Two Survivor Pathways That Allow Growth in the Absence of Telomerase Are Generated by Distinct Telomere Recombination Events (original) (raw)
Abstract
Yeast cells can survive in the absence of telomerase RNA, TLC1, by recombination-mediated telomere elongation. Two types of survivors, type I and type II, can be distinguished by their characteristic telomere patterns. RAD52 is essential for the generation of both types of survivors. Deletion of both RAD50 and RAD51 produces a phenotype similar to that produced by deletion of RAD52. Here we examined the effects of the RAD50 and the RAD51 epistasis groups as well as the RAD52 homologue, RAD59, on the types of survivors generated in the absence of telomerase. rad59 mutations completely abolished the ability to generate type II survivors, while rad50 mutations decreased the growth viability of type II survivors but did not completely eliminate their appearance. Mutations in RAD51, RAD54, and RAD57 had the converse affect: they eliminated the ability of cells to generate type I survivors in a tlc1 strain. The triple mutant, tlc1 rad51 rad59, was not able to generate survivors. Thus either type I or type II recombination pathways can allow cells to survive in the absence of telomerase; however, elimination of both pathways in a telomerase mutant leads to the inability to elongate telomeres and ultimately cell death.
Telomere integrity is essential for stable chromosome transmission and for cell viability. In Saccharomyces cerevisiae, telomeres contain a characteristic pattern of repeated sequences. The most terminal telomere sequences consist of an irregular repeat of TG1–3 sequences (25). Just internal to the TG1–3 repeats, there are more complex telomere-associated repeats, termed Y′ elements and X elements. All yeast telomeres have one X element, whereas Y′ elements are found only at some telomeres (3). Y′ elements are often found in multiple copies, and in some cases there are TG1–3 repeats between adjacent Y′ elements (3). Two classes of Y′ elements have been defined, Y′-long and Y′-short, which are 6.7 and 5.2 kb in length, respectively (17, 18).
Telomere function depends on a number of specific proteins that bind to the terminal TG1–3 repeats (reviewed in reference 6). In wild-type yeast cells, the overall length of the TG1–3 sequence is maintained at an equilibrium of around 300 bp, by activities that remove and those that add TG1–3 telomeric repeat sequences. Telomere repeats are added by the enzyme telomerase (8), which contains an essential RNA component (TLC1) (27) and a catalytic protein component (EST2) (16). In the absence of telomerase, telomeres shorten progressively due to incomplete end replication and possibly also specific nuclease activity, and after a lag of about 60 cell divisions, the growth potential of the culture decreases (19, 21, 27). This suggests that after a certain number of TG1–3 repeats have been lost from yeast chromosomes, telomere function is lost.
Although telomerase is the major pathway for telomere elongation, in the absence of telomerase, recombination can efficiently elongate telomeres and lead to the survival of a population of cells (19). In a tlc1 culture, there is a progressive decline in the fraction of growing cells to less than 1%, followed by the selection for a small population of cells that maintains telomeres via a recombination-mediated pathway (19). If the major recombination gene, RAD52, is deleted, these “survivor” cells are not generated. Thus, telomere elongation in the survivors is thought to occur via a recombinational mechanism (19).
Break-induced replication (BIR) is a gene conversion mechanism that can allow telomere healing. A centromere-proximal double-stranded break can be healed by the broken end invading a homologous region on another chromosome (20, 23). A replication fork can be established, and the entire chromosome arm can be copied, resulting in the duplication of a large portion of the chromosome arm (12). The mechanism proposed for the generation of survivors in the absence of telomerase is similar to the mechanism proposed for BIR (5, 19, 29).
Two apparently distinct types of telomere elongation events can occur via recombination in the absence of telomerase (19, 29). The two types of survivors can be distinguished on a Southern blot by the characteristic telomere bands that appear. Type I survivors show amplification of the telomere-associated Y′ elements and have very short TG1–3 repeat tracts on the ends. Type II survivors show a variable pattern of long tracts of TG1–3 repeats and only modest Y′ amplification (29). In a tlc1 mutant, there is a distribution of the two survivor types. The exact percentage of each type is strain dependent (19, 29). Type II survivors grow faster than type I survivors. Thus, if a continuous liquid culture is grown, only type II will be seen at the end of a long growth period. To determine the distribution of survivor types, single colonies from plates where there was no growth competition must be assayed (29).
Two independent pathways defined by RAD50 and RAD51 are both capable of generating survivors (15). The RAD51 pathway involves RAD51, RAD54, and RAD57 (15). These genes are known to interact with each other genetically, and the protein products interact physically (11). The RAD50 pathway involves RAD50, XRS2, and MRE11. The protein products from these genes interact to form a complex that participates in both homologous recombination and DNA end-joining pathways (reviewed in reference 10). Deletion of either RAD50 or RAD51 alone does not abolish the ability of yeast cells to generate survivors. However, deletion of RAD50 and RAD51 prevents the generation of survivors in the absence of telomerase (15).
To explore these two recombination pathways in more detail, we examined the role of RAD59 in the generation of survivors. RAD59 was identified as a gene that when mutated decreased recombination in a rad51 background (1). RAD59 has homology to RAD52, and overexpression of RAD52 will rescue a RAD59 mutant but not vise versa. Deletion of RAD59 has little effect on its own in most recombination assays; however, in a rad51 background, rad59 significantly decreases spontaneous intrachromosomal recombination (1). This synergistic effect of the two mutations suggests that rad59 plays a role in a recombination pathway separate from the rad51 pathway. In other assays, however, such as HO cleavage-induced recombination, there is little synergy between the rad59 and rad51 mutations (28).
