A Sampling of the Yeast Proteome (original) (raw)
Abstract
In this study, we examined yeast proteins by two-dimensional (2D) gel electrophoresis and gathered quantitative information from about 1,400 spots. We found that there is an enormous range of protein abundance and, for identified spots, a good correlation between protein abundance, mRNA abundance, and codon bias. For each molecule of well-translated mRNA, there were about 4,000 molecules of protein. The relative abundance of proteins was measured in glucose and ethanol media. Protein turnover was examined and found to be insignificant for abundant proteins. Some phosphoproteins were identified. The behavior of proteins in differential centrifugation experiments was examined. Such experiments with 2D gels can give a global view of the yeast proteome.
The sequence of the yeast genome has been determined (9). More recently, the number of mRNA molecules for each expressed gene has been measured (27, 30). The next logical level of analysis is that of the expressed set of proteins. We have begun to analyze the yeast proteome by using two-dimensional (2D) gels.
2D gel electrophoresis separates proteins according to isoelectric point in one dimension and molecular weight in the other dimension (21), allowing resolution of thousands of proteins on a single gel. Although modern imaging and computing techniques can extract quantitative data for each of the spots in a 2D gel, there are only a few cases in which quantitative data have been gathered from 2D gels. 2D gel electrophoresis is almost unique in its ability to examine biological responses over thousands of proteins simultaneously and should therefore allow us a relatively comprehensive view of cellular metabolism.
We and others have worked toward assembling a yeast protein database consisting of a collection of identified spots in 2D gels and of data on each of these spots under various conditions (2, 7, 8, 10, 23, 25). These data could then be used in analyzing a protein or a metabolic process. Saccharomyces cerevisiae is a good organism for this approach since it has a well-understood physiology as well as a large number of mutants, and its genome has been sequenced. Given the sequence and the relative lack of introns in S. cerevisiae, it is easy to predict the sequence of the primary protein product of most genes. This aids tremendously in identifying these proteins on 2D gels.
There are three pillars on which such a database rests: (i) visualization of many protein spots simultaneously, (ii) quantification of the protein in each spot, and (iii) identification of the gene product for each spot. Our first efforts at visualization and identification for S. cerevisiae have been described elsewhere (7, 8). Here we describe quantitative data for these proteins under a variety of experimental conditions.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae W303 (MATa ade2-1 his3-11,15 leu2-3, 112 trp1-1 ura3-1 can1-100) was used (26). −Met YNB (yeast nitrogen base) medium was 1.7 g of YNB (Difco) per liter, 5 g of ammonium sulfate per liter, and adenine, uracil, and all amino acids except methionine; −Met −Cys YNB medium was the same but without methionine or cysteine. Medium was supplemented with 2% glucose (for most experiments) or with 2% ethanol (for ethanol experiments). Low-phosphate YEPD was described by Warner (28).
Isotopic labeling of yeast and preparation of cell extracts.
Yeast strains were labeled and proteins were extracted as described by Garrels et al. (7, 8). Briefly, cells were grown to 5 × 106 cells per ml. at 30°C; 1 ml of culture was transferred to a fresh tube, and 0.3 mCi of [35S]methionine (e.g., Express protein labeling mix; New England Nuclear) was added to this 1-ml culture. The cells were incubated for a further 10 to 15 min and then transferred to a 1.5-ml microcentrifuge tube, chilled on ice, and harvested by centrifugation. The supernatant was removed, and the cell pellet was resuspended in 100 μl of lysis buffer (20 mM Tris-HCl [pH 7.6], 10 mM NaF, 10 mM sodium pyrophosphate, 0.5 mM EDTA, 0.1% deoxycholate; just before use, phenylmethylsulfonyl fluoride was added to 1 mM, leupeptin was added to 1 μg/ml, pepstatin was added to 1 μg/ml, tosylsulfonyl phenylalanyl chloromethyl ketone was added to 10 μg/ml, and soybean trypsin inhibitor was added to 10 μg/ml).
The resuspended cells were transferred to a screw-cap 1.5-ml polypropylene tube containing 0.28 g of glass beads (0.5-mm diameter; Biospec Products) or 0.40 g of zirconia beads (0.5-mm diameter; Biospec Products). After the cap was secured, the tube was inserted into a MiniBeadbeater 8 (Biospec Products) and shaken at medium high speed at 4°C for 1 min. Breakage was typically 75%. Tubes were then spun in a microcentrifuge for 10 s at 5,000 × g at 4°C.
With a very fine pipette tip, liquid was withdrawn from the beads and transferred to a prechilled 1.5-ml tube containing 7 μl of DNase I (0.5 mg/ml; Cooper product no. 6330)–RNase A (0.25 mg/ml; Cooper product no. 5679)–Mg (50 mM MgCl2) mix. Typically 70 μl of liquid was recovered. The mixture was incubated on ice for 10 min to allow the RNase and DNase to work.
Next, 75 μl of 2× dSDS (2× dSDS is 0.6% sodium dodecyl sulfate [SDS], 2% mercaptoethanol, and 0.1 M Tris-HCl [pH 8]) was added. The tube was plunged into boiling water, incubated for 1 min, and then plunged into ice. After cooling, the tube was centrifuged at 4°C for 3 min at 14,000 × g. The supernatant was transferred to a fresh tube and frozen at −70°C. About 5 μl of this supernatant was used for each 2D gel.
2D polyacrylamide gels.
2D gels were made and run as described elsewhere (6–8).
Image analysis of the gels.
The Quest II software system was used for quantitative image analysis (20, 22). Two techniques were used to collect quantitative data for analysis by Quest II software. First, before the advent of phosphorimagers, gels were dried and fluorographed. Each gel was exposed to film for three different times (typically 1 day, 2 weeks, and 6 weeks) to increase the dynamic range of the data. The films were scanned along with calibration strips to relate film optical density to disintegrations per minute in the gels and analyzed by the software to obtain a linear relationship between disintegrations per minute in the spots and optical densities of the film images. The quantitative data are expressed as parts per million of the total cellular protein. This value is calculated from the disintegrations per minute of the sample loaded onto the gel and by comparing the film density of each data spot with density of the film over the calibration strips of known radioactivity exposed to the same film. This yields the disintegrations per minute per millimeter for each spot on the gel and thence its parts-per-minute value.
After the advent of phosphorimaging, gels bearing 35S-labeled proteins were exposed to phosphorimager screens and scanned by a Fuji phosphorimager, typically for two exposures per gel. Calibration strips of known radioactivity were exposed simultaneously. Scan data from the phosphorimager was assimilated by Quest II software, and quantitative data were recorded for the spots on the gels.
Measurements of protein turnover.
Cells in exponential phase were pulse-labeled with [35S]methionine, excess cold Met and Cys were added, and samples of equal volume were taken from the culture at intervals up to 90 min (in one experiment) or up to 160 min (in a second experiment). Incorporation of 35S into protein was essentially 100% by the first sample (10 min). Extracts were made, and equal fractions of the samples were loaded on 2D gels (i.e., the different samples had different amounts of protein but equal amounts of 35S). Spots were quantitated with a phosphorimaging and Quest software.
The software was queried for spots whose radioactivity decreased through the time course. The algorithm examined all data points for all spots, drew a best-fit line through the data points, and looked for spots where this line had a statistically significant negative slope. In one of the experiments, there was one such spot. To the eye, this was a minor, unidentified spot seen only in the first two samples (10 and 20 min). In the other experiment, the Quest software found no spots meeting the criteria. Therefore, we concluded that none of the identified spots (and all but one of the visible spots) represented proteins with long half-lives.
Centrifugal fractionation.
Cells were labeled, harvested, and broken with glass beads by the standard method described above except that no detergent (i.e., no deoxycholate) was present in the lysis buffer. The crude lysate was cleared of unbroken cells and large debris by centrifugation at 300 × g for 30 s. The supernatant of this centrifugation was then spun at 16,000 × g for 10 min to give the pellet used for Fig. 6B. The supernatant of the 16,000 × g, 10-min spin was then spun at 100,000 × g for 30 min to give the supernatant used for Fig. 6A.
FIG. 6.
Fractionation by centrifugation. (A) Proteins in the supernatant of a 100,000 × g, 30-min spin; proteins in the pellet of a 16,000 × g, 10-min spin. Supernatant fractions examined in multiple experiments done over a wide range of g forces looked similar to each other, as did the pellet fractions.
Protein abundance calculations.
