Cell-Free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers Within Hydrogels (original) (raw)
. Author manuscript; available in PMC: 2019 Jan 25.
Summary:
Eukaryotic cells contain assemblies of RNAs and proteins termed RNA granules. Many proteins within these bodies contain KH or RRM RNA binding domains as well as low complexity (LC) sequences of unknown function. Exposure of cell or tissue lysates to a biotinylated isoxazole (b-isox) chemical precipitated hundreds of RNA binding proteins with significant overlap to the constituents of RNA granules. The LC sequences within these proteins are both necessary and sufficient for b-isox-mediated aggregation, and these domains can undergo a concentration-dependent phase transition to a hydrogel-like state in the absence of the chemical. X-ray diffraction and EM studies revealed the hydrogels to be composed of uniformly polymerized amyloid-like fibers. Unlike pathogenic fibers, the LC sequence-based polymers described here are dynamic and accommodate heterotypic polymerization. These observations offer a framework for understanding the function of LC sequences as well as an organizing principle for cellular structures that are not membrane invested.
Introduction
Microscopically visible aggregations of RNA-enriched granules have been observed in a wide spectrum of cell types and organisms. These include germ cell P granules of C. elegans embryos (Strome and Wood, 1982), polar granules of D. melanogaster embryos (Mahowald, 1968), stress granules that appear in cultured yeast and mammalian cells upon nutrient deprivation or other forms of metabolic stress (Buchan et al., 2011; Kedersha et al., 1999), and neuronal granules that help to shuttle mRNAs along dendrites for localized translation at or near synapses (Knowles et al., 1996). Recent studies of nematode embryos have emphasized the dynamic nature of the assembly and disassembly of P granules (Brangwynne et al., 2009; Gallo et al., 2010). The molecular components of these various forms of RNA granules, including both protein and mRNA constituents, have been studied extensively. Prototypical protein constituents of RNA granules are RNA binding proteins endowed with hnRNP-K homology (KH) or RNA recognition motif (RRM) domains that facilitate specific interaction between RNA binding proteins and their mRNA targets.
The mRNA constituents of RNA granules have been studied on a case-by-case basis. For example, the mRNA encoding the nanos transcription factor vital for establishing the anterior:posterior patterning of fruit fly embryos has long been known to be localized to the posterior pole of the embryo by virtue of its incorporation into polar granules (Lehmann and Nusslein-Volhard, 1991; Wang and Lehmann, 1991). Likewise, the mRNAs encoding certain synaptic proteins have been found to be incorporated into neuronal granules – presumably to facilitate dendritic transport for localized translation proximal to synapses (Burgin et al., 1990; Garner et al., 1988). Conceptually, it is understood that mRNAs achieve appropriate association with RNA granules via RNA binding proteins that must satisfy two tasks. First, the RNA binding protein must be capable of selective binding to the appropriate mRNA. In many cases it is well established how KH or RRM domains allow an RNA binding protein to recognize, in a sequence-specific manner, the 3’ untranslated region (3’UTR) of its target mRNA. Second, the RNA binding protein must contain regulatory information necessary for it to be addressed to RNA granules. How specific RNA binding proteins are chosen for inclusion in RNA granules remains unclear.
Here we report the perplexing observation that RNA granule-like structures can be reversibly aggregated and disaggregated in a soluble, cell-free system in response to a small molecule. The ability to perform such experiments in test tube reactions has allowed definition of the polypeptide determinants required for RNA binding proteins to reversibly enter and exit the aggregated state. Surprisingly, the determinant both necessary and sufficient for this reversible process of phase transition has proven to be low complexity (LC) polypeptide sequences, which are known to be regions/domains in proteins with little diversity in their amino acid composition. Extensive studies have shown that these LC sequences exist in a disordered state when their attendant protein is soluble (Huntley and Golding, 2002; Uversky, 2002). The distribution of the LC sequences within the proteomes of eukaryotic cells is far from random, with significant enrichment in RNA- and DNA-binding proteins of regulatory significance (Michelitsch and Weissman, 2000). By expressing, purifying and manipulating the concentration of the LC domain of the fused in sarcoma (FUS) RNA binding protein, we have been able to demonstrate the reversible formation of hydrogel droplets. Electron microscopic and X-ray diffraction studies of the FUS hydrogel give evidence of morphologically uniform amyloid-like fibers, consistent with the notion that phase transition from soluble to hydrogel state is a simple reflection of polymer formation. Hydrogel droplets formed from the LC sequence of FUS are shown to be capable of retaining proteins containing either the FUS LC sequence (homotypic trapping) or the LC sequence derived from other RNA binding proteins (heterotypic trapping). In both cases, gel trapping is inferred to reflect the incorporation of monomer test molecules into existing polymeric fibers within the FUS hydrogel. These studies offer a means of considering the fundamental biological utility of the enigmatic LC sequences commonly found in eukaryotic regulatory proteins.
Results
A high throughput drug screen conducted in search of chemicals capable of prompting mouse embryonic stem (ES) cells to differentiate towards the cardiomyocyte lineage led to the identification of an isoxazole designated 5-aryl-isoxazole-3-carboxyamide (Sadek et al., 2008). This compound is further endowed with the capacity to trigger differentiation of neuronal progenitor cells into morphologically mature neurons (Schneider et al., 2008). In efforts to discover the cellular target of this isoxazole, a biotinylated derivative was prepared (Experimental Procedures). Hereafter we will refer to this biotinylated isoxazole as b-isox.
Cytoplasmic lysates were prepared from two mouse tissues (brain and testis) and two mouse cell lines (NIH 3T3 cells and mouse embryonic stem cells) and sequentially incubated with b-isox and streptavidin beads on ice. Irrespective of tissue or cell type origin, lysates incubated with the 30uM and 100uM levels of the b-isox chemical revealed a complex pattern of precipitated proteins (Figure 1A). Analysis of control reactions revealed the unexpected observation that precipitation of the same distribution of proteins was effected in the absence of the streptavidin beads. Visual inspection of the reactions revealed a flocculent, white precipitate occurring within minutes subsequent to addition of the chemical to the cytoplasmic lysate. Subsequent centrifugation readily separated the precipitant from soluble components of the lysate, and SDS gel analysis revealed the same pattern of polypeptides as observed with combined exposure to both b-isox and streptavidin beads. The precipitation reaction was observed to be reversible. When the pelleted material was resuspended in the same buffer used to prepare the cytoplasmic lysate and warmed to 37°C, the proteins formerly in the precipitated fraction were rendered soluble (Figure 1B). As will be shown in a subsequent section of this study, the b-isox chemical forms a micro-crystalline precipitate on its own when administered at the 30-100uM level to the lysis buffer at 4°C. Moreover, the b-isox alone precipitate is solubilized when warmed to 37°C. As such, we hypothesize that the micro-crystalline precipitate itself co-precipitates the proteins visualized in lanes 6 and 7 of the four gels shown in Figure 1A.
Figure 1. Selective precipitation of proteins by the b-isox chemical.
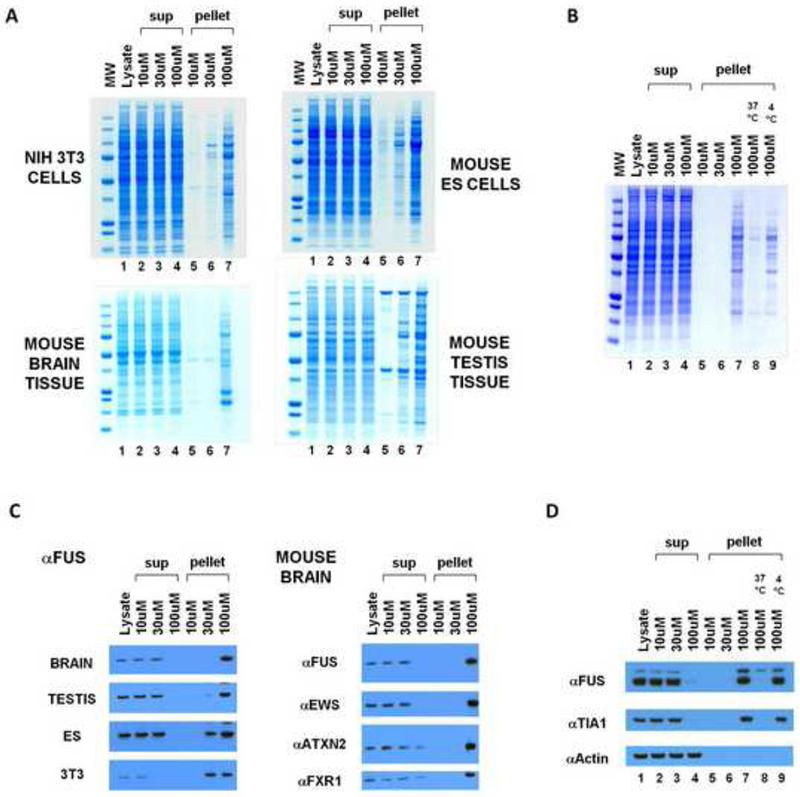
(A) B-isox-mediated precipitation of proteins from mouse 3T3 cells, ES cells, brain tissue, and testis tissue. Lysates were incubated with 10, 30, or 100uM b-isox, pelleted by centrifugation, resuspendend and resolved by SDS-PAGE and Coomassie staining. Precipitated proteins at 100uM b-isox were identified by mass spectroscopy (Figure S1).
