Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4 (original) (raw)
. Author manuscript; available in PMC: 2019 Mar 17.
Published in final edited form as: Nat Med. 2018 Sep 17;24(10):1599–1610. doi: 10.1038/s41591-018-0158-8
Abstract
Cancer cells rely on dysregulated gene expression. This establishes specific transcriptional addictions that may be therapeutically exploited. Yet, the mechanisms ultimately responsible for these addictions are poorly understood. Here we investigated the transcriptional dependencies of transformed cells to transcription factors YAP and TAZ. YAP/TAZ physically engage the general coactivator BRD4, dictating the genome-wide association of BRD4 to chromatin. YAP/TAZ flag a large set of enhancers with super-enhancer-like functional properties. YAP/TAZ-bound enhancers mediate recruitment of BRD4 and Pol II at YAP/TAZ-regulated promoters, boosting expression of a host of growth-regulating genes. Treatment with small molecule inhibitors of BRD4 blunts YAP/TAZ pro-tumorigenic activity in several cell/tissue contexts, causes regression of pre-established, YAP/TAZ-addicted neoplastic lesions, and reverts drug resistance. This work sheds light on essential mediators, mechanisms and genome-wide regulatory elements responsible for transcriptional addiction in cancer and lays the groundwork for a rational use of BET inhibitors according to YAP/TAZ biology.
An emerging paradigm in cancer biology relates to the concept of "transcriptional addiction": it posits that, to support their uncontrolled proliferation or other needs, tumor cells set high demands on transcriptional regulators, including chromatin regulators and even the basal transcriptional machinery1,2. The molecular mechanisms underlying the transcriptional dependency of cancer cells are poorly understood. Yet, it is an appealing concept, as general chromatin regulators/transcriptional cofactors are amenable to inhibition with small molecules2. The emblematic example is the antitumor activity of BET inhibitors in various xenograft model systems and clinical trials3–6. BET inhibitors oppose the activity of BET (Bromodomain and Extraterminal)-coactivators (that is, BRD4 and its related factors BRD2 and BRD3)5. Although BET proteins have been proposed to serve as general regulators of RNA polymerase II (Pol II)-dependent transcription, genome-wide studies have instead shown that BET inhibitors display selective effects on gene expression5,7. In particular, BET inhibitors have been reported to have disproportional effect on a set of highly expressed genes associated with super-enhancers5,7. The molecular basis of the transcriptional addiction associated to super-enhancers in cancer cells, as well as the determinants of the selectivity of BET inhibitors remain undefined8.
The transcription coactivators YAP/TAZ are ideal candidates to mediate cancer-specific transcriptional addictions. In fact, YAP/TAZ are genetically dispensable for homeostasis in many adult tissues9–17 while YAP/TAZ activation is a hallmark of many human malignancies13,17–19. Here we show that tumor transcriptional dependencies in fact overlap with tumor reliance on YAP/TAZ.
Results
BRD4 interacts with YAP/TAZ
With this background in mind, we started this investigation by carrying out ChIP-MS for endogenous YAP/TAZ, a procedure that allows studying the composition of the native protein complexes entertained by YAP/TAZ, and in particular nuclear interactions20. We detected some well-known nuclear partners of YAP/TAZ, including TEAD (the main YAP/TAZ DNA interacting partner) and Activator Protein 1 family members13 and several subunits of the Swi/Snf complex21. YAP/TAZ protein complexes were also enriched in chromatin readers/modifiers, such as BRD4, histone acetyltransferases (p300, p400) and the histone methyltransferase KMT2D/MLL2 (Table 1). The roles of p300, SWI/SNF and the H3K4 methyltransferase complexes in the context of YAP-dependent transcription have been previously described21–23. The association with BRD4 attracted our attention, as this hinted to a connection between YAP/TAZ regulated gene expression and the transcriptional addiction of cancer cells.
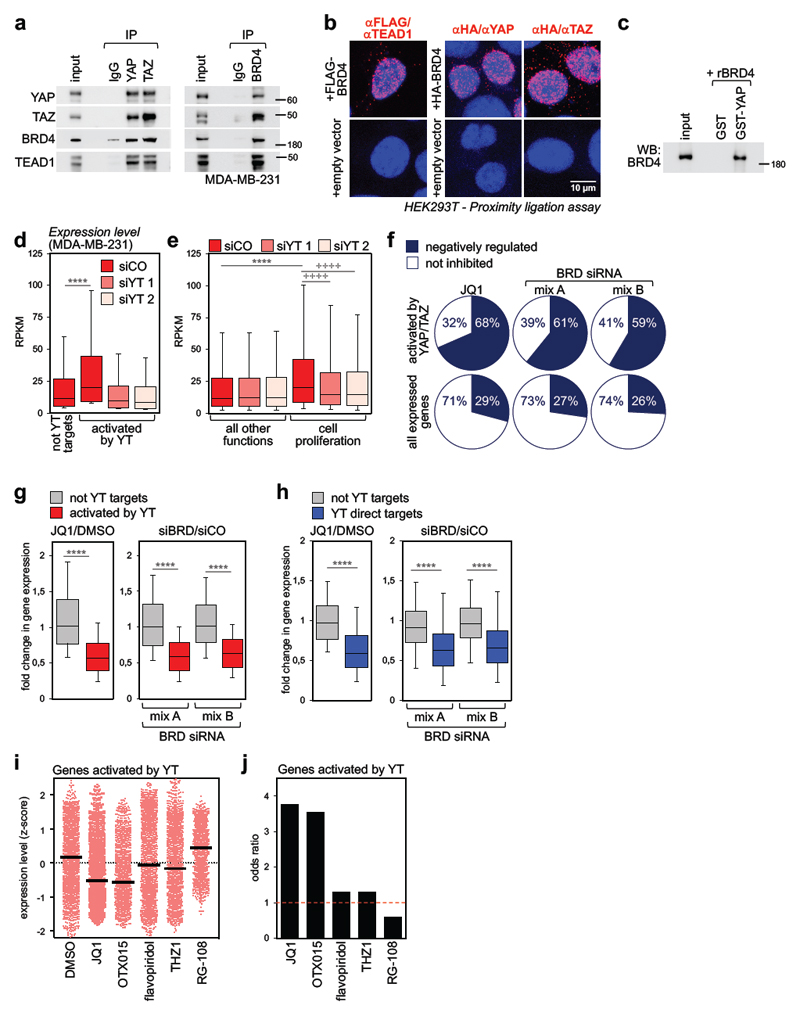
In order to validate the interactions detected by Chip-MS, we performed co-immunoprecipitation (Co-IP) of endogenous proteins, revealing the presence of BRD4 and TEAD1 in YAP and TAZ immunocomplexes, and of YAP, TAZ and TEAD1 in BRD4 immunocomplexes (Fig. 1a). By proximity ligation assays (PLA), we validated that this interaction occurs in the nucleus (Fig. 1b). Furthermore, by Co-IP, transfected FLAG-tagged YAP copurified endogenous BRD4 and BRD2 (Supplementary Fig. 1a). Importantly, the association between YAP or TAZ and BRD4 is direct, as attested by the interactions between purified recombinant proteins (Fig. 1c and Supplementary Fig. 1b). By using progressive C-terminal deletion constructs, we mapped the minimal region sufficient for association with BRD4 between aa 108-175 of mouse TAZ (Supplementary Fig. 1b-c); notably, this region includes the WW domain24. However, removal of the sole WW domain from full-length TAZ did not impair its ability to associate with BRD4 (Supplementary Fig. 1d), indicating that at least another determinant for BRD4 association exists in the C-terminal Transactivation Domain. Overall, data indicate that YAP, TAZ, TEAD1 and BET proteins are part of the same nuclear multiprotein complex.
Figure 1. BRD4 associates to YAP/TAZ and is a required cofactor for YAP/TAZ transcriptional activity.
a) Interaction of endogenous YAP/TAZ, TEAD1 and BRD4 in MDA-MB-231 cells. Each co-IP experiment was performed three times with similar results.
b) Endogenous YAP, TAZ or TEAD1 and exogenous FLAG- or HA-BRD4 interact in the nuclei of HEK293T cells, as shown by PLA signal (red fluorescent dots). Nuclei are counterstained with DAPI (blue). No dots could be detected in the nuclei of cells transfected with empty vector, confirming the specificity of interactions. Similar results were obtained in two additional experiments.
c) Recombinant BRD4 is pulled-down by GST-YAP fusion protein. GST-pulldown was repeated three times with similar results.
d) Genes activated by YAP/TAZ (n=2073) display higher expression levels compared to genes not activated by YAP/TAZ (not YT targets, n=8026) in MDA-MB-231 cells. Expression values (in RPKM) were determined by RNA-seq and are presented as box-and-whiskers plots (whiskers extend from the 10th to the 90th percentile; the box extends from the 25th to the 75th percentile; the line within the box represents the median). **** p<10-10 (one-tailed Mann-Whitney U test). See also Supplementary Fig. 1g.
e) Box-and-whiskers plots of expression values of genes involved in cell proliferation (n=1449) vs. genes associated to all other functions (n=8650) according to GO annotation. Data are presented as in d. **** p<10-10 (one-tailed Mann-Whitney U test); ++++ p<10-10 (one-tailed Wilcoxon matched-pairs signed rank test)
f) The fraction of genes activated by YAP/TAZ which are inhibited by JQ1 or BRD2/3/4 siRNAs is larger than the fraction of all expressed genes downregulated by the same treatments. See also Supplementary Fig. 1i.
g) Fold change in gene expression of not-YAP/TAZ targets (n=8026) vs. genes activated by YAP/TAZ (n=2073) upon treatment with JQ1 (left) or BET proteins depletion (right). The y axis shows the fold change in transcript levels versus DMSO-treated cells or cells transfected with control siRNA. Data are presented as box-and-whiskers plots, as in d. **** p<10-10 (one-tailed Mann-Whitney U test)
h) Fold change in gene expression of high-confidence YAP/TAZ direct targets (n=616) vs. not-YAP/TAZ targets (n=771) upon treatment with JQ1 (left) or BET proteins depletion (right). The group of not YT targets represents genes not significantly affected by YAP/TAZ depletion (FDR>0.05) in our RNA-seq dataset. Data are presented as box-and-whiskers plots, as in d. **** p<10-10 (one-tailed Mann-Whitney U test)
i) Expression level of all YAP/TAZ activated genes (n=2073) in MDA-MB-231 cells treated with DMSO (vehicle), BET inhibitors (JQ1, OTX015), CDKs inhibitors (flavopiridol, THZ1) or RG-108 (a DNA methyltransferase inhibitor, here used as negative control to assess the effect of a compound targeting an epigenetic function not related to transcription). Expression levels were determined by RNA-seq and are presented as z-scores. Individual genes and their mean (black line) are presented.
j) Odds ratio plot: genes activated by YAP/TAZ (n=2073) are more likely to be inhibited by BET inhibitors than not-YAP/TAZ target genes. CDK inhibitors and RG-108 do not display such property (see Methods).
YAP/TAZ mediate cancer transcriptional addiction
To study the connection between YAP/TAZ and transcriptional addiction in cancer, we used MDA-MB-231 cells, a well-established model of triple-negative breast cancer (TNBC), a tumor type requiring high-levels of uninterrupted transcription of large set of genes to sustain its particularly aggressive nature25,26. Are YAP/TAZ causal to these dependencies? By comparing the transcriptional profiles (obtained by RNA-seq) of control and YAP/TAZ-depleted cells (Supplementary Figs. 1e-f), we found that genes whose expression depends on YAP/TAZ were significantly more expressed than all the other genes (Fig. 1d); this conclusion was confirmed also when we restricted our analyses to high-confidence direct YAP/TAZ target genes, that is, genes dependent on YAP/TAZ whose promoters/enhancers contain YAP/TAZ binding sites as assessed by ChIP-Seq13 (Supplementary Fig. 1g). Moreover, genes whose biological function is associated with cell proliferation (~1500 genes, as determined by Gene Ontology (GO) annotation) were transcribed at higher levels compared to the bulk of expressed genes (Fig. 1e); silencing YAP/TAZ with siRNAs led to a global downregulation of such “growth program” (Fig. 1e), in line with the previous report that MDA-MB-231 cells depleted of YAP/TAZ undergo growth arrest13. Indeed, 37% of the growth genes actively transcribed in these cells are YAP/TAZ targets (541/1449); their transcripts were particularly abundant, displaying higher level of expression than non-YAP/TAZ targets associated to the same biological function (Supplementary Fig. 1h). Thus, the activation of essential growth genes in MDA-MB-231 cells relies on YAP/TAZ.
Vulnerability of YAP/TAZ activity to BET inhibitors
To assess if the interaction with BRD4 is causal for the activation of YAP/TAZ transcriptional targets, we performed RNA-seq in cells treated with JQ1, the most established BET inhibitor; JQ1 occupies the bromodomain pockets of BET proteins in a manner that is competitive with the binding to acetylated histone tails, causing their displacement from chromatin4. Most YAP/TAZ regulated genes (68%) displayed exquisite sensitivity to JQ1 (Fig. 1f and Supplementary Fig. 1i), and vice versa, genes most effectively downregulated by JQ1 were in fact YAP/TAZ-dependent (Supplementary Fig. 1j). Indeed, treatment with JQ1 selectively decreased the transcript abundance of YAP/TAZ-dependent genes, compared to all other active genes (Fig. 1g). The disproportional effect of JQ1 was confirmed when we restricted our analyses to high-confidence direct YAP/TAZ target genes (Fig. 1h).
The bias of JQ1 towards inhibition of YAP/TAZ-dependent genes (including direct targets) was also evident when restricting the analysis to genes regulating cell proliferation (Supplementary Figs. 1k-l): BET-inhibition affected the expression of 604 genes associated to GO terms linked to cell proliferation, and 428 of these (71%) were regulated by YAP/TAZ. Thus, sensitivity of a broad number of growth-regulating genes to BET inhibition relies on YAP/TAZ. Effects similar to those of JQ1 were obtained with another BET inhibitor (OTX015, Figs. 1i-j and Supplementary Figs. 1l and 1o) and by knocking down BRD2/3/4 with two independent combinations of siRNAs (Figs. 1g-h). Moreover, depletion of the sole BRD4 was sufficient, at least in part, to downregulate YAP/TAZ target genes (Supplementary Fig. 1m). We also found that endogenous YAP/TAZ remained nuclear upon treatment with BET inhibitors (Supplementary Fig. 1n), excluding the possibility that the compounds would indirectly cause YAP/TAZ cytoplasmic relocalization.