A haploid yeast cell contains about 4 × 10−12 g of protein (1, 15). Assuming a mean protein mass of 50 kDa, there are about 50 × 106 molecules of protein per cell. There are about 1.8 methionines per 10 kDa of protein mass, which implies 4.5 × 108 molecules of methionine per cell (neglecting the small pool of free Met). We measured (i) the counts per minute in each spot on the 2D gels, (ii) the total number of counts on each gel (by integrating counts over the entire gel), and (iii) the total number of counts loaded on the gel (by scintillation counting of the original sample). Thus, we know what fraction of the total incorporated radioactivity is present in each spot. After correcting for the methionine (and cysteine [see below]) content of each protein, we calculated an absolute number of protein molecules based on the fraction of radioactivity in each spot and on 50 × 106 total molecules per cell.
The labeling mixture used contained about one-fifth as much radioactive cysteine as radioactive methionine. Therefore, the number of cysteine molecules per protein was also taken into account in calculating the number of molecules of protein, but Cys molecules were weighted one-fifth as heavily as Met molecules.
mRNA abundance calculations.
For estimation of mRNA abundance, we used SAGE (serial analysis of gene expression) data (27) and Affymetrix chip hybridization data (29a, 30). The mRNA column in Table 1 shows mRNA abundance calculated from SAGE data alone. However, the SAGE data came from cells growing in YEPD medium, whereas our protein measurements were from cells growing in YNB medium. In addition, SAGE data for low-abundance mRNAs suffers from statistical variation. Therefore, we also used chip hybridization data (29a, 30) for mRNA from cells grown in YNB. These hybridization data also had disadvantages. First, the amounts of high-abundance mRNAs were systematically underestimated, probably because of saturation in the hybridizations, which used 10 μg of cRNA. For example, the abundance of ADH1 mRNA was 197 copies per cell by SAGE but only 32 copies per cell by hybridization, and the abundance of ENO2 mRNA was 248 copies per cell by SAGE but only 41 by hybridization. When the amount of cRNA used in the hybridization was reduced to 1 μg, the apparent amounts of mRNA were similar to the amounts determined by SAGE (29a, 29b). However, experiments using 1 μg of cRNA have been done for only some genes (29a). Because amounts of mRNA were normalized to 15,000 per cell, and because the amounts of abundant mRNAs were underestimated, there is a 2.2-fold overestimate of the abundance of nonabundant mRNAs. We calculated this factor of 2.2 by adding together the number of mRNA molecules from a large number of genes expressed at a low level for both SAGE data and hybridization data. The sum for the same genes from hybridization data is 2.2-fold greater than that from SAGE data.
TABLE 1.
Quantitative dataa
| Function | Name | CAI | mRNA | Adjusted mRNA | Protein (Glu) (103) | Protein (Eth) (103) | E/G ratio |
|---|---|---|---|---|---|---|---|
| Carbohydrate metabolism | Adh1 | 0.810 | 197 | 197 | 1,230 | 972 | 0.79 |
| Adh2 | 0.504 | 0 | 0 | 963 | >20 | ||
| Cit2 | 0.185 | 1 | 2.8 | 23 | 288 | 12 | |
| Eno1 | 0.870 | No Nla | 410 | 974 | 2.4 | ||
| Eno2 | 0.892 | 248 | 248 | 650 | 215 | 0.33 | |
| Fba1 | 0.868 | 179 | 179 | 640 | 608 | 0.95 | |
| Hxk1,2 | 0.500 | 13 | 10.5 | 62 | 46 | ||
| Icl1 | 0.251 | 0 | 0 | 671 | >20 | ||
| Pdb1 | 0.342 | 5 | 5 | 41 | 33 | ||
| Pdc1 | 0.903 | 226 | 226 | 280 | 205 | 0.73 | |
| Pfk1 | 0.465 | 5 | 5 | 75 | 53 | 0.71 | |
| Pgi1 | 0.681 | 14 | 14 | 160 | 120 | 0.75 | |
| Pyc1 | 0.260 | 1 | 0.7 | 37 | 34 | ||
| Tal1 | 0.579 | 5 | 5 | 110 | 35 | ||
| Tdh2 | 0.904 | 63 | 63 | 430 | 876 | NR | |
| Tdh3 | 0.924 | 460 | 460 | 1,670 | 1,927 | NR | |
| Tpi1 | 0.817 | No Nla | No Met | No Met | |||
| Protein synthesis | Efb1 | 0.762 | 33 | 16.5 | 358 | 362 | |
| Eft1,2 | 0.801 | 26 | 26 | 99 | 54 | 0.55 | |
| Prt1 | 0.303 | 4 | 0.7 | 12 | 6 | ||
| Rpa0 | 0.793 | 246 | 246 | 277 | 100 | 0.36 | |
| Tif1,2 | 0.752 | 29 | 29 | 233 | 106 | 0.46 | |
| Yef3 | 0.777 | 36 | 36 | 14 | ND | ||
| Heat shock | Hsc82 | 0.581 | 2 | 2.9 | 112 | 75 | 0.67 |
| Hsp60 | 0.381 | 9 | 2.3 | 35 | 82 | 2.3 | |
| Hsp82 | 0.517 | 2 | 1.3 | 52 | 135 | 2.6 | |
| Hsp104 | 0.304 | 7 | 7 | 70 | 161 | 2.3 | |
| Kar2 | 0.439 | 5 | 10.1 | 43 | 102 | 2.4 | |
| Ssa1 | 0.709 | 2 | 4.3 | 303 | 421 | 1.4 | |
| Ssa2 | 0.802 | 10 | 5 | 213 | 324 | 1.5 | |
| Ssb1,2 | 0.850 | 50 | 50 | 270 | 85 | ||
| Ssc1 | 0.521 | 2 | 2.6 | 68 | 80 | 1.2 | |
| Sse1 | 0.521 | 8 | 8 | 96 | 48 | ||
| Sti1 | 0.247 | 1 | 1.1 | 25 | 44 | 1.7 | |
| Amino acid synthesis | Ade1 | 0.229 | 4 | 4 | 14 | 27 | |
| Ade3 | 0.276 | 2 | 1.7 | 12 | 9 | ||
| Ade5,7 | 0.257 | 2 | 1.4 | 14 | 4 | ||
| Arg4 | 0.229 | 1 | 8.1 | 41 | 41 | ||
| Gdh1 | 0.585 | 10 | 27 | 148 | 55 | ||
| Gln1 | 0.524 | 11 | 11 | 77 | 104 | 1.3 | |
| His4 | 0.267 | 3 | 3 | 15 | 23 | 1.5 | |
| Ilv5 | 0.801 | 6 | 6 | 152 | 109 | 0.7 | |
| Lys9 | 0.332 | 4 | 4 | 32 | 17 | 0.52 | |
| Met6 | 0.657 | No Nla | 22 | 190 | 80 | 0.42 | |
| Pro2 | 0.248 | 3 | 3 | 30 | 12 | ||
| Ser1 | 0.258 | 2 | 1.2 | 15 | 8 | ||
| Trp5 | 0.319 | 5 | 5 | 28 | 12 | ||
| Miscellaneous | Act1 | 0.710 | 54 | 54 | 205 | 164 | 0.78 |
| Adk1 | 0.531 | No Nla | 47 | 43 | |||
| Ald6 | 0.520 | 3 | 3 | 181 | 159 | ||
| Atp2 | 0.424 | 1 | 4.1 | 76 | 109 | 1.4 | |
| Bmh1 | 0.322 | 46 | 46 | 191 | 137 | 0.72 | |
| Bmh2 | 0.384 | 1 | 1.4 | 134 | 147 | ||
| Cdc48 | 0.306 | 2 | 2.4 | 32 | 26 | ||
| Cdc60 | 0.299 | 2 | 0.86 | 6 | 2 | ||
| Erg20 | 0.373 | 5 | 5 | 92 | 39 | ||
| Gpp1 | 0.603 | 16 | 5 | 234 | 158 | ||
| Gsp1 | 0.621 | 3 | 3 | 115 | 39 | 0.34 | |
| Ipp1 | 0.620 | 4 | 4 | 254 | 147 | 0.58 | |
| Lcb1 | 0.173 | 0.3 | 0.8 | 19 | 40 | ||
| Mol1 | 0.423 | 0 | 0.45 | 20 | 16 | ||
| Pab1 | 0.488 | 3 | 3 | 41 | 19 | 0.47 | |
| Psa1 | 0.600 | 15 | 15 | 148 | 56 | ||
| Rnr4 | 0.497 | 6 | 6 | 44 | 37 | ||
| Sam1 | 0.494 | 5 | 5 | 59 | 21 | ||
| Sam2 | 0.497 | 3 | 15 | 63 | 20 | ||
| Sod1 | 0.376 | 36 | 36 | 631 | 618 | ||
| Uba1 | 0.212 | 2 | 2 | 14 | 20 | ||
| YKL056 | 0.731 | 62 | 62 | 253 | 112 | 0.44 | |
| YLR109 | 0.549 | 21 | 21 | 930 | |||
| YMR116 | 0.777 | 41 | 41 | 184 | 40 | 0.20 |
To take into account these difficulties, we compiled a list of “adjusted” mRNA abundance as follows. For all high-abundance mRNAs of our identified proteins, we used SAGE data. For all of these particular mRNAs, chip hybridization suggested that mRNA abundance was the same in YEPD and YNB media. For medium-abundance mRNAs, SAGE data were used, but when hybridization data showed a significant difference between YEPD and YNB, then the SAGE data were adjusted by the appropriate factor. Finally, for low-abundance mRNAs, we used data from chip hybridizations from YNB medium but divided by 2.2 to normalize to the SAGE results. These calculations were completed without reference to protein abundance.