(B) B-isox precipitant, when resuspended in fresh buffer and warmed to 37°C, is solubilized and can be re-precipitated upon repeated exposure to the 100uM level of the b-isox chemical.
(C) Western blot assays showing behavior of FUS, ataxin 2, EWS and FXR1 proteins in response to exposure to the b-isox chemical. Quantitative precipitation of FUS was observed in all four lysates at the 100uM compound concentration. EWS was also quantitatively precipitated from the mouse brain lysate at the 100uM level of the b-isox chemical. For ataxin 2 and FXR, nearly complete precipitation was observed at the 100uM level of the b-isox chemical.
(D) Western blot assays showing that b-isox-mediated precipitation of FUS and TIA1 proteins is reversible. Mouse brain lysate was exposed to 100uM levels of the b-isox chemical, and the precipitation was seperated by SDS-PAGE and subjected to western blotting to identify the FUS and TIA1 polypeptides (lane 7). After resuspension in fresh buffer and warming to 37°C, both FUS and TIA1 became soluble (lane 8). Upon re-exposure to fresh b-isox at 100uM and incubation on ice, both proteins were re-precipitated (lane 9).
By comparing the pattern of proteins left in the soluble fraction after precipitation with the highest level of b-isox (Figure 1A, lanes 4) with the initial lysate (lanes 1), no discernable difference was observed. As such, it was concluded that the b-isox-precipitated proteins (lanes 6 and 7) constituted a reasonably rare group of polypeptides invisible in the Coomassie staining pattern of the initial lysate. Therefore, we pursued identification of these precipitated proteins by mass spectrometry and found them to be substantially enriched in RNA binding proteins. Prominent among b-isox-precipitated proteins were translation initiation factors and polypeptides containing K-homology (KH) domains, RNA recognition motif (RRM) domains, and DEAD box helicase domains. We additionally observed b-isox-mediated precipitation of numerous proteins implicated in neurodegenerative diseases and mental retardation, including FMR1, FUS, TDP43 and ataxin 2. Western blotting assays confirmed the identities of these proteins and further established that they were quantitatively precipitated in response to 100uM of the b-isox compound (Figure 1C), and that precipitation reaction was reversible (Figure 1D).
Many of these proteins are constituents of neuronal granules, stress granules, processing bodies or polar granules (Anderson and Kedersha, 2006; Buchan and Parker, 2009; Voronina et al., 2011). As such, we adopted the provisional hypothesis that the b-isox compound might be capable of triggering the formation of aggregates related to RNA granules in a cell-free lysate. In order to pursue this hypothesis, we initially evaluated the overlap in proteins precipitated from the four lysates. The Venn diagram shown in Figure S1A reveals that 162 proteins were precipitated by the b-isox compound in all four cell/tissue types. After subtracting 54 ribosomal proteins, the remaining 106 proteins were compared with a data set of all proteins reported in the literature to be associated with stress granules, neuronal granules, processing bodies and polar granules (Buchan and Parker, 2009; Elvira et al., 2006; Kanai et al., 2004). Of the 106 core proteins, 53 were observed to be included within the list of 150, canonical RNA granule proteins (Figure S1B). The statistical probability of this match reveals a p-value of < 0.0001. As such, we conclude that the b-isox compound is capable of causing aggregation of a set of proteins that overlap significantly with proteins previously reported to be constituents of RNA granules.
mRNA localization via RNA granules has been intensively studied in embryos of the fruit fly, Drosophila melanogaster (Micklem, 1995; Palacios and St Johnston, 2001). Forward genetic studies have led to the discovery of numerous mRNAs localized to polar granules, as well as many RNA binding proteins that facilitate polar granule localization by binding to the 3’ untranslated regions of mRNA destined for localized storage and/or translation. Given the rich history of study of mRNA localization in Drosophila, we prepared b-isox-mediated precipitation from cytoplasmic extracts of cultured Drosophila S2 cells, and identified the proteins in the precipitation by mass spectrometry. In parallel with the enrichment pattern observed for mammalian cells and tissues, the b-isox compound led to the selective precipitation of translation initiation factors, poly-A binding proteins, mRNA cap-binding proteins, DEAD box helicases, hnRNP proteins and RNA binding proteins that contain KH domains and RRM domains (Table S1). Included in the list were the pumilio protein known to recognize nanos response elements in the 3’UTRs of mRNAs (Murata and Wharton, 1995), the squid protein required for gurken mRNA localization (Kelley, 1993; Neuman-Silberberg and Schupbach, 1993), the fly orthologs of the mammalian FMR1, TDP43 and ataxin 2 proteins, and Nup98, a nuclear pore protein known to be associated with C. elegans P granules (Voronina and Seydoux, 2010).
Sequence Determinants Required for B-isox-mediated Precipitation
RNA binding proteins containing an RRM or KH domain constituted a particularly prevalent category in the lists of b-isox-precipitated proteins from all lysate sources. In addition to these defined domains, many of these RNA binding proteins also contain long stretches of low-complexity (LC) sequence. To determine what functional domains were required for the RNA binding proteins to enter the b-isox precipitate, GFP-fused recombinant versions of a representative protein - TIA1 - were subjected to b-isox-mediated precipitation. As shown in Figure 2, the intact protein was precipitated by the b-isox chemical. Removal of three RRM domains required for direct RNA binding failed to affect b-isox-mediated precipitation. By contrast, removal of the C-terminal LC sequence prevented b-isox-mediated precipitation. Having observed that the well-folded GFP protein was itself not precipitated, we conclude that the LC domain of TIA1 is both necessary and sufficient for b-isox-mediated precipitation. Similar results were obtained with the LC domains of FUS, CIRBP, RBM3, hnRNPA1, hnRNPA2 and the yeast Sup35 protein (data not shown). The amino acid sequences of the seven LC domains investigated as purified proteins in this study are shown in Figure S2A.
Figure 2. Low complexity sequence of TIA1 is necessary and sufficient for b-isox-mediated precipitation.
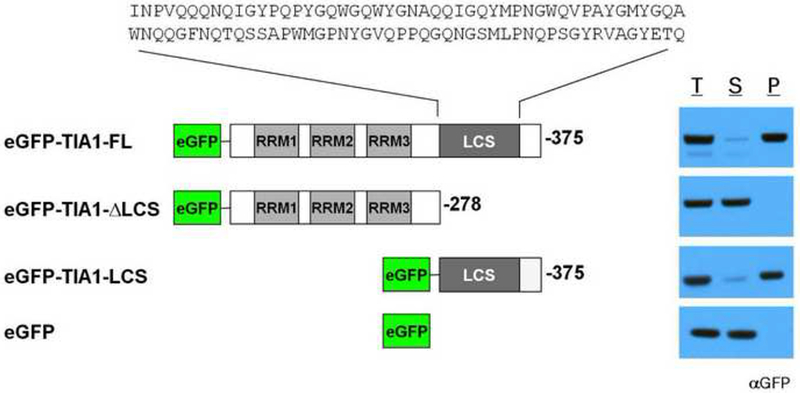
Recombinant, purified GFP-tagged TIA1 proteins were subjected to 100uM b-isox chemical. Supernatant and precipitant fractions were resolved by SDS-PAGE followed by western blot assays with a GFP antibody. RRM: RNA recognition motif, LCS: low-complexity sequence. T: total lysate, S: supernatant, P: precipitate. The sequence of the TIA1 LC domain is shown in Figure S2.
We then performed computational studies to assess the frequency of the LC domains in our collection of b-isox-precipitated proteins, and in the collection of known RNA granule proteins, as compared with the entire mouse proteome (Figure S2B). Both the b-isox-precipitated group of proteins and the literature-cited RNA granule proteins were substantially enriched in the LC domains, with a high degree of statistical significance, as compared with the entire mouse proteome.
Phase-transition of the Low Complexity Sequence Domain of FUS into a Hydrogel
While working with concentrated forms of the recombinant, full length FUS protein linked to a GST tag (∼10mg/ml), it was noted that the protein adopted a gel-like state when stored at low temperature. Hydrogel formation was observed in the absence of the b-isox chemical. Unless otherwise specifically stated, all additional experiments reported herein were conducted in the complete absence of the b-isox chemical. Recombinant derivatives of GST-FUS lacking the C-terminal RNA binding domains retained gel forming capacity. By contrast, N-terminal truncations lacking the LC sequence were incapable of phase transitioning into a hydrogel-like state. When produced as chimeric proteins with GST, mCherry or GFP, the FUS LC domain (residues 2-214, ∼50mg/ml) yielded clear, red and green colored hydrogels respectively (Figure 3A).
Figure 3. Hydrogels formed by the LC domain of FUS bind and retain the GFP-tagged LC domains of RNA binding proteins.
