Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies (original) (raw)
. Author manuscript; available in PMC: 2019 Aug 9.
SUMMARY
Eukaryotic cells contain large assemblies of RNA and protein, referred to as ribonucleoprotein (RNP) granules, which include cytoplasmic P-bodies, stress granules, neuronal and germinal granules, as well as nuclear paraspeckles, Cajal bodies and RNA foci formed from repeat expansion RNAs. Recent evidence argues that intermolecular RNA-RNA interactions play a role in forming and determining the composition of certain RNP granules. We hypothesize that intermolecular RNA-RNA interactions are favored in cells yet are limited by RNA-binding proteins, helicases, and ribosomes, thereby allowing normal RNA function. An overabundance of intermolecular RNA-RNA interactions may be toxic since perturbations that increase RNA-RNA interactions such as long repeat expansion RNAs, arginine-containing dipeptide repeat polypeptides, and sequestration or loss of abundant RNA-binding proteins can contribute to degenerative diseases.
INTRODUCTION
Eukaryotic cells contain a growing number of non-membrane bound organelles consisting of RNA and protein, which we will generically refer to as RNP granules. RNP granules include stress granules, P-bodies, germ granules, and neuronal granules in the cytoplasm as well as paraspeckles, the nucleolus and Cajal bodies in the nucleus (Buchan, 2014; Kiebler and Bassell, 2006; Voronina et al., 2012; Fox et al., 2002; Gall, 2000). RNP granules are ubiquitous and conserved in eukaryotes. Understanding the assembly mechanisms of these non-membrane bound organelles offers opportunity for new insight into cellular organization and regulation.
Historically, studies aimed at determining the assembly mechanisms of RNP granules have focused on proteins. In numerous cases mutating or deleting key RNP granule proteins reduces RNP granule formation (reviewed in Buchan, 2014; Protter and Parker, 2016). For example, the Edc3 protein strongly enhances P-body formation in Saccharomyces cerevisiae (Decker et al., 2007), the G3BP protein is required for stress granule formation in mammalian cells during oxidative stress (Kedersha et al., 2016; Tourrière et al., 2003), and the MEG1 and MEG3 proteins are required for P-granule formation in C. elegans (Wang et al., 2014). These proteins all bind RNA and are thought to connect individual RNPs into larger assemblies through protein-protein interactions.
Four different types of protein-protein interactions promote RNP granule formation. Some are stereospecific interactions between well-folded domains (Figure 1A), such as the dimerization of G3BP in stress granule assembly (Tourrière et al., 2003) or Edc3 in P-body assembly (Ling et al., 2008). Some granule-promoting interactions occur through intrinsically disordered regions (IDRs) of proteins, which are enriched in RNP granules (Decker et al., 2007; Kato et al., 2012; Reijns et al., 2008). Granule assembly can be promoted by the interactions of conserved short linear motifs (SLiMs) within IDRs with the surface of other well-folded protein domains (Figure 1B, reviewed in Jonas and Izaurralde, 2013). Moreover, short repeated sequences in IDRs containing tyrosine, referred to as low-complexity aromatic-rich kinked segments (LARKs), are enriched in RNP granule components and may form local structures that interact with other LARK containing IDRs, thereby providing additional interactions (Figure 1C, Hughes et al., 2018; Murray et al., 2017). Finally, IDRs can also interact with other proteins in a promiscuous manner (Figure 1D), perhaps through cation-π or π-π interactions (Vernon et al., 2018), and by being tethered to a specific interaction module can enhance the assembly of RNP granules (Protter et al., 2018).
Figure 1. Protein-protein interactions promote RNP granule formation.
Proteins can interact in four ways that contribute to the multivalency of RNP granules. A) Classical stereospecific interactions between well-folded domains on proteins. B) Specific proteins bind short linear motifs (SLiMs), conserved sequences within intrinsically disordered regions (IDRs). Often, structure emerges from the IDR upon binding. C) IDRs can also interact specifically with other IDRs through interaction domains with key amino acid characteristics. For example, LARKs are short repeated sequences in IDRs that contain tyrosine and can interact weakly with other LARKs on neighboring proteins. D) IDRs can also provide promiscuous interactions, potentially through π-π or cation-π interactions, which enhance assembly once components are at high-local concentrations.
Additional evidence that protein-protein interactions contribute to RNA granule formation is that many purified RNP granule proteins, and/or their IDRs, can undergo self-association in vitro in processes referred to as liquid-liquid phase separation (LLPS) and/or hydrogel formation (Appendix Table I). Although many proteins can undergo LLPS in vitro, in only a few cases have these assemblies been shown to correlate with RNP granule formation within cells. For example, the interactions of Dcp2, Pdc1, and Edc3 have been shown to promote P-body formation in S. pombe, and also promote LLPS in vitro (Fromm et al., 2014). The self-partitioning of RNP granule components in vitro is consistent with these protein-protein interactions contributing to RNP granule formation.
RNAs are also required for the formation of some RNP granules. For example, the transcription, and presence, of NEAT1 lncRNA is required for paraspeckle formation (Clemson et al., 2009; Mao et al., 2010). Similarly, stress granules and P-bodies require non-translating mRNAs for their formation (Liu et al., 2005; Sheth and Parker, 2003; Teixeira et al., 2005; Pillai et al., 2005) since inhibiting translation initiation increases P-body and stress granule assembly while trapping mRNAs in polysomes reduces P-body and stress granule assembly (Buchan et al., 2008; Teixeira, 2005; Kedersha et al., 1999; Kedersha et al., 2000). Similarly, the formation of RNA foci from repeat expansion RNAs only occurs when the RNA reaches a critical length (Lee et al., 2013; Wojciechowska and Krzyzosiak, 2011). A common model to explain how RNAs promote RNP granule formation is that RNAs provide scaffolds for multivalent RNA-binding proteins, which then, through homotypic or heterotypic protein-protein interactions, connect individual RNPs to form higher-order assemblies (Figure 2A).