To investigate the role of RAD59 and the RAD50 and RAD51 epistasis groups in telomere maintenance, we combined mutations in the telomerase RNA, TLC1, with single and double mutations in these pathways. The tlc1 rad50 and tlc1 rad59 mutants generated predominantly type I survivors, while the tlc1 rad51, tlc1 rad54, and tlc1 rad57 mutants generated only type II survivors. This suggests that the two genetic pathways, which we previously defined, can be distinguished by their different physical effects at telomeres. Both RAD50 and RAD59 play a role in a _RAD51_-independent survivor pathway. Recent experiments that demonstrate a _RAD51_-independent BIR pathway involving RAD59 and the RAD50-MRE11-XRS2 complex suggested that BIR may be the mechanism that allows the generation of survivors in the absence of telomerase (L. Signon, A. Malkova, M. Naylor, H. Klein, and J. E. Haber, unpublished data).
MATERIALS AND METHODS
Yeast strains and plasmid constructs.
All yeast strains used in this study are summarized in Table 1. The isogenic strains with mutations in several RAD genes and TLC1 gene have been described previously (22). Strains JHUY564 and JHUY563 (tlc1::URA3/TLC1 rad50::hisG′/RAD50 rad59::kanMX4/RAD59 and tlc1::URA3/TLC1 rad51::LEU2/RAD51 rad59::kanMX4/RAD59, respectively) were constructed by PCR-mediated gene disruption (2) of the RAD59 gene in strains CSHY92 and CSHY91, respectively. The rad59::kanMX4 deletion fragment was PCR amplified from pRS400 (2) with oligonucleotides AY23F (5′AGTTTAGCACATGCTTTGGACCATTAAAGGGTTACGTAGAGATTGTACTGAGAGTGCAC 3′) and AY24R (5′ATCAACGATACTGTTGATAAAGGTAAGTCGATACCTGTTCTGTGCGGTATTTCACACCG 3′). The yeast strains CSHY91 and CSHY92 were transformed using the lithium acetate method (24). The rad59 deletions were confirmed by Southern blot analysis. The genomic DNA was digested with _Bst_EII and probed with a DNA sequence produced by PCR using the oligonucleotides AY25F (5′ CATACCATTCCAAGGTAA 3′) and AY26R (5′ TAGCAGGCGACGAAGAAT 3′) by random hexamer primed DNA synthesis with the Klenow fragment in the presence of 32P-labeled dATP and dGTP (7). Deletion of RAD59, TLC1, and RAD50 in all diploid strains was confirmed by Southern blot analysis. The pRAD59 plasmid was constructed by cloning the 1.2-kb _Sca_I-_Bgl_II fragment of the RAD59 gene (1) by PCR amplification with oligonucleotides QC22F (5′CGGGATCCCGGCATCACCCATAATTG) and QC23R (5′GCTCTAGAGCCCATGCCTTCGTTACC). The resulting fragment was cloned into the vector pRS315 (26) to make pRAD59. The full 1.2-kb _Sca_I-_Bgl_II DNA fragment was sequenced to verify its accuracy. To generate the appropriate haploid strains for the cell density assay, triple- or double-mutant diploid cells were sporulated, tetrads were dissected, and spore clones were tested for the appropriate markers or by Southern blot analysis.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| CSHY91 | MATa/α Δ_hml_::ADE1/Δ_hml_::ADE1 Δ_hmr_::ADE1/Δ_hmr_::ADE1 ade1/ade1 leu2/leu2 lys5/lys5 ura3/ura3 rad51::LEU2/RAD51 tlc1::URA3/TLC1 |
| CSHY92 | MATa/α Δ_hml_::ADE1/Δ_hml_::ADE1 Δ_hmr_::ADE1/Δ_hmr_::ADE1 ade1/ade1 leu2/leu2 lys5/lys5 ura3/ura3 rad50::hisG′/RAD50 tlc1::URA3/TLC1 |
| CSHY94 | MATa/α Δ_hml_::ADE1/Δ_hml_::ADE1 Δ_hmr_::ADE1/Δ_hmr_::ADE1 ade1/ade1 leu2/leu2 lys5/lys5 ura3/ura3 rad57::LEU2/RAD57 tlc1::URA3/TLC1 |
| CSHY95 | MATa/α Δ_hml_::ADE1/Δ_hml_::ADE1 Δ_hmr_::ADE1/Δ_hmr_::ADE1 ade1/ade1 leu2/leu2 lys5/lys5 ura3/ura3 rad54::LEU2/RAD54 tlc1::URA3/TLC1 |
| JHUY563 | CSHY91 with rad59::kanMX4/RAD59 |
| JHUY564 | CSHY92 with rad59::kanMX4/RAD59 |
Cell viability assay.
Cells of the appropriate genotype were picked up from a fresh dissecting plate and grown in yeast extract-peptone-dextrose (YPD) medium to saturation (1 × 108 to 2 × 108 cells/ml). Every 24 h the cell density was measured by counting cells in a hemocytometer, and then the culture was diluted with fresh YPD liquid medium to a density of 105 cells/ml (15, 27). This cycle was repeated for 10 to 16 days. At various time points during growth, cells were plated to examine for possible contamination and some samples were collected and frozen for telomere length analysis.
