Ubiquitin signaling and autophagy (original) (raw)
Abstract
Ubiquitination is a widespread post-translational modification that controls multiple steps in autophagy, a major lysosome-mediated intracellular degradation pathway. A variety of ubiquitin chains are attached as selective labels on protein aggregates and dysfunctional organelles, thus promoting their autophagy-dependent degradation. Moreover, ubiquitin modification of autophagy regulatory components is essential to positively or negatively regulate autophagy flux in both non-selective and selective pathways. We review the current findings that elucidate the components, timing, and kinetics of the multivalent role of ubiquitin signals in control of amplitude and selectivity of autophagy pathways as well as their impact on the development of human diseases.
Introduction
A living cell is a complex and dynamic system, which quickly responds and adapts to environmental changes and stress situations. Likewise, proteins play a key role in all physiological and pathological cellular functions; therefore, understanding the mechanisms related to their synthesis, modification, and degradation is of great importance.
Autophagy is a highly dynamic catabolic process able to resolve a stress situation within minutes. This is possible because a multitude of different signaling pathways converge on the autophagy core machinery (1). The fast timing implies, at least in a first instance, the involvement of signaling cascades mainly regulated by post-translational modifications (PTMs)2 rather than transcriptional events. Initially, the focus of studies was predominantly on the regulative kinases and their phosphorylation activity. More recently, ubiquitin has emerged as a central regulator of all molecular steps of the autophagy flux, from the nucleation of the double membrane to the shutdown of the entire process after resolution of the stress situation. Moreover, phosphorylation and ubiquitination processes seem to cross-talk and influence each other to lead the entire degradative flux (2). In general, ubiquitination is the most relevant PTM employed by the ubiquitin-proteasome (UPS) and the autophagy-lysosome (AL) systems to label their substrates. In the cytosol, UPS and AL pathways act simultaneously, share components of their molecular machineries, and constantly influence each other's activity (3–5). Multiple reports indicate the autophagy receptor p62/sequestosome-1 as the principal molecule that regulates the cross-talk between the two systems (3, 6, 7); however, the strongest commonality between the two degradative systems is the small globular protein ubiquitin (Ub).
Ubiquitin is covalently conjugated to lysine (Lys) residues of a substrate protein and mediates its degradation or modulates its biological function acting as a signaling molecule, respectively. Ubiquitin conjugation is a multistep reaction that involves subsequent action of three types of enzymes: E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; and E3, ubiquitin ligases. The whole process begins with the ATP-dependent activation of the C-terminal glycine residue of the ubiquitin molecule and is terminated by the conjugation of ubiquitin to the substrate protein (8, 9). A single ubiquitin (mono-ubiquitination) or several Ub molecules (multiple mono-ubiquitinations) can be ligated to a Lys residue of the target protein. Moreover, by linking several ubiquitin moieties through one of their seven lysine residues or the N-terminal methionine residue (M1) different chains are formed (10, 11). Finally, substrate-conjugated Ub molecules can also be modified by other PTMs, such as SUMOylation, phosphorylation, and acetylation, increasing the level of complexity of the ubiquitin code (12, 13). Of note, ubiquitination is a reversible process. The linkage between ubiquitin molecules themselves as well as between ubiquitin and their substrates can be hydrolyzed through the action of deubiquitinating enzymes (DUBs) (14). Taken together, ubiquitination displays a broad variety. Mono- or multi-Ub and the exact Ub chain composition determine the fate of the tagged substrates as well as the type of signaling pathway that is involved in vivo (10).
Ubiquitin-like systems in autophagy
The autophagy core machinery harbors two evolutionarily conserved ubiquitin-like conjugation systems, which are recruited to the autophagosomal membranes during their formation, maturation, and expansion (15). The first one involves ATG7 and ATG10 and is responsible for the formation of the ATG5–ATG12–ATG16L complex; the second one includes ATG3, ATG4, and ATG7 and catalyzes the activation and membrane conjugation of the autophagy ubiquitin-like modifiers (ATG8s). The large family of mammalian ATG8 proteins (mATG8s), which can be subdivided into MAP1LC3s (LC3A, LC3B, and LC3C) and GABARAPs (GABARAP, GABARAP-L1, and GABARAP-L2), are known as ubiquitin-like modifiers for their homology with the Ub molecule and the ubiquitin-like enzymatic cascade that mediates their covalent conjugation to the lipid phosphatidylethanolamine (PE), known as “lipidation.” The conjugation process starts with the protease ATG4, which cleaves the ATG8 proteins and exposes a C-terminal glycine for the subsequent conjugation reaction. Afterward, the cascade proceeds with ATG7, the E1 enzyme; ATG8s are then taken over by the E2 enzyme ATG3; and the ATG5–ATG12–ATG16L complex mediates ATG8 linkage to PE functioning as an E3 ligase complex (Fig. 1A). Similar to ubiquitin, the ATG8 conjugation process to PE is reversible. From yeast studies, later confirmed in mammals, Atg4 also functions as the de-conjugating enzyme that releases Atg8 from the membranes (16–18). Interestingly, active mATG8s have not been reported yet to bind other entities than lipid membranes even if their exposed C-terminal glycine, after ATG4 cleavage, makes them suitable for linkage to a lysine on a target protein similarly to the Ub-conjugation mechanism. Because of the close relation between the UPS and AL systems and the similarity between the Ub molecule and the mATG8s, it is not surprising that ubiquitination influences the course of autophagy. However, it is still not fully elucidated how the Ub code regulates autophagy in terms of altering protein signaling and targeting regulatory proteins to the proteasome, respectively. Non-degradative Ub signaling stimulates autophagy induction, whereas degradative Ub signaling is needed to restore the basal autophagy flux once the stress situation is resolved (19). The complexity of the Ub signaling in autophagy is reflected by a fine cross-talk between E3 ligases, which add Lys-63 or Lys-48 chains to the components of the regulatory complexes and DUBs that counteract their actions (Fig. 1) (7, 20).