Figure 2. RNA contributes to RNP granule formation.
A) RNA can serve as a scaffold for multivalent RNA-binding proteins. These proteins can then interact with each other as described in Figure 1. B) RNAs can interact non-specifically with each other through Watson-Crick base-pairing, non-canonical base-pairing, and helical stacking. C) Molecular crowding has a greater effect on the effective concentration of larger molecules, where the available solvent is much more reduced for larger molecules. This is visualized by the two panels; the left illustrates the accessible solvent to a 55 kDa protein (dark red area) and the right a 7.5 kb RNA (dark blue area). Darker coloring denotes accessible locations of the center of the protein or RNA, respectively. D) The association of larger molecules and complexes is favored by depletion attraction, which is a force only exerted in crowded conditions. The association of two larger molecules decreases the excluded volume and increases the entropy of the smaller macromolecules also in solution. Counterintuitively, the aggregation of larger complexes is entropically favored in crowded environments like the cell.
Herein, we review evidence suggesting intermolecular RNA-RNA interactions directly promote the assembly of RNP granules, leading to a combinatorial model in which RNP granule formation is the result of a summation of protein-protein, protein-RNA and RNA-RNA interactions. This has implications for the intracellular conditions that affect RNP granule formation, how these assemblies are regulated, and the steady state nature of RNP complexes.
Intermolecular RNA-RNA interactions contribute to RNP granule formation
One argument for intermolecular RNA-RNA interactions promoting RNP granule formation is the robust self-assembly of RNA in vitro (Appendix Table II). Specifically, multiple RNAs, or mixtures of RNAs, are capable of protein-free self-assembly, including all four RNA homopolymers (Aumiller et al., 2016; Van Treeck et al., 2018), total yeast RNA (Van Treeck et al., 2018), specific mRNAs (Bounedjah et al., 2012; Langdon et al., 2018), or RNAs corresponding to repeat expansion RNAs (Jain and Vale, 2017). The self-assembly of RNA can occur at concentrations as low as 2 mg/ml (Appendix Table II), which is lower than typical concentrations used to demonstrate LLPS of RNA-binding proteins (0.5-10 mg/ml; Appendix Table I). Moreover, the concentration of exposed mRNA open reading frames during a stress response, when ribosomes run-off mRNAs and stress granules form, is estimated to be ~150-800 μg/mL in yeast and 80-300 μg/mL in human cells, which are concentrations at which RNAs will robustly self-assemble in vitro under physiological salt and polyamine concentrations (Van Treeck et al., 2018).
Three observations suggest that RNA self-assembly in vitro is relevant to in vivo formation of certain RNP granules. First, protein-free yeast RNAs assembled in vitro under physiologically relevant salt and crowding conditions largely recapitulate the yeast stress granule transcriptome (Van Treeck et al., 2018), which is biased towards longer RNAs (Khong et al., 2017). Although it remains possible that there is an unrecognized code in these self-assembling RNAs, the simplest interpretation is that longer RNAs have more sites for intermolecular RNA-RNA interactions, and therefore have enhanced self-association both in cells and in vitro. Second, pathogenic repeat expansion RNAs, are more prone to self-partitioning in vitro than length-matched counterparts (Jain and Vale, 2017). This is consistent with repeat expansion RNAs forming highly structured hairpins (Sobczak et al., 2003), the base-pairs of which could easily be rearranged to form between RNAs, creating an RNA network (Jain and Vale, 2017). Moreover, the ability of repeat expansion RNAs to self-assemble in vitro correlates with their ability to form RNA foci in cells in terms of their length requirements and sensitivity to increased ammonium acetate or doxorubicin, a nucleic acid intercalator (Jain and Vale, 2017). Third, there is a correlation of mRNA self-assembly in vitro with the localization of mRNAs in specific RNP granules in the filamentous fungus Ashbya gossypii. Specifically, the SPA2 and BNI1 mRNAs, which are enriched in an RNP granule at the growth-tip preferentially self-assemble together in vitro, while CLN3 mRNA, which is found in a nuclear associated RNP granule, preferentially assembles with itself (Langdon et al., 2018).
Additional observations are also consistent with the formation of stress granules, and potentially other RNP granules, being partially driven by promiscuous RNA-RNA interactions. Specifically, when naked RNA is injected into the cytosol, it triggers the formation of stress granules (Mahadevan et al., 2013). Similarly, transfection of the luciferase mRNA (Bounedjah et al., 2012) or short RNAs prone to forming G-quadruplexes into cells promotes stress granule formation (Fay et al., 2017). Notably, even electroporated ssDNA will nucleate stress granules, which could be explained by ssDNA base-pairing with RNAs, or ssDNA serving as a scaffold for granule-promoting RNA-binding proteins that can also bind ssDNA (Bounedjah et al., 2014). In addition, stress granule formation is sensitive to the osmotic strength of the cell in a manner that correlates with RNA-RNA interactions and not protein-protein interactions. Specifically, hyper-osmotic stress, which increases intracellular salt concentrations and enhances RNA-RNA interactions, promotes stress granule formation (Bounedjah et al., 2012). In contrast, hypo-osmotic stress, which lowers intracellular salt and diminishes shielding of the RNA’s negative backbone, results in rapid stress granule disassembly (Bounedjah et al., 2012). In contrast, LLPS of RNA-binding proteins or their IDRs in vitro is typically increased by lower salt, and decreased by higher salt concentrations (Appendix Table I). Together, these observations suggest that intermolecular RNA-RNA interactions can contribute to RNP granule assembly.