YAP/TAZ transcriptional control has been connected to CDK9-induced elongation of nascent transcripts by Pol II27. Here we find that, in stark contrast with BET inhibitors, inhibition of transcriptional CDKs with flavopiridol or THZ1 failed to display any bias towards inhibition of YAP/TAZ transcriptional targets (Figs. 1i-j and Supplementary Fig. 1o). Collectively, the data indicate that the physical association between YAP/TAZ and BRD4 is functionally relevant; BRD4 is a required cofactor for YAP/TAZ, conferring to YAP/TAZ target genes a specifically high dependency on BRD4 and vulnerability to BET inhibitors.
BRD4 is recruited to chromatin by YAP/TAZ
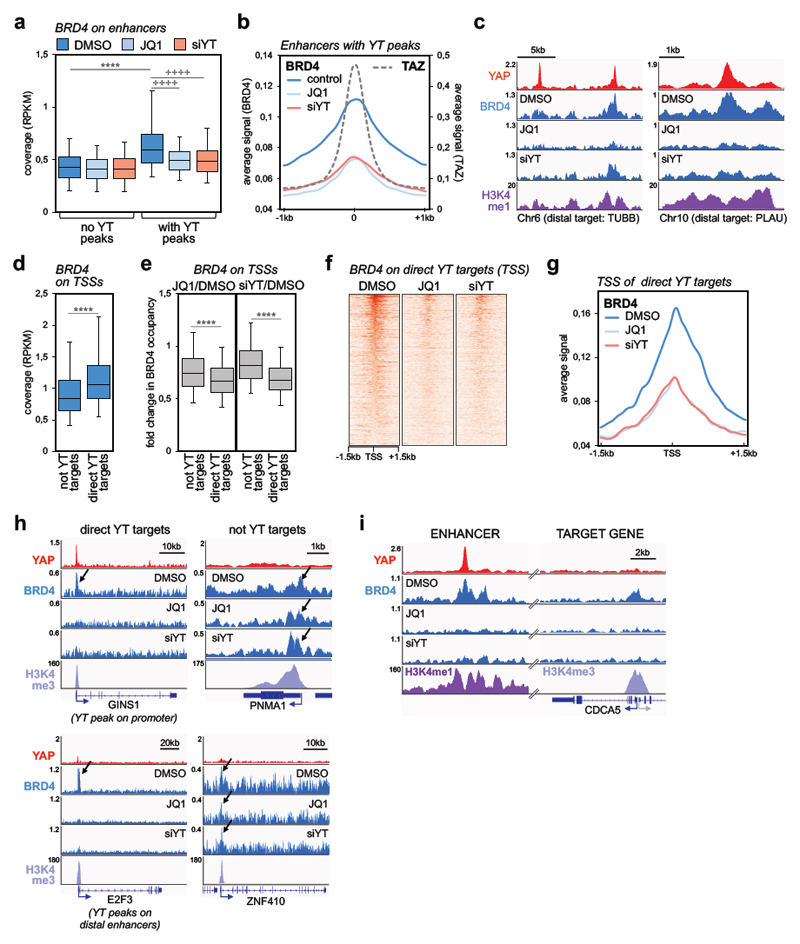
We next asked what underlies the disproportionate sensitivity of YAP/TAZ targets to BET inhibitors. To gain mechanistic insights into this connection, we performed ChIP-seq experiments to compare BRD4 and YAP/TAZ binding to chromatin. YAP/TAZ bind almost exclusively to enhancers 13,23,27, whereas BRD4 binds both active enhancers and active promoters (Supplementary Figs. 2a-b; see Materials and Methods for the definition of enhancers and promoters). We started our analysis from enhancer elements and found that BRD4 coverage was higher on enhancers containing YAP/TAZ binding sites when compared to active enhancer not occupied by YAP/TAZ (Fig. 2a). We reasoned that differential BRD4 loading might correspond to differential responsiveness to JQ1; to verify this assumption, we performed BRD4 ChIP-seq in cells treated with JQ1. We found that JQ1 induced a preferential loss of BRD4 from YAP/TAZ-occupied enhancers, compared to active enhancers without YAP/TAZ binding sites (Fig. 2a and Supplementary Fig. 2c). Thus, the presence of YAP/TAZ peaks defines enhancers enriched of BRD4 and highly sensitive to BET inhibitors on the genome-wide scale. Do these elements correspond to super-enhancers? In fact, 80% of super-enhancers in MDA-MB-231 cells do contain YAP/TAZ peaks; yet, the vast majority (85%) of YAP/TAZ-occupied enhancers are by definition typical enhancers (Supplementary Figs. 2f-g). However, we observed that genes connected to YAP/TAZ-bound typical enhancers displayed sensitivity to JQ1 strikingly similar to the much more restricted number of genes associated with super-enhancers (Supplementary Fig. 2h).
Figure 2. YAP/TAZ are required for BRD4 recruitment to chromatin.
a) Comparison of BRD4 ChIP-seq signal (expressed as normalized read density, RPKM) in active enhancers with or without YAP/TAZ peaks (n=5169 and n=30281, respectively) in MDA-MB-231 cells treated with DMSO or JQ1 (1µM, 6h), or transfected with YAP/TAZ siRNAs (48h). Data are presented as box-and-whiskers plots (whiskers extend from the 5th to the 95th percentile; the box extends from the 25th to the 75th percentile; the line within the box represents the median). **** p<10-10 (one-tailed Mann-Whitney U test); ++++ p<10-10 (one-tailed Wilcoxon matched-pairs signed rank test)
b) Average signal of BRD4 ChIP-seq reads in enhancers with YAP/TAZ peaks (n=5169) in a window of ±1 kb centered on the summit of YAP/TAZ peaks.
c) Genome browser view of YAP, BRD4 and H3K4me1 binding profiles at representative active enhancers in MDA-MB-231 cells. Both JQ1 and YAP/TAZ siRNA induce a strong decrease in BRD4 binding.
d) Box plot of BRD4 ChIP-seq signal (RPKM) comparing promoters of genes not activated by YAP/TAZ (n=8026) or of YAP/TAZ target genes (YT targets, n=616) in MDA-MB-231 cells (treated with DMSO). Data are presented as in a. **** p<10-10 (one-tailed Mann-Whitney U test)
e) Treatment with JQ1 (left) and YAP/TAZ depletion (right) induce preferential loss of BRD4 at promoters of YAP/TAZ target genes (n=616) vs. promoters of not YAP/TAZ targets (n=8026). Fold change in BRD4 binding is calculated as RPKM(JQ1 or siYT)/RPKM(DMSO). Data are presented as box-and-whiskers plots (whiskers extend from the 5th to the 95th percentile; the box extends from the 25th to the 75th percentile; the line within the box represents the median). **** p<10-10 (one-tailed Mann-Whitney U test)
f) Heatmap showing BRD4 binding on the promoters of YAP/TAZ targets in MDA-MB-231 cells, in a window of ±1.5kb centered on the transcription start site (TSS).
g) Average signal of BRD4 ChIP-seq reads on the promoters of YAP/TAZ target genes (n=616) in MDA-MB-231 cells, in a window of ±1.5 kb centered on TSS.
h) YAP, BRD4 and H3K4me3 binding profiles at representative promoters of YAP/TAZ target genes or not-YAP/TAZ targets. Arrows indicate BRD4 enrichment at the TSS. JQ1 and siYAP/TAZ induce a strong decrease in BRD4 binding on YAP/TAZ targets, whereas there is no overt variation of BRD4 binding on the TSS of not YAP/TAZ targets. GINS1 exemplifies direct YAP/TAZ target genes with YAP/TAZ binding sites in both enhancers and the TSS; E2F3 exemplifies direct YAP/TAZ target genes regulated by YAP/TAZ exclusively from distal enhancers.
i) YAP, BRD4 and H3K4me1/H3K4me3 binding profiles on a distal enhancer and on CDCA5 promoter. JQ1 (1µM, 6h) and siYAP/TAZ (48h) induce a strong decrease in BRD4 binding both on the enhancer, containing YAP/TAZ peak, and on TSS of CDCA5.
We then assessed whether the presence of YAP/TAZ was required for the engagement of BRD4 to chromatin, by performing BRD4 ChIP-seq in MDA-MB-231 cells depleted of YAP/TAZ. As shown by the average BRD4 binding profile and some representative enhancers in Figs. 2b-c, BRD4 recruitment to YAP/TAZ-containing enhancers was heavily reduced upon YAP/TAZ depletion, to an extent similar to JQ1 (see also Supplementary Figs. 2c-e). The determinants that drive the selectivity of BRD4 association to specific chromatin sites are unclear. BRD4 was reported to bind some acetylated TFs through the bromodomain5; however, arguing against this possibility in the case of YAP/TAZ, we found that their biochemical association with BRD4 is not affected by the presence of JQ1 or by mutations in the BRD4-bromodomains (Supplementary Fig. 2i). Thus, the comparable impoverishment of BRD4 recruitment to chromatin detected genome-wide after YAP/TAZ depletion or JQ1 treatment reflects the need of a dual association of BRD4 to YAP/TAZ and acetylated histones to keep BRD4 anchored to a large set of enhancers and superenhancers, as such providing selectivity to BRD4 function.
We then surmised that the disproportionate sensitivity of YAP/TAZ targets to inhibition by JQ1 should be ultimately explained at the level of YAP/TAZ-regulated promoters. Focusing on high-confidence direct YAP/TAZ target genes13 (see Table 2), we found that: i) the TSS of these genes exhibited higher BRD4 occupancy when compared to the TSS of genes not activated by YAP/TAZ (Fig. 2d); ii) YAP/TAZ were required for BRD4 accrual on the promoters of their targets, while marginally affecting the promoters of non-YAP/TAZ targets (Figs. 2e-g; see also Figs. 2h-i for representative individual gene tracks, and Supplementary Fig. 2j for validation by qPCR); iii) JQ1 caused preferential loss of BRD4 from YAP/TAZ-regulated promoters (Figs. 2e-g), matching the effects of JQ1 on gene expression. Importantly, as exemplified in Fig. 2h, JQ1 had only minor effects on BRD4 coverage on the promoters of genes not activated by YAP/TAZ, where it was in fact insufficient to induce a general downregulation of transcription (Fig. 1g-h). Thus, BRD4 levels at promoters closely reflect the dynamic of YAP/TAZ-mediated engagement of BRD4 at distant enhancers.
Mechanisms of transcriptional addiction
Data presented above suggest a model whereby YAP/TAZ bound to enhancers promote BRD4 overload on their target promoters, establishing higher expression levels of essential genes, and – concomitantly – their vulnerability to BET inhibitors. In agreement with this prediction, treatment with BET inhibitors does not alter YAP/TAZ recruitment to chromatin (Supplementary Fig. 3a); yet, BET inhibitors block downstream gene expression, even in conditions in which YAP is overexpressed, consistently with YAP/TAZ acting upstream of BRD4 (Supplementary Fig. 3b). In line, in the absence of YAP/TAZ, even overexpressed BRD4 could not promote expression of YAP/TAZ target genes (Supplementary Fig. 3c).
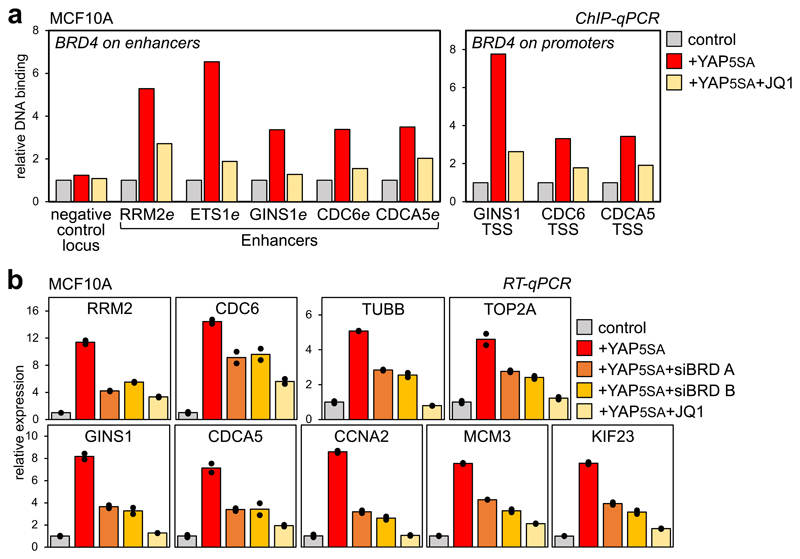
Next, we validated the functional interdependency between YAP/TAZ and BET proteins by overexpressing YAP5SA (a constitutively-active version of YAP) in mammary epithelial cells (MCF10A), which normally display low YAP/TAZ activity. By ChIP, exogenous YAP is recruited at its cognate chromatin sites (Supplementary Fig. 3d); in turn, this leads to BRD4 recruitment at the same enhancer sites and associated promoters (Fig. 3a). BET inhibitors block BRD4 recruitment to these cis-regulatory elements, supporting the requirement of BRD4 binding to histones along with YAP/TAZ. By gene expression, exogenous YAP turns on its targets, but not in cells treated with JQ1 or depleted of BET-proteins (Fig. 3b and Supplementary Fig. 3e), indicating that BRD4 operates downstream of YAP/TAZ. The same conclusion applies when we experimentally activated endogenous YAP/TAZ through inactivation of the Hippo pathway (Supplementary Fig. 3f).
