Genetic Basis of Mitochondrial Function and Morphology in Saccharomyces cerevisiae (original) (raw)
Abstract
The understanding of the processes underlying organellar function and inheritance requires the identification and characterization of the molecular components involved. We pursued a genomic approach to define the complements of genes required for respiratory growth and inheritance of mitochondria with normal morphology in yeast. With the systematic screening of a deletion mutant library covering the nonessential genes of Saccharomyces cerevisiae the numbers of genes known to be required for respiratory function and establishment of wild-type-like mitochondrial structure have been more than doubled. In addition to the identification of novel components, the systematic screen revealed unprecedented mitochondrial phenotypes that have never been observed by conventional screens. These data provide a comprehensive picture of the cellular processes and molecular components required for mitochondrial function and structure in a simple eukaryotic cell.
INTRODUCTION
Mitochondria are essential organelles of eukaryotic cells. They carry out a variety of metabolic processes including reactions of the tricarboxylic acid cycle, iron/sulfur cluster assembly, and biosynthesis of many cellular metabolites (Scheffler, 2000). Their most prominent function, however, is to supply the cell with energy generated by oxidative phosphorylation (Saraste, 1999). The cellular role of mitochondria is reflected by their structure. They are complex double membrane-bounded organelles with a characteristic morphology and intracellular distribution. Inheritance and morphogenesis depend on active transport along the cytoskeleton and continuous membrane fission and fusion events (Bereiter-Hahn and Vöth, 1994; Yaffe, 1999; Griparic and van der Bliek, 2001). The understanding of the processes underlying mitochondrial function and inheritance requires the identification and characterization of the molecular components involved. During the past few decades many of the proteins required for respiratory growth (Tzagoloff and Dieckmann, 1990; Contamine and Picard, 2000) and establishment and maintenance of mitochondrial structure (Hermann and Shaw, 1998; Jensen et al., 2000; Boldogh et al., 2001b) have been identified in yeast. The advent of the postgenomic era allows us to conduct systematic genome-wide screens to define whole complements of genes associated with particular functions (Winzeler et al., 1999; Vidan and Snyder, 2001).
The fact that Saccharomyces cerevisiae is a facultative anaerobic yeast capable of satisfying its energy requirements with ATP generated by fermentation is the reason why only relatively few mitochondrial proteins are essential for cell viability. These are restricted to a handful of factors essential for import, processing, and folding of precursor proteins, iron/sulfur cluster assembly, and flavin mononucleotide synthesis. This makes budding yeast an ideal organism for dissecting the molecular processes required for maintenance of respiratory-competent mitochondria. Its mitochondrial genome encodes eight proteins that are all essential for oxidative phosphorylation. Despite the capacity of mitochondria to encode and synthesize proteins, a vast array of genes located in the nucleus is required for respiratory competence. Mutants in these genes are commonly referred to as nuclear petite or pet mutants (Tzagoloff and Dieckmann, 1990). The Munich Information Center for Protein Sequences (MIPS; Mewes et al., 2000) currently lists 171 yeast open reading frames (ORFs) that are required for respiratory growth (http://mips.gsf.de/proj/yeast/catalogues/phenotype/fc42_25_20.html).
Mitochondria are amazingly dynamic organelles. Their morphology and distribution reflects the energy requirements of the cell (Bereiter-Hahn, 1990; Warren and Wickner, 1996; Hermann and Shaw, 1998; Yaffe, 1999). In S. cerevisiae, mitochondria form a branched tubular network below the cell cortex (Hoffmann and Avers, 1973; Stevens, 1981), the continuity of which is maintained by active actin-dependent transport and balanced membrane fusion and fission events (Nunnari et al., 1997; Hermann and Shaw, 1998). Morphological screens of randomly mutagenized yeast samples using mitochondria-specific fluorescent dyes have proven to be very successful for the identification of some key components required for maintenance of this complex structure (McConnell et al., 1990; Burgess et al., 1994; Hermann et al., 1997; Sesaki and Jensen, 1999).
