Cheng B-C. Attenuating heat-induced cellular autophagy, apoptosis and damage in H9c2 cardiomyocytes by pre-inducing HSP70 with heat shock preconditioning. Int. J. Hyperth. Off. J. Eur. Soc. Hyperthermic Oncol. North Am. Hyperth. Group. 2013;29:239–247. doi: 10.3109/02656736.2013.777853. [DOI] [PubMed] [Google Scholar]
- Hu BR, Martone ME, Jones YZ, Liu CL. Protein aggregation after transient cerebral ischemia. J. Neurosci. Off. J. Soc. Neurosci. 2000;20:3191–3199. doi: 10.1523/JNEUROSCI.20-09-03191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Frohman MA. Visualizing mitochondrial lipids and fusion events in mammalian cells. Methods Cell Biol. 2012;108:131–145. doi: 10.1016/B978-0-12-386487-1.00007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Gottschling DE. An Early-Age Increase in Vacuolar pH Limits Mitochondrial Function and Lifespan in Yeast. Nature. 2012;492:261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MB, Jasper H. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 2014;20:214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Usson Y, Morimoto RI. Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6769–6774. doi: 10.1073/pnas.96.12.6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajisto P, Döhla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348:340–343. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayani AC, Morton JP, McArdle A. The exercise-induced stress response in skeletal muscle: failure during aging. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2008;33:1033–1041. doi: 10.1139/H08-089. [DOI] [PubMed] [Google Scholar]
- Kenny TC, Hart P, Ragazzi M, Sersinghe M, Chipuk J, Sagar MaK, Eliceiri KW, LaFramboise T, Grandhi S, Santos J, et al. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPR(mt) to promote metastasis. Oncogene. 2017 doi: 10.1038/onc.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A, Ackermann B, Clement AM, Duerk H, Behl C. HSF1-controlled and age-associated chaperone capacity in neurons and muscle cells of C. elegans. PloS One. 2010;5:e8568. doi: 10.1371/journal.pone.0008568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- Kim H-E, Grant AR, Simic MS, Kohnz RA, Nomura DK, Durieux J, Riera CE, Sanchez M, Kapernick E, Wolff S, et al. Lipid Biosynthesis Coordinates a Mitochondrial-to-Cytosolic Stress Response. Cell. 2016;166:1539–1552.e16. doi: 10.1016/j.cell.2016.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim Y-N, Kim SS, Kim DH, Hur KY, Kim HK, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Codogno P. The mechanism and physiological function of macroautophagy. J. Innate Immun. 2013;5:427–433. doi: 10.1159/000351979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HS, Uehara T, Nomura Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J. Biol. Chem. 2002;277:35386–35392. doi: 10.1074/jbc.M203412200. [DOI] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Nikoletopoulou V, Tavernarakis N. Small heat-shock proteins protect from heat-stroke-associated neurodegeneration. Nature. 2012;490:213–218. doi: 10.1038/nature11417. [DOI] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Kreft SG. Protein quality and quantity control at the yeast ER. Oncotarget. 2015;6:16818–16819. doi: 10.18632/oncotarget.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Kumsta C, Chang JT, Schmalz J, Hansen M. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat. Commun. 2017;8:14337. doi: 10.1038/ncomms14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurapati R, Passananti HB, Rose MR, Tower J. Increased hsp22 RNA levels in Drosophila lines genetically selected for increased longevity. J. Gerontol. A. Biol. Sci. Med. Sci. 2000;55:B552–559. doi: 10.1093/gerona/55.11.b552. [DOI] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. The Biology of Proteostasis in Aging and Disease. Annu. Rev. Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Nunnari J. Small molecule inhibitors of mitochondrial division: tools that translate basic biological research into medicine. Chem. Biol. 2010;17:578–583. doi: 10.1016/j.chembiol.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanphere KR, Schneider S, Mermier C, Zuhl M, Dokladny K, Moseley P. Effects of Aging on the Heat Shock Response and Autophagy in Human Peripheral Blood Mononuclear Cells. FASEB J. 2013;27:994.5–994.5. [Google Scholar]