Biochemical nature of intermolecular RNA interactions
The biochemical interactions that drive self-assembly of RNA in vitro, and in cells, are of three types (Figure 2B-D). First, both Watson-Crick and non-Watson-Crick interactions between bases promote both intra- and intermolecular RNA-RNA interactions. Second, base stacking, either between single-stranded regions, or coaxial stacking of helices can also promote trans RNA-RNA interactions (Zanchetta et al., 2008). Finally, since RNAs are roughly an order of magnitude larger than their encoded proteins, RNA self-assembly may be enhanced by the crowded cellular environment (Ellis, 2001), which has a greater effect on the self-association of larger molecules or assemblies (Figure 2C, D, Marenduzzo et al., 2006)
These observations suggest that whenever there are high concentrations of exposed RNA, intermolecular RNA-RNA interactions can form, therefore contributing to higher-order RNA assemblies. Such events could occur during a stress response when large amounts of open reading frames are freed from ribosomes, or locally at sites of proliferative transcription. Given the degenerative nature of RNA-RNA base pairing, and the size of RNAs, such interactions need not be specific. For example, the average length of an mRNA in a mammalian stress granule is 7.5 kb (Khong et al., 2017), and by a sliding window contains ~7500 hexamers, thereby allowing (7500)2 possible six base-pair interactions with another 7.5 kb RNA. Since one out of 4096 hexamers can perfectly basepair, any two random 7.5 kb RNAs in stress granules would therefore be predicted to have approximately 14,000 possible six base interactions of perfect complementarity simply by chance. Even if ~99% of these possible sites of interaction are hidden by secondary structures or bound proteins, two random 7.5 kb mRNAs would be expected to have over 100 potential sites of interaction. This ignores the possible interactions from shorter helices, partial matches, non-canonical base-pairs, base stacking and crowding effects. Since large RNAs can have multiple interactions, any given individual interaction between RNAs can be weak and transient and still, through a summation of multiple weak interactions, form a stable assembly (Banani et al., 2016).
A gradient of promiscuous to specific RNA-RNA interactions
Cells appear to utilize both promiscuous and specific RNA-RNA interactions in the formation of RNP granules. Stress granules, which form rapidly during a stress response and are heavily biased towards longer mRNAs (Khong et al., 2017), may simply form through random associations between RNAs (Figure 3A, Van Treeck et al., 2018). Similarly, the retention of long mRNAs in the germ plasm of Drosophila is proposed to occur, at least in part, through essentially random base-pairing to multiple piRNAs that reside within the germ plasm simply because longer RNAs have more sites for possible interactions with the piRNAs (Vourekas et al., 2016). In contrast, specific base-pairing in trans between oskar mRNAs, or bicoid mRNAs, is required for their recruitment to RNP granules during Drosophila oocyte development (Figure 3B, Jambor et al., 2011; Ferrandon et al., 1997). mRNAs may also have structures that limit trans intermolecular interactions and thereby impart specificity to the assembly of RNP granules. This latter possibility is suggested by the observation that a mutation altering the structure of CLN3 mRNA in Ashbya gossypii increases its interaction with SPA2 and BNI1 mRNAs in vitro and the colocalization of the mRNAs in cells (Langdon et al., 2018).
Figure 3. RNA-RNA interactions can be promiscuous or specific.
A) Promiscuous interactions may be prevalent in granules containing a diverse set of RNAs in a high local concentration. Here, RNAs are predicted to assemble with a variety of interactions facilitated by the interaction capabilities of an RNA with any other RNA. An example of this may be the formation of stress granules, in which cells experience a large influx of free RNA following ribosomal run-off. B) RNA-RNA interactions contributing to assembly can also be specific. Two examples have been described in Drosophila development in which specific RNA-RNA homodimers of either bicoid or oskar RNAs are important for RNP granule assembly. C) Transcription sites may be a common location of RNP granule assembly, driven in part by the high-local concentration of RNA. As NEAT1 is transcribed, for example, a high local concentration of NEAT1 RNA is achieved. We hypothesize that newly transcribed NEAT1 RNA is capable of forming interactions with neighboring NEAT1 RNAs. As transcripts are released from transcription, a paraspeckle remains. Mature paraspeckles have a clear orientation with the middle of the RNA and certain proteins found in the center, and RNA ends oriented on the outside.
Are RNA-RNA interactions a general feature of diverse RNP granules?
The prevalence and stability of RNA-RNA interactions raise the possibility that intermolecular RNA-RNA interactions contribute to multivalent assemblies whenever such assemblies contain a high local concentration of RNA molecules. For example, several observations suggest that the formation of paraspeckles, a nuclear RNP granule containing approximately 50 copies of the long isoform of NEAT1 lncRNA (Chujo and Hirose, 2017), may be instigated by intermolecular RNA-RNA interactions that occur during transcription (Figure 3C). First, paraspeckles form during the transcription of NEAT1, require NEAT1 for their assembly, and are sensitive to RNase once formed (Fox et al., 2005; Prasanth et al., 2005). Based on the volume of a paraspeckle, 50 copies of the 23 kb NEAT1 RNA within paraspeckles (Chujo et al., 2017) equates to a NEAT1 concentration of approximately 1 mg/mL, which is well above the concentrations for RNA-based self-assembly under physiological conditions in vitro (Appendix Table II). Interestingly, 1,6 hexanediol, an aliphatic alcohol thought to disrupt some weak protein-protein interactions, causes the loss of the NONO protein from paraspeckles while NEAT1 foci remain intact, indicating the formation of NEAT1 RNA foci is independent of NONO (Yamazaki et al., 2018). Finally, numerous RNA-RNA interactions have been identified within the NEAT1 RNA both in vivo (Lu et al., 2016) and in vitro (Lin et al., 2018), and although these interactions are assumed to be intramolecular, in high local concentrations, such as the site of transcription or within paraspeckles, these interactions may also be intermolecular and contribute to paraspeckle formation.