Here, we report on the systematic screening of a yeast deletion mutant library covering the nonessential genes to define the sets of genes involved in mitochondrial structure and function. A total number of 341 ORFs was identified to be required for respiratory growth, 38 of which encode unknown proteins. Mutants with aberrant mitochondrial morphology include 5 known genes that were not previously implicated in mitochondrial morphogenesis and 10 genes encoding novel components. These data provide a comprehensive picture of the cellular processes and molecular components required for respiratory growth and inheritance of mitochondria with a normal morphology in yeast.
MATERIALS AND METHODS
Yeast Genetic Methods
Cultivation of yeast was according to standard procedures (Sherman et al., 1986). The homozygous diploid knock out library was constructed by an international consortium of yeast laboratories (Winzeler et al., 1999). It was obtained from Research Genetics (Huntsville, AL). A list of strains present in the library is available at the company's website (ftp://ftp.resgen.com/pub/deletions/Homo_diploids_061101.txt) or from the authors upon request.
Screening for pet Mutants
After completely thawing 96-well plates, cells were transferred to yeast extract/peptone/glucose (YPD) and yeast extract/peptone/glycerol (YPG) plates using a sterile pinning tool. Plates were incubated at 30°C for 2 (YPD) or 3 (YPG) days before the growth behavior was examined.
To obtain an estimate of the saturation of the genome-wide screen, we compared our results with the published information about yeast mitochondrial ribosomal proteins. Seventy-one nuclear genes encoding mitochondrial ribosomal proteins are known, according to the Yeast Proteome Database (Costanzo et al., 2001). Deletions of 62 of these genes are present in the library, and the observed growth phenotypes of 61 of these mutants correspond to published phenotypes (for detailed information see supplemental Table 1). From these numbers we estimate that the knock out library is 80–90% saturating for all nonessential yeast ORFs. In addition, some genes encoding very small proteins of <100 amino acid residues—which are only partially covered by the library—and genes encoding proteins that perform redundant functions might have been missed during the screen.
Screening for mdm Mutants
Logarithmically growing yeast cultures in YPD medium were stained with 0.1 μM rhodamine B hexyl ester (Molecular Probes, Eugene, OR) and inspected by standard fluorescence microscopy (Prokisch et al., 2000). Screening was repeated for strains that showed aberrant mitochondrial morphology or that did not stain well because of a lack of mitochondrial membrane potential. The second round of screening was performed by a different individual and included a number of wild-type strains as a control. Mutant strains (n = 268) that reproducibly did not exhibit wild-type mitochondria were transformed with plasmid pVT100U-mtGFP expressing mitochondria-targeted GFP (Westermann and Neupert, 2000). All transformants were screened at least two more times for mutants with aberrant mitochondrial morphology. All rounds of screening were performed without reference to strain identity. For all clearly identified mutants correct gene disruption was confirmed by PCR, and the mitochondrial phenotype was confirmed in a haploid genetic background (haploid deletants were obtained from EUROSCARF, Frankfurt, Germany; http://www.uni-frankfurt.de/fb15/mikro/euroscarf/). Strains that for different reasons failed to give a clear result are listed in supplemental Table 2.
RESULTS
Genes Needed for Respiratory Growth
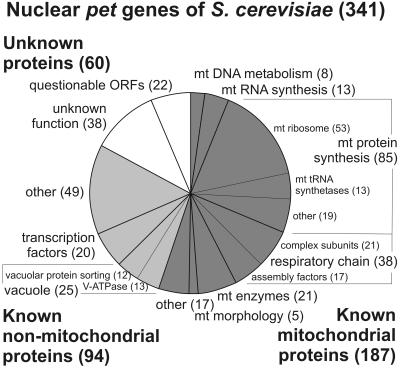
To define the molecular basis of respiratory competence, we systematically screened a library containing deletion mutants of 4794 nonessential yeast genes for strains that are respiratory deficient. Yeast knock out strains were plated on media containing the fermentable carbon source glucose or the nonfermentable carbon source glycerol and scored for mutants unable to grow on glycerol. A total number of 341 ORFs were identified to be required for respiratory growth (Figure 1; supplemental Table 3). More than half of the identified pet genes encode known mitochondrial proteins, the majority of which is devoted to replication, transcription, and translation of the mitochondrial genome or assembly of the respiratory chain. A large fraction of the pet genes encoding known nonmitochondrial proteins is associated either with vacuolar functions or encodes nuclear transcription factors. The remainder is associated with a variety of different cellular functions, and failure to grow on glycerol-containing medium might be due to cumulative effects of a compromised general cell physiology. Seventeen percent of the pet mutants contain deletions in ORFs of unknown function. We classified 22 of these as questionable ORFs because they overlap with other genes that encode known or conserved proteins. Thirty-eight ORFs encode so far unknown proteins that presumably play important roles in the maintenance of respiratory-competent mitochondria.