Figure 3. YAP/TAZ are instrumental for BRD4 recruitment to chromatin.
a) ChIP-qPCR showing increased BRD4 binding on enhancers and promoters of YAP/TAZ targets upon YAP5SA overexpression in MCF10A cells, but not in the presence of JQ1 (1µM, 6h). ChIP with pre-immune IgG displayed background signal (which was comparable in all samples). DNA enrichment was calculated as fraction of input and is presented as fold vs. BRD4 binding in control cells.
b) RT-qPCR for representative YAP/TAZ target genes showing upregulation upon YAP5SA overexpression in MCF10A cells, but not in the presence of JQ1 (1µM, 24h) or upon depletion of BRD2/3/4 (siBRD mix A and B). Data are presented as individual data points (n=2 biologically independent samples) + average (bar), from one out of three experiments, all producing similar results. BET inhibition does not impair the expression of exogenous YAP (see Supplementary Fig. 3e).
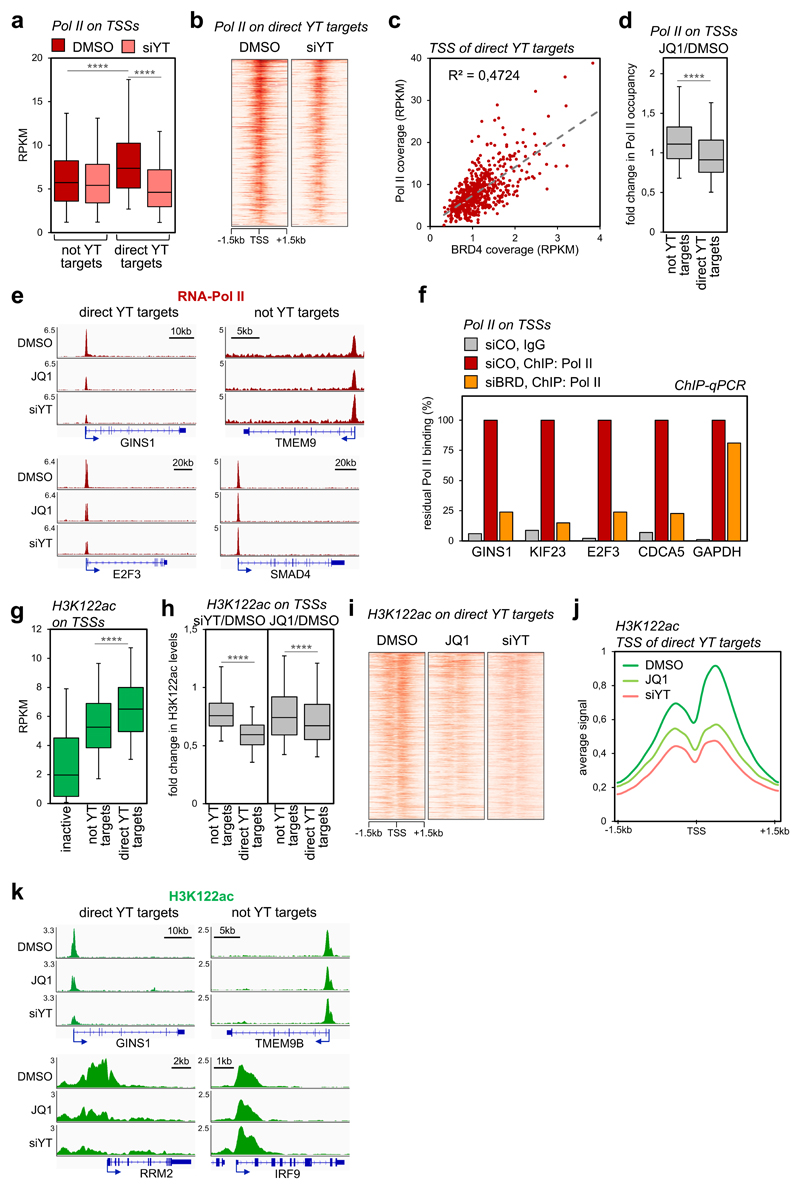
We next investigated the mechanisms by which BRD4 accrual regulates transcription initiated at YAP/TAZ-target promoters. The role of BRD4 for transcriptional activation is best understood in terms of promotion of elongation through recruitment of P-TEFb5. If so, we should expect that, upon YAP/TAZ depletion or JQ1 treatment, Pol II should remain paused, if not accumulate, on the promoters of YAP/TAZ targets. In contrast to expectations, by ChIP-seq, Pol II loading was selectively decreased on the promoters of YAP/TAZ targets in YAP/TAZ-depleted cells (Figs. 4a-b, Supplementary Fig. 4a-c). This implies that YAP/TAZ promote the recruitment of RNA Pol II. Indeed, we detected an association between YAP and Pol II in endogenous complexes by Co-IP (Supplementary Fig. 4d); intriguingly, this interaction was lost after experimental depletion of BRD4, at least suggesting that the latter serves as key element in connecting YAP/TAZ-bound cis-regulatory elements with the transcriptional apparatus assembled on cognate promoters. In line, Pol II coverage on the TSSs of YAP/TAZ target genes was on average higher compared to all other expressed genes (Fig. 4a), proportional to BRD4 binding (Fig. 4c), and selectively reduced by JQ1 treatment (Fig. 4d). Representative gene tracks showing comparable effects of JQ1 and YAP/TAZ depletion on Pol II loading on the TSS of YAP/TAZ target genes (but no substantial effects on not-YAP/TAZ targets) are presented in Fig. 4e and Supplementary Fig. 4e. Similar results were obtained after siRNA-mediated depletion of BRD2/3/4, as revealed by ChIP-qPCR at sampled promoters (Fig. 4f). This suggests that YAP/TAZ recruit Pol II by inducing BRD4 accumulation at TSS.
Figure 4. YAP/TAZ and BRD4 regulate Pol II loading and H3K122 acetylation on TSSs.
a) Box-and whiskers plots showing the distribution of RNA-Pol II ChIP-seq signal (expressed as normalized read density, RPKM) comparing promoters of genes not activated by YAP/TAZ (n=8026) or of YAP/TAZ target genes (n=616) in control (DMSO) or YAP/TAZ depleted cells. The box includes values within the 25th and 75th percentile (with the median highlighted by the line in the middle) and whiskers extend from the 5th to the 95th percentile. **** p<10-10 (one-tailed Mann-Whitney U test)
b) Heatmap showing RNA-Pol II loading on the promoters of YAP/TAZ targets in MDA-MB-231 cells, in a window of ±1.5kb centered on the transcription start site (TSS).
c) Linear correlation between BRD4 and RNA-Pol II occupancy (both expressed in RPKM) on the TSS of YAP/TAZ target genes (n=616). r2 was calculated using linear regression analysis (F-test p-value<0.0001).
d) Box-and whiskers plots (defined as in a) showing the change in RNA-Pol II promoter occupancy in JQ1-treated cells vs. control cells (DMSO), comparing promoters of genes not activated by YAP/TAZ (n=8026) with promoters of YAP/TAZ target genes (n=616). **** p<10-10 (one-tailed Mann-Whitney U test)
e) Genome browser view of RNA-Pol II binding profiles at representative promoters of YAP/TAZ target genes or not-YAP/TAZ targets. Pol II binding is reduced upon JQ1 treatment or YAP/TAZ depletion on the TSS of YAP/TAZ targets.
f) ChIP-qPCR verifying RNA-Pol II binding to promoters of established YAP/TAZ targets upon depletion of BET proteins. GAPDH promoter represents a non-YAP/TAZ target. ChIP with pre-immune IgG displayed background signal (which was comparable in all samples). DNA enrichment was calculated as fraction of input and is presented as % of RNA-Pol II binding in control cells (siCO).
g) Box-and whiskers plots of H3K122ac ChIP-seq signal (RPKM) showing its enrichment on YAP/TAZ target genes (n=616) in comparison with inactive promoters (n=4618) and not-YAP/TAZ targets (n=8026). Data are presented as in a. **** p<10-10 (one-tailed Mann-Whitney U test)
h) Box-and whiskers plots showing the change in H3K122ac promoter levels in YAP/TAZ-depleted (left) or JQ1-treated cells (right), both compared to control cells (DMSO), showing preferential loss of H3K122ac at promoters of YAP/TAZ target genes (n=616) vs. promoters of not YAP/TAZ targets (n=8026). Data are presented as in a. **** p<10-10 (one-tailed Mann-Whitney U test)
i) Heat map showing acetylation of H3K122 on the promoters of YAP/TAZ targets in MDA-MB-231 cells, in a window of ±1.5kb centered on the transcription start site (TSS).
j) Average ChIP-seq profile of H3K122ac on the promoters of YAP/TAZ target genes (n=616) in MDA-MB-231 cells, in a window of ±1.5 kb centered on TSS.
k) Genome browser view of H3K122ac levels at representative promoters of YAP/TAZ target genes vs. not-YAP/TAZ targets.
BRD4 has been recently reported, and we have repeated, to display an intrinsic acetyltransferase activity, leading to acetylation of K122 in the globular domain of H328. Intriguingly, H3K122 acetylation is associated with Pol II loading on promoters and transcriptional activation29; consistently, we discovered that the HAT domain of BRD4 is indeed crucial for expression of YAP/TAZ target genes, as an HAT-defective BRD4 (ΔHAT) cannot substitute wild type BRD4 (Supplementary Fig. 4f). We measured the levels of H3K122ac by ChIP-seq in control, YAP/TAZ-depleted or JQ1-treated cells. Strikingly, H3K122ac levels were significantly higher on the promoters of YAP/TAZ target genes (Fig. 4g), in line with the higher coverage of BRD4 (Supplementary Fig. 4g) and Pol II, and with transcriptional activation. This differential enrichment of H3K122ac was dependent on YAP/TAZ, as robust loss in H3K122 acetylation was observed in YAP/TAZ targets upon YAP/TAZ depletion (Figs. 4h-k and Supplementary Fig. 4h). Paralleling BRD4 occupancy, acetylation of H3K122 on the promoters of YAP/TAZ targets was especially sensitive to JQ1-treatment (Figs. 4h-k and Supplementary Fig. 4h). We thus propose that YAP/TAZ promote transcriptional activation of their target genes by favouring BRD4 overload on their promoters, thus favouring Pol II recruitment through H3K122 acetylation and association to Pol II.
BET inhibition opposes YAP/TAZ pro-tumorigenic functions
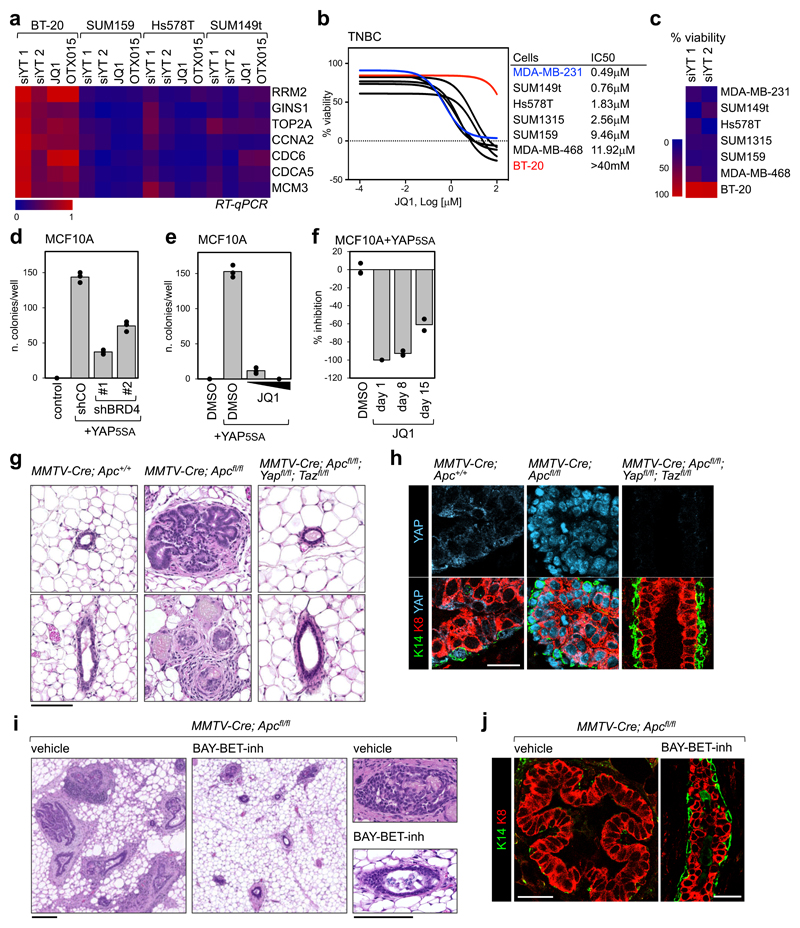
To expand on the generality of the YAP/TAZ-BRD4 connection, we then asked whether YAP/TAZ transcriptional activity is especially sensitive to BET inhibitors in TNBC cell lines other than MDA-MB-231. For this, we measured the effects of YAP/TAZ depletion or JQ1 treatment on the expression level of a set of YAP/TAZ target genes and on cell proliferation. As shown in Figs. 5a-c, TNBC cells that are YAP/TAZ-addicted are also sensitive to JQ1. Interestingly, we also found one cell line, BT20, that is not YAP/TAZ-addicted and also resistant to BET inhibitors.
