BCR/ABL Directly Inhibits Expression of SHIP, an SH2-Containing Polyinositol-5-Phosphatase Involved in the Regulation of Hematopoiesis (original) (raw)
Abstract
The BCR/ABL oncogene causes chronic myelogenous leukemia (CML), a myeloproliferative disorder characterized by clonal expansion of hematopoietic progenitor cells and granulocyte lineage cells. The SH2-containing inositol-5-phosphatase SHIP is a 145-kDa protein which has been shown to regulate hematopoiesis in mice. Targeted disruption of the murine SHIP gene results in a myeloproliferative syndrome characterized by a dramatic increase in numbers of granulocyte-macrophage progenitor cells in the marrow and spleen. Also, hematopoietic progenitor cells from SHIP−/− mice are hyperresponsive to certain hematopoietic growth factors, a phenotype very similar to the effects of BCR/ABL in murine cells. In a series of BCR/ABL-transformed hematopoietic cell lines, Philadelphia chromosome (Ph)-positive cell lines, and primary cells from patients with CML, the expression of SHIP was found to be absent or substantially reduced compared to untransformed cell lines or leukemia cells lacking BCR/ABL. Ba/F3 cells in which expression of BCR/ABL was under the control of a tetracycline-inducible promoter showed rapid loss of p145 SHIP, coincident with induction of BCR/ABL expression. Also, an ABL-specific tyrosine kinase inhibitor, CGP57148B (STI571), rapidly caused reexpression of SHIP, indicating that BCR/ABL directly, but reversibly, regulates the expression of SHIP protein. The estimated half-life of SHIP protein was reduced from 18 h to less than 3 h. However, SHIP mRNA also decreased in response to BCR/ABL, suggesting that SHIP protein levels could be affected by more than one mechanism. Reexpression of SHIP in BCR/ABL-transformed Ba/F3 cells altered the biological behavior of cells in culture. The reduction of SHIP due to BCR/ABL is likely to directly contribute to the pathogenesis of CML.
BCR/ABL is generated by the t(9,22) (q34;q11) Philadelphia chromosome (Ph) translocation and is the transforming protein in chronic myelogenous leukemia (CML) (5, 26, 47, 56). There is clonal expansion of progenitor cells of several different hematopoietic lineages, and patients characteristically have high leukocyte counts due to accumulation in the blood of immature cells of the granulocytic lineage (7). Primary CML cells demonstrate reduced apoptosis (4), altered adhesion to fibronectin (64), and hypermotility (48). Although immature hematopoietic CML cells are traditionally felt to be dependent on hematopoietic growth factors for viability (45), recent studies have shown that CD34+ lineage cells can be factor independent, possibly through an autocrine mechanism (31).
The exact mechanism of transformation by BCR/ABL is not known. The ABL tyrosine kinase activity is required for transformation by BCR/ABL (38), and there are also several other signaling pathways which are believed to contribute to transformation. For example, binding of the GRB2/SOS complex to tyrosine 177 in BCR is important for p21_RAS_ activation and is believed to contribute to transformation (1, 42, 44). BCR/ABL also phosphorylates a number of other signaling proteins, including STAT5 (8, 30, 57), SHC (37), c-CBL (3, 39, 51), and paxillin (49). However, none of the known signaling events clearly explains the myeloproliferative phenotype of BCR/ABL in CML. Additionally, it has been very difficult to link activation of any individual signaling pathway by BCR/ABL, such as p21_RAS_, to a specific biological abnormality, such as factor independence. In particular, identification of a pathway regulated by BCR/ABL which directly causes myeloproliferation would be of great value in understanding the pathogenesis of CML.
SHIP is a 145-kDa protein originally identified because of its interaction with SHC (14, 35). SHIP was found to be an SH2-containing inositol phosphatase which selectively hydrolyzes the 5′-phosphate from inositol-1,3,4,5-tetraphosphate [Ins(1,3,4,5)P4] and phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3]. SHIP is transiently tyrosine phosphorylated by growth factor stimulation and activation of immune receptors (9, 14, 40). The SHIP-related and more widely expressed SHIP2 (27, 43) is also tyrosine phosphorylated by growth factors and in BCR/ABL-transformed cells (24, 66). SHIP2 has, like SHIP, PtdIns(3,4,5)P3-specific 5′-phosphatase activity (24, 66) but may not hydrolyze Ins(1,3,4,5)P4 (66).
Mice with a disruption of the SHIP gene were found to be viable and fertile but failed to thrive, developing a myeloproliferative disorder with extensive infiltration of myeloid cells in the lung. Of further interest, marrow progenitor cells were hyperresponsive to hematopoietic growth factors (28). This is of special interest since this myeloproliferative phenotype is similar to that of BCR/ABL transformation in mice (13, 18, 25, 32). Although the mechanism whereby loss of SHIP expression might lead to a myeloproliferative disease is not known, it has been suggested that SHIP functions in part to modify a signaling pathway which is initiated by activation of phosphatidylinositol 3-kinase (PI3K) (2, 29), a lipid kinase already known to be activated and important for BCR/ABL transformation (58). For example, SHIP would be expected to metabolize the PI3K lipid product, PtdIns(3,4,5)P3, to PtdIns(3,4)P2 (14). However, it is not clear how such changes in lipid metabolism result in a myeloproliferative state.
In this study, we show that SHIP protein levels are decreased by BCR/ABL through a reversible mechanism that requires ABL kinase activity. Reexpression of SHIP in BCR/ABL-transformed cells was found to alter at least one characteristic of BCR/ABL transformation. Since loss of SHIP by gene targeting leads to a myeloproliferative syndrome, these studies further implicate phosphatidylinositol pathways as critical in the pathogenesis of CML.
MATERIALS AND METHODS
Cell culture.
The murine hematopoietic line Ba/F3 was grown in RPMI 1640 medium with 10% (vol/vol) fetal calf serum (FCS) and 10% (vol/vol) WEHI-3B conditioned medium (as a source of murine interleukin-3). Ba/F3 cell lines transfected with a TEL/ABL cDNA (BaF3/TEL-ABL) and a BCR/ABL cDNA (BaF3/p190 and BaF3/p210) were grown in RPMI 1640 medium with 10% (vol/vol) FCS. A Ba/F3 cell line transfected with a BCR/ABL cDNA under the control of a tetracycline-inducible promoter (33) (BaF3/TonB210.1; kindly provided by G. Q. Daley, Massachusetts Institute of Technology, Cambridge) and a Ba/F3 cell line expressing a kinase-dead form of p190 BCR/ABL (BaF3/p190-k.d.; kindly provided by A.-M. Pendergast, Duke University, Durham, N.C.) was grown in RPMI 1640 medium with 10% (vol/vol) FCS and 10% (vol/vol) WEHI conditioned medium. The expression of BCR/ABL in BaF3/TonB210.1 cells was induced by treatment with doxycycline (1 μg/ml). In some experiments, Ba/F3 and BaF3/TonB210.1 cells were deprived of growth factors for 18 h in RPMI 1640 medium containing 0.5% (wt/vol) bovine serum albumin. In addition, some cells were treated with LY294002 (Sigma, St. Louis, Mo.), wortmannin (Sigma), or the ABL-specific tyrosine kinase inhibitor CGP57148B (STI571; kindly provided by Novartis, Basel, Switzerland) (17). The Ph-positive cell lines BV173, K562, and Ku812, and the Ph-negative lymphoid cell lines Blin-1, Molt-4, Nalm-6, and REH were grown in RPMI 1640 medium with 10% (vol/vol) FCS. Hematopoietic progenitor cells from CML and acute lymphoblastic leukemia (ALL) patients were obtained from bone marrow aspirates with informed consent, using Dana-Farber Cancer Institute-approved protocols.
