MdmX Protects p53 from Mdm2-Mediated Degradation (original) (raw)
Abstract
The p53 tumor suppressor protein is stabilized in response to cellular stress, resulting in activation of genes responsible for either cell cycle arrest or apoptosis. The cellular pathway for releasing normal cells from p53-dependent cell cycle arrest involves the Mdm2 protein. Recently, a p53-binding protein with homology to Mdm2 was identified and called MdmX. Like Mdm2, MdmX is able to bind p53 and inhibit p53 transactivation; however, the ability of MdmX to degrade p53 has yet to be examined. We report here that MdmX is capable of associating with p53 yet is unable to facilitate nuclear export or induce p53 degradation. In addition, expression of MdmX can reverse Mdm2-targeted degradation of p53 while maintaining suppression of p53 transactivation. Using a series of MdmX deletions, we have determined that there are two distinct domains of the MdmX protein that can stabilize p53 in the presence of Mdm2. One domain requires MdmX interaction with p53 and results in the retention of both proteins within the nucleus and repression of p53 transactivation. The second domain involves the MdmX ring finger and results in stabilization of p53 and an increase in p53 transactivation. The potential basis for stabilization and increased p53 transactivation by the MdmX ring finger domain is discussed. Based on these observations, we propose that the MdmX protein may function to maintain a nuclear pool of p53 protein in undamaged cells.
Following genotoxic stress, the p53 tumor suppressor protein is induced and depending on the cell type elicits either a cell cycle arrest or apoptosis (1). These cellular effects of p53 are mediated predominately by the ability of p53 to transactivate specific genes. For example, p53-induced G1 and G2 arrests require the ability of p53 to activate genes such as p21 and 14-3-3ς (7, 11). In contrast, the ability of p53 to trigger apoptosis has been linked to activation of genes such as bax-1 (20), Killer/DR5 (34), and genes involved in oxygen free radical production (25).
While many human tumors evade the growth constraints of p53 by harboring p53 genetic alterations (mutation or deletion) or by inactivation of p53 protein via specific viral proteins, the cellular pathway that releases a normal cell from p53-dependent cell cycle arrest involves the Mdm2 protein. The gene encoding the Mdm2 protein was initially characterized as a proto-oncogene found amplified in a transformed 3T3DM cell line (6). Later it was discovered that Mdm2 could bind to p53, repress p53-dependent transactivation of target genes (21, 24), and promote rapid degradation of p53 through the ubiquitin-proteosome pathway (10, 16). In fact, the mdm2 gene can be activated following genotoxic stress by p53 (2, 35), providing a mechanism by which p53 can autoregulate its own function. Not surprisingly, overexpression of Mdm2 results in cellular transformation of cells possessing wild-type p53 (8, 30).
Recent studies have delineated the requirements for Mdm2-mediated degradation of p53. First, both a nuclear localization sequence and a nuclear export sequence (NES) must be present on Mdm2 in order for nucleocytoplasmic shuttling of p53-Mdm2 complexes to occur (33). Export of Mdm2-p53 complexes has been shown to occur via the CRM1-dependent nuclear export pathway since mutation of the Mdm2 NES or inhibition of CRM1 export with leptomycin B results in increased p53 protein stability (10, 16, 27). Second, cysteine 464 of the Mdm2 ring finger domain has been shown to be essential for the ability of Mdm2 to act as a ubiquitin ligase E3 for p53 in vitro (12) as well as its ability to mediate degradation of p53 in vivo (18). Third, specific regions of the p53 protein itself are required for efficient degradation. The Mdm2-binding domain and the oligomerization domain of p53 are required; additional evidence also indicates the involvement of the extreme C-terminal region of p53. Deletion of merely 16 amino acids renders p53 resistant to degradation even though the protein is still capable of oligomerization and interaction with Mdm2 (17).
In 1996, a p53-binding protein with considerable homology to Mdm2 was discovered and termed MdmX (29). Like the mdm2 gene, mdmX is found expressed in all murine and human tissues. However, while mdm2 gene expression is regulated in a p53-dependent manner (2, 35), mdmX gene expression does not modulate in response to DNA damage or p53 (29). Comparison of the p53 interaction domains of MdmX and Mdm2 by using phage-displayed peptides demonstrates nearly identical p53 binding pocket conformations for both proteins (5). In addition, the primary sequence of MdmX demonstrates a strong homology to Mdm2 within the zinc and ring finger domains. However, while Mdm2 and MdmX appear structurally similar, genetic evidence suggests that mdmX cannot substitute for mdm2 during early embryonic development (15, 22), perhaps indicating a divergence in function between these proteins. Recently it was shown that Mdm2 and MdmX form stable heterodimers through their ring finger domains and that this interaction resulted in a substantial increase in the half-life of Mdm2 (32).
In the present study, we demonstrate that MdmX can reverse Mdm2-targeted degradation of p53 while maintaining suppression of p53 transactivation. Using a series of MdmX deletions, we have determined that there are two distinct domains of MdmX that can stabilize p53 in the presence of Mdm2. Based on these observations, we propose that the MdmX protein functions to maintain a nuclear pool of transcriptionally inactive p53 protein in undamaged cells.