Single-colony streak assay.
Cells of the appropriate genotype were picked from a fresh dissecting plate and streaked onto a YPD plate. After incubation for 48 h at 30°C, single colonies were picked and restreaked on a YPD plate. This restreaking was repeated four times to allow loss of viability and appearance of survivors. Generation of survivors typically occurred after 3 to 4 days at 30°C. Single colonies from streak 5 were grown in YPD medium overnight, and cells were pelleted for the telomere length assay.
Telomere Southern blots.
Southern blot analysis was performed for examination of telomere length. Yeast genomic DNA was isolated, and ∼4 μg of DNA was digested with _Xho_I and separated on a 1% agarose gel. DNA was then transferred to a HybondN+ (Amersham, Piscataway, N.J.) membrane and UV cross-linked. The membrane was then hybridized with a random primed telomeric poly(d[GT/CA]) (Pharmacia, Piscataway, N.J.), and hybridization was detected with an ECL direct nucleotide labeling kit (Amersham).
RESULTS
RAD59 is required to generate survivors in a rad51 background.
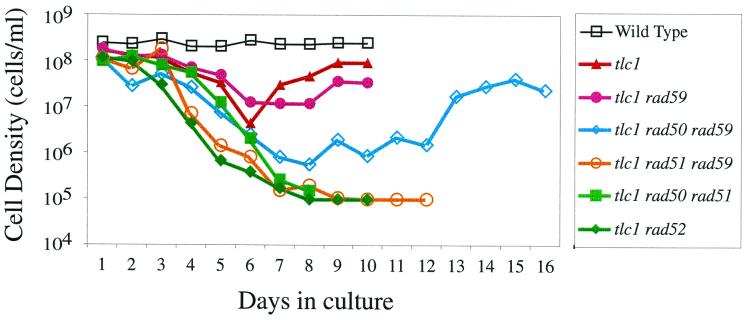
RAD59 defines a _RAD51_-independent recombination pathway (1). To examine how RAD59 affects the two previously defined telomere maintenance pathways involving RAD51 and RAD50, we compared the viabilities and telomere lengths of a set of isogenic mutants, including the tlc1, rad59, tlc1 rad59, tlc1 rad51 rad59, and tlc1 rad50 rad59 mutants. The growth of these cells and the ability to generate survivors were compared to growth and survivor generation of the wild-type, tlc1 rad52, and tlc1 rad50 rad51 cells. Cell viability was measured by diluting liquid cultures to 105 cells/ml, allowing them to grow for 24 h, and then measuring the cell density. While wild-type cells reach a density of 108 cells/ml in 24 h, mutant cells that have a loss of viability or a decreased growth rate will reach lower cell densities (15, 27). The ability to generate survivors was defined as the ability of the culture to recover after reaching a minimum growth rate after about 6 days in culture. The tlc1 rad59 double mutant showed a decline in its growth rate similar to that of the single tlc1 mutant, and survivors were generated. In contrast, the tlc1 rad51 rad59 triple mutant showed an accelerated decline in growth similar to the accelerated rate of decline seen in tlc1 rad52 (Fig. 1) (15, 19). As with the tlc1 rad52 and tlc1 rad50 rad51 mutants, no survivors were generated in the tlc1 rad51 rad59 mutant culture. These results indicate that RAD59 plays a role in generation of survivors and that RAD59 and RAD51 are in separate pathways for the generation of survivors in the absence of telomerase.
FIG. 1.
RAD59 is required to generate survivors in a rad51 background. Cells were grown to saturation in YPD medium and then diluted to 105 cells/ml every 24 h with fresh YPD medium. Cells were counted every 24 h with a hemocytometer. The curves shown are the average of results for four independent clones from each genotype.
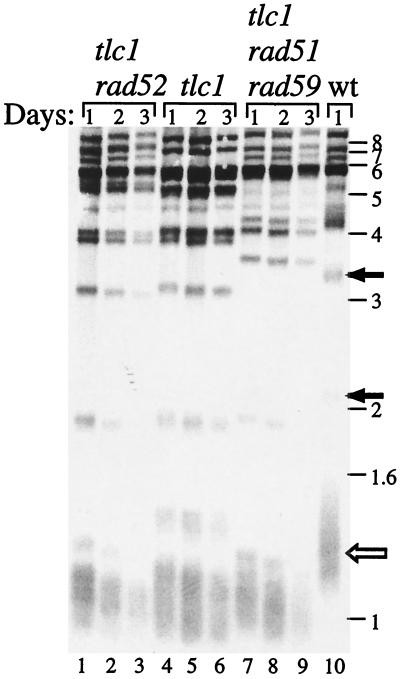
To determine whether the accelerated decline in the growth rate for the tlc1 rad51 rad59 mutant resulted from a more rapid telomere shortening, we examined telomere lengths in three mutants, tlc1, tlc1 rad52, and tlc1 rad51 rad59. There was no evidence for a more rapid telomere shortening in the tlc1 rad51 rad59 or tlc1 rad52 mutants than in the tlc1 mutant in the first 3 days of culturing (Fig. 2). Thus the accelerated decline in the growth rate of tlc1 rad51 rad59 mutants is not due to a faster decrease in telomere length.
FIG. 2.