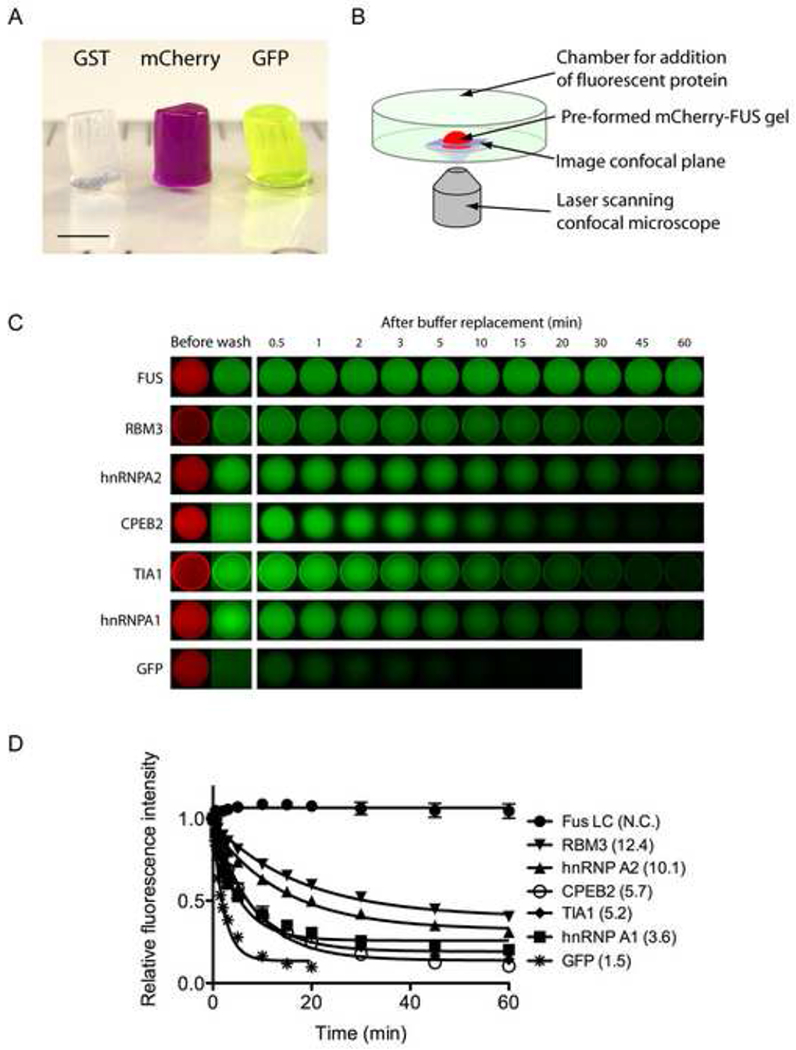
(A) Hydrogels formed by concentrated proteins linking the N-terminal LC domain of FUS to three different protein tags (GST, mCherry, and GFP) were squeezed out from silicon tubes. The hydrogels retained the cylindrical shape of the silicon tube and exhibit the respective colors of the tags (GST: clear, mCherry: red, GST: green). Scale bar at the lower left corner corresponds to 3mm.
(B) Schematic representation of fluorescence microscopic hydrogel binding assay.
(C) Hydrogel retention assays measuring the binding of GFP and GFP-fusion LC domains from RNA-binding proteins in the list of b-isox-precipitate to hydrogel droplets composed of mCherry:FUS LC. Soluble test proteins were incubated with mCherry:FUS hydrogel droplets (Before wash). Immediately after the test protein solutions were removed and replaced with buffer, retained GFP signal levels were scanned at indicated time points. Binding of four additional proteins (FMRP, CIRBP, TDP43 and yeast Sup35) were shown in Figure S3.
(D) Half-lives of the above decayed GFP signals in minutes were obtain from least-squares fitting by the Prism program (GrafPad software, La Jolla CA) and indicated in parentheses.
Hydrogel droplets of the mCherry:FUS LC domain of roughly 500um in diameter were formed on glass-bottomed microscope dishes to perform a simple, microscopic binding assay (Figure 3B). When the hydrogel droplet was exposed to GFP solution (1uM), the GFP molecules penetrated the hydrogel but - upon washing - were not selectively retained by the gel (Figure 3C). By contrast, a GFP derivative linked to the FUS LC domain was avidly retained by the mCherry:FUS LC hydrogel. We also tested the LC domains of other RNA binding proteins precipitated by the b-isox chemical. All of the LC domains tested were observed to adhere to the mCherry:FUS hydrogel, yet the decay rate varied from protein to protein (Figure 3C and 3D). Homotypic trapping of the GST:FUS LC by the mCherry:FUS LC hydrogel was sufficiently avid that GFP signal hardly decayed over the hour-long washout period. The LC domains of hnRNPA2 and RBM3 RNA binding proteins adhered to the hydrogel with moderate apparent avidity. The hnRNPA1, TIA1 and CPEB2 LC domains bound to the hydrogel with more modest avidity. Binding of the LC domains of four additional test samples (FMRP, CIRBP, TDP43 and yeast Sup35) was also observed (Figure S3A). In all nine cases, retention of these GFP fusion proteins to the hydrogel was attributed to the LC domain alone (data not shown).
Hydrogel formation was also observed for the LC domain of hnRNPA2 linked to either GFP or mCherry. The hydrogel droplets of mCherry:hnRNPA2 LC were exposed to the same proteins tested above. All the proteins, except GFP, bound to the hydrogel, but with a different hierarchy from that of mCherry:FUS LC hydrogel (Figure S3B). GFP:hnRNPA2 LC was most avidly retained, giving evidence of homotypic trapping of the test protein by the hydrogel droplet. The mCherry:hnRNPA2 hydrogel also avidly trapped the LC domains of hnRNPA1, CIRBP and RBM3. Moderate retention was observed for yeast Sup35. By contrast, considerably less avid trapping was observed for FUS, TIA1, TDP43, FMRP or CPEB2. We conclude that the phenomenon of phase transition between soluble and hydrogel-like state may be a common feature of the LC domains associated with b-isox-precipitated RNA binding proteins, yet that the hierarchy of how different hydrogels trap heterologous LC sequences are qualitatively distinct.
Tyrosine Residues Flanked by Either Glycine or Serine Specify Hydrogel Formation
Inspection of the LC sequences located in the gel-forming, N-terminal domain of FUS revealed 27 variants of the tripeptide sequence GYG, GYS, SYG or SYS (hereafter designated [G/S]Y[G/S] repeats) (Figure S4A). Speculating that these repeats might play some role in hydrogel formation, unbiased computational studies were performed on the mouse proteome in search of proteins enriched in these repeats (Figure S4B). A list of the 190 mouse proteins having the highest density of the [G/S]Y[G/S] repeats was cross-referenced with the core group of the b-isox-precipitated proteins. Of the 106 b-isox protein core, 27 were found on the computationally defined list of proteins enriched in the [G/S]Y[G/S] motif. This degree of overlap was of high statistical significance, yielding a p-value of < 2.2 × 10−16. Statistically significant overlap was also observed between these 190 mouse proteins enriched with the [G/S]Y[G/S] motifs and RNA granules proteins (p-value of <4.1 × 10−10).
Given these unusually high level of statistical significance, we prepared mutated derivatives of GFP:FUS LC wherein varying numbers of tyrosine residues in the [G/S]Y[G/S] motifs were mutated to serine. The S1 mutant changed 5 of the relevant tyrosine residues in the LC domain of FUS to serine. The S2 and S3 variants mutated 9 and 15 of the tyrosines to serine respectively, and the allS variant mutated all 27 tyrosines to serine (Figure S4A). Upon purification and concentration, none of the mutants were capable of hydrogel formation (data not shown). When these proteins were assayed for binding to mCherry:FUS LC hydrogel, moderate retention was observed for the S1 mutant, weaker retention was observed for S2, and little or no retention was observed for S3 and allS (Figure 4A and 4B).
Figure 4. Mutation of tyrosine residues within the FUS LC domain correlatively affect hydrogel retention and stress granule association.
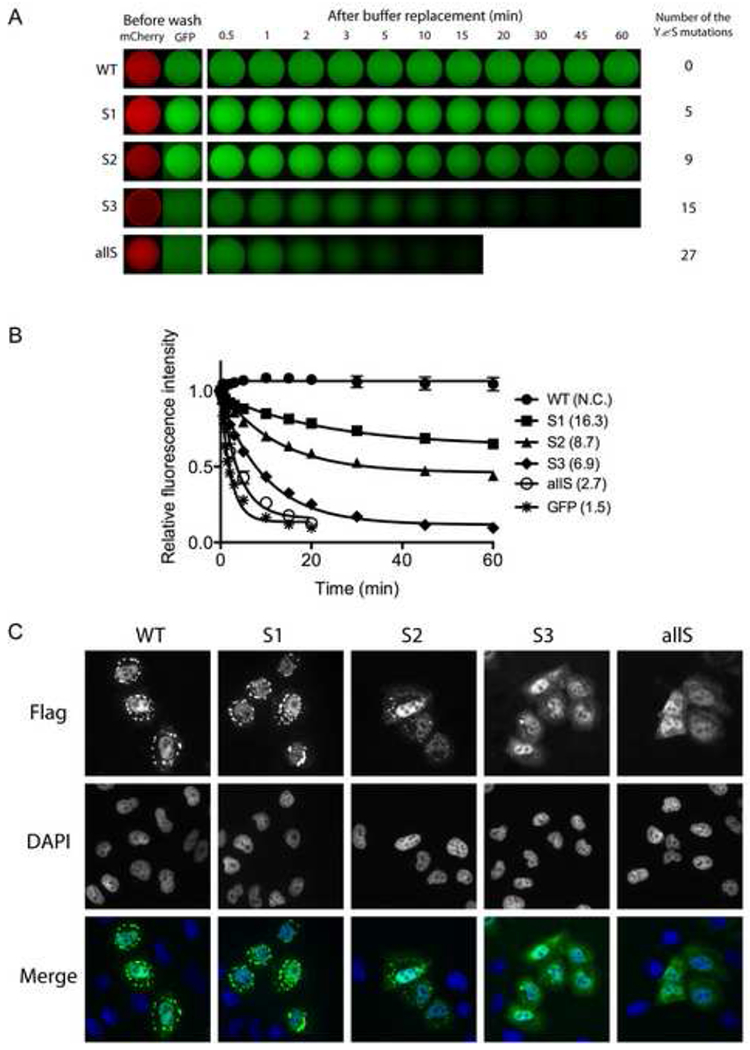
(A) Hydrogel retention assays measuring the binding of tyrosine mutants of GFP:FUS LC to mCherry:FUS LC hydrogel. Out of 27 [G/S]Y[G/S] triplet motifs in the FUS LC domain, the indicated number of tyrosines were randomly selected and replaced with serine. The resulting GFP:FUS tyrosine mutants were incubated with mCherry:FUS hydrogel droplets in chamber slides (Before wash). Immediately after the test protein solutions were removed and replaced with buffer, retained GFP signal levels were scanned at indicated time points. The sequences of all the mutants are shown in Figure S4.