Figure 1.
Distribution of functional classes of known and newly identified yeast pet genes using criteria from the Yeast Proteome Database (Costanzo et al., 2001). Numbers indicate the number of pet genes falling into each functional class. An annotated list of all identified pet genes catalogued according to their cellular function can be found in supplemental Table 3. mt, mitochondrial.
Genes Needed for Mitochondrial Distribution and Morphology
To obtain a more complete understanding of the molecular machinery determining mitochondrial behavior, we conducted a systematic genome-wide screen for genes important for mitochondrial distribution and morphology (MDM; McConnell et al., 1990). Yeast knock out strains (n = 4794) were stained with mitochondria-specific probes and screened for mutants with aberrant mitochondrial morphology. The isolated mdm mutants were grouped into three classes. Class I mutant genes encode proteins that are essential for establishing wild-type mitochondrial morphology. For these mutants, cells with wild-type-like mitochondria were never observed (see below). Mitochondria of class II and class III mutants were often fragmented or aggregated; however, a certain subfraction showed wild-type-like morphology (supplemental Table 4). Thus, these genes are considered as not being essential for establishment of normal mitochondrial structure.
Class II mutants are respiratory competent. This class includes strains that appear to exhibit mitochondrial morphology defects only under certain conditions, such as clu1, mdm20, ptc1, and yme1 (Campbell et al., 1994; Hermann et al., 1997; Fields et al., 1998; Roeder et al., 1998). Interestingly, three genes involved in biosynthesis of ergosterol, ERG6, ERG24, and ERG28, fall into this class. This is consistent with a role of ergosterol in membrane fusion that was recently demonstrated by a similar approach aimed at the identification of vacuolar inheritance components (Kato and Wickner, 2001).
Class III mutants display a pet phenotype in addition to their mitochondrial morphology defect. It has been known for a long time that loss of respiratory function often results in the loss of inner membrane cristae (Pon and Schatz, 1991). Because processes of mitochondrial morphogenesis are intimately linked to connection of the mitochondrial outer and inner membranes (Aiken Hobbs et al., 2001; Fritz et al., 2001) it is conceivable that changes in the structure of the inner membrane may affect the global structure of the organelle (Wong et al., 2000).