CAI.
The codon adaptation index (CAI) was taken from the yeast proteome database (YPD) (13), for which calculations were made according to Sharp and Li (24). Briefly, the index uses a reference set of highly expressed genes to assign a value to each codon, and then a score for a gene is calculated from the frequency of use of the various codons in that gene (24).
Statistical analysis.
The JMP program was used with the aid of T. Tully. The JMP program showed that neither mRNA nor protein abundances were normally distributed; therefore, Spearman rank correlation coefficients (rs) were calculated. The mRNA (adjusted and unadjusted) and protein data were also transformed so that Pearson product-moment correlation coefficients (rp) could be calculated. First, this was done by a Box-Cox transformation of log-transformed data. This transformation produced normal distributions, and an rp of 0.76 was achieved. However, because the Box-Cox transformation is complex, we also did a simpler logarithmic transformation. This produced a normal distribution for the protein data. However, the distribution for the mRNA and adjusted mRNA data was close to, but not quite, normal. Nevertheless, we calculated the rp and found that it was 0.76, identical to the coefficient from the Box-Cox transformed data. We therefore believe that this correlation coefficient is not misleading, despite the fact that the log(mRNA) distribution is not quite normal.
RESULTS
Visualization of 1,400 spots on three gel systems.
Yeast proteins have isoelectric points ranging from 3.1 to 12.8, and masses ranging from less than 10 kDa to 470 kDa. It is difficult to examine all proteins on a single kind of gel, because a gel with the needed range in pI and mass would give poor resolution of the thousands of spots in the central region of the gel. Therefore, we have used three gel systems: (i) pH “4 to 8” with 10% polyacrylamide; (ii) pH “3 to 10” with 10% polyacrylamide; and (iii) nonequilibrium with 15% polyacrylamide (7, 8). Each gel system allows good resolution of a subset of yeast proteins.
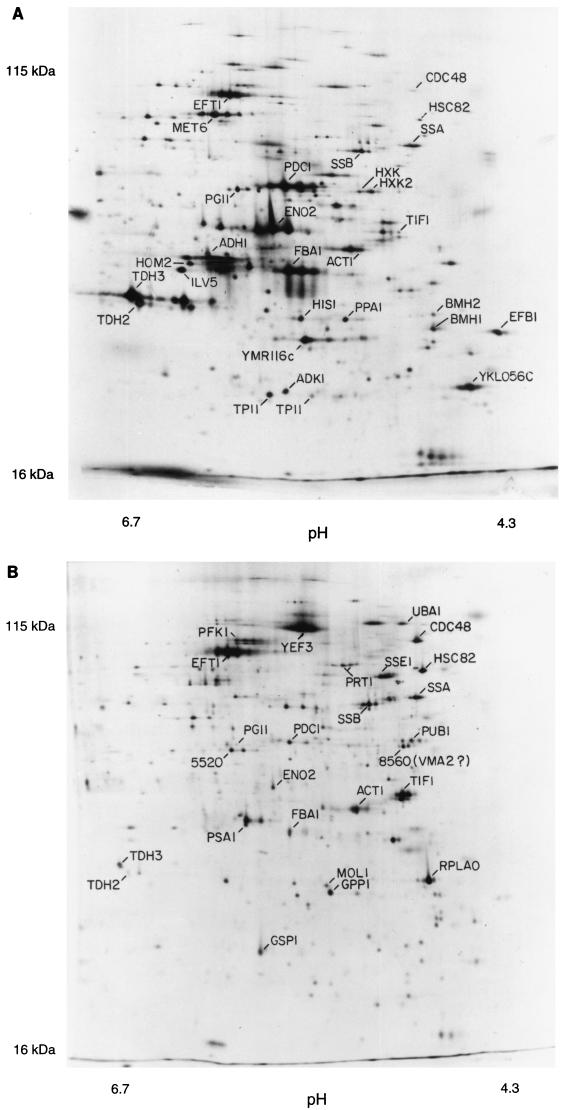
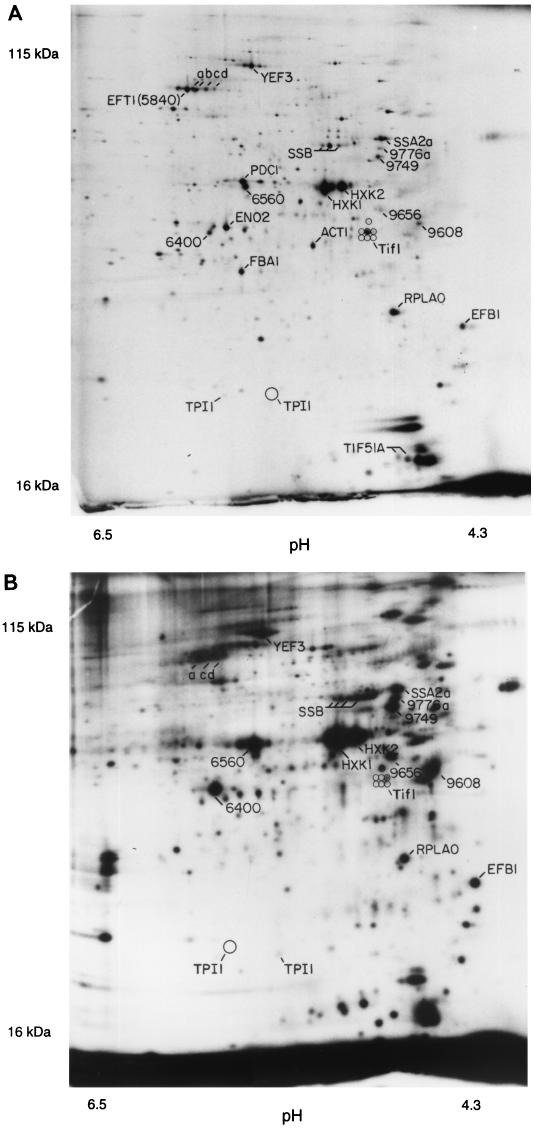
Figure 1 shows a pH 4–8, 10% polyacrylamide gel. The pH at the basic end of the isoelectric focusing gel cannot be maintained throughout focusing, and so the proteins resolved on such gels have isoelectric points between pH 4 and pH 6.7. For these pH 4–8 gels, we see 600 to 900 spots on the best gels after multiple exposures.
FIG. 1.
2D gels. The horizontal axis is the isoelectric focusing dimension, which stretches from pH 6.7 (left) to pH 4.3 (right). The vertical axis is the polyacrylamide gel dimension, which stretches from about 15 kDa (bottom) to at least 130 kDa (top). For panel A, extract was made from cells in log phase in glucose; for panel B, cells were grown in ethanol. The spots labeled 1 through 6 are unidentified proteins highly induced in ethanol.
The pH 3–10 gels (not shown) extend the pI range somewhat beyond pH 7.5, allowing detection of several hundred additional spots. Finally, we use nonequilibrium gels with 15% acrylamide in the second dimension. These allow visualization of about 100 very basic proteins and about 170 small proteins (less than 20 kDa). In total, using all three gel systems, about 1,400 spots can be seen. These represent about 1,200 different proteins, which is about one-quarter to one-third of the proteins expressed under these conditions (27, 30). Here, we focus on the proteins seen on the pH 4–8 gels.
Although nearly all expressed proteins are present on these gels, the number seen is limited by a problem we call coverage. Since there are thousands of proteins on each gel, many proteins comigrate or nearly comigrate. When two proteins are resolved, but are close together, and one protein spot is much more intense than the other, a problem arises in visualizing the weaker spot: at long exposures when the weak signal is strong enough for detection, the signal from the strong spot spreads and covers the signal from the weaker spot. Thus, weak spots can be seen only when they are well separated from strong spots.