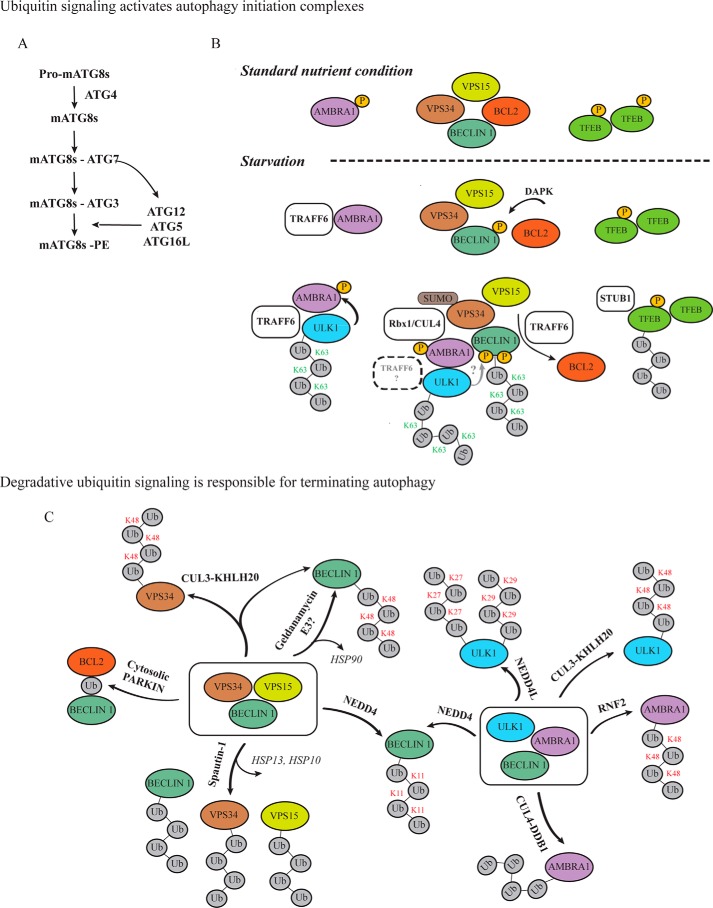
Figure 1.
Involvement of ubiquitin signaling in autophagy initiation and termination. Upon a stress stimulus, autophagy machinery is immediately activated. A, two conserved ubiquitin-like systems, involving ATG4–ATG3–ATG7 and ATG7-ATG5–ATG12–ATG16L, are responsible for the mATG8s processing. B, ULK1 and PI3K complexes are activated via Lys-63 polyubiquitin chains labeling ULK1 and BECLIN 1 proteins. Proteasome elimination of cytosolic phospho-TFEB promotes lysosome biogenesis and autophagy gene transcription. C, once the stress situation is resolved, autophagy is shut down to avoid excessive cellular catabolism. Degradative-ubiquitin signals determine proteasome elimination of the autophagy regulative proteins.
Autophagy induction via Lys-63 ubiquitination
In mammalian cells, the ULK1 and PI3K–III kinase complexes are the two major protein structures responsible for autophagy initiation and autophagosome formation. Both are regulated by phosphorylation, mainly due to mTORC1 and AMPK kinases (21–23), and ubiquitination, which involves a network of cross-talks and loops among different E3 ligases and DUBs (7). For example, TRAF6 E3 ligase positively influences both complexes by adding Lys-63 poly-Ub chains. Recent data showed that TRAF6 assembles Ub chains on ULK1 promoting its stabilization, self-association, and autophagy functions in HeLa cells. Their interaction is not direct but is mediated by AMBRA1, which acts as a bridging protein. In standard growing conditions, mTORC1 negatively regulates AMBRA1 function via its direct phosphorylation. Upon starvation, mTORC1 is inactive resulting in de-phosphorylated AMBRA1, which can bind TRAF6 and bring the E3 ligase in proximity to ULK1. Lys-63 poly-Ub chains activate ULK1, which consequently phosphorylates AMBRA1, initiating a positive regulatory loop and promoting AMBRA1 autophagy functions. Of note, ULK1 and mTORC1 phosphorylate AMBRA1 on different sites (24, 25). Besides, BECLIN 1, which forms the catalytic core of the PI3K–III complex together with VSP34 and VPS15, is also a substrate of TRAF6 in macrophages. The binding of lipopolysaccharides to TLR4 triggers a signaling cascade, which disrupts the binding between BECLIN 1 and BCL2. As a consequence, TRAF6 catalyzes the formation of Lys-63 poly-Ub chains on Lys-117 in the BH3 domain of BECLIN 1. The Ub chain generates an impediment for BCL2 binding to BECLIN 1; therefore, it promotes BECLIN 1-mediated autophagy induction. Of note, the DUB A20 hydrolyzes the Lys-63 poly-Ub chain of BECLIN 1 restoring the binding with BCL2 (26–29). Interestingly, also DAPK phosphorylation of BECLIN 1 in its BH3 domain abrogates the association with BCL2 (30). Hence, these two PTMs accomplish the same biological function, and it could be possible that BECLIN 1 phosphorylation also entails its ubiquitination, meaning that the TRAF6 E3 ligase senses the phosphorylation as a recruitment priming factor (31). Moreover, BECLIN 1 PTMs represent a cross-talking point between the ULK1 and PI3K–III complexes. Upon nutrient deprivation in mammalian cells and murine animal models, AMBRA1 directly interacts with BECLIN 1 priming its ubiquitination on Lys-437. This Lys-63-polyubiquitination is mediated by the E3 Rbx1/Cul4-ligase, which forms a complex with AMBRA1 to target BECLIN 1 under starvation. When standard conditions are restored, this reaction is reversed by the Wiskott-Aldrich syndrome protein and the SCAR homologue WASH, which is an endosomal protein belonging to the WASP protein family (32). Moreover, ULK1 can phosphorylate BECLIN 1 increasing the PI3K–III complex activity (33). The reported cross-talk between the two complexes raises the question whether AMBRA1 could also be involved in TRAF6-mediated BECLIN 1 Lys-63 ubiquitination in macrophages as a bridging protein, and whether this ubiquitination is trigged by ULK1 phosphorylation of both BECLIN 1 and AMBRA1. Moreover, although AMBRA1 and BECLIN 1 are involved in ubiquitin-dependent regulatory processes, VPS34 and VPS15 have so far been considered only for their phosphorylation capability. However, KAP1 SUMO-ligase mediates SUMO1 conjugation to VPS34. As a result, VPS34 interacts stronger with the PI3K–III complex that promotes the formation of the phagophore (34).
The role of the autophagosome is to engulf the cytosolic cargo, whereas degradation of the material enclosed occurs after fusion with a lysosome. For this reason, lysosome function and biogenesis are as important as those of autophagosomes. TFEB is the master transcriptional regulator of genes involved in autophagosome and lysosome biogenesis (35). So far, TFEB regulation was known to occur mainly by its phosphorylation via mTORC1. Phosphorylated TFEB is inactive and sequestered in the cytosol. Upon autophagy induction and mTORC1 inactivation, TFEB is dephosphorylated and can translocate, as dimer, into the nucleus where it acts as a transcription factor (35, 36). Recently, it has been shown that phosphorylated TFEB is further modified by STUB1 E3 ubiquitin ligase by analyzing _Stub1_−/− mice and further confirming in HeLa cells. STUB1 binding to TFEB is mediated by the chaperone protein HSP70 and, even if the specific Lys residue and the type of ubiquitin chain are not yet known, the degradation of Ub-modified TFEB increases the cytosolic amount of de-phosphorylated protein, which can form homodimers and translocate into the nucleus. Of note, heterodimers of phosphorylated and dephosphorylated TFEB do not shuttle to the nucleus and function as transcription factors (37). Interestingly, TFEB is the first evidence of a Ub degradative signal on an autophagy master regulator that mediates activation of autophagy (Fig. 1B).