Immunoprecipitation and Western blotting.
Immunoprecipitation and Western blotting using a chemiluminescence technique were performed as described elsewhere (50). Tyrosine-phosphorylated proteins were detected by using monoclonal antibody 4G10 (kindly provided by B. Druker, Oregon Health Science University, Portland). A mouse monoclonal antibody against SHIP (P1C1) and polyclonal rabbit antisera against SHIP (5340) and SHP2 (Santa Cruz Biotechnology, Santa Cruz, Calif.) were used for Western blotting or immunoprecipitation.
Northern blotting.
The level of SHIP mRNA in BV173, K562, and Ku812 cells was analyzed by Northern blotting using standard methods. cDNA probes from the 3′ of murine SHIP (bases 2703 to 3573) and human G3PDH (glyceraldehyde-3-phosphate dehydrogenase) (53) were used. The cDNA probes were labeled with [32P]dCTP by using Klenow fragment (High Prime kit; Boehringer, Mannheim, Germany) and purified with ProbeQuant G-50 microcolumns (Amersham Pharmacia Biotech, Piscataway, N.J.). Total RNA was isolated with Trizol reagent (Life Technologies, GibcoBRL, Gaithersburg, Md.) and used to prepare mRNA (Message Maker; Life Technologies, GibcoBRL) to evaluate gene expression. Bound probe was analyzed by phosphorimaging analysis (FLA-2000 fluorescent image analyzer; Fuji Photo Film Corp., Stamford, Conn.).
Southern blotting.
Genomic DNA was isolated from Blin-1, BV173, K562, and Ku812 cells by using a Wizard genomic DNA purification kit (Promega) according to the manufacturer’s directions. For Southern analysis, 10 μg of genomic DNA was digested with restriction enzymes _Pst_I, _Eco_RI, and _Xba_I (New England Biolabs, Beverly, Mass.) or restriction enzyme _Hin_dIII (Life Technologies, GibcoBRL), and the sample was separated on a 0.7% agarose gel by using standard methods. cDNA probes from the 5′ end of murine SHIP (bases 4 to 486) and from the 3′ end of murine SHIP (bases 2703 to 3573) were used. The cDNA probes were labeled and purified as described above, and bound probe was analyzed by autoradiography.
[35S]methionine labeling and pulse-chase analysis.
The half-life of SHIP protein was estimated by 35S labeling of cellular proteins and subsequent pulse-chase. Logarithmically growing cells were washed twice in methionine-free RPMI 1640 medium (Gibco BRL, Grand Island, N.Y.) and starved in methionine-free RPMI 1640 medium with 10% (vol/vol) dialyzed FCS (Gibco BRL) at 10 × 106 cells/ml for 1 h. Methionine-starved cells were incubated for 1 h with 200 μCi of [35S]methionine (NEN Life Science, Boston, Mass.) per ml, then washed twice in RPMI 1640 medium supplemented with 150 μg of methionine per ml, and resuspended in regular growth medium supplemented with methionine. Aliquots of cells were removed at different time points after labeling, the cells were lysed, and the cellular lysate was used for immunoprecipitation as described above, using SHIP monoclonal antibodies. The samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). The dried membranes were sprayed with En3Hance (NEN) and exposed to BioMax film (Eastman Kodak Company, Rochester, N.Y.) at −80°C.
Expression constructs.
A murine SHIP cDNA (35) was subcloned into the _Eco_RI site of the pPINCO expression vector (23) (kindly provided by P. G. Pelicci, European Institute of Oncology, Milan, Italy) and expressed under the control of the Moloney virus long terminal repeat promoter. This retroviral expression vector also expresses an enhanced green fluorescence protein (EGFP) under the control of a separate cytomegalovirus promoter. Ecotropic retroviruses for infection of Ba/F3 cells were generated in the Phoenix-Eco packaging cell line (kindly provided by G. P. Nolan, Stanford University, School of Medicine, Stanford, Calif.) as described elsewhere (41). In addition, a murine SHIP cDNA was subcloned into the _Eco_RI site of the pTRE expression vector (Clontech Laboratories, Palo Alto, Calif.) and used for transfection into Ba/F3 cells that were stably transfected with pTet-On (Clontech Laboratories) (pTet-On transfected cells were kindly provided by G. Q. Daley).
Transwell migration assay.
The membranes of transwell chambers (8-μm-pore-size polycarbonate membrane; Corning Costar Corp., Cambridge, Mass.) were coated with 10 μg of fibronectin (Life Technologies, GibcoBRL) per ml for 18 h. Cells were counted in a Coulter counter (Coulter Counter Z2; Beckman Coulter, Fullerton, Calif.), and 0.2 × 106 cells in 100 μl were transferred to the upper chamber in AIM V medium (Life Technologies, GibcoBRL) containing different stimuli. After 5 h, cells in the lower compartment were concentrated by centrifugation and living cells counted by trypan blue exclusion. The spontaneous transwell migration of cells was expressed as a migration index (number of migrating cells treated or infected with a pPINCO-SHIP retrovirus divided by the number of migrating cells left untreated or infected with the pPINCO control virus). The standard error of the mean was calculated from the migration indices of independently performed experiments.
Proliferation and viability assays.
The number of viable cells was determined by trypan blue exclusion or annexin V (Boehringer Mannheim, Indianapolis, Ind.) staining, and the cell number was determined with a Coulter particle counter (Coulter Counter Z2).
RESULTS
BCR/ABL kinase activity is required for the downregulation of SHIP protein levels.
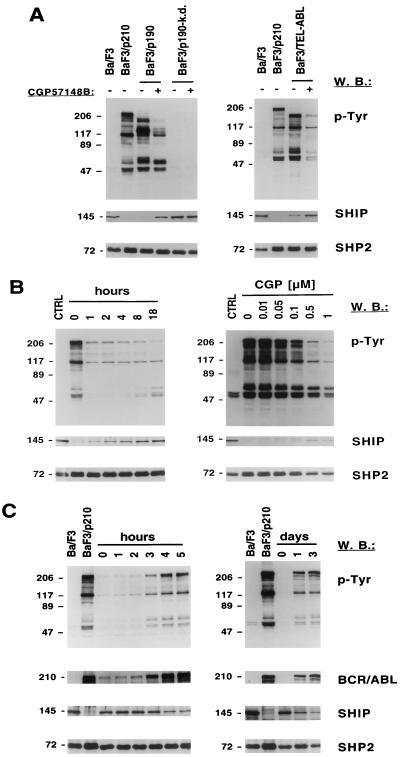
In a preliminary study, we noted that the expression of SHIP protein was reduced in a Ba/F3 cell line transformed with p210 BCR/ABL compared to the parent cell line (52). To determine if downregulation of SHIP was a general phenomenon associated with BCR/ABL transformation or only clonal variation of two cell lines, we measured SHIP protein levels in Ba/F3 cells expressing wild-type p190 BCR/ABL and in Ba/F3 cells expressing a kinase-dead mutant of p190 BCR/ABL. SHIP protein was reduced in cells transformed by p190 BCR/ABL and was increased when the p190 BCR/ABL kinase activity was inhibited by the ABL-specific tyrosine kinase inhibitor CGP57148B (STI571) (Fig. 1A, left). In contrast, in cells expressing the kinase-dead mutant, the SHIP protein was present and treatment with CGP57148B did not alter SHIP levels. Ba/F3 cells transformed with TEL/ABL (21) have detectable, but low, SHIP levels compared to untransformed cells, and SHIP protein was increased following treatment with CGP57148B (Fig. 1A, right). These results indicate that transformation by three different forms of ABL oncogenes was associated with decreased expression of SHIP protein, and that this effect was reversible with a small-molecule kinase inhibitor.
