The Proneural Gene Mash1 Specifies an Early Population of Telencephalic Oligodendrocytes (original) (raw)
Abstract
The bHLH (basic helix-loop-helix) transcription factor Mash1 is best known for its role in the regulation of neurogenesis. However, Mash1 is also expressed in oligodendrocyte precursors and has recently been shown to promote the generation of oligodendrocytes in cell culture, suggesting that it may regulate oligodendrogenesis as well. Here, we show that in the developing ventral forebrain, Mash1 is expressed by a subset of oligodendrocyte precursors (OPCs) as soon as they are generated in the ventricular zone. Using reporter mice, we demonstrate that a subset of OPCs in both the embryonic and postnatal forebrain originate from Mash1-positive progenitors, including a large fraction of adult NG2-positive OPCs. Using Mash1 null mutant mice, we show that Mash1 is required for the generation of an early population of OPCs in the ventral forebrain between embryonic day 11.5 (E11.5) and E13.5, whereas OPCs generated later in embryonic development are not affected. Overexpression of Mash1 in the dorsal telencephalon induces expression of PDGFRα (platelet-derived growth factor receptor alpha) but not other OPC markers, suggesting that Mash1specifies oligodendrogenesis in cooperation with other factors. Analysis of double-mutant mice suggests that Olig2 is one of the factors that cooperate with Mash1 for generation of OPCs. Together, our results show for the first time that Mash1 cooperates in vivo with Olig2 in oligodendrocyte specification, demonstrating an essential role for Mash1 in the generation of a subset of oligodendrocytes and revealing a genetic heterogeneity of oligodendrocyte lineages in the mouse forebrain.
Keywords: proneural, bHLH, cell lineage, glia, Olig2, Mash1
Introduction
Glial cells play an essential role in the formation and activity of the CNS. New functions for astrocytes and oligodendrocytes have recently emerged, including in regulation of synaptic transmission and in adult neurogenesis (Doetsch, 2003; Newman, 2003; Lin and Bergles, 2004). However, our understanding of the origin and heterogeneity of glial cells, and of the mechanisms that control their generation, is still very limited.
In the mouse, oligodendrocyte precursors (OPCs) are first identified in the ventral spinal cord at ∼12.5 d of embryonic development [embryonic day 12.5 (E12.5)], and at approximately E13.5 in the ventral forebrain, by the expression of the specific markers platelet-derived growth factor receptor alpha (_PDGFR_α), Olig1, and Sox10 (Pringle and Richardson, 1993; Tekki-Kessaris et al., 2001). OPCs are generated in the ventricular zone (VZ) and migrate extensively in the surrounding parenchyma while continuing to proliferate, upregulating later OPC markers such as NG2 and O4 (for review, see Rowitch, 2004). Mature OPCs eventually leave the cell cycle and upregulate myelinating proteins such as the myelin basic protein (MBP), CNPase, and proteolipid protein (PLP) (for review, see Rowitch, 2004). OPCs are also found in the adult CNS, where they have a more complex morphological appearance. These cells appear to be arrested at a precursor stage and constitute the main dividing cell population outside of the neurogenic regions (Dawson et al., 2003).
Oligodendrocytes are generated at multiple locations by progenitors with different potentials (Parnavelas, 1999; He et al., 2001; Marshall and Goldman, 2002; Marshall et al., 2003; Zerlin et al., 2004). In the forebrain, OPCs are initially produced from a ventral territory extending from the medial ganglionic eminence (MGE) to the anterior entopeduncular area (AEP) (Pringle and Richardson, 1993; Spassky et al., 1998; Tekki-Kessaris et al., 2001). A later source of embryonic OPCs has recently been identified in the lateral and/or caudal ganglionic eminences (LGE/CGE) (Kessaris et al., 2006), whereas most OPCs generated at postnatal stages have been shown to originate from the dorsal telencephalon (Gorski et al., 2002; Kessaris et al., 2006).
Important progress has been made in the characterization of the molecular mechanisms controlling the specification and differentiation of oligodendrocytes, with the discovery of the bHLH (basic helix-loop-helix) transcription factors Olig1 and Olig2. Mice mutant for Olig2 completely lack OPCs in the embryonic spinal cord whereas a few OPCs remain in the brain because of the compensatory activity of Olig1 (Lu et al., 2002; Zhou and Anderson, 2002). Mice lacking Olig1 present defects in the differentiation of OPCs into mature oligodendrocytes (Xin et al., 2005) and in axon remyelination after lesions (Arnett et al., 2004). Another bHLH protein, Mash1, has also been implicated in oligodendrogenesis. Mash1 is broadly expressed by brain and spinal cord progenitors and plays a major role in neurogenesis (Casarosa et al., 1999; Helms et al., 2005). However, Mash1 is also expressed in OPCs in the optic nerve and postnatal telencephalon (Kondo and Raff, 2000; Wang et al., 2001; Parras et al., 2004), and in vitro studies have indicated that Mash1 can cooperate with Olig2 to activate the MBP promoter (Gokhan et al., 2005). Furthermore, Mash1 null mutant mice lack some OPCs in the olfactory bulb at birth (Parras et al., 2004). However, the exact function of Mash1 in oligodendrogenesis in vivo has remained to be addressed. Here, we have directly examined in vivo the role of Mash1 in oligodendrogenesis in the embryonic forebrain. We demonstrate that Mash1 is expressed in ventricular OPCs at the time of their specification and is required for the generation of an early population of OPCs, and we show that this function involves a genetic interaction with Olig2.
Materials and Methods
Trangenic mice.
A Mash1::Cre transgene was generated from the bacterial artificial chromosome (BAC) RP24–130P7 (Children's Hospital Oakland Research Institute, Oakland, CA) with the Mash1 coding sequence replaced by an IRES-EGFP (internal ribosomal entry site-enhanced green fluorescent protein)-Cre fusion cassette from the plasmid pIGCN21 (see Materials and Methods and supplemental Fig. 3, available at www.jneurosci.org as supplemental material) by homologous recombination in bacteria as described in Lee et al. (2001). This BAC contains 70 kb of sequence upstream and 100 kb of sequence downstream of the Mash1 coding sequence. The Mash1::Cre transgenic line was generated by pronuclear injection of the modified BAC. Transgenic embryos were genotyped by PCR; the upper primer was ATCCGAAAAGAAAACGTTGA, and the lower primer was ATCCAGGTTACGGATATAGT, with 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The following transgenic lines were used and genotyped using the protocols described in the references: _Mash1_KO (Parras et al., 2002), _Olig2_KO (Lu et al., 2002), _Mash1_KINgn2 (Parras et al., 2002), _Ngn2_KIMash1 (Fode et al., 2000), Mash1::GFP (Gong et al., 2003) (newborns were genotyped by GFP expression under a UV microscope), and _Rosa26_stop-YFP (Srinivas et al., 2001). Mash1;Olig2 double mutant embryos were generated from double heterozygous mice.
Immunohistochemistry.
