Role of DBP in the Circadian Oscillatory Mechanism (original) (raw)
Abstract
Transcript levels of DBP, a member of the PAR leucine zipper transcription factor family, exhibit a robust rhythm in suprachiasmatic nuclei, the mammalian circadian center. Here we report that DBP is able to activate the promoter of a putative clock oscillating gene, mPer1, by directly binding to the mPer1 promoter. The mPer1 promoter is cooperatively activated by DBP and CLOCK-BMAL1. On the other hand, dbp transcription is activated by CLOCK-BMAL1 through E-boxes and inhibited by the mPER and mCRY proteins, as is the case for mPer1. Thus, a clock-controlled dbp gene may play an important role in central clock oscillation.
Most eukaryotes and some prokaryotes have circadian (∼24-h) rhythms governed by endogenous oscillators that control daily rhythms in physiology and behavior. Recent molecular dissections in cyanobacteria, Neurospora, Drosophila, and mice have revealed that oscillations in the transcription of specific clock genes play a central role in the generation of circadian rhythms and that negative feedback loops, in which certain gene products suppress their own transcription, form central elements of the mechanism of the circadian oscillator conserved across species (5, 17).
In mammals, the suprachiasmatic nucleus (SCN) is known as the anatomical locus of a dominant mammalian pacemaker for circadian behavior and hormonal rhythms (16, 21). Following the initial discovery of mPer1 in the SCN (31) (Sun et al. reported the same gene, RIGUI, independently [28]), the first mammalian homologue of the Drosophila clock gene per (4, 13), many noticed that this gene is one of three genes of the mammalian period gene family; Per2 (1, 26, 29) and Per3 (30, 36) are also abundantly expressed in neurons of the SCN.
Among mammalian per homologues, evidence has accumulated that at least mPer1 is a component constituting the central oscillatory mechanism. First, transcripts of mPer1 show a very robust rhythm in the SCN in mice (28, 31) and rats (35), even in extended constant dark (DD) conditions (27). Second, the auto-negative feedback loop seems to be closed for the mPer1 gene, as with the Drosophila per gene product PER, which negatively regulates the expression of its own gene (per) (11). The transcription of mPer1 was shown to be activated by the binding of the CLOCK-BMAL1 complex to the E-boxes (CACGTG) in the promoter region of the mPer1 gene (9), and this activation is specifically inhibited by PER1 protein and other negative elements, including PER2, PER3, TIM, CRY1, and CRY2 (14, 18, 23, 24). Third and perhaps more important is the finding that mPer1 expression parallels the behavioral rhythm (27). mPer1 is rapidly induced by light in a time-of-day and tissue-specific manner that correlates well with the resetting behavior of the overt rhythm in locomotor activity. Moreover, the photic thresholds and dose responses for the two processes are quantitatively very similar, and each shows reciprocity between light intensity and duration (27).
Besides the clock genes constituting this core oscillatory loop, a transcription factor, DBP (named for albumin gene D-site binding protein), is expressed in the SCN with clear rhythm in light-dark (LD) and DD conditions (20). We recently found that DBP is endogenously highly rhythmic, with an amplitude comparable to that of mPer1, and its expression is not influenced by environmental light (34). Short-duration light exposure does not alter the dbp mRNA levels at any circadian time. DBP belongs to the PAR leucine zipper transcription factor family, which includes thyrotroph embryonic factor (TEF) and hepatic leukemia factor (HLF), and is known to increase the transcription of several genes in the liver (22). Although DBP knockout mice are still rhythmic, they display significant differences in circadian locomotor activity. However, DBP protein is not required for the circadian expression of its own gene (20). Although these findings had previously been interpreted to suggest that DBP is located downstream of the clock, in the present study we present evidence that DBP functions upstream of the clock. During the analysis of the mPer1 promoter region, we noticed that the nucleotide sequence located between −28 and −37 of the mPer1 gene (+1 indicates the transcription initiation site) shows homology with the consensus DBP-binding sequence. Here we demonstrate that DBP directly binds to the mPer1 promoter and thereby increases transcription. Furthermore, we show that DBP protein can be detected in the nuclei of pacemaker cells in the SCN concomitantly with a rise in mPer1 RNA levels. We also show that dbp transcription is regulated by CLOCK-BMAL1 and PER/CRY as well as mPer1. Therefore, a clock-controlled dbp gene may play an important role in the central oscillatory mechanism.
MATERIALS AND METHODS
Genomic library screening and 5′-RACE analysis of mPer1 gene.
A mouse genomic λ phage library (Stratagene) was screened with a 32P-labeled probe derived from the 5′ portion of the mPer1 cDNA (encoding nucleotide residues 26 to 551 of RIGUI [GenBank accession number AF022992]) by PCR. Approximately 1.6 × 106 phage clones were screened as previously described (33), and 18 hybridization-positive clones were isolated. The overlapping regions of the independent clones were found to have an identical restriction map. Four representative clones were subcloned and subjected to further analysis. The entire 12.8-kbp region upstream of the 3′ end of exon 2 was sequenced.
To identify the transcription initiation site of the mPer1 gene, 5′-RACE (rapid amplification of cDNA ends) analysis using a 5′-Full RACE core set (TaKaRa, Tokyo, Japan) was carried out according to the manufacturer's instructions. Total RNA (5 μg) prepared from the hypothalamus of mice sacrified at circadian time (CT) 4 (where CT0 is subjective dawn and CT12 is subjective dusk) was used for reverse transcription with 200 pmol of 5′-phosphorylated primer R1 (5′-ATGAGTTCTTTCTGG-3′, complementary to 641 to 627 of RIGUI) representing the 5′ portion of the mPer1 coding region. Since this 5′-RACE analysis was carried out using inverse PCR, the cDNAs were circularized or concatenated with each other by T4 RNA ligase and then used as a template. Primer R2 (5′-GTCCTGCTCTGAGCTCGCACTCAGG-3′, complementary to nucleotide residues 576 to 552 of RIGUI) and primer F1 (5′-AACCCATCTACCAGTGGCTGCAGCA-3′, residues 577 to 601 of RIGUI), both representing the further 5′ upstream portion of the mPer1 coding region, were used for the first PCR (25 cycles of 94°C for 30 s, 65°C for 30 s, and 68°C for 30 s). Primer R3 (5′-AGGCTGTAGGCAATGGAGCTGCTGG-3′, complementary to nucleotide residues 551 to 527 of RIGUI) and primer F2 (5′-GTGAACAGTCAGCTCGAGCCAGGAC-3′, residues 602 to 626 of RIGUI) were employed for the second nested PCR (27 cycles of 94°C for 30 s, 65°C for 30 s, and 68°C for 30 s). The resultant products were electrophoresed on a 0.8% SeaKem GTG agarose gel (FMC Corp.). The specific 5′-RACE product of approximately 550 bp was extracted from the gel and subcloned into pCR2.1-TOPO (Invitrogen) for sequence determination.