FIG. 1.
BCR/ABL kinase activity is required for the downregulation of SHIP protein levels. Tyrosine phosphorylation of cellular proteins was determined in total cell lysate by antiphosphotyrosine (p-Tyr) Western blotting (W.B.). The blots were stripped and reprobed for antibodies against SHIP, SHP2, or ABL (BCR/ABL). Lysates of Ba/F3 or BaF3/p210 cells were used as controls (CTRL), shown on the left side of each panel; molecular masses of the proteins are indicated in kilodaltons on the left. (A) BaF3/p190 and BaF3/p190-k.d. (expressing a kinase-dead BCR/ABL protein) were treated with 1 μM CGP57148B (STI571) or were left untreated (left). BaF3/TEL-ABL and were treated with 1 μM CGP57148B or were left untreated (right). (B) BaF3/p210 cells were treated with 1 μM CGP57148B for 0 to 18 h (left) or with 0 to 1 μM CGP57148B for 18 h (right) as indicated. (C) BCR/ABL expression was induced by doxycycline treatment of BaF3/TonB210.1 cells for 0 to 5 h (left) or 0 to 3 days (right) as indicated.
To further evaluate the relationship of SHIP levels to BCR/ABL kinase activity, kinetics and dose-response assays for CGP57148B on SHIP protein levels were performed. Total-cell lysate of BaF3/p210 cells treated for 0 to 18 h with CGP57148B was analyzed by antiphosphotyrosine and SHIP Western blotting (Fig. 1B, left). Total-cell tyrosine phosphorylation was reduced within 1 h of CGP57148B treatment. Increased expression of SHIP protein was time and dose dependent (Fig. 1B).
To confirm the inverse relationship between BCR/ABL kinase activity and SHIP protein expression, BCR/ABL was expressed under the control of a tetracycline-inducible promoter (Fig. 1C). The induction of BCR/ABL protein correlated directly with increased cellular tyrosine phosphorylation and decreased SHIP protein expression. BCR/ABL protein was significantly induced within 3 h of doxycycline treatment, and the level of SHIP protein was reduced within an additional hour.
These results indicate that the inhibitory effect of BCR/ABL on SHIP is both rapid and reversible. These effects were also specific for SHIP, since expression of a variety of other signaling proteins, including c-ABL, SHP2, c-CBL, PI3K, VAV, SHC, and paxillin, was not found to be changed (data not shown).
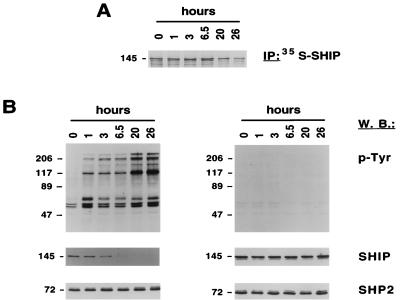
The half-life of SHIP is reduced in BCR/ABL-transformed Ba/F3 cells.
To determine if the reduction in SHIP levels was due to accelerated metabolism of the SHIP protein in BCR/ABL-transformed cells, SHIP protein half-life was measured. The half-life of SHIP was estimated at 17 h through a pulse-chase experiment in Ba/F3 cells, using a 1-h pulse with [35S]methionine (Fig. 2A). This value is consistent with a previous report of 10 h in DA-ER cells (15). In BCR/ABL-transformed cells, pulse-chase experiments were unsuccessful due to the extremely low level of SHIP protein. We therefore used a different technique to estimate SHIP half-life. BaF3/p210 cells were treated with CGP57148B for 18 h to induce SHIP expression, and then CGP57148B was washed out (Fig. 2B, left), resulting in rapid reactivation of BCR/ABL kinase activity. The subsequent decrease of SHIP protein was then measured by Western blotting. SHIP protein levels were reduced by more than half within 3 h of CGP57148B removal (Fig. 2B), suggesting that the actual half-life of SHIP in BCR/ABL cells was 3 h or less. The blots in Fig. 2 were stripped and reprobed for SHP2 to demonstrate equal loading of protein. Treatment with the ABL kinase inhibitor CGP57148B did not alter cellular tyrosine phosphorylation and did not affect SHIP levels of untransformed Ba/F3 cells (Fig. 2B, right). Therefore, the half-life of SHIP in BCR/ABL-transformed cells is likely to be significantly reduced compared to untransformed cells, although we were unable to use identical methods to compare transformed and untransformed cells. A change in SHIP protein half-life may be due to the activation of protein degradation pathway; however, specific inhibitors such as MG132 (46), lactacystin (16), and proteasome inhibitor I (62) did not alter SHIP levels in BCR/ABL-transformed cells (data not shown).
FIG. 2.
The half-life of SHIP is reduced in BCR/ABL-transformed Ba/F3 cells. (A) Ba/F3 cells labeled with [35S]methionine were lysed at 0 to 26 h after [35S]methionine removal as indicated. SHIP protein was immunoprecipitated (IP) from cell lysate of Ba/F3 cells labeled with [35S]methionine and visualized by autoradiography. (B) Ba/F3 cells and BaF3/p210 cells were treated with CGP57148B (STI571) for 18 h and lysed at 0 to 26 h after CGP57148B removal as indicated. Total cell lysate was analyzed by antiphosphotyrosine (p-Tyr), SHIP, and SHP2 Western blotting (W.B.). Molecular masses of the proteins are indicated in kilodaltons on the left of each panel.
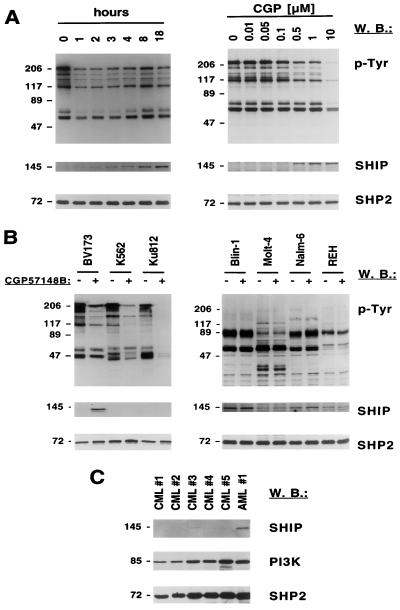
SHIP protein expression is downregulated in Ph-positive cells.
To determine if BCR/ABL alters SHIP protein expression in cell lines derived from patients with Ph-positive CML in addition to transfected cell lines, the Ph-positive CML cell line BV173 was treated with CGP57148B for 0 to 18 h. The tyrosine phosphorylation of multiple cellular proteins was reduced within 1 h of CGP57148B treatment, and SHIP protein increased within 1 h of treatment but was not maximal until 18 h (Fig. 3A, left). A significant reduction of cellular tyrosine phosphorylation and induction of SHIP protein was observed with 0.5 μM CGP57148B (Fig. 3A, right). We examined two other Ph-positive cell lines for SHIP expression and found that K562 and Ku812 cells had undetectable levels of SHIP (Fig. 3B, left). In contrast to BV173 cells, however, CGP57148B did not induce SHIP expression. We also tested the BCR/ABL-negative and Ph-negative human leukemia cell lines Blin-1, Molt-4, Nalm-6, and REH and evaluated their responses to CGP57148B (Fig. 3B, right). The ABL kinase inhibitor did not alter cellular tyrosine phosphorylation or SHIP protein expression in any of these Ph-negative cell lines. However, the ratio of SHIP protein to SHP2 protein was variable. These data suggest that SHIP protein expression may vary considerably among human leukemia cell lines, even when they do not express BCR/ABL.
