Transcriptional Repression by Drosophila Methyl-CpG-Binding Proteins (original) (raw)
Abstract
C methylation at genomic CpG dinucleotides has been implicated in the regulation of a number of genetic activities during vertebrate cell differentiation and embryo development. The methylated CpG could induce chromatin condensation through the recruitment of histone deacetylase (HDAC)-containing complexes by methyl-CpG-binding proteins. These proteins consist of the methylated-DNA binding domain (MBD). Unexpectedly, however, several studies have identified MBD-containing proteins encoded by genes of Drosophila melanogaster, an invertebrate species supposed to be void of detectable m5CpG. We now report the genomic structure of a Drosophila gene, dMBD2/3, that codes for two MBD-containing, alternatively spliced, and developmentally regulated isoforms of proteins, dMBD2/3 and dMBD2/3Δ. Interestingly, in vitro binding experiments showed that as was the case for vertebrate MBD proteins, dMBD2/3Δ could preferentially recognize m5CpG-containing DNA through its MBD. Furthermore, dMBD2/3Δ as well as one of its orthologs in mouse, MBD2b, could function in human cells as a transcriptional corepressor or repressor. The activities of HDACs appeared to be dispensable for transcriptional repression by dMBD2/3Δ. Finally, dMBD2/3Δ also could repress transcription effectively in transfected Drosophila cells. The surprisingly similar structures and characteristics of the MBD proteins as well as DNA cytosine (C-5) methyltransferase-related proteins in Drosophila and vertebrates suggest interesting scenarios for their roles in eukaryotic cellular functions.
In vertebrates, including the mammals, the chromosomal DNAs are modified by C methylation at a limited number of CpG dinucleotides, resulting in m5CpG, methylated at position 5 of the C residues. This methylation at CpG has been implicated in the regulation of a number of genetic activities during mammalian cell differentiation and embryo development. These activities include tissue-specific gene transcription, X chromosome inactivation, genomic imprinting, cellular defense against viral agents, and tumorigenesis (references 1, 2, 11, 13, 15, and 34 and references therein). The level of m5CpG in the vertebrate cells is presumably balanced by the combined actions of DNA cytosine (C-5) methyltransferases (CpG MTases) (2, 26) and DNA demethylases (3).
It is generally accepted that regulation of the different cellular activities by DNA methylation is modulated mainly through the m5CpG residues. Indeed, m5CpG could be recognized by a specific class of proteins that consist of the methylated-DNA binding domain (MBD) (21). These “MBD proteins,” in particular MeCP2 and MBD2/3, could preferentially bind to m5CpG-containing DNA, recruit histone deacetylase (HDAC)-containing complexes, and thus cause chromatin condensation in the vicinity of m5CpG (12, 22, 23, 33). This is very likely one major, but not the only, scheme involved in the functioning of vertebrate DNA methylation.
Unlike the vertebrates, several invertebrate species, including Drosophila melanogaster (27, 31), do not have apparent DNA methylation in their genomes. Nor has CpG MTase been reported to occur in these invertebrate animals. It was thus surprising to find mammalian CpG MTase-related proteins as well as MBD proteins expressed in Drosophila. A Drosophila protein, DmMTR1, is characteristically similar to the vertebrate maintenance CpG MTase, dnmt1, in several aspects (9). In particular, DmMTR1 molecules are located outside the nuclei during interphase of the syncytial Drosophila embryos, as is dnmt1 in the mouse blastocyst (19). However, DmMTR1 molecules appear to be rapidly transported into and then out of the nuclei again, as the embryos undergo the mitotic waves (9). Immunofluorescence data further indicate that DmMTR1 molecules “paint” the whole set of condensed Drosophila chromosomes throughout the mitotic phase, suggesting that it may play an essential role in the cell cycle-regulated condensation of the Drosophila chromosomes. In addition to DmMTR1, another Drosophila polypeptide, DmMT2, exhibiting high sequence homology to mammalian dnmt2, a relatively short homolog of dnmt1 but without detectable methylation activities (25), has been identified through a database search (9, 30). Expression of DmMT2 in Drosophila is developmentally regulated (9).
Interestingly, several recent reports have also noted the identification of an MBD protein encoded by the Drosophila genome (9, 30, 33). Like the mammalian MBD (12, 22, 23, 33), the Drosophila MBD protein, which was termed the “Drosophila MBD-like sequence” (33) or dMBD2/3 (30), interacts with HDAC in vitro and in vivo (30). However, it did not appear to recognize m5CpG (30). Here we report the complete genomic structure of the Drosophila dMBD2/3 gene, which encodes two alternatively spliced and developmentally regulated MBD proteins. We also provide the first evidence that the Drosophila MBD protein(s) could recognize m5CpG-containing DNA, albeit with a lower affinity than the mammalian MBD proteins. Remarkably, the protein(s) could also function as a corepressor via an HDAC-independent pathway.
MATERIALS AND METHODS
Recombinant clones.
The three P1 clones containing the dMBD2/3 gene were identified by the hybridization of filter-containing Drosophila P1 genomic clones (Genome Systems Inc.) with an expression sequence tag (EST) clone, LD03777. The nucleotide sequences of the P1 clones, LD03777, and another EST clone, LD46808, were determined by automated sequencing of both strands.
All recombinant procedures were done by the methods of Sambrook et al. (29). The cDNAs of dMBD2/3Δ and mouse MBD2b were cloned in frame into pGEX-4T-2 and pGEX-4T-1 (Amersham Pharmacia Biotech), respectively. To generate the plasmid pGEX-dMBD2/3Δ (36-226), encoding the glutathione transferase (GST) fusion protein of dMBD2/3Δ without the MBD, the cDNA of _dMBD2/3_Δ was cut with _Hae_II and blunt ended, and the 3′ fragment was cloned into pGEX-4T-1.