Embryos and dissected embryonic brains were fixed for 30 min in 4% paraformaldehyde (Sigma, St. Louis, MO) at 4°C and cryoprotected in 20% sucrose. Samples were embedded in octopamine (BDH Chemicals, Poole, UK) and sectioned at 10 μm sections with a cryostat (Leica, Nussloch, Germany) and then processed for immunohistochemistry or stored at −80°C. Postnatal and adult mice were perfused with chilled 4% paraformaldehyde. Brains were dissected out, processed as described above, sectioned at 30–40 μm with a cryostat, and labeled by immunohistochemistry as floating sections. Sections were incubated in a blocking solution [PBS plus 10% normal goat serum (Vector Laboratories, Burlingame, CA) and 0.1% Tween 20 or Triton X-100] and then with primary antibodies overnight at 4°C. The following primary antibodies were used: mouse monoclonal antibodies to adenomatus polyposis coli (APC; 1:200; Calbiochem, La Jolla, CA), Mash1 (1:10; a gift from D. J. Anderson, California Institute of Technology, Pasadena, CA), 2′3′-cyclic nucleotide 3′-phosphodiesterase (CNPase; 1:500; Millipore, Bedford, MA), GFP (1:500; Invitrogen, San Diego, CA), Ngn2 (1:10; gift from D. J. Anderson), and Olig2 (1:400) (Ligon et al., 2006); rabbit polyclonal antibodies to Caspase3 (1:1000; R & D Systems, Minneapolis, MN), GFP (1:2000; Invitrogen), Gsh2 (1:3000; gift from K. Campell, Children's Hospital Medical Center, Cincinnati, OH), NG2 (1:1000; Millipore), Olig1 (1:1000) (Mizuguchi et al., 2001), and Olig2 (1:1000; Millipore); and rat monoclonal antibody to PDGFRα (1:800; BD Biosciences, Franklin Lakes, NJ). Secondary antibodies were incubated 1 h at room temperature and included the following: goat anti-mouse Alexa Fluor 488, goat anti-mouse Alexa Fluor 568, goat anti-rabbit Alexa Fluor 488, goat anti-rabbit Alexa Fluor 568, goat anti-rat Alexa Fluor 568, (1:1000; all from Invitrogen), and Cy5 conjugated goat anti-rat (1:500; Jackson ImmunoResearch, West Grove, PA). DAPI (4,6,diamidino-2-phenylindole; 1:5000) was used to label DNA, and sections were mounted in Aquapolymount medium (Polysciences, Warrington, PA). Immunofluorescence was visualized with a Leica TC SP1 confocal microscope. Images were treated using an imaging software package created by J. L. Vonesh (Institute of Genetics and Molecular and Cellular Biology, Strasbourg, France) and Adobe Photoshop.
Cell counting.
Because of the important variability in OPC numbers at different rostrocaudal levels of the forebrain, the data for Figures 4, 5, and 6 were obtained by counting one series of sections covering the whole forebrain. The total number of PDGFRα+ cells in the forebrain was estimated by multiplying the number obtained from counting one series by the number of series performed (two for E12.5, three for E13.5, and four for E14.5).
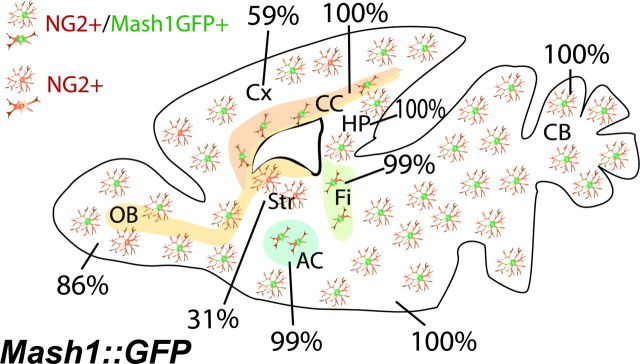
Figure 4.
Mash1::GFP is expressed in many adult NG2+ OPCs. Schematic representation of a sagittal brain section from a 6-week-old Mash1::GFP mouse. The ratio of GFP-positive (brown/green) to GFP-negative (brown) cells among NG2+ cells varies in different gray matter regions, whereas most NG2+ cells express GFP in white matter regions. AC, Anterior commissure; CC, corpus callosum; CB, cerebellum; Cx, cortex; Fi, fimbria; HP, hippocampus; OB, olfactory bulb; Str, striatum.
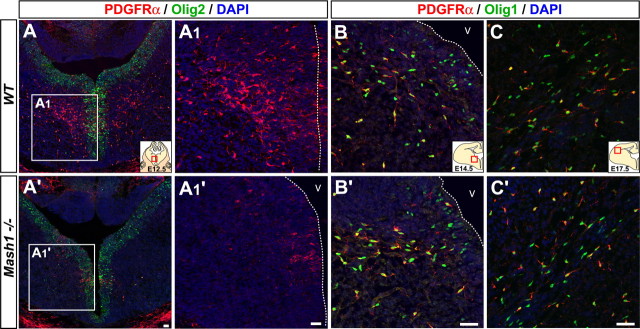
Figure 5.
Mash1 is required for the generation of early born OPCs in embryonic telencephalon. Telencephalic coronal sections of wild-type (A–C) and _Mash1_−/− embryos (A′–C′). PDGFRα+ cells found in VZ and SVZ at E12.5 in wild-type embryos (A, A1) are mostly missing in _Mash1_−/− embryos (A′, A1′). At E14.5 both Olig1+,PDGFRα+ OPCs and Olig1+,PDGFRα− cells are present in the AEP of wild-type (B) and Mash1 mutant (B′) embryos. At E17.5, no difference is found in the distribution of Olig1+,PDGFRα+ OPCs migrating into the cortex in wild-type (C) and _Mash1_−/− (C′) embryos. The dotted line marks the ventricular surface. V, Ventricle. Scale bars, 20 μm.
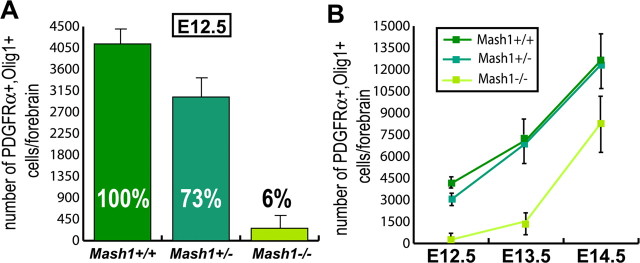
Figure 6.
Quantification of the requirement for Mash1 in the generation of early born OPCs in embryonic telencephalon. Quantification of PDGFRα+,Olig1+ cells in wild-type, Mash1+/−, and _Mash1_−/− embryos at E12.5 (A) and at different embryonic stages (B). Series of forebrain sections were counted to estimate the total number OPCs in the forebrain (n = 3). Error bars indicate SEM.
Results
To begin to address the role of Mash1 in the specification of oligodendrocytes in the embryonic telencephalon, we first characterized the distribution of OPCs in this brain region and examined their expression of Mash1, using antibodies against PDGFRα, Olig1, and Olig2, the earliest known markers for OPCs.
Olig1 and PDGFRα were coexpressed in a subset of cells in the VZ and subventricular zone (SVZ) of the ventral forebrain, mostly confined to a domain extending continuously from the posterior part of the MGE rostrally, to the AEP and the hypothalamic area of the diencephalon caudally (Fig. 1A–D) (Olivier et al., 2001; Spassky et al., 2001; Tekki-Kessaris et al., 2001; Kessaris et al., 2006). From E11.5, when they were first detected, to E13.5, all Olig1+ cells coexpressed PDGFRα in the VZ and the SVZ, and reciprocally, most PDGFRα+ cells coexpressed Olig1 (Fig. 1A). From E14.5 onwards, however, only a fraction of Olig1+ cells, both in the VZ and outside of it, coexpressed PDGFRα (Fig. 1B,C). Olig1+,PDGFRα+ cells coexpressed NG2 but Olig1+,PDGFRα− cells did not (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), suggesting that this later population is not engaged in oligodendrogenesis. At these early stages, expression of other early markers of oligodendritic lineage such as Sox10 and Nkx2.2, could not be detected at the protein level in forebrain OPCs (data not shown).
Figure 1.