(B) Decay rates of hydrogel binding of the four tyrosine-to-serine mutants, compared with the intact GFP:FUS LC domain. The curve fittings were done as described in Figure 3D.
(C) In vivo stress granule recruitment assay for the tyrosine mutants of FUS. Mammalian expression vectors were prepared wherein the full-length FUS was fused to a Flag epitope tag at its N-terminus and the C-terminal nuclear localization sequence was deleted. In the mutant vectors, the same mutations as those of S1, S2, S3, and allS described in (A) were introduced. After human U2OS cells were transfected with each expression vector, stress granule formation was induced by 0.5mM sodium arsenite. Wild-type FUS was observed to enter cytoplasmic puncta, presumed to reflect stress granules, which was co-stained with antibodies specific to TIA1 (Figure S4C).
The FUS protein has been observed to partition to stress granules when expressed in mammalian cells, particularly when lacking its C-terminal nuclear localization sequence (Bosco et al., 2010). We therefore prepared mammalian expression vectors to monitor the propensity of FUS to enter stress granules as a function of the status of the [G/S]Y[G/S] repeats. As shown in Figures 4C and S4C, the version of FUS containing 27 intact [G/S]Y[G/S] repeats localized to stress granules, as did the S1 mutant lacking 5 of the relevant tyrosine residues. By contrast, the S2 mutant lacking 9 tyrosines could barely be detected in stress granules, and no stress granule co-localization whatsoever was observed for the S3 and allS mutants.
The FUS and hnRNPA2 Hydrogels are Composed of Amyloid-like Fibers
To initiate morphological and structural studies of the nature of the mCherry:FUS LC hydrogel, we first gathered transmission electron microscope (TEM) images of a pre-formed hydrogel composed of either mCherry:FUS LC or a His6-tag version of the FUS LC domain (Figure 5A and 5B). Both the mCherry:FUS and His:FUS preparations revealed morphologically uniform amyloid-like fibers. The dimensions of the fibers were roughly 500nm in length, with the mCherry:FUS fibers showing a thickness (30nm) slightly greater than that of the His:FUS fibers (20nm).
Figure 5. Morphological and structural properties hydrogel droplets and b-isox precipitates.
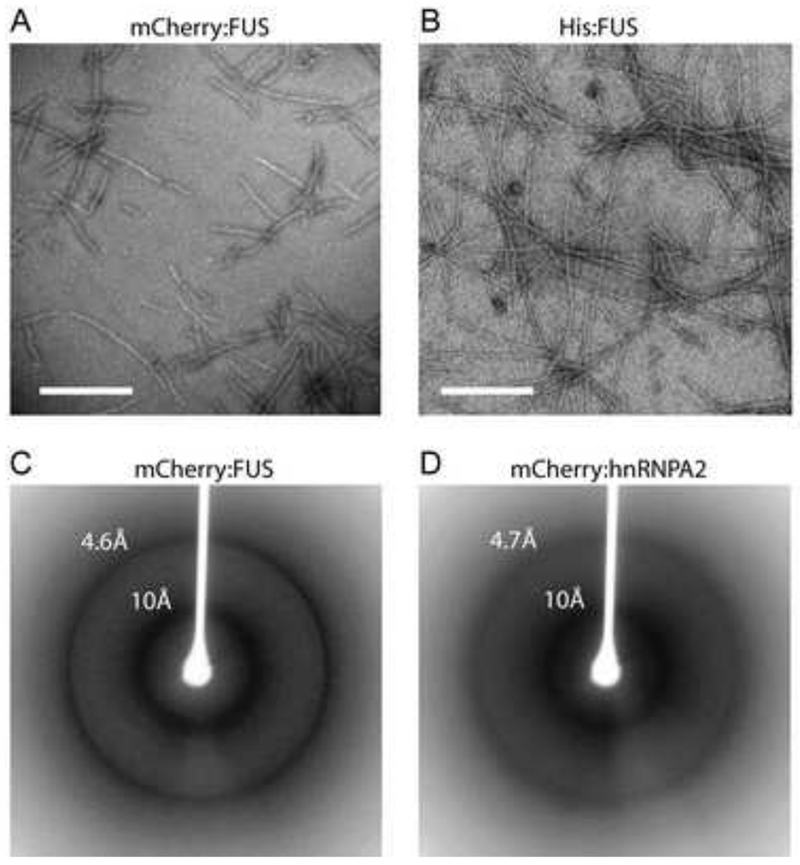
(A and B) Materials derived from mCherry:FUS LC (A) and His-tagged:FUS LC (B) hydrogel droplets were visualized by transmission electron microscopy. Both samples revealed uniformly similar, polymeric, amyloid-like fibers. White scale bar indicates 500nm for (A) and 320nm for (B).
(C and D) Materials from mCherry:FUS LC (C) and mCherry:hnRNPA2 LC (D) hydrogel droplets were subjected to the X-ray diffraction. Clear X-ray reflections were observed at 4.6Å and 10Å for the mCherry:FUS material, and 4.7Å and 10Å for the mCherry:hnRNPA2 material.
To assess the presence of cross-β properties in the filaments, hydrogels of mCherry:FUS LC and mCherry:hnRNPA2 LC were subjected to X-ray diffraction analysis. As shown in Figure 5C and 5D, both materials yielded a diffraction pattern showing prominent, circular reflections at 4.6-4.7Å and 10Å. These reflections are prototypic of cross-β structure (Astbury and Dickinson, 1935; Geddes et al., 1968; Sunde and Blake, 1997). The X-ray diffraction pattern of mCherry alone did not reveal these reflections, nor were they seen in soluble preparations of mCherry:FUS LC or mCherry:hnRNPA2 LC (data not shown). In combination with the TEM studies, these X-ray diffraction images give strong evidence of the presence of amyloid-like polymers as the structural basis of hydrogel architecture.
Relative Fragility of mCherry:FUS and mCherry:hnRNPA2 Amyloid-like Fibers
Extreme stability is a hallmark property of pathogenic amyloid fibers (Dobson, 2003; McKinley et al., 1983; Meersman and Dobson, 2006; Smith et al., 2006; Wille and Prusiner, 1999). Lindquist and colleagues recently described dozens of new, ultra-stable amyloids formed in yeast, many derived from the LC sequences associated with transcription factors and RNA binding proteins (Alberti et al., 2009). A uniform property of these prion-like amyloid fibers is insensitivity to the solubilizing effects of sodium dodecyl sulfate (SDS). To examine the SDS sensitivity:resistance of the mCherry:FUS LC and mCherry:hnRNPA2 LC fibers, heavily polymerized preparations (Figure 6A) were filtrated by a membrane allowing passage of monomeric proteins, but not fibers. As shown in Figure 6B, little or no protein passed through the filter when the fibers were diluted in standard buffer and filtrated immediately. By contrast, exposure to 2% SDS at 37°C for 10 min allowed the vast majority of UV-adsorbing material to pass into the filtrate. Such results indicate that the fibers were depolymerized into monomers by SDS treatment. To visualize fiber disassembly, we utilized semi-denaturating detergent agarose gel electrophoresis (SDD-AGE) to compare the stability of our fibers with those generated from the yeast Sup35 protein (Wickner, 1994). Sample preparations were incubated at 37°C for 10 min in the presence of no SDS, 0.5%, 1% and 2% SDS then applied to an agarose gel for electrophoretic separation. The Sup35 fibers were almost completely resistant to even the highest level of SDS tested (2%). By contrast, mCherry:FUS LC fibers were almost fully depolymerized in the no SDS control. Likewise, the hnRNPA2 LC fibers were fully depolymerized upon incubation at 37°C in the absence of SDS (Figure 6C). It thus appears that despite sharing morphological similarities and common evidence of the presence of cross-β structural properties, the mCherry:FUS and mCherry:hnRNPA2 fibers must be fundamentally different from the prion-like, irreversible fibers described broadly throughout the literature.
Figure 6. Amyloid-like fibers composed of mCherry:FUS and mCherry:hnRNPA2 de-polymerize upon exposure to SDS.