Figure 5. Treatment with BET inhibitor blunts YAP/TAZ-addicted breast tumors.
a) Heat map showing the regulation of YAP/TAZ target genes in triple negative breast cancer cells after YAP/TAZ depletion (siYT1, siYT2) or treatment with BET inhibitors (1µM, 24h). Expression values are normalized to cells transfected with control (siRNA) and to GAPDH.
b) Left: viability curves of TNBC cells treated with increasing doses of JQ1 (1nM to 100µM). Data are mean of n=8 independent wells (independently treated and evaluated). Right: IC50 of listed cell lines.
c) Proliferation of BT20 cells is not impaired by YAP/TAZ depletion, whereas all cells sensitive to JQ1 are also affected by YAP/TAZ depletion.
d) BRD4 downregulation by shRNAs impairs colony formation by YAP5SA-overexpressing MCF10A cells in soft agar. Data are presented as individual data points (n=3 independent samples) + average (bar), from one of three experiments providing similar results. Similar results were obtained in MDA-MB-231 cells, whose colony-forming capacity depends on endogenous YAP/TAZ (see Supplementary Fig. 5b).
e) Quantification of colonies formed by YAP5SA-overexpressing MCF10A cells in soft agar, upon treatment with 0,1µM or 1µM JQ1 for the entire experiment. Data are presented as in d. Similar results were obtained in MDA-MB-231 cells (see Supplementary Fig. 5c).
f) Inhibition of the growth of colonies initiated by YAP5SA-overexpressing MCF10A cells in soft agar upon addition of JQ1 (1µM) to culture medium 8 or 15 days after seeding (treatment with JQ1 at day1 is presented as reference for maximal inhibition). Data are presented as in d.
g) Representative hematoxylin and eosin (H&E) staining of sections of mammary glands from MMTV-Cre;Apc+/+, MMTV-Cre;Apcfl/fl, or MMTV-Cre;Apcfl/fl;Yapfl/fl;Tazfl/fl mice. Scale bar is 0.1 mm. The same phenotype was observed in at least 4 mice per each experimental group.
h) Representative immunofluorescence (IF) pictures of mammary glands from the indicated mice, showing YAP accumulation in the nuclei of epithelial cells, expansion of luminal cells (K8-positive) and discontinuities in the basal layer of K14-positive cells in MMTV-Cre;Apcfl/fl. Ducts of MMTV-Cre;Apcfl/fl;Yapfl/fl;Tazfl/fl mice display a normal morphology. Scale bar is 25 µm. IF was performed on sections derived from 4 mice per each genotype.
i) Representative H&E staining of sections of mammary glands from MMTV-Cre;Apcfl/fl mice, treated with vehicle (n=5) or BAY-BET inhibitor (n=5) for 6 weeks. All scale bars are 0.1mm. See BAY-BET-inhibitor has no effect on the histological appearance of mammary glands of Apcfl/fl (_Cre_-negative) littermates (see Supplementary Fig. 5g).
j) Representative IF pictures of mammary glands from MMTV-Cre;Apcfl/fl mice, treated with vehicle (n=5) or BAY-BET inhibitor (n=5) for 6 weeks, showing that treatment with BET inhibitor restores normal distribution of the luminal marker K8 and the basal marker K14 in the mammary ducts. Scale bars are 25 µm. See Supplementary Fig. 5h for normal K8/K14 staining in Apcfl/fl (_Cre_-negative) mice treated with BAY-BET-inhibitor.
Next, we evaluated the effect of BRD4 silencing on YAP-induced cell transformation. For this, we performed a colony formation assay in soft agar with MCF10A cells, which are per se unable to seed colonies, but acquire this capacity upon overexpression of YAP5SA. Downregulation of BRD4 (with shRNAs) reduced the number of colonies initiated by YAP-overexpressing cells (Fig. 5d and Supplementary Fig. 5a). In line, addition of JQ1 to culture medium potently impaired initial colony formation (Fig. 5e), as well as growth of established colonies (Fig. 5f), in agreement with the effects of JQ1 on the transcriptional and epigenetic effects of YAP expression.
Extending the translational significance of these findings, we assayed whether inhibition of BET proteins could blunt the growth of, if not cure, YAP/TAZ-addicted mammary tumors in vivo. Constitutive activation of the Wnt cascade in the mammary epithelium has been shown to induce TNBC-like tumors in mice30, a finding that parallels the high frequency of APC epigenetic silencing in human TNBC31. YAP/TAZ are potently activated by aberrant Wnt signalling9 and, consistently, MMTV-Cre;Apcfl/fl mice exhibited massive YAP stabilization (Fig. 5h). By 8 weeks of age, MMTV-Cre;Apcfl/fl mice displayed massive overgrowth of the mammary epithelium, with panductal and panlobular atypical hyperplasia and fibrosis (Fig. 5g and Supplementary Fig. 5d), expansion of the luminal layer and large discontinuities in the basal/myoepithelial layer (Figs. 5h, 5j and Supplementary Fig. 5e), collectively configuring a preneoplastic/early neoplastic scenario. Strikingly, all these lesions did not develop in MMTV-Cre;Apcfl/fl mice also bearing Yap and Taz conditional alleles (Figs. 5g-h), indicating that YAP/TAZ are required for epithelial overgrowth and development of these mammary lesions. We next tested whether established, already grown neoplastic lesions in MMTV-Cre;Apcfl/fl mice could be treated by administration of BET inhibitors. For this, we treated 8-week-old female mice (i.e., with an overt mammary gland phenotype) with a potent BET inhibitor (BAY-123809732, 75 mg/kg/week) for 6 weeks. Strikingly, at the end of treatment, lesions had greatly regressed due to cell death and epithelial remodelling with few remaining signs of mammary hyperplasia or fibrosis to an extent that the main mammary ducts returned to a normal appearance (Figs. 5i-j and Supplementary Fig. 5f). As a control, treatment of Apcfl/fl siblings (i.e., lacking Cre expression) was overtly well tolerated, and inconsequential for mammary gland homeostasis (Supplementary Figs. 5g-h). Thus, BET inhibition defines a vulnerability for YAP/TAZ-driven mammary tumors, in line with the results obtained in human TNBC cell lines.
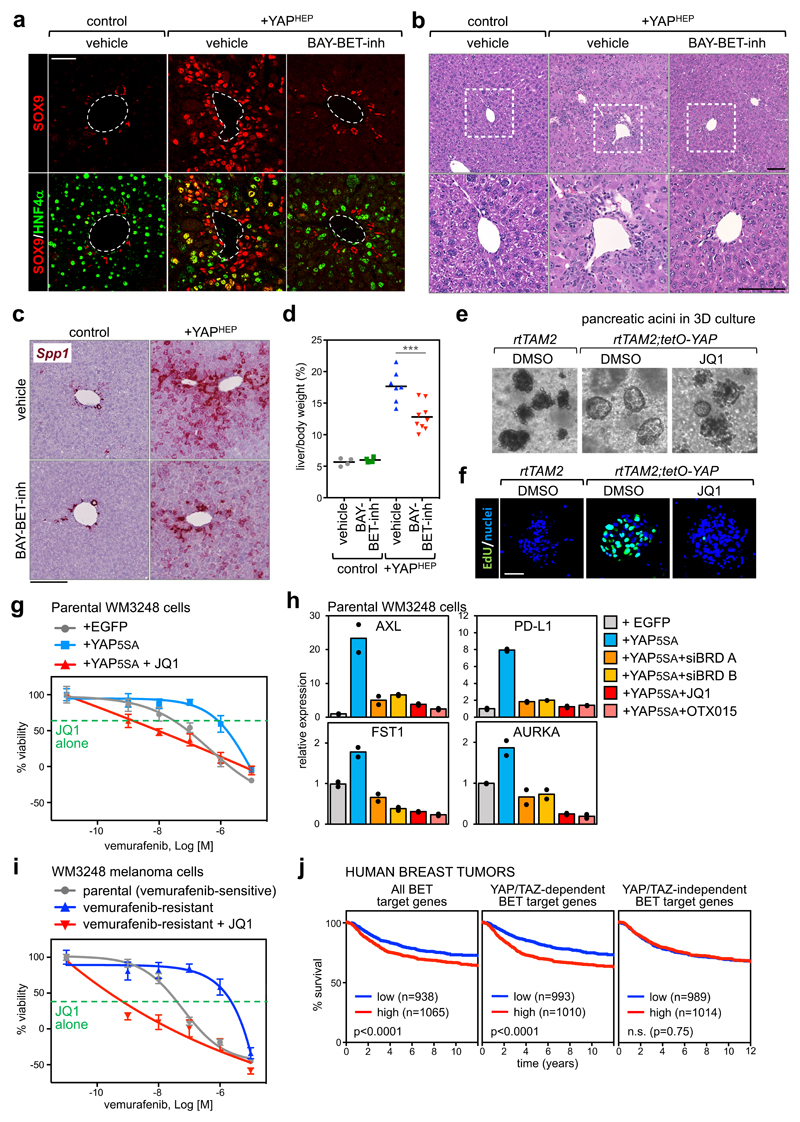
We next explored the functional dependence of YAP/TAZ on BET proteins in tissues other than the mammary gland. For this, we opted for the mouse liver, a classic model system for the study of YAP/TAZ function in vivo33,34. As previously reported35, YAP activation in hepatocytes of adult Albumin-CreERT2; R26-LSL-rtTA; tet-O-YAPS127A mice (Supplementary Fig. 6a-b) promotes a pre-neoplastic condition, whereby differentiated hepatocytes transdifferentiate into liver progenitor cells labelled by SOX9, a YAP/TAZ direct target in the liver (Fig. 6a); these cells then contribute to the generation of a "ductular reaction", defined by small ("oval") cells infiltrating the liver parenchyma (Fig. 6b). Remarkably, treatment with BET inhibitor (BAY-1238097) abolishes the appearance of both transdifferentiating cells (captured "in transition" by the co-expression of SOX9 and of the hepatocyte-specific HNF4α) and of ductular reactions (Fig. 6a-b and Supplementary Fig. 6c-d), without affecting the expression of the transgene (Supplementary Fig. 6b). By RT-qPCR and in situ hybridization, the oval-cell marker Osteopontin (Spp1) is induced in YAP transgenic livers but suppressed by concomitant treatment with BET inhibitor (Fig. 6c and Supplementary Fig. 6e). Histologically, BET inhibitor remarkably prevented the otherwise massive changes in the architecture of the liver parenchyma and reduced the appearance of proliferating Ki-67+ hepatocytes that are typically observed in vehicle-treated YAP transgenic livers (Fig. 6b and Supplementary Fig. 6h). Finally, liver overgrowth induced by YAP expression is inhibited by BET inhibitor (Fig. 6d).
Figure 6. Treatment with BET inhibitors blunts YAP/TAZ-driven responses in vivo.
a) Representative immunofluorescence (IF) for SOX9 and HNF4α in sections of control mouse livers or livers with hepatocyte-specific overexpression of YAPS127A (+YAPHEP), treated with vehicle or BAY-BET-inhibitor. Quantification of double positive cells is in Supplementary Fig. 6c. Scale bar is 50µm. IF was performed in n=4 control mice, n=4 +YAPHEP mice treated with vehicle, and n=4 +YAPHEP mice treated with BAY-BET-inhibitor.
b) Representative hematoxylin and eosin (H&E) staining of liver sections from control mice + vehicle (n=4), +YAPHEP mice + vehicle (n=7), +YAPHEP mice + BAY-BET-inh (n=9). Lower panels are magnifications of the portal area. Scale bars are 100μm. Administration of BET-inhibitor to control mice had no overt consequences on liver morphology or molecular features (see Supplementary Fig. 6f-g).
c) RNA in situ hybridization on liver tissues for Osteopontin (Spp1). Scale bar is 200µm. The experiment was performed in liver sections from 2 mice per each experimental group with similar results.
d) BAY-BET inhibitor impairs liver overgrowth induced by YAP expression. Data are liver/body weight ratios in all examined mice (control mice + vehicle, n=4; control mice + BAY-BET-inh, n=4; +YAPHEP mice + vehicle, n=7; +YAPHEP mice + BAY-BET-inh, n=9). Lines represent the mean of each group. ***p=0.00098 (unpaired t-test, two-tailed)
e) Representative images of pancreatic acini in 3D culture, derived from the indicated mice, after 3 days of culture in the presence of doxycycline to activate YAP expression. Treatment with BET inhibitor opposes YAP-induced ADM in organoids (see quantification in Supplementary Fig. 6i). The experiment was repeated four times with similar results.
f) EdU staining showing as treatment with BET inhibitor impairs cell proliferation. Scale bar is 50 μm. The experiment was repeated two times with similar results.
g) Viability curves of parental WM3248 cells (per se vemurafenib-sensitive) transduced with EGFP or YAP5SA, treated with increasing doses of vemurafenib (1nM to 10µM) with or without JQ1(1µM). The green line shows the effect of JQ1 alone (1µM). Data are mean + SD of n=8 independent wells (independently treated and evaluated). One representative experiment is shown; similar results were obtained in two additional independent experiments.
h) RT-qPCR for YAP/TAZ target genes showing upregulation upon YAP5SA overexpression in WM3248 cells and downregulation upon treatment with BET inhibitors (1µM, 24h) or depletion of BRD2/3/4 (siBRD mix A and B). Data are presented as individual data points (n=2 independent samples) + average (bar).
i) Viability curves of parental (vemurafenib-sensitive) WM3248 and vemurafenib-resistance WM3248, treated with increasing doses of vemurafenib (1nM to 10µM) with or without JQ1(1µM). The green line shows the effect of JQ1 alone (1µM). Data are presented as in g. One of three independent experiments (all with similar results) is shown.
j) Kaplan–Meier graph representing the probability of metastasis-free survival in breast cancer patients. Survival curves are significantly different when patients are stratified according to high or low expression of all BET target genes and common YAP/TAZ/BET target genes, but not when patients are stratified according to BET-only targets (Log-rank Mantel Cox Test).
Next, we focused on another endoderm-derived tissue, the pancreas, testing the functional interdependency of YAP/TAZ and BRD4 in early event of pancreatic tumor formation, that is acinar-to-ductal metaplasia (ADM)36. As recapitulated by ex vivo organoid cultures, transgenic expression of YAP in pancreatic acinar cells induces ADM, initially by turning acinar cells into ductal progenitors that only then start to proliferate18. Treatment with JQ1 opposes YAP-induced ADM in organoids and impairs ensuing cell proliferation (Figs. 6e-f and Supplementary Fig. i), as also validated by the expression levels of the ductal marker Krt19 and of Ccnd1 (Supplementary Fig. 6j). These data complement the data shown above for the mammary gland and expand the generality of the YAP/TAZ-BRD4 interplay to diverse gene expression programs and in distinct tissue contexts both in vitro and in vivo.