We suggest that intermolecular RNA-RNA interactions should be expected to occur in any biological context with high local concentrations of RNA, unless actively limited by the cell (see below). Such situations would include transcription sites with the propensity for intermolecular interactions increased by high rates of transcription, longer transcripts, and specific sequences prone to intermolecular interactions. Other possible contexts where intermolecular RNA-RNA interactions could be important for subcellular assemblies include the nucleolus, which is dependent on rRNA transcription for its formation (Falahati et al., 2016), Barr bodies, where the formation of an XIST RNA-protein complex coating the inactive X chromosome could be promoted by intermolecular interactions between XIST molecules (Lu et al., 2016; da Rocha and Heard, 2017), and viral RNA synthesis factories, which have a high local concentration of RNA (Nikolic et al., 2017).
One intriguing hypothesis is that RNA-RNA interactions could also play a role in the function of eRNAs, which are produced at both enhancers and super-enhancers, are important for enhancer function, and stimulate transcription proportional to their abundance (Hnisz et al., 2017). However, how eRNAs enhance transcription is not clear. A recent model for enhancer and super-enhancer function is that the concentration of transcription factors and eRNAs at these sites allows for the formation of a local phase separation, which leads to downstream transcriptional activation (Hnisz et al., 2017). Given the propensity of RNAs to interact in trans, one prediction is that eRNA-eRNA interactions might play a role in the assembly of this phase transition at enhancers and thereby trigger downstream transcriptional activation.
A combinatorial model for RNP granule assembly
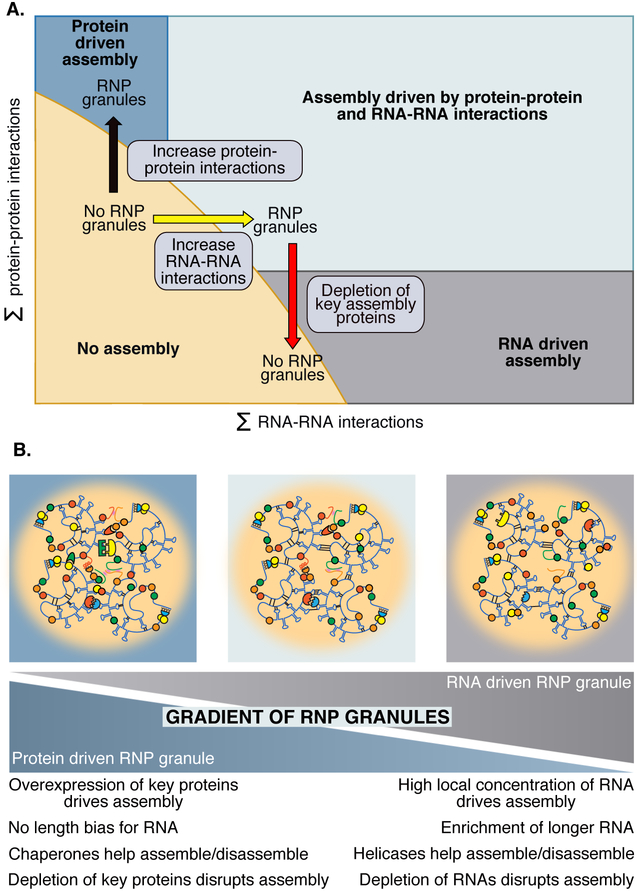
Given the diversity of interactions promoting RNP granule assembly, we suggest a “four-phase” model wherein RNP granules form when the summation of protein-protein, protein-RNA and RNA-RNA interactions increase over a threshold for assembly (Figure 4A). An assembly diagram containing four unique regions illustrates this model. Increasing protein-protein interactions leads to RNP granule assembly as the cellular environment moves up on the Y-axis, to a position in which protein-protein interactions are sufficient to drive assembly. Conversely, increasing RNA-RNA interactions, through rapid influxes of RNA, or the production of RNAs with increased propensity to assemble can lead to formations primarily driven by RNA. This model also provides context for why disruption of protein-protein interactions could disrupt formation of a primarily RNA-driven assembly.
Figure 4. A four-phase model of RNP granule assembly incorporates protein-protein and RNA-RNA interactions and has specific implications.
A) RNP granules can form through either RNA or protein dominated assembly pathways, or through combinations of these pathways. Increasing key protein-protein interactions can shift monomeric RNPs upwards into an RNP granule regime (black arrow). Increasing RNA-RNA interactions (yellow arrow) can stimulate assembly, but the depletion of assembly RBPs (red arrow) can prevent assembly, even in conditions where RNA-RNA interactions are increased. B) RNP granules can have different requirements for assembly. The relative contributions of RNA-RNA or protein-protein interactions is expected to vary from one type of RNP granule to the next. Protein-driven granules are expected to be highly influenced by the overexpression or deletion of key protein components. These granules are also expected to be regulated by post-translational modifications and chaperones. In contrast, granules primarily driven by RNA-RNA interactions are predicted to have a high local concentration of RNA, with an enrichment for long RNAs or RNAs with stable, specific interactions. In addition, enzymes acting on RNAs, such as helicases, would be expected to modulate the dynamics of RNA-based assemblies. Most granules will reside somewhere in the middle, with some characteristics properties from both sides of the spectrum.