Autophagy termination is regulated via degradative ubiquitination
Once the stress situation is resolved, for example when the standard nutrient conditions are restored, or when the degradative process is markedly extending in time, the autophagy machinery needs to be shut down to avoid the excessive degradation of cellular components that will lead to apoptosis. Once more, the cross-talk between phosphorylation and ubiquitination signals on the ULK1 and PI3K–III complexes primes the process (19). The degradative ubiquitin signal, which involves the Lys-48 Ub chain's labeling of the target protein followed by its subsequent proteasomal degradation, reduces the protein level of the autophagy regulators restoring a physiological basal autophagy flux. The HECT-type E3 ligase NEDD4 mediates Lys-11-linked ubiquitination of BECLIN 1, which is subsequently degraded. The reduced level of BECLIN 1 destabilizes the VPS34–BECLIN 1 complex affecting its biological function and consequently blocking autophagy (38). NEDD4L, an E3 ligase highly homologous to NEDD4, negatively modulates ULK1 protein level during autophagy progression boosting its degradation. NEDD4L directs ULK1 to the proteasome via Lys-27- and Lys-29-linked polyubiquitination (39). Moreover, in the specific case of selenite-induced mitophagy in HeLa cells, ULK1 becomes a substrate for the mitochondrial outer membrane (MOM) E3 ligase MUL1 (40). Recently it has been described that ULK1, BECLIN 1, and VPS34 are also substrates of CULLIN3–KHLH20 ligase, which mediates their degradation via Lys-48 poly-Ub chains (41).
Interestingly, BECLIN 1 and AMBRA1 play a pivotal role in both autophagy induction and termination. Therefore, their protein level is finely controlled by several E3 ligases and DUBs. Under prolonged stress conditions such as long starvation periods in HeLa cells, the CULLIN4–DDB1 complex shuts down autophagy promoting AMBRA1 degradation via the proteasome (42). Moreover, RNF2 E3 ligase can modify Lys-45 of AMBRA1 with a Lys-48–linked Ub chain, which targets the substrate directly to the proteasome (43). However, understanding BECLIN 1 regulation is probably the most challenging endeavor. Besides its direct interaction with NEDD4, BECLIN 1 function is indirectly regulated also by the E3 ligase PARKIN. In HeLa cells, the interaction between BECLIN 1 and BCL2 is strengthened by PARKIN-mediated mono-ubiquitination of BCL2, with the consequent result of impairing BECLIN 1 function in autophagy (44). The exact mechanism of how mono-ubiquitination contributes to the stability of the complex has remained unclear so far. However, the dual function of PARKIN reported, depending on its cellular localization, bears interesting starting points for further, deeper studies. On mitochondria, it promotes Lys-63 and Lys-48 poly-Ub chain reactions to induce mitophagy, although when PARKIN is cytosolic, it has an inhibitory effect on autophagy relayed through mono-ubiquitination of BCL2. Last, but not less important, is the DUB-mediated regulation of BECLIN 1 protein levels. Here, the DUBs USP13 and USP10 normally protect BECLIN 1 from ubiquitination. Upon their inhibition by cellular treatment with the chemical compound Spautin-1, a still unknown E3 ligase rapidly tags BECLIN 1 with degradative Ub chains and directs it to the proteasome (45). Importantly, a similar scenario is pictured by geldanamycin, which displaces the chaperone protein HSP90 from BECLIN 1 with the final result of Lys-48 polyubiquitin chain formation on BECLIN 1 (46). Also, in the case of geldanamycin, the E3 ligase involved is still not characterized (Fig. 1C).
It is worth speculating that the autophagy system employs E3 ligases in a cascade-like regulation of different steps in the induction, procedure, and termination of the flux. Moreover, the existence of such a thorny E3 ligase network is extremely challenging when characterizing its details, and obviously the cross-talk between proteasome and autophagy represents a future frontier for the biomedicine.
Role of ubiquitin in selective autophagy
Selective autophagy can be considered a surgical instrument used by the cell to eliminate protein aggregates and damaged organelles without affecting any other cytosolic components. The degradative process is driven and controlled by specialized molecules: the autophagy receptors, which physically link the autophagosomal membranes with the cargo for clearance. Selective autophagy can be classified as ubiquitin-dependent or -independent. In both cases, receptors harbor an LIR motif to bind the ATG8s, which are linked to the expanding autophagosomal membranes. In the case of ubiquitin-dependent autophagy, receptors directly bind the ubiquitin chains, present on the cargo surface, through their ubiquitin-binding domain. Of note, the two types of selective autophagy are not exclusive, frequently both kind of receptors are found present on the same cargo (47–49). Several types of selective autophagy have been observed and named accordingly to the substrate they target for degradation (47, 48). Technically, all ubiquitin-labeled cellular components are suitable for lysosomal degradation; however, autophagy can distinguish its specific cargos leaving the other cellular components unaffected, which are also labeled with poly-Ub chains, or directing them to the proteasome. One possible explanation is the presence of ubiquitin clusters, which increase the avidity for the autophagy receptors on the cargo surface. An additional level of regulation could be the quality of the Ub chains. Lys-63 chains seem to have a preferential affinity for the autophagy receptors (50, 51), whereas proteins decorated with Lys-48-, Lys-27-, Lys-11-linked ubiquitin chains undergo proteasomal degradation (52). Nevertheless, in autophagy-deficient mice displaying compromised autophagy flux, all ubiquitin chain types are enriched (53). Therefore, the relative contribution of the different chains is still unclear and a matter of investigation.
