Establishment of a high-resolution 3D modeling system for studying pancreatic epithelial cell biology in vitro (original) (raw)
Abstract
Objective
Translation of basic research from bench-to-bedside relies on a better understanding of similarities and differences between mouse and human cell biology, tissue formation, and organogenesis. Thus, establishing ex vivo modeling systems of mouse and human pancreas development will help not only to understand evolutionary conserved mechanisms of differentiation and morphogenesis but also to understand pathomechanisms of disease and design strategies for tissue engineering.
Methods
Here, we established a simple and reproducible Matrigel-based three-dimensional (3D) cyst culture model system of mouse and human pancreatic progenitors (PPs) to study pancreatic epithelialization and endocrinogenesis ex vivo. In addition, we reanalyzed previously reported single-cell RNA sequencing (scRNA-seq) of mouse and human pancreatic lineages to obtain a comprehensive picture of differential expression of key transcription factors (TFs), cell–cell adhesion molecules and cell polarity components in PPs during endocrinogenesis.
Results
We generated mouse and human polarized pancreatic epithelial cysts derived from PPs. This system allowed to monitor establishment of pancreatic epithelial polarity and lumen formation in cellular and sub-cellular resolution in a dynamic time-resolved fashion. Furthermore, both mouse and human pancreatic cysts were able to differentiate towards the endocrine fate. This differentiation system together with scRNA-seq analysis revealed how apical-basal polarity and tight and adherens junctions change during endocrine differentiation.
Conclusions
We have established a simple 3D pancreatic cyst culture system that allows to tempo-spatial resolve cellular and subcellular processes on the mechanistical level, which is otherwise not possible in vivo.
Keywords: Three dimensional (3D), Pancreatic progenitors, scRNA-seq, Endocrinogenesis, Cell polarity, Cell–cell adhesion
1. Introduction
The pancreas comprises of exocrine and endocrine compartments responsible for secretion of nutrient-digestive enzymes and blood glucose-regulating hormones, respectively. Malfunction of the exocrine pancreas is associated with diseases, such as pancreatitis and pancreatic cancer. On the other hand, loss or progressive dysfunction of insulin-producing β-cells leads to hyperglycemia and diabetes mellitus. Currently, there is no treatment that can stop or reverse diabetes progression. Replacing the lost β-cells by generating in vitro differentiated β-cells and triggering endogenous mechanisms of repair hold great promise. Yet, accomplishing these therapeutic goals requires a better understanding of how pancreatic lineages, specifically β-cells, are formed during human embryonic development [1].
In mouse, pancreas organogenesis involves complex events of tissue patterning and cell differentiation [2]. First, pancreatic epithelial buds are formed from the foregut endoderm that consist of multipotent pancreatic progenitors (MPCs). In the central region of the buds, some MPCs become polarized and contribute to the formation of central microlumens [3], [4]. Subsequent fusion of the microlumens together with the patterning of the epithelial buds into the central trunk and peripheral tip domains gradually produce a single-layered epithelial network at embryonic day (E) 15.5 [5]. During these epithelial remodeling processes, MPCs become progressively lineage restricted and segregate into three main pancreatic lineages, namely acinar, ductal, and endocrine cells. Among these, endocrine cells are differentiated from bipotent ductal/endocrine progenitors located within the pancreatic epithelium [2], [6]. First, bipotent progenitors express low levels of the TF neurogenin3 (Neurog3, Ngn3) to become Ngn3low progenitors. Then, these progenitors increase the expression levels of Ngn3 and generate Ngn3high precursors, which differentiate into hormone−/Fevhigh population. Finally, Fevhigh cells generate fully differentiated hormone+ endocrine cells [7], [8], which cluster into islets of Langerhans and regulate blood glucose homeostasis through producing and secreting hormones, such as insulin and glucagon [1], [6].
Over the past decade, our understanding of human pancreas development has steadily increased [9], [10], [11], [12], [13]. This is partially due to the recent advancements in differentiation of human pluripotent stem cells (hPSCs) into pancreatic islet-like clusters (ILCs) [14], [15], [16], [17]. Although this in vitro differentiation system has uncovered detailed gene regulatory networks and a roadmap of human endocrinogenesis [17], [18], it cannot address the impact of tissue morphogenesis on endocrine cell differentiation. Therefore, understanding the molecular details of coupling epithelial dynamics, cell polarity, cell–cell and cell–matrix adhesion to pancreatic differentiation programs requires high-resolution spatial and temporal modeling systems [4], [19], [20], [21].
3D organoids are complex structures consisting of a polarized epithelial layer with a central lumen and carry great potential to study human development and organ-specific diseases, which are otherwise not assessable. In terms of organogenesis, these epithelial-based structures are unique systems that address developmental processes regulating niche signals and lineage decision, cell–cell interactions as well as tissue morphogenesis and patterning [22], [23], [24], [25]. Several groups have previously investigated pancreatic lineage decision or cell plasticity using organoids derived from embryonic or adult pancreatic cells, respectively [26], [27], [28], [29], [30], [31], [32], [33]. Among these, a pioneering work by Greggio et al. has generated 3D organoids that faithfully resembles mouse embryonic pancreas and allows in vitro lineage expansion and differentiation [26]. However, the complex epithelial structure of organoids deteriorates their potential to investigate dynamic regulation of cell polarity, adhesion, and differentiation in a temporal fashion. In contrast, 3D epithelial cysts or spheres are circular and polarized epithelial structures with a central lumen that present simple cell-type composition and allow for high-resolution cellular and subcellular analyses over time that are not possible in vivo. These structures have been generated from mouse and human pancreatic epithelial cells [27], [28]. Yet, their application to study early pancreatic formation and patterning during endocrinogenesis has not been thoroughly examined.
To study dynamic changes in pancreatic epithelial morphology during endocrine cell differentiation, we developed a simple high-resolution ex vivo 3D cyst culture from pancreatic progenitors (PPs). We generated polarized pancreatic epithelial cysts (PECs) originated from mouse primary PPs or human iPSCs-derived PPs that present similar molecular characteristics to the pancreatic epithelium in vivo. To obtain molecular details of cell biological changes during endocrinogenesis, we first reanalyzed single-cell RNA sequencing (scRNA-seq) of mouse embryonic pancreatic epithelial cells [7] and in vitro human endocrine cell differentiation [17], [18], indicating changes in expression levels of key TFs, cell–cell adhesion molecules and cell polarity components. To support this finding in a dynamic time-resolved culture system, we next differentiated PECs into endocrine cells and found remodeling of cell adhesion molecules and loss of apical-basal (AB) polarity during endocrine cell differentiation. Overall, establishment of a simple and reproducible PEC culture provided a high-resolution modeling system that not only allows for studying pancreas development in a dynamic temporal fashion but also enables comparing pancreatic epithelial biology across species and genotypes.
2. Material and methods
2.1. Mouse lines
Mouse lines were kept at the central facilities at Helmholtz Center Munich (HMGU) and animal experiments were performed in accordance with the German animal welfare legislation with the approved guidelines of the Society of Laboratory Animals (GV-SOLAS) and of the Federation of Laboratory Animal Science Associations (FELASA). Post-mortem examination of organs was not subject to regulatory authorization. The following mouse lines were used in the study: C57BL/6J, 129/SvJ and Tg(Neurog3-cre)C1Able/J (Ngn3Cre) [34] crossed with Gt(ROSA)26mTmG [35] to generate Ngn3Cre;mTmG line.
2.2. Cell source
An episomal reprogrammed iPSC line (HMGUi001-A) was generated in our laboratory from healthy donors (Gibco Human Episomal iPSC, Cat. No: A18945, Life Technologies) [36]. The cell line has been authenticated by Cell Line Genetics (Madison, WI) and confirmed to be mycoplasma-free by using the Lonza MycoAlert Mycoplasma Detection Kit (Lonza, Cat. No: LT07-418).
2.3. 3D culture of mouse embryonic pancreatic epithelial cells
8-well uncoated Ibidi chambers (Ibidi, Cat. No: 80821) were coated with 100% growth factor-reduced Matrigel (BD Biosciences, Cat. No: 356231) (5 μl/well) for 15 min at 37 °C before adding the cell suspension. Mouse embryonic pancreata were dissected from E14.5 embryos and incubated in 0.25% trypsin–EDTA (Invitrogen, Cat. No: 25200-056) for ≤30 min on ice followed by 5 min incubation at 37 °C. The trypsin was replaced by culture medium (medium A or B) and single-cell suspension was obtained by 30 times pipetting up and down of the samples. Cells were cultured on 100% Matrigel pre-coated chambers supplemented with 2% Matrigel in the culture medium. Each E14.5 pancreata was split in 2–3 wells of μ-slide 8-well Ibidi chambers.
Medium A: High glucose DMEM including l-Glutamine and 25 mM HEPES (21063-029), 1:100 MEM nonessential amino acids solution (NEAA) (100X) (Gibco, Cat, No: 11140-035), 1:100 Penicillin/Streptamycin (Gibco, Cat. No: 15140-122), 0.1 mM 2-Mercaptoethanol (Gibco, Cat. No: 31350010), 1:100 Insulin (SITE liquid Media 100x including Transferrin and Prostaglandin E1) (Sigma, Cat. No: S4920-5ML) and 0.33 ng/ml recombinant mouse hepatocyte growth factor (HGF) (PromoKine, Cat. No: D-64530).
Medium B: The medium was prepared by modification of a previously published protocol [37]. High glucose DMEM including l-Glutamine and 25 mM HEPES (21063-029), 2.4 mg/ml AlbuMAX II (Invitrogen, 11021-029), 1:100 Insulin-Transferrin-Selenium-G (100X) (Invitrogen, Cat. No: 41400-045), 1:100 Penicillin/Streptomycin (Gibco, Cat. No: 15140-122), 1:100 MEM nonessential amino acids solution (NEAA) (100X) (Gibco, Cat, No: 11140-035), 0.1 mM 2-Mercaptoethanol (Gibco, Cat. No: 31350010) and 0.33 ng/ml recombinant mouse HGF (PromoKine, Cat. No: D-64530).
For endocrine differentiation of mouse pancreatic cysts, cells were cultured for 1 day in basal medium B and the media was replaced by the differentiation medium consisting of basal medium B and the following components: 10 nM Exendin-4 (Sigma, Cat. No: E7144), 10 μM ALK5 inhibitor II (Axxora - Enzo Life Sciences, Cat. No: ALX-270-445-M005), 2 μM retinoic acid (Sigma, Cat. No: R2625-100MG), 100 ng/ml recombinant mouse Noggin (R&D Systems, Cat. No: 1967-NG-025), 10 mM Nicotinamide (Sigma, Cat. No: N0636-100G), 1.25 μM Tetrabenazine (TOCRIS, Cat. No: 2175), and 1.25 μM Dibutyryl-cAMP (dBu-cAMP) (Enzo Life Sciences, Cat. No: BML-CN125-0030).