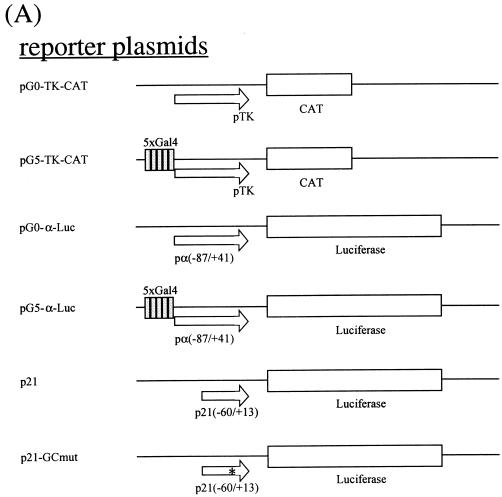
The maps of the recombinant plasmids used in transfection are depicted in Fig. 4A. pSG424-dMBD2/3Δ contains the sequence for amino acids (aa) 1 to 226 of dMBD2/3Δ cloned in frame in the expression vector pSG424 (28). pSG424-MBD2b contains the sequence for aa 1 to 226 of the mouse MBD2b protein (8). pSG424-p45 was a gift from N. Gavva at the University of California at Davis. pCI-Gal4 harbors the sequence for the Gal4 DNA binding domain downstream of the cytomegalovirus (CMV) promoter in pCI (Promega Biotech). pCI-dMBD2/3Δ contains the sequence for aa 1 to 226 of dMBD2/3Δ cloned in frame with Gal4 of pCI-Gal4. Similarly, pCI-Gal4-MBD2b contains the sequence for aa 1 to 262 of the mouse MBD2b protein. To obtain pCI-Gal4-dMBD2/3Δ (36-226) without the MBD, the respective cDNA was released from pGEX-dMBD2/3Δ (36-226) by _Eco_RI/_Xho_I digestion and cloned into _Eco_RI/Sal_I-cut pCI-Gal4. pG5-TK-CAT was a gift from M. Ptashne. It contains five Gal4-binding sites in front of the thymidine kinase (TK) basic promoter. pG0-TK-CAT is the derivative without Gal4-binding sites. pG0-α-Luc contains the proximal human α1 globin promoter (−87 to +41) upstream of the luciferase gene of pGL3-basic (Promega Biotech). To create pG5-α-Luc, five Gal4-binding sites were cloned upstream of the α1 globin promoter in pG0-α-Luc. p21 (35) contains the proximal promoter region (−60 to +13) of the murine p21_waf1 gene upstream of the luciferase gene in pGL3-basic. p21-GCmut is a derivative containing a mutated GC box. pPacSp1 was a generous gift from Guntram Suske (Institute of Molecular Biology and Tumor Research, Philipps University, Marburg, Germany). pAc5.1/V5-His/lacZ was obtained from Invitrogen.
FIG. 4.
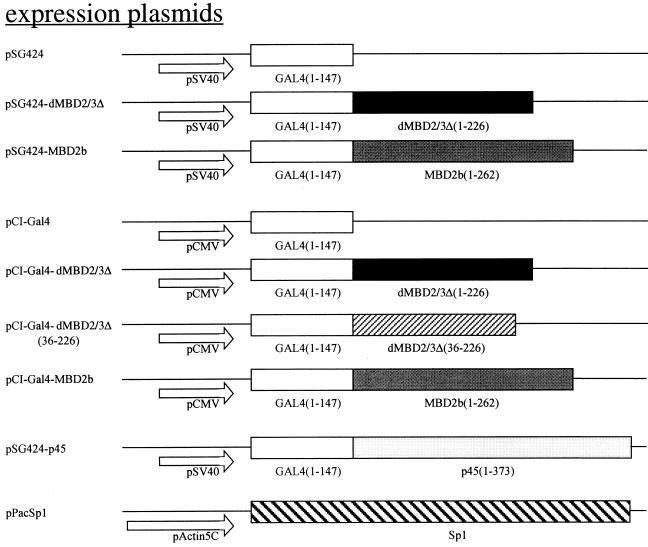
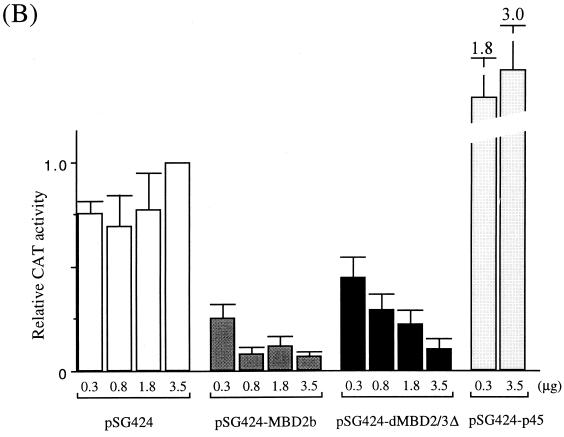
Transcription repression by Drosophila and mammalian MBD proteins in human cells. (A) Maps of reporter and expression plasmids used in the cotransfection assays. For more details of the plasmids, see Materials and Methods. (B) Relative CAT activities from pG5-TK-CAT in human 293T cells cotransfected with increasing amounts of different recombinant expression plasmids. The CAT activities were standardized with the β-galactosidase activity from cotransfected pCMV-β-gal. Activity with 3.5 μg of cotransfected pSG424 is defined as 1. The bars represent standard deviations of two sets of transfection carried out in duplicate. (C) Relative luciferase activities from pG5-α-Luc or pG0-α-Luc in human 293T cells cotransfected with increasing amounts of different recombinant expression plasmids. The activation from pG5-α-Luc or pG0-α-Luc was measured and standardized with the β-galactosidase activity from cotransfected pCMV-β-gal. The luciferase activity from pG5-α-Luc without cotransfected expression plasmid is defined as 1 (lane 1). Note that in contrast with activity shown in panel B, the expression of the Gal fusions in this set of experiments was directed by a CMV promoter in the pCI vector.