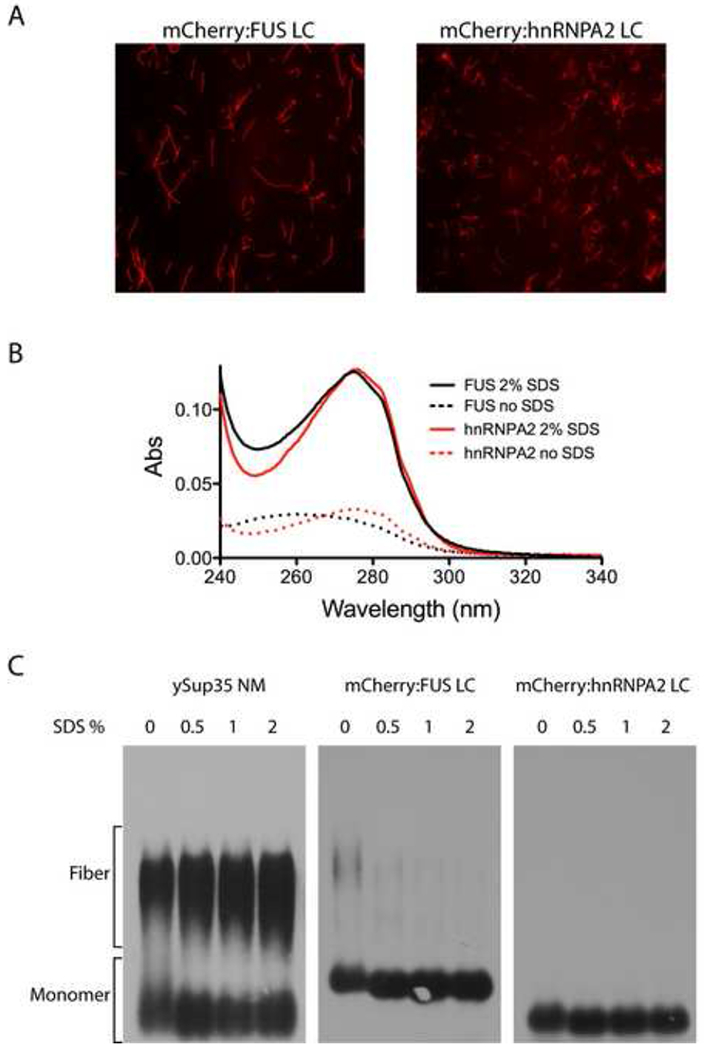
(A) Fluorescence microscope images of amyloid-like fibers observed upon incubation of concentrated solutions (10mg/ml) of mCherry:FUS LC and mCherry:hnRNPA2 LC domains.
(B) UV adsorption of filtrate of amyloid-like fibers shown in (A) before (dotted lines) and after (solid lines) exposure to 2% SDS and 10 min incubation at 37°C. Little or none of the mCherry:FUS or mCherry:hnRNPA2 material passed through the filter without exposure to SDS. After SDS treatment, essentially all protein samples passed through the filter.
(C) SDS gel electrophoresis assays of migration of yeast Sup35 NM protein aggregates, mCherry:FUS LC aggregates and mCherry:hnRNPA2 LC aggregates. 10ug samples of each protein were warmed to 37°C for 10 min in the absence (0) or presence of varying amounts of SDS (0.5%, 1% or 2%). Following incubation samples were loaded onto and electrophoresed through an agarose gel, transferred to a nitrocellulose filter and western blotted with antibodies to either ySup35 (left panel) or the His epitope tag (middle and right panels). SDS exposure did not substantially affect ySup35 NM aggregates. mCherry:FUS LC aggregates were almost fully de-polymerized in the zero SDS sample, and fully de-polymerized in samples containing 0.5%, 1% and 2% SDS (middle panel). mCherry:hnRNPA2 aggregates were fully de-polymerized under all conditions (right panel).
A Fiber Polymerization Model for Hydrogel Retention
Having observed FUS hydrogels to be composed of amyloid-like fibers, it became possible to consider various mechanisms by which exogenously-supplied proteins could be trapped and retained by the gel. The network of existing polymers could, for example, be decorated by Velcro-like appendages constituting lateral sites for the binding of either homotypic or heterotypic LC domains. Alternatively, when a monomeric GFP-LC domain protein is applied to the mCherry:FUS LC hydrogel, it is possible that the high density of fiber ends within the gel might seed incorporation of the test monomer into the polymeric network of the gel. As a test of these hypotheses, mCherry:FUS LC fibers were polymerized and sonicated to form fibrous seeds to which monomeric GFP variants could be exposed. When such seeds were exposed to GFP alone, no evidence of attachment – either lateral to the fiber or extending from either end – was observed (data not shown). By contrast, the red mCherry:FUS LC seeds were extended in length when exposed to soluble GFP:FUS LC (Figure 7A). The extension was observed from both ends of the seed, yet growth in one direction typically exceeded the other. In all cases, polymeric extension from the seeds was almost exclusively green in color.
Figure 7. Co-polymerization of GFP:FUS, GFP:FUS tyrosine-to-serine mutants, and GFP:hnRNPA1 to mCherry:FUS seed fibers.
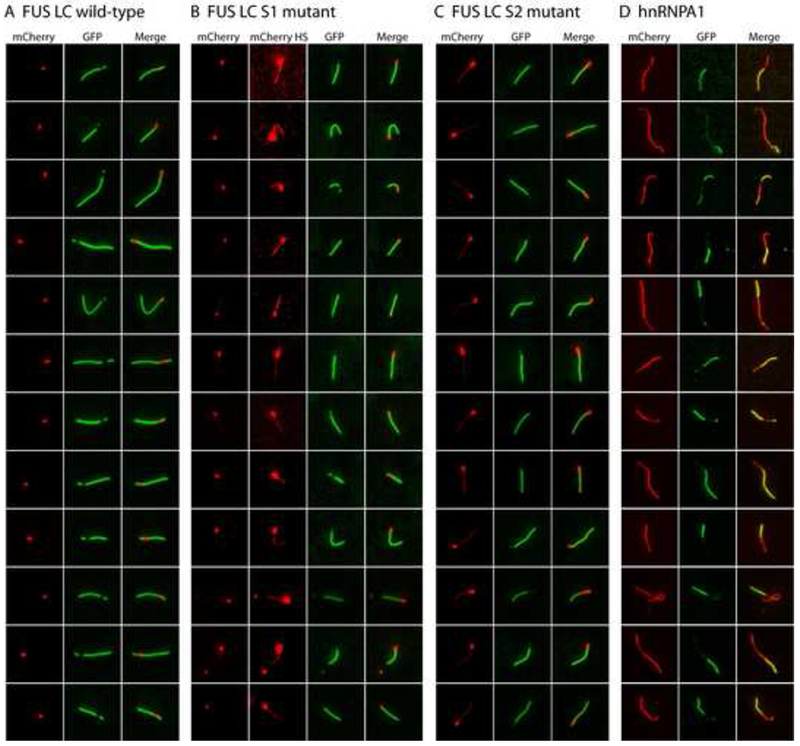
(A) Extensions of pre-formed mCherry:FUS LC fiber seeds by GFP-FUS LC monomers. mCherry:FUS seeds were mixed with monomeric GFP:FUS LC domain (wild-type) at an approximately 5-fold ratio of GFP:FUS to mCherry:FUS seeds. After incubation for 2 hr, fluorescent images for extended fibers were taken in a standard epifluorescent mode on an Olympus TIRF microscope (Experimental Procedures).
(B) Co-extension of mCherry:FUS seeds by monomeric GFP:FUS LC S1 mutant. Higher contrast mCherry images (mCherry HS) revealed that the monomeric S1 mutant (green color) co-polymerized with mCherry:FUS monomer from the seeds.
(C) Co-extension of mCherry:FUS seeds by monomeric GFP-FUS S2 mutant. mCherry signals in the co-polymerization region was stronger than that of (B) and visible in standard contrast images.
(D) Heterotypic co-polymerization of mCherry:FUS and GFP:hnRNPA1. mCherry:FUS seeds were mixed with monomeric GFP:hnRNPA1 LC at equal ratios. Red regions are interpreted to be mCherry:FUS seeds that extended homomeric polymerization during the reaction, green regions are interpreted to reflect heteromeric polymerization of GFP:hnRNPA1 into the mCherry:FUS seeds. Merged images reveal regions of yellow color interpreted to reflect co-polymerization of mCherry:FUS and GFP:hnRNPA1. Additional examples of heterotypic co-polymerization were provided in Figure S5.
The S1 and S2 mutants of the FUS LC domain lack the capability to form hydrogels but partially retain their ability to bind to the mCherry:FUS LC hydrogel and to associate with stress granules in living cells (Figure 4). As such, we assessed the ability of these mutants to grow from the mCherry:FUS LC seeds. As shown in Figure 7B and 7C, both of these proteins were capable of growing unidirectionally from the seeds. Close inspection of the images revealed that the extended regions of fiber growth were composed of both red and green color. In contrast to the green-only extension observed for the native FUS LC domain, the S1 and S2 mutants appear to have facilitated fiber extension via some form of copolymerization with mCherry:FUS LC. Finally, no evidence of seed extension was observed with either of the S3 or alls variants (data not shown).