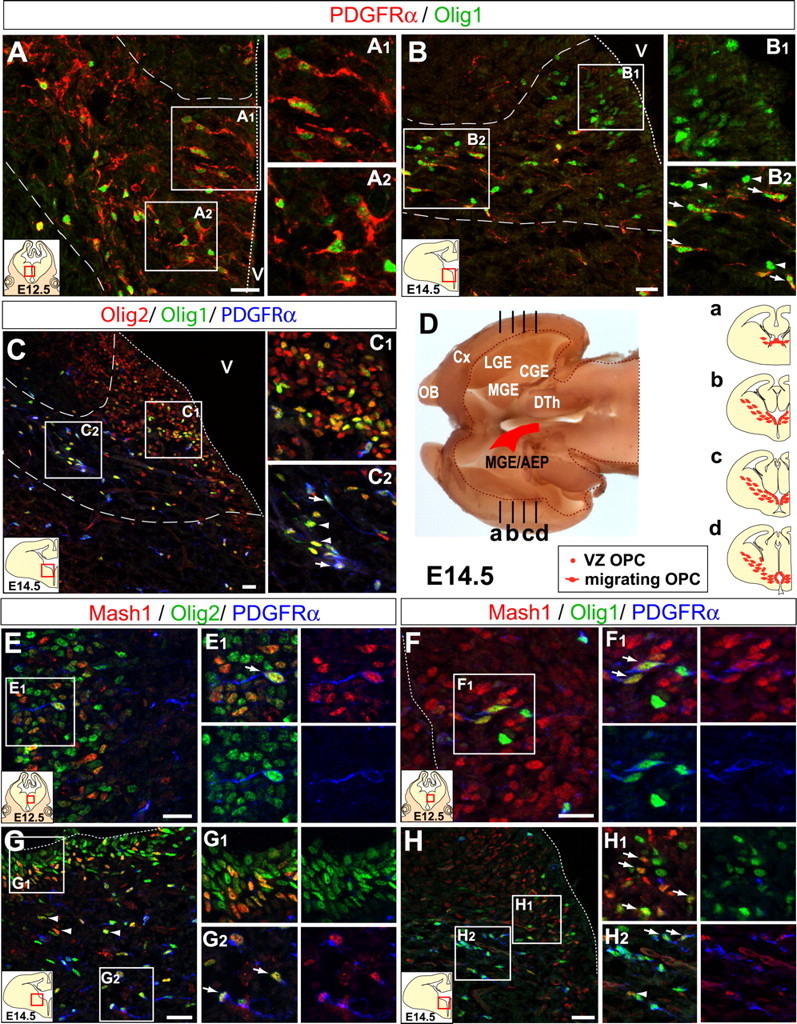
Mash1 is expressed in VZ and SVZ OPCs. A, At E12.5 in mouse telencephalic coronal sections, PDGFRα and Olig1 are coexpressed in a ventral territory (AEP) in both VZ cells (A1) and cells migrating into the SVZ (A2). A1 and A2 are enlargements of the areas outlined by squares in A. B, At E14.5 in the same region, many Olig1+ VZ cells also express PDGFRα (B1), whereas in the SVZ, some Olig1+ cells express PDGFRα (arrows) and others do not (B2, arrowheads). C, All Olig1+ cells in the VZ (C1) and SVZ (C2) coexpress Olig2, whereas only a fraction coexpresses PDGFRα (arrows). D, Horizontal view of the forebrain at E14.5 after removal of dorsal tissue depicting in red the region where Olig1+,PDGFRα+ cells are found in the VZ and SVZ. Da–Dd, schematic of coronal sections at different rostrocaudal levels. E, F, At E12.5, Mash1 is expressed in PDGFRα+,Olig2+ cells (E, E1) and in PDGFRα+,Olig1+ cells in the VZ of the AEP (F, F1). G, At E14.5, all Mash1+ cells coexpress Olig2 in the VZ (G1) and SVZ, and some of them coexpress PDGFRα (G2) whereas others do not (arrowheads). H, A fraction of Olig1+ cells coexpresses Mash1 in the VZ (H1) and in the SVZ (H2), and some of these cells coexpress PDGFRα (arrows) whereas others do not (arrowhead). Dotted line, ventricular surface; Dashed line, borders of the main OPC migratory stream. OB, Olfactory bulb; Cx, cortex; CGE, caudal ganglionic eminence; DTh, dorsal thalamus; V, ventricle. Scale bar, 20 μm.
Mash1 is expressed by telencephalic OPCs at the time of their specification
The proneural gene Mash1 is expressed by a large fraction of VZ and SVZ progenitors throughout the embryonic ventral telencephalon (Fode et al., 2000). We asked whether Mash1 is expressed by OPCs as soon as they are generated in the VZ and after they have begun to migrate. At E12.5, most Olig1+, PDGFRα+ cells in the VZ expressed Mash1 (83 ± 2%) (Fig. 1F), suggesting that Mash1 is present in OPCs at the time of their specification. Mash1 was also expressed by a fraction of OPCs that migrated away from the VZ and SVZ (57 ± 8% of PDGFRα+ cells and 53 ± 5% of Olig1+ cells; data not shown). At E14.5, fewer ventricular Olig1+ cells expressed Mash1 (69 ± 4%) (Fig. 1H), whereas the same proportion of migrating OPCs were Mash1+ (51 ± 11% of PDGFRα+, Olig1+ cells) (Fig. 1H). Thus, Mash1 is expressed by many but not all early born OPCs at the time of their generation.
Mash1 was also present at E14.5 in a distinct and smaller population of OPCs located more laterally, at the border between the piriform cortex and the LGE/CGE, which coexpressed PDGFRα and the ventral telencephalic marker Gsh2 (supplemental Fig. 2_A,B_, available at www.jneurosci.org as supplemental material).
In contrast with Olig1, which appears to be restricted to the oligodendrocyte lineage from E11.5 to E13.5, Olig2 was expressed throughout the VZ and SVZ of the ventral forebrain, including in all Mash1+ cells (Fig. 1C,E,G). A small fraction of Mash1+,Olig2+ cells corresponds to PDGFRα+ OPCs (Fig. 1E,G), whereas another subset represents βIII-tubulin+ neuronal precursors, and a third population does not express cell type-specific markers and may thus correspond to multipotent progenitors (data not shown).
Mash1 expression is restricted to a subset of telencephalic OPCs
The heterogeneity of OPCs regarding Mash1 expression could reflect differences in the timing of Mash1 expression in OPCs, or alternatively, the existence of genetically distinct lineages of oligodendrocytes in the embryonic forebrain. To distinguish between these two possibilities, we asked whether all forebrain OPCs, or only a fraction of them, originated from Mash1+ cells. We used Mash1-GFP reporter mice to transiently mark the progeny of Mash1+ cells with GFP and Mash1-Cre mice to irreversibly mark all cells originating from Mash1+ progenitors. A transgenic line (named thereafter Mash1::GFP) in which GFP is inserted in the Mash1 gene in a BAC containing 210 kb of sequence around the Mash1 locus (Gong et al., 2003) expresses GFP at a high level in a pattern indistinguishable from that of Mash1 in forebrain progenitors, and expression of GFP, which is more stable than Mash1, is maintained in the progeny of Mash1+ cells that have left the VZ and SVZ (Fig. 2A) (supplemental Fig. 5_A_, available at www.jneurosci.org as supplemental material, and data not shown). At E12.5 and E13.5 in Mash1::GFP embryos, most PDGFRα+ OPCs expressed GFP, thus indicating that the first OPCs originate from Mash1+ progenitors (Fig. 2C,H). However, from E14.5 onwards, a significant and growing fraction of PDGFRα+ OPCs were GFP-negative, representing half of all OPCs found in the telencephalon toward the end of embryonic development (Fig. 2E,G,H).
Figure 2.