2.4. In vitro differentiation of human iPSCs towards pancreatic β-like cells
Human iPSCs were cultured on 1:30 diluted Geltrex (Invitrogen, U.K, Cat. No: A1413302) in StemMACS iPS-Brew medium (Miltenyi Biotec, Germany, Cat. No: 130-104-368). When 70–80% confluency was reached, cells were split by washing the cells with 1x DPBS (Invitrogen, Cat. No: 14190094) followed by incubation with 0.05 mM EDTA (Applichem, Cat. No: 12604-021) for 3 min at 37 °C. Next StemMACS iPS-Brew medium containing Y-27632 (10 μM; Sigma–Aldrich, Cat. No: Y0503) was added and cell suspension was seeded for maintenance cultures (1:10) and differentiation (2–2.5 × 105/cm2) on Geltrex-coated surfaces. The differentiation was started 24 hrs after cell seeding. For differentiation towards endocrine cells, a slightly modified version (at S2) of the β-cell differentiation protocol by Rezania et al. was used [16].
Medium S1: Definitive endoderm was induced by using MCDB131 medium supplemented with 0.5% BSA (Sigma, Cat. No: 10775835001), 1xGlutamax (Gibco, Cat. No: A12860-01), CHIR 99021 (R&D Systems, Cat. No: 4423/10) for the first day and 100 ng/ml Activin A (Peprotech, Cat. No: 120-14-300) for all the 3 days.
Medium S2: The cells were differentiated towards primitive gut tube (PGT) with MCDB131 supplemented with 0.5% BSA, 1xGlutamax, 50 ng/ml of FGF7 (Peprotech, Cat. No: 100-19-100), 1.25 μM of IWP2 (Tocris, Cat. No: 3533-10), and 0.25 mM Vit. C (Sigma, Cat. No: 120-14-300) for 2 days.
Medium S3: Pancreatic progenitors (PP1) were induced by MCDB131 medium supplemented with 1xGlutamax, 2% BSA, 0.25 mM Vit. C, 50 ng/ml FGF7, 0.25 μM SANT-1 (Sigma, Cat. No: S4572-5MG), 1 μM retinoic acid (Sigma, Cat. No: R2625-50MG), 100 nM LDN193189 (Sigma, Cat. No: 04–0074), 1:200 ITS-X (Gibco, Cat. No: 51500-056), and 200 nM TPB (Merck Millipore, Cat. No: 565740-1MG) for 2 days.
Medium S4: PP1 were then differentiated towards pancreatic progenitors 2 (PP2) with MCDB131 supplemented with 1xGlutamax, 10 mM final glucose concentration, 2% BSA, 0.25 mM Vit.C, 2 ng/ml FGF7, 0.25 μM SANT-1, 0.1 μM retinoic acid, 200 nM LDN193189, 1:200 ITS-X, and 100 nM TPB for 3 days.
Medium S5: For endocrine induction, cells were exposed to MCDB131 medium supplemented with 1xGlutamax, 20 mM final glucose concentration, 2% BSA, 0.25 μM SANT-1, 0.05 μM retinoic acid, 100 nM LDN193189, 1:200 ITS-X, 1 μM T3 (Sigma, Cat#T6397-100MG), 10 μM ALK5 Inhibitor II (Enzo Life Sciences, Cat. No: ALX-270-445-M005), 10 μM zinc sulphate (Sigma, Cat. No: SI Z0251-100G), and 10 μg/ml heparin (Sigma, Cat. No: H3149) for 3 days.
Medium S6: Hormone synthesizing cells were induced with MCDB131 supplemented with 1xGlutamax, 20 mM final glucose concentration, 2% BSA, 100 nM LDN193189, 1:200 ITS-X, 1μM T3, 10 mM ALK5 Inhibitor II, 10 μM zinc sulphate, 100 nM gamma secretase inhibitor XX (Merck, Cat. No: 565789) for 7 days.
2.5. 3D culture of human pancreatic progenitors
For cyst generation, 8-well Ibidi chambers were coated with 100% Matrigel (5 μl/well) and kept for 15 min at 37 °C to polymerize the Matrigel. Human differentiated iPSCs at different stages (DE, PGT, PP1, PP2 and EN) were trypsinized with 1:1 0.05% Trypsin–EDTA (Cat. No: 25300054) and DPBS for 3 min at 37 °C. Subsequently the enzymatic reaction was neutralized by StemMACS iPS-Brew, spun for 3 min at 1200 rpm and seeded in differentiation medium containing 10 μM Y-27632 and 5% Matrigel. The following day the medium was changed with the appropriate differentiation medium.
2.6. Immunostaining and imaging
2.6.1. Preparation of embryonic pancreatic sections and staining
Embryonic pancreata were dissected and fixed with 4% paraformaldehyde (PFA) for 2 hrs at 4 °C before subsequent merging in 7.5, 15 and 30% sucrose-PBS solutions (2 hrs incubation for each solution) at RT. Next, samples were embedded in cryoblocks using tissue-freezing medium (Leica 14020108926) and sections of 20 μm thickness were prepared and used for immunostaining described as follow.
First, samples were permeabilized (0.1% Triton, 100 mM Glycine) for 15 min and were subjected to the blocking solution (10% FCS, 0.1% BSA, 3% Donkey serum and 0.1% Tween-20 in PBS) for 1 hr at RT. Then, primary antibodies (Supplementary Table 1) that were diluted in the blocking solution were added overnight at 4 °C. Samples were then extensively washed with PBST (0.1% Tween-20 in PBS), and were incubated with secondary antibodies for 3–5 hrs at RT. The samples were then incubated with DAPI and embedded in the commercial medium (Life Tech., ProLong Gold).
2.6.2. Immunostaining of mPECs
Mouse PECs were fixed with 4% PFA for 10–15 min at 37 °C and were permeabilized (100 mM Glycine and 0.2% Triton X-100) for 10 min at RT. Next, fixed samples were incubated with blocking solution (10% FCS, 0.1% BSA, 3% Donkey serum and 0.1% Tween-20 in PBS) for 30 min at RT. Subsequently, mPECs were incubated with primary antibodies (diluted in blocking solution) (Supplementary Table 1) for 1–2 hrs at RT. After washing with PBST, samples were incubated with secondary antibodies for 1 hr at RT followed by addition of the mounting medium.
2.6.3. Immunostaining of hPECs
iPSC-derived hPECs were rinsed once with 1 x PBS before samples were fixed with 4% PFA for 15 min at RT, then washed once with PBS and permeabilized with 0.1% Triton X-100 and 0.1 M Glycine in PBS for 20 min at RT. The hPECs were blocked with the blocking solution (10% FCS, 0.1% BSA, 3% Donkey serum and 0.1% Tween-20 in PBS) for 1 hr at RT on a shaker. Next, hPECs were exposed to the primary antibodies (Supplementary Table 1) for 1 hr at RT and then at 4 °C overnight. The following day the antibody solution was removed and washed 3 x with PBS for 10 min followed by incubation with secondary antibodies for 3 hrs at RT. The hPECs were washed 3 x with PBS for 10 min and then kept in PBS for direct imaging or addition of Elvanol for long term storage.
2.6.4. Confocal microscopy imaging
Imaging of mPECs and pancreatic sections was performed using a laser scanning confocal microscopy. Images were acquired with a Leica microscope of the type DMI 6000 and a Leica TCS SP5 confocal laser scanning setup. hPECs were imaged using Zeiss LSM 800 confocal laser microscope.
2.6.5. Time-lapse imaging
Time-lapse movies were captured using a Leica microscope of the type DMI 6000 and a Leica TCS SP5 confocal laser scanning setup. E14.5 primary pancreatic cells were culture in 3D Matrigel and imaged after 4 or 24 hrs at 37 °C in a controlled chamber supported by O2 and CO2.
2.7. Single-cell data analysis
All analyses were performed using Python 3.5 and the Single Cell Analysis in Python (Scanpy) (v1.3.2) (https://github.com/theislab/scanpy) [38].
2.7.1. Re-analysis of murine pancreatic endocrinogenesis
Processed, normalized, and annotated single cell RNA sequencing data were downloaded from GEO (accession number GSE132188). The original data contained cells from the pancreatic epithelium analyzed at four time points (E12.5-E15.5) [7]. For the scope of this manuscript, we used a subset of the data that contained from time points E13.5–15.5 ductal, endocrine progenitor-precursor cells (_Ngn3_low and _Ngn3_high) as well as Fev+/hormone- and hormone+ endocrine cells (Fev+-Beta, Fev+-Alpha, Beta, Alpha, Fev+-Delta, Delta, Fev+-Epsilon, Epsilon) according to the clustering in Figures 6 and 7 of the original publication [7]. For visualization, Uniform Manifold Approximation and Projection (UMAP) [39] was newly calculated on the subset of the data by recomputing the single cell neighborhood graph (kNN-graph) on the first 50 principal components using 15 neighbors. As input data was subset to the highly variable genes as identified in the original publication.
2.7.2. Re-analysis of human in vitro stem cell differentiation
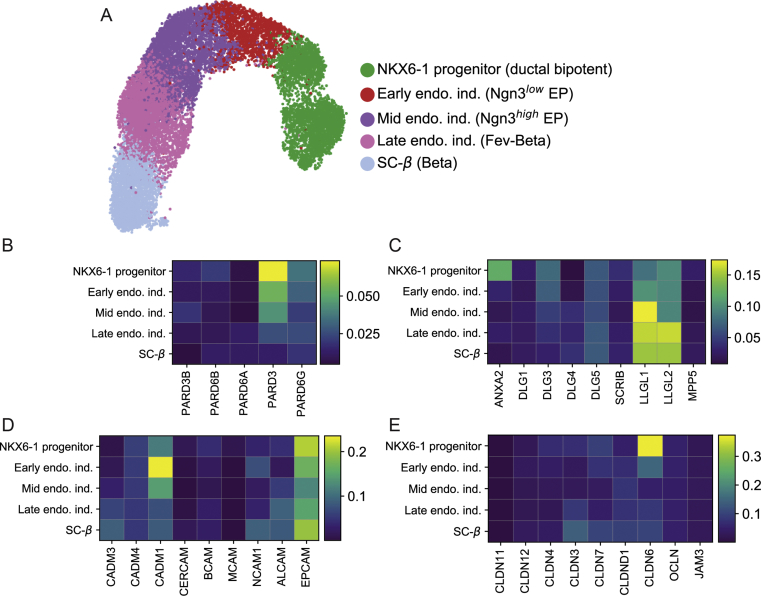
Processed single cell RNA sequencing data and annotations were downloaded from GEO (accession number GSE132188). The data of stage 5 cells from the original publication [17] were used (GSE114412_Stage_5.all.processed_counts.tsv and GSE114412_Stage_5.all.cell_metadata.tsv) and subset to endocrine progenitors and the β-cell lineage according to the annotation of the original publication in Figure 5 (clusters NKX6-1 progenitor, Early endocrine induction, Mid endocrine induction, Late endocrine induction, SC-β). A cluster of SC-β cells that did not express insulin (annotated as early SC-β cells) was excluded. The data were normalized to total counts per cell using the pp.normalize_per_cell function in scanpy with default parameters and log-transformed (pp.log1p). Genes expressed in less than 10 cells were filtered. For visualization, a UMAP was calculated by computing the single cell neighborhood graph (kNN-graph) on the first 50 principal components using 15 neighbors. As input, the data were subset to the top 3000 highly variable genes selected based on normalized dispersion using the pp.filter_genes_dispersion with default parameters.