Transcriptional assay.
HepG2 cells were grown in Dulbecco's modified Eagle's medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (Sigma). Cells were plated at ca. 6.0 × 105 cells per well in six-well plates 24 h before transfection. Cells were transfected with LipofectAmine-Plus reagent (Gibco; LipofectAmine, 8 μl; Plus reagent, 6 μl) according to the manufacturer's instructions. Unless otherwise noted, cells in each well were transfected with 1 μg (total) of expression plasmids with the indicated inserts in pcDNA3 (Invitrogen), 10 ng of reporter plasmids, and 0.01 to 0.1 ng of internal control plasmids. The total amount of DNA per well was adjusted by adding pcDNA3 vector. After 48 h, cells in each well were extracted with 200 μl of passive lysis buffer (Promega), and 20 μl of the extracts was taken for assays of firefly luciferase and Renilla luciferase by luminometry. For statistical analysis, a two-sample t test was applied.
For the transcriptional assay of the mPer1 gene, reporter constructs and internal control plasmid were made as follows. A 1.3-kbp fragment derived from the 5′-flanking region of the mPer1 gene (−1296 to +32; +1 indicates the transcription initiation site) was ligated to the Renilla luciferase reporter gene (1,212-bp _Hin_dIII-_Xba_I fragment of the pRL-TK vector [Promega]) and the simian virus 40 (SV40) polyadenylation signal (3,092-bp _Xba_I-_Mlu_I fragment of the pGL3-Basic vector [Promega]). PCR-based mutagenesis was used to construct the reporter plasmid containing the same 1.3-kb fragment of the mPer1 promoter with the DBP-binding site mutated to TCGCCATGGC.
A 60-bp construct in which three copies of a 20-bp sequence centered on the DBP-binding site were linked in tandem was made by annealing oligonucleotides 5′-GATCTCTGGCATTATGCAACCCGCCCTGGCATTATGCAACCCGCCCTGGCATTATGCAACCCGCCA-3′ and 5′-GATCTGGCGGGTTGCATAATGCCAGGGCGGGTTGCATAATGCCAGGGCGGGTTGCATAATGCC AGA-3′ (binding sites are underlined). The annealed oligonucleotides were inserted into the _Bgl_II site of the pRL-TK vector. A 60-bp construct in which three DBP-binding sites were mutated was constructed with oligonucleotides 5′-GATCTCTGGCTCACCCGGCTCCGCCCTGGCTCACCCGGCTCCG CCCTGGCTCACCCGGCTCCGCCA-3′ and 5′-GATCTGGCGGAGCCGGG TGAGCCAGGGCGGAGCCGGGTGAGCCAGGGCGGAGCCGGGTGAG CCAGA-3′ (mutated binding sites are underlined). These annealed oligonucleotides were also subcloned into the _Bgl_II site of the pRL-TK vector.
As the firefly luciferase gene of the pGL3-luciferase vectors (Promega) contains a sequence with 9 of 10 bases identical to the DBP consensus sequence (and is activated by transfection of DBP), we made the following internal control plasmid and used a small amount (0.01 to 0.02 ng). The sequence 5′-GTTATGTAAA-3′ 1,204 nucleotide residues downstream from the translation initiation site was altered to 5′-GCTATGTGAA-3′ without changing the deduced amino acid sequence, and then the coding region was ligated to a 3,132-bp _Xba_I-_Nhe_I fragment derived from vector pRL-CMV (Promega), allowing expression from the cytomegalovirus (CMV) promoter.
For transcriptional assay of the dbp gene, reporter plasmids were made as follows. A 300-bp fragment containing the two E-boxes derived from the second intron of the mouse dbp gene (GenBank accession number U29762; nucleotide residues 2900 to 3200) was amplified from genomic DNA by PCR and subcloned into the pGL3-promoter vector (Promega). PCR-based mutagenesis was used to construct the reporter plasmids containing mutated E-boxes. The upstream and downstream E-boxes were replaced with TCGCTC and GCTAGT, respectively. The pRL-CMV vector (Promega) was used as an internal control for transcriptional assay of the dbp gene. The sequences of all reporter constructs were confirmed.
Expression constructs were made as follows. The coding regions of mouse dbp (GenBank accession number U29762), mPer1 (AB002108), mPer2 (AF035830), mPer3 (AB013605), mTim (AB019001), mCry1 (AB000777), and human BMAL1b (AB000813) were obtained by reverse transcription-PCR and used after confirming their sequences. The plasmids containing the human Clock (AB002332; KIAA0334 gene) and partial human Cry2 (AB014558; KIAA0658 gene) were generously provided by Takahiro Nagase, KAZUSA DNA Research Institute. As the KIAA0658 gene lacks the 5′ end of the human Cry2 coding region, an upstream 13-bp fragment was added by PCR according to the sequence of an expressed sequence tag (EST) clone (accession number AL040215). All coding regions were ligated into the pcDNA3 vector.
Gel shift assay.
A double-stranded oligonucleotide (top strand, 5′-GGCAGGGCCTGGCATTATGCAACCCGCCTCC-3′; bottom strand, 5′-GGGAGGCGGGTTGCATAATGCCAGGCCCTGC-3′) including the DBP-responsive site derived from the mPer1 promoter with its flanking sequences, was labeled at both ends with Klenow enzyme in the presence of [α-32P]dCTP and used as a probe. The AP2 consensus binding probe was labeled in the same way using an annealed oligonucleotide (top strand, 5′-GGTGGAAAGTCCCCAGGCTGTGAATCC-3′; bottom strand, 5′-GGGATTCACAGCCTGGGGACTTTCCAC-3′). Nuclear extracts were prepared as described (25), and 8 μg was incubated with a radiolabeled probe. For the control experiment, 30 ng of human AP2 (Promega) was added. Binding reactions and electrophoresis were performed as described before (15). Competitors containing the PAR protein recognition sequence or the mutated sequence were made by annealing the oligonucleotides 5′-GTTCTTGGTTACGTAATCTCCAATGGTTCTT-3′ (top strand) and 5′-AAGAACCATTGGAGATTACGTAACCAAGAAC-3′ (bottom strand) and 5′-GTTCTTGTCGCCATGGCCTCCAATGGTTCTT-3′ (top strand) and 5′-AAGAACCATTGGAGGCCATGGCGACAAGAAC-3′ (bottom strand), respectively. For the supershift assays, 0.25 μl of DBP antiserum was added to DNA-protein complexes, and the incubation was continued for 15 min on ice.