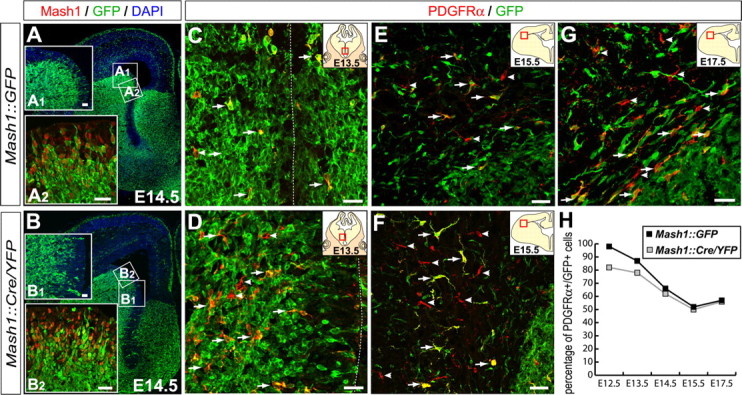
Mash1 progenitors generate a subpopulation of OPCs. Coronal sections through the telencephalon of Mash1::GFP (A, C, E, G) and Mash1::Cre/YFP (B, D, F) embryos stained with anti-GFP antibody. A, B, GFP is detected in many ventrally born cells, including dorsally migrating cortical interneurons. GFP and YFP proteins are detected in most Mash1+ VZ cells in Mash1::GFP (A2) and Mash1::Cre/YFP (B2) embryos, respectively. Insets in A1 and B1 are enlargements of the areas outlined by squares in A and B, respectively. Many PDGFRα+ cells express GFP (arrows) close to the region where they are generated, whereas a fraction that increases with time is GFP-negative (arrowheads) in both Mash1::GFP (C, E, G) and Mash1::Cre/YFP (B, D, F) embryos, as shown in particular for PDGFRα+ cells invading the cortex. H, Percentage of PDGFRα+ cells coexpressing GFP or YFP at different stages demonstrating that the fraction of OPCs that do not originate from Mash1+ progenitors increases with time. The dotted line marks the ventricular surface. Scale bars, 20 μm.
To rule out the possibility that GFP-negative OPCs in Mash1::GFP embryos have downregulated GFP, we used a transgenic line in which the Cre recombinase has been inserted in the Mash1 gene in a BAC containing 170 kb of sequence around the Mash1 locus (Mash1::Cre) (see Materials and Methods and supplemental Fig. 3_A_, available at www.jneurosci.org as supplemental material). The Mash1::Cre line was crossed with the reporter line Rosa26stop-YFP (Srinivas et al., 2001) and analysis of double transgenic embryos (thereafter called Mash1::Cre/YFP) revealed a pattern of yellow fluorescent protein (YFP) expression in the telencephalon very similar to that of GFP in Mash1::GFP embryos (Fig. 2B,D,F). At E12.5 and E13.5, most PDGFRα+ cells expressed YFP (suggesting a rate of Cre recombination of ∼80%) (Fig. 2D,H), whereas an increasing fraction of YFP-negative OPCs was found at E14.5 and E15.5 (Fig. 2F,H). Given the high recombination rate calculated at early stages, the YFP-negative OPC population identified subsequently is not likely because of inefficient recombination by the Mash1::Cre line. Together with the Mash1::GFP data, it suggests instead that the embryonic telencephalon contains two genetically distinct lineages of OPCs, one produced by Mash1+ progenitors, which predominates initially, and another produced by _Mash1_-negative progenitors that expands subsequently. The contribution of the Mash1+ lineage to OPC populations varied in different telencephalic regions toward the end of embryonic development (septum, ∼35%; striatum, ∼50%; cortex, ∼65%) (Fig. 2G and data not shown).
Mash1+ progenitors generate a subset of adult forebrain OPCs
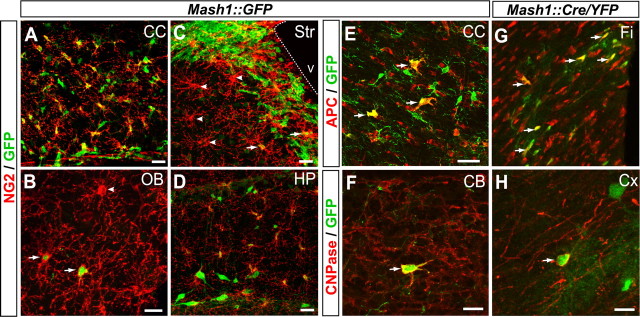
OPCs are generated in the telencephalon not only during embryogenesis but also at postnatal stages (Ivanova et al., 2003; Kessaris et al., 2006). OPCs at postnatal stages are thought to give rise to both myelinating oligodendrocytes and quiescent OPCs (NG2+ cells) (Ligon et al., 2006; Nishiyama et al., 1996). We asked whether Mash1+ progenitors contributed to the generation of OPCs in the adult forebrain. In the brain of 6 week-old Mash1::GFP mice, almost all NG2+ cells in the white matter, including the corpus callosum, fimbria, and anterior commissure, were GFP+ (Figs. 3A, 4 and data not shown). A smaller fraction of NG2+ cells in the gray matter expressed GFP (31% in the striatum, 52% in ventral SVZ, 59% in the cerebral cortex, 86% in the olfactory bulb, 100% in hippocampus) (Fig. 3B–D). Using APC as a marker of premyelinating oligodendrocytes (Bhat et al., 1996), we found that a smaller fraction of APC+ cells expressed GFP in both white (fimbria, 34%; corpus callosum, 43%; anterior commissure, 41%) (Fig. 3E and data not shown) and gray matters (cortex, 16%; striatum, 29%; olfactory bulb, 11%; data not shown). We also found GFP+ cells expressing the myelinating marker CNPase (Fig. 3F). Similar results were obtained with the Mash1::Cre/YFP mice (Fig. 3G,H). These data therefore suggest that Mash1+ progenitors give rise to a subset of both adult NG2+ OPCs and myelinating oligodendrocytes and that these populations are likely to be genetically heterogeneous.
Figure 3.
Mash1 progenitors generate both adult NG2+ OPCs and myelinating oligodendrocytes. Sagittal brain sections from 6-week-old Mash1::GFP (A–F) and Mash1::Cre/YFP (G, H) mice. All NG2+ cells in white matter regions express the Mash1::GFP transgene, including in the corpus callosum (A) (see also Fig. 4). Different proportions of NG2+ cells are GFP-positive (arrows) and GFP-negative (arrowheads) in gray matter regions, including in the olfactory bulb (B), striatum (C), and hippocampus (D). In both Mash1::GFP (E, F) and Mash::Cre/YFP (G, H) brains, a fraction of maturing oligodendrocytes expressing the premyelinating marker APC (E, G) or the myelinating marker CNPase (F, H) coexpresses GFP (arrows) in both white matter (E, corpus callosum; H, fimbria) and gray matter regions (F, cerebellum; H, cortex). The dotted line marks the ventricular surface. CC, Corpus callosum; CB, cerebellum; Cx, cortex; Fi, fimbria; OB, olfactory bulb; HP, hippocampus; Str, striatum. V, ventricle. Scale bars, 20 μm.