Beyond controlling tumour initiation and growth, YAP/TAZ endow cancer cells with the capacity to acquire resistance to chemotherapeutics and molecularly targeted drugs17. Melanoma cells bearing BRAF activating mutations are a point in case. It has been recently shown that resistance to BRAF inhibitors (such as PLX4032/vemurafenib) is rapidly installed in a YAP/TAZ-dependent manner in melanoma cells37,38. In line, YAP overexpression in BRAF-mutant but still vemurafenib-sensitive cells is sufficient to install chemoresistance37. We hypothesized that JQ1 could be used to revert YAP-induced drug resistance. Indeed, the growth of YAP-overexpressing BRAF-mutant melanoma cells was strongly inhibited by the combined exposure to vemurafenib and JQ1, which was per se poorly active (Fig. 6g and Supplementary Fig. 6k). Depletion of BET proteins in YAP-overexpressing cells or treatment with BET inhibitors impaired the expression of YAP direct target genes (Fig. 6h and Supplementary Fig. 6l); these genes include AXL13, which has been reported to be a pillar in resistance to BRAF inhibitors39, and the immune checkpoint ligand PD-L140. Next, we wanted to verify if JQ1 could re-sensitize cells that have spontaneously acquired resistance to vemurafenib after chronic exposure. We found that JQ1 could inhibit the activity of the TEAD luciferase reporter in vemurafenib-resistant cells (Supplementary Fig. 6m), and, in viability assays, the combined treatment with JQ1 sensitized resistant cells to low doses of vemurafenib, and impaired tumour cell viability to an extent that neither vemurafenib nor JQ1 could achieve when used individually (Fig. 6i and Supplementary Fig. 6n). The effect of JQ1 was phenocopied by combination of vemurafenib with two independent mixes of BRD siRNAs (Supplementary Fig. 6o). Collectively, these experiments indicate the requirement of BET proteins in maintaining YAP/TAZ-induced resistance to vemurafenib in BRAF mutant melanoma cells, and suggest that BET inhibitors might indeed prove useful to revert YAP/TAZ-dependent drug resistance in melanoma cells.
Our data imply that, in human tumors, a relevant fraction of the oncogenic functions of BRD4, and its associated transcriptional dependencies, may be in fact associated to the biology of YAP/TAZ. To put this idea to the test, we analysed a large dataset of human breast cancer patients41 containing clinical annotations and transcriptional profiling. We stratified patients according to the expression levels of BET-dependent genes, identified in MDA-MB-231 at the beginning of this study. Patients with a higher expression of this signature had a worse prognosis, as expected from the known oncogenic functions of BRD4 (Fig. 6j). Remarkably, however, when we split the list of BET target genes into YAP/TAZ-dependent and independent signatures, we found that only common YAP/TAZ/BET target genes retained predictive value, whereas BET targets that were not YAP/TAZ-dependent did not (Fig. 6j). This implies that BRD4 oncogenic properties substantially rest upon YAP/TAZ transcriptional responses. Similarly, analysis of a dataset of human hepatocellular carcinomas42 with signatures of shared YAP/TAZ/BET targets vs. BET targets that were not YAP/TAZ targets (derived from analyses of YAP/TAZ-depleted or JQ1-treated HepG2 cells, see details in methods), indicated that only the shared signature was meaningful for tumor aggressiveness (Supplementary Fig. 6p).
Finally, in a cohort of BRAF mutant melanoma patients treated with vemurafenib43, only expression of a signature of shared YAP/TAZ/BET-target genes in the primary tumor was associated to early disease progression (Supplementary Fig. 6q).
Discussion
A large body of evidence from several investigators has addressed various means by which BRD4 interacts with transcriptional regulators to nuance transcription44. The present discovery of a YAP/TAZ-BRD4 axis advances on this paradigm and may suggest new therapeutic opportunities for cancer and other diseases relying on YAP/TAZ transcriptional programs.
We have advanced on the molecular mechanisms underlying transcriptional addiction in tumor cells, identifying YAP/TAZ as relevant players in this phenomenon. The underlying molecular event is the physical and functional association between YAP/TAZ and BRD4: YAP/TAZ-bound enhancers recruit BRD4, leading to BRD4 accrual on their target promoters. We show that the YAP/TAZ/BRD4 complex confers a transcriptional advantage to a broad number of YAP/TAZ target genes; such transcriptional "edge" can be targeted by BET inhibitors in different cellular and tissue contexts with tumor preventive and suppressive effects.
The present results advance on the molecular definition of enhancer elements that are responsible for transcriptional dysregulations in cancer. Major emphasis in this respect has recently been placed on super-enhancers, although the molecular identity of the key TFs underlying the properties of these regulatory elements remains mysterious8. We found that super-enhancers largely consist of YAP/TAZ-occupied enhancers; this raises questions on the fact that a number of cancer-specific super-enhancers may simply represent the more noticeable "tip of the iceberg" of a larger set of YAP/TAZ-bound enhancers, that nonetheless display super-enhancer-like properties, as defined by strong enrichment of BRD4, higher expression level of regulated target genes and higher than average sensitivity to BET inhibitors.
We show that association of BRD4 to chromatin requires the dual interaction with YAP/TAZ and with acetylated histones. This is reminiscent of the previously proposed requirement of both bromodomain-dependent and -independent roles for BRD4 function5,25; such complex formation on chromatin likely involves cooperative associations akin to those postulated to stabilize distinct TFs bound to joined motifs at their modular cis-regulatory elements. We postulate that new therapeutics may be designed around the BET-YAP/TAZ interaction surfaces, including the YAP/TAZ WW-domain, the so far poorly understood YAP/TAZ transactivation domains and the YAP/TAZ-interacting domains of BRD4, that remain here unexplored.
Drugging YAP/TAZ is clearly a very challenging yet exciting goal for cancer research19, given the widespread and pervasive functions of YAP/TAZ in cancer cells, contrasting their dispensability for healthy tissues; BET inhibitors may start to fulfil this unmet need. From the other perspective, BET inhibitors are promising anticancer drugs, although drug resistance and identification of responsive patient subpopulations remain critical open issues3. Our results collectively indicate that the oncogenic effects of BET proteins are in close association to YAP/TAZ biology, potentially offering new perspectives on how to select patients that are more likely to receive benefit from BET inhibitors, alone or in combination with other drugs. Molecularly annotated datasets of patient cohorts treated with BET inhibitors are not yet available; however, here we show that patients stratified according to YAP/TAZ classifiers might in fact display differential sensitivity to these drugs, as the oncogenic potential of BET proteins, as inferred from gene expression signatures, appears to be essentially contained within genes addicted to BRD4 through YAP/TAZ.
YAP/TAZ are critical for inducing cell-fate plasticity in normal and tumor cells alike17. For example, they reprogram normal/differentiated mammary cells into mammary stem cells18, or more differentiated tumor cells into cancer stem cells45. The nature of the epigenetic barriers controlling these transitions remains unknown, but it is tempting to speculate that BRD4 availability, and potentially other factors assembled by YAP/TAZ on chromatin, may link YAP/TAZ function to permissive vs. restrictive chromatin states, as such guiding cell reprogramming or barring it. Thus, the YAP/TAZ-BRD4 connection may hold relevance in contexts other than cancer in which YAP/TAZ play essential roles, such as heart repair and tissue regeneration.
Online Methods
Reagents and plasmids
Doxycycline, OTX015 (SML1605), flavopiridol (F3055), human insulin, hydrocortisone and cholera toxin were from Sigma. Human EGF was from Peprotech. Vemurafenib-PLX4032 (A3004) and THZ1 (A8882) were from Apex Bio. JQ1 was from BPS Bioscience (27402). RG 108 (ab141013) was from Abcam.
pCDNA-FLAG-YAP vectors (wild-type or 5SA, siRNA insensitive) were described in Ref. 46. FLAG-YAP 5SA and FLAG-YAP wild type were subcloned in pBABE- retroviral plasmids. pBABE-blasti retroviral vectors was generated by replacing the puromycin resistance gene with the blasticidin resistance gene in pBABE-puro (Addgene plasmid #1764, a gift of H. Land, J. Morgenstern and R. Weinberg). Annealed oligos for shRNAs (shCO, shBRD4#1, shBRD4#2) were cloned into Tet-pLKO-puro inducible lentiviral vector.
pFlag-CMV2-BRD4 was from Addgene (#22304, a gift from Eric Verdin47). BRD4 CDS with a N-ter HA tag was subcloned in CSII-CMV-MCS-IRES2-Puro lentiviral plasmid. CSII-CMV-MCS-IRES2-Puro vector was generated by replacing the blasticidin resistance gene with the puromycin resistance gene in CSII-CMV-MCS-IRES2-Bsd, kindly provided by H. Miyoshi (RIKEN BSI, #RDB04385). pcDNA5-Flag-BRD4-WT (Addgene plasmid # 90331) and pCDNA5-Flag-BRD4-BD (Addgene plasmid # 90005) were a gift from Kornelia Polyak25.
The coding sequence of HAT-deficient mBRD4 was a gift from Dinah S. Singer28. Wild-type and HAT-deficient mBRD4 were subcloned in CSII-CMV-MCS-IRES2-Bsd lentiviral plasmid. GFP from PL-SIN-EOS-C(3+)-EiP (Addgene plasmid # 21313, a gift from James Ellis48) and FLAG-YAP 5SA were subcloned in pCW57.1 lentiviral plasmid to establish stable cell lines. pCW57.1 was a gift from David Root (Addgene plasmid # 41393) RSV-Flag-Brd2 (Addgene plasmid # 86614) was a gift from Mario García-Domínguez49. For GST-pull down experiments WT full-length YAP was cloned in pGEX-4T-3. GST-TAZ constructs were described in Ref. 50. 8xGTIIC-lux was described in Ref. 51; pCMV-LacZ was from Clontech. All constructs were confirmed by sequencing.
Cell lines, treatments and transfections
MDA-MB-231 cells were from ICLC. MDA-MB-231 cells were cultured in DMEM/F12 (Life Technologies) supplemented with 10% FBS, glutamine and antibiotics. For YAP overexpression, cells were transduced with pBABE-puro-FLAG-YAP. For BRD4 overexpression, cells were transduced with CSII-CMV-MCS-IRES2-Puro-HA-hBRD4, or CSII-CMV-MCS-IRES2-Bst-FLAG-mBRD4 WT or HAT-deficient (or empty vector as control). For BRD4 knockdown, cells were transduced with Tet-pLKO-puro-shBRD4 (#1 or #2) or Tet-pLKO-puro-shCO. Interfering sequences were: shCO (CCTATTTATGAGGCGACGGAA), shBRD4#1 (GCCTGGAGATGACATAGTCTTA), shBRD4#2 (ACAGTGACAGTTCGACTGATGA). After infection with doxycycline-inducible vectors, MDA-MB-231 cells were maintained in media supplemented with Tet-approved FBS (Clontech), to reduce background expression of the transgene in the absence of doxycycline.
MCF10A cells (from ATCC) were cultured in DMEM/F12 (Life Technologies) with 5% horse serum, glutamine and antibiotics, freshly supplemented with insulin, human EGF, hydrocortisone, and cholera toxin. For gene expression and ChIP experiments, MCF10A cells were seeded at high density (75.000 cells/cm2) and harvested after 48 hours. For YAP overexpression, cells were transduced with pBABE-blasti-FLAG-YAP5SA (or empty vector as control). For BRD4 knockdown, cells were further transduced with Tet-pLKO-puro-shBRD4 (#1 or #2) or Tet-pLKO-puro-shCO.
HEK293T cells (from ATCC) were maintained in DMEM supplemented with 10% FBS (Life Technologies), glutamine and antibiotics. MDA-MB-231, MCF10A and HEK293T were authenticated by DSMZ service.
BT-20 cells were obtained from the ATCC (HTB-19) and cultured in DMEM supplemented with 10%FBS (Life Technologies), glutamine and antibiotics. SUM149PT and SUM-159PT were kindly provided by Dr. S. Ethier and cultured in F12 with 5%FBS (Life Technologies), glutamine and antibiotics, freshly supplemented with 5μg/ml Insulin and 1μg/ml hydrocortisone. Hs578T cells were obtained from ICLC and mainteined in DMEM with 10%FBS (Life Technologies), glutamine and antibiotics, freshly supplemented with 10μg/ml Insulin.
M229 and M229-R5 cells (a gift from JC Marine, Leuven Center for Cancer Biology) were cultured in DMEM (Life Technologies), 10 % FBS, glutamine and antibiotics. SKMEL28 and SKMEL28-R2 cells (a gift from J.Kim, KAIST) were cultured in MEM (Life Technologies), 10%FBS, glutamine and antibiotics. WM3248 and WM3248-R6 cells (a gift from J.Kim, KAIST) were cultured in MCBD (Sigma), 2%FBS, 1.68mM CaCl2. M229-R5 were cultured in the presence of 1 µm PLX4032, SKMEL28-R2 and WM3248-R6 were maintained in 2 µm PLX4032. For YAP overexpression, SKMEL28 and WM3248 were transduced with pCW57.1-Flag-YAP5SA (or EGFP as control) and maintained in presence of doxycycline. All cell lines were routinely tested for mycoplasma contamination and were negative. None of the cell lines used in this study is present in the database of commonly misidentified cell lines.
Drugs were resuspended in DMSO and used at a final concentration of 1μM for 24h, unless differently specified. DNA transfections were performed with TransitLT1 (Mirus Bio) according to manufacturer instructions. siRNAs were transfected with Lipofectamine RNAi-MAX (Life Technologies) in antibiotics-free medium according to manufacturer instructions. Cells were harvested 48h after transfection with YAP/TAZ siRNAs and 72h after transfection with siRNAs targeting BET-proteins. YAP/TAZ siRNA mix 1 (siYT1) contained the following siRNAs: GACAUCUUCUGGUCAGAGAdTdT + ACGUUGACUUAGGAACUUUdTdT; YAP/TAZ siRNA mix 2 (siYT2): CUGGUCAGAGAUACUUCUUdTdT + AGGUACUUCCUCAAUCACAdTdT. BRD2/3/4 siRNA mix A (siBRD A) contained the following siRNAs: GUAGCAGUGUCACGCCUUAdTdT + CCUGCCGGAUUAUCAUAAAdTdT + GAGGACAAGUGCAAGCCUAdTdT; BRD2/3/4 siRNA mix B (siBRD B): GUAGCAGUGUCACGCCUUAdTdT + GCCCGUGGACGCAAUCAAAdTdT + GCGUUUCCACGGUACCAAAdTdT. Negative control siRNA was purchased from Qiagen (cat. 1027280, AllStars Negative Control siRNA).