MATERIALS AND METHODS
Cell lines and antibodies.
2KO cells are embryo fibroblasts derived from mice lacking both p53 and mdm2 genes (generously provided by Guillermina Lozano). H1299 cells are a non-small-cell lung carcinoma line devoid of p53. HeLa and NIH 3T3 cells were obtained from the American Type Culture Collection. All cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and 10 μg of gentamicin per ml. p53-specific antibodies FL 1-393 (Santa Cruz Biotechnology, Inc.) and Ab421 (Oncogene) were used. MdmX and Mdm2 polyclonal rabbit antibodies were generated by using bacterially expressed recombinant protein and do not show cross-reactivity (data not shown). The MdmX antibody was purified by using an Immunopure immunoglobulin G purification kit (Pierce). The polyclonal Mdm2 antibody 2A10 was a kind gift from Gerald Zambetti, St. Jude Research Hospital. A horseradish peroxidase-conjugated secondary antibody (Promega) was used for chemiluminescence detection of proteins.
Plasmids.
Cytomegalovirus (CMV)-based constructs CMVp53, CMVmdmX, CMVhdm2, and CMVmdm2 were generated by cloning each wild-type cDNA (including the stop codons) into pcDNA3.1mychis2 (Invitrogen) such that the c-Myc epitope and six-histidine tag are not in frame with the cDNAs. CMVmdm2ΔRF was generated by cloning the mdm2 cDNA encoding the first 467 amino acids in frame with the c-Myc epitope and six-histidine tag. Enhanced green fluorescent protein (EGFP)-expressing constructs CMVmdmX-EGFP and CMVmdm2-EGFP were constructed by cloning the respective cDNA into pEGFP-N1 (Clontech). MdmX deletion mutants were constructed by PCR using Pfu DNA polymerase. All cDNA constructs were confirmed by DNA sequencing. CMVHPV16-E6 was a gift from Kathleen R. Cho. PG13CAT and MG15CAT contain 13 copies of the p53 consensus sequence and 15 copies of a mutated p53 consensus sequence, respectively. Both PG13CAT and MG15CAT were gifts from Bert Vogelstein. pGL3Basic and CMVRLuc (both from Promega) were used for normalization of transfection efficiency.
Transfections.
H1299, HeLa, and MEF 2KO cells were transiently transfected with the indicated amounts of plasmid by using Lipofectamine (Gibco BRL). After a 5-h incubation in DMEM lacking serum and antibiotics, the cells were refed with DMEM containing serum and antibiotics, and whole-cell extracts were made 24 h later. Whole-cell extracts were made by incubating frozen cell pellets in a 2× volume of either single lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1.0% NP-40, 1.0 μg of aprotinin per ml, 100 μg of phenylmethylsulfonyl fluoride per ml) or PBSA (phosphate-buffered saline [PBS] containing 5 mM EDTA and 0.5% Triton X-100). In all degradation assays, transfection efficiency was normalized to that of either pGL3Basic or CMVRLuc and quantified from whole-cell extracts with a luciferase assay system (Promega).
Protein analysis.
The immunoprecipitation for Fig. 2 was performed by incubating each extract with 250 μl of LSAB (100 mM NaCl, 100 mM Tris [pH 8.0], 0.5% NP-40, 10 mM dithiothreitol, 100 μg of phenylmethylsulfonyl fluoride per ml) with or without 4.0 μg of Ab421 per ml for 1 h. Protein G-agarose (25 μl of a 50% PBS slurry) was added to each reaction and incubated an additional 1 h. The protein G-agarose–antibody complexes were washed three times in LSAB and then resuspended in a sodium dodecyl sulfate loading dye. The immunoprecipitated proteins were analyzed via Western analysis using a polyclonal antibody for p53 (FL 1-393) or MdmX. For Fig. 6, the cells were lysed in 300 μl of PBSA and the cell lysate was immunoprecipitated with 15 μl of polyclonal MdmX antibody for 12 h. For Western analysis, proteins were resolved on a sodium dodecyl sulfate–10% polyacrylamide gel followed by transfer of proteins to a polyvinylidene difluoride membrane (Millipore) by using a Transblot system (Bio-Rad). Immunoblotting was performed as described elsewhere (3), using primary antibodies at a 1:3,000 dilution and secondary antibodies (goat anti-mouse or goat anti-rabbit conjugated to horseradish peroxidase) at a 1:5,000 dilution. Filters were then exposed to a chemiluminescent reagent and exposed to X-ray film. For Fig. 4, 1 μg of bacterially produced p53 was mixed with equivalent radioactivity counts of in vitro-translated 35S-labeled MdmX or MdmXΔp53 in 250 μl of LSAB containing 4.0 μg of Ab421 per ml for 1 h. The remaining steps of the immunoprecipitation were performed as described above.
FIG. 2.