Mash1 is required for the generation of a subpopulation of embryonic OPCs in the telencephalon
We next asked whether Mash1 is required for the generation of OPCs in the embryonic telencephalon. PDGFRα,Olig1 and PDGFRα,Olig2 double-labeling experiments revealed that most early born OPCs were absent from Mash1 null mutant (Mash1_−/−) embryos at E12.5 and E13.5 (Figs. 5A,A′, 6A,B) (supplemental Fig. 5_A,A′, available at www.jneurosci.org as supplemental material). This was not because of premature death, because activated Caspase3 labeling did not reveal any elevated apoptosis (data not shown), suggesting instead that Mash1 is required for the generation of most OPCs at early stages of oligodendrogenesis. Strikingly, the defect in OPC production was transient. Quantifying OPCs at later stages revealed that similar numbers of new OPC were added in _Mash1_−/− and wild-type embryos between E13.5 and E14.5 (∼6500 and 5000 new OPCs, respectively) (Figs. 5B,B′, 6B). The production of a similar number of OPCs during this period from OPC populations that have very different sizes at E13.5 (∼1500 in _Mash1_−/− embryos and 7500 wild-type embryos), suggests that OPCs in _Mash1_−/− embryos proliferate at a much higher rate than their wild-type counterparts or, more likely, that a greater number of OPCs are specified de novo from uncommitted progenitors in mutant embryos. Indeed, BrdU incorporation experiments have not revealed significant differences in the rate of proliferation of PDGFRα+ cells between Mash1 mutant and wild-type embryos (data not shown). Examination of PDGFRα+,Olig1+ cells in the forebrain at E17.5 showed no significant difference between wild-type and _Mash1_−/− embryos (Fig. 4C,C′), thus confirming that the requirement for Mash1 function in embryonic oligodendrogenesis is confined to the earliest born population of OPCs.
We attempted to track the fate of the subset of OPCs transcribing the Mash1 gene in a Mash1 null background by generating _Mash1_−/− embryos carrying the _Mash1::_Cre-recombined YFP transgene. However, this transgene was abnormally regulated in the absence of Mash1, with virtually all cells in the ventral telencephalon expressing GFP (supplemental Fig. 5, available at www.jneurosci.org as supplemental material). This is likely because of a loss of Notch signaling in _Mash1_−/− embryos, resulting in uniform activation of the Mash1 locus in ventral telencephalic progenitors (Casarosa et al., 1999). We were therefore unable to specifically mark Mash1+ cells and their progeny in the absence of Mash1 function.
Mash1 function in oligodendrogenesis is independent from its role in neurogenesis
The loss of OPCs in _Mash1_−/− embryos could reflect a direct role of Mash1 in the generation of OPCs, or a more indirect role because of its involvement in the generation of telencephalic neurons (Casarosa et al., 1999; Horton et al., 1999), which could themselves be required for OPC specification. To distinguish between these two possibilities, we examined mice carrying a knock-in allele of Mash1 in which the coding sequence of Mash1 has been replaced by that of another proneural gene, Neurogenin2 (Mash1KINgn2) (Parras et al., 2002). In Mash1KINgn2/Ngn2 embryos, ectopic expression of Ngn2 in the VZ of the ventral telencephalon (Fig. 7A,A′) rescues the loss of neuronal precursors and of correctly specified postmitotic neurons resulting from the deletion of Mash1 (Parras et al., 2002). Analysis of PDGFRα expression in Mash1KINgn2/Ngn2 embryos revealed that the deficit in OPCs was as severe as in _Mash1_−/− embryos (Fig. 7B–D). Moreover, the normal number of OPCs observed in Mash1KINgn2/+ embryos indicates that Ngn2 expression is not inhibitory for oligodendrogenesis in the ventral forebrain. The lack of OPCs in Mash1KINgn2/Ngn2 embryos is thus because of the loss of Mash1 and not to the ectopic expression of Ngn2 (Fig. 7C,D). The role of Mash1 in oligodendrogenesis is therefore independent of the level of neurogenesis, arguing for a direct role of Mash1 in the generation of OPCs.
Figure 7.
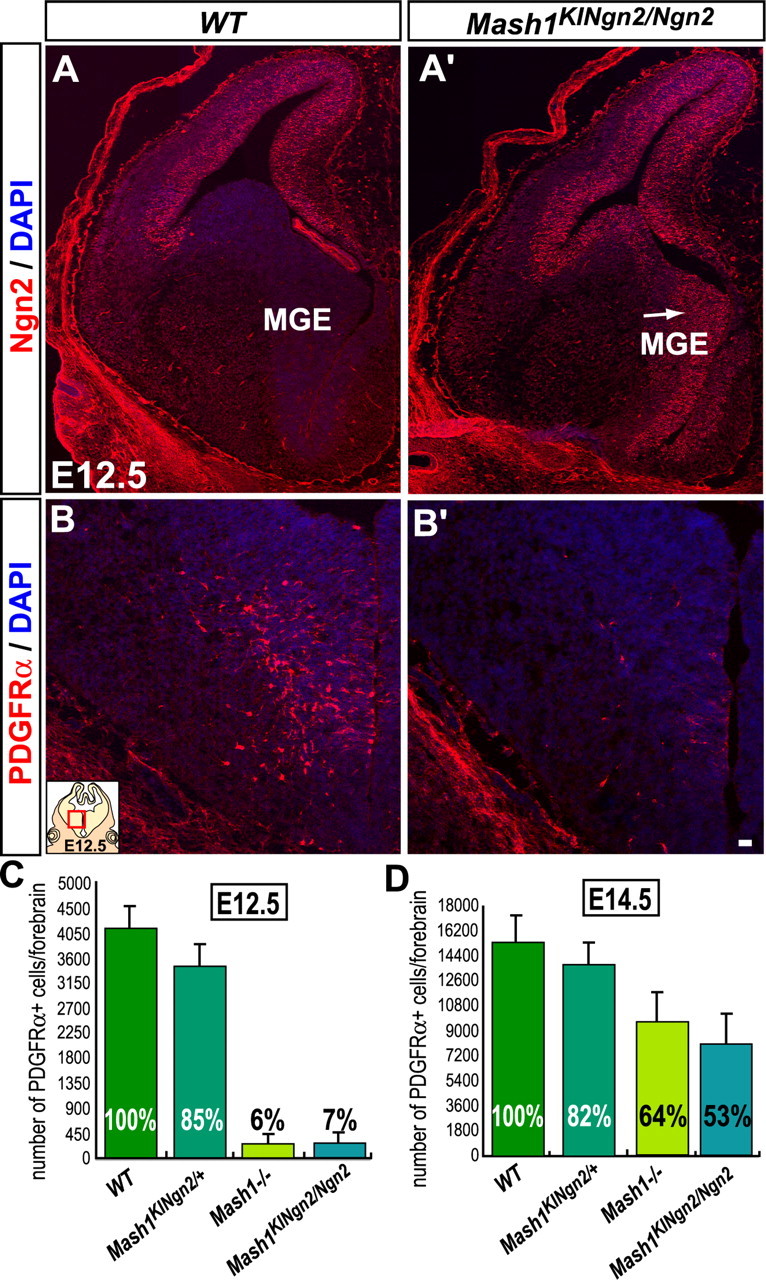
Mash1 requirement for the generation of OPCs is independent of neurogenesis. Telencephalic coronal sections of wild-type (A, B) and Mash1KINgn2/Ngn2 embryos (A′, B′) at E12.5. (A, A′) Expression of Ngn2 protein in the dorsal telencephalon of wild-type embryos (A) and ectopic Ngn2 expression in the ventral domain in Mash1KINgn2/Ngn2 embryos (A′). The MGE develops normally (arrow) when Ngn2 is expressed instead of Mash1. (B, B′) PDGFRα+ cells found in VZ and SVZ of the MGE in wild-type embryos at E12.5 (B) are mostly missing in Mash1KINgn2/Ngn2 embryos (B′). Quantification of PDGFRα+ cells in wild-type, _Mash1KINgn2/+, Mash1_−/− and Mash1KINgn2/Ngn2 mutant embryos at E12.5 (C) and at E14.5 (D) showing no difference in numbers of PDGFRα+ cells between wild-type and Mash1KINgn2/+ embryos and between _Mash1_−/− and Mash1KINgn2/Ngn2 embryos (n = 3), indicating that Ngn2 does not inhibit the OPC fate nor rescue Mash1 function in OPC specification. WT, Wild-type. Scale bars, 20 μm. Error bars indicate SEM.