Northern blot analysis.
Total Drosophila RNAs were isolated from different stages using the TRIZOL reagent (Life Technologies). Poly(A) RNAs were then isolated with a kit from Qiagen. One microgram of the RNAs was loaded in each lane, blotted onto nitrocellulose filter paper, and hybridized with 32P-labeled LD03777 probe. α1 tubulin RNA patterns were used as the internal control for the Northern blot analysis.
EMSA.
The sequence of the β-TATA oligonucleotide used in the electrophoretic mobility shift assay (EMSA) is 5′-GATCTCACTGTGTCGACCTTGGGCATAAAAGGCAGAGCACTGGAGCTGCTGCTTAA-3′ (complement, 3′-CTAGAGTGACACAGCTGGAACCCGTATTTTCCGTCTCGTGACCTC GACGACGAATT-5′). To obtain methylated DNA, the oligonucleotide or pBluescript II KS (pBSKS; Stratagene) was fully methylated with _Sss_I methylase according to the instructions of the manufacturer (New England Biolabs). The oligonucleotides were labeled with 32P at their 5′ ends and gel purified prior to EMSA. The GST fusion proteins were all generated from recombinant pGEX-4T plasmids (Amersham Pharmacia Biotech) transformed into Escherichia coli BL21 cells. The proteins were purified with glutathione-Sephadex beads (Amersham Pharmacia Biotech). The expression plasmid pGST-ankyrin was a gift from Xin Chen. For EMSA, different amounts of the fusion proteins were preincubated with 100 ng of pBSKS and 8 μg of bovine serum albumin at room temperature for 5 min in 20 mM HEPES (pH 7.9)–3 mM MgCl2–1 mM EDTA–10 mM β-mercaptoethanol–0.1% Triton X-100–10% glycerol in a total volume of 20 μl. The labeled oligonucleotides (40,000 cpm, approximately 1 ng) were then added, and the samples were further incubated for 20 min at room temperature. To separate the protein-DNA complexes, the reaction mixtures were loaded onto a running nondenaturing 4% polyacrylamide gel, which had been prerun in 0.5× Tris-borate-EDTA buffer for 30 min at 4°C and 200 V. Electrophoresis was further carried out at 4°C and 200 V for 2 1/2 h. Gels were dried and exposed on X-ray film at −80°C. For competition experiments, a 100-fold molar excess of the competitor DNAs was included in the binding reactions.
Transient DNA transfection.
The human kidney epithelial cell line 293T was grown in Dulbecco modified Eagle medium–10% fetal calf serum and transfected at 20% confluence in 2-cm “Cell Wells.” The DNA samples for transfection were introduced as a calcium phosphate coprecipitate consisting of 1 μg of a reporter plasmid (pG0-TK-CAT, pG5-TK-CAT, pG0-α-Luc, pG5-α-Luc, p21, or p21-GCmut), 0.3 μg of pCMV-β-gal (18), 10 μg of salmon sperm DNA as the carrier, and different amounts of an expression plasmid [pSG424-dMBD2/3Δ, pSG424-MBD2b, pCI-Gal4, pCI-Gal4-dMBD2/3Δ, pCI-Gal4-dMBD2/3Δ (36-226), pCI-Gal4-MBD2b, or pSG424-p45]. pSG424 and/or pCI-Gal4 was used as the control plasmid(s), and either was also added as required to keep the total amount of DNA used in each transfection the same. Sixteen hours after transfection, the medium was changed and cells were incubated for another 48 h. The chloramphenicol acetyltransferase (CAT), luciferase, and β-galactosidase assays were performed as described previously.
For transfection of the Drosophila SL2 cell line, the cells were maintained at 23.5°C in a modified M3 medium (Invitrogen) supplemented with 10% fetal bovine serum and penicillin and streptomycin (both from Gibco BRL). At 18 h before the transfection, 2 × 106 cells were plated out onto individual 10-cm petri dishes. On the following day, the cells were harvested and resuspended in medium without the serum and antibiotics. Then 7.5 × 105 cells/well were seeded onto a 24-well plate, and DNA transfection was carried out using Lipofectin. Each liposome-DNA mixture contained 1 μg of the reporter plasmid pG5-α-Luc or pG0-α-Luc, different amounts of an MBD expression plasmid or its corresponding vector, 0.2 μg of pPac-Sp1, and 0.3 μg of pAc5.1/V5-His/lacZ. The transfected cells were kept in antibiotic-free and serum-free medium at 23.5°C for 6 h before replacement with normal serum. The luciferase and β-galactosidase activities were assayed after another 72 h of incubation.