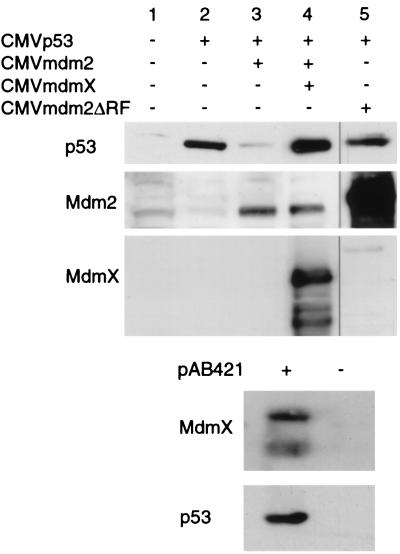
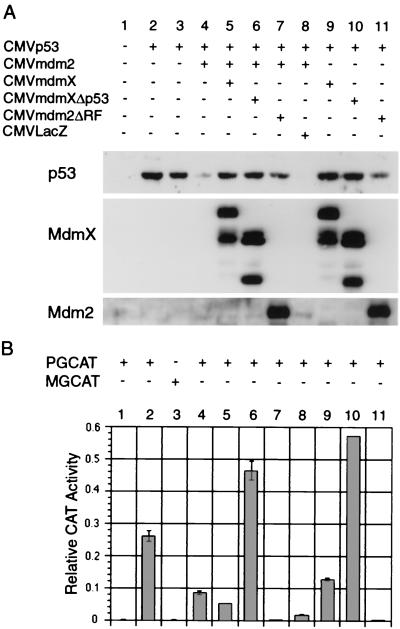
MdmX reverses Mdm2-mediated degradation of p53. H1299 cells were cotransfected with 0.5 μg of CMVp53, 1 μg of CMVmdm2, and 7.0 μg of either CMV, CMVmdmX, or CMVmdm2ΔRF. Cell extracts were prepared 24 h after transfection, and equal luciferase units were analyzed via Western analysis. The recovery of p53 protein levels from Mdm2-mediated degradation is specific to MdmX, as both CMVLacZ (Fig. 5, lane 8) and CMVLacINLS (data not shown) have been used at the same ratios with no recovery in p53 levels. Also, titration experiments demonstrated that beginning at a 3.5:1 ratio of mdmX to mdm2, MdmX was able to protect p53 (data not shown). Immunoprecipitation of p53 from lane 4 transfection was performed with p53-specific monoclonal antibody Ab421 to demonstrate the presence of MdmX proteins associated with p53.
FIG. 6.
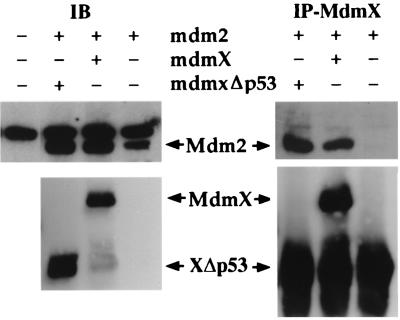
Mdm2 interacts with MdmX and MdmXΔp53. H1299 cells were transfected with 5.0 μg of human mdm2 and 2.5 μg of the indicated mdmX plasmids. Whole-cell extracts were immunoblotted (IB) with either a monoclonal Mdm2 antibody (2A10) or a polyclonal MdmX antibody. Mdm2 was coimmunoprecipitated (IP) with a polyclonal MdmX antibody. In the absence of MdmX, no Mdm2 protein was coprecipitated with MdmX, indicating that there is no cross-reactivity with the polyclonal antibody used. Both full-length MdmX and MdmXΔp53 coprecipitated with Mdm2, demonstrating the direct interaction previously reported (32).
FIG. 4.
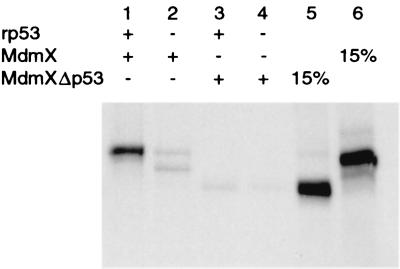
MdmXΔp53 is unable to interact with p53. An MdmX deletion mutant lacking the first 127 amino acids (MdmXΔp53) and wild-type MdmX were produced by using a coupled in vitro transcription-translation system in the presence of [35S]methionine. The resulting proteins were incubated with Ab421 in the presence (+ lanes) or absence (− lanes) of recombinant bacterially produced p53 protein.
Cellular localization studies.