FIG. 3.
CGP57148B (STI571) differentially regulates the SHIP protein expression in Ph-positive cell lines and is differentially expressed in human cell lines. (A and B) Tyrosine phosphorylation of cellular proteins was determined in total cell lysate by antiphosphotyrosine (p-Tyr) Western blotting (W.B.). The blots were stripped and reprobed for antibodies against SHIP and SHP2. Molecular masses of the proteins are indicated in kilodaltons on the left of each panel. (A) BV173 cells were treated with 1 μM CGP57148B for 0 to 18 h (left) or with 0 to 10 μM CGP57148B for 18 h (right) as indicated. (B) BV173, K562, and Ku812 (left) or Blin-1, Molt-4, Nalm-6, and REH (right) cells were treated with CGP57148B for 18 h with 1 μM CGP57148B or left untreated as indicated. (C) SHIP protein expression was determined by Western blotting in total cell lysate of primary cells from patients with Ph-positive CML and one patient with acute myeloblastic leukemia (AML). The blots were stripped and reprobed for antibodies against PI3K and SHP2.
Finally, the expression of SHIP was measured in bone marrow samples of patients with CML or a control leukemia, ALL (Ph negative), and compared to expression of other cellular proteins, including PI3K and SHP2 (Fig. 3C). In CML samples, SHIP was found to be not expressed or expressed at very low levels compared to the ALL sample. In contrast, all samples showed comparable levels of p85 PI3K and SHP2 expression, suggesting that primary CML cells also have reduced SHIP levels.
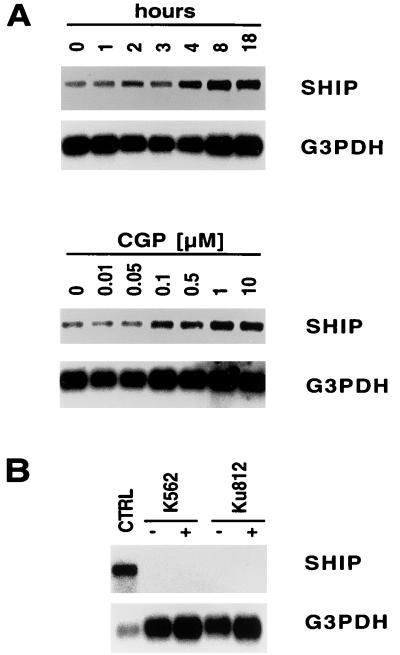
CGP57148B (STI571) induces upregulation of SHIP mRNA in BV173 but not K562 or Ku812 cells.
To determine if BCR/ABL affects expression of the SHIP gene, BV173 cells were treated for 0 to 18 h with CGP57148B, and SHIP mRNA levels were evaluated by Northern blotting (Fig. 4A, top). SHIP mRNA increased modestly with maximal induction at 8 h of CGP57148B treatment. A dose-response study of CGP57148B treatment indicates that optimal induction of SHIP mRNA was at 1 μM CGP57148B (Fig. 4A, bottom). These data suggest that increased levels of SHIP protein following CGP57148B treatment may in part be due to an increase in SHIP mRNA. In contrast, K562 cells and Ku812 cells did not show any detectable levels of SHIP mRNA before or after CGP57148B treatment (Fig. 4B). A Southern analysis was performed to determine if the SHIP gene in these two cell lines is deleted or has major structural abnormalities. The SHIP gene was found to be present in K562 and Ku812 cells and restriction digests showed banding patterns similar to that of the SHIP expressing Blin-1 cell line (data not shown).
FIG. 4.
BCR/ABL regulates the level of SHIP mRNA in BV173 cells. (A and B) Expression of SHIP mRNA and G3PDH mRNA was determined by Northern blotting using specific probes. (A) BV173 cells were treated with CGP57148B (STI571) for 0 to 18 h (top) or with 0 to 10 μM CGP57148B for 18 h (bottom) as indicated. (B) K562 and Ku812 cells were left untreated (control [CTRL]) or treated with CGP57148B for 18 h with 1 μM CGP57148B as indicated.
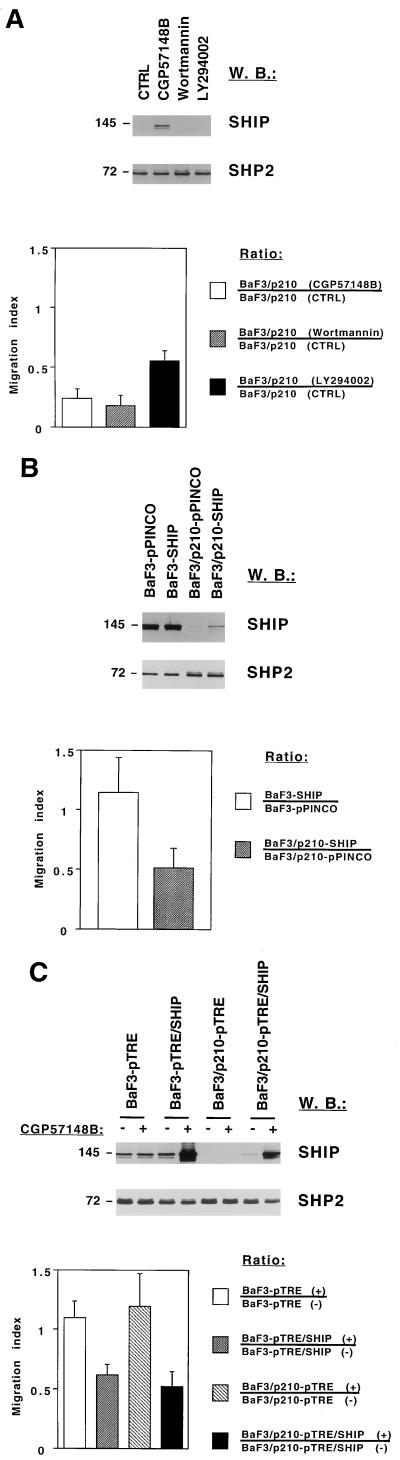
SHIP inhibits spontaneous transwell migration in BaF3/p210 cells.
We reexpressed SHIP in BaF3/p210 cells by using a retroviral expression vector or a doxycycline-inducible expression system (Fig. 5B, top; Fig. 5C, top). Compared to vector-only transfected control BaF3/p210 cells, reexpression of SHIP at the levels achieved here did not reduce growth rates, alter factor independence, or reduce viability. In addition, although SHIP has been shown to regulate AKT activation in B cells (2), the levels of phospho-AKT were not changed in BCR/ABL transformed Ba/F3 cells by reexpression of SHIP (data not shown).
FIG. 5.
SHIP regulates spontaneous transwell migration in BCR/ABL-transformed cells. SHIP and SHP2 protein expression was determined in total-cell lysate by Western blotting (W.B.). Molecular masses of the proteins are indicated in kilodaltons on the left of each panel. (Top) Cells were used for a transwell migration assay and the number of viable cells in the lower chamber was determined after 5 or 24 h (BaF3/p210-pTRE and BaF3/p210-pTRE/SHIP cells only) by trypan blue exclusion. The error bars indicate the standard error of the mean (bottom). (A) BaF3/p210 cells were pretreated for 2 h and subsequently incubated during the transwell migration assay (n = 2) with dimethyl sulfoxide (control [CTRL]), 1 μM CGP57148B (STI571), 50 nM wortmannin, and 4 μM LY294002 or used for Western blotting after 7 h of treatment. (B) Ba/F3 and BaF3/p210 cells were infected with retroviruses by using an empty vector (pPINCO) or the pPINCO vector containing the SHIP cDNA (SHIP) and used for Western blotting or transwell migration (n = 4). (C) Ba/F3 and BaF3/p210 cells transfected with a control vector (pTRE) or a SHIP expression vector (pTRE-SHIP) were either left untreated (−) or treated with doxycycline (+) and used for Western blotting or transwell migration (n = 3).