Quantitative in situ hybridization using radiolabeled probes. (i) Animals.
Male BALB/c mice (Japan Animal Company, Osaka, Japan) purchased 5 weeks postpartum were exposed to 2 to 4 weeks of complete light (fluorescent light, 300 lux)-dark (LD) cycles and then kept in complete darkness for 2 days as a continuation of the dark phase of the last LD cycle. The expression profiles of mPer1 and dbp mRNA were examined in the second dark-dark (DD) cycle every 2 h (n = 5 at each time point) starting at CT0. The experimental protocol of the current research was approved by the Committee for Animal Research at Kobe University School of Medicine.
(ii) In situ hybridization.
In situ hybridization histochemistry using the free-floating sections was performed according to the method detailed previously (2, 27). We used 33P-radiolabeled complementary RNA (cRNA) probes for mPer1 and dbp for the present in situ hybridization studies (27, 34). The radioactivity of each SCN on BioMax film (Kodak) was analyzed using a microcomputer interfaced to an image-analyzing system after conversion into the relative optical densities produced by the [14C]acrylic standards. The intensities of the optical density of the 10 sections of the SCN were then summed. The results are expressed as means ± standard error of the mean (SEM). For statistical analysis, one-way analysis of variance (ANOVA) followed by Sheffe's multiple comparisons were applied. The peak value was adjusted to 100 (CT4 for dbp, and CT6 for mPer1), and relative RNA abundance was used.
Immunocytochemistry of DBP. (i) Antiserum to DBP.
The total coding region of mouse dbp was subcloned into vector pGEX6P-1 (Pharmacia). The full length of DBP fused with glutathione-_S_-transferase (GST) was expressed in Escherichia coli and purified through a glutathione-Sepharose 4B column (Pharmacia). New Zealand White rabbits received a subcutaneous injection of purified antigen containing 50% Freund's complete adjuvant. The animals were boosted three times with purified antigen containing 50% Freund's incomplete adjuvant at 2-week intervals. Two weeks after the last injection, blood was collected. To remove antibodies which cross-react with other PAR proteins, antiserum was incubated with HLF-GST fusion protein bound to glutathione-Sepharose 4B. Antibodies were checked by Western blots of nuclear extracts of mouse liver and DBP expression plasmid-transfected HepG2 cells. In both cases, we detected a single band lying at a relative molecular mass of approximately 46 kDa, corresponding to the size reported previously (8).
(ii) Immunocytochemistry.
Under deep ether anesthesia, DD-housed male BALB/c mice were examined for DBP expression in the second DD cycle every 2 h (n = 4 at each time point) starting at CT0.
Immunocytochemistry of DBP was performed using the avidin-peroxidase method as described previously (2). Immunoreactions were visualized with diaminobenzidine (DAB). The specificity was confirmed by dilution tests and absorption tests; 10 μg of purified antigen expressed in E. coli completely abolished the positive reaction. Quantitative analysis was performed in DBP-immunoreactive sections at each time point. We counted the number of immunoreactive nuclei in three sections in the middle portion of the SCN. We repeated the examination in four animals. Statistical analysis of the data was done by one-way ANOVA followed by Scheffe's multiple comparisons.
Double-labeling histochemical method for mPer1 and dbp.
To find out if cells express both mPer1 and dbp, we used two double-labeling histochemical methods. Colocalization of two types of mRNAs was detected by double-labeling in situ hybridization using [35S]CTP (New England Nuclear)-labeled dbp probe and digoxigenin-labeled mPer1 probe in CT6 animals (n = 3). We made digoxigenin-labeled antisense mPer1 cRNA probes using digoxigenin-UTP (Boehringer, Mannheim, Germany) following the manufacturer's instructions. Tissue preparation, prehybridization, hybridization, and posthybridization washing were the same as for isotope probe hybridization except that we used 20-μm-thick sections. For digoxigenin-labeled probes, sections were processed for immunocytochemistry with the nucleic acid detection kit (Boehringer).
We also tried to detect DBP protein and mPer1 mRNA in single cells by double-labeling immunocytochemistry and in situ hybridization. Tissue fixation was performed at CT6 (n = 3) as for the in situ hybridization described above. The sections were processed for immunocytochemistry of DBP as described above. After the brown DAB reaction, sections were processed for prehybridization, hybridization, and posthybridization washes as described above. Perikaryal digoxigenin-labeled mPer1 mRNA signals were stained blue with the nucleic acid detection kit (Boehringer).
Characterization of the mPer1 antisense probe (nucleotide positions 538 to 1752) and dbp antisense probe (595 to 1100) used in this study was precisely described in previous reports (27, 34). The specificities of these probes were confirmed by RNase-digested sections and competition experiments.
RESULTS
DBP activates mPer1 promoter.
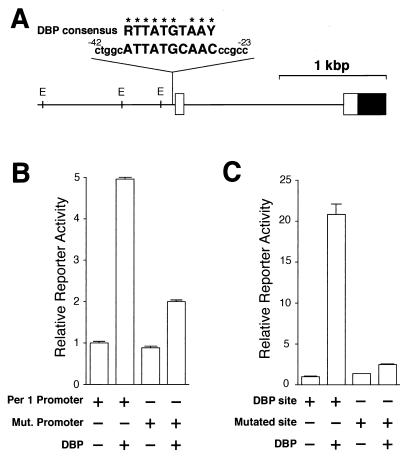
Since the nucleotide sequence located between −28 and −37 in the mPer1 gene shows homology with the consensus DBP-binding sequence (Fig. 1A) (9 of 10 bases match), we tested the ability of DBP to drive mPer1 transcription through the putative DBP-binding site. A 1.3-kbp fragment of the 5′-flanking region of the mPer1 gene containing the endogenous promoter including this potential site was subcloned into a promoterless reporter vector for use in transcriptional analysis in HepG2 cells. Acting through this promoter, DBP produced an increase in transcriptional activity (5.0-fold; P < 0.001) (Fig. 1B); this activation was dependent on the putative DBP-binding site, because mutation of this site reduced the transcriptional activation by DBP from 5.0-fold to 2.0-fold (Fig. 1B). To confirm that this putative binding site is actually responsive to DBP, we next examined the transcriptional activity of a construct in which three copies of a 20-bp sequence centered on that site were linked in tandem and subcloned into a reporter vector containing the herpes simplex virus thymidine kinase (HSV-TK) minimal promoter. DBP produced a substantial activation through this tandem repeat (20.8-fold; P < 0.001) (Fig. 1C), and this activation was reduced from 20.8-fold to 2.5-fold when the three putative DBP-binding sites were mutated (Fig. 1C). Thus, we conclude that DBP is able to activate the mPer1 promoter through this site in vitro.
FIG. 1.