Heterokaryon assays were performed as previously described (27, 31). Briefly, 3 μg of CMVmdmX-EGFP or CMVmdm2-EGFP was transfected into HeLa cells as described above. Transfected HeLa cells (105) were plated with 1.5 × 105 murine NIH 3T3 fibroblasts onto Lab-Tek II chamber slides (Nalge Nunc International). After 18 h, the cells were treated with cycloheximide (100 μg/ml) for 15 min, fused with 50% polyethylene glycol 3350-PBS for 4 min, and incubated in complete medium containing cycloheximide for an additional hour. In an additional experiment, H1299 cells were used in place of HeLa cells and the postfusion incubation time was extended from 1 h to 3 h (Fig. 3A, panels g to i). The cells were then fixed in 3% paraformaldehyde in PBS for 10 min, permeabilized with 1% Triton X-100 in PBS for 15 min, and blocked with PBS containing 10% goat serum, 0.5% bovine serum albumin, and 0.05% Tween 20. Cells were then incubated with a monoclonal antibody to β-actin (Sigma) at a 1:250 dilution and a secondary antibody conjugated to Texas red (Jackson Laboratories) at a 1:500 dilution. The cells were treated with Hoechst 33342 (50 μg/ml) for 10 min and mounted with Gel Mount (Biomeda Corp.) The cells were analyzed by confocal fluorescence microscopy.
FIG. 3.
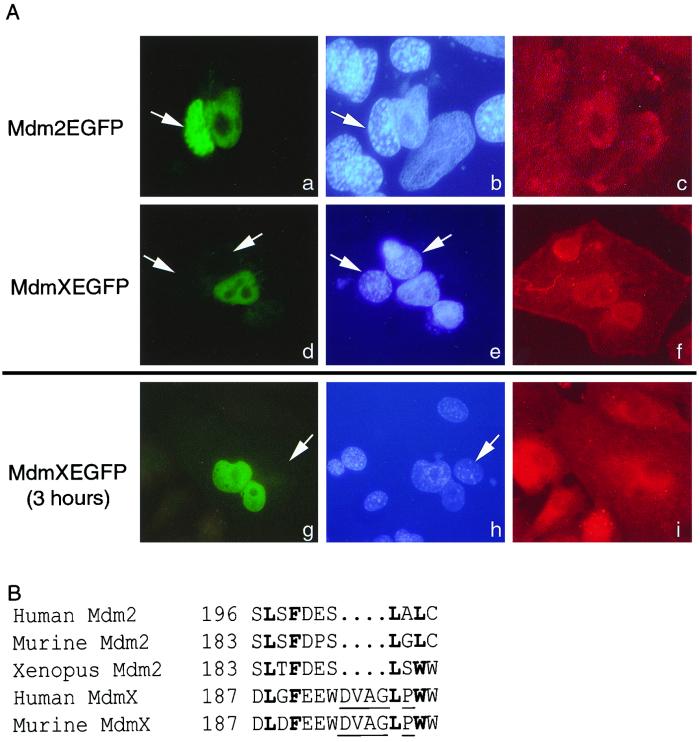
MdmX is unable to undergo nuclear export. (A) Fluorescence of Mdm2-EGFP (a) or MdmX-EGFP (d and g) is observed in three heterokaryons. The human and murine nuclei (b, e, and h, white arrows) are visualized by Hoechst dye. Murine nuclei are indicatively stained with a punctate pattern distinct from that of the human nuclei. A β-actin antibody followed with a Texas red-conjugated secondary antibody allowed confirmation that the nuclei are within heterokaryons (c, f, and i). Mdm2-EGFP serves as a positive control demonstrating that the EGFP fusion does not inhibit nucleocytoplasmic shuttling. The data presented are representative of more than 20 separate heterokaryons. (B) Comparison of the Mdm2 NES with the putative MdmX NES. Boldfaced amino acids represent conserved hydrophobic amino acids; underlined amino acids produce an altered secondary structure within the MdmX NES.
CAT assays.
Chloramphenicol acetyltransferase (CAT) activity was measured by incubating each extract (equal luciferase units) with 0.25 μCi of [14C]chloramphenicol and 25 μg of _n_-butyl coenzyme A in 125 μl of 0.25 M Tris-Cl (pH 8.0). Acetylated products were purified by extraction with mixed xylenes followed by three back extractions with 0.25 M Tris-Cl (pH 8.0). The mixed xylenes were then transferred to a vial containing liquid scintillation fluid for quantification of acetylated [14C]chloramphenicol. Each assay was performed in duplicate, and the bar graphs (Fig. 5B and 7C) represent the average deviation of the duplicate samples.
FIG. 5.
MdmX maintains nuclear pools of p53 and blocks p53 transactivation. H1299 cells were transfected with 0.25 μg of CMVp53 (lanes 2 to 8), 0.75 μg of CMVmdm2 (lanes 4 to 8), and 5.25 μg of CMVmdmX, CMVmdmXΔp53, CMVLacZ, or CMVmdm2ΔRF. Each transfection also contained 0.5 μg of pGL3Luc and 1 μg of the indicated reporter plasmid. Cell extracts were prepared 24 h after transfection, and equal luciferase units were analyzed via Western analysis (A) or CAT assay (B). Error bars represent difference between duplicates of the same extracts. Similar results have been observed in three independent experiments.