The interactions that allow an RNP granule to assemble should be expected to vary between conditions and granule types. Specifically, one expects that the relative importance of protein-protein to RNA-RNA interactions to vary between different types of RNP granules (Figure 4B). For example, stress granules that form from a rapid loss of mRNAs from translation and preferentially recruit long mRNAs may utilize a significant amount of RNA-RNA interactions in their formation. Similarly, RNA foci formed from repeat expansion RNAs with a high propensity to base pair would be expected to be primarily driven by RNA-RNA interactions. In contrast, RNP granules with a low concentration of RNAs, and highly efficient self-assembling proteins, would be predicted to be more dependent on protein interactions. Moreover, the specific protein-protein interactions that contribute to RNP granule assembly can vary. For example, P-body assembly in yeast can be driven by different combinations of proteins depending on the genetic context (Rao and Parker, 2017). Similarly, the deletion of key stress granule proteins in mammalian cells can abrogate stress granule assembly in some stresses and not in others (Kedersha et al., 2016). Thus, one anticipates that the underlying proteome and transcriptome of individual cells allows varying sets of interactions to drive RNP granule assembly in different cell types and biological contexts.
The self-assembly of RNA and RNA-binding proteins, as well as their interactions with each other predicts a complex set of conditions whereby the concentrations, size, and valency of RNA and RNA-binding proteins will influence the formation and properties of larger assemblies (Appendix Table III, Figure 5). For example, the addition of high concentrations of RNA can inhibit the self-assembly of RNA-binding proteins (Banerjee et al., 2017; Schwartz et al., 2013, Maharana et al., 2018), perhaps because the RNA competes with the protein-protein interaction surfaces. However, RNA can also enhance the assembly of RNA-binding proteins (Lin et al., 2015; Molliex et al., 2015; Patel et al., 2015) at lower concentrations or if in conditions that allow RNA-RNA interactions to contribute (Figure 5B). Similarly, the ratios and valency of RNA-binding proteins can affect the nature of assemblies driven by RNA-RNA interactions (Figure 5A).
Figure 5. Concentrations and properties of granule components influence the formation and properties of granules themselves.
A) Effects of monovalent and multivalent RNA-bp on assemblies formed by RNA-RNA interactions. At low concentrations of protein, both monovalent and multivalent RNA-bp are recruited to RNA assemblies, but do not dramatically change the assembly (top left and right) (Van Treeck et al., 2018). In contrast, at high concentrations a monovalent RNA-bp can inhibit assembly by competing for RNA-RNA interactions (bottom left), while a multivalent RNA-bp can enhance assembly by providing additional cross-linking interactions between RNA molecules (bottom right) (Bounedjah et al., 2014). B) Effects of RNAs on self-assemblies of RNA-binding proteins. The addition of low concentrations of short or long RNAs should result in the RNA being effectively recruited to the assembly (top left and right). At high concentrations, both types of RNAs can also inhibit assembly by competing with the protein-protein interaction surface (bottom) (Lin et al., 2015; Schwartz et al., 2013; Maharana et al., 2018). However, RNA can also enhance the assembly of RNA-binding proteins (Banerjee et al., 2017; Molliex et al., 2015; Patel et al., 2015) by either providing a scaffold to increase the valency of RNA-protein complex interactions (Figure 2A), by triggering a conformational change in the protein that promotes assembly, which can occur with short RNAs (middle left), or through RNA-RNA interactions that promote assembly, which is favored for longer RNAs (middle right).
Regulation of the equilibrium between monomeric and multimeric RNPs
Given the robust self-assembly properties of RNA, we hypothesize that cellular RNAs are in a continual exchange between monomeric, oligomeric, and multimeric assemblies large enough to be seen in the light microscope and identified as RNP granules. Evidence for intermolecular interactions at scales smaller than RNP granules includes smFISH of the mRNA DYNC1H1, which does not show random distribution in the cytoplasm, but concentrates into clusters of 3-7 mRNAs (Pichon et al., 2016). In addition, mapping of RNA-RNA interactions in vivo identified over 990 diverse mRNA-mRNA interactions with a substantial fraction being between two different mRNAs (Aw et al., 2016; Lu et al., 2016; Sharma et al., 2016; Gong et al., 2018). Moreover, the imperfect binding of Alu-containing lncRNAs to mRNAs has been suggested to promote Staufen-dependent mRNA decay (Gong and Maquat, 2011). Although more work needs to be done to achieve a more comprehensive and reproducible summary of RNA-RNA interactions in the cell, 170 mRNAs have already emerged as potential interaction hubs with more than 100 documented RNA interaction partners (Gong et al., 2018).
Since the proper functioning of RNAs frequently requires a monomeric RNP, we hypothesize that cells utilize multiple mechanisms to modulate the formation of RNA-based assemblies (Figure 6). For example, ribosomes or monovalent RNA-binding proteins can limit RNA-RNA interactions by binding RNA sequences and restricting their availability for RNA-RNA interactions, while multivalent proteins may increase RNP aggregation (Figure 5A, Appendix Table III). This principle has been demonstrated both in vitro and in vivo, where increased concentrations of the monovalent RNA-binding protein YB1 inhibits stress granule formation in cells, and limits the formation of an RNA-TIA1 protein assembly in vitro (Bounedjah et al., 2014). Similarly, knockdown of the abundant RNA-binding protein TDP-43 in human cells, or its ortholog in C. elegans, leads to the accumulation of dsRNAs in either cytoplasmic or nuclear foci (Saldi et al., 2014). This suggests that the binding of proteins to RNA can limit the formation of dsRNA and intermolecular interactions between RNAs that result in RNA foci.
Figure 6. A model for the modulation of interactions between RNPs.