The tlc1 rad51 rad59 mutant does not show a higher rate of telomere shortening. tlc1, tlc1 rad52, and tlc1 rad51 rad59 cells were grown for five successive days in liquid culture, and telomere length was determined on Southern blots for the first 3 days. The tlc1 rad52 and tlc1 rad51 rad59 mutants reached a minimum growth rate at day 4. Solid arrows indicate X telomeres and the open arrow indicates Y′ telomeres in the wild-type strain.
In contrast to results for the tlc1 rad51 rad59 triple mutant, the tlc1 rad50 rad59 triple mutant did generate survivors (Fig. 1). However, the recovery for the tlc1 rad50 rad59 triple mutant was slower than that for tlc1. This slow recovery is similar to the slow recovery of tlc1 rad50 mutants (15) and may be due to the low growth rate of rad50 mutants (14). The ability of the tlc1 rad50 rad59 mutants to generate survivors as do the tlc1 rad50 and tlc1 rad59 mutants suggests that RAD50 and RAD59 are in the same pathway for telomere maintenance in the absence of telomerase. The failure of tlc1 rad51 rad59 to generate survivors supports the conclusion that RAD59 and RAD51 are in different pathways.
tlc1 rad59 mutants generate only type I survivors.
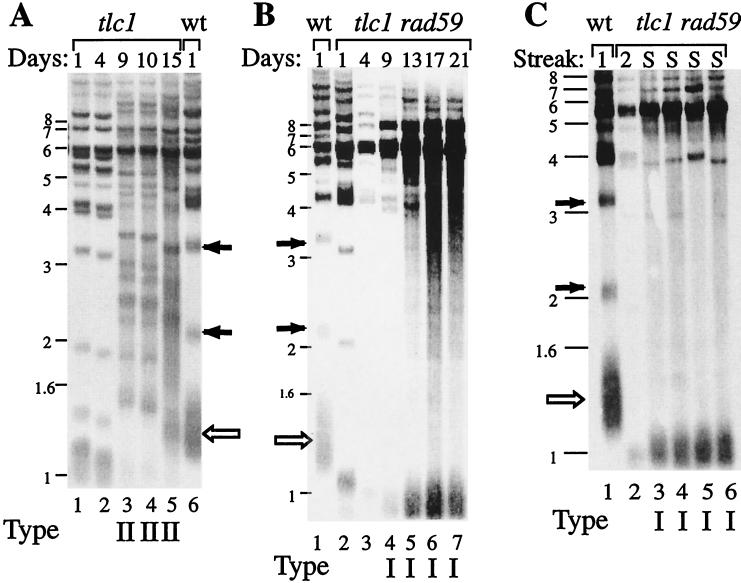
In the absence of telomerase, at least two distinct types of survivors are generated (19, 29). Type I and type II survivors can be distinguished by the pattern of telomere restriction fragments on Southern blots. The telomere bands in type II survivors have long tracts of telomeric TG1–3 sequence and some amplification of Y′ elements (Fig. 3) (29). In these survivors, the multiple distinct telomere bands between 1 and 6 kb represent individual chromosomes with different lengths of TG1–3 repeats. The major _Xho_I band near 1 kb, representing telomeres containing Y′ elements, is no longer prominent, since these short telomeres are all elongated. In contrast, type I survivors have a short _Xho_I band just below 1 kb, lack telomere bands between 1 and 6 kb, and show strong amplification of the 5.2- and 6.7-kb Y′ elements. Since type II survivors have a growth advantage over type I survivors, type II survivors are expected to predominate when cells are grown in liquid culture (29). Consistent with this, the Southern analysis of tlc1 survivors grown in liquid culture showed type II survivors. In contrast, the liquid cultures of tlc1 rad59 mutants showed only type I survivors (Fig. 3). Thus the absence of RAD59 inhibits type II survivor formation.
FIG. 3.
tlc1 rad59 double mutants generate only type I survivors. Southern blots of tlc1 and tlc1 rad59 mutants are shown. The numbers at the top of the lane represent the number of days that cells were grown in liquid culture. The letter S at the top of the lanes represents survivor cells that were streaked out at least two times after survivors were generated. The numbers on the side indicate the DNA molecular size markers in kilobases. The survivor type is indicated below each lane as type I or type II. Solid arrows indicate X telomeres and open arrows indicate Y′ telomeres in the wild-type strain. (A) tlc1 mutants were grown in liquid culture, and telomeres were measured on Southern blots at days 1 and 4, before the generation of survivors (lanes 1 and 2). At days 9, 10, and 15, type II survivors were apparent (lanes 3 to 5). Wild-type telomeres are shown for comparison in lane 6. (B) The telomere patterns of tlc1 rad59 double mutants are shown before the generation of survivors at days 1, 4, and 9 (lanes 2 to 4) and after type I survivors were generated at days 13, 17, and 21 (lanes 5 to 7). (C) The single-colony assay was used for tlc1 rad59 cells. Wild-type cells (lane 1), a presurvivor colony at the second streak-out (lane 2), and four independent type I tlc1 rad59 survivor colonies are shown.