3. Results
3.1. Establishment of 3D mouse pancreatic cyst culture
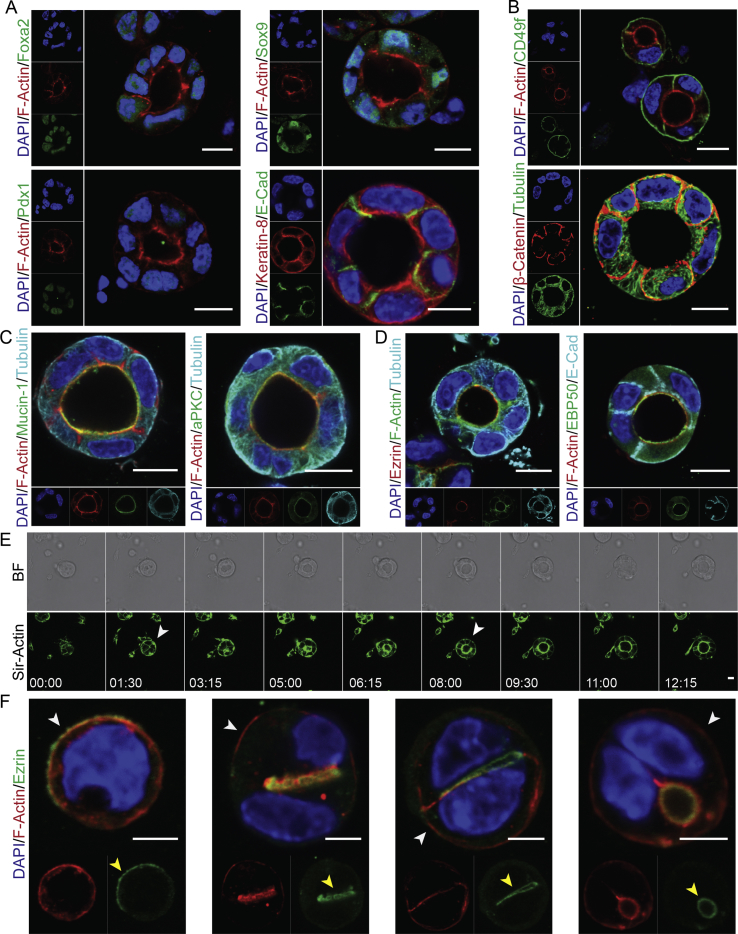
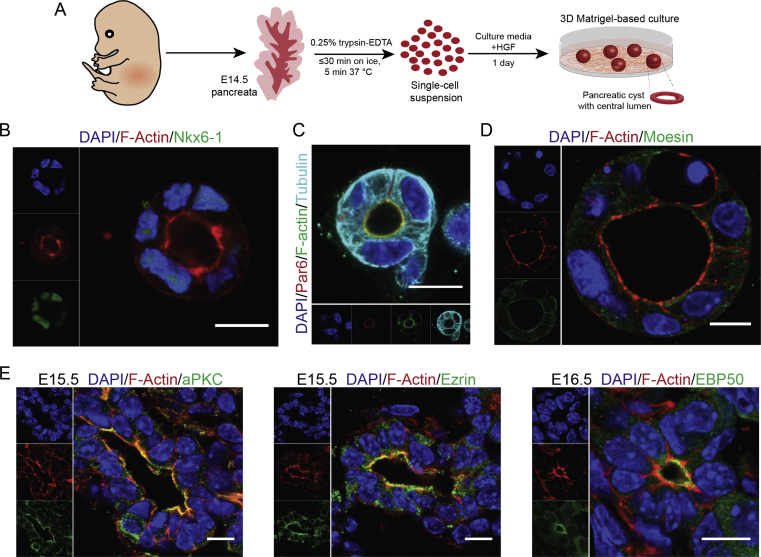
To temporally resolve cell biological and molecular processes occurring in the pancreatic epithelium during expansion and differentiation, we established an in vitro mouse polarized PEC culture. Therefore, we used single cell suspensions of primary pancreatic cells at E14.5 cultured in a Matrigel-based 3D condition using two different culture media containing hepatocyte growth factor (HGF) (Suppl. Figure 1A). Although, the initial cell suspension included all pancreatic and non-pancreatic cells (mesenchymal, epithelial and endothelial), our culture conditions allowed only pancreatic progenitors to aggregate and gradually form a polarized epithelial cyst with a central lumen during one day of culture (Suppl. Movie 1). Alternatively, cyst formation was initiated by division of a single progenitor followed by establishment of a central lumen at 2-cell stage (Figure 1F). Further maintenance of PECs in culture resulted in their growth and expansion (Suppl. Movie 2). Following this, we stained mouse PECs (mPECs) for pancreatic ductal epithelial markers. We found that mPECs express several pancreatic progenitor TFs, such as Sox9, low levels of Foxa2, Pdx1, and Nkx6-1 as well as the ductal marker Cytokeratin-8 (CK8) (Figure 1A and Suppl. Figure 1B). These results indicate that the PP-derived mPECs represent the molecular signatures of pancreatic ductal epithelium in vivo.
Figure 1.
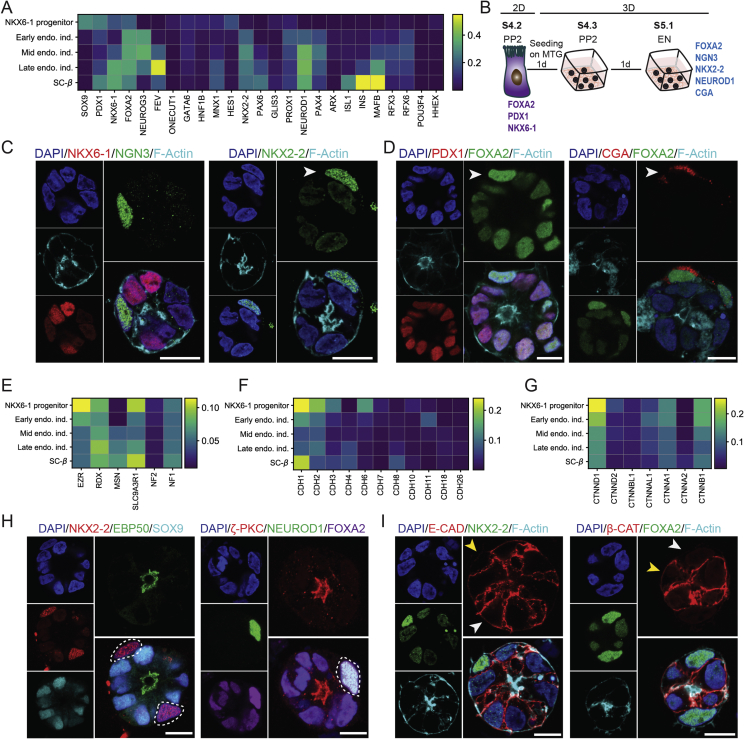
Establishment and characterization of 3D mouse PECs culture. (A) mPECs express key TFs of embryonic pancreatic epithelium including Foxa2, Sox9, and Pdx1 as well as the pancreatic ductal marker Keratin-8. (B) mPECs express apicobasal polarity markers including F-Actin (apical), β-catenin (lateral) and CD49f (basal). (C,D) Expression of different polarity proteins including Mucin-1, aPKC, Ezrin, and EBP50 at the apical domain of mPECs. (E) Time-lapse imaging of mPECs formation over 12 hrs. mPECs were labeled with 200 mM F-Actin probe (SiR-actin) 4 hrs before starting the course of imaging. White arrows indicate the levels of F-Actin at the cyst periphery. (F) Different stages of polarity establishment and central lumen formation in pancreatic epithelial cells. White arrows indicate the levels of F-Actin at the cyst periphery. Yellow arrows show the apical-like domain (single-cell stage), AMIS and central lumen formation (from two-cell stage onwards). Scale bar: 10 μm (A–E); 5 μm (F).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.molmet.2019.09.005
The following are the supplementary data related to this article:
Next, we analyzed if mPECs recapitulate AB polarity of embryonic pancreatic epithelium. Immunostaining analysis revealed the basal expression of the cell–matrix adhesion molecule CD49f (Integrin α6) and basolateral staining of the cell–cell adhesion molecule β-Catenin (β-Cat) in mPECs (Figure 1B). Furthermore, staining of different apical markers including cortical plasma membrane (PM) filamentous Actin (F-Actin), Mucin-1, atypical protein kinase C (aPKC), and partitioning defective 6 homolog alpha (Par6) indicated the orientation of apical domain towards the central lumen (Figure 1C and Suppl. Figure 1C). In epithelial cells, F-Actin is tethered to the nearby PM mediated by the ERM (Ezrin, Radixin, Moesin) proteins that are differentially expressed in distinct tissues. Accordingly, we found the localization of Ezrin and its related protein, Ezrin-Radixin-Moesin-binding phosphoprotein 50 (EBP50/NHERF1), in the apical domain of these structures (Figure 1D). Among these, the expression of EBP50 has not been reported in embryonic pancreatic epithelium before. Of note, we could not detect specific staining for another ERM member Moesin in the apical domain of mPECs (Suppl. Figure 1D). Moreover, the apical expression of aPKC, Ezrin, and EBP50 was confirmed in embryonic pancreatic sections (Suppl. Figure 1E), further indicating the similar expression of AB polarity markers between in vitro generated mPECs and in vivo pancreatic epithelial progenitors.