We hypothesize RNPs may be more prone to associate when their components are in high local concentrations, they are composed of longer RNAs, or contain multivalent RNA-binding proteins or RNAs with increased interaction propensity with other RNAs (such as in repeat expansion diseases). Disassembly may be promoted by increased recruitment of monovalent RNA-binding proteins or short RNAs that limit associations between RNPs. Active remodelers, like chaperones, can also disassemble RNP granules (reviewed in Protter and Parker 2016). In addition, helicases and ribosomes can unwind intermolecular RNA-RNA interactions and block potential RNA-RNA and RNA-protein interactions, respectively.
RNA helicases should be expected to disassemble RNA-RNA interactions and thereby limit RNP granule formation. Strikingly, the loss of the RNA helicase CGH-1 in C. elegans generates solid sheets of other P-granule components (Hubstenberger et al., 2013) and the ATPase activity of CGH-1 is necessary for P granule disassembly following extrusion from cells (Smith and Seydoux, 2018). Similarly, ATP hydrolysis mutants in the DEAD-box helicases Dhh1 or Ded1/Ddx3, results in constitutive P-bodies or stress granules, respectively (Mugler et al., 2016, Hilliker et al., 2011). The disassembly role of helicases may be difficult to demonstrate in some cases as RNA helicases can also play roles in RNP granule assembly by nucleating protein-protein interactions (e.g. Hilliker et al., 2011).
Cells can also limit RNP granule formation by degrading RNAs; evidence for this effect comes from the increase in P-bodies seen in mammalian or yeast cells when mRNA decapping is blocked (Cougot et al., 2004; Sheth and Parker, 2003).
RNA-RNA interactions and disease
Several genetic perturbations linked to human disease may function by tipping the equilibrium between monomeric and multimeric RNAs (Figure 6). The prevalence of RNA foci in repeat expansion disease is a clear example of a toxic RNA being produced that has the potential for multiple intermolecular interactions and thereby formation of a multimeric RNA assembly in the cell (Jain and Vale, 2017). Moreover, many repeat expansion RNAs, such as ALS-linked G4C2 expansions in the C9orf72 gene, can produce polypeptides composed of dipeptide repeats through a process referred to as repeat associated non-AUG (RAN) translation (Zu et al., 2010). The most toxic of these dipeptides are either (RG)n or (PR)n, which can essentially function as polyamines and promote intermolecular RNA-RNA interactions (Van Treeck et al., 2018), although whether this is the basis of their toxicity remains to be established. Finally, a decrease in the functional pool of abundant hnRNP proteins appears to promote intermolecular RNA-RNA interactions. For example, the sequestration of abundant RNA-binding proteins such as TDP-43 into cytoplasmic foci in numerous degenerative diseases may contribute to toxicity by allowing RNA-RNA interactions, and a stress response to dsRNA (Saldi et al., 2014).
RNA self-assembly: Perspectives and Retrospectives
One predicts that RNA-RNA interactions will continue to emerge as key players in cellular mechanisms and assemblies. While base-pairing interactions will be easiest to find, important RNA-RNA interactions that are short and transient may remain more elusive. The ubiquity of RNA-RNA interactions argues that such interactions will be prevalent, but also carefully modulated by the cell. As such, mis-regulation of RNA-RNA interactions could result in pathogenic phenotypes. An important area of research will be in defining the range of RNA-RNA interactions in cells, how cells utilize such interactions for normal function, and how they are modulated by cellular machineries.
Retrospectively, it is self-evident that the robust and degenerate self-assembly of RNA would have been an ideal crucible for the origins of self-replicating RNA. Given an abiotic source of oligonucleotides at a high enough concentration and with abundant counter-ions, essentially any collection of oligonucleotides can self-assemble into a higher-order structure with an increased local concentration of oligonucleotides (Aumiller et al., 2016; Van Treeck et al., 2018). Moreover, because such assemblies concentrate RNA molecules, they can increase the formation of catalytic moieties by increasing interactions between molecules that promote chemical reactions (Strulson et al., 2012). Such RNA-based assemblies may have preferentially retained longer RNAs while, due to the absence of a membrane, allowed facile entry of oligonucleotide precursors for continued rounds of replication.
Supplementary Material
Appendix Tables I-III
Table I. Conditions used for protein self-assembly in vitro. This table serves as a representative list of in vitro protein assemblies and the conditions used for each experiment. Each entry lists the protein identity used for assembly, the concentration of the protein component, the buffer conditions used for assembly, extra comments and the reference.
Table II. Conditions used for RNA self-assembly in vitro. This table covers the in vitro conditions used for the self-assembly of RNA. Each entry lists the RNA used, the concentration of the RNA, the buffer conditions used for assembly, extra comments, and the reference.