Transcriptional regulation of the mPer1 promoter by DBP. (A) A long vertical line indicates the site that matches the consensus DBP-binding sequence in the 5′-flanking region of the mPer1 gene. The sequence of that site and 5 bp of flanking sequence (lowercase letters) on each side and the consensus DBP-binding sequence (R, purine; Y, pyrimidine) are noted (identical bases are shown by asterisks). The numbers of the nucleotide residues indicate the distance from the transcription start site. The solid and open boxes show the protein-coding region and the 5′ untranslated region, respectively. The locations of the E-box sites (E) are also indicated by short vertical lines. (B) Transcriptional activation of the reporter plasmid including the mPer1 promoter. Reporter plasmids containing a 1.3-kbp fragment including the putative DBP-binding site (ATTATGCAAC) (Per1 promoter) or a mutated site (TCGCCATGGC) (Mut. promoter) were used for the transcriptional assay. (C) Transcriptional activation of an HSV-TK-driven reporter plasmid containing the putative DBP-binding site. A 60-bp construct in which three copies of either the putative DBP-binding site (DBP site) or a mutated site (TCACCCGGCT) (Mutated site) and flanking sequence were linked in tandem was subcloned into the HSV-TK-driven reporter plasmid. (B and C) Presence (+) or absence (−) of reporter and DBP expression plasmids (750 ng) is noted. Each value is the mean + SEM of three replicates for a single assay. The results shown are representative of at least three independent experiments.
DBP directly binds to mPer1 promoter.
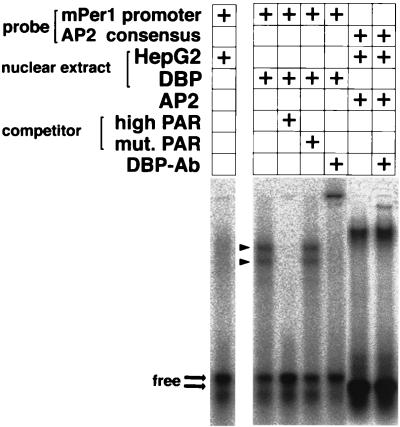
The optimal core binding motif for DBP, RTTATGTAAY (where R is a purine and Y is a pyrimidine), is known to be bound by the other members of the PAR and C/EBP families of basic leucine zipper proteins (7). Thus, we used a gel shift assay to examine whether transcriptional activation by DBP is due to direct binding of DBP to the mPer1 promoter or to indirect mechanisms, including other regulatory proteins. When a radiolabeled 32-bp probe encompassing the putative DBP-binding site was incubated with nuclear extract from untransfected HepG2 cells, only weak smeared bands corresponding to endogenous proteins were observed (Fig. 2). In contrast, nuclear extracts from cells transfected with the DBP expression vector gave rise to two intense bands. These intense bands were abolished by a 100-fold excess of an unlabeled competitor containing PAR protein high-affinity sequence; on the other hand, they were not affected by a 100-fold excess of competitor containing a mutated sequence (Fig. 2). Thus, these intense bands represent specific protein-DNA complexes.
FIG. 2.
Binding of DBP to the mPer1 promoter. Gel shift assays with a probe encompassing the putative DBP-binding site derived from the mPer1 promoter were performed. Nuclear extracts from untransfected (HepG2) or DBP expression plasmid-transfected (DBP) HepG2 cells were incubated with the probe. For the competition experiment, a 100-fold excess of a double-stranded oligonucleotide containing the PAR protein recognition sequence GTTACGTAAT (high PAR) or containing the mutated sequence TCGCCATGGC (mut.PAR) was added. For the supershift experiment, 0.25 μl of DBP antiserum was added. The complex of human AP2 and an AP2 consensus binding probe was not affected by adding the DBP antiserum. The presence (+) of each probe, nuclear extract, AP2, competitor, and anti-DBP antibody is noted. Arrowheads indicate two bands representing complexes of DBP and the probe. Thick arrows point to the locations of the free probes.
To characterize the proteins involved in the protein-DNA complexes, DBP antiserum was added to the binding reaction for the nuclear extract from the DBP-expressing cells. The resultant protein-DNA complexes showed a supershifted band, and the two intense bands previously observed were diminished (Fig. 2). This is a specific effect of the antiserum with DBP, since a complex of AP2 transcription factor with its consensus binding probe was not affected by adding DBP antiserum (Fig. 2). Thus, the two intense bands contain the DBP proteins. Although DBP can form heterodimers with HLF and TEF (8), the endogenous expression levels of these proteins in HepG2 cells are low (data not shown). Therefore, each of the intense bands may represent different posttranslational modifications or proteolytic products.
Taken together with the ability of DBP to drive mPer1 in the transcriptional assay, it is suggested that DBP positively regulates mPer1 gene expression by directly binding to the mPer1 promoter.
DBP and CLOCK-BMAL1 cooperatively activate the mPer1 promoter.
Since the CLOCK-BMAL1 complex is already known to activate transcription through E-boxes in the mPer1 promoter (9), we next tested for a possible interaction with DBP. The 1.3-kbp mPer1 promoter reporter plasmid includes the three E-boxes and the DBP-responsive site (Fig. 1A), and it was cotransfected with CLOCK, BMAL1, and DBP expression plasmids.
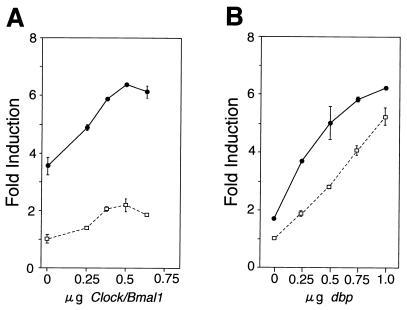
The mPer1 promoter responded in a dose-dependent manner to increasing amounts of transfected CLOCK and BMAL1 expression vectors in the absence of the DBP expression vector (Fig. 3A). The small increase in transcriptional activity (maximum 2.3-fold; P < 0.01) that we observed might be due to the HepG2 cells used in this assay, since a more substantial increase was observed when other cell lines, such as NIH 3T3 cells, were used (data not shown). Coexpressing DBP at any dose of CLOCK and BMAL1 expression plasmids further increased the transcriptional activation (maximum 6.6-fold; P < 0.001) (Fig. 3A). This response is additive rather than synergistic, because the activation induced by both DBP and CLOCK-BMAL1 is close to the sum of the effects of each working alone.
FIG. 3.