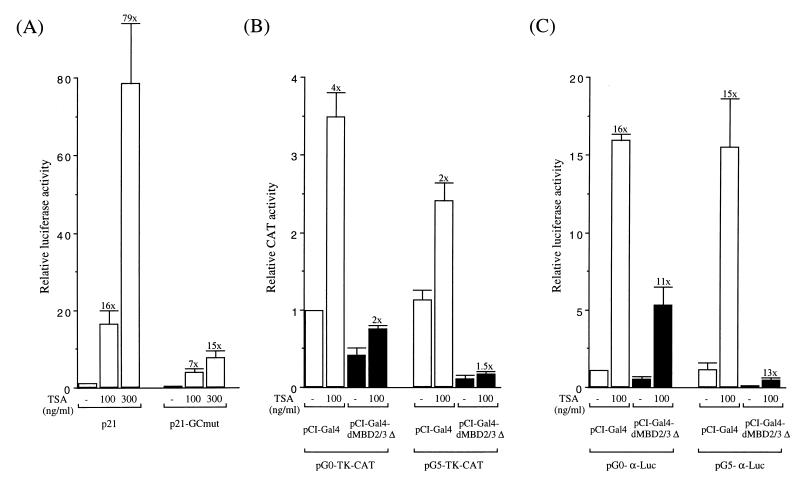
For trichostatin A (TSA) treatment, 1 μg of the reporter plasmid(s) and 50 ng of the expression plasmid(s) were used in each cotransfection. Twenty hours after transfection, the transfected 293T cells were treated with TSA at various concentrations (see Fig. 5) for 24 h. Luciferase or CAT activities were then assayed and normalized to the extract protein concentrations. In keeping with previous studies (16), we observed a general increase of the promoter activities after the TSA treatment, e.g., six- to eightfold for the CMV promoter and five- to ninefold for the Rous sarcoma virus promoter (data not shown).
FIG. 5.
Effects of TSA treatment on the repression function of dMBD2/3Δ. (A) Activity increases of the p21_waf1_ and p21-GCmut promoters after treatment with the indicated concentrations of TSA. Increases relative to the promoter activity in the absence of TSA treatment are indicated above the bars. (B) Activity increases of the TK promoter by TSA treatment. The blank bars represent the TK promoter activities in the presence of exogenously expressed Gal4 protein, and the black bars represent activities with the Gal4 fusion of dMBD2/3Δ. (C) Activity increases of the human α globin promoter caused by TSA treatment.
RESULTS AND DISCUSSION
Genomic organization of dMBD2/3, a Drosophila gene encoding two MBD-containing polypeptides.
Multiple EST clones were identified by Blast search using the mammalian MBD2b sequence (8) as the bait. One of the clones (LD03777) originating from a Drosophila embryonic cDNA library was first sequenced. Its insert is 865 bp long, including the poly(A) tail. LD03777 bears an open reading frame corresponding to 226 aa, which has been referred to previously as the “Drosophila MBD-like sequence” (33) or dMBD2/3Δ (30). The protein contains an MBD-like sequence in the N-terminal domain and a coiled-coil motif near the C terminus. It is apparently the ortholog of the vertebrate MBD2/MBD3 family (8, 30, 33).
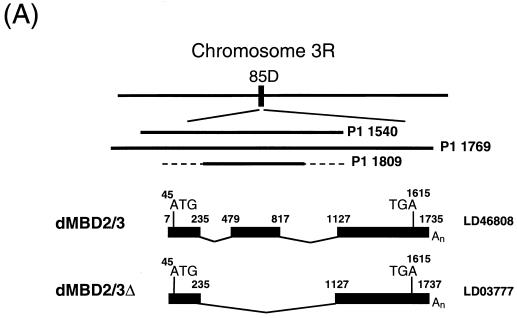
_dMBD2/3_Δ is the product of alternative splicing. First, blot hybridization of a P1 Drosophila genomic filter with LD03777 identified three P1 clones, 1540, 1769, and 1809 (Fig. 1A). The in situ data located clones 1540 and 1769 to 85D20-85D24 and 85D18-85D27, respectively, but no in situ information was available for clone 1809. Comparison of the nucleotide sequences of LD03777 and the P1 clones revealed the existence of an 891-bp intron (bp 236 to 1126 [Fig. 1B]) downstream of the MBD of dMBD2/3Δ. Second, we found another EST clone, LD46808, 1,198 bp long, which contains an extra exon of 339 bp derived from the 891-bp intron (bp 479 to 817 [Fig. 1A and B]). These data indicate that the primary transcript of the dMBD2/3 gene shown in Fig. 1B is alternatively spliced to generate two mRNAs coding for dMBD2/3Δ and a longer protein, dMBD2/3 (Fig. 1A).
FIG. 1.
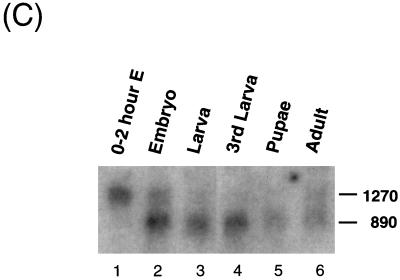
Structure and expression of the Drosophila dMBD2/3 gene. (A) Genomic and cDNA organization of dMBD2/3. The chromosomal location (85D) of the gene and the three P1 clones spanning the region are indicated. Shown below the P1 clones are the cDNAs for dMBD2/3 and dMBD2/3Δ, which were derived from the cDNA clones LD46808 and LD03777, respectively. The base numbering of the two cDNAs corresponds to that of the genomic sequence of dMBD2/3 shown in panel B. (B) Genomic sequence of dMBD2/3. The sequence of the dMBD2/3 gene was deduced from the three P1 clones. The putative site of transcriptional initiation, as inferred from the sequences of the two cDNA clones, is designated +1. The two introns and the regions upstream and downstream of the gene are shown with lowercase letters. The corresponding amino acids are also shown, with those homologous to the conserved amino acids among the vertebrate MBDs indicated by bold letters. The putative TATA box in the upstream promoter region and the poly(A) signal, AATAAA, are underlined. (C) Expression of dMBD2/3 and _dMBD2/3_Δ mRNA. Drosophila poly(A) RNAs isolated from different stages of development were analyzed by Northern blot hybridization with the cDNA clone LD03777 as the probe. The two transcripts (1,270 and 890 nt long) are indicated.