- Lautenschläger J, Prell T, Ruhmer J, Weidemann L, Witte OW, Grosskreutz J. Overexpression of human mutated G93A SOD1 changes dynamics of the ER mitochondria calcium cycle specifically in mouse embryonic motor neurons. Exp. Neurol. 2013;247:91–100. doi: 10.1016/j.expneurol.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ZY, Prouteau M, Gotta M, Barral Y. Compartmentalization of the endoplasmic reticulum in the early C. elegans embryos. J. Cell Biol. 2016;214:665–676. doi: 10.1083/jcb.201601047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Garrido-Urbani S, Ginestier C, Fournier G, Esterni B, Monville F, Adélaïde J, Geneix J, Xerri L, Dubreuil P, et al. Correlated break at PARK2/FRA6E and loss of AF-6/Afadin protein expression are associated with poor outcome in breast cancer. Oncogene. 2007;26:298–307. doi: 10.1038/sj.onc.1209772. [DOI] [PubMed] [Google Scholar]
- Li D, Yallowitz A, Ozog L, Marchenko N. A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis. 2014;5:e1194. doi: 10.1038/cddis.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in Stress, Development, and Organismal Health. Trends Cell Biol. 2017 doi: 10.1016/j.tcb.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lin Y-F, Schulz AM, Pellegrino MW, Lu Y, Shaham S, Haynes CM. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature. 2016;533:416–419. doi: 10.1038/nature17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chang A. Heat shock response relieves ER stress. EMBO J. 2008a;27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chang A. Heat shock response relieves ER stress. EMBO J. 2008b;27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, Nyström T. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedeke C, Frei SB, Sbalzarini I, Schwarz H, Spang A, Barral Y. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 2005;169:897–908. doi: 10.1083/jcb.200412143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan S, Oku M, Tsuda M, Hoseki J, Sakai Y. Mitochondrial impairment triggers cytosolic oxidative stress and cell death following proteasome inhibition. Sci. Rep. 2014;4 doi: 10.1038/srep05896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Brady OA, Puertollano R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 2016;35:479–495. doi: 10.15252/embj.201593428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Høj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur. J. Biochem. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 2010;12:260–272. doi: 10.1016/j.cmet.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10:885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melentijevic I, Toth ML, Arnold ML, Guasp RJ, Harinath G, Nguyen KC, Taub D, Parker JA, Neri C, Gabel CV, et al. C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature. 2017;542:367–371. doi: 10.1038/nature21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell. 2016;165:1209–1223. doi: 10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J-N, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene. 2007;26:5086–5097. doi: 10.1038/sj.onc.1210317. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi S, Ohkuma A, Talim B, Karahashi M, Koumura T, Aoyama C, Kurihara M, Quinlivan R, Sewry C, Mitsuhashi H, et al. A congenital muscular dystrophy with mitochondrial structural abnormalities caused by defective de novo phosphatidylcholine biosynthesis. Am. J. Hum. Genet. 2011;88:845–851. doi: 10.1016/j.ajhg.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima N, Tsutsumi S, Sourbier C, Beebe K, Mollapour M, Rivas C, Yoshida S, Trepel JB, Huang Y, Tatokoro M, et al. The HSP90 inhibitor ganetespib synergizes with the MET kinase inhibitor crizotinib in both crizotinib-sensitive and -resistant MET-driven tumor models. Cancer Res. 2013;73:7022–7033. doi: 10.1158/0008-5472.CAN-13-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D, Pilz G, Araúzo-Bravo M, Barral Y, Jessberger S. A mechanism for the segregation of age in mammalian neural stem cells. Science. 2015;349:1334–1338. doi: 10.1126/science.aac9868. [DOI] [PubMed] [Google Scholar]