Mash1 interacts genetically with Olig2 for the generation of telencephalic OPCs
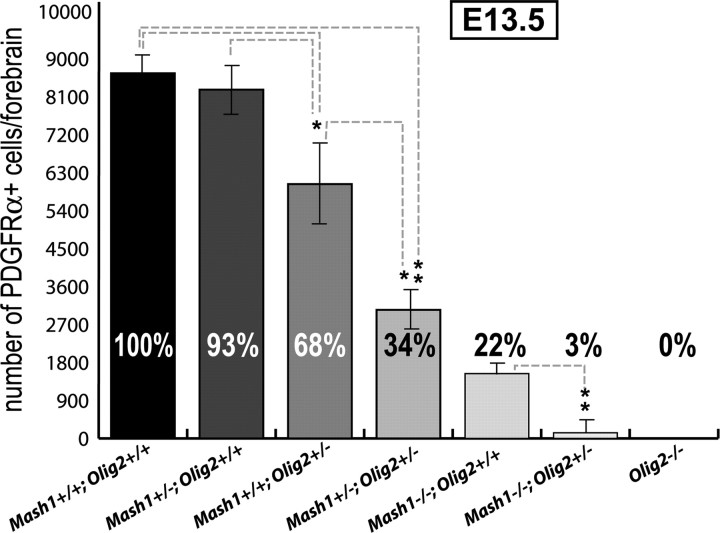
In addition to Mash1, the transcription factor Olig2 has also been implicated in the generation of OPCs in the embryonic telencephalon (Lu et al., 2002). To determine whether Mash1 and Olig2 regulate OPC production independently or through a common pathway, we examined whether the two factors interact to produce OPCs. Embryos carrying mutant alleles of Mash1 and/or Olig2 were generated and examined for PDGFRα expression at E13.5, a stage when the generation of most OPCs depends on Mash1 function (Fig. 8). As expected, Olig2−/− embryos showed a complete absence of OPCs (Fig. 8) (Lu et al., 2002). Loss of one wild-type copy of Olig2 leads to a 32% reduction in number of OPCs, whereas heterozygosity at the Mash1 locus did not affect OPC production. Embryos with one dose of Mash1 and one dose of Olig2 had a further reduction in number of OPCs (66%). Moreover, loss of one copy of Olig2 also significantly aggravated the OPC defect seen in _Mash1_−/− embryos (Fig. 8). These data demonstrate that Mash1 and Olig2 interact genetically and suggest that the two genes act in the same pathway to specify early born OPCs in the ventral telencephalon.
Figure 8.
Genetic interaction between Mash1 and Olig2. Quantification of PDGFRα+ cells in the forebrain of E13.5 embryos carrying different combinations of Mash1 and Olig2 alleles. Loss of one wild-type copy of Mash1 enhances the defect in PDGFRα+ OPCs observed in Olig2+/− embryos, and loss of one wild-type copy of Olig2 enhances the defect in OPCs observed in _Mash1_−/− embryos. Percentages represent the ratio of forebrain PDGFRα+ cells observed in embryos of indicated genotypes over wild-type embryos (n = 3). *p < 0.05; **p < 0.01; Student's t test. Error bars indicate SEM.
Overexpression of Mash1 is not sufficient to specify OPCs
To further address the role of Mash1 in specification of OPCs, we asked whether ectopic expression of Mash1 could promote the generation of OPCs or at least the expression of OPC markers at ectopic locations. For this, we examined a mouse line in which the Mash1 coding sequence replaces the coding sequence of Ngn2, resulting in ectopic expression of Mash1 in progenitors of the dorsal telencephalon (Ngn2KIMah1 mice) (Fode et al., 2000; Parras et al., 2002) (Fig. 9B). PDGFRα was expressed ectopically in the SVZ of the lateral cortex in Ngn2KIMash1/Mash1 embryos at E15.5 (Fig. 9A′,B). We also examined other OPC markers but were unable to detect expression of Olig2, Sox10, Nkx2.2, or NG2 in this region. Only a few Mash1+,PDGFRα+ cells also expressed Olig1 in the dorsal telencephalon of Ngn2KIMash1/Mash1 embryos (Fig. 9C and data not shown). Thus, Mash1 can induce expression of PDGFRα but is not sufficient to activate a full program of oligodendrogenesis when ectopically expressed in dorsal telencephalic progenitors.
Figure 9.
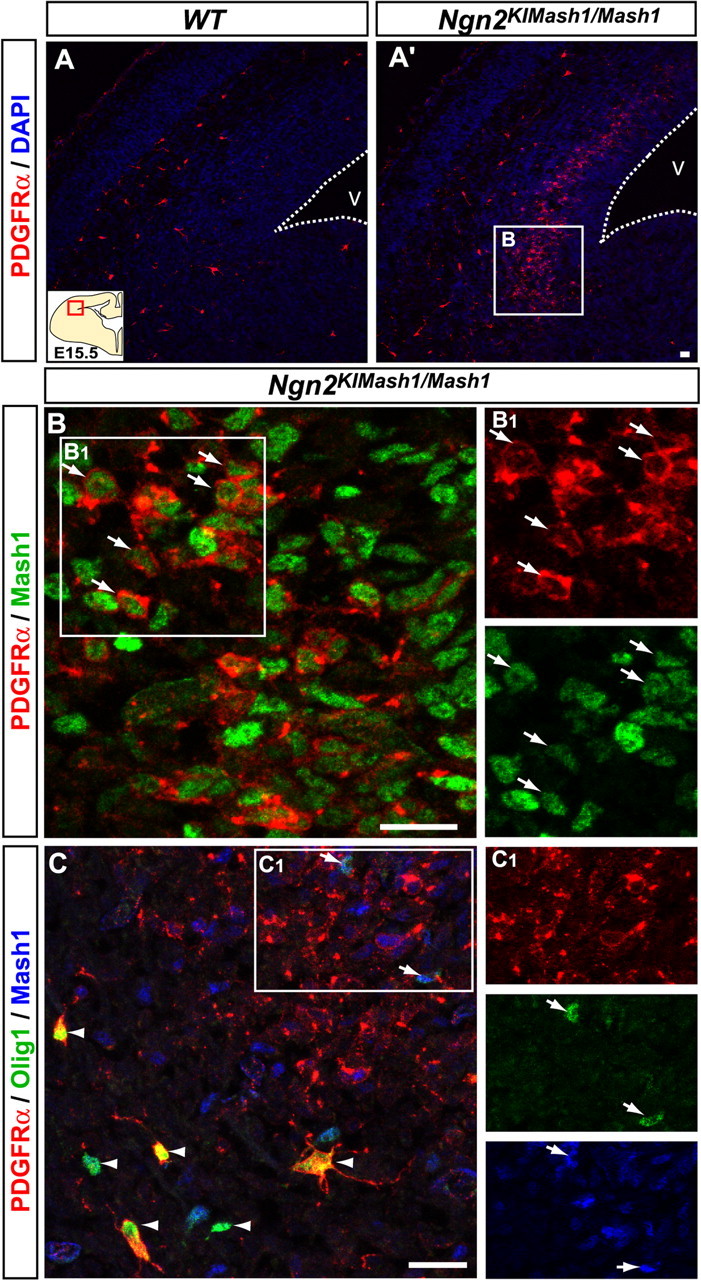
Mash1 can induce some traits of OPC specification in the absence of Ngn2. Telencephalic coronal sections of wild-type (A) and Ngn2KIMash1/Mash1 (A′, B, C) embryos at E15.5 are shown. In wild-type embryos, a few PDGFRα+ cells begin to invade the intermediate zone of the lateral cortex (A). In Ngn2KIMash1/Mash1 embryos, abundant PDGFRα+ cells are ectopically located in the SVZ of the lateral cortex (A′). B, Ectopic PDGFRα+ cells coexpress Mash1 (arrows). C, A few ectopic PDGFRα+,Mash1+ cells express low level of Olig1 (arrows) compared with high level of Olig1 expression in ventrally derived OPCs (arrowheads). The dotted line marks the ventricular surface. The territory outlined by a square in A' is enlarged in B'. Insets B1 and C1 are enlargements of the areas outlined by squares in B and C, respectively. V, Ventricle. Scale bars, 20 μm.