Growth in liquid culture for many population doublings provides a strong selection for the fastest-growing cells. Thus we used a single-colony assay to determine the distribution of survivor types in mutant strains. We plated for single colonies and assayed for type I or type II telomere length patterns on Southern blots. The tlc1 and tlc1 rad59 cells were each streaked five successive times on YPD plates until survivors were generated (approximately 125 cell divisions). Single colonies were then inoculated into liquid culture for overnight growth, and genomic DNA was isolated. Of 39 single tlc1 colonies assayed, 24 (62%) showed a type I pattern and 15 (38%) showed a type II telomere pattern (Table 2 and Fig. 5A). This ratio of type I to type II is similar to that found in other strain backgrounds (19, 29). In contrast, of the 22 tlc1 rad59 single colonies analyzed, all 22 showed type I survivors (Table 2 and Fig. 3C). This result further supports the conclusion that RAD59 is required for the formation of type II survivors.
TABLE 2.
Distribution of survivor types
| Genotype | No. of colonies (% of total) | ||
|---|---|---|---|
| Total studied | Type I | Type II | |
| tlc1 | 39 | 24 (62) | 15 (38) |
| tlc1 rad51 | 28 | 0 (0) | 28 (100) |
| tlc1 rad54 | 21 | 0 (0) | 21 (100) |
| tlc1 rad57 | 21 | 0 (0) | 21 (100) |
| tlc1 rad59 | 22 | 22 (100) | 0 (0) |
| tlc1 rad50 | 20 | 18 (90) | 2 (10) |
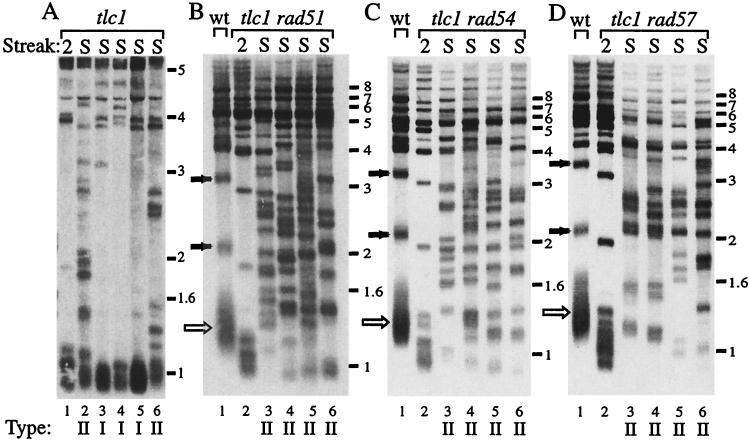
FIG. 5.
tlc1 rad51 mutants generate only type II survivors in the single-colony assay. tlc1 and tlc1 rad51 cells were streaked out repeatedly until survivors were generated (see Material and Methods). DNA samples from either presenescence cells (streak 2) or survivor cells (S) were used for telomere length pattern analysis. (A) For tlc1, both type I (lanes 3 to 5) and type II (lanes 2 and 6) survivors were detected. (B) For tlc1 rad51, of four independent survivors, all were type II (lanes 3 to 6). (C) For tlc1 rad54, of four independent survivors, all were type II (lanes 3 to 6). (D) For tlc1 rad57, of four independent survivors, all were type II (lanes 3 to 6). The survivor type is indicated below each lane. Solid arrows indicate X telomeres and open arrows indicate Y′ telomeres in the wild-type strain.
tlc1 rad50 mutants have a reduced percentage of type II survivors.
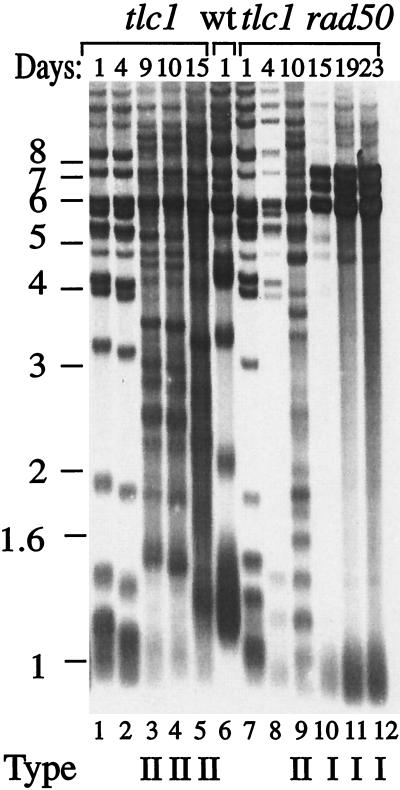
The specific requirement for RAD59 for the generation of type II survivors suggested that the two different survivor pathways defined by the RAD50 and RAD51 epistasis groups might independently affect the two survivor types. We thus assayed whether deletion of genes in these groups would also affect the distribution of survivor types. In a liquid culture assay, the cell density of the tlc1 rad50 double mutant declined continuously until survivors were generated, and then the cell density increased (data not shown) (15). Telomere length in the tlc1 rad50 double mutant shortened progressively, and in two of three independent experiments only type I survivors were found. In the third experiment, type II survivors were detected at day 10, but they were no longer present at days 15, 19, and 23 (Fig. 4). The cell growth rate from day 10 to 23 was similar to that of wild-type cells (data not shown). This suggested that type II survivors were initially formed but that they were not maintained during continued growth.
FIG. 4.
Type II survivors that are generated in tlc1 rad50 cells are not maintained during continuous culturing. tlc1 cells were cultured for 15 days, and telomeres were examined at days 1, 4, 9, 10, and 15 (lanes 1 to 5). Type II survivors were detected at days 9, 10, and 15 (lanes 3 to 5). tlc1 rad50 cells were grown for 23 days, and telomeres were examined at days 1, 4, 10, 15, 19, and 23 (lanes 8 to 12). Type II survivors were detected at day 10 but not at days 15, 19, and 23. The survivor type is indicated below each lane as type I or type II.