Aggrephagy
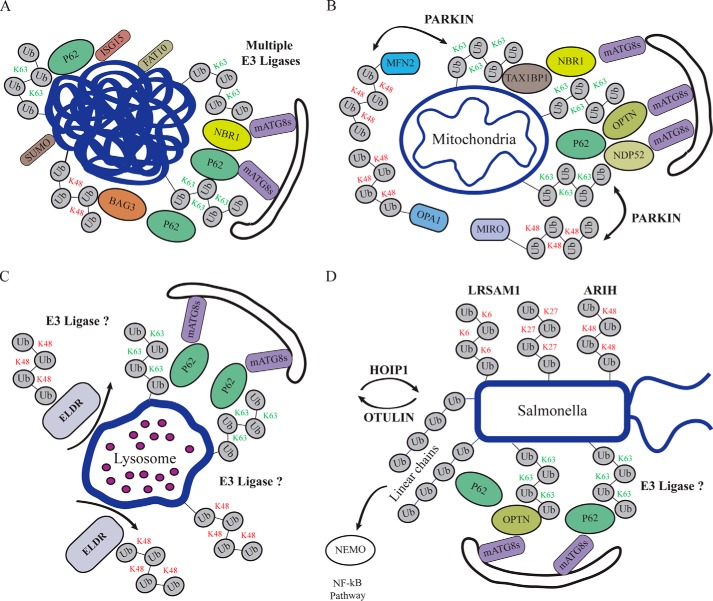
So far, almost 20 different types of selective autophagy have been described (47, 48), and nearly half of them are ubiquitin-driven. In this context, the first one to be described was aggrephagy, probably because it represents one of the best examples of cross-talk between the UPS- and AL-degradative systems (Fig. 2A). Therefore, it is not surprising that many components of the ubiquitin system are shared between the UPS and aggrephagy. Generally spoken, the proteasome mainly degrades proteins linked to Lys-48 poly-Ub chains, whereas the aggrephagy machinery is stimulated by Lys-63 poly-Ub chains, which recruit the autophagy receptors p62 and NBR1 (54, 55). As part of the reversible signaling networks, de-ubiquitination is involved in aggrephagy, too. The USP9X protease removes mono-ubiquitin from the toxin α-synuclein, which is then preferentially disposed in the lysosome rather than by the proteasome (56). The proteasome subunit RPN11, a DUB, generates unanchored free Ub chains, which bind and activate HDAC6 that favors protein aggregate clearance supporting the autophagosome and cargo movement along microtubules (57, 58). Autophagy receptors and Lys-63 poly-Ub chains are essential for aggrephagy, and their activity is maintained also by other Ub-related molecules. The ubiquitin-like proteins SUMO-1 and FAT10 bind and contribute to the formation of protein aggregates (59, 60), whereas ISG15 binds to HDAC6 and p62 promoting lysosomal clearance of aggregates (61). Finally, BAG family members BAG1 and BAG3 compete for the HSP-bound polyubiquitinated substrates. Although BAG1 delivers chaperon-recognized misfolded proteins to the proteasome, BAG3 directly interacts with p62 and, at the same time, binds to Lys-48 poly-Ub chains pulling through the lysosome proteins initially addressed to the proteasome (62).
Figure 2.
Autophagy receptors bind to Lys-63 polyubiquitin chains and recruit phagophores to the cargo. A, in aggrephagy, multiple E3 ligases are involved in the protein aggregate ubiquitination. p62 and NBR1 receptors are recruited via Lys-63 chains. SUMO, FAT10, and ISG15 are other ubiquitin-like proteins, which label the protein aggregate surface. ISG15 interacts with p62, whereas BAG3 binds simultaneously Lys-48 Ub chains and p62 redirecting Lys-48 substrates to lysosomes. B, during mitophagy, PARKIN translocates to the mitochondria, where it mediates the ubiquitination of a plethora of mitochondrial proteins. OMM proteins, which are involved in the mitochondrial fusion process and anchorage to the cytoskeleton, are degraded via proteasome. Isolated single organelles are then recognized by autophagy receptors via Lys-63 polyubiquitin chains. C, damaged lysosomes are eliminated via autophagy, too. To recruit LC3 on the lysosome surface, Lys-48 polyubiquitin chains must be removed by the ELDR complex. Lys-63 chains are unaffected by ELDR, so they promote autophagy receptor recruitment. The E3 ligases involved in lysosome ubiquitination are so far unknown. D, upon infection, the Salmonella surface is tagged by several E3 ligases as a defense mechanism. LRSAM1 and ARIH add, respectively, Lys-6–Lys-27 and Lys-48 poly-Ub chains. Linear Ub chains are formed and regulated by the HOIP1 and OTULIN (LUBAC complex), whereas the E3 ligase responsible for Lys-63 chains is unknown. The autophagy machinery is recruited by Lys-63 and linear chains.
Mitophagy
Mitochondrial clearance is another relevant example of collaboration between the autophagy and proteasome ubiquitin signals (Fig. 2B). Exhausted mitochondria are eliminated via both Ub-dependent and Ub-independent autophagy. The E3 ligase PARKIN mediates the ubiquitination of mitochondrial proteins in order to promote mitophagy. Conversely, FUNDC1, BNIP3, and NIX are specific autophagy receptors, which lead to Ub-independent mitophagy (63, 64). Independently of the type of mitophagy, mitochondria must go through a fission process to be enclosed in an autophagosome. The dysfunctional organelle is quarantined from the whole mitochondrial network, and its anchorage from the microtubules is relieved. These biological processes are all finely regulated by ubiquitin at different levels. Mitochondrial outer membrane (MOM) proteins MFN1/2, FIS1, DRP1, and OPA1, involved in mitochondrial fission and fusion, are ubiquitinated by E3 ligases and then removed via the proteasome, boosting a re-arrangement of the mitochondrial network morphology (65–68). Degradation of MOM fusion proteins (MFN2 and OPA1) unbalances the mitochondrial dynamic to fission, which is necessary to isolate the single damaged organelle. At the same time, MIRO clearance ensures the release of the mitochondria from their motor complex and makes them suitable for engulfment into autophagosomes (69). In contrast, during amino acid starvation healthy mitochondria escape degradation promoting their elongation to preserve ATP production and avoid mitophagy (70, 71). In this context, Lys-48 polyubiquitination of DRP1, MFN1, and MiD49 eliminates the fission protein regulators and therefore supports mitochondrial fusion and elongation (66, 72). Isolated and dysfunctional mitochondria are then ready to be engulfed by double membranes and eliminated. The core mitophagy process is led by the kinase PINK1, which phosphorylates the E3 ligase PARKIN and the poly-Ub chains on the mitochondrial surface, generating a positive feedback loop. Phospho-Ub chains display a stronger affinity for PARKIN, which is consequently more efficiently recruited from the cytosol to the mitochondria (73–77). Moreover, PARKIN phosphorylation is an activation signal for this E3 ligase itself, which is responsible for the ubiquitination of a plethora of MOM proteins. The massive PARKIN ubiquitination favors the binding of the autophagy receptors (p62, OPTN, NPD2, TAX1BP1, and NBR1), which bridge the autophagy membranes. Furthermore, TBK1 kinase mediates the phosphorylation of all five mitophagy receptors, generating a positive signaling loop (78, 79). On the surface of mitochondria, PARKIN also interacts with AMBRA1. The nature and the meaning of this interaction is not completely clear as yet (80, 81). Potentially, AMBRA1 functions as an adaptor to recruit other E3 ligases like TRAF6 and CUL4–DDB1 to mitochondria. As part of the PARKIN-mediated mitophagy, the USP30 DUB controls PARKIN activity by removing poly-Ub chains from the mitochondrial surface (82).