Transcriptional activation of the mPer1 promoter by DBP and CLOCK-BMAL1. (A) Dose-response curve of CLOCK-BMAL1-mediated transcriptional activation in the presence and absence of DBP. The reporter plasmid which contains the 1.3-kbp mPer1 promoter, including the three E-boxes and the DBP-responsive site, was used. Increasing doses of CLOCK and BMAL1 expression plasmids were transfected with (solid line) or without (dashed line) 750 ng of DBP expression plasmid. The total amount of DNA (2 μg) per well was adjusted by adding pcDNA3 vector. (B) Dose-response curve of DBP-mediated transcriptional activation in the presence and absence of CLOCK-BMAL1. Increasing doses of DBP expression plasmid were transfected with (solid line) or without (dashed line) 500 ng each of CLOCK and BMAL1 expression plasmids. (A and B) Each value is the mean ± SEM of duplicate for a single assay. A similar pattern of activation was reproduced in another experiment.
This additive effect is also observed at any dose of DBP expression plasmid when 0.5 μg each of CLOCK and BMAL1 expression plasmids were cotransfected (Fig. 3B). These observations indicate that DBP and CLOCK-BMAL1 cooperatively activate the mPer1 transcription in an additive fashion.
Circadian expression of DBP is in phase with the transcription of mPer1 in the SCN.
mPer1 transcripts exhibit a striking circadian oscillation in the SCN, peaking during the subjective morning (31). If DBP protein increases the transcription of mPer1 in the SCN, as indicated by the transcriptional assay for the mPer1 promoter, DBP protein should be expressed in advance of or at least in phase with mPer1 transcription in the SCN. To test this hypothesis, we examined the precise circadian profiles in the mouse SCN of dbp mRNA and DBP protein and compared them to that of mPer1 mRNA.
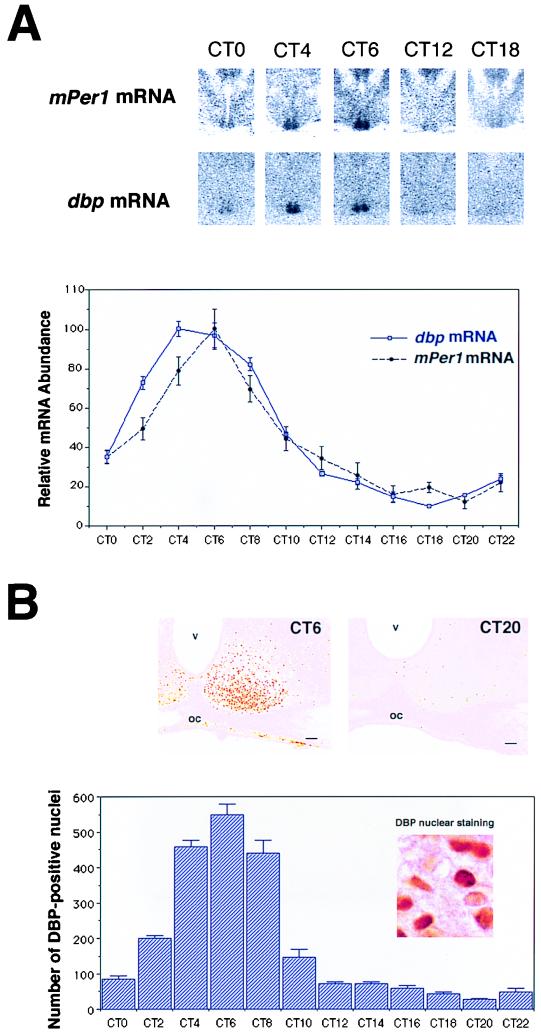
For detecting circadian changes in mRNA in the SCN, we adopted a well-established quantitative in situ hybridization with free-floating sections and examined DD-housed mice perfused for fixation every 2 h (27) (Fig. 4A). dbp mRNA levels were highest in the subjective morning at CT4 (CT0, subjective light on; CT12, subjective light off) and lowest in the subjective early night (CT18) (P < 0.001). The amplitude of this oscillation under DD conditions was over 10-fold.
FIG. 4.
Circadian expression of dbp and mPer1 mRNA and DBP protein in the SCN. (A) Quantitative analysis of dbp (open square with blue line) and mPer1 (solid circle with dotted black line) mRNA expressed in the SCN in DD conditions (n = 5, mean ± SEM). Relative dbp and mPer1 mRNA abundance was determined by quantitative in situ hybridization using isotope-labeled probes with the mean peak values adjusted to 100. Representative in situ hybridization autoradiograms at specific time points are shown on the top panels. (B) Circadian expression of DBP immunoreactivity in the SCN. The photomicrographs show immunoreactivity at CT6 and CT20. Cell counts of SCN nuclei staining positive for DBP (per section) are shown. Values are means + SEM for four animals. oc, optic chiasma; v, third ventricle. Bar, 70 μm.
Next, DBP protein expression was examined in the SCN by immunocytochemistry using anti-DBP serum (Fig. 4B). Analysis of the brains of mice sampled at CT6 showed DBP only in the nuclei (not cytoplasm) in the SCN (Fig. 4B, inset) as in other brain regions, including caudate-putamen and piriform cortex (data not shown). This characteristic of DBP nuclear antigenicity has already been reported in hepatocytes (22). The majority of SCN cells (>80%) appeared to be immunoreactive for the antigen tested, and the immunoreactivity is specific, being completely blocked by preincubation with affinity-purified DBP antigen (data not shown). In contrast, the SCN sampled at CT20 contained only a few weakly stained DBP-immunoreactive nuclei. Quantitative analysis of the number of immunoreactive nuclei in the SCN sampled at 2-h intervals over 24 h in DD showed a clear circadian variation (Fig. 4B). The abundance of proteins was low at subjective dawn (CT0) and rapidly increased to the highest level at CT6, and subsequently there was a progressive decline during subjective afternoon, producing trough levels in the subjective night (P < 0.001). In contrast, expression of DBP in other areas did not exhibit appreciable circadian variation (data not shown), consistent with the constitutive expression of mPER1 proteins in regions of the brain other than the SCN (12).
Thus, in the SCN, we found that the peak time of dbp mRNA accumulation was CT4 and that of DBP protein was CT6, and the time lag between the two was about 2 h. This is consistent with prior observations in the liver, where the appearance of dbp mRNA precedes DBP protein by about 2 h (6). The rapid translation and nuclear accumulation of DBP contrast strikingly with mPER1, which accumulates with a delay of about 6 h (12).
Next we compared the circadian profile of DBP protein with that of mPer1 mRNA, which showed a peak at CT6 and a trough at CT20. Strikingly, the profiles of DBP protein accumulation and mPer1 mRNA are the same (Fig. 4A and B). These data indicate that DBP is expressed at the right time to activate mPer1 transcription in the SCN.