The molecular details of early events during pancreatic epithelial development are not well-characterized. In Madin–Darby canine kidney (MDCK) cell line, epithelial lumen formation starts with the appearance of an apical-like domain on one side of the single cell that after cell division translocates to the area between two cells to form the apical membrane initiation site (AMIS) [40], [41]. Further growth of epithelial cysts relies on the expansion of this domain and increase in cell division process. To explore how the central lumen and epithelial polarity are established, we analyzed the time-lapse imaging of pancreatic cyst formation. We found that at early stages F-Actin network is enriched at the cell periphery but it gradually localizes centrally at later stages (Figure 1E and Suppl. Movie 1). Moreover, staining of pancreatic epithelial cells revealed the existence of an apical-like domain at single-cell stage (marked by Ezrin) and the appearance and extension of an AMIS at the two-cell stage that further generated a central lumen and established the epithelial AB polarity (Figure 1F). Thus, our primary cell culture model allows detailed analysis of a cell biological and molecular process that has only been shown to exist in immortalized MDCK cells before.
3.2. ScRNA-seq analysis reveals molecular details of cell biological processes during endocrinogenesis
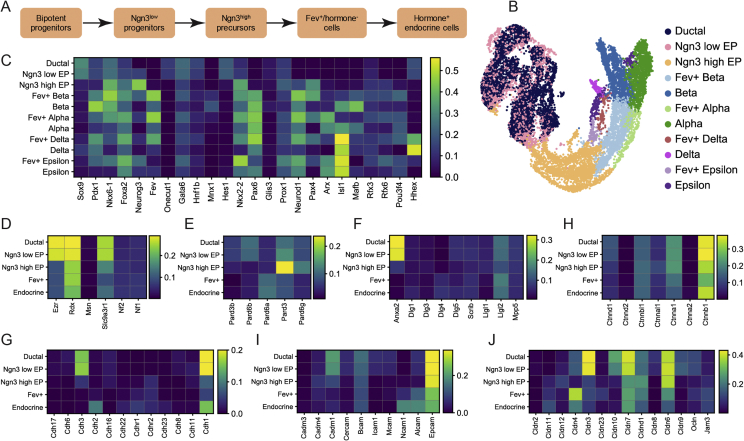
After successful generation of PECs, we used the model system to study the molecular details of endocrinogenesis ex vivo. To compare and have a blueprint of cell differentiation programs during endocrine cell differentiation in vivo, we first analyzed transcriptional profiles of mouse embryonic pancreatic cells at single-cell-level resolution using our recently published dataset [7] (Figure 2A,B). As expected, TFs Sox9, Gata6, Hes1, and Hhex were expressed higher in ductal cluster and _Ngn3_low progenitors, but their expression were strikingly lower or almost absent in other clusters. At _Ngn3_high precursor state, there was an upregulation of Ngn3, Foxa2, _Nkx6_-1, _Nkx2_-2, and Pax4 expression. In contrast, the expression levels of Pdx1, Pax6, and Neurod1 were only increased at _Fev_high state. The differentiated α-cells expressed higher levels of Arx and Pou3f4, while the differentiated β-cells were marked by increased expression levels of Pdx1, _Nkx6_-1, Mnx1, and Ins2 (Figure 2C). Thus, this analysis provides a comprehensive picture of changes in expression levels of key pancreatic TFs during mouse endocrinogenesis.
Figure 2.
ScRNA-seq analysis of cell–cell adhesion and polarity molecules during endocrinogenesis. (A) Endocrine lineage formation during mouse embryonic development. (B) UMAP plot of 11 pancreatic epithelial cell types including ductal, _Ngn3_low progenitors, _Ngn3_high precursors, Fev+ beta, β-cells, Fev+ alpha, α-cells, Fev+ delta, δ-cells, Fev+ epsilon, and ε-cells from E13.5, 14.5 and 15.5 mouse embryonic pancreas. Each cell cluster is shown with a different color code. (C) ScRNA-seq analysis of E13.5–15.5 mouse pancreatic epithelial cells indicating changes in the expression levels of key pancreatic TFs during endocrine cell differentiation. (D–F) Heatmap showing the mean expression levels of apical polarity molecules including members of ERM proteins (D), Par proteins (E) and other known epithelial apical markers (F) in ductal, _Ngn3_low progenitors, _Ngn3_high precursors, Fev+ and endocrine cells. Compared to part B, Fev+ beta, Fev+ alpha, Fev+ delta, and Fev+ epsilon as well as β-, α-,δ-cells and ε-cells have been combined and shown as Fev+ and endocrine clusters, respectively. (G–J) Heatmap showing the mean expression levels of major epithelial cell–cell adhesion molecules including different members of cadherins (G), catenins (H), cell adhesion molecules (CAMs) (I), and claudins (J) in ductal, _Ngn3_low progenitors, _Ngn3_high precursors, Fev+ and endocrine cells. Ocln and Jam3 are shown within the claudins heatmap. Expression values are normalized and scaled to range from 0 to 1.
Upon differentiation, endocrine cells change morphology, leave the ductal epithelium in a so-called delamination process, and cluster within the surrounding mesenchyme to form proto-islets [6], [42], [43]. The cell biological and molecular details of this dynamic process are largely unknown. Therefore, we hypothesized that changes in AB polarity as well as resolving and remodeling of adherens and tight junctions (AJs/TJs) underlie this process. Towards this end, we analyzed the relative expression levels of key AB polarity molecules including members of the ERM and Par families. Among the ERM molecules, Ezrin (Ezr) was expressed higher in ductal and _Ngn3_low progenitors, but its expression levels strikingly decreased from _Ngn3_high precursor state onward. In comparison, the expression levels of Moesin (Msn) were extremely low in pancreatic lineages, while the expression of EBP50 (Slc9a3rl) were reduced from _Ngn3_high precursor state (Figure 2D). From the Par molecules, Pard6b was expressed higher in ductal and _Ngn3_low progenitors, while Pard6a was expressed higher in _Fev_high and endocrine cells. Interestingly, the expression levels of Pard3 and Pard6g were transiently increased at _Ngn3_high precursor state but decreased in _Fev_high cells (Figure 2E). Among other apical domain molecules, we found higher expression levels of Annexin A2 (Anxa2) in ductal and _Ngn3_low progenitors that was reduced from _Ngn3_high precursor state. Additionally, the expression level of Lethal giant larvae protein homolog 2 (Llgl2) was transiently reduced in _Ngn3_high precursors (Figure 2F).
Next, we analyzed the relative changes in the expression levels of AJs/TJs molecules during endocrinogenesis. Amongst cell–cell adhesion molecules, we analyzed the expression levels of cadherins, catenins, cell adhesion molecules (CAMs) and claudins. The major members of cadherins that were expressed in pancreatic cells were E-Cadherin (Cdh1), N-Cadherin (Cdh2), and P-Cadherin (Cdh3). Cdh1 was expressed higher in ductal cluster and _Ngn3_low progenitors, but its levels decreased in _Ngn3_high precursors and _Fev_high cells and increased again in endocrine cells. In comparison, Cdh2 was only expressed in endocrine cells (Figure 2G). Catenin α-1 (Ctnna1), β-Catenin (Ctnnb1), Ctnnbl1, and p120 catenin (Ctnnd1) were the major catenins that were expressed during endocrinogenesis. Similar to Cdh1, the expression levels of Ctnnb1 were also reduced in _Ngn3_high precursors and _Fev_high cells but increased in endocrine cells. In addition, the expression levels of Ctnnd1 were reduced during endocrinogenesis as it has been reported previously [44] (Figure 2H). Among the CAMs, Cadm1 and Basal cell adhesion molecule (Bcam) were downregulated during endocrinogenesis, while the expression levels of Neural cell adhesion molecule 11 (Ncam) and CD166 antigen (Alcam) were increased in endocrine cells (Figure 2I). Finally, we found the transient downregulation of Cldn4 in _Ngn3_high precursors that was strikingly restored in _Fev_high cells (Figure 2J). Altogether, scRNA-seq analysis indicates loss of AB polarity components and dynamic changes in expression levels of AJs/TJs molecules during mouse endocrinogenesis.
3.3. 3D mPECs enable studying cell biological processes during endocrinogenesis
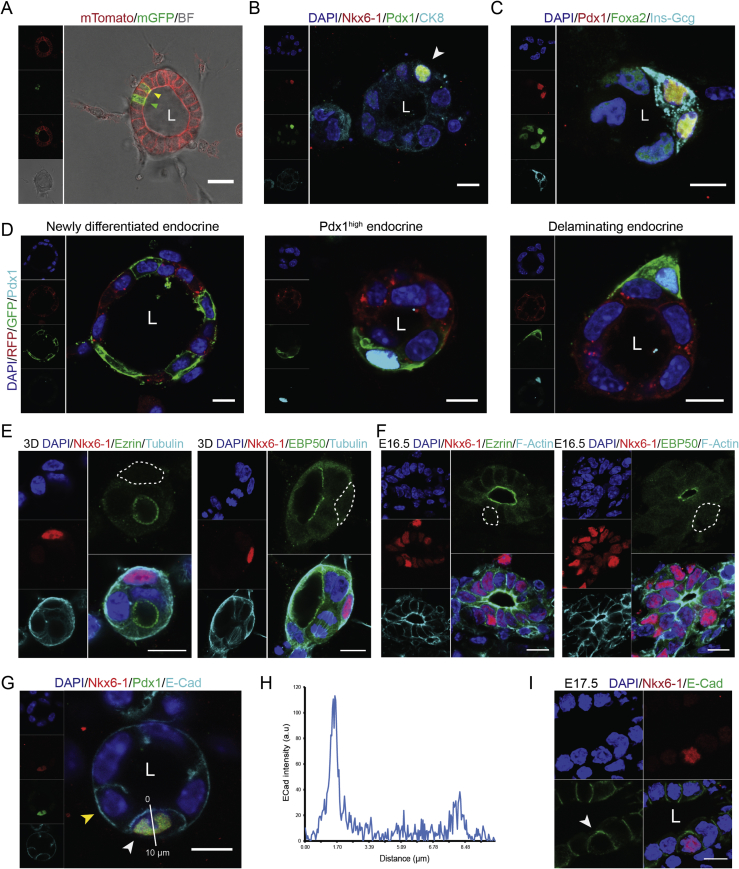
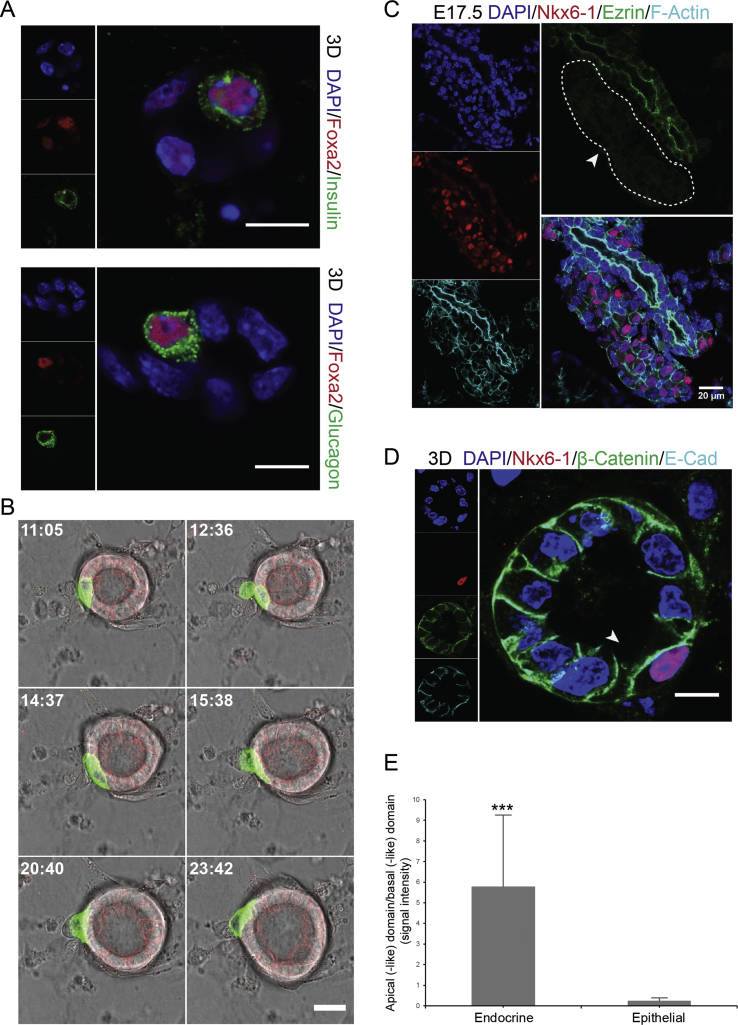
To confirm the cell biological changes during endocrinogenesis that were predicted by the scRNA-seq analysis, we triggered endocrine cell fate in mPECs using previously reported mouse and human endocrine differentiation factors (see M&M). To monitor endocrinogenesis, we used the mTmG reporter mouse line, in which membranous Tomato (mTomato) is expressed ubiquitously [35]. Crossing of this mouse model with Ngn3Cre line [34] resulted in generation of Ngn3Cre; mTmG mice, in which Cre-mediated excision of the membrane Tomato (mT) reporter gene switches the expression to membrane GFP (mGFP) expression. Thus, endocrine cells first appeared in yellow (expressing mTomato and mGFP) and remained green afterwards (expressing only mGFP) (Figure 3A and Suppl. Movie 3). It should be noted that in this system, endocrine induction occurs with low frequency; however, even low number of differentiated cells allowed morphological analysis. Immunostaining analysis confirmed the appearance of endocrine cells within the mPECs that were marked by high expression levels of Foxa2, Pdx1, and Nkx6-1 (Figure 3B,C), as predicted by our scRNA-seq data (Figure 2). Furthermore, immunostaining revealed the emergence of hormone-producing cells from the 3D mPECs (Figure 3C). Although, we found the presence of insulin- and glucagon-positive cells within the mPECs (Suppl. Figure 2A), the rate of generated mono- and poly-hormonal cells in this system needs to be analyzed in future studies. Overall, these data indicate that primary mPEC culture mimics endogenous activation of endocrine TF programs during endocrinogenesis.