Developmental regulation of the alternative splicing of dMBD2/3.
The two dMBD2/3 mRNAs are differentially expressed during Drosophila development, as shown by Northern blot analysis using LD03777 as the hybridization probe (Fig. 1C). In overnight embryos and in adult flies, two mRNAs of lengths similar to those of dMBD2/3Δ (890 nucleotides [nt]) and dMBD2/3 (1,270 nt), respectively, were present (Fig. 1C, lanes 2 and 6). On the other hand, only the long transcript was expressed in embryos younger than 2 h (Fig. 1C, lane 1), and the short mRNA was the predominant species at the other stages of development (Fig. 1C, lanes 3 through 5). The Northern blotting data suggest that the alternative splicing scheme generating dMBD2/3 and dMBD2/3Δ mRNAs is developmentally regulated and that the two proteins play different roles during development. dMBD2/3 is likely an essential gene for the early development of Drosophila, since a line with P-element insertion in the putative promoter region of dMBD2/3 is lethal to homozygous embryos (M.-S. Hung, unpublished data).
dMBD2/3Δ could preferentially recognize m5CpG-containing DNA.
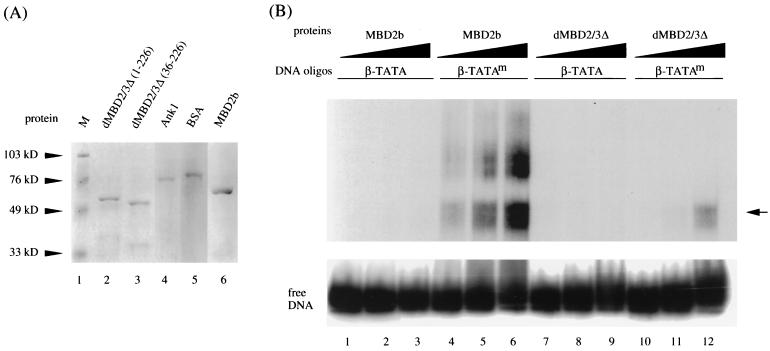
Whether the Drosophila MBD protein binds to m5CpG is an interesting question. All vertebrate MBD proteins identified thus far could recognize and preferentially bind to DNA containing m5CpG (reference 4 and reference therein). Close sequence examination indicates that the MBD in dMBD2/3 is similar to those in vertebrates but with many amino acid differences, including relatively long deletions (Fig. 1B) (30, 33). Recombinant GST fusions of different MBD proteins (Fig. 2A) were thus compared for their abilities to bind methylated or unmethylated DNA (Fig. 2B). As expected from the previous studies (8), little binding of mouse MBD2b to unmethylated probe could be detected (Fig. 2B, lanes 1 through 3). On the other hand, it bound to the methylated DNA probe with relatively high affinity (Fig. 2B, compare lanes 4 through 6 to lanes 1 through 3).
FIG. 2.
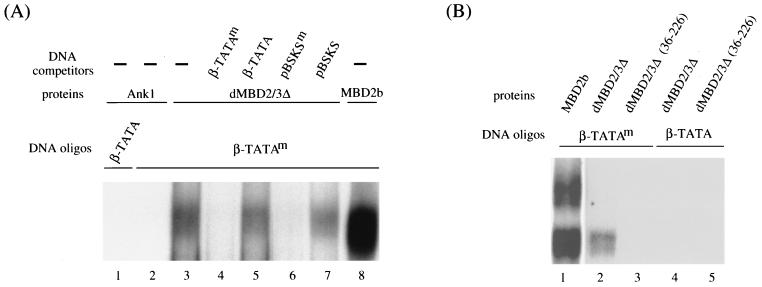
Preferential binding of Drosophila and mammalian MBD proteins to m5CpG-containing DNA. (A) Coomassie blue staining patterns of recombinant GST fusion polypeptides. One microgram each of GST fusions with the intact dMBD2/3Δ (lane 2), dMBD2/3Δ (36-226) (lane 3), a mouse ankyrin polypeptide (lane 4), and mouse MBD2b (lane 6) was electrophoresed on a denaturing sodium dodecyl sulfate-polyacrylamide gel and stained with Coomassie blue. Also run on the gel were molecular mass markers (lane 1) and bovine serum albumin (lane 5). (B) EMSA of recombinant GST fusions. The abilities of the Drosophila dMBD2/3Δ and mouse MBD2b proteins to recognize m5CpG were compared by EMSA. A 56-bp DNA probe (β-TATA) containing the human β globin TATA box with a single CpG was electrophoresed after incubation with competitor DNA and increasing amounts of proteins (2, 20, and 200 ng). Another probe, β-TATAm, which contains the same sequence as β-TATA but with the CpG site methylated at position 5 of the C residues, was analyzed in the same way. The oligonucleotides β-TATA and β-TATAm were used as the probes in an EMSA after binding with increasing amounts (2, 20, and 200 ng) of recombinant GST fusions of MBD2b or dMBD2/3Δ. The arrow points to the complex formed between the β-TATAm oligonucleotide and both MBD2b and dMBD2/3Δ. The GST protein did not form a complex either with β-TATA or with β-TATAm (data not shown).