The aforementioned studies give evidence that the high density of fiber ends in the hydrogel may homotypically trap GFP:FUS LC proteins. Next, we asked whether the mCherry:FUS LC fiber seeds might also prompt heterotypic co-polymerization with the LC domains of distinct RNA binding proteins. As shown in Figure 7D, when the mCherry:FUS seeds were incubated with soluble preparations of GFP:hnRNPA1 LC, fiber extension was observed to emanate from the red seeds. As in the cases of the S1 and S2 mutants of the FUS LC domain, fiber extensions with GFP:hnRNPA1 LC were observed to be of mixed color (green and red). We hypothesize that these extensions represent co-polymers composed of both the red, mCherry:FUS LC hybrid and the green, GFP:hnRNPA1 LC hybrid. Co-polymerization was similarly observed for: (i) GFP:RBM3 protein with mCherry:hnRNPA2 LC fiber seeds; (ii) GFP:hnRNPA1 LC with mCherry:hnRNPA2 seeds; and (iii) GFP:CIRBP with mCherry:FUS LC seeds (Figure S5). In the latter cases, particularly for GFP:CIRBP with mCherry:FUS seeds, the fibers were observed to alternate repeatedly in the intensity of green, red or mixed fluorescent signal as if homotypic fiber growth might be favored over heterotypic polymerization.
X-ray Structure of B-isox Crystals
To examine possible mechanisms for b-isox-mediated precipitation of the selective proteins, the b-isox-precipitated materials from cellular lysates of human U20S were visualized by light microscopy. Careful inspection of the precipitate revealed amorphous materials intermixed with micro-crystals (Figure 8A). This amorphous materials was positively stained with ethidium bromide as visualized by fluorescence microscopy, consistent with the presence of nucleic acids in the b-isox precipitate (Figure 8B) (Han et al., 2012). When the b-isox chemical was alone dissolved in the lysis buffer, star-like micro-crystals having multiple, needle-like projections were observed to the exclusion of any amorphous precipitate (Figure 8C and 8D). These micro-crystals were not stained with ethidium bromide (data not shown). Thus, we concluded that the b-isox chemical itself forms micro-crystals that selectively co-precipitate RNA-binding proteins containing LC domains. A central focus of the accompanying manuscript (Han et al., 2012) deals with molecular characterization of the RNA species selectively precipitated by the b-isox microcrystals.
Figure 8. X-ray structure of b-isox crystal.
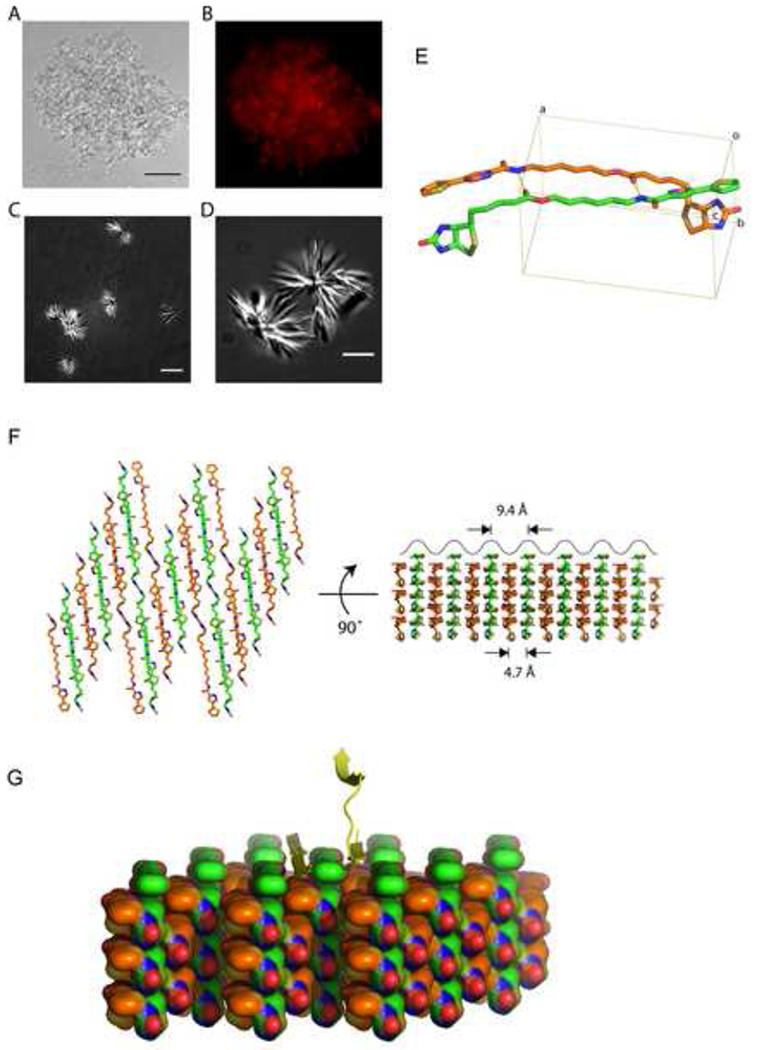
(A and B) Light and fluorescence microscopic images of precipitated materials observed upon exposure of b-isox chemical to lysate prepared from human U2OS cells. Prior to b-isox compound addition, lug/ml of ethidium bromide was added to the lysate. Precipitated aggregates revealed amorphous materials intermixed with b-isox micro-crystals (A). Black scale bar indicates 50um. Aggregated granules stained positively with ethidium bromide as imaged by fluorescence microscopy (B).
(C and D) Light microscopic images of precipitated materials observed upon exposure of b-isox chemical to lysate buffer alone. Precipitated material was photographed with differential interference contrast mode on a Deltavision fluorescent microscope. Precipitated material was exclusively composed of starshaped micro-crystals having multiple, needle-like projections. White scale bar indicates 10um for (C) and 5um for (D).
(E) Structure of the b-isox chemical. Two crystallographically independent molecules in the asymmetric unit are shown as a stick model. Atom color codes are: red – oxygen, blue – nitrogen, yellow – sulfur, orange and green – carbon. Hydrogen bonds are represented by yellow dot lines. Rectangular lines define unit cell. An averaged distance between the aliphatic linkers of the two molecules is 4.7Å. Statistics for X-ray data collection and structure refinement were provided in Table S2.
(F) Assembly of the symmetry-related molecules in the b-isox crystal. Left: top view of the assembly, right: side view as rotated by 90°. Two independent molecules (orange and green) antiparallely aligned and alternately form respective layers. The upper surface reveals a wave of line corresponding to the visible surface of the left view. Individual valleys are sites for addition of b-isox molecules when the crystal grows. Since the width of the valley is 9.4Å, it provides reasonable access sites for a β-strand polypeptide.
(G) Hypothetical model showing the binding of β-strand polypeptide within the valleys of the surface of the b-isox crystal. A tandem array of β–strands are hypothesized on the left side. One β-strand of a cross-β structure is hypothesized on the right side.
How might the b-isox micro-crystals selectively precipitate LC domain-containing proteins? In pursuit of this question, we determined the structure of the b-isox compound in a crystalline form prepared in the same buffer used for all biochemical experiments performed herein. The b-isox chemical in the crystals exists in an extended structure wherein the isoxazole and biotin moieties are maximally separated by the aliphatic linker, and two molecules in the asymmetric unit interact in an anti-parallel manner (Figure 8E). Two hydrogen bonds facilitate linker interaction and the distance between linkers is 4.7Å (average for the nine paired atoms). Symmetry-related molecules generated far beyond the unit cell formed respective layers, which stacked alternatively (Figure 8F). As a result, one surface of the crystal generates a wavy, repetitive terrain, with valleys representing sites for the docking of additional b-isox molecules for crystal growth. Since the interaction of two b-isox molecules in the crystal match the dimensions of β-strand – β-strand interaction (4.7Å), we hypothesize that the valleys on the wavy surface of the b-isox crystal represent docking sites for β-strands as modeled in Figure 8G. If correct, this concept offers the simple prediction that the surface of b-isox crystals offers a site for random coil polypeptides (LC sequences) to transform to extended β-strand conformation templated by the ordered and repetitive valleys on the crystal surface. Properly folded proteins would not be expected to display sufficiently extensive segments of random coil to allow conversion to extended β-strand structure and tight interaction with the crystalline surface. Finally, dissolution of the b-isox crystals, which takes place upon warming to 37°C, would be expected to allow bound proteins to re-convert back to random coil and solubility as observed experimentally (Figure 1).
Discussion
The central thesis evolving from the present study is that LC sequences associated with regulatory proteins can reversibly transform from solubility to a polymeric, amyloid-like state. We hypothesize that the latter state may form the organizing principle for sub-cellular structures that are not membrane bound, including various forms of RNA granules that have been recognized from cytological studies for more than a century. Ample evidence has accumulated showing that LC sequences can form pathological aggregates, particularly in the context of neurodegenerative disease (Ross and Poirier, 2004; Shastry, 2003). By contrast, the underlying biological utility of LC sequences remains enigmatic. Here we offer the potentially unifying hypothesis that LC sequences allow the movement of regulatory proteins in and out of organized, sub-cellular domains via reversible polymerization into dynamic, amyloid-like fibers. These may include cytologically visible domains such as RNA granules, Cajal bodies, nuclear speckles, and nuclear “factories” that organize events including RNA splicing and gene transcription. We emphasize, however, that a continuum likely ranges from the sub-microscopic use of dynamically operative, fibrous organizing centers all the way to readily visualized RNA granules.