Figure 3.
Application of mPEC system to study endocrinogenesis at cellular and subcellular resolution levels. (A) Differentiation of mPECs derived from Ngn3Cre;mTmG pancreata towards endocrine cell fate. Newly generated endocrine cell expressing both mTomato and mGFP is shown by yellow arrow and the fully differentiated endocrine cell expressing mGFP is shown by green arrow. (B) Endocrine cell differentiation (white arrow) in mPECs is marked by expression of high levels of Pdx1 and Nkx6-1 compared to the nearby epithelial cells. (C) Emergence of hormone-positive cells within the mPECs. Samples are stained with antibodies against insulin and glucagon derived from similar species, and therefore stained with a single secondary antibody. (D) Newly generated endocrine cells (mGFP+) that are located within the epithelium do not express high levels of Pdx1. Cells expressing high levels of Pdx1 are not in direct contact with the central lumen but rather are located with the second epithelial layer. Anti-RFP was used to stain mTomato in fixed samples. (E,F) The apical proteins Ezrin and EBP50 are downregulated in the endocrine cells differentiated in the mPECs (E) and in vivo condition (F). Dashed circles indicate endocrine cells. (G) Differentiated endocrine cells exhibit a distinct expression pattern of E-Cad compare to the nearby epithelial cells. White and yellow arrows indicate the expression of E-cad at the basal side of epithelial and endocrine cell, respectively. (H) Plot profile of expression pattern of E-Cad in endocrine cells. Higher expression level of E-Cad is evident at the domain towards the central lumen. (I) Expression pattern of E-Cad in a differentiated endocrine cell in E17.5 embryonic pancreatic section. White arrow shows expression of E-Cad at the domain towards the central lumen. L, lumen; BF, bright-field. Scale bar: 20 μm (A); 10 μm (B–I).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.molmet.2019.09.005
The following is the supplementary data related to this article:
To explore endocrine cell dynamics in the PEC model system, we performed time-lapse imaging of mPECs derived from Ngn3Cre;mTmG pancreata. We found that GFP+ cells have a tendency to cluster together within the epithelial layer (Suppl. Movie 4) and due to their highly active movements, they ultimately evaginated from the epithelial plane (Suppl. Figure 2B and Suppl. Movie 5). Moreover, the Pdx1 TF was expressed at low levels in GFP+ cells that were fully located within the epithelium, but the expression levels were increased upon endocrine cell delamination (Figure 3D). These data indicate that the delamination process requires an activation of further downstream TF cascades during endocrine cell development.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.molmet.2019.09.005
The following are the supplementary data related to this article:
Based on our scRNA-seq analyses, we next investigated how endocrine cell morphology is changed upon cell differentiation in mPECs. ScRNA-seq analyses showed striking downregulation of epithelial AB polarity proteins including ERM members during endocrinogenesis. Therefore, we stained differentiated mPECs for AB polarity markers Ezrin and EBP50 and found the loss of these proteins in the differentiated endocrine cells upon and after delamination process (Figure 3E). These data were also supported by staining of E16.5 pancreatic sections (Figure 3F and Suppl. Figure 2C).
Single-cell mRNA profiling also indicated striking changes in the expression levels of several cell–cell adhesion molecules (Figure 2G–J). Among these, we analyzed the expression of two major adhesion proteins E-Cad and β-Cat in mPECs that exhibited distinct expression pattern in epithelial and endocrine cells. Whereas epithelial cells expressed both proteins at the basolateral sides where adherens junctions are localized, the differentiated endocrine cells exhibited a reversed localization of these proteins, in which E-Cad and β-Cat were detected in the lateral and the domain towards the central lumen (apical-like domain) (Figure 3G,H and Suppl. Figure 2D,E). A similar expression pattern of E-Cad was also found in E17.5 pancreatic tissue (Figure 3I). Altogether, in vitro and in vivo analysis indicate that the sequence of events of cell biological processes occurring during endocrine induction and differentiation can be fine-staged to the specific progenitor cell type e.g. _Ngn_3high precursors and Fev+ cells.
3.4. Establishment of 3D human pancreatic cyst culture
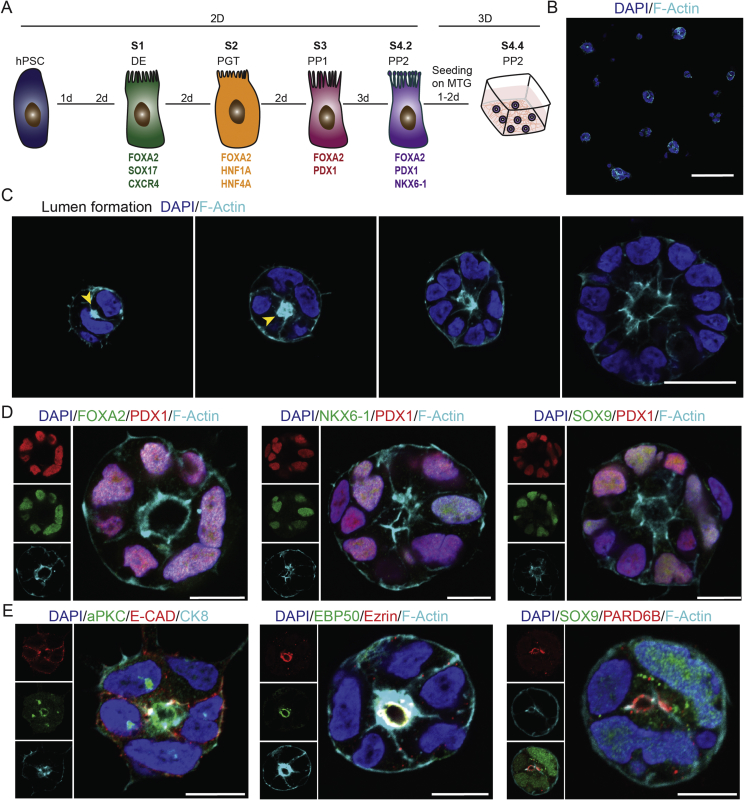
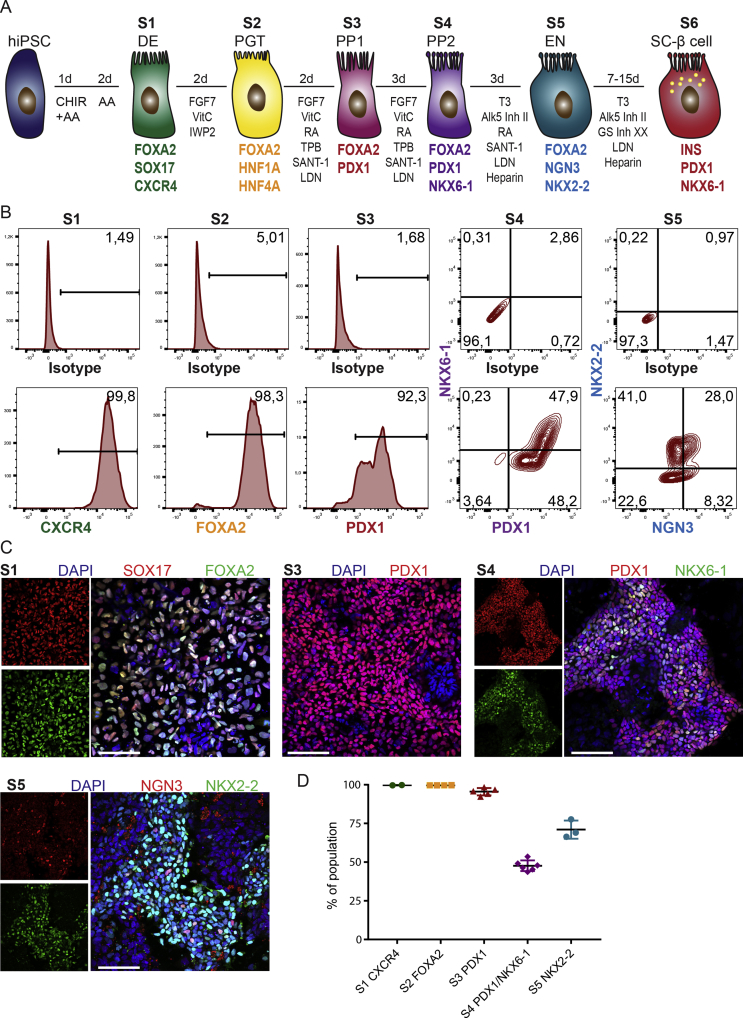
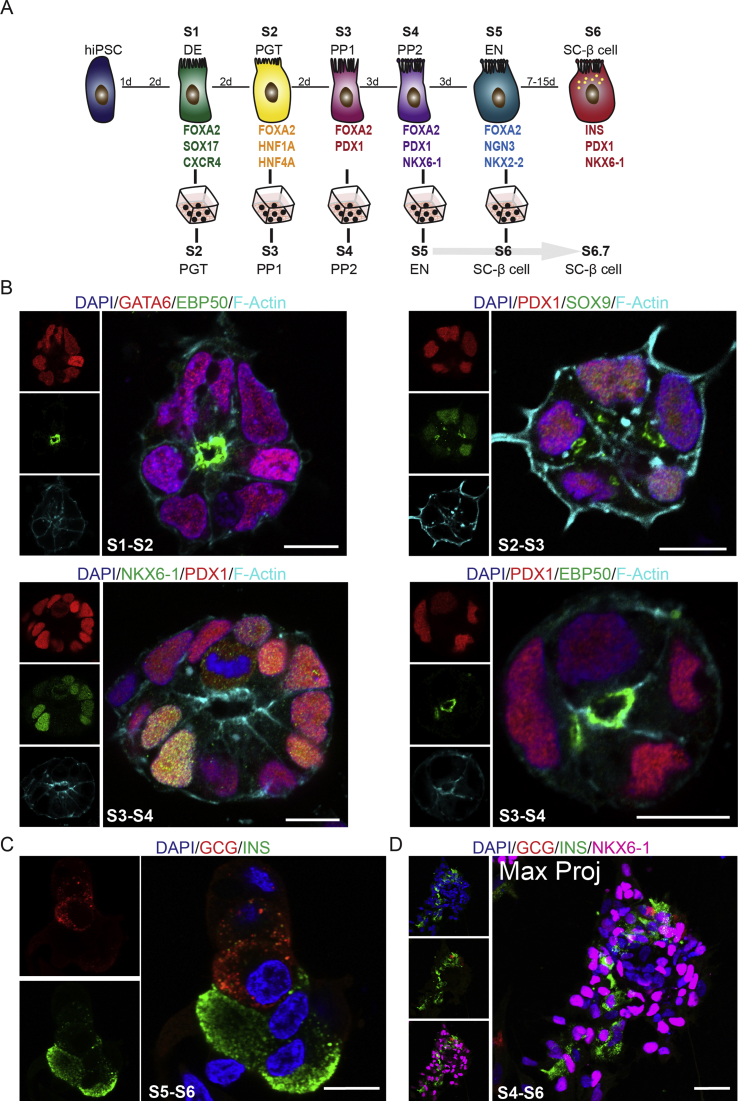
To translate the in vitro PEC platform from mouse to human, we established a 3D culture of hiPSC-derived pancreatic epithelial cells. We used our recently generated hiPSC line (HMGUi001-A) and differentiated them towards endocrine lineage in adherent 2D cultures [36]. The differentiation procedure included definitive endoderm (DE, stage 1 (S1)), primitive gut tube (PGT, stage 2 (S2)), pancreatic progenitors 1 (PP1s, stage 3 (S3)), pancreatic progenitors 2 (PP2s, stage 4 (S4)) marked by expression of both PDX1 and NKX6-1 TFs and endocrine lineage (S5) expressing NGN3 and NKX2-2 [16], [45] (Suppl. Figure 3).
To test the potential of human pancreatic lineages to form human PECs (hPECs), we harvested single cells at stages S1, S2, S3, S4, and S5 from 2D and further cultured them under 3D condition. We noticed that although in all stages cells were capable of forming polarized cysts, the PP2 cells exhibited a higher tendency to form 3D cysts (qualitative observation) (Figure 4A,B and Suppl. Figure 4B). The higher potential for hPEC generation might be due to the right niche factors within the culture media and bipotency of PP2 cells to generate endocrine or ductal epithelial lineages. Notably, at S5 stage, cells were not able to generate polarized cysts in 3D condition but rather accumulated as small aggregates that were mainly composed of NKX6-1high cells and hormone+ endocrine cells (Suppl. Figure 4C,D). These data highlight the reduction in self-renewal and expansion potential as well as loss of epithelial characteristics of endocrine cells that hampers their ability to form cysts with a central lumen.
Figure 4.