Retroviral and lentiviral infections were carried out as in Ref. 50,52.
ChIP-MS
Cells were cross-linked with 1% formaldehyde (Sigma) in culture medium for 10min at room temperature followed by 5min treatment with 0.125M Glycine/PBS. Cells were harvested and incubated in Lysis Buffer 1 (50mM HEPES, pH7.5, 10mM NaCl, 1mM EDTA, 10% Glycerol, 0.5% NP-40, 0.25% Triton X-100, with protease inhibitors; 20 min at 4°C), then in Lysis Buffer 2 (10mM Tris-HCl pH8, 200 mM NaCl, 1mM EDTA, 0.5 mM EGTA, plus protease inhibitors; 10 min at RT); finally nuclei were resuspended in Lysis Buffer 3 (10mM Tris-HCl pH8, 200 mM NaCl, 1mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, plus protease inhibitors) and sonicated using a Branson Sonifier 4500D (5x1min pulse, duty cycle 0.5, 30% amplitude). Immunoprecipitation was performed by incubating cleared extracts (corresponding to 2x106 cells) with 20 µg of antibody overnight at 4°C (anti-YAP: EP1674Y, Abcam; anti-TAZ: HPA007415, Sigma; pre-immune rabbit IgGs: I5006, Sigma). Antibody/antigen complexes were recovered with ProteinG-Dynabeads (Invitrogen, 5 μL Dynabeads/1 μg antibody) for 3h at 4°C. The precipitates were washed twice in low salt wash buffer (20mM Tris-HCl pH8, 150mM NaCl, 0.1 % SDS, 0.5% Triton X-100 and 2mM EDTA), twice in high salt wash buffer (20mM Tris-HCl pH8, 2mM EDTA, 300mM NaCl, 0.1% SDS and 0.5% TritonX), and once with 100 mM Tris-HCl. Precipitates were eluted in 7.5%SDS, 200mM DTT and incubated at 37°C for 30min to revert crosslinks. Upon alkylation with iodoacetamide (IAA), proteins were purified with SP3 beads as previously described (PMID 25358341), resuspended in 50mM ammonium bicarbonate and digested with 300 ng trypsin 16h at 37°C. Peptides were subjected to SP3 cleanup and they were eluted in 0.1% TFA. Samples were analyzed on an Orbitrap Fusion mass spectrometer (Thermo Fisher).
Co-immunoprecipitation of endogenous nuclear proteins
Cells were rinsed twice with ice-cold HBSS and incubated with ice-cold hypotonic buffer (2 x 1min, 20mM HEPES, 20% Glycerol, 10mM NaCl, 1.5mM MgCl2, 0.2mM EDTA, 0.1% NP40, freshly supplemented with 1 mM DTT, and protease and phosphatase inhibitor cocktails). Nuclei were then harvested by scraping in hypertonic buffer (hypotonic buffer + 500mM NaCl, 400 µl/60 cm2) and disrupted by sonication in a water-bath sonicator. Nuclear lysates were cleared by centrifugation and quantified by Bradford. For immunoprecipitation, extracts were diluted to 140mM NaCl and incubated 4h at 4°C with magnetic beads (Dynabeads Protein A or G, Invitrogen) preloaded with specific primary antibodies. Immunocomplexes were then washed in binding buffer four times; finally, beads were resuspended in SDS sample buffer.
Antibodies used for immunoprecipitation: anti-BRD4 (E2A7X, CST); anti-YAP1 (13584-1-AP, Proteintech); anti-WWTR1 (HPA007415, Sigma); anti-FLAG (clone M2, A8592, Sigma); normal rabbit IgG (I5006, Sigma).
GST Pull-Down
GST-YAP and GST-TAZ were produced in E.coli and immobilized on glutathione sepharose 4B (GE/Sigma). Resins were blocked in 5%BSA/PBS and then incubated with full lenght recombinant BRD4 (cat. RD-21-153, Cambridge Bioscience, 500ng/reaction) in binding buffer (20mM Hepes KOH, 20% glycerol, 100mM KCl, 1.5mM MgCl2, 0.2mM EDTA, 0.1% NP-40, 1mM DTT) o.n. at 4°C. Resins were then washed in binding buffer (4 x 5min) and resuspended in SDS sample buffer for subsequent analysis)
Western Blot
Cells were harvested in Lysis Buffer (20 mM HEPES (pH 7.8), 100 mM NaCl, 5% Glycerol, 0.5% NP40, 5mM EDTA, 1 mM DTT, phosphatase and protease inhibitors) and lysed by sonication.
About 10mg of liver were mechanically disrupted, resuspended in 500µl of Lysis Buffer and lysed by sonication. Extracts were quantified with Bradford method. Samples were run in 4-12% Nupage-MOPS acrylamide gels (ThermoFisher) and transferred onto PVDF membranes by wet electrophoretic transfer. Blots were blocked with 0.5% non-fat dry milk and incubated overnight at 4°C with primary antibodies. Secondary antibodies were incubated 1 hour at room temperature, and then blots were developed with chemiluminescent reagents. Images were acquired with Image Quant LAS 4000 (GE healthcare). In vitro experiments were performed three times with similar results.
Antibodies used for Western blot: anti-YAP/TAZ (sc-101199), anti-HA (Y-11, sc-805), and anti-BRD2 (D-2, sc-514102) from Santa Cruz; anti-GAPDH (MAB347, Millipore); anti-TEF1 (clone 31, 610923, BD Biosciences); horseradish-peroxidase-conjugated anti-FLAG (clone M2, A8592) and anti-BRD4 (HPA015055) from Sigma; anti-H3 (ab1791) and anti-RNA polymerase II CTD repeat YSPTSPS antibody (ab817) from Abcam.
Immunofluorescence of cultured cells
Cells were cultured on glass slides and treated with 1µM JQ1 or OTX015 for 24h. Cells were fixed 10 min at room temperature with 4% PFA in PBS, permeabilized 10 min at RT with PBS+0.3% Triton X-100, blocked in 10% Goat Serum (GS) in PBS + 0.1% Triton X-100 (PBST) for 1h, and then incubated with anti-YAP/TAZ (sc-101199; SantaCruz) primary antibody diluted in 2% GS in PBST, overnight at 4°C. After four washes in PBST, samples were incubated with secondary antibody (Alexa 488, 1:200 in 2% GS in PBST) for 2h at room temperature. Nuclei were counterstained with ProLong-DAPI (Molecular Probes, Life Technologies). Images were acquired with a Leica TCS SP5 confocal microscope equipped with a CCD camera.
In situ proximity ligation assay
HEK293T cells were seeded on fibronectin-coated glass chamber slides and transfected with pFlag-CMV2-BRD4, pCS2-HA-BRD4 or empty pCS2+ as negative control. After 24 hours, cells were fixed in 4% PFA for 10 min at RT. In situ PLA was performed with DuoLink In Situ Reagents (Sigma) according to manufacturer’s instructions. Primary antibodies used in the PLA are: mouse anti-HA (F-7, sc-7392, SantaCruz), mouse anti-TEF1 (610923; BD Biosciences), rabbit anti-FLAG (F-7425; Sigma), rabbit anti-YAP1 (EP1674Y, abcam), rabbit anti-WWTR1 (HPA007415, Sigma). Images were acquired with a Leica TCS SP5 confocal microscope equipped with a CCD camera; for each field, a Z-stack was acquired; images were processed using Volocity software (PerkinElmer). We verified that the fraction of nuclei with positive PLA signal corresponded to the fraction of transfected cells (determined by immunofluorescence for FLAG or HA).
Luciferase reporter assays
Luciferase assays were performed in M229 and M229-R5 cells with the TEAD reporter 8xGTIIC-Lux. Luciferase reporter (25 ng/cm2) was transfected together with CMV-β-gal (25 ng/cm2) to normalize for transfection efficiency with CPRG (Roche) colorimetric assay. DNA content in all samples was kept uniform by adding pBluescript plasmid up to 250 ng/cm2. Cells were plated at 20% confluence (day 0) and the following day (day1) transfected with DNA. After 6h, cells were treated with different doses of JQ1 (1nM, 10nM, 0.1µM and 1µM). Cells were harvested after 24h of treatment (day2). Firefly luciferase activity was measured with an Infinite F200PRO plate reader (TECAN), using Luciferin (Sigma) as substrate. Data are presented as firefly/β-gal activity. Each sample was transfected in duplicate and each experiment was repeated at least three times independently with similar results.
Viability assays
Cells were seeded in 96-well plates (4000 cells/well) one day before treatment with drugs or transfection with siRNAs. Cells fixed after 72h with a crystal violet solution (0.05% w/v Crystal violet, 1% formaldehyde, 1% methanol in PBS) for 20 min at RT; stained cells were washed with water until a clear background was visible, and air-dried. Crystal violet was extracted with 1% SDS (w/v in ddH2O, 100 μl/well) and absorbance at λ=595 nm was measured with an Infinite F200PRO plate reader (TECAN). 8 wells were analyzed for each experimental condition; data are presented as mean + SD. Data are presented as % viability compared to control cells (treated with DMSO or transfected with siCO), where absorbance at the beginning of treatment was set as 0%, and absorbance at the end of experiment was set as 100%. Each experiment was performed at least twice, with similar results.
Colony formation assay in soft agar
104 MCF10A cells and 3x104 MDA-MB-231 cells were resuspended in complete growth medium with 0.35% agarose (Invitrogen) and were layered onto 0.5% agar beds in six-well plates. Complete medium was added on top of cells and was replaced with fresh medium twice a week 3 weeks. Complete medium contained 1µg/ml doxycycline to activate the expression of shRNAs, where necessary. The indicated doses of JQ1 were added to culture starting 24h after seeding (unless differently specified). Assays were conducted in triplicate. For MCF10A cells, all colonies in a well were counted. For MDA-MB-231 cells, colonies were counted in 6 fields of each samples and the average number of colonies/field was calculated for each sample. Experiments contained three independent samples for each condition and were performed three times, with similar results.
Isolation and culture of pancreatic acini
Primary pancreatic acini were isolated as described in Panciera et al. 201618 from rtTAM2; colYAP mice, or from rtTAM2 littermates as control (both male and females, 6-8 weeks old). Acini were seeded in neutralized rat tail collagen type I (Cultrex)/acinar culture medium (1:1)53 and overlaid with acinar culture medium (Waymouth’s medium [Life Technologies] supplemented with 0.1% FBS [Life Technologies], 0.1% BSA, 0.2 mg/ml soybean trypsin inhibitor [SBTI], 1× insulin-transferrin-selenium-ethanolamine [ITS-X] [Life Technologies], 50 μg/ml bovine pituitary extract [BPE] [Life Technologies], 1μg/ml dexamethasone [Sigma], and antibiotics) supplemented with 0.5μg/ml doxycycline and DMSO or 10μM JQ1, as indicated. ADM events were counted 2-4 days after seeding. For EdU incorporation, 20μM EdU was added to culture medium for 90 min, then collagen cushions containing acini/ducts were extensively washed in PBS and fixed in 4% PFA for 20 min at RT. EdU staining was performed with Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermo Fisher Scientific), according to manufacturer instructions. Total RNA was extracted with TRIzol Reagent (Invitrogen).
Mice
Animal experiments were performed adhering to our institutional guidelines as approved by OPBA (University of Padua) and the Italian Ministry of Health. All experimental mice used in this study were mixed strains and more than 6 weeks old; for mammary gland experiments we used exclusively female mice. Transgenic lines used in the experiments were kindly provided by: Duojia Pan54 (Yap1fl/fl); Alan R. Clarke55 (Apcfl/fl); F. Camargo33 (tetO-YAPS127A); Pierre Chambon56 (Albumin-CreERT2). Tazfl/fl and double Yapfl/fl; Tazfl/fl conditional knock-out mice were as described in Ref. 9. MMTV-Cre57 (stock #003553) and R26-LSL-rtTA58 (stock #005670) were purchased from The Jackson Laboratory. Yap, Taz and Apc conditional knockouts were intercrossed with MMTV-Cre mice to obtain the different genotypes. Mice carrying Albumin-CreERT2, R26-LSL-rtTA and tetO-YAPS127A alleles were intercrossed to obtain Albumin-CreERT2; R26-LSL-rtTA/+; tetO-YAPS127A mice. Albumin-CreERT2; R26-LSL-rtTA/+ littermates were used as control. Animals were genotyped with standard procedures and with the recommended set of primers.
Control (Apcfl/fl, n=5) or MMTV-Cre; Apcfl/fl (n=5) mice were administered BAY-BET-inh (BAY-1238097) by intraperitoneal injection for 6 weeks, starting at 8 weeks of age (15mg/kg, 5inj/week). Control mice were injected with vehicle (0.9% NaCl, pH 4). Harvesting, processing and stainings on mammary glands were performed as in Ref. 18.
For the induction of the recombination in the liver, control (Albumin-CreERT2; R26-LSL-rtTA/+) mice and Albumin-CreERT2; R26-LSL-rtTA/+; tetO-YAPS127A mice received 1 intraperitoneal injection per day of 3 mg Tamoxifen (Sigma) dissolved in corn oil (Sigma) during 2 consecutive days. After 2 weeks, mice were administered doxycycline in drinking water for 10 days, during which they also received BAY-BETinh (15 mg/kg, 5 inj/week) or vehicle by intraperitoneal injections, as indicated in the corresponding Figures.