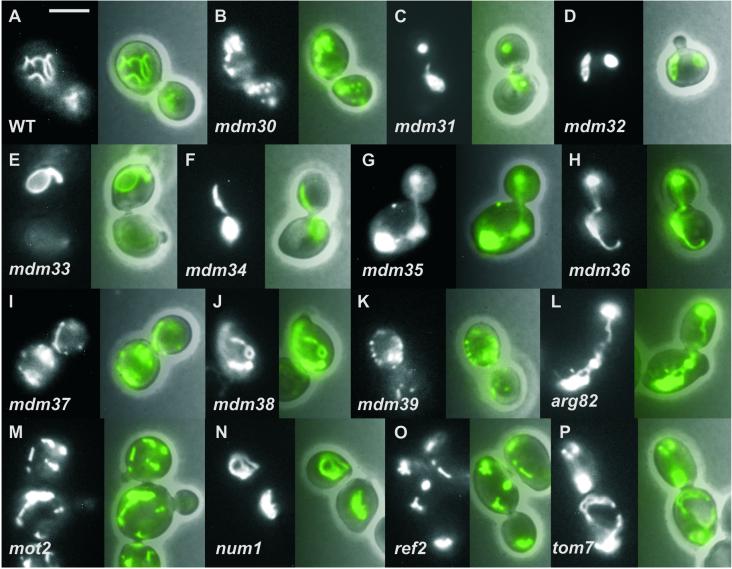
Among the class I mutants are eight genes that are known to encode key components of the machinery of mitochondrial morphology. In addition, we identified 10 previously uncharacterized genes as being essential for the establishment of normal mitochondrial morphology (Table 1). Four of these genes, MDM31, MDM32, MDM37, and MDM38, encode proteins of unknown function that are predicted to be located in the mitochondrial inner membrane. Mutants of the closely related mdm31 and mdm32 genes contain compact mitochondrial aggregates (Figure 2, C and D). In contrast, mdm37 mutant cells harbor mainly fragmented mitochondria with very few short tubules (Figure 2I) resembling mutants defective in mitochondrial fusion (Hermann et al., 1998; Rapaport et al., 1998; Fritz et al., 2001; Sesaki and Jensen, 2001). Mitochondria of cells lacking the evolutionarily conserved Mdm38 protein appear enlarged with very few branches and often form rings or lariat-like structures, a phenotype that was never reported before (Figure 2J).
Table 1.
Genes essential for maintenance of normal mitochondrial morphology in yeast
| Gene name (standard/systematic) | TM | Presequence | Mitochondrial morphology | Reference |
|---|---|---|---|---|
| DNM1/YLL001W | — | — | Net-like | Otsuga et al., 1998; Bleazard et al., 1999; Sesaki and Jensen, 1999 |
| FIS1/YIL065C | OM | — | Net-like | Mozdy et al., 2000 |
| FZO1/YBR179C | OM | — | Fragmented | Hermann et al., 1998; Rapaport et al., 1998 |
| MDM10/YAL010C | OM | — | Giant spherical | Sogo and Yaffe, 1994 |
| MDM12/YOL009C | OM | — | Giant spherical | Berger et al., 1997 |
| MDV1/YJL112W | — | — | Net-like | Fekkes et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001 |
| MGM1/YOR211C | ? | e | Fragmented/aggregated | Guan et al., 1993; Shepard and Yaffe, 1999; Wong et al., 2000 |
| MMM1/YLL006W | OM | — | Giant spherical | Burgess et al., 1994 |
| MDM30/YLR368W | — | — | See Fig. 2B | This study |
| MDM31/YHR194W | p | 0.9986 | See Fig. 2C | This study |
| MDM32/YOR147W | p | 0.9989 | See Fig. 2D | This study |
| MDM33/YDR393W | p | 0.5117 | See Fig. 2E | This study |
| MDM34/YGL219C | — | — | See Fig. 2F | This study |
| MDM35/YKL053C-A | — | — | See Fig. 2G | This study |
| MDM36/YPR083W | — | — | See Fig. 2H | This study |
| MDM37/YGR101W | p | 0.9943 | See Fig. 2I | This study |
| MDM38/YOL027C | p | 0.9302 | See Fig. 2J | This study |
| MDM39/YGL020C | p | — | See Fig. 2K | This study |
| ARG82/YDR173C | — | — | See Fig. 2L | This study |
| MOT2/YER068W | — | — | See Fig. 2M | This study |
| NUM1/YDR150W | — | — | See Fig. 2N | This study |
| REF2/YDR195W | — | — | See Fig. 2O | This study |
| TOM7/YNL070W | OM | — | See Fig. 2P | This study |
Figure 2.
Mitochondrial morphology of newly identified mdm mutants. Strains expressing mitochondria-targeted GFP were grown in glucose-containing YPD medium at 30°C to logarithmic growth phase and subjected to fluorescence microscopy. Left, mitochondrial morphology of representative cells; right, overlay with the corresponding phase contrast image. WT, wild-type. Bar, 5 μm.
Two genes, MDM33 and MDM39, encode predicted membrane proteins lacking a clear mitochondrial presequence. Most mdm33 mutant cells exhibit giant ring-like mitochondrial structures, an unprecedented phenotype (Figure 2E), whereas the mdm39 mutant shows fragmented mitochondria (Figure 2K).