Hematopoietic cells spontaneously migrate on surfaces coated with extracellular matrix proteins when stimulated with hematopoietic growth factors, a process which can be in part quantified by a transwell migration assay (53). BCR/ABL-transformed Ba/F3 cells demonstrate a significant level of spontaneous migration which can be reduced by the ABL kinase inhibitor CGP57148B or the PI3K inhibitors wortmannin (50 nM) and LY294002 (4 μM) (Fig. 5A). Treatment of BaF3/p210 cells with CGP57148B but not with wortmannin or LY294002 increased protein expression of SHIP (Fig. 5A, top). In a typical experiment 0.45 × 105 out of 2 × 105 untreated BaF3/p210 cells migrated within 5 h through the transwell membrane.
To determine if overexpressing SHIP in BaF3/p210 cells altered migration, cells were infected with a retrovirus (pPINCO) expressing SHIP under the control of a long terminal repeat promoter and GFP under the control of a separate cytomegalovirus promoter. All cell lines had comparable GFP expression, with 55 to 62% positive cells by fluorescence-activated cell sorting analysis. Increased SHIP expression was detected in BaF3/p210 cells infected with the pPINCO-SHIP virus but not the control virus (Fig. 5B, top). The increase in SHIP protein did not reach levels present in the parent Ba/F3 cells. Migration of cells containing the empty vector (BaF3-pPINCO and BaF3/p210-pPINCO) or the pPINCO-SHIP vector (BaF3-SHIP and Ba3/p210-SHIP) was determined in a transwell migration chamber and expressed as a migration index. The mean (n = 4) of the migration indices of BaF3-SHIP compared to BaF3-pPINCO cells was 1.15, and that of BaF3/p210-SHIP compared to BaF3/p210-pPINCO cells was 0.51 (Fig. 5B, bottom), indicating that SHIP significantly decreased the transwell migration of BaF3/p210 cells but not of Ba/F3 cells.
The effect of SHIP on transwell migration was also investigated in the BaF3-pTRE/SHIP and BaF3/p210-pTRE/SHIP cells, where SHIP was under the control of a tetracycline-inducible promoter. Doxycycline treatment resulted in expression of SHIP severalfold higher than in the untreated cells or the parent BaF3-pTRE and BaF3/p210-pTRE cells (Fig. 5C, top). However, there was already some increased expression of SHIP in doxycycline-untreated BaF3/p210-pTRE/SHIP cells compared to untreated BaF3/p210-pTRE cells, likely indicating that the promoter is leaky. It can also be appreciated that overexpression of the SHIP protein in Ba/F3 as well as BaF3/p210 cells leads to the appearance of additional bands that are immunoreactive with the SHIP antibody. Most of these bands have a molecular weight lower than that of full-length SHIP, suggesting these might be degraded forms, consistent with previous reports (15).
Increased expression of SHIP decreased spontaneous migration of both Ba/F3 and BaF3/p210 cells (Fig. 5C, bottom). The means (n = 3) of the migration indices were found to be 1.10 for BaF3-pTRE cells and 1.20 for BaF3/p210-pTRE cells, indicating that doxycycline treatment alone did not alter migration. In contrast, the mean of migration indices for BaF3-pTRE/SHIP was 0.62 and that for BaF3/p210-pTRE/SHIP was 0.52, indicating that when overexpressed at very high levels, SHIP reduces transwell migration in Ba/F3 as well as BaF3/p210 cells. In addition, in four independent experiments, the migration in SHIP-overexpressing cells using uncoated surfaces was reduced by 43% compared to control cells, suggesting that this effect is independent of extracellular matrix proteins.
DISCUSSION
We demonstrate here that the expression of the phosphatidylinositol-5-phosphatase SHIP is rapidly and reversibly downregulated by BCR/ABL and that this requires ABL kinase activity. SHIP levels are not altered in cells expressing a kinase-dead form of BCR/ABL, and SHIP rapidly reaccumulates in cells treated with a small-molecule ABL kinase inhibitor, CGP57148B (STI571). In addition, induction of the BCR/ABL protein in a tetracycline-inducible system correlates with downregulation of the SHIP protein. This downregulation of SHIP protein is specific since a variety of other signaling proteins, such as SHP2, PI3K, c-ABL, c-CBL, VAV, SHC, or paxillin, are not affected by BCR/ABL. The mechanism may be multifactorial since BCR/ABL not only reduces the estimated half-life of SHIP protein but also downregulates the amount of SHIP mRNA. Both processes can be rapidly blocked with the CGP57148B inhibitor. These observations are of note, since disruption of the SHIP gene by gene targeting results in a myeloproliferative disorder in mice (28) with many similarities to the myeloproliferative syndrome caused by BCR/ABL in mice (13, 18, 25, 32). Thus, the results presented here implicate SHIP in the pathogenesis of the myeloproliferative syndrome characteristic of CML.
The downregulation of SHIP reported here is unique. Although there are many known signaling proteins which are tyrosine phosphorylated by BCR/ABL, overall expression of other kinase targets is generally not affected. The protein tyrosine phosphatase PTP-1B has been reported to be upregulated by BCR/ABL; this is of particular interest since PTP1B coimmunoprecipitates with and dephosphorylates BCR/ABL at Tyr177, therefore inhibiting binding of the adapter protein GRB2 and suppressing RAS-dependent transcriptional activation (34). Thus, increased levels of PTP-1B in BCR/ABL-transformed cells should suppress transformation (34). Expression of Abi (Abl-interacting) protein has been shown to be reduced in cells transformed by BCR/ABL through activation of the ubiquitin-proteasome pathway (12). Similar to expression of SHIP, Abi expression is nearly undetectable in some cells from patients with Ph-positive leukemias. Abi proteins are negative regulators of transformation by ABL kinases, suggesting that loss of Abi proteins could contribute to transformation. To date, however, a direct role for Abi has not been shown in BCR/ABL transformation. Abi interacts with BCR/ABL through the SH3 domain of ABL. In addition, Abi proteins can also bind to ABL C-terminal sequences (11, 55). The fact that BCR/ABL downregulates Abi expression by inducing ubiquitination suggests that SHIP may also be ubiquitinated and degraded through the same mechanism. However, treatment of BaF3/p210 cells for 6 h with the proteasome pathway inhibitors MG132 (5 μM), lactacystin (10 μM), and proteasome inhibitor 1 (1 μM) did not significantly change expression or ubiquitination of SHIP protein, detected by SHIP and ubiquitin immunoblotting in SHIP immunoprecipitations (data not shown).
Additional studies are ongoing to define the role of ubiquitination, if any, in SHIP regulation. Degradation of SHIP unrelated to BCR/ABL by an unknown mechanism has been found by Damen et al. in the murine hematopoietic cell line DA-ER (15). In these cells, antibodies against the 145-kDa SHIP protein also identified several bands of smaller size. It was suggested that a calpain-like protease is involved in the degradation of SHIP in these cells and that the truncated forms could have different signaling properties. For example, a smaller 110-kDa form of SHIP still retained PtdIns(3,4,5)P3-specific 5′-phosphatase activity but, in contrast to the 145-kDa form of SHIP, was exclusively localized to the cytoskeleton and could not bind SHC after interleukin-3 stimulation (15). However, at least one of the smaller SHIP-reactive bands has been identified as a spliced form of SHIP rather than a degradation product (36). We have also observed increased levels of smaller forms of SHIP, especially when a SHIP cDNA is overexpressed in Ba/F3 and BaF3/p210 cells. Studies to determine if these smaller forms are degradation products of SHIP are under way.