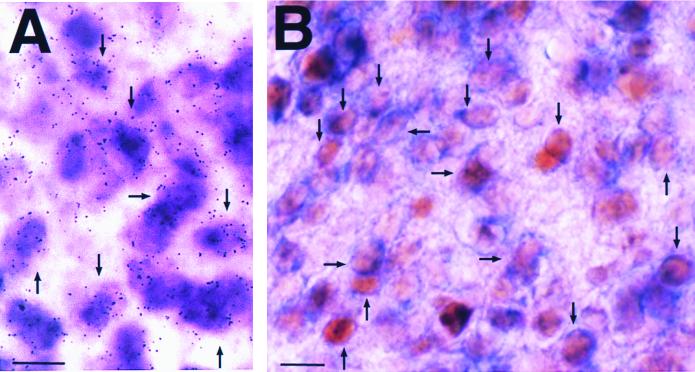
To test whether dbp and mPer1 are coexpressed in the same cells within the SCN, we performed double-labeling in situ hybridization experiments on the SCN at CT6, when levels of both dbp and mPer1 are high. We used an isotope-labeled dbp probe, which results in silver grains, and a digoxigenin-labeled mPer1 probe, which gives a purple reaction product. In the SCN, most mPer1 mRNA-positive cells also gave dbp mRNA-positive signals (Fig. 5A). This coexpression was confirmed by double-labeling using mPer1 in situ hybridization and DBP protein immunocytochemistry. By this combination, we found that the cytoplasmic blue stain of digoxigenin-labeled mPer1 mRNA surrounded the nuclear brown-stained DBP protein in many SCN cells (Fig. 5B). These findings demonstrate that DBP and mPer1 are coexpressed in the majority of SCN cells. The results therefore show that DBP is expressed in the same cells as mPer1 in the SCN and at the right time to participate in mPer1 transcription. This adds in vivo relevance to our transcriptional and DNA-binding studies. We propose that DBP may accelerate mPer1 transcription and contribute to the central oscillatory mechanism in SCN cells.
FIG. 5.
Coexpression of dbp transcripts and DBP proteins with mPer1 transcripts. (A) Double-labeling in situ hybridization using digoxigenin-labeled mPer1 probe and isotope-labeled dbp probe. Note that most cells express both mPer1 (purple) and dbp (silver grains) mRNAs. (B) Double labeling of mPer1 in situ hybridization and DBP protein immunocytochemistry. Note that the blue stain of digoxigenin-labeled mPer1 mRNA in the thin cytoplasm surrounds the brown-stained DBP protein localized in the nucleus. Arrows indicate representative double-labeled cells. All SCN sections were sampled at CT6. Bar, 10 μm.
dbp transcription is activated by CLOCK-BMAL1 and suppressed by CRY1, CRY2, and mPER3.
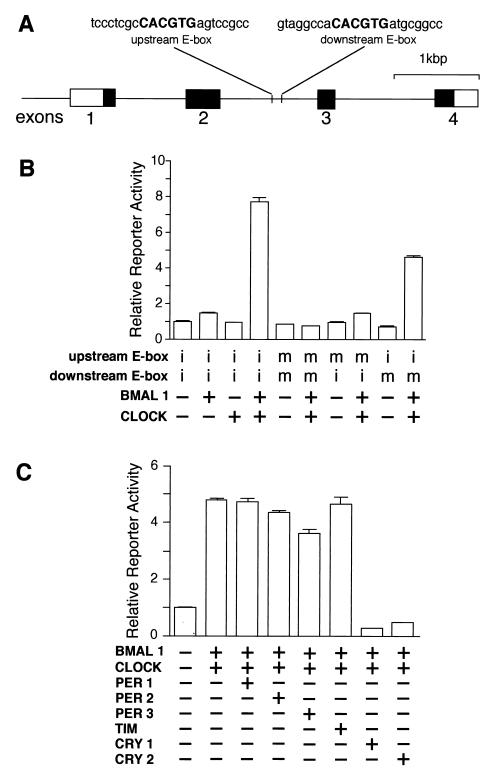
The above findings suggest that rhythmic expression of dbp plays an important role in regulating the amplitude of the mPer1 transcript rhythm. How then is the cyclic expression of dbp regulated? Circadian expression profiles indicate that transcription of dbp occurs with a similar phase to mPer1, although dbp is slightly earlier. Since mPer1 transcription is known to be activated by the CLOCK-BMAL1 heterodimer through E-boxes (CACGTG) and suppressed by mPER1, mPER2, mPER3, mTIM, CRY1, and CRY2 (9, 14, 18, 23, 24), in the following experiments we examined the possibility that similar transcriptional regulation occurs for dbp gene transcription. The mouse dbp gene is composed of four exons, and data from mice lacking exons 1 to 4 has shown that this region is essential for the circadian expression of its own gene in the SCN (20). We found two E-boxes in the second intron, ∼2.5 kbp downstream from the DBP transcription initiation site (Fig. 6A).
FIG. 6.
Transcriptional regulation of the mouse dbp gene by clock genes. (A) Structure of the mouse dbp gene and locations of the E-box sites. The solid and open boxes show the protein-coding region and the 5′ and 3′ untranslated regions, respectively. The locations of the E-box sites are indicated by vertical lines, and the sequence of each E-box with 8 bp of flanking sequence (lowercase letters) on each side is shown at the top. (B) Transactivation from the E-box sites by CLOCK-BMAL1 heterodimer. For transcriptional analysis, an SV40-driven reporter containing a 300-bp fragment derived from the mouse dbp gene centered on the two E-boxes was constructed and used (2.5 ng). The fragment was inserted immediately upstream of the SV40 promoter in the native 5′-to-3′ direction. Reporter plasmids in which one or two E-boxes were mutated were also constructed and used. Intact (i) or mutated (m) E-box of each reporter plasmid is shown, and 500 ng each of expression plasmids was used. (C) Negative regulation of CLOCK-BMAL1-induced transcription. The reporter plasmid (5 ng) containing the 300-bp fragment with the intact E-boxes was used. The amounts of the expression plasmids used were 750 ng for BMAL1 and CLOCK; 125 ng for PER1, PER2, PER3, and mTIM; and 50 ng for CRY1 and CRY2. The total amount (2 μg) of DNA was adjusted by adding pcDNA3 vector. (B and C) Presence (+) or absence (−) of the expression plasmids is shown. Each value is the mean + SEM of three replicates for a single assay. The results shown are representative of at least three independent experiments.