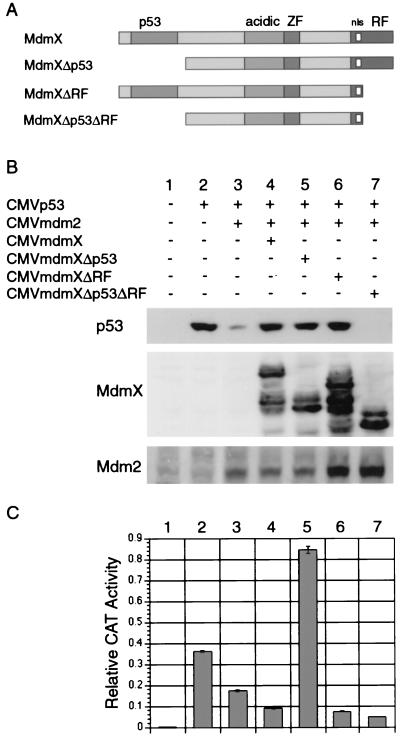
FIG. 7.
MdmX stabilizes p53 through two distinct domains. (A) MdmX deletion mutants lacking either the p53-binding domain, the ring finger, or both domains were constructed. Transfections were performed as described for Fig. 5, and cell extracts were subjected to Western analysis (B) and CAT assay (C). These results have been seen in three independent experiments.
RESULTS
MdmX can reverse Mdm2-mediated degradation.
Mdm2 inactivation of p53 function results from direct interaction with the p53 transactivation domain and nuclear export of p53 for ubiquitin-mediated degradation (27). By comparing Mdm2 and MdmX, we concluded that the Mdm2 NES, essential for transporting nuclear p53 to the cytoplasm, was altered in MdmX (Fig. 3B). Thus we decided to examine how MdmX affected the levels of p53 protein.
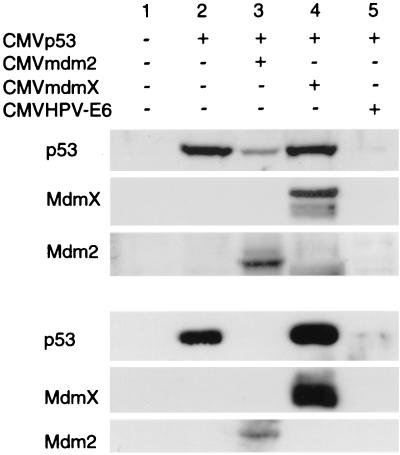
In Fig. 1, we examined the ability of MdmX to degrade p53 in either murine embryo fibroblasts devoid of p53 and Mdm2 (MEF 2KO) or human lung carcinoma cells lacking p53 (H1299). With both cell types, p53 degradation was not observed when p53 was cotransfected with MdmX (Fig. 1, lane 4). In contrast, p53 protein levels were dramatically decreased in p53 transfections containing either Mdm2 or human papillomavirus E6 expression vectors (Fig. 1, lanes 3 and 5). These results suggest a difference of function where MdmX is unable to mediate the Mdm2 activity of p53 degradation. In fact, MdmX coexpression with p53 in cells possessing endogenous Mdm2 (H1299) resulted in a slightly greater amount of p53 protein levels relative to p53 expression alone, indicating that MdmX may have a stabilizing effect on p53 protein levels in the presence of Mdm2.
FIG. 1.
MdmX is unable to mediate p53 degradation. MEF 2KO (top) and H1299 (bottom) cells were cotransfected with the CMVmdm2, CMVmdmX, or CMVHPV-E6 expression vector and a CMVp53 expression vector at a 2.5:1 (MEF 2KO) or 5:1 (H1299) ratio. Each transfection contained 0.5 μg of the luciferase expression vector, pGL3Luc (Promega), and a total DNA concentration of 7 μg. Cell extracts were prepared 24 h following transfection, and equal luciferase units were analyzed via Western analysis. We have not observed any direct effect of p53 overexpression on expression of the luciferase internal control (data not shown).
To confirm whether MdmX stabilizes p53 protein in the presence of Mdm2, we performed a second series of transfections in which MdmX was expressed in the presence of both p53 and Mdm2. Figure 2 demonstrates that while Mdm2 is capable of degrading p53, the addition of MdmX reverses Mdm2-induced p53 degradation (Fig. 2, lane 4). In a control transfection, expression of a Mdm2 mutant lacking the carboxyl-terminal ring finger (Mdm2ΔRF) was unable to induce any degradation of p53 even when cotransfected 14-fold relative to p53 (Fig. 2, lane 5). Ring finger mutants similar to Mdm2ΔRF have previously been shown to stabilize p53 (18). Immunoprecipitation of p53 from cellular extracts containing elevated p53 protein in the presence of Mdm2 and MdmX (Fig. 2, lane 4) confirmed that MdmX proteins were associated with p53. Two MdmX proteins were found to associate with p53 protein (Fig. 2, + pAB421 lane), one migrating at 70 kDa (full length) and a second, faster-migrating protein of approximately 55 kDa. This smaller MdmX protein most likely represents the N-terminal MdmX protein produced by caspase cleavage at a conserved cleavage motif present near the carboxyl terminus of MdmX (9).