Interestingly, Drosophila dMBD2/3Δ also formed a specific complex with the methylated probe but not with unmethylated DNA (Fig. 2B, lanes 7 through 12). This complex comigrates with the fast band of the mouse MBD2b-DNA complexes (Fig. 2B, compare lanes 6 and 12). Quantitation of the respective complexes by PhosphorImager analysis (Fig. 2B) and experiments using lower concentrations of the recombinant proteins (data not shown) indicated that the affinity of binding of dMBD2/3Δ to the methylated DNA probe was approximately 20-fold lower than that of the mouse MBD2b. This reduced affinity of binding of dMBD2/3Δ to methylated DNA is likely due to the relatively divergent sequence and consequently the structure of the MBD in dMBD2/3Δ. The preferential binding of dMBD2/3Δ to DNA containing m5CpG was also noted by others (A. Wolffe, personal communication).
MBD is required for dMBD2/3Δ binding to methylated DNA.
The specificity of the dMBD2/3Δ-DNA complex was further confirmed by experiments shown in Fig. 3. First, as exemplified for a GST-ankyrin fusion polypeptide (Fig. 3A, lanes 1 and 2), proteins without MBD do not bind to the probe, methylated or unmethylated. Similarly, GST alone could not form a complex with the same set of DNA oligonucleotides (data not shown). Second, the complex was effectively competed away upon the addition of a 100-fold molar excess of methylated probe or methylated pBluescript DNA (Fig. 3A, compare lane 3 to lanes 4 and 6) but not upon addition of the same amount of unmethylated competitors (Fig. 3A, lanes 5 and 7). Finally, removal of aa 1 to 35 from dMBD2/3Δ, which comprise a major portion of the MBD-like sequence, abolished its ability to bind methylated DNA, as demonstrated by EMSA (Fig. 3B, compare lanes 2 and 3) and by Southwestern analysis (data not shown). It should be noted here that Tweedie et al. (30) have found no methyl-CpG-binding activity for either dMBD2/3 or dMBD2/3Δ in their EMSA. However, in those cases, a different DNA oligonucleotide was used. It is likely that the recognition of m5CpG by these two Drosophila MBD proteins is dependent on the DNA sequence context.
FIG. 3.
(A) Oligonucleotide competition EMSA of dMBD2/3Δ. β-TATA and β-TATAm oligonucleotides were allowed to form complexes with 200 ng of the GST fusion of dMBD2/3Δ, 20 ng of the GST fusion of MBD2b, or 200 ng of the GST fusion of a mouse ankyrin polypeptide. The binding reactions were carried out without (−) or with the inclusion of 100 ng of competitor DNA oligonucleotide β-TATA, β-TATAm, plasmid pBSKS, or pBSKS methylated at all of the CpG sites (pBSKSm). The reaction products were then analyzed by EMSA. (B) Requirement of the MBD of dMBD2/3Δ for complex formation. The oligonucleotides were allowed to form a complex with 200 ng of the GST fusion of intact dMBD2/3Δ or with 200 ng of the GST fusion of a truncated dMBD2/3Δ.
dMBD2/3Δ and mouse MBD2b could function as transcription corepressors or repressors in human 293T cells.
The mammalian MBD2a protein repressed promoter activity in a cotransfection assay (23). To examine whether the Drosophila MBD protein(s) could also act as a transcriptional corepressor, we first carried out side-by-side comparison of the effects of MBD2b, which is an isoform of mouse MBD2a, and of Drosophila dMBD2/3Δ on the expression in human 293T cells of a cotransfected TK promoter with five Gal4-binding sites (Fig. 4B). The Gal4 fusions of either protein significantly repressed the TK promoter in a dosage-dependent manner (Fig. 4B, see data for pSG424-MBD2b and pSG424-dMBD2/3Δ) when being recruited to the promoter via the upstream Gal4-binding sites in pG5-TK-CAT. In contrast, expression of only the Gal4 (Fig. 4B, see data for pSG424) had no obvious effect and that of a fusion polypeptide between Gal4 and the activation domain of p45, a subunit of the erythroid enriched factor NF-E2, stimulated the TK promoter activity (Fig. 4B, see data for pSG424-p45).
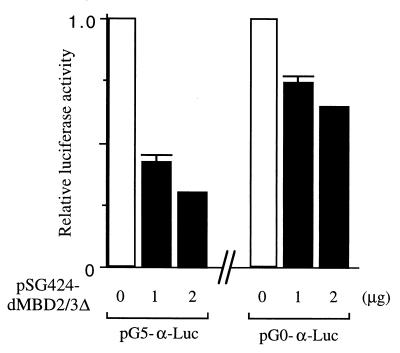
The strong repression effects by the MBD proteins were also observed in a separate set of experiments. In this case, a minimal promoter of the human α globin gene with five Gal4-binding sites (Fig. 4A, see map) was used as the reporter. The expression of either Gal4-dMBD2/3Δ (Fig. 4C, lanes 2 through 4) or Gal4-MBD2b (Fig. 4C, lanes 6 through 8) significantly repressed, by more than 90%, the luciferase activity of pG5-α-Luc. Interestingly, removal of the MBD from dMBD2/3Δ did not affect its function as a transcription corepressor (Fig. 4C, compare lanes 4 and 5). When a reporter plasmid without the Gal4-binding sites, pG0-α-Luc, was used, the two MBD proteins could still repress the promoter activity but to a much smaller extent (approximately 50%) (Fig. 4C, compare lanes 10 through 16 and 2 through 8). This is consistent with our finding that, like for MeCP2 (20) but unlike for MBD1 (24), the repression effect of dMBD2/3Δ is dependent on its distance from the promoter (K. Roder, unpublished data).
The data in Fig. 4 indicate that in human cells, both Drosophila dMBD2/3Δ and mouse MBD2b could function as transcriptional corepressors of at least two different basic promoters. Furthermore, as exemplified for dMBD2/3Δ, the m5CpG recognition and transcriptional repression by these MBD proteins are exerted through two separate domains.
TSA treatment could not inhibit the repressor or corepressor function of dMBD2/3Δ.