Four components, Mdm30p, Mdm34p, Mdm35p, and Mdm36p, do not have predicted presequences or transmembrane domains. The mdm30 mutant displays many fragmented or aggregated mitochondria with only very few short tubules (Figure 2B). Mutants with deletions in the mdm34 gene (Figure 2F) or the overlapping questionable ORF ygl218w (not shown) have spherical mitochondria, similar to mdm10, mdm12, and mmm1 mutants (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997), and sometimes single tubules extending through the cell. Mitochondria of cells lacking the small, highly conserved Mdm35 protein are spherical (Figure 2G), whereas the organelles of the mdm36 mutant are mostly aggregated at one side of the cell (Figure 2H).
Five genes were identified that encode known proteins that were previously not implicated in mitochondrial morphogenesis (Table 1). The tom7 mutant displays aggregated, often fenestrated mitochondria that are unevenly distributed in the cell (Figure 2P). Tom7p is a component of the general preprotein import complex of mitochondria and plays an important role in the insertion of proteins into the outer membrane (Hönlinger et al., 1996). We propose that Tom7p is required for the insertion of one or several morphogenesis factor(s) into the outer membrane. A reduced import of this factor(s) would consequently lead to an abnormal mitochondrial morphology. Deletion mutants of num1 (Figure 2N) and the overlapping questionable ORF ydr149c display highly aggregated mitochondria. Num1p is a cell cortex-associated protein involved in nuclear migration (Kormanec et al., 1991; Heil-Chapdelaine et al., 2000). Our data assign to Num1p an additional function in positioning of mitochondria. Arg82p, a nuclear inositol 1,4,5-trisphosphate kinase, Mot2p, a global transcriptional repressor, and Ref2p, an RNA processing factor, each influence the expression of a large number of different genes (Cade and Errede, 1994; Dubois and Messenguy, 1994; Irie et al., 1994; Russnak et al., 1995; Odom et al., 2000). The pronounced mitochondrial phenotypes of these mutants (Figure 2, L, M, and O) can be sufficiently explained by the assumption that expression of at least one crucial protein is affected.
DISCUSSION
With the completion of a comprehensive genome-wide screen the number of genes implicated in mitochondrial function and morphology has been more than doubled. The systematic approach enabled the identification of a number of components that have been missed by classical approaches that typically were based on the screening of collections of temperature-sensitive mutants generated by random mutagenesis (Hermann and Shaw, 1998). Most notably, the systematic screening of a deletion mutant library revealed a number of mdm mutants lacking an obvious growth defect (e.g., mdm30, mdm33, mdm35, mdm36, mdm38, mdm39) that presumably could not be easily identified by conventional genetic screens. This illustrates the power of the genomic approach that should turn out to be equally fruitful also for the study of many other cellular processes.
What might be the cellular roles of the newly discovered MDM genes? Only in two cases the predicted protein sequence is suggestive of a function. MDM30 encodes a protein of unknown function that contains an F-box, a motif involved in targeting of proteins to ubiquitin-dependent proteolysis (Patton et al., 1998). Furthermore, high-throughput two-hybrid analysis (Uetz et al., 2000) identified Mdm30p as a possible interaction partner of Cdc53p and Skp1p, two core components of the SCF (Skp1p-cullin-F-box) complexes, which target proteins for ubiquitin-dependent degradation (Skowyra et al., 1997). Because it is known that protein ubiquitination is important for mitochondrial inheritance (Fisk and Yaffe, 1999), we propose that Mdm30p is a novel factor involved in this process. MDM38 encodes a protein that shares homology with a mitochondrial protein of unknown function of Drosophila, the CG4589 gene product (Caggese et al., 1999). This protein is a calcium-binding protein that contains two EF hand calcium binding domains. It will be interesting to see whether Mdm38p plays a role in calcium homeostasis of mitochondria and thereby influences organellar morphology.