For a given gel, the number of detectable spots initially rises with exposure time. However, beyond an optimal exposure, the number of distinguishable spots begins to decrease, because signals from strong spots cover signals from nearby weak spots. At long exposures, the whole autoradiogram turns black. Thus, there is an optimum exposure yielding the maximum number of spots, and at this exposure the weakest spots are not seen.
Largely because of the problem of coverage, the proteins seen are strongly biased toward abundant proteins. All identified proteins have a CAI of 0.18 or more, and we have identified no transcription factors or protein kinases, which are nonabundant proteins. Thus, this technology is useful for examining protein synthesis, amino acid metabolism, and glycolysis but not for examining transcription, DNA replication, or the cell cycle.
Spot identification.
The identification of various spots has been described elsewhere (7, 8). At present, 169 different spots representing 148 proteins have been identified. Many of these spots have been independently identified (2, 10, 23, 25). The main methods used in spot identification have been analysis of amino acid composition, gene overexpression, peptide sequencing, and mass spectrometry.
Pulse-chase experiments and protein turnover.
Pulse-chase experiments were done to measure protein half-lives (Materials and Methods). Cells were labeled with [35S]methionine for 10 min, and then an excess of unlabeled methionine was added. Samples were taken at 0, 10, 20, 30, 60, and 90 min after the beginning of the chase. Equal amounts of 35S were loaded from each sample; 2D gels were run, and spots were quantitated. Surprisingly, almost every spot was nearly constant in amount of radioactivity over the entire time course (not shown). A few spots shifted from one position to another because of posttranslational modifications (e.g., phosphorylation of Rpa0 and Efb1). Thus, the proteins being visualized are all or nearly all very stable proteins, with half-lives of more than 90 min. Gygi et al. (10) have come to a similar conclusion by using the N-end rule to predict protein half-lives. This result does not imply that all yeast proteins are stable. The proteins being visualized are abundant proteins; this is partly because they are stable proteins.
Protein quantitation.
Because all of the proteins seen had effectively the same half-life, the abundance of each protein was directly proportional to the amount of radioactivity incorporated during labeling. Thus, after taking into account the total number of protein molecules per cell, the average content of methionine and cysteine, and the methionine and cysteine content of each identified protein, we could calculate the abundance of each identified protein (Tables 1 and 2; Materials and Methods). About 1,000 unidentified proteins were also quantified, assuming an average content of Met and Cys.
TABLE 2.
Functions of proteins listed in Table 1
| Namea | YPD title linesb |
|---|---|
| Adh1 | Alcohol dehydrogenase I; cytoplasmic isozyme reducing acetaldehyde to ethanol, regenerating NAD+ |
| Adh2 | Alcohol dehydrogenase II; oxidizes ethanol to acetaldehyde, glucose repressed |
| Cit2 | Citrate synthase, peroxisomal (nonmitochondrial); converts acetyl-CoA and oxaloacetate to citrate plus CoA |
| Eno1 | Enolase 1 (2-phosphoglycerate dehydratase); converts 2-phospho-d-glycerate to phosphoenolpyruvate in glycolysis |
| Eno2 | Enolase 2 (2-phosphoglycerate dehydratase); converts 2-phospho-d-glycerate to phosphoenolpyruvate in glycolysis |
| Fba1 | Fructose bisphosphate aldolase II; sixth step in glycolysis |
| Hxk1 | Hexokinase I; converts hexoses to hexose phosphates in glycolysis; repressed by glucose |
| Hxk2 | Hexokinase II; converts hexoses to hexose phosphates in glycolysis and plays a regulatory role in glucose repression |
| Icl1 | Isocitrate lyase, peroxisomal; carries out part of the glyoxylate cycle; required for gluconeogenesis |
| Pdb1 | Pyruvate dehydrogenase complex, E1 beta subunit |
| Pdc1 | Pyruvate decarboxylase isozyme 1 |
| Pfk1 | Phosphofructokinase alpha subunit, part of a complex with Pfk2p which carries out a key regulatory step in glycolysis |
| Pgi1 | Glucose-6-phosphate isomerase, converts glucose-6-phosphate to fructose-6-phosphate |
| Pyc1 | Pyruvate carboxylase 1; converts pyruvate to oxaloacetate for gluconeogenesis |
| Tal1 | Transaldolase; component of nonoxidative part of pentose phosphate pathway |
| Tdh2 | Glyceraldehyde-3-phosphate dehydrogenase 2; converts d-glyceraldehyde 3-phosphate to 1,3-dephosphoglycerate |
| Tdh3 | Glyceraldehyde-3-phosphate dehydrogenase 3; converts d-glyceraldehyde 3-phosphate to 1,3-dephosphoglycerate |
| Tpi1 | Triosephosphate isomerase; interconverts glyceraldehyde-3-phosphate and dihydroxyacetone phosphate |
| Efb1 | Translation elongation factor EF-1β; GDP/GTP exchange factor for Tef1p/Tef2p |
| Eft1 | Translation elongation factor EF-2; contains diphthamide which is not essential for activity; identical to Eft2p |
| Eft2 | Translation elongation factor EF-2; contains diphthamide which is not essential for activity; identical to Eft1p |
| Prt1 | Translation initiation factor eIF3 beta subunit (p90); has an RNA recognition domain |
| Rpa0 (RPPO) | Acidic ribosomal protein A0 |
| Tif1 | Translation initiation factor 4A (eIF4A) of the DEAD box family |
| Tif2 | Translation initiation factor 4A (eIF4A) of the DEAD box family |
| Yef3 | Translation elongation factor EF-3A; member of ATP-binding cassette superfamily |
| Hsc82 | Chaperonin homologous to E. coli HtpG and mammalian HSP90 |
| Hsp60 | Mitochondrial chaperonin that cooperates with Hsp10p; homolog of E. coli GroEL |
| Hsp82 | Heat-inducible chaperonin homologous to E. coli HtpG and mammalian HSP90 |
| Hsp104 | Heat shock protein required for induced thermotolerance and for resolubilizing aggregates of denatured proteins; important for [psi−]-to-[PSI+] prion conversion |
| Kar2 | Heat shock protein of the endoplasmic reticulum lumen required for protein translocation across the endoplasmic reticulum membrane and for nuclear fusion; member of the HSP70 family |
| Ssa1 | Cytoplasmic chaperone; heat shock protein of the HSP70 family |
| Ssa2 | Cytoplasmic chaperone; member of the HSP70 family |
| Ssb1 | Heat shock protein of HSP70 family involved in the translational apparatus |
| Ssb2 | Heat shock protein of HSP70 family, cytoplasmic |
| Ssc1 | Mitochondrial protein that acts as an import motor with Tim44p and plays a chaperonin role in receiving and folding of protein chains during import; heat shock protein of HSP70 family |
| Sse1 | Heat shock protein of the HSP70 family; multicopy suppressor of mutants with hyperactivated Ras/cyclic AMP pathway |
| Sti1 | Stress-induced protein required for optimal growth at high and low temperature; has tetratricopeptide repeats |
| Ade1 | Phosphoribosylamidoimidazole-succinocarboxamide synthase: catalyzes the seventh step in de novo purine biosynthesis pathway |
| Ade3 | C1 tetrahydrofolate synthase (trifunctional enzyme), cytoplasmic |
| Ade5,7 | Phosphoribosylamine-glycine ligase plus phosphoribosylformylglycinamidine cyclo-ligase; bifunctional protein |
| Arg4 | Argininosuccinate lyase; catalyzes the final step in arginine biosynthesis |
| Gdh1 | Glutamate dehydrogenase (NADP+); combines ammonia and α-ketoglutarate to form glutamate |
| Gln1 | Glutamine synthetase; combines ammonia to glutamate in ATP-driven reaction |
| His4 | Phosphoribosyl-AMP cyclohydrolase/phosphoribosyl-ATP pyrophosphohydrolase/histidinol dehydrogenase; 2nd, 3rd, and 10th steps of his biosynthesis pathway |
| Ilv5 | Ketol-acid reductoisomerase (acetohydroxy, acid reductoisomerase) (alpha-keto-β-hydroxylacyl) reductoisomerase); second step in Val and Ilv biosynthesis pathway |
| Lys9 | Saccharopine dehydrogenase (NADP+, l-glutamate forming) (saccharopine reductase), seventh step in lysine biosynthesis pathway |