Establishment and characterization of 3D human PEC culture. (A) Schematic picture presenting different steps of in vitro differentiation of hPCSs into PPs and subsequent culturing of PPs in 3D Matrigel-based culture. (B) Culturing S4 PPs in 3D condition results in formation of hPECs after 1–2 days. (C) hPECs are formed by the formation of AMIS (marked by F-Actin) and subsequent increase in cell number along with expansion of the central lumen located at the apical domain. Yellow arrows show the AMIS. (D) The expression of key TFs of embryonic pancreatic epithelium including PDX1, FOXA2, SOX9, and NKX6-1 in hPECs. (E) hPECs are highly polarized epithelial structures expressing apical polarity markers aPKC, Ezrin, EBP50, and PARD6B. Scale bar: 100 μm (B) 20 μm (C) 10 μm (D–E).
To assess how pancreatic epithelial morphogenesis occurs in human 3D cysts, we monitored different stages of epithelial polarization and central lumen formation. We found that upon culturing in 3D condition, human PPs aggregated and formed an AMIS at two-cell stage that was defined by the accumulation of F-Actin at the interface area (Figure 4C). AMIS development in hPECs mimicked the AMIS formation observed in mPECs showing that this is a very reproducible and stereotypic process occurring in polarized human and mouse epithelial cells upon polarization and lumen formation. Increased cell number with elongation of the central apical domain resulted in growth and expansion of hPECs (Figure 4C). Next, we examined if hPECs express human embryonic pancreatic epithelial markers. Immunostaining analysis revealed the expression of TFs, such as PDX1, FOXA2, NKX6-1, and SOX9 in hPECs (Figure 4D). This expression profile is similar to what has been previously reported for the human embryonic pancreatic epithelial cells [11]. Furthermore, comparable to mPECs, the expression of the intermediate filament Cytokeratin-8 (CK8), as a pancreatic epithelial marker was detected in hPECs (Figure 4E). Finally, we detected the expression of AB polarity markers aPKC, Ezrin and EBP50 in the apical domain of hPECs (Figure 4E). Collectively, these data show that mouse and human PPs are able to generate polarized pancreatic cysts in 3D condition that follow similar building principles of polarity establishment, central lumen formation and cyst expansion.
3.5. 3D hPECs enable studying cell biology of human endocrinogenesis
To explore whether our 3D model system allows studying human endocrinogenesis ex vivo, we first obtained a blueprint of cell differentiation programs using a recently reported scRNA-seq data set from in vitro human endocrine cell differentiation [17]. We analyzed the transcriptional profiles of five major clusters involved in β-cell differentiation (Suppl. Figure 5A) that revealed the expression pattern of key TFs during lineage segregations towards endocrine fate (Figure 5A). For endocrine differentiation of hPECs, we harvested PP2 cells from 2D and cultured them in 3D for 1 day in S4 medium to form hPECs. Next, we switched the medium to endocrine differentiation condition and further cultured the hPECs for another day (Figure 5B). This method resulted in induction and differentiation of endocrine cells marked by the expression of TFs NGN3, NKX2-2 and NEUROD1 (Figure 5C,H). Additionally, we found the appearance of cells expressing high levels of FOXA2 that were positive for pan-endocrine marker chromogranin A (CGA) (Figure 5D). Interestingly, both scRNA-seq data and hPEC staining indicated upregulation of FOXA2 levels during endocrine differentiation. In comparison PDX1 levels were not increased in endocrine cells compared to the pancreatic progenitors (Figure 5A,D), contrary to what was observed in mouse system. Notably, we observed that cells expressing NKX2-2 and high levels of FOXA2 were not in direct contact to the apical lumen but rather were located in the second layer below the cyst epithelium (Figure 5C,D). This observation likely reflects the delamination process of human endocrine cells similar to what we observed in the mouse primary progenitor cell system.
Figure 5.
ScRNA-seq analysis and hPEC differentiation enable studying cellular processes during human endocrinogenesis. (A) ScRNA-seq analysis of in vitro differentiated human pancreatic lineages indicating changes in the expression levels of key pancreatic TFs during endocrinogenesis. Expression values are normalized and scaled to range from 0 to 1. (B) Schematic picture showing differentiation of endocrine cells in hPEC system. (C) Expression of NGN3 and NKX2-2 (white arrow) indicate endocrinogenesis process in hPECs. (D) Endocrine differentiation in hPECs is marked by expression of high levels of FOXA2 and CGA (white arrows). (E–G) Heatmap showing the mean expression levels of different members of ERM molecules (E), cadherins (F) and catenins (G) during in vitro human endocrinogenesis. Expression values are normalized and scaled to range from 0 to 1. (H) The apical proteins EBP50 and aPKC are downregulated in the endocrine cells differentiated in the hPECs. Dashed circles indicate endocrine cells. (I) Different expression pattern of E-CAD and β-CAT in human epithelial and endocrine cells. White and yellow arrows indicate the expression of E-CAD and β-CAT at the basal side of epithelial and endocrine cell, respectively. Scale bar: 10 μm.
To explore whether changes in AB polarity and remodeling of AJs/TJs are evolutionary conserved mechanisms during endocrine cell differentiation, we first analyzed the relative expression levels of apical polarity components and AJs/TJs molecules in the scRNA-seq data from human pancreatic lineages (Figure 5E–G and Suppl. Figure 5B-E). Among apical polarity molecules, the expression levels of Ezrin were reduced during endocrinogenesis, while the expression levels of EBP50 (SLC9A3Rl) were transiently downregulated in endocrine progenitors (Figure 5E). Interestingly and differently with the mouse scRNA-seq, the expression levels of Moesin (MSN) were increased during human endocrinogenesis. Similar to the mouse system, the expression levels of E-CAD and β-CAT were transiently downregulated in endocrine progenitors and were increased in differentiated endocrine cells (Figure 5F,G). Through immunostaining analysis, we confirmed these findings in the hPEC system, that revealed loss of expression of AB polarity markers aPKC and EBP50 in the differentiated endocrine cells compared to the nearby epithelial cells (Figure 5H). Moreover, we found a differential expression pattern of E-CAD and β-CAT in human endocrine and epithelial cells. Similar to mouse system, both proteins were expressed at the basolateral sides of epithelial cells but were not present on the basal sides of differentiated endocrine cells (Figure 5I). Overall, combining scRNA-seq analysis with hPEC differentiation was able to identify similarities and differences of cell differentiation programs and cellular processes during endocrinogenesis in mouse and human.