For validation of the Albumin-CreERT2 driver (Figure S6A), Albumin-CreERT2; R26-LSL-YFP/+ mice were injected with 3 mg Tamoxifen (Sigma) per day dissolved in corn oil (Sigma) during 5 consecutive days and were sacrificed after 2 weeks.
Immunostainings and immunofluorescences of liver tissue
Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded tissue sections as described in 45. Primary anti-Ki67 polyclonal antibody (clone SP6; M3062, Spring Bioscience) was from Spring Bioscience.
Immunofluorescences on PFA-fixed paraffin embedded tissue slices was performed as in 45. Primary antibodies were anti-cytokeratin (wide spectrum screening, ZO622; Dako), anti-HNF4α (sc-6556; Santa Cruz Biotechnology), anti-SOX9 (AB5535; Millipore) and anti-GFP (ab13970; abcam). Samples were counterstained with ProLong-DAPI (Molecular Probes, Life Technologies) to label cell nuclei. Confocal images were obtained with a Leica TCS SP5 equipped with a CCD camera. Bright field images were obtained with a Nanozoomer Scanner 2.0RS (Hamamatsu).
RNA in situ hybridization
Tissue sections (formalin-fixed paraffin-embedded) were processed for RNA in situ detection using the RNAscope Duplex Detection Kit (Chromogenic) according to the manufacturer’s instructions (Advanced Cell Diagnostics). RNAscope probe used was Spp1 (Osteopontin; NM_001204201.1, region 2-1079), which was detected using the Fast Red detection reagent.
RT-qPCR
Total RNA extraction from cells and tissue was performed with NucleoSpin 8 RNA Core Kit (Macherey-Nagel, REF.740465.4) using an automated system (Freedom EVO, Tecan). Contaminant DNA was removed by rDNase Set (Macherey-Nagel, REF.740963). Pancreatic acini were harvested in TriPure (Roche) for total RNA extraction.cDNA synthesis was carried out with oligo(dT)-primed MuMLV Reverse Trascriptase (Invitrogen).
Gene expression analyses by quantitative real-time PCR (RT-qPCR) were carried out with QuantStudio 5 thermal cycler (ThermoFisher). Experiments were performed at least three times. Expression levels are calculated relative to GAPDH.
Human primer pairs are:
AURKA: GCCCTCTGGGTAAAGGAAAG, GCCGAAGGTGGGACTGTAT;
AXL: CACCAGCAAGAGCGATGTGT, CGGTCCTGGGGATTTAGCTC;
BRD4: GAGGCAGACCAACCAACTGC, CAGGGAGGTTCAGCTTGACG;
CCNA2: TTTGATAGATGCTGACCCATACC, ATGCTGTGGTGCTTTGAGGT;
CDC6: CGCAAAGCACTGGATGTTT, CAACCCTCTTGGGAATCAGA;
CDCA5: CCTGAAATCTGGCCGAAGAC, CTCCTGCGAGGTGATTGGAC;
E2F3: GAACAAGGCAGCAGAAGTGC, CCCCCATCCTCAGACAGACT;
FST1: CCGGTGTTCCCTCTGTGATG, TCCTCTTCCTCGGTGTCTTCC;
GAPDH: CTCCTGCACCACCAACTGCT, GGGCCATCCACAGTCTTCTG;
GINS1: TCAACGAGGATGGACTCAGA, AAGCAAGCGGTCATACAGGT;
KIF23: AGTTCAGGCTCCCTTGGATG, TCTGTTCCCTTCTGCTCTGGTC;
MCM3: TGGGTTGTGCCGAGAGAGTT, CCAACATTCCTCGCCTTCAG;
PD-L1: GGTGCCGACTACAAGCGAAT, GGTGACTGGATCCACAACCAA;
PLAU: CGCCACACACTGCTTCATT, CAAACTTCATCTCCCCTTGC;
RRM2: TGGCTCAAGAAACGAGGACTG, TGAACATCAGGCAAGCAAAATC;
TOP2A: CGCCGCAAAAGGAAGCCATC, TTTTGCCCGAGGAGCCACAG;
TUBB1: GTGGCCTCAAGATGGCAGTC, TCTCAGCCTCGGTGAACTCC.
Mouse primer pairs are:
Gapdh: ATCCTGCACCACCAACTGCT, GGGCCATCCACAGTCTTCTG;
Rn18s: TGTCTCAAAGATTAAGCCATGC, GCGACCAAAGGAACCATAAC;
Sox9: AGGCCACGGAACAGACTCAC, CCCCTCTCGCTTCAGATCAA;
Spp1: CTGGTGCCTGACCCATCTCA, TCATCCGAGTCCACAGAATCC;
YAP S127A: ACAGCATGTTCGAGCTCATG, TGTGACGTTCATCTGGGACA;
Krt19: GGTCAGTGTGGAGGTGGATTC, GACTTCGGTCTTGCTTATCTGGA;
Ccnd1: GACCTTTGTGGCCCTCTGTG, AAAGTGCGTTGTGCGGTAGC.
RNA-seq
Cells were harvested by RNeasy Mini Kit (Qiagen) for total RNA extraction and contaminant DNA was removed by RNase-Free DNase Set (Qiagen).
RNA-seq libraries for deep-sequencing were prepared with the Illumina TruSeq Standard Total RNA with Ribo-Zero GOLD kit, and sequencing was performed with Illumina HiSeq2500. About 40M reads/sample were obtained. Raw reads were aligned using TopHat59 (version 2.0.5) to build version hg19 of the human genome. Counts for UCSC annotated genes were calculated from the aligned reads using HTSeq60 (version 0.6.0). Normalization and differential analysis were carried out using edgeR package61 and R (version 3.0.0). Raw counts were normalized to obtain Counts Per Million mapped reads (CPM) and Reads Per Kilobase per Million mapped reads (RPKM). Only genes with a RPKM greater than 1 in at least 2 samples were retained for differential analysis. Genes were considered differentially expressed with a Benjamini-Hochberg FDR less than or equal to 1% and a fold change equal or lower than 0.75. The 10th percentile, first quartile, median, third quartile and 90th percentile are plotted in box and whiskers graphs. Fold changes were calculated as the ratio of RPKM.
Gene Ontology Analysis
Gene Ontology (GO) analyses were performed on all the genes expressed in MDA-MB-231 cells (RNA-seq RPKM >= 1) using Enrichr (http://amp.pharm.mssm.edu/Enrichr/) 62,63. Genes annotated with GO terms related to cell cycle (e.g. GO:0000278~mitotic cell cycle, GO:0008283~cell proliferation), DNA replication and repair (e.g. GO:0006260~DNA replication, GO:0006281~DNA repair), mitosis (e.g. GO:0000279~M phase and GO:0007067~mitosis), cytokinesis (e.g. GO:0000281~mitotic cytokinesis and GO:0051301~cell division), microtubule cytoskeleton (e.g. GO: 0007020~microtubule nucleation and GO:0007051~spindle organization), and telomere maintenance (e.g. 0000723~telomere maintenance) were included in a single category named “cell proliferation”.
ChIP-seq and ChIP-qPCR
ChIP was performed as previously described13. Briefly, cells were crosslinked with 1% formaldehyde (Sigma) in culture medium for 10 min at room temperature, and chromatin from lysed nuclei was sheared to 200–600 bp fragments using a Branson Sonifier 4500D.
For ChIP-seq, ~200 μg of chromatin were incubated with 10 μg of antibody overnight at 4°C (anti-BRD4: A301-985A, Bethyl-Lab; anti-PolII: ab817, abcam; normal rabbit IgG: Sigma; normal mouse IgG: Santa Cruz). For ChIP-seq of H3K122ac, ~50 μg of chromatin were incubated with 10 μg of anti-Histone H3 (acetyl K122) (ab33309, abcam). Antibody/antigen complexes were recovered with ProteinA-Dynabeads (Invitrogen, 5 μl Dynabeads/1 μg antibody) for 2h at 4°C (1h for anti-H3K122ac). The precipitates were washed and eluted in 50mM Tris-HCl pH 8, 1%SDS, 1mM EDTA for 20 min at 65°C. Chromatin was decrosslinked, treated with RNaseA and Proteinase K and DNA was purified by QIAquick PCR Purification Kit (Qiagen, 28106). Enrichment of target sequences was checked by qPCR, then libraries were generated with the Ovation Ultra Low Library Prep Kit (NuGEN) according to manufacturer's instructions. Sequencing was performed on an Illumina HiSeq 2500 platform.
For ChIP-qPCR, 100 μg of sheared chromatin and 3–5 μg of antibody were used.
For ChIPs of YAP, anti-YAP1 (ab52771) from Abcam was used. For ChIPs of H3K122ac, at least 10 μg of chromatin were incubated with 2 μg of antibody. Quantitative real-time PCR was carried out with QuantStudio 5 thermal cycler (ThermoFisher); each sample was analysed in triplicate and was presented as mean + SD. The amount of immunoprecipitated DNA in each sample was determined as the fraction of the input [amplification efficiency ^ (Ct INPUT - Ct ChIP)], and normalized to IgG control.
Duplicate experiments were performed at least twice with similar results.
Primers used are:
ANKRD1: AAAAAGGGCAGTGATGTGGTG, GAAGAGGGAGGGGAGGACAA;
CCNA2 Enh: ACAGAAGGGGAGCGACTGG, CCCACCGTTTTCACTTTTTC;
CDC6 Enh: GCTGGGCATCACAGTCTTGG, GGCATGGCTGGGTGACTC;
CDC6 TSS: CAAGGCGAAAGGCTCTGTGA, CAAGCCCCTGAACAAACTGC;
CDCA5 Enh: AGTGCTGCTCCCCCACACTA, CCTGCAAGGAAAGAGCTGGA;
CDCA5 TSS: GCGTTCGCCTCCCAGACATA, TTCCGCTTCCTTTCCCGCAG;
CYR61: CACACACAAAGGTGCAATGGAG, CCGGAGCCCGCCTTTTATAC;
ETS1 Enh: CCCTTGTCCCAACACACACA, AAAACTGTCTCCACCTCCTAATGC;
E2F3 TSS: GCGTAAACCGTATCCCTTCA, CAAAAATAATCGGGGCTCTGG;
FOSL1 TSS: TACACGGCTGCTGGGTTC, GGTGGAGCCTGGAGGTGAC;
GAPDH TSS: TCGCTCTCTGCTCCTCCTGT, GTTTCTCTCCGCCCGTCTTC;
GINS1 Enh: CCCCAAAAGTGTCCATGACC, CAGGATCACCCCCATCTCAA;
GINS1 TSS: GCCGAGAGCCCAGATACCAT, CGTTGAAGGCAGGCAGTAG;
HBB: GCTTCTGACACAACTGTGTTCACTAGC, CACCAACTTCATCCACGTTCACC;
KIF23 TSS: TTGGCCCGTTTGAAATGCGC, ACGTTAGGACCGGCAGCAAG;
MCM3 Enh: AGTTGGGATAGGCGGAGACC, GCAGGTGGGGCTTGTTTAGG;
MCM3 TSS: TCCCGCCACCAAAGGTTAC, AGCGGAAAACCCGAAGAAGA;
PLAU Enh: GCTGGCTTCACCCTTCACAC, ATGGGGCAGACGGACTCTTC;
PLAU TSS: CCTCAGTCCAGACGCTGTTG, CTCCCTCCCCTGTCTTGCAG;
RRM2 Enh: AGGGCTGTTGCTCACCTCTTG, GCATTCTTCCTGGCTCTTTGTG;
RRM2 TSS: TTAAAGGCTGCTGGAGTGAGG, CGGAGGGAGAGCATAGTGGA;
TMEM200B TSS: AAAGGGAGGGCGAGGGAGAA, CAGCGCGGTGGTTCTTTAGGA;
TOP2A Enh: CCCCACCCAGACAGGAAA, TGAGGCAGGGCAGTTTAGAA;
TUBB Enh: ACTGGCTTCGGCTGTGTCTT, AATAAAGGATGTGGGGAGCA;
TUBB TSS: TTCTTGGCAGGCACATTTTG, GACCGTTTCCGCATCTCTCT.
Analysis of ChIP-seq data
Raw reads were aligned using Bowtie64 (version 0.12.7) to build version hg19 of the human genome retaining only uniquely mapped reads. Redundant reads were removed using SAMtools.
The lists of H3K4me1-, H3K4me3-, H3K27ac-, YAP- and TAZ-enriched regions (peaks) in MDA-MB-231 cells were already described in Ref. 13.
The overlap of peaks from different ChIP-seq experiments was determined using the BEDTools2 suite 65.
Definition of promoters and enhancers
To define promoter regions in the genome of MDA-MB-231 cells, we first defined a list of 2 kb-wide regions centered on each transcription start site (TSS) mapped in the build version hg19 of the human genome (downloaded from the UCSC genome browser, Ref. 66). We then obtained a list of promoter regions by including only the TSSs overlapping with H3K4me3-enriched regions.
Active enhancers were defined as non-promoter regions displaying enrichment for H3K4me1 and H3K27ac. For this, we first defined a list of enhancers based on H3K4me1 peaks, purged of those overlapping with promoter regions. The width of each enhancer was set to the same of the corresponding H3K4me1 peak. From this list we generated a list of active enhancers, by including only the enhancer elements overlapping with peaks for H3K27ac.
Annotation of active enhancers to target genes
Active enhancers were annotated using the chromatin interactions reported in Supplementary data 2 of Jin et al. 201367, derived from a high-resolution Hi-C experiment; the data sheets report the genomic locations of all target peaks interacting with more than 10,000 anchors located at gene promoters. Active enhancers overlapping with these target peaks were assigned to the corresponding interacting promoter region.
Annotation of YAP/TAZ binding regions to enhancers and target gene promoters
YAP/TAZ-bound enhancers were defined as active enhancers overlapping with both YAP and TAZ peaks. Similarly, YAP/TAZ-bound promoters were defined as promoter regions (as defined above) overlapping with both YAP and TAZ peaks. Gene promoters associated with YAP/TAZ-bound enhancers through DNA looping were defined as promoter regions associated with at least one YAP/TAZ-bound enhancer on the bases of Hi-C data (as in Ref. 13). YAP/TAZ direct target genes were defined as those whose promoters are associated with YAP/TAZ-bound enhancers or are directly binding YAP/TAZ.