The instability of type II survivors in tlc1 rad50 cells was also seen in single-colony assays. In the first streak after survivor generation, 2 of 20 individual tlc1 rad50 survivor colonies assayed showed the type II pattern (Table 2). However, after streaking an additional three times, only type I survivors were detected (data not shown). This is consistent with the appearance of type II survivors in tlc1 rad50 mutants seen in earlier work (15) when cells were assayed soon after survivor generation. These data suggest that type II survivors are generated at a low frequency in tlc1 rad50 mutants but cannot be maintained during subsequent growth. Thus RAD50 facilitates the formation and maintenance of type II survivors.
tlc1 rad51, tlc1 rad54, and tlc1 rad57 mutants generate only type II survivors.
RAD51 defines a pathway that includes RAD54 and RAD57 and is separate from the RAD50 pathway that is able to maintain telomere length in the absence of telomerase (15). We used Southern analysis to determine the type of survivors that are formed in tlc1 rad51, tlc1 rad54, and tlc1 rad57 double-mutant cells. In liquid culture, all survivors showed a type II pattern (15) (data not shown). Since type II survivors have a growth advantage and will take over a liquid culture, we assayed single colonies to examine the distribution of survivor types. tlc1 rad51, tlc1 rad54, and tlc1 rad57 double mutants were streaked five times on YPD plates to allow the generation of survivors, and single colonies were examined by Southern blot analysis. Each of the double mutants tlc1 rad51, tlc1 rad54, and tlc1 rad57 generated only type II survivor clones (Table 2 and Fig. 5B, C, and D). This differs significantly from the tlc1 single-mutant survivors in the same genetic background, where only 38% of the survivors were type II (Table 2 and Fig. 5A). Thus RAD51, RAD54, and RAD57 are required for the formation of type I survivors in the absence of telomerase.
Reintroduction of the RAD59 gene into late tlc1 rad59 survivors does not reestablish a wild-type distribution of type I and type II survivors.
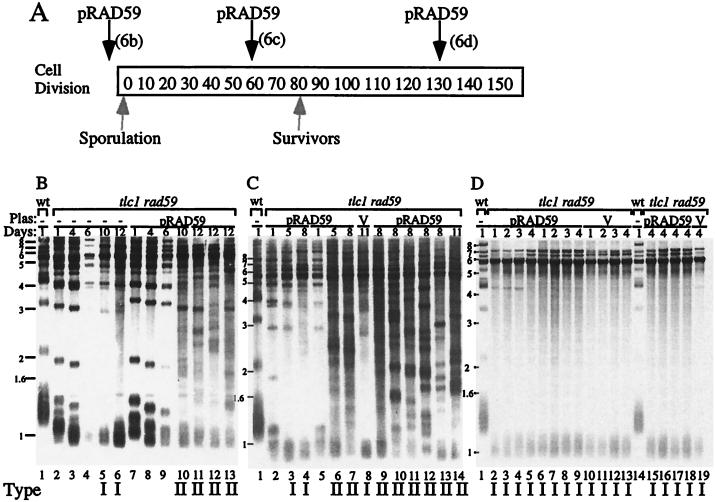
To examine the role that RAD59 might play in the generation of type II survivors, we tested whether reintroduction of the wild-type gene at three different points during survivor generation would rescue the ability to generate type II survivors (Fig. 6A). A plasmid containing RAD59 was transformed into a diploid heterozygous for tlc1 and rad59. The diploid was sporulated, and haploid spore clones were selected. tlc1 rad59 spore clones without the pRAD59 plasmid generated only type I survivors in liquid culture (Fig. 6B). However, tlc1 rad59 spore clones with the pRAD59 plasmid generated type II survivors, showing that the plasmid can complement the loss of rad59 from the genome (Fig. 6B) (see Materials and Methods). As an independent control, we tested the methyl methanesulfonate (MMS) sensitivity of the spore clones. The cells without pRAD59 were sensitive to MMS, but the cells with pRAD59 were not (data not shown). We next tested whether transformation of the pRAD59 plasmid into tlc1 rad59 cells just before or just after survivors were generated would affect the distribution of survivor types. Transformation into a culture that had undergone approximately 60 cell divisions (before survivors were generated) restored the ability to generate type II survivors in 10 of the 18 transformants examined. The remaining 8 transformants showed the type I pattern (Fig. 6C and data not shown). Transformation of a control vector plasmid generated only type I survivors, indicating that the transformation itself did not restore type II survivors. Strikingly, transformation of the pRAD59 plasmid into cells that were initially grown for 130 cell divisions (after survivors were generated) did not restore the appearance of type II survivors (Fig. 6D). Telomeres were examined for five independent transformants at days 1, 2, 3, and 4 during growth of the culture, and an additional seven transformants were assayed at day 4 only. None of these transformants showed type II survivors (Fig. 6d and data not shown). Thus the restoration of RAD59 function after the establishment of type I survivors does not allow conversion to type II despite the type II survivor growth advantage.
FIG. 6.