MdmX is maintained within the nucleus.
Since p53 was found complexed to MdmX (Fig. 2), we next tested whether the stabilization of p53 was due to a lack of nucleocytoplasmic shuttling of MdmX. Although the hydrophobic amino acids necessary for nuclear export are conserved between MdmX and Mdm2, there are additional amino acids and the presence of a nonconserved proline residue that may alter the ability of MdmX to interact with the nuclear export machinery (Fig. 3B). To test this, a heterokaryon assay was performed. HeLa cells were transfected with either a CMVmdm2-EGFP or CMVmdmX-EGFP expression vector and then fused with murine NIH 3T3 fibroblasts by methods previously used to illustrate nuclear export of Mdm2 (27, 31). Figure 3A contains three heterokaryons, one expressing Mdm2-EGFP (panels a to c) and two expressing MdmX-EGFP (panels d to i). The nuclei were viewed by using Hoechst dye in which murine nuclei (white arrows) are present with a punctate pattern distinct from that of human nuclei (panels b, e, and h). Fused cells were confirmed by β-actin localization (panels c, f, and i). The heterokaryon expressing Mdm2-EFGP contains the fusion protein in both human and murine nuclei, demonstrating the functionality of the assay. In contrast, both heterokaryons expressing MdmX-EFGP failed to demonstrate any nucleocytoplasmic shuttling irrespective of whether the cells were incubated for 1 or 3 h following fusion. Thus, we conclude that while MdmX was capable of binding to p53, even in the presence of Mdm2, it lacks nuclear export capability.
MdmX blocks p53 transactivation and degradation.
To determine the effect of MdmX-mediated p53 stabilization on its ability to transactivate, H1299 cells were transfected as before but with inclusion of a p53 reporter gene. In the MdmX and Mdm2ΔRF transfections containing Mdm2, p53 protein was stabilized yet p53 transactivation remained repressed at levels comparable to those for transfection with only Mdm2 (see Fig. 5B, lanes 5 and 7). Although these forms of Mdm2 and MdmX are defective in steps of the p53 degradation pathway, both are able to bind p53 and therefore were able to inhibit p53 transactivation. These results differed dramatically with those seen with an MdmX deletion mutant lacking the first 127 amino acids (MdmXΔp53). MdmXΔp53 was in vitro translated in the presence of [35S]methionine and shown to be unable to associate with p53 (Fig. 4). It was expected since MdmXΔp53 could not bind to p53, it would be unable to stabilize p53 in the presence of Mdm2. However, MdmXΔp53 coexpressed with Mdm2 and p53 was able to stabilize p53 protein to levels comparable to those for wild-type MdmX (Fig. 5A, compare lanes 5 to 6). Even more interesting were the results obtained when p53 transactivation was monitored in the presence of MdmXΔp53. MdmXΔp53 produced a 2-fold (in the presence of Mdm2 [Fig. 5B, lane 6]) or a 2.5-fold (with p53 alone [Fig. 5B, lane 10]) increase in p53 transactivation. These results suggested the existence of a second domain on MdmX capable of stabilizing p53 without direct interaction with p53.
MdmX stabilizes p53 through two distinct domains.
MdmX and Mdm2 were recently reported to heterodimerize through their ring finger domains (32). Using an immunoprecipitation-Western blot experiment, MdmX and MdmXΔp53 were shown to associate with Mdm2 (Fig. 6). Based on these findings, we focused on addressing whether formation of an MdmX-Mdm2 heterodimer inhibited p53 degradation. To test this hypothesis in our assay system, we constructed two additional MdmX deletion mutants (Fig. 7A). MdmXΔRF lacks the last 45 amino acids, including four cysteine residues that comprise the ring finger, and is therefore unable to bind to Mdm2 (data not shown). MdmXΔp53ΔRF lacks both the p53 and ring finger domains and is therefore unable to bind to either p53 or Mdm2 (data not shown). Using the transfection assay described for Fig. 5, MdmXΔp53 and MdmXΔRF were both shown to be capable of stabilizing p53 in the presence of Mdm2 (Fig. 7B, lanes 5 and 6), while MdmXΔp53ΔRF could not recover p53 from Mdm2-mediated degradation (Fig. 7B, lane 7). In addition, MdmXΔRF also maintained suppression of p53 transactivation function whereas MdmXΔp53 stimulated this aspect of p53 function (Fig. 7C).
DISCUSSION
The MdmX protein represents yet another cellular protein capable of regulating p53 transactivation. Although MdmX possesses significant homology to Mdm2, evidence to date suggests that MdmX most likely functions in a biological fashion distinct from that of Mdm2. For example, while mdmX (14) and mdm2 are both expressed independent of cell proliferation (23) and differentiation (3, 19), the mdmX gene, unlike mdm2, is not induced in response to DNA damage (28, 29). Thus, it is unlikely that MdmX plays any significant role in regulating p53 function during or following a genotoxic stress. Furthermore, even though the role for MdmX during development has not been addressed, MdmX clearly is unable to compensate for the early embryonic lethality observed in Mdm2 null mice (15, 22).