During the past decade, several genetic and morphological screens have revealed a number of important components involved in mitochondrial inheritance, yet many processes determining mitochondrial behavior are not well understood. These include the machinery connecting mitochondria to the actin cytoskeleton (Simon et al., 1995; Boldogh et al., 1998, 2001a), proteins cooperating with Fzo1p (Hermann et al., 1998; Rapaport et al., 1998; Fritz et al., 2001) or Ugo1p (Sesaki and Jensen, 2001) in mediating mitochondrial membrane fusion, and components shaping the internal structure of mitochondria (Wong et al., 2000). The proteins encoded by the majority of the newly identified MDM genes do not share homology to other known proteins. Their varied and striking mutant phenotypes suggest that they are important players in a number of different processes contributing to the morphogenesis of mitochondria. The relatively small number of newly identified components suggests that the screen was rather specific. It is important to note, however, that our results were obtained with null alleles. Thus, it cannot be excluded that some of the newly identified genes perform primary functions not directly related to mitochondrial morphogenesis and that the mitochondrial phenotypes might be due to indirect effects accumulating in the deletion strains. It will be a major challenge for the future to establish the molecular functions of the newly discovered proteins and to unravel the interactions among the components of mitochondrial behavior. These studies will certainly improve our understanding of the mechanisms that shape this complex double membrane-bounded organelle.
The results of the screen suggest that most of the proteins constituting the core machinery of mitochondrial morphogenesis uniquely affect this organelle. With the exception of the dynamin-related proteins Dnm1p and Mgm1p, none of the key components of mitochondrial inheritance shares homology with any other known protein involved in membrane trafficking events of other organelles. Vice versa, besides Num1p, none of the components involved in biogenesis of other organelles was found to be essential for normal mitochondrial structure. It appears that Nature invented an entirely new machinery of organelle maintenance after the endosymbiotic ancestors of mitochondria entered the eukaryotic cell.
Supplementary Material
MBC Supplemental Material
ACKNOWLEDGMENTS
The authors thank Gabriele Ludwig for excellent technical assistance and Johannes Herrmann and William Wickner for many helpful discussions. This work was supported by the Deutsche Forschungsgemeinschaft through grants WE 2174/2–1 (to B.W.), Sonderforschungsbereich 413 Teilprojekt B3 (to B.W. and W.N.), and by the BMBF through grant MITOP (to W.N.).
Footnotes
REFERENCES
- Aiken Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Vöth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Berger KL, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw J. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I, Vojtov N, Karmon S, Pon LA. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol. 1998;141:1371–1381. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Yang H-C, Nowakowski WD, Karmon SL, Hays LG, Yates III JR, Pon LA. Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc Natl Acad Sci USA. 2001a;98:3162–3167. doi: 10.1073/pnas.051494698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Yang H-C, Pon LA. Mitochondrial inheritance in budding yeast. Traffic. 2001b;2:368–374. doi: 10.1034/j.1600-0854.2001.002006368.x. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade RM, Errede B. MOT2 encodes a negative regulator of gene expression that affects basal expression of pheromone-responsive genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3139–3149. doi: 10.1128/mcb.14.5.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggese C, Ragone G, Perrini B, Moschetti R, De Pinto V, Caizzi R, Barsanti P. Identification of nuclear genes encoding mitochondrial proteins: isolation of a collection of D. melanogaster cDNAs homologous to sequences in the Human Gene Index database. Mol Gen Genet. 1999;261:64–70. doi: 10.1007/s004380050942. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Tanaka N, White KH, Thorsness PE. Mitochondrial morphology and functional defects caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol Biol Cell. 1994;5:899–905. doi: 10.1091/mbc.5.8.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny KL, McCaffery JM, Jensen RE. Division of mitochondria requires a novel DNM1-interacting protein, Net2p. Mol Biol Cell. 2001;12:309–321. doi: 10.1091/mbc.12.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, et al. YPDTM, PombePDTM and WormPDTM: model organism volumes of the BioKnowledgeTM Library, an integrated resource for protein information. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E, Messenguy F. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol Gen Genet. 1994;243:315–324. doi: 10.1007/BF00301067. [DOI] [PubMed] [Google Scholar]