A 300-bp fragment centered on these E-boxes was subcloned into a reporter vector containing the SV40 promoter for use in transcriptional analysis in HepG2 cells. We found that CLOCK and BMAL1 together produced a large increase in transcriptional activity through this region (7.7-fold; P < 0.001) (Fig. 6B). Only a negligible increase was detected when either CLOCK or BMAL1 was examined alone. CLOCK-BMAL1 heterodimers apparently act through the E-box elements, because when both E-boxes were mutated, this activation was completely abolished. Unexpectedly, this CLOCK-BMAL1 activation was mainly dependent on the upstream E-box, because mutation of this site reduced the activation by CLOCK-BMAL1 from 7.7-fold to 1.5-fold, while mutation of the downstream E-box resulted in a small reduction (4.6-fold) (Fig. 6B). Interestingly, when compared to the reported sequence of the human dbp gene, the upstream E-box and the surrounding sequence are seen to be well conserved, but the downstream E box was replaced (CACGTG to GACGTG) in the human genome.
Although the E-boxes are located within an intron, they are thought to act as an enhancer which functions regardless of direction or location. In fact, in both cases when the fragment was inserted in the reverse 5′-to-3′ direction immediately upstream of the SV40 promoter or in the native 5′-to-3′ direction immediately downstream of the luciferase reporter gene, CLOCK-BMAL1 produced a large increase in transcriptional activity (10.1-fold [P < 0.001] and 8.8-fold [P < 0.001], respectively). Thus, the data indicate that CLOCK-BMAL1 has the ability to regulate DBP transcription through the E-boxes.
To investigate the negative elements which may contribute to DBP regulation, we examined whether mPER, mTIM, or CRY could individually inhibit CLOCK-BMAL1-induced transcription. Among them, mPER3 significantly reduced (24.3%; P < 0.001) and CRY1 and CRY2 completely abolished CLOCK-BMAL1-mediated transcription (Fig. 6C). Any possible combination of mPERs and mTIM expression plasmids did not inhibit more effectively than when they were transfected alone (data not shown). These results do not exclude the possibility that mPERs and mTIM are important for negative regulation, because endogenous expression of these genes is observed in the cell lines used (data not shown). Although the precise interactions among negative elements requires further analysis, the data imply that the dbp gene is regulated by central clock components, including CLOCK-BMAL1 and PER3, CRY1, and CRY2.
DISCUSSION
Molecular dissection of the mammalian clock oscillating system has been advanced using Drosophila as a model, because many genes structurally homologous to Drosophila clock genes are found in mammals. The first and most completely characterized clock gene in the animal kingdom is the Drosophila per gene. Three mammalian structural homologues were isolated from mouse and human (1, 25, 27, 29, 30), and among them mPer1, the first identified mouse period gene, attracted intense attention because it resembled the Drosophila per gene in many aspects (5). Transcription of mPer1 is activated by binding of the CLOCK-BMAL1 heterocomplex, both of which are basic helix-loop-helix (bHLH)-PAS proteins, to the E-boxes in the promoter region of mPer1. The negative limb of the feedback loop includes mPER1, mPER2, mPER3, CRY1, CRY2, and mTIM, and these are thought to form a multimeric protein complex with CLOCK-BMAL1, similar to the fruit fly PER, TIM, and CLOCK (19), and to suppress transcription.
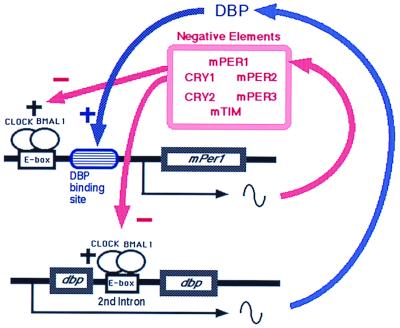
In the present study, we have integrated the dbp gene and DBP protein into this negative feedback loop (Fig. 7). In this model, at the phase of increasing mPer1 transcription in the subjective morning, when negative elements cease their suppression of CLOCK-BMAL1-induced transactivation, CLOCK-BMAL1 begins to promote the transcription of mPer1. At the same time, CLOCK-BMAL1 binds to E-boxes in the second intron of the dbp gene and activates dbp transcription. Rapidly produced DBP proteins go into the nucleus and directly bind to the DBP-binding site of the mPer1 promoter. There, DBP cooperatively increases the transcription rate of mPer1. During the subjective afternoon, when negative elements strongly suppress the CLOCK-BMAL1-induced transcription, the transcripts of mPer1 and dbp decrease. The decrease in dbp transcripts is followed by the immediate decrease in DBP protein. Because mPer1 is regulated by CLOCK-BMAL1, PER, CRY, and DBP, decreased DBP protein facilitates the decrease in mPer1 transcripts. Thus, the time-specific appearance and disappearance of DBP may help to increase the amplitude of mPer1 transcripts.
FIG. 7.
Schematic representation of the role of DBP in the circadian oscillatory mechanism of the SCN. In addition to the negative autoregulatory feedback loop of mPer1, a DBP-mediated loop exists. As DBP protein positively regulates the mPer1 promoter, DBP amplifies the circadian oscillation of mPer1 and thereby influences the circadian oscillator.
DBP as well as mTIM, mPER1, mCRY1, and mCRY2 (12, 18) is found in the nucleus of clock-oscillating SCN cells. These genes show time-specific expression in SCN cells except for mTIM, which is constantly expressed at all times examined (12). Interestingly, the peaks of CRY1, CRY2, and mPER1 proteins occur at subjective dusk (CT12) in the SCN (12, 18). However, the circadian expression profile of DBP was completely different from these and showed a peak in the subjective morning (CT6). Since DBP is a positive element and mPER1, mCRY1, and mCRY2 are negative elements, it is possible that these transcription factors are expressed according to their role in mPer1 transcription and in the clock.
In addition to its role in core clock oscillation, DBP may also be involved in the circadian output system. It was recently shown that circadian oscillation of vasopressin mRNA levels is directly regulated by the central loop of the mammalian clock. The vasopressin gene is activated by CLOCK-BMAL1 heterodimers and repressed by the mPER, mTIM, and mCRY proteins through an E-box in the promoter region (14, 18). Therefore, output genes that may be responsible for changes in the physiology and behavior of the animal can be directly linked to the central clock mechanism. This type of clock gene regulation will fit genes having an E-box. The consensus binding site for DBP is different from the CLOCK-BMAL1 E-box binding site. Therefore, DBP, which is tightly linked to core clock oscillation, could potentially regulate a totally different set of output genes. In fact, DBP activates the transcription of some genes in the liver by directly binding to their promoters, such as the albumin, cholesterol 7α hydroxylase, and cytochrome P450 (CYP2C6) genes (7). Although the target genes in SCN neurons have not been elucidated at present, DBP may diversify the circadian output in the SCN.