| Met6 | Homocysteine methyltransferase; (5-methyltetrahydropteroyl triglutamate-homocysteine methyltransferase), methionine synthase, cobalamin independent |
| Pro2 | γ-Glutamyl phosphate reductase (phosphoglutamate dehydrogenase), proline biosynthetic enzyme |
| Ser1 | Phosphoserine transaminase; involved in synthesis of serine from 3-phosphoglycerate |
| Trp5 | Tryptophan synthase, last (5th) step in tryptophan biosynthesis pathway |
| Act1 | Actin; involved in cell polarization, endocytosis, and other cytoskeletal functions |
| Adk1 | Adenylate kinase (GTP:AMP phosphotransferase), cytoplasmic |
| Ald6 | Cytosolic acetaldehyde dehydrogenase |
| Atp2 | Beta subunit of F1-ATP synthase; 3 copies are found in each F1 oligomer |
| Bmh1 | Homolog of mammalian 14-3-3 protein; has strong similarity to Bmh2p |
| Bmh2 | Homolog of mammalian 14-3-3 protein; has strong similarity to Bmh1p |
| Cdc48 | Protein of the AAA family of ATPases; required for cell division and homotypic membrane fusion |
| Cdc60 | Leucyl-tRNA synthetase, cytoplasmic |
| Erg20 | Farnesyl pyrophosphate synthetase; may be rate-limiting step in sterol biosynthesis pathway |
| Gpp1 (Rhr2) | dl-Glycerol phosphate phosphatase |
| Gsp1 | Ran, a GTP-binding protein of the Ras superfamily involved in trafficking through nuclear pores |
| Ipp1 | Inorganic pyrophosphatase, cytoplasmic |
| Lcb1 | Component of serine C-palmitoyltransferase; first step in biosynthesis of long-chain base component of sphingolipids |
| Mol1 (Thi4) | Thiamine-repressed protein essential for growth in the absence of thiamine |
| Pab1 | Poly(A)-binding protein of cytoplasm and nucleus; part of the 3′-end RNA-processing complex (cleavage factor I); has 4 RNA recognition domains |
| Psa1 | Mannose-1-phosphate guanyltransferase; GDP-mannose pyrophosphorylase |
| Rnr4 | Ribonucleotide reductase small subunit |
| Sam1 | _S_-Adenosylmethionine synthetase 1 |
| Sam2 | _S_-Adenosylmethionine synthetase 2 |
| Sod1 | Copper-zinc superoxide dismutase |
| Uba1 | Ubiquitin-activating (E1) enzyme |
| YKL056 | Resembles translationally controlled tumor protein of animal cells and higher plants |
| YLR109 (Ahp1) | Alkyl hydroperoxide reductase |
| YMR116 (Asc1) | Abundant protein with effects on translational efficiency and cell size, has two WD (WD-40) repeats |
Many proteins give multiple spots (7, 8). The contribution from each spot was summed to give the total protein amount. However, many proteins probably have minor spots that we are not aware of, causing the amount of protein to be underestimated.
When the proteins on a pH 4–8 gel were ordered by abundance, the most abundant protein had 8,904 ppm, the 10th most abundant had 2,842 ppm, the 100th most abundant had 314 ppm, the 500th most abundant had 57 ppm, and the 1,000th most abundant (visualized at greater than optimum exposure) had 23 ppm. Thus, there is more than a 300-fold range in abundance among the visualized proteins. The most abundant 10 proteins account for about 25% of the total protein on the pH 4–8 gel, the most abundant 60 proteins account for 50%, and the most abundant 500 proteins account for 80%. Since it seems likely that the pH 4–8 gels give a representative sampling of all proteins, we estimate that half of the total cellular protein is accounted for by fewer than 100 different gene products, principally glycolytic enzymes and proteins involved in protein synthesis.
Correlation of protein abundance with mRNA abundance.
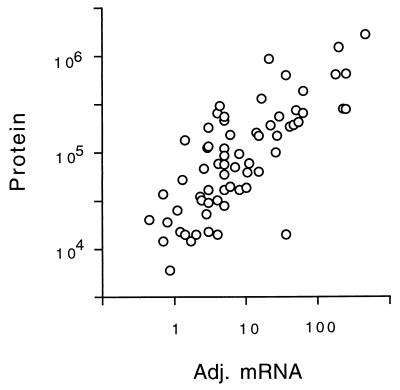
Estimates of mRNA abundance for each gene have been made by SAGE (27) and by hybridization of cRNA to oligonucleotide arrays (30). These two methods give broadly similar results, yet each method has strengths and weaknesses (Materials and Methods). Table 1 lists the number of molecules of mRNA per cell for each gene studied. One measurement (mRNA) uses data from SAGE analysis alone (27); a second incorporates data from both SAGE and hybridization (30) (adjusted mRNA) (Table 1; Materials and Methods). We correlated protein abundance with mRNA abundance (Fig. 2). For adjusted mRNA versus protein, the Spearman rank correlation coefficient, rs, was 0.74 (P < 0.0001), and the Pearson correlation coefficient, rp, on log transformed data (Materials and Methods) was 0.76 (P < 0.00001). We obtained similar correlations for mRNA versus protein and also for other data transformations (Materials and Methods). Thus, several statistical methods show a strong and significant correlation between mRNA abundance and protein abundance. Of course, the correlation is far from perfect; for mRNAs of a given abundance, there is at least a 10-fold range of protein abundance (Fig. 2). Some of this scatter is probably due to posttranscriptional regulation, and some is due to errors in the mRNA or protein data. For example, the protein Yef3 runs poorly on our gels, giving multiple smeared spots. Its abundance has probably been underestimated, partly explaining the low protein/mRNA ratio of Yef3. It is the most extreme outlier in Fig. 2.
FIG. 2.
Correlation of protein abundance with adjusted mRNA abundance. The number of molecules per cell of each protein is plotted against the number of molecules per cell of the cognate mRNA, with an rp of 0.76. Note the logarithmic axes. Data for mRNA were taken from references 27 and 30 and combined as described in Materials and Methods.
These data on mRNA (27, 30) and protein abundance (Table 1) suggest that for each mRNA molecule, there are on average 4,000 molecules of the cognate protein. For instance, for Act1 (actin) there are about 54 molecules of mRNA per cell and about 205,000 molecules of protein. Assuming an mRNA half-life of 30 min (12) and a cell doubling time of 120 min, this suggests that an individual molecule of mRNA might be translated roughly 1,000 times. These calculations are limited to mRNAs for abundant proteins, which are likely to be the mRNAs that are translated best.
A full complement of cell protein is synthesized in about 120 min under these conditions. Thus, 4,000 molecules of protein per molecule of mRNA implies that translation initiates on an mRNA about once every 2 s. This is a remarkably high rate; it implies that if an average mRNA bears 10 ribosomes engaged in translation, then each ribosome completes translation in 20 s; if an average protein has 450 residues; this in turn implies translation of over 20 amino acids per s, a rate considerably higher than estimated for mammalians (3 to 8 amino acids per s) (18). These estimates depend on the amount of mRNA per cell (11, 27).
The large number of protein molecules that can be made from a single mRNA raises the issue of how abundance is controlled for less abundant proteins. Many nonabundant proteins may be unstable, and this would reduce the protein/mRNA ratio. In addition, many nonabundant proteins may be translated at suboptimal rates. We have found that mRNAs for nonabundant proteins usually have suboptimal contexts for translational initiation. For example, there are over 600 yeast genes which probably have short open reading frames in the mRNA upstream of the main open reading frame (17a). These may be devices for reducing the amount of protein made from a molecule of mRNA.
Correlation of codon bias with protein abundance.
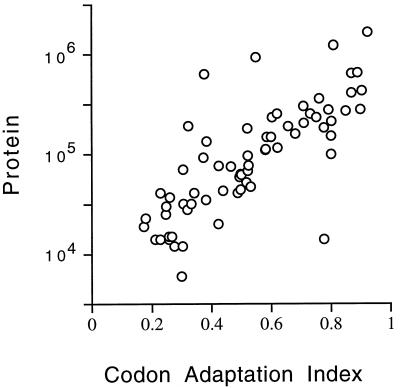
The mRNAs for highly expressed proteins preferentially use some codons rather than others specifying the same amino acid (14). This preference is called codon bias. The codons preferred are those for which the tRNAs are present in the greatest amounts. Use of these codons may make translation faster or more efficient and may decrease misincorporation. These effects are most important for the cell for abundant proteins, and so codon bias is most extreme for abundant proteins. The effect can be dramatic—highly biased mRNAs may use only 25 of the 61 codons.