Discussion
In this study, we have analyzed the role of Mash1 in the specification of OPCs, focusing on the embryonic forebrain. Our data support a role of Mash1 in the specification of a subpopulation of OPCs from multipotent progenitors: (1) Mash1 is extensively coexpressed with Olig2 in the VZ that contains multipotent stem cells, and it is expressed in OPCs as soon as they are identified in the VZ by expression of PDGFRα and Olig1; (2) a large subset of OPCs that appear in the ventral forebrain between E11.5 and E13.5 and express Mash1 are not generated in Mash1 null mutant embryos; (3) analysis of double-heterozygous embryos for Mash1 and Olig2 reveals an interaction between the two genes, suggesting that they both participate to the same genetic program of oligodendrogenesis; (4) forced expression of Mash1 in the embryonic cortex induces ectopic expression of the OPC marker PDGFRα, although it is not sufficient to induce a full program of oligodendrogenesis; (5) a subset of OPCs and oligodendrocytes in the embryonic, early postnatal, and adult forebrain, including most adult NG2+ OPCs, originate from Mash1+ progenitors. We discuss below the role of Mash1 in oligodendrocyte specification and the heterogeneity of oligodendrocyte lineages with respect to Mash1 expression.
Mash1 function in specification of OPCs
Olig2 and Mash1 are coexpressed in the VZ and SVZ of the ventral telencephalon at the time when OPCs are specified and a fraction of Olig2+,Mash1+ cells express the early OPC markers, PDGFRα and Olig1. Olig2 and Mash1 are also coexpressed by neuronal precursors, marked by expression of βIII-tubulin, as well as by cells that do not express cell type-specific markers and are presumably multipotent progenitors. Coexpression of Olig2 and Mash1 therefore does not appear to be sufficient to commit progenitors to either a neuronal or an oligodendroglial fate. Overexpression experiments of Mash1 in the dorsal telencephalon (present study; Lu et al., 2001) also indicate that expression of Mash1 alone is not sufficient to induce oligodendrogenesis in the telencephalon, and suggesting that the role of this factor in this lineage is context dependent.
The transcriptional mechanisms that determine the choice between neuronal and oligodendrocyte fates in ventral telencephalic progenitors remain unclear. Other factors may act in combination with Olig2 and Mash1 to specify OPCs. Gain of function experiments have suggested that Olig2 must interact with Nkx2.2 to promote oligodendrogenesis in the chick spinal cord (Zhou et al., 2001). However, analysis of mice mutant for Nkx2.1, the paralog of Nkx2.2 in the forebrain, demonstrates that this gene is not absolutely required for oligodendrogenesis in the ventral forebrain (Tekki-Kessaris et al., 2001), and the broad expression of Nkx2.1 in the medial part of the ventral telencephalon, including in neuronal and OPCs, indicates that the combination of Olig2 with Nkx2.1 is not sufficient to determine the oligodendrocyte fate. Olig1 is the earliest transcription factor whose expression is restricted to the oligodendroglial lineage in the early ventral forebrain. Indeed, Olig1 expression is mostly induced in VZ cells that already express PDGFRα and are therefore already engaged in the oligodendroglial lineage. This, together with the analysis of Olig1 mutant mice, rules out Olig1 as an important determinant of the oligodendrocyte lineage in the embryonic forebrain, involved in the specification of OPCs from multipotent progenitors. In contrast, the Sox9 gene, which is expressed broadly in the VZ throughout the CNS and is required for the specification of oligodendrocytes and astrocytes in the ventral spinal cord (Stolt et al., 2003) could work in combination with Olig2 and/or Mash1 to specify oligodendrocytes in the forebrain.
Oligodendrogenesis in _Mash1_−/− embryos is significantly affected only between E11.5 and E13.5. Thereafter, OPCs are produced at an accelerated rate to compensate for the early deficit. This could reflect a vastly accelerated rate of proliferation of the OPCs present in _Mash1_−/− embryos at E13.5, or an immigration of OPCs generated from more caudal Mash1-negative regions, or, more likely, an increase in the rate of specification of OPCs from uncommitted progenitors, reflecting the activity of genes capable of compensating, albeit with a delay, the loss of Mash1 function in OPC specification. Such genes may normally be involved in OPC generation together with Mash1, or may be involved only in the Mash1 mutant context (see Discussion on a similar issue in Pattyn et al., 2006). The finding that Mash1 and Olig2 interact genetically suggests that the two genes act in the same pathway for the generation of OPCs. One possibility is that in the absence of Mash1, this pathway is not completely blocked, and Olig2 activity is only partially impaired, with, for example, a lower level of expression of common targets of Mash1 and Olig2. Addressing this possibility will require the characterization of the molecular mechanisms underlying the interaction of Mash1 with Olig2 in specification of OPCs.
OPCs have been shown to originate not only from the MGE but also from the LGE/CGE and the dorsal telencephalon, at late embryonic and postnatal stages, respectively (Gorski et al., 2002; Kessaris et al., 2006). We found that OPCs located in the SVZ of the LGE during embryogenesis and in the cortical SVZ and subcortical white matter at birth and at postnatal stages, also express Mash1 (present study and Parras et al., 2004; and data not shown). A recent study proposed, on the contrary, that Mash1 is not expressed in OPCs of the cortical SVZ at birth and is present in the cytoplasm of OPCs from the subcortical white matter at postnatal day 14 (Gokhan et al., 2005), probably because of a lower sensitivity and higher background of the staining procedure used in that study. In addition, analysis of Mash1::GFP reporter mice suggests that a large fraction of OPCs present in the forebrain at birth and at adult stages originate from Mash1-positive progenitors (present study). These data, together with the earlier demonstration of a deficit in OPCs in the olfactory bulb of newborn Mash1 mutant mice (Parras et al., 2004), suggests that Mash1 may be involved in the specification of oligodendrocyte lineages at postnatal stages as well. Addressing this possibility will require the analysis of mice with a conditional ablation of Mash1.
Subsets of OPCs originating either from Mash1+ or from Mash1− progenitors
Our study reveals that OPCs are heterogeneous with respect to Mash1 expression. This heterogeneity reflects in part the transient nature of Mash1 expression, which is initiated in OPCs as they are generated in the VZ and is turned off in OPCs migrating away from the VZ and SVZ toward (e.g., the cortex). However, characterization of OPCs at the time of their generation in the VZ and analysis of Mash1::GFP and Mash1::Cre mice to mark the progeny of Mash1+ cells also shows the existence of several OPC lineages that originate from Mash1+ and Mash1− progenitors respectively, and that coexist in the brain during embryogenesis as well as in newborns and adults. Heterogeneity of embryonic OPCs with respect to expression of the markers _PDGFR_α and PLP/dm20 has been reported previously (Spassky et al., 1998, 2001), but there is no apparent correlation between expression of Mash1 and that of PDGFRα or PLP/dm20, because Mash1+ progenitors give rise to only a subset of PDGFRα+ cells and PLP/dm20 is not expressed in the embryonic telencephalon (Spassky et al., 1998, 2001).