Whether the corepressor and repressor functions of dMBD2/3Δ or MBD2b are mediated through HDAC(s) has been tested with the use of TSA. In general, TSA treatment could increase the activities of many endogenous and transfected promoters (10, 32). We have observed the same phenomenon. As shown in Fig. 5A, the activity of p21, the promoter of the murine p21_waf1_ gene, was greatly increased in a TSA concentration-dependent manner. This is consistent with the finding by Xiao et al. (35). Furthermore, a mutant promoter, p21-GCmut, with a mutation in the proximal GC box of p21, exhibited a much smaller magnitude of TSA inducibility (Fig. 5A). Similar to p21_waf1_, the activities of the TK promoter in plasmid pG0-TK-CAT or pG5-TK-CAT (Fig. 5B) and the α globin promoter in pG0-α-Luc or pG5-α-Luc (Fig. 5C) were all increased after TSA treatment, 2- to 4-fold and 15- to 16-fold, respectively. However, it is obvious from Fig. 5B and C that TSA treatment could not relieve the repression by dMBD2/3Δ, whether or not the reporter plasmids contained the Gal4-binding sites.
These results together indicated that dMBD2/3Δ and MBD2b could function in vivo as transcription corepressors without the involvement of HDAC. However, dMBD2/3Δ could associate with HDAC in vitro and in vivo (30). Thus, like mammalian MBD2a (23), Drosophila MBD proteins such as dMBD2/3Δ are also capable of functioning as transcription corepressors through either HDAC-independent or HDAC-dependent repression pathways in a promoter context-specific manner. This utilization of two alternative pathways to repress transcription has been implicated for at least one other Drosophila corepressor, Groucho (5).
dMBD2/3Δ also could effectively repress transcription in Drosophila cells.
To test whether dMBD2/3Δ could function as a corepressor and/or repressor in a Drosophila cellular environment, we carried out DNA transfection in an SL2 cell culture. As in those experiments performed for Fig. 4B and C, pG5-α-Luc or pG0-α-Luc was cotransfected with pSG424 or pSG424-dMBD2/3Δ. In addition, pPacSp1 was also cotransfected since high-level activities of both the α globin promoter in pG5-α-Luc or pG0-α-Luc and the simian virus 40 promoter in pSG424 require the binding of Sp1 factor, which Drosophila cells lack (6).
Indeed, as shown in Fig. 6, dMBD2/3Δ could effectively repress the α globin promoter in pG5-α-Luc. Furthermore, as was true for 293T cells, dMBD2/3Δ also repressed transcription in the absence of the five Gal4-binding sites, although the magnitude of repression was smaller (Fig. 6). Experiments using pCI-Gal4-dMBD2/3Δ instead of pSG424-dMBD2/3Δ gave similar results (data not shown).
FIG. 6.
Transcriptional repression by dMBD2/3Δ in Drosophila SL2 cells. The relative luciferase activities from pG5-α-Luc or pG0-α-Luc in SL2 cells cotransfected with pSG424 or pSG424-dMBD2/3Δ were calculated. Activity with 1 μg of cotransfected pSG424 is defined as 1. The α globin promoter and simian virus 40 promoter in pG0-α-Luc and pG5-α-Luc and in pSG424, respectively, were driven by Sp1 factor expressed by cotransfected pPacSp1. Indeed, the luciferase activities were at least 10 times lower without cotransfection with pPacSp1 (data not shown).
Conservation of Drosophila proteins related to factors involved in the vertebrate DNA methylation program.
As already mentioned, the end product of the DNA methylation reaction, m5CpG, has been recognized as the key determinant responsible for the functioning of CpG MTases and DNA demethylases in vertebrate cells (reference 13 and reference therein). Among the possible mechanisms, m5CpG serves as the binding site for MBD proteins, many of which appear to reside within chromatin remodeling complexes that contain HDACs (12, 22, 23, 30). The identification of Drosophila proteins (DmMTR1 [9] and DmMT2 [9, 30]) related to two of the vertebrate CpG MTase family members and of the Drosophila MBD proteins (9, 30, 33) has thus raised interesting questions regarding the mechanisms of functioning of these proteins in vertebrates as well as in Drosophila (9, 30). Two scenarios could be envisioned to explain the evolutionary conservation of the vertebrate CpG MTase-related proteins in Drosophila. First, DNA methylation-demethylation processes actually occur in Drosophila but only transiently at specific stages of the development or cell cycle, thereby escaping previous methods of detection (27, 31). Alternatively, these proteins and their mammalian counterparts carry out currently unknown functions not requiring the enzymatic reactions. Our data on dMBD2/3Δ have interesting implications for the above models.
Previously, it was shown that ectopically expressed mammalian MBD proteins, in particular MBD1, could effectively repress transfected promoters in Drosophila cells, if the promoter DNA was methylated at CpGs prior to the transfection (7, 24). Furthermore, Lyko et al. (17) have reported that ectopic expression of mammalian CpG MTases caused genomic DNA methylation and embryonic lethality in Drosophila. The lethality is most likely due to the silencing of essential genes during Drosophila development, which in turn, as suggested by the authors, could be caused either by steric inhibition of transcription factor binding or through changes in the chromatin structure near the m5CpG residues. The capability of dMBD2/3Δ to recognize and preferentially bind m5CpG-containing DNA (Fig. 2 and 3) lends support to the first scenario outlined above.