Although DBP knockout mice are rhythmic with a slightly shortened period (20), this finding is consistent with the suggestion that DBP is an important component of the circadian clock. However, interpretation of this rhythmicity and short period length should be done with extreme discretion, since DBP is one of the PAR (proline and acidic amino acid-rich) leucine zipper transcription factors, as well as HLF and TEF (8). HLF and TEF may interact with DBP and share the possible binding sites (6). This kind of redundancy and complexity were found in mCry genes, composed of mCry1 and mCry2. Although mCry1 mCry2 double-knockout mice were completely arrhythmic, mCry1 or mCry2 single-knockout mice were rhythmic (31). Moreover, mCry1 and mCry2 single-knockout mice showed a shorter and longer free-running period, respectively (31), in spite of the indistinguishable actions of mCRY1 and mCRY2 proteins on the mPer1 promoter (18). Further study of the potential influence of the other PAR leucine zipper transcription factors would thus be interesting.
Recently, the Drosophila vri (vrille) gene, which showed strong homology in its DNA-binding domain to DBP (10), was demonstrated to also be expressed in pacemaker cells (3). VRI also has a role in circadian locomotor activity: the gene dosage of vri affects the period of the circadian rhythm (3). VRI apparently lacks a PAR domain, which is conserved among DBP, HLF, and TEF. However, there is a set of proteins, including mammalian E4BP4, Drosophila Giant, and Caenorhabditis elegans CES2 (10), that have DNA-binding domains closely related to that of DBP but also lack a PAR domain. Interestingly, the E4BP4, Giant, and CES2 proteins all have the ability to repress transcription. Therefore, VRI may play a suppressive role in the Drosophila clock, in contrast to DBP's activating role. Indeed, recent data have shown that continuous vri expression decreases PER protein and tim mRNA levels (3). It is possible that the activating PAR leucine zipper proteins and the related repressing transcription factors make good complements to each other for regulating the clock.
In summary, our data indicate that DBP is part of the central oscillatory clock in mammals and has an important role in ensuring a precise 24-h rhythm. This finding will provide a new viewpoint for the circadian oscillatory system and suggests that this family of transcription factors may have roles in the clock in many diverse organisms.
ACKNOWLEDGMENTS
We thank K. Taguchi, S. Takekida, Y. Sumi, and H. Onishi for technical assistance; T. Nagase for providing human Clock and Cry2 clones; K. Okubo for donating HepG2 cells; and J. Blau, M. Young, and Y. Shigeyoshi for useful discussions and comments on the manuscript.
This work was supported in part by grants from the Special Coordination Funds of the Science and Technology Agency of Japan, the Grant-in-Aid for the Scientific Research on Priority Areas of the Ministry of Education, Science, Sports and Culture of Japan, Mitsubishi Foundation, and SRF.
REFERENCES
- 1.Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 2.Ban Y, Shigeyoshi Y, Okamura H. Development of circadian VIP rhythm in the rat suprachiasmatic nucleus. J Neurosci. 1997;17:3920–3931. doi: 10.1523/JNEUROSCI.17-10-03920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blau J, Young M W. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 4.Citri Y, Colot H V, Jacquier A C, Yu Q, Hall J C, Baltimore D, Rosbach M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- 5.Dunlap J C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 6.Falvey E, Fleury-Olela F, Schibler U. The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falvey E, Marcacci L, Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- 8.Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- 9.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Tahahashi J S, Weitz C J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 10.George H, Terracol R. The vrille gene of Drosophila is a maternal enhancer of decapentaplegic and encodes a new member of the bZIP family of transcription factors. Genomics. 1997;146:1345–1363. doi: 10.1093/genetics/146.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardin P E, Hall J C, Rosbash M. Feedback of the drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 12.Hastings M H, Field M D, Maywood E S, Weaver D R, Reppert S M. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;11:1–7. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson F R, Bargiello T A, Yun S H, Young M W. Product of per locus of Drosophila shares homology with proteoglycans. Nature. 1986;320:185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- 14.Jin X, Shearman L P, Weaver D R, Zylka M J, de Vries G J, Reppert S M. A molecular mechanism regulating rhythmic output from the suprachiasmatic nucleus. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 15.Kageyama R, Sasai Y, Nakanishi S. Molecular characterization of transcription factors that bind to the cAMP responsive region of the substance P precursor gene: cDNA cloning of a novel C/EBP-related factor. J Biol Chem. 1991;266:15525–15531. [PubMed] [Google Scholar]
- 16.Klein D C, Moore R Y, Reppert S M. Suprachiasmatic nucleus: the mind's clock. New York, N.Y: Oxford University Press; 1991. p. 467. [Google Scholar]
- 17.Kondo T, Ishiura M. The circadian clocks of plants and cyanobacteria. Trends Plant Sci. 1999;4:171–176. doi: 10.1016/s1360-1385(99)01410-7. [DOI] [PubMed] [Google Scholar]
- 18.Kume K, Zylka M J, Sriram S, Shearman L P, Weaver D H, Jin X, Maywood E, Hastings M H, Reppert S M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 19.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore R Y. The suprachiasmatic nucleus and the organization of a circadian system. TINS (Trends Neurosci) 1982;5:404–407. [Google Scholar]
- 22.Mueller C R, Maire P, Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 1990;61:279–291. doi: 10.1016/0092-8674(90)90808-r. [DOI] [PubMed] [Google Scholar]
- 23.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers J H, van der Horst G T. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 24.Sangoram A M, Saez L, Antoch M P, Gekakis N, Staknis D, Whiteley A, Fruechte E M, Vitaterna M H, Shimomura K, King D P, Young M W, Weitz C J, Takahashi J S. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 25.Schreiber E, Matthias P, Mueller M, Schffner W. Rapid detection of octamer binding proteins with “mini-extracts” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F J, Reppert S M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 27.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros J J, Dunlap J C, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 28.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 29.Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, Sakakida Y, Okumura K, Takashima N, Okamura H. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells. 1998;3:167–176. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- 30.Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, Yagita K, Yan L, Young M L, Okamura H. A light independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 32.van der Horst G T, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkert A, Eker A P, van Leenen D, Buijs R, Bootsma D, Hoeijmakers J H, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi S, Nakanishi S. Regional expression and regulation of alternative forms of mRNAs derived from two distinct transcription initiation sites of the rat mGluR5 gene. J Neurochem. 1998;71:60–68. doi: 10.1046/j.1471-4159.1998.71010060.x. [DOI] [PubMed] [Google Scholar]
- 34.Yan, L., S. Miyake, and H. Okamura. 1999. Distribution and circadian expression of dbp in SCN and extra-SCN areas in the mouse brain. J. Neurosci. Res., in press. [DOI] [PubMed]
- 35.Yan L, Takekida S, Shigeyoshi Y, Okamura H. Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience. 1999;94:141–150. doi: 10.1016/s0306-4522(99)00223-7. [DOI] [PubMed] [Google Scholar]
- 36.Zylka M J, Shearman L P, Weaver D R, Reppert S M. Three period homolog in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]