In addition to the changes in SHIP protein levels, we found an increase of SHIP mRNA in CGP57148B-treated BV173 cells. SHIP mRNA was already present at significant levels in untreated cells, and the increase in mRNA after CGP57148B treatment did not reflect the more substantial increase in protein levels. This suggests that BCR/ABL-induced regulation of SHIP mRNA alone is probably not sufficient to account for all of the change in altered SHIP protein levels in these cells. BCR/ABL has been shown to regulate the expression of several genes and the activity of transcription factors such as STAT5 (8, 30, 57). Some of the genes that are known to be regulated by BCR/ABL, including c-myc (60) and c-jun, are involved in cell cycle regulation (6). Other genes, such as the inosine 5′-monophosphate dehydrogenase gene (20), involved in de novo guanylate synthesis, or several ribosomal genes (10) are likely to be regulated as a result of increased proliferation.
In contrast to BV173 cells, the two Ph-positive cell lines K562 and Ku812 had undetectable levels of SHIP mRNA and protein, and the complete loss of SHIP was irreversible when BCR/ABL kinase activity was inhibited by CGP57148B treatment. Southern blot analysis revealed that at least one copy of the SHIP gene is present in these two cell lines but did not reveal rearrangement of the gene. It is possible that there are smaller mutations which inactivate expression. The identification of mutations which reduce SHIP expression or function in these Ph-positive cell lines would provide additional evidence that SHIP functions as a tumor suppressor gene in CML.
The mechanism whereby loss of SHIP expression causes a myeloproliferative disease is unknown. A reasonable hypothesis, however, is that SHIP normally modulates levels of inositol lipids formed in response to activation of PI3K and related enzymes. PI3K activity is important for normal hematopoietic cells (19, 54, 61) and for transformation by BCR/ABL (58). Recently the expression and tyrosine phosphorylation of the SHIP-related protein SHIP2 in CML cells had been described (66). SHIP2, like SHIP, has PtdIns(3,4,5)P3-specific 5′-phosphatase activity (24, 66), but in contrast, the SHIP2 protein levels are not changed in BCR/ABL-transformed cells (66). It is unlikely that the activities of SHIP2 and SHIP are redundant, since SHIP−/− mice have a severe hematopoietic phenotype.
Vollenweider et al. have reported that expression of SHIP after nuclear microinjection inhibits membrane ruffling induced by insulin, insulin-like growth factor I, and platelet-derived growth factor in 3T3-L1 adipocytes (65). This effect of SHIP on the cytoskeleton was restricted since growth factor-induced stress fiber breakdown was not affected (65). This finding further supports the notion that SHIP substrates are likely to play an important role in cytoskeletal rearrangements. Interestingly, disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes in yeast with similarities to SHIP results in cytoskeletal abnormalities (59). Nevertheless, we did not detect a significant difference in F-actin content in BaF3/p210 cells overexpressing SHIP compared to wild-type cells (data not shown).
We have investigated the effects of overexpressing SHIP in BCR/ABL-transformed cells by transfection using two different approaches. Overexpressing SHIP at moderate levels with a retrovirus led to a decrease in spontaneous transwell migration of BCR/ABL-transformed Ba/F3 cells. However, with a tetracycline-inducible promoter to express SHIP at very high levels, there was also a significant reduction in transwell migration in untransformed cells. Abnormal cytoskeletal function has previously been linked to the transformed phenotype in CML cells. We have previously shown that BCR/ABL induces cytoskeletal abnormalities that affect morphology, motility, and adhesion (48). Also, CML progenitor cells were shown to have diminished ability to adhere to stromal cells (22) and reduced long-term adhesion to fibronectin (64). It has been suggested that altered cytoskeletal function leads to premature release of CML cells from the marrow and accumulation of myeloid cells in the blood. The regulation of cytoskeletal function is equally important for the regulation of normal hematopoiesis (63). Downregulation of SHIP by BCR/ABL and increased cell migration may therefore contribute to an increased release of cells from the bone marrow.
Considering the results presented here, it will be of interest to further compare the mechanism inducing the myeloproliferative disease in SHIP knockout mice with that of BCR/ABL. By identifying critical signaling pathways affected by loss of SHIP expression, it may be possible that disease-specific, rational drug design can take place. In any case, it is likely that further characterization of the myeloproliferative phenotype in the SHIP knockout mice will help to understand the mechanism of transformation by BCR/ABL in CML.
ACKNOWLEDGMENTS
This work was supported by Leukemia Foundation fellowship FIJC-95/INT (M.S.) and NIH grant DK50654 (J.D.G.).
REFERENCES
- 1.Afar D E, Goga A, McLaughlin J, Witte O N, Sawyers C L. Differential complementation of BCR-ABL point mutants with c-Myc. Science. 1994;264:424–426. doi: 10.1126/science.8153630. [DOI] [PubMed] [Google Scholar]
- 2.Aman M J, Lamkin T D, Okada H, Kurosaki T, Ravichandran K S. The inositol phosphatase SHIP inhibits Akt/PKB activation in B cells. J Biol Chem. 1998;273:33922–33928. doi: 10.1074/jbc.273.51.33922. [DOI] [PubMed] [Google Scholar]
- 3.Andoniou C E, Thien C, Langdon W Y. Tumour induction by activated Abl involves tyrosine phosphorylation of the product of the Cbl oncogene. EMBO J. 1994;13:4515–4523. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedi A, Zehnbauer B A, Barber J P, Sharkis S J, Jones R J. Inhibition of apoptosis by BCR-ABL in chronic myeloid leukemia. Blood. 1994;83:2038–2044. [PubMed] [Google Scholar]
- 5.Ben-Neriah Y, Daley G Q, Mes-Masson A M, Witte O N, Baltimore D. The chronic myelogenous leukemia-specific p210 protein is the product of the BCR/ABL hybrid gene. Science. 1986;233:212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- 6.Burgess G S, Williamson E A, Cripe L D, Litz-Jackson S, Bhatt J A, Stanley K, Stewart M J, Kraft A S, Nakshatri H, Boswell H S. Regulation of the c-jun gene in p210 BCR-ABL transformed cells corresponds with activity of JNK, the c-jun N-terminal kinase. Blood. 1998;92:2450–2460. [PubMed] [Google Scholar]
- 7.Canellos G. Diagnosis and treatment of chronic granulocytic leukemia. In: Wiernik P H, Canellos G P, Kyle R A, et al., editors. Neoplastic disease of the blood. New York, N.Y: Churchill Livingstone; 1991. pp. 61–76. [Google Scholar]
- 8.Carlesso N, Frank D A, Griffin J D. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by BCR/ABL. J Exp Med. 1996;183:811–820. doi: 10.1084/jem.183.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chacko G W, Tridandapani S, Damen J E, Liu L, Krystal G, Coggeshall K M. Negative signaling in B lymphocytes induces tyrosine phosphorylation of the 145-kDa inositol polyphosphate 5-phosphatase, SHIP. J Immunol. 1996;157:2234–2238. [PubMed] [Google Scholar]