Lysophagy
Damaged lysosomes are themselves a target for selective autophagy, and different types of poly-Ub chains have been detected on their surface (83–85). Yet, only recently has the significance of the poly-Ub chains that label lysosomes been partially elucidated (Fig. 2C). Damaged lysosomes are initially tagged with Lys-63 poly-Ub chains, which recruit p62, and in the second instance, only a subpopulation of dysfunctional organelles is decorated by Lys-48 poly-Ub chains. Lys-48 ubiquitination is the signal for p97 recruitment on the damaged lysosomes, where it cooperates with the co-factors UBXD1, PLAA, and YOD1 (endo-lysosomal damage response (ELDR) complex) to specifically remove the Lys-48 poly-Ub chains. Both ATPase activity of p97 and the de-ubiquitinase function of YOD1 are essential to remove Lys-48 polyubiquitin. Additionally, the whole integrity of the ELDR complex is required to extract Lys-48 poly-Ub chains. Interestingly, although ELDR does not affect p62 recruitment, because it leaves the Lys-63 chains unaffected, removal of Lys-48 poly-Ub chains favors the arrival of mATG8s on damaged lysosome surfaces (86). This situation is reminiscent of mitochondria during mitophagy, so it will be extremely interesting to understand which are the Lys-48 target proteins and why they need to be eliminated to recruit LC3. The clearance of Lys-48 chains could be functional to unbalance the poly-Ub chain population, in favor of the Lys-63 ones, or to degrade specific substrates that may inhibit or mask the still unknown autophagy receptors on the lysosomal surface. Furthermore, the identity of the E3 ligases that mediate lysosomal Lys-63 and Lys-48 ubiquitination still remains an open question.
Xenophagy
Another interesting example of Ub-dependent selective autophagy is xenophagy of Salmonella typhimurium or other intracellular pathogens such as Mycobacterium (Fig. 2D) (87–89). Bacterial infection triggers a dynamic change in the global ubiquitinome in both the host cell and bacteria itself (90). In the host cell, the Salmonella surface is promptly tagged with multiple types of Ub chains, which activate the cellular defense mechanism to constrain pathogen growth (87, 89). The E3 ligases responsible for the pathogen ubiquitination are LRSAM1 for Lys-6 and Lys-27 chains, ARIH for Lys-48 chains, and HOIP1 for the linear ubiquitination (91–93). Curiously, the E3 ligase responsible for the Lys-63 chains' decoration of bacteria is still unknown. Likewise, the E3 ligase RNF166 also localizes to Salmonella, but it mediates Lys-29 and Lys-33 polyubiquitination of p62 and not of pathogen proteins (94). Of note, there is a significant increase in ubiquitination sites of linear poly-Ub chains upon infection (M1-linked), assembled by the LUBAC complex, which induces two different types of signaling response: selective autophagy induction and NF-κB–signaling activation. Moreover, different poly-Ub chains are clustered in small, distinct foci on the bacterial surface, where even some hybrid poly-Ub chains are detected (95). M1 and Lys-63 polyubiquitin chains are recognized by p62, NDP52, and OPTN, which mediate the recruitment of the autophagy machinery around the pathogen. In particular, OPTN is fundamental to restrict Salmonella infection via xenophagy. OPTN affinity for LC3 is then further increased by TBK1, which mediates OPTN phosphorylation in its LIR domain (89, 90). Additionally, linear chains function as a platform for NF-κB signaling activation. M1-Ub chains on Salmonella trigger the local recruitment of NEMO, activation of IKKα/IKKβ, and consequently NF-κB, which in turn promotes the secretion of pro-inflammatory cytokines to restrict bacterial proliferation (92, 95). Interestingly, the only DUB that has been identified so far to counteract Salmonella infection is OTULIN, which is a key component of the LUBAC complex (95).
Overall, ubiquitin signaling is involved in several types of selective autophagy and regulates the process at different stages. The combination of Lys-48, Lys-63, M1, and likely other poly-Ub chain signaling potentially mediates cargo morphology changing and its cytosolic isolation, the recognition by the autophagy receptors, and eventually the activation of other signaling pathways. A further level of complexity is then achieved with the presence of additional PTMs on poly-Ub chains, like phosphorylation (12).
Concluding remarks
The autophagy field has been extensively investigated during the course of the past years. However, how the whole pathway is activated in response to stress stimuli still represents a challenge. The molecular signaling looks like an intricate combination of PTMs, which activate and inhibit the regulatory complexes generating a domino effect. Unraveling the cross-relations among the different PTMs is of great interest. The combined impact of phosphorylation, ubiquitination, and acetylation allows regulatory proteins to rapidly activate and assemble as well as to self-inhibit their action (19, 96). Further work is still required to elucidate the ubiquitination reaction and the identity of the E3 ligases responsible for activating or inhibiting the autophagic processes. The list of E3 ligases involved in the regulative steps is constantly growing. The enzymes can act together, during the same phase of the process, or work sequentially at different stages, and they can also counteract each other. Most important, the nature of the generated poly-Ub chains on the target proteins determines their fate. Lys-63 chains are used to positively regulate the system, whereas Lys-48 poly-Ub chains have an inhibitory effect. Interestingly, linear ubiquitin chains still do not have a precise identity in the signaling cascade. Until now, their role has been described only during xenophagy; nevertheless, they may be involved in other types of selective autophagy as well as in the regulative signaling. Of great relevance is also the Ub signal that modifies the dynamics of the targeted organelles to prepare them for the selective autophagy. Only mitochondria have been studied so far; however, the endoplasmic reticulum (ER) morphology can also be affected during bacterial infection by a novel type of phosphoribosyl-dependent serine ubiquitination (97, 98). The two ER-phagy receptors FAM134B and RTN3 harbor the intrinsic property to shape ER membranes, so their relation to ubiquitin could further explain their molecular mechanism of action in ER-phagy (99, 100). Moreover, the elucidation of the dynamic regulatory network is important to understand the cross-talks among the different components in the occurrence of the plethora of autophagy-linked diseases.
Acknowledgments
We thank Lina Herhaus and Heide Genau for critical reading of the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 1177, European Research Council (ERC) Grant Ub-BAC, Cluster of Excellence “Macromolecular Complexes” Grant (EXC 115), LOEWE Grant Ub-Net, LOEWE Centre for Gene and Cell Therapy Frankfurt (CGT) (to I. D.), and COFUND, Goethe International Postdoc Programme (GO-IN) Grant 291776 (to P. G.). This is the third article in the Thematic Minireview series “Autophagy.” The authors declare that they have no conflicts of interest with the contents of this article.