- Moreno JA, Halliday M, Molloy C, Radford H, Verity N, Axten JM, Ortori CA, Willis AE, Fischer PM, Barrett DA, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 2013;5:206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton EA, Lamitina T. Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock. Aging Cell. 2013;12:112–120. doi: 10.1111/acel.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch C, Harper JW. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature. 2016;534:710–713. doi: 10.1038/nature18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow L, Debnath J. Autophagy as a Stress-Response and Quality-Control Mechanism: Implications for Cell Injury and Human Disease. Annu. Rev. Pathol. Mech. Dis. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Master M, Zhu Y, Pack AI. Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J. Neurosci. Off. J. Soc. Neurosci. 2008;28:6539–6548. doi: 10.1523/JNEUROSCI.5685-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I, Hayashi JI. Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med. 2001;7:934–940. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Fujimoto M, Fukushima S, Nakamura A, Hayashida N, Takii R, Takaki E, Nakai A, Muto M. Heat shock factor 1 is required for migration and invasion of human melanoma in vitro and in vivo. Cancer Lett. 2014;354:329–335. doi: 10.1016/j.canlet.2014.08.029. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen D-F, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Mol. Cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Jaeger AM, Gomez-Pastor R, Willmund F, Frydman J, Thiele DJ. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 2014;9:955–966. doi: 10.1016/j.celrep.2014.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivon M, Richet E, Codogno P, Arrigo A-P, Kretz-Remy C. Autophagy activation by NFkappaB is essential for cell survival after heat shock. Autophagy. 2009;5:766–783. doi: 10.4161/auto.8788. [DOI] [PubMed] [Google Scholar]
- Nuss JE, Choksi KB, DeFord JH, Papaconstantinou J. Decreased enzyme activities of chaperones PDI and BiP in aged mouse livers. Biochem. Biophys. Res. Commun. 2008;365:355–361. doi: 10.1016/j.bbrc.2007.10.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberley TD, Swanlund JM, Zhang HJ, Kregel KC. Aging results in increased autophagy of mitochondria and protein nitration in rat hepatocytes following heat stress. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2008;56:615–627. doi: 10.1369/jhc.2008.950873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrodnik M, Salmonowicz H, Brown R, Turkowska J, Średniawa W, Pattabiraman S, Amen T, Abraham A, Eichler N, Lyakhovetsky R, et al. Dynamic JUNQ inclusion bodies are asymmetrically inherited in mammalian cell lines through the asymmetric partitioning of vimentin. Proc. Natl. Acad. Sci. 2014;111:8049–8054. doi: 10.1073/pnas.1324035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostberg JR, Kaplan KC, Repasky EA. Induction of stress proteins in a panel of mouse tissues by fever-range whole body hyperthermia. Int. J. Hyperth. Off. J. Eur. Soc. Hyperthermic Oncol. North Am. Hyperth. Group. 2002;18:552–562. doi: 10.1080/02656730210166168. [DOI] [PubMed] [Google Scholar]
- Östling P, Björk JK, Roos-Mattjus P, Mezger V, Sistonen L. Heat Shock Factor 2 (HSF2) Contributes to Inducible Expression of hsp Genes through Interplay with HSF1. J. Biol. Chem. 2007;282:7077–7086. doi: 10.1074/jbc.M607556200. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim J-M, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Pawelec G, Solana R. Immunosenescence. Immunol. Today. 1997;18:514–516. doi: 10.1016/s0167-5699(97)01145-6. [DOI] [PubMed] [Google Scholar]
- Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams DJ, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C, Valtorta M, Tedeschi G, Galbusera E, Pastori V, Bigi A, Nonnis S, Grassi E, Fusi P. Study of subcellular localization and proteolysis of ataxin-3. Neurobiol. Dis. 2008;30:190–200. doi: 10.1016/j.nbd.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, Morimoto RI. Regulation of the Cellular Heat Shock Response in Caenorhabditis elegans by Thermosensory Neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printsev I, Curiel D, Carraway KL. Membrane Protein Quantity Control at the Endoplasmic Reticulum. J. Membr. Biol. 2016 doi: 10.1007/s00232-016-9931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao A, Jin X, Pang J, Moskophidis D, Mivechi NF. The transcriptional regulator of the chaperone response HSF1 controls hepatic bioenergetics and protein homeostasis. J. Cell Biol. 2017;216:723–741. doi: 10.1083/jcb.201607091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana A, Schwindling C, Wenning AS, Becherer U, Rettig J, Schwarz EC, Hoth M. T cell activation requires mitochondrial translocation to the immunological synapse. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14418–14423. doi: 10.1073/pnas.0703126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabek JP, Boylston WH, Papaconstantinou J. Carbonylation of ER chaperone proteins in aged mouse liver. Biochem. Biophys. Res. Commun. 2003;305:566–572. doi: 10.1016/s0006-291x(03)00826-x. [DOI] [PubMed] [Google Scholar]
- Ramadasan-Nair R, Gayathri N, Mishra S, Sunitha B, Mythri RB, Nalini A, Subbannayya Y, Harsha HC, Kolthur-Seetharam U, Srinivas Bharath MM. Mitochondrial alterations and oxidative stress in an acute transient mouse model of muscle degeneration: implications for muscular dystrophy and related muscle pathologies. J. Biol. Chem. 2014;289:485–509. doi: 10.1074/jbc.M113.493270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A, Oliveira MP, Khamoui AV, Aparicio R, Rera M, Rossiter HB, Walker DW. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat. Commun. 2017;8:448. doi: 10.1038/s41467-017-00525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reyniès A, Assié G, Rickman DS, Tissier F, Groussin L, René-Corail F, Dousset B, Bertagna X, Clauser E, Bertherat J. Gene expression profiling reveals a new classification of adrenocortical tumors and identifies molecular predictors of malignancy and survival. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009;27:1108–1115. doi: 10.1200/JCO.2008.18.5678. [DOI] [PubMed] [Google Scholar]
- Ritossa F. A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia. 1962;18:571–573. [Google Scholar]
- Ruan L, Zhou C, Jin E, Kucharavy A, Zhang Y, Wen Z, Florens L, Li R. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature. 2017;543:443–446. doi: 10.1038/nature21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Rujano MA, Bosveld F, Salomons FA, Dijk F, Waarde MAWH, van Want JJL, van der Vos RAI, de Brunt ER, Sibon OCM, Kampinga HH. Polarised Asymmetric Inheritance of Accumulated Protein Damage in Higher Eukaryotes. PLOS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Félix AA, Williams EG, Jha P, Lo Sasso G, Huzard D, et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 2016;22:879–888. doi: 10.1038/nm.4132. [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Caudron F, Prasad R, Moreno DF, Bolognesi A, Aldea M, Barral Y. Compartmentalization of ER-Bound Chaperone Confines Protein Deposit Formation to the Aging Yeast Cell. Curr. Biol. CB. 2017;27:773–783. doi: 10.1016/j.cub.2017.01.069. [DOI] [PubMed] [Google Scholar]
- Sandqvist A, Björk JK, Åkerfelt M, Chitikova Z, Grichine A, Vourc’h C, Jolly C, Salminen TA, Nymalm Y, Sistonen L. Heterotrimerization of Heat-Shock Factors 1 and 2 Provides a Transcriptional Switch in Response to Distinct Stimuli. Mol. Biol. Cell. 2009;20:1340–1347. doi: 10.1091/mbc.E08-08-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Amon A. Aneuploidy triggers a TFEB-mediated lysosomal stress response. Autophagy. 2015;11:2383–2384. doi: 10.1080/15548627.2015.1110670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Yoshida H. Organelle autoregulation-stress responses in the ER, Golgi, mitochondria and lysosome. J. Biochem. (Tokyo) 2015;157:185–195. doi: 10.1093/jb/mvv010. [DOI] [PubMed] [Google Scholar]
- Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- Scharf K-D, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta. 2012;1819:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nyström T, Osiewacz HD. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 2007;9:99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- Schindler AJ, Schekman R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17775–17780. doi: 10.1073/pnas.0910342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F, Baker D, Chambers MG, Su H, Li D, et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature. 2017 doi: 10.1038/nature23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 2014;127:4078–4088. doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz AM, Haynes CM. UPR(mt)-mediated cytoprotection and organismal aging. Biochim. Biophys. Acta. 2015;1847:1448–1456. doi: 10.1016/j.bbabio.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, Min C, Jaskelioff M, Xiao Y, Wu C-J, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20:92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo K, Choi E, Lee D, Jeong D-E, Jang SK, Lee S-J. Heat shock factor 1 mediates the longevity conferred by inhibition of TOR and insulin/IGF-1 signaling pathways in C. elegans. Aging Cell. 2013;12:1073–1081. doi: 10.1111/acel.12140. [DOI] [PubMed] [Google Scholar]
- Shao L-W, Niu R, Liu Y. Neuropeptide signals cell non-autonomous mitochondrial unfolded protein response. Cell Res. 2016;26:1182–1196. doi: 10.1038/cr.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- Siegelin MD, Dohi T, Raskett CM, Orlowski GM, Powers CM, Gilbert CA, Ross AH, Plescia J, Altieri DC. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J. Clin. Invest. 2011;121:1349–1360. doi: 10.1172/JCI44855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh IS, Gupta A, Nagarsekar A, Cooper Z, Manka C, Hester L, Benjamin IJ, He J-R, Hasday JD. Heat shock co-activates interleukin-8 transcription. Am. J. Respir. Cell Mol. Biol. 2008;39:235–242. doi: 10.1165/rcmb.2007-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton KD, Wischmeyer PE. Effects of HSP70.1/3 gene knockout on acute respiratory distress syndrome and the inflammatory response following sepsis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;290:L956–961. doi: 10.1152/ajplung.00466.2005. [DOI] [PubMed] [Google Scholar]
- Solís EJ, Pandey JP, Zheng X, Jin DX, Gupta PB, Airoldi EM, Pincus D, Denic V. Defining the Essential Function of Yeast Hsf1 Reveals a Compact Transcriptional Program for Maintaining Eukaryotic Proteostasis. Mol. Cell. 2016;63:60–71. doi: 10.1016/j.molcel.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo KY, Atkin JD, Horne MK, Nagley P. Recruitment of mitochondria into apoptotic signaling correlates with the presence of inclusions formed by amyotrophic lateral sclerosis-associated SOD1 mutations. J. Neurochem. 2009;108:578–590. doi: 10.1111/j.1471-4159.2008.05799.x. [DOI] [PubMed] [Google Scholar]
- Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- Stauch KL, Purnell PR, Fox HS. Aging synaptic mitochondria exhibit dynamic proteomic changes while maintaining bioenergetic function. Aging. 2014;6:320–334. doi: 10.18632/aging.100657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Górska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Sugiura A, Yonashiro R, Fukuda T, Matsushita N, Nagashima S, Inatome R, Yanagi S. A mitochondrial ubiquitin ligase MITOL controls cell toxicity of polyglutamine-expanded protein. Mitochondrion. 2011;11:139–146. doi: 10.1016/j.mito.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science. 2011;332:729–732. doi: 10.1126/science.1203411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Liu Y, Aballay A. Organismal regulation of XBP-1-mediated unfolded protein response during development and immune activation. EMBO Rep. 2012;13:855–860. doi: 10.1038/embor.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmström KM, Fergusson MM, Yoo YH, Combs CA, et al. Measuring In Vivo Mitophagy. Mol. Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A, Elo JM, Pietiläinen KH, Hakonen AH, Sevastianova K, Korpela M, Isohanni P, Marjavaara SK, Tyni T, Kiuru-Enari S, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 2011;10:806–818. doi: 10.1016/S1474-4422(11)70155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Várnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, Sheng M. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr. Biol. CB. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen D-F, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Uehara T, Nomura Y. Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J. Biol. Chem. 2000;275:10388–10393. doi: 10.1074/jbc.275.14.10388. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Yoshida H. TFE3, HSP47, and CREB3 Pathways of the Mammalian Golgi Stress Response. Cell Struct. Funct. 2017;42:27–36. doi: 10.1247/csf.16023. [DOI] [PubMed] [Google Scholar]
- Tatum MC, Ooi FK, Chikka MR, Chauve L, Martinez-Velazquez LA, Steinbusch HWM, Morimoto RI, Prahlad V. Neuronal serotonin release triggers the heat shock response in C. elegans in the absence of temperature increase. Curr. Biol. CB. 2015;25:163–174. doi: 10.1016/j.cub.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink N, van der Sanden MHM, Houweling M, Helms JB, Vaandrager AB. Depletion of phosphatidylcholine affects endoplasmic reticulum morphology and protein traffic at the Golgi complex. J. Lipid Res. 2009;50:2182–2192. doi: 10.1194/jlr.M800660-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt) Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissières A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J. Mol. Biol. 1974;84:389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- Toma-Jonik A, Widlak W, Korfanty J, Cichon T, Smolarczyk R, Gogler-Piglowska A, Widlak P, Vydra N. Active heat shock transcription factor 1 supports migration of the melanoma cells via vinculin down-regulation. Cell. Signal. 2015;27:394–401. doi: 10.1016/j.cellsig.2014.11.029. [DOI] [PubMed] [Google Scholar]
- Tran JR, Tomsic LR, Brodsky JL. A Cdc48p–associated factor modulates endoplasmic reticulum-associated degradation, cell stress, and ubiquitinated protein homeostasis. J. Biol. Chem. 2011;286:5744–5755. doi: 10.1074/jbc.M110.179259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta AP, Chipuk JE. Mitochondrial dynamics as regulators of cancer biology. Cell. Mol. Life Sci. 2017;74:1999–2017. doi: 10.1007/s00018-016-2451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiri EN, Trougakos IP. International Review of Cell and Molecular Biology. Elsevier; 2015. The Amazing Ubiquitin-Proteasome System: Structural Components and Implication in?Aging; pp. 171–237. [DOI] [PubMed] [Google Scholar]
- Tsao W, Jeong YH, Lin S, Ling J, Price DL, Chiang P-M, Wong PC. Rodent models of TDP-43: recent advances. Brain Res. 2012;1462:26–39. doi: 10.1016/j.brainres.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulapurkar ME, Almutairy EA, Shah NG, He J, Puche AC, Shapiro P, Singh IS, Hasday JD. Febrile-range hyperthermia modifies endothelial and neutrophilic functions to promote extravasation. Am. J. Respir. Cell Mol. Biol. 2012;46:807–814. doi: 10.1165/rcmb.2011-0378OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkilä S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjøsund KE, Tulkki V, et al. Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vance JE. Molecular and cell biology of phosphatidylserine and phosphatidylethanolamine metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003;75:69–111. doi: 10.1016/s0079-6603(03)75003-x. [DOI] [PubMed] [Google Scholar]
- Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan AJ, Pao W, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat. Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez JM, Lindquist S. hsp70: nuclear concentration during environmental stress and cytoplasmic storage during recovery. Cell. 1984;36:655–662. doi: 10.1016/0092-8674(84)90345-3. [DOI] [PubMed] [Google Scholar]