We asked whether the correlation of codon bias with abundance continues for medium-abundance proteins. There are various mathematical expressions quantifying codon bias; here, we have used the CAI (24) (Materials and Methods) because it gives a result between 0 and 1. The rs for CAI versus protein abundance is 0.80 (P < 0.0001), similar to the mRNA-protein correlation, confirming a strong correlation between CAI and protein abundance (Fig. 3). The relationship between CAI and protein abundance is log linear from about 1,000,000 to about 10,000 molecules per cell. We have no data for rarer proteins.
FIG. 3.
Correlation of protein abundance with CAI. The number of molecules per cell of each protein is plotted against the CAI for that protein. Note the logarithmic scale on the protein axis. Data for the CAI are from the YPD database (13).
It is not clear whether CAI reflects maximum or average levels of protein expression. The proteins used for the CAI-protein correlation included some proteins which were not expressed at maximum levels under the condition of the experiment (Hsc82, Hsp104, Ssa1, Ade1, Arg4, His4, and others). When these proteins were removed from consideration and the correlation between CAI and the remaining (presumably constitutive) proteins was recalculated, the rs was essentially unchanged (not shown).
The equation describing the graph in Fig. 3 is log (protein molecules/cell) = (2.3 × CAI) + 3.7. Thus, under certain conditions (a CAI of 0.3 or greater; a constitutively expressed gene), a very rough estimate of protein abundance can be made by raising 10 to the power of [(2.3 × CAI) + 3.7].
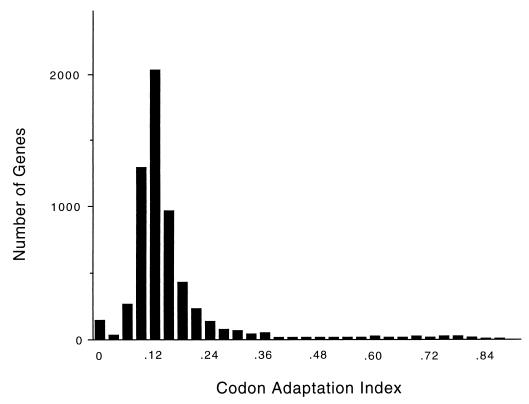
The distribution of CAI over the genome (Fig. 4) consists of a lower, bell-shaped distribution, possibly indicating a region where there is no selection for codon bias, and an upper, flat distribution, starting at a CAI of about 0.3, possibly indicating a region where there is selection for codon bias. Almost all of the proteins whose abundance we have measured are in the upper, flat portion of the distribution. In the lower, bell-shaped region, we do not know whether there is a correlation between CAI and protein abundance.
FIG. 4.
Distribution of CAI over the whole genome, shown in intervals of 0.030 (i.e., there are 150 genes with a CAI between 0.000 and 0.030, inclusive; 31 genes with a CAI between 0.031 and 0.060; 269 genes with a CAI between 0.061 and 0.090; 1,296 genes with a CAI between 0.091 and 0.120; etc.). The distribution peaks with 2,028 genes with a CAI between 0.121 and 0.150.
Changes in protein abundance in glucose and ethanol.
A comparison of cells grown in glucose (Fig. 1A) with cells grown in ethanol (Fig. 1B) is shown in Table 1. As is well known, some proteins are induced tremendously during growth on ethanol. Two striking examples are the peroxisomal enzymes Icl1 (isocitrate lyase) and Cit2 (citrate synthase), which are induced in ethanol by more than 100- and 12-fold, respectively (Fig. 1; Table 1). These enzymes are key components of the glyoxylate shunt, which diverts some acetyl coenzyme A (acetyl-CoA) from the tricarboxylic acid cycle to gluconeogenesis. S. cerevisiae requires large amounts of carbohydrate for its cell wall; in ethanol medium, this carbohydrate comes from gluconeogenesis, which depends on the glyoxylate shunt and on the glycolytic pathway running in reverse. The need for gluconeogenesis also explains why glycolytic enzymes are abundant even in ethanol medium. Thus, 2D gel analysis shows the prominence of the glycolytic and glyoxylate shunt enzymes in cells grown on ethanol, emphasizing that gluconeogenesis, presumably largely for production of the cell wall, is a major metabolic activity under these conditions.
During gluconeogenesis, substrate-product relationships are reversed for the glycolytic enzymes. One might expect that not all glycolytic enzymes would be well adapted to the reverse reaction. Indeed, 2D gels show that in ethanol, Adh2 (alcohol dehydrogenase 2) is strongly induced (16), while its isozyme Adh1 is not greatly affected. Adh1 and Adh2 each interconvert acetaldehyde and ethanol. Adh1 has a relatively high Km for ethanol (17 mM), while Adh2 has a lower Km (0.8 mM) (5). Thus, it is thought that Adh1 is specialized for glycolysis (acetaldehyde to ethanol), while Adh2 is specialized for respiration (ethanol to acetaldehyde) (5, 29). Similarly, Eno1 (enolase 1) is induced in ethanol, while its isozyme Eno2 (enolase 2) decreases in abundance (Table 1) (4, 19). Eno1 is inhibited by 2-phosphoglycerate (the glycolytic substrate), while Eno2 is inhibited by phosphoenolpyruvate (the gluconeogenic substrate) (4). Perhaps Eno1 has a lower Km for phosphoenolpyruvate than does Eno2, though to our knowledge this has not been tested. Thus, the 2D gels distinguish isozymes specialized for growth on glucose (Adh1 and Eno2) from isozymes specialized for ethanol (Adh2 and Eno1).
Many heat shock proteins (e.g., Hsp60, Hsp82, Hsp104, and Kar2) were about twofold more abundant in ethanol medium than in glucose medium. This is consistent with the increased heat resistance of cells grown in ethanol (3).
Enzymes involved in protein synthesis (Eft1, Rpa0, and Tif1) were about twice as abundant in glucose medium as in ethanol medium. This may reflect the higher growth rate of the cells in glucose.
Phosphorylation of proteins.
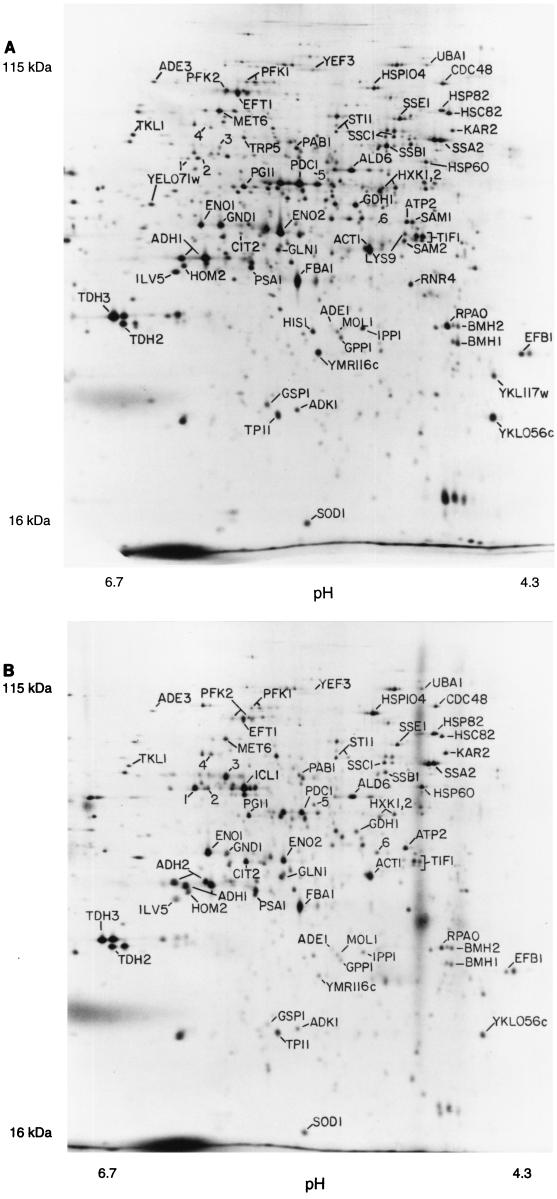
To examine protein phosphorylation, we labeled cells with 32P and ran 2D gels to examine phosphoproteins. About 300 distinct spots, probably representing 150 to 200 proteins, could be seen on pH 4–8 gels (Fig. 5B). We then aligned autoradiograms of three gels, each with a different kind of labeled protein (32P only [Fig. 5B], 32P plus 35S [Fig. 5A], and 35S only [not shown, but see Fig. 1 for example]). In this way, we made provisional identification of some of the 32P-labeled spots as particular 35S-labeled spots. All such identifications are somewhat uncertain, since precise alignments are difficult, and of course multiple spots may exactly comigrate. Nevertheless, we believe that most of the provisional identifications are probably correct. Among the major 32P-labeled proteins are the hexokinases Hxk1 and Hxk2, the acidic ribosome-associated protein Rpa0, the translation factors Yef3 and Efb1, and probably Hsp70 heat shock proteins of the Ssa and Ssb families. Rpa0 and Efb1 are quantitatively monophosphorylated.