2
The abbreviations used are:
PTM
post-translational modification
UPS
ubiquitin-proteasome
AL
autophagy-lysosome
Ub
ubiquitin
DUB
deubiquitinating enzyme
ELDR
endo-lysosomal damage response
ER
endoplasmic reticulum
PE
phosphatidylethanolamine
MOM
mitochondrial outer membrane.
References
- 1.Botti-Millet J., Nascimbeni A. C., Dupont N., Morel E., and Codogno P. (2016) Fine-tuning autophagy: from transcriptional to post-translational regulation. Am. J. Physiol. Cell Physiol. 311, C351–C362 10.1152/ajpcell.00129.2016 [DOI] [PubMed] [Google Scholar]
- 2.McEwan D. G., and Dikic I. (2011) The three musketeers of autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 21, 195–201 10.1016/j.tcb.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dikic I. (2017) Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 10.1146/annurev-biochem-061516-044908 [DOI] [PubMed] [Google Scholar]
- 4.Korolchuk V. I., Mansilla A., Menzies F. M., and Rubinsztein D. C. (2009) Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell 33, 517–527 10.1016/j.molcel.2009.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall R. S., Li F., Gemperline D. C., Book A. J., and Vierstra R. D. (2015) Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 58, 1053–1066 10.1016/j.molcel.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto G., Wada K., Okuno M., Kurosawa M., and Nukina N. (2011) Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289 10.1016/j.molcel.2011.07.039 [DOI] [PubMed] [Google Scholar]
- 7.Reidick C., El Magraoui F., Meyer H. E., Stenmark H., and Platta H. W. (2014) Regulation of the tumor-suppressor function of the class III phosphatidylinositol 3-kinase complex by ubiquitin and SUMO. Cancers 7, 1–29 10.3390/cancers7010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458, 422–429 10.1038/nature07958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Husnjak K., and Dikic I. (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 10.1146/annurev-biochem-051810-094654 [DOI] [PubMed] [Google Scholar]
- 11.Kirisako T., Kamei K., Murata S., Kato M., Fukumoto H., Kanie M., Sano S., Tokunaga F., Tanaka K., and Iwai K. (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 10.1038/sj.emboj.7601360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herhaus L., and Dikic I. (2015) Expanding the ubiquitin code through post-translational modification. EMBO Rep. 16, 1071–1083 10.15252/embr.201540891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seeler J. S., and Dejean A. (2017) SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184–197 10.1038/nrc.2016.143 [DOI] [PubMed] [Google Scholar]
- 14.Clague M. J., Barsukov I., Coulson J. M., Liu H., Rigden D. J., and Urbé S. (2013) Deubiquitylases from genes to organism. Physiol. Rev. 93, 1289–1315 10.1152/physrev.00002.2013 [DOI] [PubMed] [Google Scholar]
- 15.Nakatogawa H., Ichimura Y., and Ohsumi Y. (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- 16.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., and Ohsumi Y. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 17.Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., and Ohsumi Y. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276 10.1083/jcb.151.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., and Yoshimori T. (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812 10.1242/jcs.01131 [DOI] [PubMed] [Google Scholar]
- 19.Antonioli M., Di Rienzo M., Piacentini M., and Fimia G. M. (2017) Emerging mechanisms in initiating and terminating autophagy. Trends Biochem. Sci. 42, 28–41 10.1016/j.tibs.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 20.Abrahamsen H., Stenmark H., and Platta H. W. (2012) Ubiquitination and phosphorylation of Beclin 1 and its binding partners: tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett. 586, 1584–1591 10.1016/j.febslet.2012.04.046 [DOI] [PubMed] [Google Scholar]
- 21.Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., Asara J. M., Fitzpatrick J., Dillin A., Viollet B., Kundu M., et al. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J., and Guan K. L. (2013) AMPK connects energy stress to PIK3C3/VPS34 regulation. Autophagy 9, 1110–1111 10.4161/auto.24877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan H. X., Russell R. C., and Guan K. L. (2013) Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 9, 1983–1995 10.4161/auto.26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nazio F., Strappazzon F., Antonioli M., Bielli P., Cianfanelli V., Bordi M., Gretzmeier C., Dengjel J., Piacentini M., Fimia G. M., and Cecconi F. (2013) mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15, 406–416 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- 25.Di Bartolomeo S., Corazzari M., Nazio F., Oliverio S., Lisi G., Antonioli M., Pagliarini V., Matteoni S., Fuoco C., Giunta L., D'Amelio M., Nardacci R., Romagnoli A., Piacentini M., Cecconi F., and Fimia G. M. (2010) The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 191, 155–168 10.1083/jcb.201002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashkenazi A., Bento C. F., Ricketts T., Vicinanza M., Siddiqi F., Pavel M., Squitieri F., Hardenberg M. C., Imarisio S., Menzies F. M., and Rubinsztein D. C. (2017) Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545, 108–111 10.1038/nature22078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanjuan M. A., Dillon C. P., Tait S. W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J. L., Withoff S., and Green D. R. (2007) Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- 28.Shi C. S., and Kehrl J. H. (2008) MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283, 33175–33182 10.1074/jbc.M804478200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi C. S., and Kehrl J. H. (2010) TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 3, ra42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zalckvar E., Berissi H., Mizrachy L., Idelchuk Y., Koren I., Eisenstein M., Sabanay H., Pinkas-Kramarski R., and Kimchi A. (2009) DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 10, 285–292 10.1038/embor.2008.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng H., Lopez G. Y., Kim C. K., Alvarez A., Duncan C. G., Nishikawa R., Nagane M., Su A. J., Auron P. E., Hedberg M. L., Wang L., Raizer J. J., Kessler J. A., Parsa A. T., Gao W. Q., et al. (2014) EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J. Clin. Invest. 124, 3741–3756 10.1172/JCI73093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia P., Wang S., Du Y., Zhao Z., Shi L., Sun L., Huang G., Ye B., Li C., Dai Z., Hou N., Cheng X., Sun Q., Li L., Yang X., and Fan Z. (2013) WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 32, 2685–2696 10.1038/emboj.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., and Guan K. L. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y., Fiskus W., Yong B., Atadja P., Takahashi Y., Pandita T. K., Wang H. G., and Bhalla K. N. (2013) Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. U.S.A. 110, 6841–6846 10.1073/pnas.1217692110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., and Ballabio A. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M. C., Facchinetti V., Sabatini D. M., and Ballabio A. (2012) A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sha Y., Rao L., Settembre C., Ballabio A., and Eissa N. T. (2017) STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 36, 2544–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platta H. W., Abrahamsen H., Thoresen S. B., and Stenmark H. (2012) Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 441, 399–406 10.1042/BJ20111424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nazio F., Carinci M., Valacca C., Bielli P., Strappazzon F., Antonioli M., Ciccosanti F., Rodolfo C., Campello S., Fimia G. M., Sette C., Bonaldo P., and Cecconi F. (2016) Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J. Cell Biol. 215, 841–856 10.1083/jcb.201605089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Qi W., Chen G., Feng D., Liu J., Ma B., Zhou C., Mu C., Zhang W., Chen Q., and Zhu Y. (2015) Mitochondrial outer-membrane E3 ligase MUL1 ubiquitinates ULK1 and regulates selenite-induced mitophagy. Autophagy 11, 1216–1229 10.1080/15548627.2015.1017180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C. C., Lin Y. C., Chen Y. H., Chen C. M., Pang L. Y., Chen H. A., Wu P. R., Lin M. Y., Jiang S. T., Tsai T. F., and Chen R. H. (2016) Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol. Cell 61, 84–97 10.1016/j.molcel.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 42.Antonioli M., Albiero F., Nazio F., Vescovo T., Perdomo A. B., Corazzari M., Marsella C., Piselli P., Gretzmeier C., Dengjel J., Cecconi F., Piacentini M., and Fimia G. M. (2014) AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev. Cell 31, 734–746 10.1016/j.devcel.2014.11.013 [DOI] [PubMed] [Google Scholar]
- 43.Xia P., Wang S., Huang G., Du Y., Zhu P., Li M., and Fan Z. (2014) RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 24, 943–958 10.1038/cr.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen D., Gao F., Li B., Wang H., Xu Y., Zhu C., and Wang G. (2010) Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J. Biol. Chem. 285, 38214–38223 10.1074/jbc.M110.101469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J., Xia H., Kim M., Xu L., Li Y., Zhang L., Cai Y., Norberg H. V., Zhang T., Furuya T., Jin M., Zhu Z., Wang H., Yu J., Li Y., et al. (2011) Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234 10.1016/j.cell.2011.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu C., Liu J., Hsu L. C., Luo Y., Xiang R., and Chuang T. H. (2011) Functional interaction of heat shock protein 90 and Beclin 1 modulates Toll-like receptor-mediated autophagy. FASEB J. 25, 2700–2710 10.1096/fj.10-167676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khaminets A., Behl C., and Dikic I. (2016) Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6–16 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- 48.Rogov V., Dötsch V., Johansen T., and Kirkin V. (2014) Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167–178 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 49.Stolz A., and Dikic I. (2014) PINK1-PARKIN interplay: down to ubiquitin phosphorylation. Mol. Cell 56, 341–342 10.1016/j.molcel.2014.10.022 [DOI] [PubMed] [Google Scholar]
- 50.Linares J. F., Duran A., Yajima T., Pasparakis M., Moscat J., and Diaz-Meco M. T. (2013) K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol. Cell 51, 283–296 10.1016/j.molcel.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olzmann J. A., Li L., Chudaev M. V., Chen J., Perez F. A., Palmiter R. D., and Chin L. S. (2007) Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178, 1025–1038 10.1083/jcb.200611128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins G. A., and Goldberg A. L. (2017) The logic of the 26S proteasome. Cell 169, 792–806 10.1016/j.cell.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riley B. E., Kaiser S. E., Shaler T. A., Ng A. C., Hara T., Hipp M. S., Lage K., Xavier R. J., Ryu K. Y., Taguchi K., Yamamoto M., Tanaka K., Mizushima N., Komatsu M., and Kopito R. R. (2010) Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 191, 537–552 10.1083/jcb.201005012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirkin V., Lamark T., Sou Y. S., Bjorkoy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Overvatn A., Ishii T., Elazar Z., et al. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- 55.Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L., Kleinert R., Prinz M., Aguzzi A., and Denk H. (2002) p62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160, 255–263 10.1016/S0002-9440(10)64369-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rott R., Szargel R., Haskin J., Bandopadhyay R., Lees A. J., Shani V., and Engelender S. (2011) α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc. Natl. Acad. Sci. U.S.A. 108, 18666–18671 10.1073/pnas.1105725108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., and Yao T. P. (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- 58.Lee J. Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y. S., Pandey U. B., Kaushik S., Tresse E., Lu J., Taylor J. P., Cuervo A. M., and Yao T. P. (2010) HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 29, 969–980 10.1038/emboj.2009.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho S. J., Yun S. M., Jo C., Lee D. H., Choi K. J., Song J. C., Park S. I., Kim Y. J., and Koh Y. H. (2015) SUMO1 promotes Abeta production via the modulation of autophagy. Autophagy 11, 100–112 10.4161/15548627.2014.984283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalveram B., Schmidtke G., and Groettrup M. (2008) The ubiquitin-like modifier FAT10 interacts with HDAC6 and localizes to aggresomes under proteasome inhibition. J. Cell Sci. 121, 4079–4088 10.1242/jcs.035006 [DOI] [PubMed] [Google Scholar]
- 61.Nakashima H., Nguyen T., Goins W. F., and Chiocca E. A. (2015) Interferon-stimulated gene 15 (ISG15) and ISG15-linked proteins can associate with members of the selective autophagic process, histone deacetylase 6 (HDAC6) and SQSTM1/p62. J. Biol. Chem. 290, 1485–1495 10.1074/jbc.M114.593871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gamerdinger M., Kaya A. M., Wolfrum U., Clement A. M., and Behl C. (2011) BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 12, 149–156 10.1038/embor.2010.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., Huang L., Xue P., Li B., Wang X., Jin H., et al. (2012) Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185 10.1038/ncb2422 [DOI] [PubMed] [Google Scholar]
- 64.Novak I., Kirkin V., McEwan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Lohr F., Popovic D., Occhipinti A., Reichert A. S., Terzic J., Dotsch V., Ney P. A., and Dikic I. (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., and Joazeiro C. A. (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3, e1487 10.1371/journal.pone.0001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakamura N., Kimura Y., Tokuda M., Honda S., and Hirose S. (2006) MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 7, 1019–1022 10.1038/sj.embor.7400790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narendra D., Tanaka A., Suen D. F., and Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yonashiro R., Ishido S., Kyo S., Fukuda T., Goto E., Matsuki Y., Ohmura-Hoshino M., Sada K., Hotta H., Yamamura H., Inatome R., and Yanagi S. (2006) A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 25, 3618–3626 10.1038/sj.emboj.7601249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y. L., Selkoe D., Rice S., Steen J., LaVoie M. J., and Schwarz T. L. (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 10.1016/j.cell.