Reintroduction of the RAD59 gene into late-generation tlc1 rad59 survivor cultures does not reestablish a wild-type distribution of survivor types. (A) The pRAD59 plasmid was transformed into tlc1 rad59 cells at various times during the generation of survivor cells as indicated. (B) tlc1 rad59 spore clones that did not contain the pRAD59 plasmid (lanes 2 to 6) and those that did (lanes 7 to 11) were analyzed after 12 days of growth in liquid culture. Two additional spore clones containing the pRAD59 plasmid were also analyzed at day 12. (C) pRAD59 was transformed into tlc1 rad59 cells at generation ∼60, and independent transformants were examined for their survivor type distribution by Southern blot analysis. Two independent transformants containing pRAD59 (lanes 2 to 4 and 5 to 7) were examined at days 1, 5, and 8. Control cells transformed with the pRS315 vector only are shown in lane 8. Additional independent transformants containing the pRAD59 plasmid were assayed at day 8 (lanes 9 to 13) or day 11 (lane 14). (D) pRAD59 or the vector pRS315 was transformed into tlc1 rad59 cells at generation ∼130 (see Material and Methods), and independent transformants were cultured for 4 days to test survivor types. Two transformants containing pRAD59 (lanes 2 to 5 and 6 to 9) and one with the pRS315 vector (lanes 10 to 13) were assayed for telomere pattern. Additional independent colonies containing pRAD59 (lanes 15 to 18) or vector (lane 19) at day 4 were assayed. Numbers on the side are molecular size markers in kilobases. Lanes marked wt contain wild-type DNA as a control. Plas, plasmid.
DISCUSSION
Two types of survivors are generated via recombination-mediated telomere elongation in the absence of telomerase. Specific genes mediate the pathways that allow generation of the survivors; the RAD51, RAD54, and RAD57 genes are involved in the generation of type I survivors, and both RAD59 and RAD50 mediate the generation of type II survivors (Fig. 7). When both survivor generation pathways are eliminated, as in the tlc1 rad50 rad51 mutants or the tlc1 rad51 rad59 mutants, no survivors are generated.
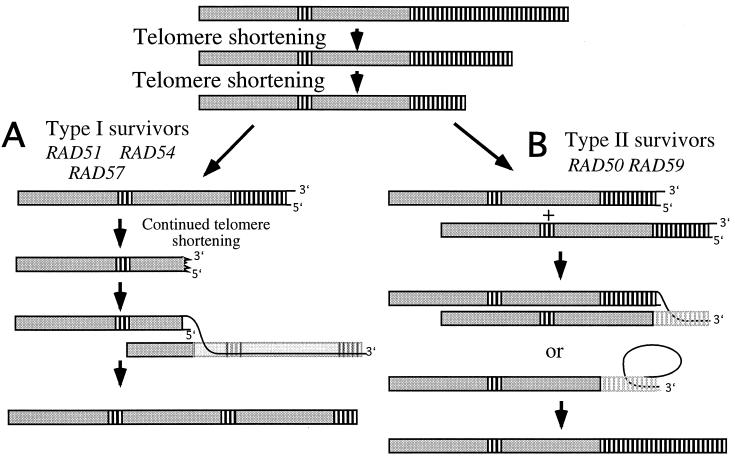
FIG. 7.
Two types of survivors are generated by two distinct genetic pathways. A telomere containing two tandem Y′ elements (large gray boxes) separated by TG1–3 repeats (small white boxes) is represented at the top. Telomere shortening occurs in the absence of telomerase. Survivors can be generated via two different mechanisms. (A) The _RAD51_-, _RAD54_-, and _RAD57_-dependent pathway generates type I survivors. Telomere shortening continues into the Y′ element, exposing single-stranded 3′ overhangs. The single-stranded DNA invades a homologous region in the Y′ element on some other telomere, and BIR allows duplication of the intact telomere onto the telomere which had lost the TG1–3 repeats. The telomere that is copied is shown in light gray for clarity, to reveal the 3′ end elongation of the invading strand. This kind of recombination event could also occur at X sequences on telomeres that do not contain Y′ elements (see the text). (B) Telomere shortening results in recombination before all of the telomere repeats are lost from the ends. RAD50 and RAD59 allow recombination in the irregular TG1–3 repeats to occur efficiently. Alternatively, the telomeric TG1–3 tracts self-prime DNA replication and allow extension of the telomere sequences with a rolling-circle-type mechanism.
RAD51, sequence identity-dependent strand annealing, and type I survivors.
Type I survivors involve the amplification of the telomere-adjacent Y′ elements and require the presence of RAD51. These survivors may arise via continued resecting of the telomere in the absence of telomerase until the Y′ elements are at the molecular terminus. Recombination is then initiated within the Y′ elements. If the recombination occurs with an internal Y′ element on a chromosome that has tandem Y′ elements, amplification of Y′s will occur (Fig. 7A). Alternatively, on chromosomes that have only X elements, resectioning into the X sequences may allow recombination with other X elements. This could lead to all chromosomes picking up Y′s, since many chromosomes have both X and Y′ elements. This would explain the apparent disappearance of the X-containing bands between 1 and 6 kb in the type I survivors. The requirements for RAD51 for type I survivors may be due to the fact that recombination in the Y′s requires a high level of sequence identity. RAD51 mediates strand invasion during recombination, and it is very sensitive to the mismatches in the homologous region (4). Y′ elements are very well conserved in sequence, showing only 1% sequence divergence (17), while TG1–3 telomere repeats are much more irregular in sequence. Thus, RAD51 may be able to mediate the recombination between Y′s but not be able to mediate the recombination that occurs between the less identical TG1–3 tracts. In the absence of RAD51, the recombination events between Y′ elements cannot be carried out.