FIG. 5.
Phosphorylated proteins. (A) Mixture of 32P-labeled proteins and 35S-labeled proteins. Two separate labeling reactions were done, one with 32P and one with 35S, and extracts were mixed and run on a 2D gel. Spots marked with numbers rather than gene names represent spots noted on 35S gels but unidentified. Spots labeling with 32P were identified by (i) increased labeling compared to the 35S-only gel (not shown); (ii) the characteristic fuzziness of a 32P-labeled spot; and (iii) the decay of signal intensity seen on exposures made 4 weeks later (not shown). A minor form of Tpi1 and at least six minor forms of Tif1 have been noted in overexpression experiments (see also Fig. 6B); positions of the minor forms are indicated by circles. (B) 32P-only labeling. The major form of Tpi1, which is not labeled with 32P, is indicated by a large circle; positions of seven forms of Tif1 are indicated by smaller circles.
Many yeast proteins resolve into multiple spots on these 2D gels (7). Yef3 has five or more spots, at least four of which comigrate with 32P. Tpi1 has a major spot showing no 32P labeling and a minor, more acidic spot which overlaps with some 32P label. Tif1 has at least seven spots (7); two of these overlap with some 32P label, but five do not (Fig. 5). Eft1 has at least three spots (7), and none of these overlap with 32P, although there are three nearby, unidentified 32P-labeled spots (a, c, and d in Fig. 5). Spots that seem to be extra forms of Met6, Pdc1, Eno2, and Fba1 can be seen in Fig. 6A, but there is little 32P at these positions in Fig. 5. Thus, phosphorylation explains some but not all of the different protein isoforms seen.
The cell cycle is regulated in part by phosphorylation. We compared 32P-labeled proteins from cells synchronized in G1 with α-factor, in cells synchronized in G1 by depletion of G1 cyclins, and in cells synchronized in M phase with nocodazole. Only very minor differences were seen, and these were difficult to reproduce. The cell cycle proteins regulated by phosphorylation may not be abundant enough for this technique to be applied easily.
Centrifugal fractionation.
We fractionated 35S-labeled extracts by centrifugation (Materials and Methods). Figure 6A shows the proteins in the supernatant of a high-speed (100,000 × g, 30 min) centrifugation, while Fig. 6B shows the proteins in the pellet of a low-speed (16,000 × g, 10 min) centrifugation. Many proteins are tremendously enriched in one fraction or the other, while others are present in both. Most glycolytic enzymes (e.g., Tdh2, Tdh3, Eno2, Pdc1, Adh1, and Fba1) are enriched in the supernatant fraction. The only exception is Pfk1 (not indicated), which is found in both pellet and supernatant fractions. Many proteins involved in protein synthesis (Eft1, Yef3, Prt1, Tif1, and Rpa0) are in the pellet, possibly because of the association of ribosomes with the endoplasmic reticulum. However, Efb1 is in the supernatant, as is a substantial portion of the Eft1. Perhaps surprisingly, several mitochondrial proteins (Atp2 [not shown] and Ilv5) are largely in the supernatant. Perhaps glass bead breakage of cells releases mitochondrial proteins. The nuclear protein Gsp1 is in the pellet fraction. The enrichment produced by centrifugation makes it possible to see minor spots which are otherwise poorly resolved from surrounding proteins. Figure 6B shows that the previously identified Tif1 spot is surrounded by as many as six other spots that cofractionate. We observed six identical or very similar additional spots when we overexpressed Tif1 from a high-copy-number plasmid (not shown). Signal overlaps only one or two of these spots in 32P-labeling experiments (Fig. 5), and so the different forms are not mainly due to different phosphorylation states.
DISCUSSION
Our experience with developing a 2D gel protein database for S. cerevisiae is summarized here. With current technology, we can see the most abundant 1,200 proteins, which is about one-third to one-quarter of the proteins expressed. The remaining proteins will be difficult to see and study with the methods that we have used, not because of a lack of sensitivity but because weak spots are covered by nearby strong spots.
Of the 1,200 proteins seen, we have identified 148, with a bias toward the most abundant proteins. Steady application of the methods already used would allow identification of most of the remaining proteins. Gene overexpression will be particularly useful, since it is not affected by the lower abundance of the remaining visible proteins.
2D gels of the kind that we have used are not suitable for visualization of rare proteins. However it will be possible to study on a global basis metabolic processes involving relatively abundant proteins, such as protein synthesis, glycolysis, gluconeogenesis, amino acid synthesis, cell wall synthesis, nucleotide synthesis, lipid metabolism, and the heat shock response.
Gygi et al. (10) have recently completed a study similar to ours. Despite generating broadly similar data, Gygi et al. reached markedly different conclusions. We believe that both mRNA abundance and codon bias are useful predictors of protein abundance. However, Gygi et al. feel that mRNA abundance is a poor predictor of protein abundance and that “codon bias is not a predictor of either protein or mRNA levels” (10). These different conclusions are partly a matter of viewpoint. Gygi et al. focus on the fact that the correlations of mRNA and codon bias with protein abundance are far from perfect, while we focus on the fact that, considering the wide range of mRNA and protein abundance and the undoubted presence of other mechanisms affecting protein abundance, the correlations are quite good.
However, the different conclusions are also partly due to different methods of statistical analysis and to real differences in data. With respect to statistics, Gygi et al. used the Pearson product-moment correlation coefficient (rp) to measure the covariance of mRNA and protein abundance. Depending on the subset of data included, their rp values ranged from 0.1 to 0.94. Because of the low rp values with some subsets of the data, Gygi et al. concluded that the correlation of mRNA to protein was poor. However, the rp correlation is a parametric statistic and so requires variates following a bivariate normal distribution; that is, it would be valid only if both mRNA and protein abundances were normally distributed. In fact, both distributions are very far from normal (data not shown), and so a calculation of rp is inappropriate. There was no statistical backing for the assertion that codon bias fails to predict protein abundance.
We have taken two statistical approaches. First, we have used the Spearman rank correlation coefficient (rs). Since this statistic is nonparametric, there is no requirement for the data to be normally distributed. Using the rs, we find that mRNA abundance is well correlated with protein abundance (rs = 0.74), and the CAI is also well correlated with protein abundance (rs = 0.80) (and also with mRNA abundance [data not shown]). For the data of Gygi et al. (10), we obtained similar results, though with their data the correlation is not as good; rs = 0.59 for the mRNA-to-protein correlation, and rs = 0.59 for the codon bias-to-protein correlation.
In a second approach, we transformed the mRNA and protein data to forms where they were normally distributed, to allow calculation of an rp (Materials and Methods). Two transformations, Box-Cox and logarithmic, were used; both gave good correlations with our data [e.g., rp = 0.76 for log(adjusted RNA) to log(protein)]. We were not able to transform the data of Gygi et al. to a normal distribution.
Finally, there are also some differences in data between the two studies. These may be partly due to the different measurement techniques used: Gygi et al. measured protein abundance by cutting spots out of gels and measuring the radioactivity in each spot by scintillation counting, whereas we used phosphorimaging of intact gels coupled to image analysis. We compared our data to theirs for the proteins common between the studies (but excluding proteins whose mRNAs are known to differ between rich and minimal media, and excluding Tif1, which was anomalous in differing by 100-fold between the two data sets). The rs between the two protein data sets was 0.88 (P < 0.0001). Although this is a strong correlation, the fact that it is less than 1.0 suggests that there may have been errors in measuring protein abundance in one or both studies. After normalizing the two data sets to assume the same amount of protein per cell, we found a systematic tendency for the protein abundance data of Gygi et al. to be slightly higher than ours for the highest-abundance proteins and also for the lowest-abundance proteins but slightly lower than ours for the middle-abundance proteins. These systematic differences suggest some systematic errors in protein measurement. Although we do not know what the errors are, we suggest the following as a reasonable speculation. For the highest-abundance proteins, we may have underestimated the amount of protein because of a slightly nonlinear response of the phosphorimager screens. For the lowest-abundance proteins, Gygi et al. may have overestimated the amount of protein because of difficulties in accurately cutting very small spots out of the gel and because of difficulties in background subtraction for these small, weak spots. The difference in the middle abundance proteins may be a consequence of normalization, given the two errors above.