In this study, we have also demonstrated for the first time that Drosophila MBD proteins could function as transcription corepressors or repressors for unmethylated promoters in mammalian as well as Drosophila cells (Fig. 4 and 6). This is remarkably similar to the behavior of the human MBD1 proteins in transfected mammalian and Drosophila cell cultures (7). Furthermore, TSA inhibition experiments suggested that repression by dMBD2/3Δ was mostly HDAC independent (Fig. 5), as previously observed for repression of certain promoters by the mammalian MBD proteins (23, 36). In this regard, it should be noted that certain HDACs, such as the Sir2 HDAC, could not be inhibited by TSA (9a). Since dMBD2/3Δ could associate with several types of HDACs (30), it may also repress transcription through HDAC-dependent pathways. In any case, our data also provide strong support for the second scenario, i.e., that Drosophila CpG MTase- and MBD protein-related factors, as well as their mammalian homologs, carry out functions, e.g., repression of transcription, not requiring the presence of m5CpG residues.
In summary, we have shown that Drosophila MBD proteins such as dMBD2/3Δ could recognize m5CpG through the MBD motif. dMBD2/3Δ also acts as a potent repressor in both Drosophila and mammalian cells. These data together with the previous reports indicate that a nearly full complement of factors related to those involved in the vertebrate DNA methylation program are expressed in Drosophila. Interestingly, as already mentioned above, the functional conservation of the MBD proteins is further supported by the finding that a Drosophila strain carrying a P-element insertion in the dMBD2/3 promoter is lethal to homozygous embryos (Hung, unpublished). Molecular genetic analysis of these proteins in Drosophila will provide interesting and essential insights into the mechanistic aspects of the functions of CpG MTases, MBD proteins, and their related factors in eukaryotic cells.
ACKNOWLEDGMENTS
The first four authors contributed equally to this work.
We are grateful to Shigeo Hayashi at the Genetic Strain Research Center, National Institute of Genetics, Japan, for providing the P1 genomic clones and to Alan Wolffe for communicating unpublished results. We also thank Duen-wei Hsu, Xin Chen, Mark Ptashne, Narender Gavva, Bon-chu Chung, and Guntram Suske for some of the vectors and clones used in the study.
K. Roder is the recipient of a DFG postdoctoral fellowship (Ro 2121/1-1). This research was supported by the Academia Sinica, the National Science Council, and the Foundation of Biomedical Sciences, Taipei, Taiwan, Republic of China.
REFERENCES
- 1.Baylin S B. Tying it all together: epigenetics, genetics, cell cycle, and cancer. Science. 1997;277:1948–1949. doi: 10.1126/science.277.5334.1948. [DOI] [PubMed] [Google Scholar]
- 2.Bestor T H, Verdine G L. DNA methyltransferases. Curr Opin Cell Biol. 1994;6:380–389. doi: 10.1016/0955-0674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya S K, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 4.Bird A, Wolffe A P. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 5.Chen G, Fernandez J, Mische S, Courey A J. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 7.Fujita N, Takebayashi S-I, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415–6426. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung M S, Karthikeyan N, Huang B, Koo H-C, Kiger J, Shen C-K J. Drosophila proteins related to vertebrate DNA (5-cytosine) methyltransferases. Proc Natl Acad Sci USA. 1999;96:11940–11945. doi: 10.1073/pnas.96.21.11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Imai S-I, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 10.Jin S, Scotto K W. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones P A, Gonzalgo M L. Altered DNA methylation and genome instability—a new pathway to cancer? Proc Natl Acad Sci USA. 1997;94:2103–2105. doi: 10.1073/pnas.94.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 13.Jost J P, Saluz H P, editors. DNA methylation: molecular and biological significance. Basel, Switzerland: Birkhäuser Verlag; 1993. [Google Scholar]
- 14.Kudo S. Methyl-CpG-binding protein MeCP2 represses SP1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol Cell Biol. 1998;18:5492–5499. doi: 10.1128/mcb.18.9.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laird P W, Jaenisch R. The role of DNA methylation in cancer genetic and epigenetics. Annu Rev Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- 16.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 17.Lyko F, Ramsahoye B H, Kashevsky H, Tudor M, Mastrangelo M-A, Orr-Weaver T L, Jaenisch R. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat Genet. 1999;23:363–366. doi: 10.1038/15551. [DOI] [PubMed] [Google Scholar]
- 18.MacGregor G R, Caskey C T. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 1989;17:2365. doi: 10.1093/nar/17.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mertineit C, Yoder J A, Taketo T, Laird D W, Trasler J M, Bestor T H. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- 20.Nan X, Campoy F J, Bird A. MeCp2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1987;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 21.Nan X, Meehan R R, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 23.Ng H-H, Zhang Y, Hendrich B, Johnson C A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 24.Ng H-H, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26:2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okano M, Bell D W, Haber D A, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 27.Patel C V, Gopinathan K P. Determination of trace amounts of 5-methylcytosine in DNA by reverse-phase high-performance liquid chromatography. Anal Biochem. 1987;164:164–169. doi: 10.1016/0003-2697(87)90381-2. [DOI] [PubMed] [Google Scholar]
- 28.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Tweedie S, Ng H-H, Barlow A L, Turner B M, Hendrich B, Bird A. Vestiges of a DNA methylation system in Drosophila melanogaster? Nat Genet. 1999;23:389–390. doi: 10.1038/70490. [DOI] [PubMed] [Google Scholar]
- 31.Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982;146:148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- 32.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 33.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 34.Walsh C P, Bestor T H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao H, Hasegawa T, Isobe K-I. Both Sp1 and Sp3 are responsible for p21waf1 promoter activity induced by histone deacetylase inhibitor in NIH3T3 cells. J Cell Biochem. 1999;73:291–302. [PubMed] [Google Scholar]
- 36.Yu F, Thiesen J, Stratling W H. Histone-deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 2000;28:2201–2206. doi: 10.1093/nar/28.10.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]