- Verma P, Pfister JA, Mallick S, D’Mello SR. HSF1 protects neurons through a novel trimerization- and HSP-independent mechanism. J. Neurosci. Off. J. Soc. Neurosci. 2014;34:1599–1612. doi: 10.1523/JNEUROSCI.3039-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Ly CV, Venken KJT, Koh T-W, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Vevea JD, Garcia EJ, Chan RB, Zhou B, Schultz M, Di Paolo G, McCaffery JM, Pon LA. Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev. Cell. 2015;35:584–599. doi: 10.1016/j.devcel.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues APC, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- Voelker DR. Genetic and biochemical analysis of non-vesicular lipid traffic. Annu. Rev. Biochem. 2009;78:827–856. doi: 10.1146/annurev.biochem.78.081307.112144. [DOI] [PubMed] [Google Scholar]
- Volovik Y, Maman M, Dubnikov T, Bejerano-Sagie M, Joyce D, Kapernick EA, Cohen E, Dillin A. Temporal requirements of heat shock factor-1 for longevity assurance. Aging Cell. 2012;11:491–499. doi: 10.1111/j.1474-9726.2012.00811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang D, Wang J, Bonamy GMC, Meeusen S, Brusch RG, Turk C, Yang P, Schultz PG. A small molecule promotes mitochondrial fusion in mammalian cells. Angew. Chem. Int. Ed Engl. 2012;51:9302–9305. doi: 10.1002/anie.201204589. [DOI] [PubMed] [Google Scholar]
- Wang J, Farr GW, Hall DH, Li F, Furtak K, Dreier L, Horwich AL. An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000350. doi: 10.1371/journal.pgen.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GDJ, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Weir HJ, Yao P, Huynh FK, Escoubas CC, Goncalves RL, Burkewitz K, Laboy R, Hirschey MD, Mair WB. Dietary Restriction and AMPK Increase Lifespan via Mitochondrial Network and Peroxisome Remodeling. Cell Metab. 2017;0 doi: 10.1016/j.cmet.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Merging mitochondria matters: cellular role and molecular machinery of mitochondrial fusion. EMBO Rep. 2002;3:527–531. doi: 10.1093/embo-reports/kvf113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta BBA - Bioenerg. 2012;1817:1833–1838. doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Meisinger C, Pfanner N. Cell biology. Connecting organelles. Science. 2009;325:403–404. doi: 10.1126/science.1178016. [DOI] [PubMed] [Google Scholar]
- Williams KW, Liu T, Kong X, Fukuda M, Deng Y, Berglund ED, Deng Z, Gao Y, Liu T, Sohn J-W, et al. Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metabolism. 2014;20:471–482. doi: 10.1016/j.cmet.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P, van Montfort R. EML4-ALK fusions: propelling cancer but creating exploitable chaperone dependence. Cancer Discov. 2014;4:642–645. doi: 10.1158/2159-8290.CD-14-0409. [DOI] [PubMed] [Google Scholar]
- Wu X, Li L, Jiang H. Doa1 targets ubiquitinated substrates for mitochondria-associated degradation. J. Cell Biol. 2016;213:49–63. doi: 10.1083/jcb.201510098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Tang WK, Ye Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene. 2016;583:64–77. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol. Biol. Cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Calay ES, Fan J, Arduini A, Kunz RC, Gygi SP, Yalcin A, Fu S, Hotamisligil GS. METABOLISM. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science. 2015;349:500–506. doi: 10.1126/science.aaa0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yee C, Yang W, Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157:897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M, Miyatake T, Attardi G. Complementation of mutant and wild-type human mitochondrial DNAs coexisting since the mutation event and lack of complementation of DNAs introduced separately into a cell within distinct organelles. Mol. Cell. Biol. 1994;14:2699–2712. doi: 10.1128/mcb.14.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zattas D, Hochstrasser M. Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit. Rev. Biochem. Mol. Biol. 2015;50:1–17. doi: 10.3109/10409238.2014.959889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Gong S, Shunmei E, Zou J. Induction of macroautophagy by heat. Mol. Biol. Rep. 2009;36:2323–2327. doi: 10.1007/s11033-009-9451-4. [DOI] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JR, Guo F, Yu Z, Mickey K, Narkar A, Ross RT, McClain M, Li R. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159:530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]