- Fekkes P, Shepard KA, Yaffe MP. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields SD, Conrad MN, Clarke M. The S. cerevisiae CLU1 and D. discoideum cluA genes are functional homologues that influence mitochondrial morphology and distribution. J Cell Sci. 1998;111:1717–1727. doi: 10.1242/jcs.111.12.1717. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Yaffe MP. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S, Rapaport D, Klanner E, Neupert W, Westermann B. Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J Cell Biol. 2001;152:683–692. doi: 10.1083/jcb.152.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L, van der Bliek AM. The many shapes of mitochondrial membranes. Traffic. 2001;2:235–244. doi: 10.1034/j.1600-0854.2001.1r008.x. [DOI] [PubMed] [Google Scholar]
- Guan K, Farh L, Marshall TK, Deshenes RJ. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr Genet. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Oberle JR, Cooper JA. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol. 2000;151:1337–1343. doi: 10.1083/jcb.151.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Shaw JM. Mitochondrial dynamics in yeast. Annu Rev Cell Dev Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H-P, Avers CJ. Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science. 1973;181:749–750. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- Hönlinger A, Bömer U, Alconada A, Eckerskorn C, Lottspeich F, Dietmeier K, Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 1996;15:2125–2137. [PMC free article] [PubMed] [Google Scholar]
- Irie K, Yamaguchi K, Kawase K, Matsumoto K. The yeast MOT2 gene encodes a putative zinc finger protein that serves as a global negative regulator affecting expression of several categories of genes, including mating-responsive genes. Mol Cell Biol. 1994;14:3150–3157. doi: 10.1128/mcb.14.5.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Hobbs AE, Cerveny KL, Sesaki H. Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc Res Tech. 2000;51:573–583. doi: 10.1002/1097-0029(20001215)51:6<573::AID-JEMT7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Kato M, Wickner W. Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO J. 2001;20:4035–4040. doi: 10.1093/emboj/20.15.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormanec J, Schaaf-Gerstenschläger I, Zimmermann FK, Perecko D, Küntzel H. Nuclear migration in Saccharomyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1 protein. Mol Gen Genet. 1991;230:277–287. doi: 10.1007/BF00290678. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, et al. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2000;28:37–40. doi: 10.1093/nar/28.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fusion is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Pon L, Schatz G. Biogenesis of yeast mitochondria. In: Broach JR, Pringle JR, Jones EW, editors. The Molecular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1991. pp. 333–406. [Google Scholar]
- Prokisch H, Neupert W, Westermann B. Role of MMM1 in maintaining mitochondrial morphology in Neurospora crassa. Mol Biol Cell. 2000;11:2961–2971. doi: 10.1091/mbc.11.9.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Roeder AD, Hermann GJ, Keegan BR, Thatcher SA, Shaw JM. Mitochondrial inheritance is delayed in Saccharomyces cerevisiae cells lacking the serine/threonine phosphatase PTC1. Mol Biol Cell. 1998;9:917–930. doi: 10.1091/mbc.9.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak R, Nehrke KW, Platt T. REF2 encodes an RNA-binding protein directly involved in yeast mRNA 3′-end formation. Mol Cell Biol. 1995;15:1689–1697. doi: 10.1128/mcb.15.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- Scheffler IE. A century of mitochondrial research: achievements and perspectives. Mitochondrion. 2000;1:3–31. doi: 10.1016/s1567-7249(00)00002-7. [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol. 2001;152:1123–1134. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Yaffe MP. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J Cell Biol. 1999;144:711–720. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks J. Methods in Yeast Genetics: A Laboratory Course. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;130:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B. Mitochondrial structure. In: Strathern EW, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1981. pp. 471–504. [Google Scholar]
- Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–365. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:601–603. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vidan S, Snyder M. Large-scale mutagenesis: yeast genetics in the genome era. Curr Opin Biotechnol. 2001;12:28–34. doi: 10.1016/s0958-1669(00)00171-3. [DOI] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Westermann B, Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wong ED, Wagner JA, Gorsich SW, McCaffery JM, Shaw JM, Nunnari J. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP. The machinery of mitochondrial inheritance and behavior. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MBC Supplemental Material