Calculating normalized read count and density at enhancers and promoters
ChIP-Seq reads aligning to each cis-regulatory region (active enhancers and promoters for BRD4 ChIP-seq experiments and promoters only for PolII and H3K122ac ChIP-seq experiments) were calculated using the BEDTools2 suite 65. Total number of reads was normalized to the total number of million mapped reads, producing normalized read counts in units of reads per million mapped reads (RPM). Normalized read density in units of reads per kilobase per million mapped reads (RPKM) was determined by dividing the total RPM count by the width of each cis-regulatory region in kilobases. Relative occupancy was calculated as the ratio of RPKM. The 5th percentile, first quartile, median, third quartile and 95th percentile are plotted in box and whiskers graphs.
Super-enhancers identification and annotation to target genes
The list of super-enhancers was obtained following the method described by Ref. 68, using total normalized reads of BRD4 from control (DMSO-treated) MDA-MB-231 cells, subtracted of normalized counts from IgG ChIP-seq (background) of the same cells. Briefly, we first ranked all active enhancers by increasing total background-subtracted normalized reads of BRD4 (x-axis), and plotted the total background-subtracted normalized counts of BRD4 in units of total rpm (y-axis) (see Figure S4F). This plot revealed a clear point in the distribution of BRD4 at active enhancers where the occupancy signal began increasing rapidly. To geometrically define this point, we found the x axis point for which a line with a slope of 1 was tangent to the curve. We define enhancers above this point to be super-enhancers, and enhancers below that point to be typical enhancers. Annotation of super-enhancers to target genes was carried out on the bases of Hi-C data as detailed before for all active enhancers. Genes were considered as associated to super-enhancers if their promoters were associated at least with one super-enhancer; all the genes whose promoters are associated with active enhancers but with no super-enhancers were labeled as genes associated with typical enhancers.
ChIP-seq heatmaps and average profiles
Heatmaps and average signal profiles were generated using a custom R script which considers a 1-kb window centered on TAZ peak summits falling on active enhancers or a 1.5 kb window centered on TSS of YAP/TAZ target genes. Normalized read density (reads per million, rpm) was calculated from pooled replicates using MACS69 (version 2.0.10) callpeak function with appropriate control samples (IgG for BRD4 and Input DNA for PolII and H3K122ac) and displayed using Integrative Genomics Viewer (IGV). Normalized reads density was calculated with a resolution of 50 bp. Each row in the heatmap represents a genomic region around a peak summit or TSS and rows are ranked according to TAZ or DMSO_BRD4 ChIP-seq signal intensity, respectively.
Generation of the signatures of BET-dependent genes
For analysis of the breast cancer dataset, we used the data generated for this study to identify a list of BET-dependent genes, defined as those whose expression was significantly downregulated (fold change equal or lower than 0.75; FDR less than or equal to 0.01) in both JQ1-treated and siBRD2/3/4-transfected MDA-MB-231 cells compared to control cells. Starting from this list, we then defined a list of common YAP/TAZ/BRD target genes, defined as BET-dependent genes that are also YAP/TAZ direct target genes whose expression is robustly downregulated (fold change less than or equal to 0.67) upon transfection with both YAP/TAZ siRNA mixes. We also defined a list of BRD-dependent but YAP/TAZ-independent genes in BC cells, composed by genes that are not downregulated (fold changes greater than 0.75) after transfection with either YAP/TAZ siRNA mixes. These two lists are of similar size, being the YAP/TAZ/BRD signature composed by 220 genes, and the BRD-dependent/ YAP/TAZ-independent signature composed by 228 genes.
For the analysis of the hepatocellular carcinoma dataset, we made use of public gene expression datasets obtained from human hepatoma HepG2 cells treated with JQ170 (GSE51143) to identify BET target genes, defined as those whose expression was significantly downregulated (fold change equal or less than 0.7; p value less than or equal to 0.05) by JQ1 treatment as compared with DMSO-treated cells. We then used data obtained by Ref. 71 (GSE49384) by transfecting YAP/TAZ siRNAs in HepG2 cells also transfected with siRNAs against NF2 and LATS2, to identify YAP/TAZ-dependent genes in these cells. We used these data to identify common YAP/TAZ/BET target genes, defined as BET target genes significantly dowregulated (fold change less than or equal to 0.75; p value less than or equal to 0.05) after YAP/TAZ knockdown; conversely, BRD-dependent genes that are not significantly downregulated (fold change greater than or equal to 1; p value greater than 0.05) after YAP/TAZ knockdown were used to define the signature of BET target genes that are YAP/TAZ-independent. These two lists are of similar size, being the YAP/TAZ/BET signature composed by 84 genes, and the BET-dependent/YAP/TAZ-independent signature composed by 89 genes.
For the analysis of the Vemurafenib-resistant BRAF-mutant melanoma dataset, YAP/TAZ/BET shared genes were defined as the JQ1-sensitive genes identified by Fontanals-Ciera et al., 201772 in two different melanoma cell lines, that were also found significantly downregulated (fold change less than or equal to 0.5; FDR less than or equal to 0.01) after YAP/TAZ-knockdown in two vemurafenib-resistant melanoma cell lines, according to the data from the dataset GSE6859937; JQ1-sensitive genes not significantly downregulated (fold change greater than or equal to 1.33; p value greater than 0.05) after YAP/TAZ knockdown were instead included in the signature of BET-dependent, YAP/TAZ independent genes in melanoma cells. These two lists are of similar size, being the YAP/TAZ/BET signature composed by 41 genes, and the BET-dependent/YAP/TAZ-independent signature composed by 47 genes.
Collection and processing of gene expression data
Breast cancer gene expression data were generated, normalized, and annotated as described in Ref. 41. Briefly, starting from a collection of 4,640 samples from 27 major data sets comprising microarray data and clinical information, we derived a compendium (meta-data set) comprising gene expression levels and clinical outcome for 3,661 unique samples from 25 independent cohorts. Gene expression data of 247 human hepatocellular carcinoma were downloaded from Gene Expression Omnibus GSE14520 series matrix files and used as is after removing non-tumor and normal samples42,73. Transcriptional profiles of BRAF-mutant melanomas were obtained from GSE5050943. Briefly, starting from non-normalized data, we generated normalized gene expression signals for melanoma tumor samples from patients before commencing treatment with dabrafenib or vemurafenib (n=27) using variance stabilization and quantile normalization of the lumi Bioconductor package74. All microarray data analyses have been performed in R version 3.4.2 with the annotation packages of Bioconductor packages of Release 3.5.
Average signature expression and signature scores
Signature scores have been obtained summarizing the standardized expression levels of signature genes into a combined score with zero mean75. Average signature expression has been calculated as the standardized average expression of all signature genes in all samples and plotted as mean ± standard error of the mean (SEM).
Kaplan–Meier survival analysis
To identify two groups of tumors with either high or low signature, we used the classification rule described in Ref. 75. Briefly, tumors were classified as ‘Low’ if the combined signature score was negative and as ‘High’ if the combined signature score was positive. This classification was applied to expression values of the breast cancer meta-dataset and of GSE14520 hepatocellular carcinomas. To evaluate the prognostic value of the BRD-dependent signatures, we estimated the probabilities that patients would remain free of metastases/survive using the Kaplan–Meier method. To confirm these findings, the Kaplan–Meier curves were compared using the log-rank (Mantel–Cox) test. P-values were calculated according to the standard normal asymptotic distribution. Survival analysis was performed in GraphPad Prism.
Statistical analysis
Statistical analysis for the generation of gene expression and DNA binding data were performed in R and are described in the above paragraphs of Methods.
Other statistical analyses were performed with GraphPad Prism 7.0. One-tailed Mann-Whitney U test was used for pairwise comparison of gene expression levels between groups of genes (from RNA-seq experiments), or DNA binding (from ChIP-seq data); one-tailed Wilcoxon matched-pairs signed rank test was used when comparing the same group of genes/binding sites in different experimental conditions. Linear regression analysis was used to study correlation. The log-rank (Mantel–Cox) test was used to compare survival curves.
For in vivo studies, mice were randomly assigned to treatment groups; sample size was not pre-determined. Two-tailed unpaired Student’s _t_-test was applied for comparison between groups, when required; all analyzed samples were included for statistical analysis.
Supplementary Material
Reporting summary
Supplementary figures
Supplementary table 1
Supplementary table 2
Acknowledgements
We are grateful to J.C. Marine (Leuven Center for Cancer Biology), J. Kim (KAIST) for gift of cell lines; D.J. Pan (University of Texas), Alan R. Clarke (Cardiff University), F. Camargo (Boston Children’s Hospital) and P. Chambon (University of Strasbourg) for gifts of mice. MMTV-Cre mice were purchased from The Jackson Laboratory, where they were kindly deposited by L. Hennighausen. This work is supported by AIRC Special Program Molecular Clinical Oncology “5 per mille”, by an AIRC PI-Grant to S.P, by Epigenetics Flagship project CNR-MIUR. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (DENOVOSTEM grant agreement No 670126).
Footnotes
Author Contributions
F.Z., M.C. and S.P. designed the study, analyzed data and wrote the manuscript. F.Z., G.B., L.F. performed experiments. M.Forcato and S.B. performed bioinformatics analysis. L.A., E.Q., D.D.B, V.G., M.Fassan performed animal experiments and histological analysis. P.L. and B.H. provided reagents and advice for animal experiments. G.S. and J.K. performed mass spectrometry. A.M. performed the initial experiments of this study.
Competing Financial Interest: BH and PL are employees of Bayer AG. SP is a consultant for and received institutional grants from Bayer AG.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Bradner JE, Hnisz D, Young RA. Transcriptional Addiction in Cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villicana C, Cruz G, Zurita M. The basal transcription machinery as a target for cancer therapy. Cancer Cell Int. 2014;14:18. doi: 10.1186/1475-2867-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrieu G, Belkina AC, Denis GV. Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today Technol. 2016;19:45–50. doi: 10.1016/j.ddtec.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stathis A, et al. Clinical Response of Carcinomas Harboring the BRD4-NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer Discov. 2016;6:492–500. doi: 10.1158/2159-8290.CD-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 9.Azzolin L, et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Bai H, et al. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology. 2012;56:1097–1107. doi: 10.1002/hep.25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai J, et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes & development. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, et al. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes & development. 2014;28:432–437. doi: 10.1101/gad.233676.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su T, et al. Two-signal requirement for growth-promoting function of Yap in hepatocytes. Elife. 2015;4 doi: 10.7554/eLife.02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Science signaling. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi K, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panciera T, et al. Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nature reviews. Drug discovery. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rafiee MR, Girardot C, Sigismondo G, Krijgsveld J. Expanding the Circuitry of Pluripotency by Selective Isolation of Chromatin-Associated Proteins. Mol Cell. 2016;64:624–635. doi: 10.1016/j.molcel.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skibinski A, et al. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep. 2014;6:1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh H, et al. Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Rep. 2014;8:449–459. doi: 10.1016/j.celrep.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein C, et al. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 2015;11:e1005465. doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu S, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell. 2015;163:174–186. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galli GG, et al. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell. 2015;60:328–337. doi: 10.1016/j.molcel.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devaiah BN, et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol. 2016;23:540–548. doi: 10.1038/nsmb.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tropberger P, et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013;152:859–872. doi: 10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 30.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swellam M, et al. Aberrant methylation of APC and RARbeta2 genes in breast cancer patients. IUBMB Life. 2015;67:61–68. doi: 10.1002/iub.1346. [DOI] [PubMed] [Google Scholar]
- 32.Bernasconi E, et al. Preclinical evaluation of the BET bromodomain inhibitor BAY 1238097 for the treatment of lymphoma. Br J Haematol. 2017 doi: 10.1111/bjh.14803. [DOI] [PubMed] [Google Scholar]
- 33.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 34.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yimlamai D, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2017;14:296–304. doi: 10.1038/nrgastro.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MH, et al. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J. 2016;35:462–478. doi: 10.15252/embj.201592081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin L, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. 2015;47:250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo Q, et al. AXL/AKT axis mediated-resistance to BRAF inhibitor depends on PTEN status in melanoma. Oncogene. 2018 doi: 10.1038/s41388-018-0205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee BS, et al. Hippo effector YAP directly regulates the expression of PD-L1 transcripts in EGFR-TKI-resistant lung adenocarcinoma. Biochemical and biophysical research communications. 2017;491:493–499. doi: 10.1016/j.bbrc.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Enzo E, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34:1349–1370. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roessler S, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizos H, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014;20:1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, Vakoc CR. Targeting Cancer Cells with BET Bromodomain Inhibitors. Cold Spring Harb Perspect Med. 2017:7. doi: 10.1101/cshperspect.a026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 46.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 47.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hotta A, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nature methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Gutierrez P, Mundi M, Garcia-Dominguez M. Association of bromodomain BET proteins with chromatin requires dimerization through the conserved motif B. Journal of cell science. 2012;125:3671–3680. doi: 10.1242/jcs.105841. [DOI] [PubMed] [Google Scholar]
- 50.Azzolin L, et al. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 51.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 52.Martello G, et al. A MicroRNA Targeting Dicer for Metastasis Control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 53.Means AL, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132:3767–3776. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 54.Zhang N, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sansom OJ, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes & development. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuler M, Dierich A, Chambon P, Metzger D. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis. 2004;39:167–172. doi: 10.1002/gene.20039. [DOI] [PubMed] [Google Scholar]
- 57.Wagner KU, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belteki G, et al. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Picaud S, et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A. 2013;110:19754–19759. doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mori M, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fontanals-Cirera B, et al. Harnessing BET Inhibitor Sensitivity Reveals AMIGO2 as a Melanoma Survival Gene. Mol Cell. 2017;68:731–744 e739. doi: 10.1016/j.molcel.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roessler S, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966 e912. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 75.Adorno M, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reporting summary
Supplementary figures
Supplementary table 1
Supplementary table 2