The low-abundance proteins in the data set of Gygi et al. have a poor correlation with mRNA abundance. We calculate that the rs is 0.74 for the top 54 proteins of Gygi et al. but only 0.22 for the bottom 53 proteins, a statistically significant difference. However, with our data set, the rs is 0.62 for the top 33 proteins and 0.56 (not significantly different) for the bottom 33 proteins (which are comparable in abundance to the bottom 53 proteins of Gygi et al.). Thus, our data set maintains a good correlation between mRNA and protein abundance even at low protein abundance. This is consistent with our speculation that protein quantification by phosphorimaging and image analysis may be more accurate for small, weak spots than is cutting out spots followed by scintillation counting. Our relatively good correlations even for nonabundant proteins may also reflect the fact that we used both SAGE data and RNA hybridization data, which is most helpful for the least abundant mRNAs. In summary, we feel that the poor correlation of protein to mRNA for the nonabundant proteins of Gygi et al. may reflect difficulty in accurately measuring these nonabundant proteins and mRNAs, rather than indicating a truly poor correlation in vivo. It is not surprising that observed correlations would be poorer with less-abundant proteins and mRNAs, simply because the accuracy of measurement would be worse.
How well can mRNA abundance predict protein abundance? With rp = 0.76 for logarithmically transformed mRNA and protein data, the coefficient of determination, (rp)2, is 0.58. This means that more than half (in log space) of the variation in protein abundance is explained by variation in mRNA abundance. When converted back to arithmetic values, protein abundances vary over about 200-fold (Table 1), and (rp)2 = 0.58 for the log data means that of this 200-fold variation, about 20-fold is explained by variation in the abundance of mRNA and about 10-fold is unexplained (but could be due partly to measurement errors). For proteins much less abundant than those considered here, we imagine the in vivo correlation between mRNA and protein abundance will be worse, and other regulatory mechanisms such as protein turnover will be more important.
Some important conclusions can be drawn from this sampling of the proteome. First, there is an enormous range of protein abundance, from nearly 2,000,000 molecules per cell for some glycolytic enzymes to about 100 per cell for some cell cycle proteins (26a). Second, about half of all cellular protein is found in fewer than 100 different gene products, which are mostly involved in carbohydrate metabolism or protein synthesis. Third, the correlation between protein abundance and CAI is log linear as far as we can see, which is from about 10,000 protein molecules per cell to about 1,000,000. This is somewhat surprising, because it implies that selective forces for codon bias are significant even at moderate expression levels. It also means that codon bias is a useful predictor of protein abundance even for moderately low bias proteins. Fourth, there is a good correlation between protein abundance and mRNA abundance for the proteins that we have studied. This validates the use of mRNA abundance as a rough predictor of protein abundance, at least for relatively abundant proteins. Fifth, for these abundant proteins, there are about 4,000 molecules of protein for each molecule of mRNA. This last conclusion raises questions as to how the levels of nonabundant proteins are regulated and suggests that protein instability, regulated translation, suboptimal rates of translation, and other mechanisms in addition to transcriptional control may be very important for these proteins.
ACKNOWLEDGMENTS
We thank Neena Sareen and Nick Bizios (CSHL 2D gel laboratory) for production of 2D gels, Tom Volpe for help with some experiments, Corine Driessens for help with calculations and statistics, and Herman Wijnen and Nick Edgington for comments on the manuscript. We especially thank Tim Tully for in-depth statistical analysis and for insightful discussions on statistical interpretations.
This work was supported by grant P41-RR02188 from the NIH Biomedical Research Technology Program, Division of Research Resources, to J.I.G., by Small Business Innovation Research grant R44 GM54110 to Proteome, Inc., by grant DAMD17-94-J4050 from the Army Breast Cancer Program to B.F., and by NIH grant RO1 GM45410 to B.F.
REFERENCES
- 1.Baroni M D, Martegani E, Monti P, Alberghina L. Cell size modulation by CDC25 and RAS2 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:2715–2723. doi: 10.1128/mcb.9.6.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucherie H, Sagliocco F, Joubert R, Maillet I, Labarre J, Perrot M. Two-dimensional gel protein database of Saccharomyces cerevisiae. Electrophoresis. 1996;17:1683–1699. doi: 10.1002/elps.1150171106. [DOI] [PubMed] [Google Scholar]
- 3.Elliott B, Futcher B. Stress resistance of yeast cells is largely independent of cell cycle phase. Yeast. 1993;9:33–42. doi: 10.1002/yea.320090105. [DOI] [PubMed] [Google Scholar]
- 4.Entian K D, Meurer B, Kohler H, Mann K H, Mecke D. Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae. Biochim Biophys Acta. 1987;923:214–221. doi: 10.1016/0304-4165(87)90006-7. [DOI] [PubMed] [Google Scholar]
- 5.Ganzhorn A J, Green D W, Hershey A D, Gould R M, Plapp B V. Kinetic characterization of yeast alcohol dehydrogenases. Amino acid residue 294 and substrate specificity. J Biol Chem. 1987;262:3754–3761. [PubMed] [Google Scholar]
- 6.Garrels J I. The Quest system for quantitative analysis of two-dimensional gels. J Biol Chem. 1989;264:5269–5282. [PubMed] [Google Scholar]
- 7.Garrels J I, Futcher B, Kobayashi R, Latter G I, Schwender B, Volpe T, Warner J R, McLaughlin C S. Protein identifications for a Saccharomyces cerevisiae protein database. Electrophoresis. 1994;15:1466–1486. doi: 10.1002/elps.11501501210. [DOI] [PubMed] [Google Scholar]
- 8.Garrels J I, McLaughlin C S, Warner J R, Futcher B, Latter G I, Kobayashi R, Schwender B, Volpe T, Anderson D S, Mesquita-Fuentes R, Payne W E. Proteome studies of S. cerevisiae: identification and characterization of abundant proteins. Electrophoresis. 1997;18:1347–1360. doi: 10.1002/elps.1150180810. [DOI] [PubMed] [Google Scholar]
- 9.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 10.Gygi S P, Rochon Y, Franza B R, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hereford L M, Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977;10:453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- 12.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodges P E, McKee A H, Davis B P, Payne W E, Garrels J I. The Yeast Proteome Database (YPD): a model for the organization of genome-wide functional data. Nucleic Acids Res. 1999;27:69–73. doi: 10.1093/nar/27.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 15.Johnston G C, Pringle F R, Hartwell L H. Coordination of growth with cell division in the yeast S. cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 16.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E, Pringle J, Broach J, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 17.Kornblatt M J, Klugerman A. Characterization of the enolase isozymes of rabbit brain: kinetic differences between mammalian and yeast enolases. Biochem Cell Biol. 1989;67:103–107. doi: 10.1139/o89-016. [DOI] [PubMed] [Google Scholar]
- 17a.Latter, G., and B. Futcher. Unpublished data.
- 18.Mathews B, Sonenberg N, Hershey J W B. Origins and targets of translational control. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 1–29. [Google Scholar]
- 19.McAlister L, Holland M J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J Biol Chem. 1982;257:7181–7188. [PubMed] [Google Scholar]
- 20.Monardo P J, Boutell T, Garrels J I, Latter G I. A distributed system for two-dimensional gel analysis. Comput Appl Biosci. 1994;10:137–143. doi: 10.1093/bioinformatics/10.2.137. [DOI] [PubMed] [Google Scholar]
- 21.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson S D, Latter G I. Evaluation of storage phosphor imaging for quantitative analysis of 2-D gels using the Quest II system. BioTechniques. 1993;15:1076–1083. [PubMed] [Google Scholar]
- 23.Sagliocco F, Guillemot J C, Monribot C, Capdevielle J, Perrot M, Ferran E, Ferrara P, Boucherie H. Identification of proteins of the yeast protein map using genetically manipulated strains and peptide-mass fingerprinting. Yeast. 1996;12:1519–1533. doi: 10.1002/(SICI)1097-0061(199612)12:15%3C1519::AID-YEA47%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 24.Sharp P M, Li W H. The Codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevchenko A, Jensen O N, Podtelejnikov A V, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 26a.Tyers, M., and B. Futcher. Unpublished data.
- 27.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 28.Warner J. Labeling of RNA and phosphoproteins in S. cerevisiae. Methods Enzymol. 1991;194:423–428. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- 29.Wills C. Production of yeast alcohol dehydrogenase isoenzymes by selection. Nature. 1976;261:26–29. doi: 10.1038/261026a0. [DOI] [PubMed] [Google Scholar]
- 29a.Wodicka, L. Personal communication.
- 29b.Wodicka, L. Unpublished data.
- 30.Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]