- 10.Daheron L, Salmeron S, Patri S, Brizard A, Guilhot F, Chomel J C, Kitzis A. Identification of several genes differentially expressed during progression of chronic myelogenous leukemia. Leukemia. 1998;12:326–332. doi: 10.1038/sj.leu.2400923. [DOI] [PubMed] [Google Scholar]
- 11.Dai Z H, Pendergast A M. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 1995;9:2569–2582. doi: 10.1101/gad.9.21.2569. [DOI] [PubMed] [Google Scholar]
- 12.Dai Z H, Quackenbush R C, Courtney K D, Grove M, Cortez D, Reuther G W, Pendergast A M. Oncogenic Abl and Src tyrosine kinases elicit the ubiquitin-dependent degradation of target proteins through a Ras-independent pathway. Genes Dev. 1998;12:1415–1424. doi: 10.1101/gad.12.10.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daley G Q, Van Etten R A, Baltimore D. Induction of chronic myelogenous leukemia in mice by the p210BCR/ABL gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 14.Damen J E, Liu L, Rosten P, Humphries R K, Jefferson A B, Majerus P W, Krystal G. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase. Proc Natl Acad Sci USA. 1996;93:1689–1693. doi: 10.1073/pnas.93.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damen J E, Liu L, Ware M D, Ermolaeva M, Majerus P W, Krystal G. Multiple forms of the SH2-containing inositol phosphatase, SHIP, are generated by C-terminal truncation. Blood. 1998;92:1199–1205. [PubMed] [Google Scholar]
- 16.Dick L R, Cruikshank A A, Destree A T, Grenier L, McCormack T A, Melandri F D, Nunes S L, Palombella V J, Parent L A, Plamondon L, Stein R L. Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272:182–188. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- 17.Druker B J, Tamura S, Buchdunger E, Ohno S, Segal G M, Fanning S, Zimmermann J, Lydon N B. Effects of a selective inhibitor of the ABL tyrosine kinase on the growth of BCR-ABL positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 18.Elefanty A G, Hariharan I K, Cory S. BCR-ABL, the hallmark of chronic myeloid leukaemia in man, induces multiple haemopoietic neoplasms in mice. EMBO J. 1990;9:1069–1078. doi: 10.1002/j.1460-2075.1990.tb08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fruman D A, Snapper S B, Yballe C M, Davidson L, Yu J Y, Alt F W, Cantley L C. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85 alpha. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 20.Gharehbaghi K, Burgess G S, Collart F R, Litz-Jackson S, Huberman E, Jayaram H N, Boswell H S. p210 BCR-ABL confers overexpression of inosine monophosphate dehydrogenase: an intrinsic pathway to drug resistance mediated by oncogene. Leukemia. 1994;8:1257–1263. [PubMed] [Google Scholar]
- 21.Golub T R, Goga A, Barker G F, Afar D, McLaughlin J, Bohlander S K, Rowley J D, Witte O N, Gilliland D G. Oligomerization of the Abl tyrosine kinase by the Ets protein Tel in human leukemia. Mol Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon M Y, Dowding C R, Riley G P, Goldman J M, Greaves M F. Altered adhesive interactions with marrow stroma of haematopoietic progenitor cells in chronic myeloid leukaemia. Nature. 1987;328:342–344. doi: 10.1038/328342a0. [DOI] [PubMed] [Google Scholar]
- 23.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, Lanfrancone L, Peschle C, Nolan G P, Pelicci P G. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid ebv/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 24.Habib T, Hejna J A, Moses R E, Decker S J. Growth factors and insulin stimulate tyrosine phosphorylation of the 51C/SHIP2 protein. J Biol Chem. 1998;273:18605–18609. doi: 10.1074/jbc.273.29.18605. [DOI] [PubMed] [Google Scholar]
- 25.Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale P K, Groffen J. Acute leukaemia in BCR/ABL transgenic mice. Nature. 1990;344:251–253. doi: 10.1038/344251a0. [DOI] [PubMed] [Google Scholar]
- 26.Heisterkamp N, Stephenson J R, Groffen J, Hansen P F, de Klein A, Bartram C R, Grosveld G. Localization of the c-Abl oncogene adjacent to a translocation breakpoint in chronic myelocytic leukaemia. Nature. 1983;306:239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 27.Hejna J A, Saito H, Merkens L S, Tittle T V, Jakobs P M, Whitney M A, Grompe M, Friedberg A S, Moses R E. Cloning and characterization of a human cDNA (INNPL1) sharing homology with inositol polyphosphate phosphatases. Genomics. 1995;29:285–287. doi: 10.1006/geno.1995.1247. [DOI] [PubMed] [Google Scholar]
- 28.Helgason C D, Damen J E, Rosten P, Grewal R, Sorensen P, Chappel S M, Borowski A, Jirik F, Krystal G, Humphries R K. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter M G, Avalos B R. Phosphatidylinositol 3′-kinase and SH2-containing inositol phosphatase (SHIP) are recruited by distinct positive and negative growth-regulatory domains in the granulocyte colony-stimulating factor receptor. J Immunol. 1998;160:4979–4987. [PubMed] [Google Scholar]
- 30.Ilaria R L, Vanetten R A. P210 and p190BCR/ABL induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 31.Jiang X, Fujisaki T, Berger M, Eaves A, Eaves C. Abst. 1041: autonomous multi-lineage differentiation in vitro of primitive Ph+ CD34+ cells from patients with chronic myelogenous leukemia (CML) Blood Suppl. 1998;90:254a. doi: 10.1038/sj.leu.2401752. [DOI] [PubMed] [Google Scholar]
- 32.Kelliher M A, McLaughlin J, Witte O N, Rosenberg N. Induction of a chronic myelogenous leukemia-like syndrome in mice with v-abl and BCR/ABL. Proc Natl Acad Sci USA. 1990;87:6649–6653. doi: 10.1073/pnas.87.17.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klucher K M, Lopez D V, Daley G Q. Secondary mutation maintains the transformed state in BaF3 cells with inducible Bcr/Abl expression. Blood. 1998;91:3927–3934. [PubMed] [Google Scholar]
- 34.Lamontagne K R, Flint A J, Franza B R, Pendergast A M, Tonks N K. Protein tyrosine phosphatase 1B antagonizes signalling by oncoprotein tyrosine kinase p210 bcr-abl in vivo. Mol Cell Biol. 1998;18:2965–2975. doi: 10.1128/mcb.18.5.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lioubin M N, Algate P A, Tsai S, Carlberg K, Aebersold R, Rohrschneider L R. p150-SHIP, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 36.Lucas D M, Rohrschneider L R. A novel spliced form of SH2-containing inositol phosphatase is expressed during myeloid development. Blood. 1999;93:1922–1933. [PubMed] [Google Scholar]
- 37.Matsuguchi T, Salgia R, Hallek M, Eder M, Druker B, Ernst T, Griffin J. SHC phosphorylation in myeloid cells is regulated by granulocyte macrophage colony-stimulating factor, interkeukin-3, and steel factor and is constitutively increased by p210BCR/ABL. J Biol Chem. 1994;269:5016–5021. [PubMed] [Google Scholar]
- 38.McWhirter J R, Galasso D L, Wang J Y. A coiled-coil oligomerization domain of BCR is essential for the transforming function of BCR-ABL oncoproteins. Mol Cell Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odai H, Sasaki K, Iwamatsu A, Hanazono Y, Tanaka T, Mitani K, Yazaki Y, Hirai H. The proto-oncogene product c-Cbl becomes tyrosine phosphorylated by stimulation with GM-CSF or EPO and constitutively binds to the SH3 domain of GRB2/ASH in human hematopoietic cells. J Biol Chem. 1995;270:10800–10805. doi: 10.1074/jbc.270.18.10800. [DOI] [PubMed] [Google Scholar]