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gomes L. C., Di Benedetto G., and Scorrano L. (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13, 589–598 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rambold A. S., Kostelecky B., Elia N., and Lippincott-Schwartz J. (2011) Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U.S.A. 108, 10190–10195 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu J., Meng Y., Zhang Z., Yan Q., Jiang X., Lv Z., and Hu L. (2017) MARCH5 RNA promotes autophagy, migration, and invasion of ovarian cancer cells. Autophagy 13, 333–344 10.1080/15548627.2016.1256520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarraf S. A., Raman M., Guarani-Pereira V., Sowa M. E., Huttlin E. L., Gygi S. P., and Harper J. W. (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kane L. A., Lazarou M., Fogel A. I., Li Y., Yamano K., Sarraf S. A., Banerjee S., and Youle R. J. (2014) PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ordureau A., Heo J. M., Duda D. M., Paulo J. A., Olszewski J. L., Yanishevski D., Rinehart J., Schulman B. A., and Harper J. W. (2015) Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl. Acad. Sci. U.S.A. 112, 6637–6642 10.1073/pnas.1506593112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., Endo T., Fon E. A., Trempe J. F., Saeki Y., Tanaka K., and Matsuda N. (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- 77.Ordureau A., Sarraf S. A., Duda D. M., Heo J. M., Jedrychowski M. P., Sviderskiy V. O., Olszewski J.L., Koerber J. T., Xie T., Beausoleil S. A., Wells J. A., Gygi S. P., Schulman B., and Harper J. W. (2014) Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell. 56, 360–375 10.1016/j.molcel.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heo J. M., Ordureau A., Paulo J. A., Rinehart J., and Harper J. W. (2015) The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 60, 7–20 10.1016/j.molcel.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richter B., Sliter D. A., Herhaus L., Stolz A., Wang C., Beli P., Zaffagnini G., Wild P., Martens S., Wagner S. A., Youle R. J., and Dikic I. (2016) Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. U.S.A. 113, 4039–4044 10.1073/pnas.1523926113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strappazzon F., Nazio F., Corrado M., Cianfanelli V., Romagnoli A., Fimia G. M., Campello S., Nardacci R., Piacentini M., Campanella M., and Cecconi F. (2015) AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 22, 517 10.1038/cdd.2014.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van Humbeeck C., Cornelissen T., Hofkens H., Mandemakers W., Gevaert K., De Strooper B., and Vandenberghe W. (2011) Parkin interacts with Ambra1 to induce mitophagy. J. Neurosci. 31, 10249–10261 10.1523/JNEUROSCI.1917-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bingol B., Tea J. S., Phu L., Reichelt M., Bakalarski C. E., Song Q., Foreman O., Kirkpatrick D. S., and Sheng M. (2014) The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375 10.1038/nature13418 [DOI] [PubMed] [Google Scholar]
- 83.Hung Y. H., Chen L. M., Yang J. Y., and Yang W. Y. (2013) Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat. Commun. 4, 2111 [DOI] [PubMed] [Google Scholar]
- 84.Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., and Yoshimori T. (2013) Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fujita N., Morita E., Itoh T., Tanaka A., Nakaoka M., Osada Y., Umemoto T., Saitoh T., Nakatogawa H., Kobayashi S., Haraguchi T., Guan J. L., Iwai K., Tokunaga F., Saito K., et al. (2013) Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 203, 115–128 10.1083/jcb.201304188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Papadopoulos C., Kirchner P., Bug M., Grum D., Koerver L., Schulze N., Poehler R., Dressler A., Fengler S., Arhzaouy K., Lux V., Ehrmann M., Weihl C. C., and Meyer H. (2017) VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 36, 135–150 10.15252/embj.201695148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Birmingham C. L., Smith A. C., Bakowski M. A., Yoshimori T., and Brumell J. H. (2006) Autophagy controls Salmonella infection in response to damage to the _Salmonella_-containing vacuole. J. Biol. Chem. 281, 11374–11383 10.1074/jbc.M509157200 [DOI] [PubMed] [Google Scholar]
- 88.Noda T., Kageyama S., Fujita N., and Yoshimori T. (2012) Three-axis model for Atg recruitment in autophagy against Salmonella. Int. J. Cell Biol. 2012, 389562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wild P., Farhan H., McEwan D. G., Wagner S., Rogov V. V., Brady N. R., Richter B., Korac J., Waidmann O., Choudhary C., Dötsch V., Bumann D., and Dikic I. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fiskin E., Bionda T., Dikic I., and Behrends C. (2016) Global analysis of host and bacterial ubiquitinome in response to Salmonella typhimurium infection. Mol. Cell 62, 967–981 10.1016/j.molcel.2016.04.015 [DOI] [PubMed] [Google Scholar]
- 91.Huett A., Heath R. J., Begun J., Sassi S. O., Baxt L. A., Vyas J. M., Goldberg M. B., and Xavier R. J. (2012) The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella typhimurium. Cell Host Microbe 12, 778–790 10.1016/j.chom.2012.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Noad J., von der Malsburg A., Pathe C., Michel M. A., Komander D., and Randow F. (2017) LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-κB. Nat. Microbiol. 2, 17063 10.1038/nmicrobiol.2017.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Polajnar M., Dietz M. S., Heilemann M., and Behrends C. (2017) Expanding the host cell ubiquitylation machinery targeting cytosolic Salmonella. EMBO Rep. 18, 1572–1585 10.15252/embr.201643851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heath R. J., Goel G., Baxt L. A., Rush J. S., Mohanan V., Paulus G. L., Jani V., Lassen K. G., and Xavier R. J. (2016) RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep. 17, 2183–2194 10.1016/j.celrep.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Wijk S. J. L., Fricke F., Herhaus L., Gupta J., Hötte K., Pampaloni F., Grumati P., Kaulich M., Sou Y. S., Komatsu M., Greten F. R., Fulda S., Heilemann M., and Dikic I. (2017) Linear ubiquitination of cytosolic Salmonella typhimurium activates NF-κB and restricts bacterial proliferation. Nat. Microbiol. 2, 17066 10.1038/nmicrobiol.2017.66 [DOI] [PubMed] [Google Scholar]
- 96.Xie Y., Kang R., Sun X., Zhong M., Huang J., Klionsky D. J., and Tang D. (2015) Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11, 28–45 10.4161/15548627.2014.984267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhogaraju S., Kalayil S., Liu Y., Bonn F., Colby T., Matic I., and Dikic I. (2016) Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167, 1636–1649.e13 10.1016/j.cell.2016.11.019 [DOI] [PubMed] [Google Scholar]
- 98.Kotewicz K. M., Ramabhadran V., Sjoblom N., Vogel J. P., Haenssler E., Zhang M., Behringer J., Scheck R. A., and Isberg R. R. (2017) A single legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell Host Microbe 21, 169–181 10.1016/j.chom.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grumati P., Morozzi G., Hölper S., Mari M., Harwardt M. I., Yan R., Müller S., Reggiori F., Heilemann M., and Dikic I. (2017) Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 6, e25555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A. K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N., Mauthe M., Katona I., Qualmann B., Weis J., Reggiori F., et al. (2015) Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]