Based on those observations and that both MdmX and Mdm2 are capable of binding p53 and inhibiting transactivation, we decided to focus on how MdmX and Mdm2 might affect p53 protein in nondamaged cells. The results presented in this study unequivocally demonstrate that while MdmX is capable of blocking p53 transactivation, it is unable to modulate p53 protein levels. Not only does MdmX coexpressed with p53 fail to reduce p53 protein levels, but MdmX coexpressed with Mdm2 and p53 can reverse the ability of Mdm2 to degrade p53. It is this latter effect that provided the first hint of a possible biological role for MdmX.
In an attempt to determine the basis for the ability of MdmX to block Mdm2-mediated degradation of p53, the following discoveries were made. First, using a heterokaryon assay, we demonstrated that the putative MdmX NES was in fact nonfunctional and that MdmX is unable to undergo nuclear export. While other domains of Mdm2 required for mediating p53 degradation may also be lacking in MdmX, the inability to export p53 could create a nuclear pool of p53 protein protected from Mdm2 degradation through its association with MdmX.
Second, the immunoprecipitation of MdmX with stabilized p53 (Fig. 2) and the MdmXΔRF mutant phenotype showing no p53 degradation (Fig. 7) confirmed that association of MdmX with p53 can protect p53 in the presence of Mdm2. Consistent with this model, expression of an MdmX containing only the first 162 amino acids was also able to stabilize p53 in the presence of Mdm2 (data not shown). However, additional MdmX mutants uncovered a second novel domain through which MdmX could also block Mdm2-mediated p53 degradation. When constructing an MdmX protein unable to bind p53 (MdmXΔp53), we anticipated observing a MdmX protein that could neither bind p53 nor protect p53 from degradation by Mdm2. Based on the results shown in Fig. 4 and 5, the prediction was only partially correct. While MdmXΔp53 would not associate with p53 in vitro (Fig. 4), it was able to stabilize the p53 protein in the presence of Mdm2 (Fig. 5A). Based on the recent report that MdmX and Mdm2 can heterodimerize via their ring finger domains (32) and the Mdm2 ring finger domain is required for p53 degradation (18), the ability of MdmXΔp53 to stabilize p53 in the presence of exogenous (Fig. 5B, lane 6) or endogenous (Fig. 5B, lane 10) Mdm2 most likely results from it directly binding to Mdm2. Determination of how association between MdmX and Mdm2 results in p53 stability and in what ratio the two proteins interact is currently under way. For example, it will be interesting to determine whether binding of merely one Mdm2 molecule is enough to mediate degradation of a p53 tetramer. Conversely, does binding of one MdmX molecule mediate recovery from Mdm2-mediated degradation?
Taken together, these results suggest that MdmX is capable of blocking p53 degradation by Mdm2. Rb (13) and p19_ARF_ (26) represent two other cellular proteins shown to be able to block Mdm2-mediated degradation of p53. Interestingly, both proteins have been characterized as tumor suppressor proteins. Based on the results presented here, it is possible that the inability to detect MdmX overexpression in human tumors may represent the fact that deregulation of MdmX does not phenotypically resemble the oncogenic activities attributed to its family member Mdm2. Clearly, more studies are required to test this hypothesis.
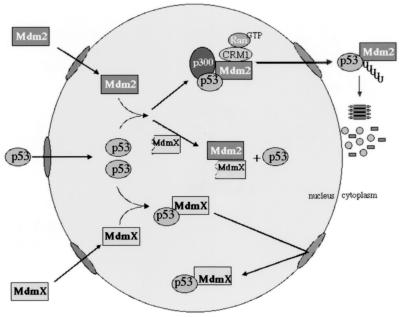
In the absence of any reported modulation of MdmX protein, we propose the following model for MdmX in normal, undamaged cells (Fig. 8). We believe that low levels of wild-type p53 protein seen in undamaged cells are maintained due to a competition for p53 binding by Mdm2 and MdmX. The Mdm2-p53 complexes are shuttled from the nucleus into the cytoplasm and degraded, while the MdmX-p53 complexes are retained in the nucleus (Fig. 8). Consistent with such a model, we have been able to modulate endogenous p53 in cell lines by transiently expressing MdmXΔp53 (data not shown). The results from the MdmXΔp53 mutant suggest that MdmX may also stabilize p53 protein by interacting with Mdm2, which based on the increase in p53 transactivation must somehow release p53 from interaction with Mdm2. Production of such a form of MdmX protein could potentially occur in two fashions. First, potential downstream translation start sites may produce MdmX proteins lacking the N-terminal region. When overexpressed, the full-length mdmX cDNA does produce smaller MdmX proteins that correlate with sizes predicted from these ATG sites. A second method could involve activation of the conserved caspase 3 cleavage of the MdmX protein. This would produce a C-terminal MdmX protein that lacked the p53 interaction domain but retained a functional ring finger.
FIG. 8.