- 40.Osborne M A, Zenner G, Lubinus M, Zhang X L, Songyang Z, Cantley L C, Majerus P, Burn P, Kochan J P. The inositol 5′-phosphatase SHIP binds to immunoreceptor signaling motifs and responds to high affinity IgE receptor aggregation. J Biol Chem. 1996;271:29271–29278. doi: 10.1074/jbc.271.46.29271. [DOI] [PubMed] [Google Scholar]
- 41.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pendergast A M, Quilliam L A, Cripe L D, Bassing C H, Dai Z, Li N, Batzer A, Rabun K M, Der C J, Schlessinger J, Gishizky M L. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell. 1993;75:175–185. [PubMed] [Google Scholar]
- 43.Pesesse X, Deleu S, Desmedt F, Drayer L, Erneux C. Identification of a second SH2-domain-containing protein closely related to the phosphatidylinositol polyphosphate 5-phosphatase SHIP. Biochem Biophys Res Commun. 1997;239:697–700. doi: 10.1006/bbrc.1997.7538. [DOI] [PubMed] [Google Scholar]
- 44.Puil L, Liu J, Gish G, Mbamalu G, Bowtell D, Pelicci P G, Arlinghaus R, Pawson T. BCR-ABL oncoproteins bind directly to activators of the Ras signalling pathway. EMBO J. 1994;13:764–773. doi: 10.1002/j.1460-2075.1994.tb06319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renshaw M W, McWhirter J R, Wang J Y. The human leukemia oncogene BCR-ABL abrogates the anchorage requirement but not the growth factor requirement for proliferation. Mol Cell Biol. 1995;15:1286–1293. doi: 10.1128/mcb.15.3.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 47.Rowley J D. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 48.Salgia R, Li J L, Ewaniuk D S, Pear W, Pisick E, Burky S A, Ernst T, Sattler M, Chen L B, Griffin J D. BCR/ABL induces multiple abnormalities of cytoskeletal function. J Clin Investig. 1997;100:46–57. doi: 10.1172/JCI119520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salgia R, Li J L, Lo S H, Brunkhorst B, Kansas G S, Sobhany E S, Sun Y P, Pisick E, Hallek M, Ernst T, Tantravahi R, Chen L B, Griffin J D. Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by p210BCR/ABL. J Biol Chem. 1995;270:5039–5047. doi: 10.1074/jbc.270.10.5039. [DOI] [PubMed] [Google Scholar]
- 50.Sattler M, Durstin M A, Frank D A, Okuda K, Kaushansky K, Salgia R, Griffin J D. The thrombopoietin receptor c-MPL activates JAK2 and TYK2 tyrosine kinases. Exp Hematol. 1995;23:1040–1048. [PubMed] [Google Scholar]
- 51.Sattler M, Salgia R, Durstin M A, Prasad K V, Griffin J D. Thrombopoietin induces activation of the phosphatidylinositol-3′ kinase pathway and formation of a complex containing p85PI3K and the protooncoprotein p120CBL. J Cell Physiol. 1997;171:28–33. doi: 10.1002/(SICI)1097-4652(199704)171:1<28::AID-JCP4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 52.Sattler M, Salgia R, Shrikhande G, Verma S, Choi J L, Rohrschneider L R, Griffin J D. The phosphatidylinositol polyphosphate 5-phosphatase SHIP and the protein tyrosine phosphatase SHP-2 form a complex in hematopoietic cells which can be regulated by BCR/ABL and growth factors. Oncogene. 1997;15:2379–2384. doi: 10.1038/sj.onc.1201422. [DOI] [PubMed] [Google Scholar]
- 53.Sattler M, Winkler T, Verma S, Byrne C H, Shrikhande G, Salgia R, Griffin J D. Hematopoietic growth factors signal through the formation of reactive oxygen species. Blood. 1999;93:2928–2935. [PubMed] [Google Scholar]
- 54.Serve H, Yee N S, Stella G, Sepp-Lorenzino L, Tan J C, Besmer P. Differential roles of PI3-kinase and Kit tyrosine 821 in Kit receptor-mediated proliferation, survival and cell adhesion in mast cells. EMBO J. 1995;14:473–483. doi: 10.1002/j.1460-2075.1995.tb07023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi Y G, Alin K, Goff S P. Abl-interactor-1, a novel SH3 protein binding to the carboxy-terminal portion of the Abl protein, suppresses v-Abl transforming activity. Genes Dev. 1995;9:2583–2597. doi: 10.1101/gad.9.21.2583. [DOI] [PubMed] [Google Scholar]
- 56.Shtivelman E, Lifshitz B, Gale R P, Canaani E. Fused transcript of Abl and Bcr genes in chronic myelogenous leukaemia. Nature. 1985;315:550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 57.Shuai K, Halpern J, ten Hoeve J, Rao X P, Sawyers C L. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene. 1996;13:247–254. [PubMed] [Google Scholar]
- 58.Skorski T, Kanakaraj P, Nieborowska-Skorska M, Ratajczak M Z, Wen S C, Zon G, Gewirtz A M, Perussia B, Calabretta B. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood. 1995;86:726–736. [PubMed] [Google Scholar]
- 59.Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy S F, Emr S, Decamilli P, Nussbaum R. Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol. 1997;74:350–360. [PubMed] [Google Scholar]
- 60.Stewart M J, Litz-Jackson S, Burgess G S, Williamson E A, Leibowitz D S, Boswell H S. Role for E2F1 in p210 BCR-ABL downstream regulation of c-myc transcription initiation. Studies in murine myeloid cells. Leukemia. 1995;9:1499–1507. [PubMed] [Google Scholar]
- 61.Suzuki H, Terauchi Y, Fujiwara M, Aizawa S, Yazaki Y, Kadowaki T, Koyasu S. Xid-like immunodeficiency in mice with disruption of the p85 alpha subunit of phosphoinositide 3-kinase. Science. 1999;283:390–392. doi: 10.1126/science.283.5400.390. [DOI] [PubMed] [Google Scholar]
- 62.Traenckner E B, Wilk S, Baeuerle P A. A proteasome inhibitor prevents activation of NF-kappa B and stabilizes a newly phosphorylated form of I kappa B-alpha that is still bound to NF-kappa B. EMBO J. 1994;13:5433–5441. doi: 10.1002/j.1460-2075.1994.tb06878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verfaillie C M, Hurley R, Lundell B I, Zhao C H, Bhatia R. Integrin-mediated regulation of hematopoiesis—do BCR/ABL-induced defects in integrin function underlie the abnormal circulation and proliferation of CML progenitors. Acta Haematol. 1997;97:40–52. doi: 10.1159/000203658. [DOI] [PubMed] [Google Scholar]
- 64.Verfaillie C M, McCarthy J B, McGlave P B. Mechanisms underlying abnormal trafficking of malignant progenitors in chronic myelogenous leukemia. Decreased adhesion to stroma and fibronectin but increased adhesion to the basement membrane components laminin and collagen type IV. J Clin Investig. 1992;90:1232–1241. doi: 10.1172/JCI115985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vollenweider P, Clodi M, Martin S S, Imamura T, Kavanaugh W M, Olefsky J M. An SH2 domain-containing 5 ′ inositolphosphatase inhibits insulin-induced GLUT4 translocation and growth factor-induced actin filament rearrangement. Mol Cell Biol. 1999;19:1081–1091. doi: 10.1128/mcb.19.2.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wisniewski D, Strife A, Swendeman S, Erdjument-Bromage H, Geromanos S, Kavanaugh W M, Tempst P, Clarkson B. A novel SH2-containing phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase (SHIP2) is constitutively tyrosine phosphorylated and associated with src homologous and collagen gene (SHC) in chronic myelogenous leukemia progenitor cells. Blood. 1999;93:2707–2720. [PubMed] [Google Scholar]