Table III. Conditions used for RNA-protein co-assemblies in vitro. This table describes the assembly conditions used for protein-RNA co-assemblies in vitro. The protein identity and concentration, RNA identity and concentration, buffer conditions, comments and relevant references are included.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aumiller WM Jr., Pir Cakmak F, Davis BW, and Keating CD (2016). RNA-Based Coacervates as a Model for Membraneless Organelles: Formation, Properties, and Interfacial Liposome Assembly. Langmuir acs.langmuir.6b02499. [DOI] [PubMed] [Google Scholar]
- Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon K-L, Tapsin S, Chan Y-S, Tan C-P, Sim AYL, et al. (2016). In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Molecular Cell 62, 603–617. [DOI] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, and Rosen MK (2016). Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PR, Milin AN, Moosa MM, Onuchic PL, and Deniz AA (2017). Reentrant Phase Transition Drives Dynamic Substructure Formation in Ribonucleoprotein Droplets. Angewandte Chemie International Edition 56, 11354–11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, and Brangwynne CP (2015). RNA transcription modulates phase transition-driven nuclear body assembly. Proceedings of the National Academy of Sciences 112, E5237–E5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, et al. (2017). Molecular Cell 65, 1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, Hamon L, Curmi PA, Guerquin-Kern JL, Pietrement O, and Pastre D (2014). Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Research 42, 8678–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O, Hamon L, Savarin P, Desforges B, Curmi PA, and Pastre D (2012). Macromolecular Crowding Regulates Assembly of mRNA Stress Granules after Osmotic Stress: New Role For Compatible Osmolytes. Journal of Biological Chemistry 287, 2446–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, and Parker R (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. The Journal of Cell Biology 183, 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR (2014). mRNP granules: Assembly, function, and connections with disease. RNA Biology 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T and Hirose T (2017). Nuclear Bodies Built on Architectural Long Noncoding RNAs: Unifying Principles of Their Construction and Function. Molecules and Cells 40, 889–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Yamazaki T, Kawaguchi T, Kurosaka S, Takumi T, Nakagawa S, and Hirose T (2017). Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. The EMBO Journal 36, 1447–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, and Lawrence JB (2009). An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Molecular Cell 33, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, and Sèraphin B (2004). Cytoplasmic foci are sites of mRNA decay in human cells. The Journal of Cell Biology 165, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, and Heard E (2017). Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nature Structural and Molecular Biology 24, 197–204. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, and Parker R (2007). Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. The Journal of Cell Biology 179, 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC-H, Eckmann CR, Myong S, and Brangwynne CP (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ (2001). Macromolecular crowding: obvious but underappreciated. TRENDS in Biochemical Sciences 26, 597–604. [DOI] [PubMed] [Google Scholar]
- Falahati H, Pelham-Webb B, Blythe S, and Wieschaus E (2016). Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Current Biology 26, 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MM, Anderson PJ, and Ivanov P (2017). ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. CellReports 21, 3573–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Koch I, Westhof E, and Nusslein-Volhard C (1997). RNA-RNA interaction is required for the formation of specific bicoid mRNA 3' UTR-STAUFEN ribonucleoprotein particles. The EMBO Journal 16, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Bond CS, and Lamond AI (2005). P54nrb Forms a Heterodimer with PSP1 That Localizes to Paraspeckles in an RNA-dependent Manner. Molecular Biology of the Cell 16, 5304–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Wah Lam Y, Leung AKL, Lyon CE, Andersen J, Mann M, and Lamond AI (2002). Paraspeckles: A Novel Nuclear Domain. Current Biology 12, 13–25. [DOI] [PubMed] [Google Scholar]
- Frey S and Görlich D (2007). A Saturated FG-Repeat Hydrogel Can Reproduce the Permeability Properties of Nuclear Pore Complexes. Cell 130, 512–523. [DOI] [PubMed] [Google Scholar]
- Fromm SA, Kamenz J, Nöldeke ER, Neu A, Zocher G, and Sprangers R (2014). In Vitro Reconstitution of a Cellular Phase-Transition Process that Involves the mRNA Decapping Machinery. Angewandte Chemie International Edition 53, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG (2000). Cajal Bodies: The First 100 Years. Annual Review of Cell and Developmental Biology 16, 1–31. [DOI] [PubMed] [Google Scholar]
- Gong C, and Maquat LE (2011). lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 39 UTRs via Alu elements. Nature 470, 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Shao D, Xu K, Lu Z, Lu ZJ, Yang YT, and Zhang QC (2018). RISE: a database of RNA interactome Nucleic Acids Research 46, D194–D200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G et al. (2012). Cell-free Formation of RNA Granules: Bound RNAs Identify Features and Components of Cellular Assemblies. Cell 149, 768–779. [DOI] [PubMed] [Google Scholar]
- Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knolt GJ, Iyer KS, Ho D, Newcombe EA, et al. (2015). Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. Journal of Cell Biology 210, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, and Parker R (2011). The DEAD-Box Protein Ded1 Modulates Translation by the Formation and Resolution of an eIF4F-mRNA Complex. Molecular Cell 43, 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, and Sharp PA (2017). A Phase Separation Model for Transcriptional Control. Cell 169, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Noble SL, Cameron C, and Evans TC (2013). Translation Repressors, an RNA Helicase, and Developmental Cues Control RNP Phase Transitions during Early Development. Developmental Cell 27, doi: 10.1016/j.devcel.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, and Eisenberg DS (2018). Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, and Vale RD (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Brunel C, and Ephrussi A (2011). Dimerization of oskar 3' UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 17, 2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, and Izaurralde E (2013). The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes & Development 27, 2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. (2012). Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell 149, 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, and Anderson P (2000). Dynamic Shuttling of TIA-1 Accompanies the Recruitment of mRNA to Mammalian Stress Granules. The Journal of Cell Biology 151, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, and Anderson P (1999). RNA-binding Proteins TIA-1 and TIAR Link the Phosphorylation of eIF-2α to the Assembly of Mammalian Stress Granules. The Journal of Cell Biology 147, 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. (2016). G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. The Journal of Cell Biology 212, 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, and Parker R (2017). The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Molecular Cell 68, 808–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, and Bassell GJ (2006). Neuronal RNA Granules: Movers and Makers. Neuron 51, 685–690. [DOI] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann C, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, et al. (2018). mRNA structure determines specificity of a polyQ-driven phase separation. Science 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, and Narlikar GJ (2017). Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-B, Chen H-J, Peres JN, Gomez-Deza J, Attig J, Štalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. (2013). Hexanucleotide Repeats in ALS/FTD Form Length-Dependent RNA Foci, Sequester RNA Binding Proteins, and Are Neurotoxic. Cell Reports 5, 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Schmidt BF, Bruchez MP, and McManus CJ (2018). Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Research 22, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, and Parker R (2015). Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Molecular Cell 60, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SHM, Decker CJ, Walsh MA, She M, Parker R, and Song H (2008). Crystal Structure of Human Edc3 and Its Functional Implications. Molecular and Cellular Biology 28, 5965–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, and Parker R (2005). MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature Cell Biology 7, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. (2016). RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 165, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann T, et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan K, Zhang H, Akef A, Cui XA, Gueroussov S, Cenik C, Roth FP, and Palazzo AF (2013). RanBP2/Nup358 Potentiates the Translation of a Subset of mRNAs Encoding Secretory Proteins. Plos Biol 11, e1001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang Bin, and Spector DL (2010). Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nature Cell Biology 13, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D, Finan K, and Cook PR (2006). The depletion attraction: an underappreciated force driving cellular organization. The Journal of Cell Biology 175, 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O'Meally R, Dignon GL, Conicella AE, Zheng W, et al. (2017). Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. The EMBO Journal 36, 2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugler CF, Hondele M, Heinrich S, Sachdev R, Vallotton P, Koek AY, Chan LY, and Weis K (2016). ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. eLife 5, e18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, and Tycko R (2017). Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 171, 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudriere-Gesbert C, Gaudin Y, and Blondel D (2017). Negri bodies are viral factories with properties of liquid organelles. Nature Communications 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, and Baldwin AJ (2015). Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Cell 57, 936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Pichon X, Bastide A, Safieddine A, Chouaib R, Samacoits A, Basyuk E, Peter M, Mueller F, and Bertrand E (2016). Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. The Journal of Cell Biology 214, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E and Filipowicz W (2005). Inhibition of Translational Initiation by Let-7 MicroRNA in Human Cells. Science 309, 1573–1576. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, and Spector DL (2005). Regulating Gene Expression through RNA Nuclear Retention. Cell 123, 249–263. [DOI] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016). Principles and Properties of Stress Granules. Trends in Cell Biology 1245, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, and Parker R (2018). Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Reports 22, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao BS, and Parker R (2017). Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences 114, E9569–E9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MAM, Alexander RD, Spiller MP, and Beggs JD (2008). A role for Q/N-rich aggregation-prone regions in P-body localization. Journal of Cell Science 121, 2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi TK, Ash PE, Wilson G, Gonzales P, Garrido Lecca A, Roberts CM, Dostal V, Gendron TF, Stein LD, Blumenthal T, et al. (2014). TDP-1, the Caenorhabditis elegans ortholog of TDP-43, limits the accumulation of double-stranded RNA. The EMBO Journal 33, 2947–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, and Cech TR (2013). RNA Seeds Higher-Order Assembly of FUS Protein. Cell Reports 5, 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E, Sterne-Weiler T, O’Hanlon D, and Blencowe BJ (2016). Global Mapping of Human RNA-RNA Interactions. Molecular Cell 62, 618–626. [DOI] [PubMed] [Google Scholar]
- Sheth U and Parker R (2003). Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J and MacRae IJ (2018). Phase Transitions in the Assembly and Function of Human miRISC. Cell 173, 946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, and Seydoux G (2018). Liquid-like P granules require ATP hydrolysis to avoid solidification. BioRxiv doi: 10.1101/245878. [DOI] [Google Scholar]
- Sobczak K, de Mezer M, Michlewski G, Krol J, and Krzyzosiak WJ (2003). RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Research 31, 5469–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strulson CA, Molden RC, Keating CD and Bevilacqua PC (2012). RNA catalysis through compartmentalization. Nature Chemistry 4, 941–946. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, and Parker R (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, and Tazi J (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. The Journal of Cell Biology 160, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, and Parker R (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proceedings of the National Academy of Sciences 115, 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon RM, Chong PA, Tsang B, Klm TH, Bah A, Farber P, Lin H, Forman-Kay JD (2018). Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Paix A, and Seydoux G (2012). The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 139, 3732–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Alexiou P, Vrettos N, Maragkakis M, and Mourelatos Z (2016). Sequence-dependent but not sequence-specificpiRNA adhesion traps mRNAs to the germ plasm. Nature 531, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen B-C, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, and Seydoux G (2014). Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3, e04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T, Commins C, et al. (2018). Tau protein liquid-liquid phase separation can initiate tau aggregation. The EMBO Journal 37, e98049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowska M, and Krzyzosiak WJ (2011). Cellular toxicity of expanded RNA repeats: focus on RNA foci. Human Molecular Genetics 20, 3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, and Hirose T (2018). Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Molecular Cell 70, 1038–1053. [DOI] [PubMed] [Google Scholar]
- Zanchetta G, Nakata M, Buscaglia M, Clark NA, and Bellini T (2008). Liquid crystal ordering of DNA and RNA oligomers with partially overlapping sequences. Journal of Physics: Condensed Matter 20, 494294. [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, and Gladfelter AS (2015). RNA Controls PolyQ Protein Phase Transitions. Molecular Cell 60, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MAC, et al. (2010). Non-ATG-initiated translation directed by microsatellite expansions. Proceedings of the National Academy of Sciences 108, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix Tables I-III
Table I. Conditions used for protein self-assembly in vitro. This table serves as a representative list of in vitro protein assemblies and the conditions used for each experiment. Each entry lists the protein identity used for assembly, the concentration of the protein component, the buffer conditions used for assembly, extra comments and the reference.
Table II. Conditions used for RNA self-assembly in vitro. This table covers the in vitro conditions used for the self-assembly of RNA. Each entry lists the RNA used, the concentration of the RNA, the buffer conditions used for assembly, extra comments, and the reference.
Table III. Conditions used for RNA-protein co-assemblies in vitro. This table describes the assembly conditions used for protein-RNA co-assemblies in vitro. The protein identity and concentration, RNA identity and concentration, buffer conditions, comments and relevant references are included.