4. Discussion
Due to the evolutionary diversities in organ formation and function, translation of clinical findings from animals to human requires better understanding of similarities and differences between these species. Thus, it is essential to design novel ex vivo modeling systems that allow time-resolved longitudinal analysis of biological processes. Here, we have established a robust and reproducible high-resolution 3D PEC platform, which is suitable to uncover mechanisms of early pancreatic epithelial development in mouse and human. The ability of this platform to perform single-cell continuous live cell imaging with high temporal and spatial resolution enabled studying cell biological processes underlying pancreatic progenitor epithelialization, endocrine differentiation and delamination. Yet, this 3D cyst model does not reflect the complex organization of pancreatic epithelium patterned into a central trunk and peripheral tip domains. However, it offers a simple modeling platform to analyze the early events of polarity establishment, pancreatic lumen formation, and endocrinogenesis. In addition, this system has the capacity for pancreatic-associated disease modelling with unprecedented resolution. The application of 3D epithelial cysts to model pancreatitis and pancreatic cancer has been already reported [46], [47], [48], [49], [50]. Therefore, generating hPECs from patient-derived hiPSCs or CRISPR/Cas9-mediated genetically modified cells will expand the application of this system to study human disorders of the endocrine and exocrine pancreas in a dish.
We showed that both mouse and human PECs have potential for differentiation towards endocrine fate. However, the rate of endocrine differentiation in our system was lower than current 2D or 3D protocols. Several groups have provided evidence that PPs cultured in 3D spheroids or aggregates (cell clusters without a central lumen) efficiently differentiate into endocrine cells [51], [52]. Additionally, a recent study has reported that EGF and R-spondin-1 increased self-renewal of PPs when cultured in 3D aggregates and that long-term maintenance of PPs did not reduce their potency for endocrine fate induction [53]. These findings support the idea that 3D aggregates are likely more efficient systems for PP expansion and differentiation than 3D cysts or organoids. Despite low endocrine differentiation efficiency, we were able to monitor cellular processes during endocrinogenesis in a time-resolved fashion. Hence, the PEC platform offers a suitable system for studying cell biological events during endocrine induction rather than expanding high numbers of endocrine progenitors or differentiated cells. Future work should identify triggering factors to increase self-renewal and endocrine differentiation potential of PECs that will extend the applications of this system for clinical translation.
ScRNA-seq analysis and PEC differentiation revealed a loss of AB polarity during endocrine induction in mouse and human. Two previous studies have shown that changes in AB polarity regulate pancreatic endocrine induction and differentiation. In an elegant study, Löf-Öhlin et al. found that activation of EGFR signaling in endocrine progenitors inhibits aPKC to reduce apical luminal size. This apical domain narrowing leads to Notch signaling inhibition and Ngn3 upregulation to promote endocrinogenesis [20]. In another study led by Christopher Wright, non-muscle myosin II (nmMyoII) was found to regulate apical narrowing, basolateral movement and Ngn3 upregulation in endocrine progenitors. In addition, nmMyoII function was necessary for abscission of the cell-rear focalized apical domain of the delaminating endocrine cells [21]. Therefore, these studies have provided mechanistic details of how changes in AB polarity impact epithelial morphogenesis and endocrine differentiation. In line with their findings, our data provide further evidence that AB polarity components are dynamically regulated and ultimately downregulated during endocrinogenesis, possibly as a part of the process that differentiated endocrine cells change their epithelial characteristics. However, whether after differentiation and delamination, endocrine cells rearrange their polarity machinery to acquire front-rear or planar cell polarity needs further investigation.
During epithelial morphogenesis, loss of apical polarity results in remodeling and/or resolving of AJs and TJs. In support of this notion, our data showed the differential expression levels and pattern of several cell–cell adhesion molecules during endocrinogenesis. Among these, the transient downregulation of E-Cad and β-Cat in _Ngn3_high precursors and _Fev_high cluster as well as the different expression pattern of these proteins in endocrine vs epithelial cells were remarkable. Such differential expression of adhesion molecules not only is required for morphogenetic events but also impacts cell differentiation programs via tissue segregation and cell sorting mechanisms [54]. In support of this idea, a recent study led by Henrik Semb identified that distinct levels of p120ctn segregate tip vs trunk as well as endocrine vs ductal fate during embryonic development [44], [55]. Therefore, understanding the spatio-temporal expression of cell–cell adhesion molecules allows efficient generation of endocrine cells from human stem cells. Moreover, a detailed knowledge of cell surface marker changes, as has been reported for CD133, CD49f [56], EpCAM [10], and CD49a [17], will help to sort progenitors from stem cell differentiation cultures to enrich for certain homogeneous cell populations to improve differentiation efficiency. Such surface markers might also help to reconstruct stem-cell derived ILCs with defined endocrine cell types in tissue engineering approaches.
We found transient or complete changes in expression levels of several cell adhesion and polarity proteins in _Ngn3_high precursors and _Fev_high population. Such findings support previous evidence that the onset of morphogenetic changes starts at Ngn3high state and continues along endocrine cell differentiation course. Along this line, time-lapse imaging of mPECs derived from Ngn3Cre;mTmG reporter mouse line allowed us to monitor endocrine cell dynamics and supported the idea that cell mobility increases during endocrinogenesis. However, the molecular details of the machinery regulating endocrine cell movement still need to be identified. It should be noted that there are differences between endocrine cell dynamics in our ex vivo system with previously reported pancreatic explant culture [21], [57]. Such differences arise in the absence of the tissues (endothelial, mesenchymal, and neuronal cells) that surround pancreatic epithelium and provide ECM components and a variety of signaling cues essential for pancreatic lineage decision, morphogenesis and patterning. Thus, it is likely that endocrine cell motility and delamination are coordinated through intrinsic programs directed by signals from the neighboring tissues. Along this line, the axonal guidance cue semaphorin 3a, secreted from surrounding mesenchyme, induces endocrine cell motility and delamination in mouse [58]. This study provides an example that how endocrine cell morphogenesis is regulated by the secreted factors from non-pancreatic cells. Therefore, future works should seek to establish 3D modeling systems derived from pancreatic lineages co-cultured with non-epithelial pancreatic tissues. Such complex multi-lineage systems will more faithfully mirror in vivo developmental programs coupling human pancreatic morphogenesis and lineage formation.
In summary, we provide a high-resolution 3D culture system that closely resembles embryonic pancreatic epithelium biology in mouse and human. This model system helps to better understand molecular details of pancreatic epithelial formation and endocrine cell differentiation during early development. Yet, the application of this system can be improved and broadened by generating epithelial cysts with homogenous size and composition co-cultured with mesenchymal, neuronal, and endothelial cells. Such improvements not only help to decipher molecular details of human endocrinogenesis but also aid to achieve a more efficient β-cell and islet cell differentiation platform to generate high numbers of glucose-responsive functional ILCs for cell-based therapy of diabetes.
Author contributions
M.B. conceptualized and designed the project, performed and analyzed experiments and wrote the manuscript. K.S. performed and analyzed experiments and helped drafting the manuscript. S.T. reanalyzed the scRNA-seq data and helped drafting the manuscript. A.B. and M.T. performed experiments. F.J.T. supervised the scRNA-seq data reanalysis. H.L. conceptualized and designed the project and wrote the manuscript.
Acknowledgements
We thank Xianming Wang, Pallavi Mahaddalkar, Anika Böttcher, Anne Theis, Jessica Jaki, Bianca Vogel, Kerstin Diemer. and Julia Beckenbauer for technical support. This work was supported by the Helmholtz-Gemeinschaft (Helmholtz Portfolio Theme ‘Metabolic Dysfunction and Common Disease) and Deutsches Zentrum für Diabetesforschung (DZD). M.B. and H.L. acknowledge financial support by the DZD Grant NEXT funding. This study was also, supported by the PancChip project funded by the German Federal Ministry of Education and Research (project number 01EK1607A).
Footnotes
Contributor Information
Mostafa Bakhti, Email: mostafa.bakhti@helmholtz-muenchen.de.
Heiko Lickert, Email: heiko.lickert@helmholtz-muenchen.de.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 1
Fig S1.
Fig S2.
Fig S3.
Fig S4.
Fig S5.
Fig S6.