When the b-isox chemical was applied to lysates prepared from a variety of cell or tissue types, it triggered the precipitation of a select group of proteins associated with RNA biogenesis (Figure 1). This unusual property of the b-isox compound is unlikely to have anything to do with the ability of the parental isoxazole to cause mouse embryonic stem cells to differentiate into cardiac myocytes (Sadek et al., 2008), or to cause neuronal stem cells to adopt the morphology of differentiated neurons (Schneider et al., 2008). This conclusion is based upon the observation that the parental isoxazole has no capacity whatsoever to cause precipitation of the spectrum of RNA binding proteins described in this study (data not shown).
We hypothesize that the b-isox micro-crystals co-precipitate the spectrum of RNA binding proteins described in this study. One possible interpretation of this unusual phenomenon is that the surface of the micro-crystals may create an appropriately organized template to trigger the conversion of the LC sequences from a soluble state to a polymerized, fiber-like state. Structural studies of the b-isox micro-crystals reveal an ordered, wavy surface (Figure 8). The valleys of this surface are of dimensions suitable for accommodation of extended β-strand, raising the possibility that the b-isox crystal surface does no more than facilitate the transition of disordered polypeptide sequences from a random coil state to an extended β-strand.
Central to our interpretation of many observations in this study is the conclusion that the LC sequences associated with many different RNA binding proteins can reversibly transform from a soluble state into polymeric, amyloid-like fibers. The LC domains are both necessary and sufficient for b-isox-mediated precipitation, for hydrogel formation, and for retention by pre-formed hydrogel droplets (Figures 2 and 3). Numerous studies have given evidence that these same LC sequences are capable of helping localize RNA binding proteins to P bodies in yeast (Decker et al., 2007; Reijns et al., 2008) and mammalian stress granules (Gilks et al., 2004). We have observed correlative effects of mutations of the [G/S]Y[G/S] triplet repeats found in the FUS LC domain on retention to a pre-formed hydrogel and attraction to stress granules in cultured U20S cells (Figure 4). As such, we hypothesize that the dynamic movement of RNA binding proteins in and out of stress granules, P bodies, polar granules and neuronal granules may be driven primarily by the reversible transition of the LC sequences from solubility to polymeric fibers. Additional data consistent with this interpretation are described in the accompanying manuscript (Han et al., 2012).
That the FUS LC domain can be polymerized into amyloid-like fibers is supported by both TEM and X-ray diffraction (Figure 5). The protein concentration required to observe fiber polymerization, roughly 10mg/ml, is clearly higher than the ambient concentrations of individual RNA binding proteins within cells. One means of considering how appropriately high, local concentrations of these proteins might be met is via recruitment to an RNA target. hnRNP proteins coat mRNAs in a manner equally dense to the coating of DNA by histone proteins. As such, it is possible that substrate binding helps achieve locally high concentrations of RNA binding proteins adequate to facilitate the conversion of the LC sequences from a disordered, random coil state to polymeric fibers.
Although the amyloid-like fibers of mCherry:FUS LC and mCherry:hnRNPA2 LC are morphologically similar to pathogenic, prion-like amyloids, and yield an X-ray diffraction pattern consistent with cross-β structure (Figure 5), a substantive difference is that the fibers reported herein are readily de-polymerized (Figure 6). It is clear that additional biophysical experiments will be required to understand the structural underpinnings of these newly-described amyloid-like fibers, as well as the dynamics controlling both polymerization and de-polymerization.
By incubating pre-formed fibrous seeds with various samples of soluble test proteins, it was possible to visualize fiber extension (Figure 7). Mutated variants of the GFP:FUS LC domain missing either 5 (S1) or 9 (S2) of the tyrosine residues centered within the [G/S]Y[G/S] repeats were capable of polymerizing from the mCherry:FUS LC seeds. Compared with the native GFP:FUS LC domain, however, these mutants tended to grow uni-directionally. It was likewise observed for these mutants that the green polymeric growth was mixed with red fluorescence, particularly in the case of the S2 mutant. We hypothesize that the mixture of red and green color reflects the fact that these extensions consist of a mixture of native and mutant subunits. We tentatively conclude that the mutated LC domains are somehow assisted into the polymeric state by the native LC domains of mCherry:FUS subunits that are themselves dynamically polymerizing and depolymerizing from the mCherry:FUS seeds originally doped into the reaction assays.
The mCherry:FUS LC hydrogel droplets were observed to trap and retain GFP fusion proteins linked to the LC domains of numerous RNA binding proteins. Whereas we do not fully understand this heterotypic trapping, evidence is provided in favor of the capacity of mCherry:FUS polymeric seeds to stimulate fiber extension with the GFP-fused LC domains of hnRNPA1 and CIRBP (Figures 7 and S5). In the former of these cases, fiber growth from the seeds appeared to be relative uniform and consist of roughly equal amounts of green and red color. More complicated patterns of co-polymerization were observed when either mCherry:FUS or mCherry:hnRNPA2 seeds were exposed to the LC domains of hnRNPA1 or RBM3 (Figure S5). Interpreted most simply, these images appear to reflect amyloid-like polymers composed of the LC domains of heterologous RNA binding proteins. The classically studied polymeric fibers of eukaryotic cells, including actin, microtubules and various forms of intermediate filaments, are homotypic in composition. As such, it is with caution that we offer the tentative conclusion that the amyloid-like fibers polymerized from the LC domains of RNA binding proteins might be hetero-polymeric. Although additional studies will be required to properly evaluate these provocative observations, the results of this study offer compelling evidence that mCherry:FUS and mCherry:hnRNPA2 hydrogel droplets are capable of trapping the LC domains of heterologous RNA binding proteins. As demonstrated by Han and colleagues in the accompanying manuscript (Han et al., 2012), heterotypic trapping can be extended to native RNA binding proteins associated, in cellular lysates, with their cognate messenger RNAs.
Hydrogels not unlike what we describe in this report have been observed upon expression, purification and concentration of the Nsp1p component of nuclear pores (Frey and Gorlich, 2007; Frey et al., 2006). Nsp1p is the yeast ortholog of the mammalian Nup62 nuclear pore protein. Solid state NMR studies of Nsp1p hydrogels revealed evidence of β-structure, favoring the conclusion that Nsp1p may – upon concentration – become organized into some sort of fibrous network. Nsp1p, like many other nuclear pore proteins, contains multiple phenylalanine-glycine (FG) repeats. Mutation of all phenylalanine residues in Nsp1p to serine blocked hydrogel formation, yet mutation of the same residues to tyrosine did not. It is possible that the network formed from the Nsp1p nuclear pore protein rich in FG repeats might be related to the amyloid-like fibers formed by the LC domain of FUS that contains 27 repeats of the tripeptide sequence [G/S]Y[G/S], It is intriguing that one nuclear pore protein, Nup98, has been reported not only to be present in polar granules of C. elegans embryos, but required for their integrity and function (Voronina and Seydoux, 2010). We make note of the fact that the mammalian and fly orthologs of Nsp1p/Nup98 were precipitated by the b-isox micro-crystals along with the diverse spectrum of proteins involved in RNA biogenesis. We close by pointing out the perplexing observation that polar granules of C. elegans are held in close proximity to clusters of nuclear pores in immature germ cells (Pitt et al., 2000). What connections might be envisioned from these observations, be they theoretical, conceptual or mechanistic, are open to speculation.
Experimental Procedures
Detailed experimental procedures are available as Supplementary Data.
Materials
Synthesis of the biotinylated isoxazole is described in the Extended Experimental Procedures. The transfection plasmids for the stress granules recruitment assay were constructed using the pcDNA3.1(+) vector for mammalian expression (Invitrogen, USA). The expression plasmids for the recombinant proteins used in all the biochemical assays were constructed using pHis-parallel vectors (Sheffield et al., 1999). The proteins were overexpressed in E. coll BL21(DE3) and purified with Ni-NTA (Qiagen, USA) or glutathione resin (GE Healthcare, USA).
Biotinylated Isoxazole-mediated Precipitation
Biotinylated isoxazole was added to cell or tissue lysates at 10, 30, or 100uM final concentration. The reaction solutions were incubated at 4°C for 1 hour and then centrifuged at 14K rpm for 15 min. The pellets and supernatants were separately analyzed by 4-12% Tris-Glycine gradient gels.
Computation Analysis of Low Complexity Sequences and [GS]Y[GS] Motifs
Low complexity sequences were identified using the SEG program with default parameter settings (Wootton and Federhen, 1996). The [GS]Y[GS] tripeptide motif was counted for each protein in the human, mouse and fly proteomes, and the proteins with four or more copies of this motif were considered to be enriched. Statistical significance was analyzed by Fisher’s Exact test using “R” software (R Development Core Team, 2010).
Stress Granule Recruitment Assay
U20S cells were transfected with 10ug of plasmids of Flag-FUSΔ32 WT or Flag-FUSΔ32 Y->S mutants. After 36 hour, stress granules were induced with 0.5mM sodium arsenite for 1 hour. The cells were fixed on coverslips with αFlag antibody and imaged by fluorescent microscopy.