Role of MdmX maintaining nuclear pools of p53. The two mechanisms for MdmX-mediated recovery of p53 in the presence of Mdm2 are illustrated. Shown are the possible influences of wild-type MdmX and MdmXΔp53. See Discussion for further details.
Finally, it did not escape our observation that MdmXΔp53 was able to stabilize p53 protein in a transcriptionally competent conformation (Fig. 5B, lanes 6 and 10). This ability of MdmXΔp53 to increase p53 transactivation above the levels seen when p53 was transfected alone suggested that the MdmX-Mdm2 complex must be unable to block p53 transactivation. Similar approaches have been used to create Mdm2 peptides capable of reactivating p53 function in vivo (4). In addition to examining the effects of this mutant on p53 cell cycle arrest and apoptosis, possible anticancer therapeutic uses for MdmX mutants like MdmXΔp53 may prove extremely valuable in tumors harboring elevated Mdm2 and wild-type p53 proteins.
ACKNOWLEDGMENTS
We thank David Cool for use of his microscope facilities, Guillermina Lozano for the MEF 2KO cells, Bert Vogelstein for the PG13-CAT and MG15-CAT reporter constructs, Kathleen Cho for the CMVHPV-E6 construct, Gerald Zambetti for the 2A10 antibody, Madhavi Kadakia for the CMVhdm2 construct, and Charlotte Slader for technical assistance and helpful discussions.
This work was supported by a grant from the NIH CA64430 (to S.J.B.).
REFERENCES
- 1.Amundson S A, Myers T G, Fornace A J., Jr Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene. 1998;17:3287–3299. doi: 10.1038/sj.onc.1202576. [DOI] [PubMed] [Google Scholar]
- 2.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berberich S J, Litteral V, Mayo L D, Tabesh D, Morris D. mdm-2 gene amplification in 3T3-L1 preadipocytes. Differentiation. 1999;64:205–212. doi: 10.1046/j.1432-0436.1999.6440205.x. [DOI] [PubMed] [Google Scholar]
- 4.Bottger A, Bottger V, Sparks A, Liu W L, Howard S F, Lane D P. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 5.Bottger V, Bottger A, Garcia-Echeverria C, Ramos Y F M, van der Eb A J, Jochemsen A G, Lane D P. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene. 1999;18:189–199. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 6.Cahilly-Snyder L, Yang-Feng T, Francke U, George D L. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet. 1987;13:235–244. doi: 10.1007/BF01535205. [DOI] [PubMed] [Google Scholar]
- 7.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein V. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–835. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 8.Finlay C A. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman D A, Wu L, Levine A J. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haupt Y, Maya R, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 11.Hermeking H, Lengauer C, Polyak K, He T C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 12.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh J K, Chan F S, O'Connor D J, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 14.Jackson M, Berberich S J. Constitutive mdmx expression during cell growth, differentiation and DNA damage. DNA Cell Biol. 1999;18:696–700. doi: 10.1089/104454999314971. [DOI] [PubMed] [Google Scholar]
- 15.Jones S N, Roe A E, Donehower L A, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 16.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 17.Kubbutat M H G, Ludwig R L, Ashcroft M, Vousden K H. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5680–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubbutat M H G, Ludwig R L, Levine A J, Vousden K H. Analysis of the degradation function of Mdm2. Cell Growth Differ. 1999;10:87–92. [PubMed] [Google Scholar]
- 19.Mayo L D, Berberich S J. Wild type p53 protein is unable to activate the mdm-2 gene during F9 cell differentiation. Oncogene. 1996;13:2315–2321. [PubMed] [Google Scholar]
- 20.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 21.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 22.Montes de Oca Luna R, Wagner D S, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 23.Mosner J, Deppert W. p53 and mdm2 are expressed independently during cellular proliferation. Oncogene. 1994;9:3321–3328. [PubMed] [Google Scholar]
- 24.Oliner J D, Peitenol J A, Thiagalingam S, Gyuris J, Kinzler K W, Voglestein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 25.Polyak K, Xia Y, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 26.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 27.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shvarts A, Bazuine M, Dekker P, Ramos Y F M, Steegenga W T, Merckx G, van Ham R C A, van der Houven van Oordt W, van der Eb A J, Jochemsen A G. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics. 1997;43:34. doi: 10.1006/geno.1997.4775. [DOI] [PubMed] [Google Scholar]
- 29.Shvarts A S, Wilma T, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham R C A, van der Houven van Oordt W, Hateboer G, van der Eb A J, Jochemsen A G. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 30.Sigalas I, Calvert A H, Anerson J J, Neal D E, Lunec J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med. 1996;2:912–917. doi: 10.1038/nm0896-912. [DOI] [PubMed] [Google Scholar]
- 31.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 33.Tao W, Levine A J. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G S, Burns T F, McDonald III E R, Jiang W, Meng R, Krantz I D, Kao G, Gan D D, Zhou J Y, Muschel R, Hamilton S R, Spinner N B, Markowitz S, Wu G, el-Deiry W S. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 35.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]