RAD59 and the establishment of type II survivors.
RAD59 is involved in a _RAD51_-independent mitotic recombination pathway mediating recombination between intrachromosomal inverted repeats (1). Consistent with this, we found that RAD59 plays a role in a _RAD51_-independent pathway in the generation of tlc1 survivors. In the absence of RAD59, all survivors were type I, indicating that RAD59 promotes the generation of type II survivors. These survivors likely arise via a recombination mechanism that is initiated within the TG1–3 repeats themselves. This recombination event could be initiated through either an interchromosomal invasion and copying of the TG1–3 repeats or perhaps intrachromosomal copying of a telomere that is looped back on itself (Fig. 7B). There is evidence that mammalian telomeres form t-loops, in which the telomere sequences are looped back on themselves and the 3′ G-rich overhang is base paired with a duplex region of repeats forming a D-loop (9). A D-loop resembles half of a replication fork and may be able to prime DNA polymerization. Perhaps such a structure would become deregulated upon telomere shortening and serve as the substrate for the generation of type II survivors (Fig. 7B).
RAD50 is required for maintenance of type II survivors.
rad50 mutants can initially generate type II survivors at a low rate, but these survivors are not maintained over time (Table 2 and Fig. 3) (15). Recently it was found that Rad50 protein is bound to human telomeres, and it was proposed that Rad50 is involved in the establishment of the t-loop structure (30). The initial appearance and subsequent loss of type II survivors in tlc1 rad50 mutants is consistent with rad50 playing a role in the efficient establishment of t-loops. If t-loops form in rad50 cells but at a lower rate than usual, those cells that were able to establish t-loops could initially generate type II survivors via priming from the 3′ end in the t-loop. However, in the absence of telomerase, there is strong pressure to maintain telomere length since many cells in the culture stop dividing when telomeres become short; perhaps the impaired t-loop formation in rad50 mutants cannot keep up. Thus the type I survivors in the culture are able to take over and become the dominant survivor type.
Break-induced replication and survivors arise through a similar mechanism.
The mechanisms outlined in Fig. 7 by which survivors are generated in the absence of telomerase may be similar to BIR. BIR is one mechanism by which a double strand break can be healed. If there is sequence homology on one side of a break and not on the other, the homologous region can invade, pair with, and copy an entire chromosome arm (20). The mechanisms proposed for the generation of survivors (5) (reviewed in references 13, 19, and 29) are very similar to that proposed for BIR. Recent experiments indicate that the genetic requirements for BIR are similar to those for survivor generation. There is a _RAD51_-independent BIR pathway involving RAD59 and the RAD50-MRE11-XRS2 complex (Signon et al., unpublished data). rad51 or rad50 single mutants do not eliminate BIR, but rad51 rad50 double mutants dramatically reduce BIR. Similarly, rad59 single mutants have little effect on BIR, yet rad51 rad59 double mutants reduce BIR significantly. These genetic requirements could reflect the two mechanisms outlined in Fig. 7 for the two genetic pathways that allow generation of survivors.
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH, GM43080, to C.W.G.
We thank Anna Malkova for testing MMS sensitivity. We thank Siyuan Le, Jennifer Hackett, and Kay Keyer-Opperman for critical reading of the manuscript. We thank James Haber for sharing data before publication and for helpful discussions.
ADDENDUM IN PROOF
While this paper was under review, Teng et al. published a paper that also showed a requirement for RAD50 for type II survivors (S. Teng, J. Chang, B. McCowan, and V. A. Zakian, Mol. Cell **6:**947–952, 2000).
REFERENCES
- 1.Bai Y, Symington L S. A Rad52 homologue is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 2.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Chan C S M, Tye B. Organization of DNA sequences and replication origins at yeast telomeres. Cell. 1983;33:563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn B, Szauter P, Pardue M L, Szostak J W. Transfer of yeast telomeres to linear plasmids by recombination. Cell. 1985;39:191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- 6.Fang G, Cech T R. Telomere proteins. In: Blackburn E H, Greider C W, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 69–105. [Google Scholar]
- 7.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 8.Greider C W, Blackburn E H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 9.Griffith J D, Comeau L, Rosenfield S, Stansel R M, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 10.Haber J E. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 11.Hays S L, Firmenich A A, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes A M, Haber J E. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 13.Kass-Eisler A, Greider C W. Recombination in telomere-length maintenance. Trends Biochem Sci. 2000;25:200–204. doi: 10.1016/s0968-0004(00)01557-7. [DOI] [PubMed] [Google Scholar]
- 14.Kironmai K M, Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 15.Le S, Moore J K, Haber J E, Greider C W. RAD51 and RAD50 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 17.Louis E J, Haber J E. The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis E J, Haber J E. The subtelomeric Y′ repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics. 1990;124:533–545. doi: 10.1093/genetics/124.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundblad V, Blackburn E H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 20.Malkova A, Ivanov E L, Haber J E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McEachern M J, Blackburn E H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 22.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrow D M, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 25.Shampay J, Szostak J W, Blackburn E H. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 28.Sugawara N, Ira G, Haber J E. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng S C, Zakian V A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X D, Kuster B, Mann M, Petrini J H, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]