Hydrogel Binding Assay
Preformed hydrogel droplets (0.5ul) of mCherry:FUS LC and mCherry:hnRNPA2 LC domains were soaked with 1uM GFP-fusion LC domains at 4 °C for up to a week. Horizontal sections of the hydrogel droplets were scanned with fluorescent microscopy. For signal decay assays, the GFP solution was replaced with fresh buffer, and GFP fluorescent images were subsequently taken at the indicated time points. A one-phase exponential decay equation was applied to calculate half lives of the GFP signal retention.
TEM Imaging and X-Ray Diffraction
Each of the mCherry:FUS and His-tagged FUS hydrogels were directly transferred to a TEM grid. The grid was washed with distilled water, stained with 3% uranyl acetate, and dried in air. TEM images were obtained at 80kV on FEI Morgagni TEM. For X-ray diffraction experiments, pre-formed mCherry:FUS and mCherry:hnRNPA2 hydrogels were dialyzed against distilled water overnight twice followed by lyophilization. The solid materials were mounted on a nylon cryoloop in 50% ethylene glycol. Diffraction images were collected at 100 K using a Rigaku FR-E generator (wavelength 1.5418 Å) equipped with a R-AXIS HTC imaging plate detector.
Fiber Extension Assays
Pre-formed fibers of mCherry:FUS LC or mCherry:hnRNPA2 LC domains were sonicated to be broken into many short fiber seeds. The seeds were mixed with 100uM of the monomeric LC domains of GFP:FUS wild-type, S1, S2, S3, or alls or 10uM of GFP:hnRNPA1, GFP:RBM3, or GFP:CIRBP. After incubation at RT or 4°C, the extended fibers were visualized with a total internal reflection fluorescent (TIRF) microscope with a 100X objective lens.
Assays of SDS Sensitivity
Pre-formed fibers of mCherry:FUS and mCherry:hnRNPA2 LC domains were passed through a O.lum spin filter to remove fiber particles with/without 2% SDS. UV absorbance was measured to monitor the amount of monomeric protein that passed through the filter. For SDD-AGE experiments, fiber solutions were briefly sonicated to break down long fibers to allow entry into the 1.5% agarose gel. Fiber samples were mixed with the indicated concentrations of SDS and 5% glycerol and bromophenol blue and incubated at 37°Cfor 10 min. Fibers and monomers were separated by agarose gel electrophoresis in TAE buffer supplemented with 0.1% SDS. In parallel, pre-formed fibers of the yeast Sup35 protein were treated under the same conditions. The separated proteins were transferred to a nitrocellulose and visualized by western blotting using a His-tag antibody for mCherry:FUS and mCherry:hnRNPA2, and an antibody specific for Sup35.
X-ray Structure Determination of B-Isox Crystals
The b-isox chemical was crystallized at 4°C by vapor diffusion under the following conditions: a compound drop (4ul) containing 0.75mM b-isox, 80% DMSO and 20% lysis buffer was diffused against a reservoir (1ml) consisting of 70% DMSO and 30% lysis buffer. X-ray diffraction data were collected at Beamline 19ID of the Structural Biology Center at Argonne Photon Source (Argonne, Chicago IL). The structure was solved by a direct method using SHELX program package (Sheldrick, 2008). Statistics for X-ray data collection and structure refinement is shown in Table S2. The coordinate and reflection files were deposited at Cambridge Crystallographic Data Center (CCDC 873064).
Supplementary Material
1
Acknowledgements
We thank Joe Ready, David Trudgian, Randal Halfmann and Tom Pollard for valuable advice; Joan Steitz, Geraldine Sedoux, Joe Gall, Ben Tu, Mike Rosen, Randal Halfmann and Phil Sharp for helpful criticisms of the manuscript; Randal Halfman for providing yeast Sup35 amyloid fibers and antibodies specific to the Sup35 NM protein; Xianbing Qi for re-synthesis of the biotinylated isoxazole; Chad A. Brautigam and Frank J. Rotella for X-ray data collection at APS; and the shared resource cores in Microscopic Imaging, Proteomics and DNA Sequencing at UTSWMC for technical support. This work was funded by an unrestricted endowment provided SLM by an anonymous donor.
References
- Alberti S, Halfmann R, King O, Kapila A, and Lindquist S (2009). A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N (2006). RNA granules. J Cell Biol 172, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astbury WT, and Dickinson S (1935). The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J 29, 2351–2360 2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ Jr., Sapp P, McKenna-Yasek D, Brown RH Jr., and Hayward LJ (2010). Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 19, 4160–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, and Hyman AA (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- Buchan JR, and Parker R (2009). Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon JH, and Parker R (2011). Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci 124, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, and Kelly PT (1990). In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci 10, 1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, and Parker R (2007). Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179, 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM (2003). Protein folding and misfolding. Nature 426, 884–890. [DOI] [PubMed] [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, et al. (2006). Characterization of an RNA granule from developing brain. Mol Cell Proteomics 5, 635–651. [DOI] [PubMed] [Google Scholar]
- Frey S, and Gorlich D (2007). A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130, 512–523. [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, and Gorlich D (2006). FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Wang JT, Motegi F, and Seydoux G (2010). Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science 330, 1685–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CC, Tucker RP, and Matus A (1988). Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature 336, 674–677. [DOI] [PubMed] [Google Scholar]
- Geddes AJ, Parker KD, Atkins ED, and Beighton E (1968). “Cross-beta” conformation in proteins. J Mol Biol 32, 343–358. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, and Anderson P (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15, 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu L, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. (2012). Cell-free Formation of RNA Granules: Bound RNAs Identify Features and Components of Cellular Assemblies. Cell **, ***-***. [DOI] [PubMed]
- Huntley MA, and Golding GB (2002). Simple sequences are rare in the Protein Data Bank. Proteins 48, 134–140. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, and Hirokawa N (2004). Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, and Anderson P (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL (1993). Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes Dev 7, 948–960. [DOI] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, and Kosik KS (1996). Translocation of RNA granules in living neurons. J Neurosci 16, 7812–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, and Nusslein-Volhard C (1991). The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 112, 679–691. [DOI] [PubMed] [Google Scholar]
- Mahowald AP (1968). Polar granules of Drosophila. II. Ultrastructural changes during early embryogenesis. J Exp Zool 167, 237–261. [DOI] [PubMed] [Google Scholar]
- McKinley MP, Bolton DC, and Prusiner SB (1983). A protease-resistant protein is a structural component of the Scrapie prion. Cell 35, 57–62. [DOI] [PubMed] [Google Scholar]
- Meersman F, and Dobson CM (2006). Probing the pressure-temperature stability of amyloid fibrils provides new insights into their molecular properties. Biochim Biophys Acta 1764, 452–460. [DOI] [PubMed] [Google Scholar]
- Michelitsch MD, and Weissman JS (2000). A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A 97, 11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem DR (1995). mRNA localisation during development. Dev Biol 172, 377–395. [DOI] [PubMed] [Google Scholar]
- Murata Y, and Wharton RP (1995). Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80, 747–756. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, and Schupbach T (1993). The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 75, 165–174. [PubMed] [Google Scholar]
- Palacios IM, and St Johnston D (2001). Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu Rev Cell Dev Biol 17, 569–614. [DOI] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, and Priess JR (2000). P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol 219, 315–333. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2010). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, and Beggs JD (2008). A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci 121, 2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, and Poirier MA (2004). Protein aggregation and neurodegenerative disease. Nat Med 10 Suppl, S10–17. [DOI] [PubMed] [Google Scholar]
- Sadek H, Hannack B, Choe E, Wang J, Latif S, Garry MG, Garry DJ, Longgood J, Frantz DE, Olson EN, et al. (2008). Cardiogenic small molecules that enhance myocardial repair by stem cells. Proc Natl Acad Sci U S A 105, 6063–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JW, Gao Z, Li S, Farooqi M, Tang TS, Bezprozvanny I, Frantz DE, and Hsieh J (2008). Small-molecule activation of neuronal cell fate. Nat Chem Biol 4, 408–410. [DOI] [PubMed] [Google Scholar]
- Shastry BS (2003). Neurodegenerative disorders of protein aggregation. Neurochem Int 43, 1–7. [DOI] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, and Derewenda Z (1999). Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif 15, 34–39. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (2008). A short history of SHELX. Acta Crystallogr A 64, 112–122. [DOI] [PubMed] [Google Scholar]
- Smith JF, Knowles TP, Dobson CM, Macphee CE, and Welland ME (2006). Characterization of the nanoscale properties of individual amyloid fibrils. Proc Natl Acad Sci U S A 103, 15806–15811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, and Wood WB (1982). Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc Natl Acad Sci U S A 79, 1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde M, and Blake C (1997). The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv Protein Chem 50, 123–159. [DOI] [PubMed] [Google Scholar]
- Uversky VN (2002). Natively unfolded proteins: a point where biology waits for physics. Protein Sci 11, 739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, and Seydoux G (2010). The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development 137, 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Seydoux G, Sassone-Corsi P, and Nagamori I (2011). RNA Granules in Germ Cells. Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, and Lehmann R (1991). Nanos is the localized posterior determinant in Drosophila. Cell 66, 637–647. [DOI] [PubMed] [Google Scholar]
- Wickner RB (1994). [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569. [DOI] [PubMed] [Google Scholar]
- Wille H, and Prusiner SB (1999). Ultrastructural studies on scrapie prion protein crystals obtained from reverse micellar solutions. Biophys J 76, 1048–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, and Federhen S (1996). Analysis of compositionally biased regions in sequence databases. Methods Enzymol 266, 554–571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1