Different subpopulations of oligodendrocytes and their precursors have been described in the adult CNS. Oligodendrocytes are very heterogeneous morphologically, which has led to their classification into distinct subtypes (Del Rio Hortega, 1942). In the spinal cord, expression of different combinations of the transcription factors Olig1, Olig2, and Nkx2.2 also defines several populations that mostly correspond to different stages of maturation along the oligodendroglial lineage but possibly also to functionally distinct classes of oligodendrocytes (Kitada and Rowitch, 2006). In the brain, two populations of oligodendroglial cells expressing the chondroitin sulfate proteoglycan NG2 have been proposed. Mitotically active and bipolar NG2+ cells are progenitors for myelinating oligodendrocytes (Dawson et al., 2003), whereas quiescent and morphologically more complex NG2+ cells have neurological functions, including in synaptic transmission and regeneration (Bergles et al., 2000; Butt and Berry, 2000). Our results suggest that both Mash1+ and Mash1− progenitors contribute to these two populations in the adult forebrain. Whether Mash1 expression defines functionally distinct subtypes of oligodendrocytes in the embryonic and adult brain remains to be determined.
Footnotes
This work was supported by a grant from the European Commission Research and Technological Development programme to F.G. and by institutional funds from the Medical Research Council (MRC). C.M.P. was supported by an MRC career development fellowship and is presently supported by an Avenir Contract from Institut National de la Santé et de la Recherche Médicale and Fondation de France. We gratefully acknowledge Charles Stiles and David Anderson for the gift of Olig2, Mash1, and Ngn2 monoclonal antibodies; Kenny Campbell for the gift of Gsh2 antibody; Neal Copeland for providing the pIGCN21 plasmid and bacterial strains used to generate the Mash1::Cre BAC; Jane Johnson for providing the Mash1::GFP line; Vicky Snowden for generating the Mash1::Cre line; and Melinda Bray for help with animal care.
References
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Bergles DE, Roberts JD, Somogyi P, Jahr CE. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature. 2000;405:187–191. doi: 10.1038/35012083. [DOI] [PubMed] [Google Scholar]
- Bhat RV, Axt KJ, Fosnaugh JS, Smith KJ, Johnson KA, Hill DE, Kinzler KW, Baraban JM. Expression of the APC tumor suppressor protein in oligodendroglia. Glia. 1996;17:169–174. doi: 10.1002/(SICI)1098-1136(199606)17:2<169::AID-GLIA8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Butt AM, Berry M. Oligodendrocytes and the control of myelination in vivo: new insights from the rat anterior medullary velum. J Neurosci Res. 2000;59:477–488. doi: 10.1002/(SICI)1097-4547(20000215)59:4<477::AID-JNR2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Dawson MR, Polito A, Levine JM, Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci. 2003;24:476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- Del Rio Hortega P. La neuroglia normal. Conceptos de neuroglioma y angioglioma. Arch de Histol Norm y Patol. 1942;1:5–71. [Google Scholar]
- Doetsch F. The glial identity of neural stem cells. Nat Neurosci. 2003;6:1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang S-L, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Ingraham C, Rising L, Goderie S, Temple S. Multipotent stem cells from the mouse basal forebrain contribute GABAergic neurons and oligodendrocytes to the cerebral cortex during embryogenesis. J Neurosci. 2001;21:8854–8862. doi: 10.1523/JNEUROSCI.21-22-08854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development. 2005;132:2709–2719. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci. 1999;14:355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- Ivanova A, Nakahira E, Kagawa T, Oba A, Wada T, Takebayashi H, Spassky N, Levine J, Zalc B, Ikenaka K. Evidence for a second wave of oligodendrogenesis in the postnatal cerebral cortex of the mouse. J Neurosci Res. 2003;73:581–592. doi: 10.1002/jnr.10717. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada M, Rowitch DH. Transcription factor co-expression patterns indicate heterogeneity of oligodendroglial subpopulations in adult spinal cord. Glia. 2006;54:35–46. doi: 10.1002/glia.20354. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Kesari S, Kitada M, Sun T, Arnett HA, Alberta JA, Anderson DJ, Stiles CD, Rowitch DH. Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci USA. 2006;103:7853–7858. doi: 10.1073/pnas.0511001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Bergles DE. Synaptic signaling between GABAergic interneurons and oligodendrocyte precursor cells in the hippocampus. Nat Neurosci. 2004;7:24–32. doi: 10.1038/nn1162. [DOI] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch DH, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Marshall CA, Goldman JE. Subpallial dlx2-expressing cells give rise to astrocytes and oligodendrocytes in the cerebral cortex and white matter. J Neurosci. 2002;22:9821–9830. doi: 10.1523/JNEUROSCI.22-22-09821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CA, Suzuki SO, Goldman JE. Gliogenic and neurogenic progenitors of the subventricular zone: who are they, where did they come from, and where are they going? Glia. 2003;43:52–61. doi: 10.1002/glia.10213. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y-i, Shimamura K, Nakafuku M. Combinatorial roles of Olig2 and Neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- Newman EA. New roles for astrocytes: regulation of synaptic transmission. Trends Neurosci. 2003;26:536–542. doi: 10.1016/S0166-2236(03)00237-6. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Lin XH, Giese N, Heldin CH, Stallcup WB. Co-localization of NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells in the developing rat brain. J Neurosci Res. 1996;43:299–314. doi: 10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Olivier C, Cobos I, Perez Villegas E, Spassky N, Zalc B, Martinez S, Thomas J. Monofocal origin of telencephalic oligodendrocytes in the anterior entopeduncular area of the chick embryo. Development. 2001;128:1757–1769. doi: 10.1242/dev.128.10.1757. [DOI] [PubMed] [Google Scholar]
- Parnavelas JG. Glial cell lineages in the rat cerebral cortex. Exp Neurol. 1999;156:418–429. doi: 10.1006/exnr.1999.7044. [DOI] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras CM, Galli R, Britz O, Soares S, Galichet C, Battiste J, Johnson JE, Nakafuku M, Vescovi A, Guillemot F. Mash1 specifies neurons and oligodendrocytes in the postnatal brain. EMBO J. 2004;23:4495–4505. doi: 10.1038/sj.emboj.7600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Guillemot F, Brunet JF. Delays in neuronal differentiation in Mash1/Ascl1 mutants. Dev Biol. 2006;295:67–75. doi: 10.1016/j.ydbio.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Pringle N, Richardson W. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117:525–533. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Spassky N, Goujet-Zalc C, Parmantier E, Olivier C, Martinez S, Ivanova A, Ikenaka K, Macklin W, Cerruti I, Zalc B, Thomas JL. Multiple restricted origin of oligodendrocytes. J Neurosci. 1998;18:8331–8343. doi: 10.1523/JNEUROSCI.18-20-08331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Heydon K, Mangatal A, Jankovski A, Olivier C, Queraud-Lesaux F, Goujet-Zalc C, Thomas JL, Zalc B. Sonic hedgehog-dependent emergence of oligodendrocytes in the telencephalon: evidence for a source of oligodendrocytes in the olfactory bulb that is independent of PDGFRalpha signaling. Development. 2001;128:4993–5004. doi: 10.1242/dev.128.24.4993. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekki-Kessaris N, Woodruff R, Hall AC, Gaffield W, Kimura S, Stiles CD, Rowitch DH, Richardson WD. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. doi: 10.1242/dev.128.13.2545. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu F-f, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerlin M, Milosevic A, Goldman JE. Glial progenitors of the neonatal subventricular zone differentiate asynchronously, leading to spatial dispersion of glial clones and to the persistence of immature glia in the adult mammalian CNS. Dev Biol. 2004;270:200–213. doi: 10.1016/j.ydbio.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]