References
- 1.Bakhti M., Böttcher A., Lickert H. Modelling the endocrine pancreas in health and disease. Nature Reviews Endocrinology. 2019;15:155–171. doi: 10.1038/s41574-018-0132-z. [DOI] [PubMed] [Google Scholar]
- 2.Pan F.C., Wright C. Pancreas organogenesis: from bud to plexus to gland. Developmental Dynamics. 2011;240(3):530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 3.Villasenor A., Chong D.C., Henkemeyer M., Cleaver O. Epithelial dynamics of pancreatic branching morphogenesis. Development (Cambridge, England) 2010;137(24):4295–4305. doi: 10.1242/dev.052993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kesavan G., Sand F.W., Greiner T.U., Johansson J.K., Kobberup S., Wu X. Cdc42-mediated tubulogenesis controls cell specification. Cell. 2009;139(4):791–801. doi: 10.1016/j.cell.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 5.Bankaitis E.D., Bechard M.E., Wright C.V.E. Feedback control of growth, differentiation, and morphogenesis of pancreatic endocrine progenitors in an epithelial plexus niche. Genes & Development. 2015;29(20):2203–2216. doi: 10.1101/gad.267914.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastidas-Ponce A., Scheibner K., Lickert H., Bakhti M. Cellular and molecular mechanisms coordinating pancreas development. Development. 2017;144(16):2873–2888. doi: 10.1242/dev.140756. [DOI] [PubMed] [Google Scholar]
- 7.Bastidas-Ponce A., Tritschler S., Dony L., Scheibner K., Tarquis-Medina M., Salinno C. Comprehensive single-cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development. 2019;146(12):173849. doi: 10.1242/dev.173849. [DOI] [PubMed] [Google Scholar]
- 8.Byrnes L.E., Wong D.M., Subramaniam M., Meyer N.P., Gilchrist C.L., Knox S.M. Lineage dynamics of murine pancreatic development at single-cell resolution. Nature Communications. 2018;9(1):1–17. doi: 10.1038/s41467-018-06176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramond C., Beydag-Tasöz B.S., Azad A., van de Bunt M., Petersen M.B.K., Beer N.L. Understanding human fetal pancreas development using subpopulation sorting, RNA sequencing and single-cell profiling. Development. 2018;145:165480. doi: 10.1242/dev.165480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramond C., Glaser N., Berthault C., Ameri J., Kirkegaard J.S., Hansson M. Reconstructing human pancreatic differentiation by mapping specific cell populations during development. ELife. 2017;6:e27564. doi: 10.7554/eLife.27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jennings R.E., Berry A.a., Kirkwood-Wilson R., Roberts N.a., Hearn T., Salisbury R.J. Development of the human pancreas from foregut to endocrine commitment. Diabetes. 2013;62(10):3514–3522. doi: 10.2337/db12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jennings R.E., Berry A.A., Strutt J.P., Gerrard D.T., Hanley N.A. Human pancreas development. Development (Cambridge, England) 2015;142(18):3126–3137. doi: 10.1242/dev.120063. [DOI] [PubMed] [Google Scholar]
- 13.Jennings R.E., Berry A.A., Gerrard D.T., Wearne S.J., Strutt J., Withey S. Laser capture and deep sequencing reveals the transcriptomic programmes regulating the onset of pancreas and liver differentiation in human embryos. Stem Cell Reports. 2017;9(5):1387–1394. doi: 10.1016/j.stemcr.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159(2):428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russ H.a., Parent A.V., Ringler J.J., Hennings T.G., Nair G.G., Shveygert M. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. The EMBO Journal. 2015;34(13):1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature Biotechnology. 2014;32(11):1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 17.Veres A., Faust A.L., Bushnell H.L., Engquist E.N., Kenty J.H.-R., Harb G. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569(7756):368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theis F.J., Lickert H. A map of β-cell differentiation pathways supports cell therapies for diabetes. Nature. 2019;569(7756):342–343. doi: 10.1038/d41586-019-01211-9. [DOI] [PubMed] [Google Scholar]
- 19.Kesavan G., Lieven O., Mamidi A., Öhlin Z.L., Johansson J.K., Li W.-C. Cdc42/N-WASP signaling links actin dynamics to pancreatic β cell delamination and differentiation. Development (Cambridge, England) 2014;141(3):685–696. doi: 10.1242/dev.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löf-Öhlin Z.M., Nyeng P., Bechard M.E., Hess K., Bankaitis E., Greiner T.U. EGFR signalling controls cellular fate and pancreatic organogenesis by regulating apicobasal polarity. Nature Cell Biology. 2017;19(11):1313–1325. doi: 10.1038/ncb3628. [DOI] [PubMed] [Google Scholar]
- 21.Bankaitis E.D., Bechard M.E., Gu G., Magnuson M.A., Wright C.V.E. ROCK-nmMyoII, Notch and Neurog3 gene-dosage link epithelial morphogenesis with cell fate in the pancreatic endocrine-progenitor niche. Development. 2018;145(18):dev162115. doi: 10.1242/dev.162115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011 doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 23.Lancaster M.A., Knoblich J.A. Organogenesisin a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 24.Huch M., Koo B.-K. Modeling mouse and human development using organoid cultures. Development. 2015;142(18):3113–3125. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- 25.Clevers H. Modeling development and disease with organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 26.Greggio C., De Franceschi F., Figueiredo-Larsen M., Gobaa S., Ranga A., Semb H. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140:4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugiyama T., Benitez C.M., Ghodasara A., Liu L., McLean G.W., Lee J. Reconstituting pancreas development from purified progenitor cells reveals genes essential for islet differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(31):12691–12696. doi: 10.1073/pnas.1304507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonfanti P., Nobecourt E., Oshima M., Albagli-Curiel O., Laurysens V., Stangé G. Ex vivo expansion and differentiation of human and mouse fetal pancreatic progenitors are modulated by epidermal growth factor. Stem Cells and Development. 2015;24(15):1766–1778. doi: 10.1089/scd.2014.0550. [DOI] [PubMed] [Google Scholar]
- 29.Jin L., Feng T., Shih H.P., Zerda R., Luo A., Hsu J. Colony-forming cells in the adult mouse pancreas are expandable in Matrigel and form endocrine/acinar colonies in laminin hydrogel. Proceedings of the National Academy of Sciences. 2013;110(10):3907–3912. doi: 10.1073/pnas.1301889110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin L., Feng T., Zerda R., Chen C.-C., Riggs A.D., Ku H.T. In vitro multilineage differentiation and self-renewal of single pancreatic colony-forming cells from adult C57Bl/6 mice. Stem Cells and Development. 2014;23(8):899–909. doi: 10.1089/scd.2013.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huch M., Bonfanti P., Boj S.F., Sato T., Loomans C.J.M., van de Wetering M. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. The EMBO Journal. 2013;32(20):2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loomans C.J.M., Williams Giuliani N., Balak J., Ringnalda F., van Gurp L., Huch M. Expansion of adult human pancreatic tissue yields organoids harboring progenitor cells with endocrine differentiation potential. Stem Cell Reports. 2018;10(3):1088–1101. doi: 10.1016/j.stemcr.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.K. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nature Protocols. 2016;11(9):1724–1743. doi: 10.1038/nprot.2016.097. [DOI] [PubMed] [Google Scholar]
- 34.Schonhoff S.E., Giel-Moloney M., Leiter A.B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Developmental Biology. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Muzumdar M.D., Tasic B., Miyamichi K., Li N., Luo L. A global double-fluorescent cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 36.Wang X., Sterr M., Burtscher I., Chen S., Hieronimus A., Machicao F. Genome-wide analysis of PDX1 target genes in human pancreatic progenitors. Molecular Metabolism. 2018;9:57–68. doi: 10.1016/j.molmet.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakano D., Shiraki N., Kume S. Pancreatic differentiation from murine embryonic stem cells. Methods in Molecular Biology. 2016;1341:417–423. doi: 10.1007/7651_2015_217. [DOI] [PubMed] [Google Scholar]
- 38.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biology. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G. Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnology. 2019 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 40.Bryant D.M., Datta A., Rodríguez-Fraticelli A.E., PeräCurrency Signnen J., Martín-Belmonte F., Mostov K.E. A molecular network for de novo generation of the apical surface and lumen. Nature Cell Biology. 2010;12(11):1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryant D.M., Roignot J., Datta A., Overeem A.W., Kim M., Yu W. A molecular switch for the orientation of epithelial cell polarization. Developmental Cell. 2014;31(2):171–187. doi: 10.1016/j.devcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gouzi M., Kim Y.H., Katsumoto K., Johansson K., Grapin-Botton A. Neurogenin3 initiates stepwise delamination of differentiating endocrine cells during pancreas development. Developmental Dynamics. 2011;240(3):589–604. doi: 10.1002/dvdy.22544. [DOI] [PubMed] [Google Scholar]
- 43.Sharon N., Chawla R., Mueller J., Vanderhooft J., Whitehorn L.J., Rosenthal B. A peninsular structure coordinates asynchronous differentiation with morphogenesis to generate pancreatic islets. Cell. 2019;176(4):790–804. doi: 10.1016/j.cell.2018.12.003. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyeng P., Heilmann S., Löf-Öhlin Z.M., Pettersson N.F., Hermann F.M., Reynolds A.B. p120ctn-Mediated organ patterning precedes and determines pancreatic progenitor fate. Developmental Cell. 2019;49:31–47. doi: 10.1016/j.devcel.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Wang X., Sterr M., Ansarullah, Burtscher I., Böttcher A., Beckenbauer J. Point mutations in the PDX1 transactivation domain impair human β-cell development and function. Molecular Metabolism. 2019;24:80–97. doi: 10.1016/j.molmet.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boj S.F., Hwang C. Il., Baker L.A., Chio I.I.C., Engle D.D., Corbo V. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J., Snyder E.R., Liu Y., Gu X., Wang J., Flowers B.M. Reconstituting development of pancreatic intraepithelial neoplasia from primary human pancreas duct cells. Nature Communications. 2017;8:1–14. doi: 10.1038/ncomms14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang L., Holtzinger A., Jagan I., Begora M., Lohse I., Ngai N. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nature Medicine. 2015;21(11):1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hohwieler M., Illing A., Hermann P.C., Mayer T., Stockmann M., Perkhofer L. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2017;66(3):473–486. doi: 10.1136/gutjnl-2016-312423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker L.A., Tiriac H., Clevers H., Tuveson D.A. Modeling pancreatic cancer with organoids. Trends in Cancer. 2016;2:176–190. doi: 10.1016/j.trecan.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nair G.G., Liu J.S., Russ H.A., Tran S., Saxton M.S., Chen R. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nature Cell Biology. 2019;21:263–274. doi: 10.1038/s41556-018-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shim J.H., Kim J.H., Han J., An S.Y., Jang Y.J., Son J. Pancreatic islet-like three-dimensional aggregates derived from human embryonic stem cells ameliorate hyperglycemia in streptozotocin-induced diabetic mice. Cell Transplantation. 2015;24:2155–2168. doi: 10.3727/096368914X685438. [DOI] [PubMed] [Google Scholar]
- 53.Konagaya S., Iwata H. Chemically defined conditions for long-term maintenance of pancreatic progenitors derived from human induced pluripotent stem cells. Scientific Reports. 2019;9(1):6–7. doi: 10.1038/s41598-018-36606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Townes P.L., Holtfreter J. Directed movements and selective adhesion of embryonic amphibian cells. Journal of Experimental Zoology. 1955;128(1):53–120. doi: 10.1002/jez.a.114. [DOI] [PubMed] [Google Scholar]
- 55.Bakhti M., Bastidas-Ponce A., Lickert H. Sorting out fate determination. Developmental Cell. 2019;49(1):1–3. doi: 10.1016/j.devcel.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 56.Sugiyama T., Rodriguez R.T., McLean G.W., Kim S.K. Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proceedings of the National Academy of Sciences. 2007;104(1):175–180. doi: 10.1073/pnas.0609490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bechard M.E., Bankaitis E.D., Hipkens S.B., Ustione A., Piston D.W., Yang Y.-P. Precommitment low-level Neurog3 expression defines a long-lived mitotic endocrine-biased progenitor pool that drives production of endocrine-committed cells. Genes & Development. 2016;30(16):1852–1865. doi: 10.1101/gad.284729.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pauerstein P.T., Tellez K., Willmarth K.B., Park K.M., Hsueh B., Efsun Arda H. A radial axis defined by semaphorin-to-neuropilin signaling controls pancreatic islet morphogenesis. Development. 2017;144(20):3744–3754. doi: 10.1242/dev.148684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1