p300 Acts as a Transcriptional Coactivator for Mammalian Notch-1 (original) (raw)
Abstract
Notch-1 belongs to a family of transmembrane receptor proteins that direct the decisions as to various cell fates. After ligand binding, a proteolytic cleavage step occurs and the intracellular part of Notch-1, Notch-1-IC, translocates into the nucleus, where it targets the DNA binding protein RBP-Jκ/CBF1. RBP-Jκ mediates repression through recruitment of a histone deacetylase-containing complex. The Notch-1-IC/RBP-Jκ complex overcomes repression and activates the transcription of Notch target genes. We have identified a novel domain in Notch-1-IC, the EP domain, which is indispensable for full transcriptional activation. This transactivation domain is localized adjacent to the ankyrin repeats of Notch-1-IC. In cotransfection experiments, Notch-1-IC-mediated transcriptional activation was inhibited by E1A12S and p53, two proteins, which interfere with the function of the common coactivator p300. Protein-protein interaction assays demonstrated the association of Notch-1-IC and the CH3 region of p300. In addition, the interaction of mammalian Notch-1-IC with p300 was destabilized after deletion of the EP domain of Notch-1-IC. Based on physical interaction with Notch-1-IC and coactivator functions of p300, we propose a model for Notch-1-mediated gene regulation via p300.
The Notch signaling pathway is a highly conserved signaling mechanism, which is believed to control cell fate decisions in multiple developmental programs (2). In vertebrates, Notch proteins comprise a family of four transmembrane receptors (Notch-1 to Notch-4) that contain multiple epidermal growth factor-like repeats followed by conserved cysteine-rich Notch/Lin12 repeats in their extracellular domain and six cdc10/ankyrin repeats in their intracellular domain. The Notch ligands (Jagged-1, Jagged-2, and Delta-1 to Delta-3) represent transmembrane proteins that, like Notch, contain multiple epidermal growth factor-like repeats in their extracellular domain (11). Ligand binding leads to a cleavage step near the transmembrane region of the C-terminal protein fragment, resulting in the release of the intracellular domain (Notch-IC) followed by its nuclear translocation (41, 46).
An important nuclear target of activated Notch-1 is the ubiquitous DNA binding protein RBP-Jκ/CBF-1, the mammalian homologue of_Drosophila melanogaster Suppressor of Hairless_ [Su(H)] (13, 15). Activated Notch interacts with RBP-Jκ/Su(H) primarily through the RAM23 domain, a sequence that was identified N-terminal to the ankyrin repeats, resulting in activation of transcription (47). Downstream targets of Notch signaling such as Enhancer of split [E(spl)] complex genes (4,28) and mammalian homologues of Hairy and E(spl) genes, HES-1 and HES-5, (18, 32) have been identified. These basic helix-loop-helix (bHLH) proteins antagonize other bHLH factors like MyoD that induce differentiation (25).
In the absence of Notch-1-IC, RBP-Jκ acts as a transcriptional repressor (9, 36). Recent data indicate that RBP-Jκ-mediated repression includes destabilization of the transcription factor IID (TFIID)-TFIIA interaction (33) and recruitment of histone deacetylase corepressor complexes (16,20).
Whereas hypoacetylated histones are implicated in gene silencing, hyperacetylated histones accumulate within transcriptionally active genes (24). Indeed, many transcription factors associate with histone acetyltransferase activity. One of these proteins, p300, belongs to a family of transcriptional coactivators that also includes the closely related cyclicAMP response element binding protein, CBP. The p300 protein associates with many classes of transcription factors including basic leucine zipper (bZIP) proteins like Jun and Fos (1), nuclear receptors (7), members of the NF-κB family (37), and bHLH proteins (53).
After association with RBP-Jκ, Notch-IC stimulates the expression of target genes by overcoming RBP-Jκ-mediated repression and activation of transcription through the presence of an endogenous transactivation domain (15, 27). In addition, recent studies by Kurooka et al. demonstrated a functional interaction of Notch-1-IC with the histone acetyltransferases P/CAF and GCN5 (26).
Here we present the identification and characterization of a novel domain within the C-terminal protein fragment of mammalian Notch-1, which we named the EP domain. Deletion of this domain did not interfere with nuclear localization but abolished Notch-1-mediated transactivation of both an artificial promoter construct and the murine HES-1 promoter. Protein-protein interaction assays demonstrated that the intracellular part of Notch-1 (Notch-1-IC) is targeted by the common coactivator p300. Coimmunoprecipitation assays indicate that deletion of the EP domain within Notch-1-IC destabilizes the interaction with p300 in vivo. Furthermore, in cotransfection experiments, mNotch-1-IC-mediated transactivation was inhibited by E1A12S and p53, two proteins that interfere with p300 function. Our results suggest that recruitment of p300 through the EP domain might be involved in Notch-1-mediated gene regulation.
MATERIALS AND METHODS
Plasmids.
The murine Notch-1-IC cDNA was isolated from pSG5mNotch1IC (15) by digestion with _Kpn_I and_Xba_I and subcloned into the expression vector pcDNA3 (InVitrogen). The C-terminal deletion constructs mNotch-1-X/XB, mNotch-1-P/XB, mNotch-1-E/XB, mNotch-1-N/XB, and mNotch-1-Eh/XB were made by digestion of pcDNA3-mNotch-1-IC with _Xba_I and_Xho_I (mNotch-1-X/XB), _Pvu_II (mNotch-1-P/XB),_Eco_RV (mNotch-1-E/XB), _Not_I (mNotch-1-N/XB), and_Ehe_I (mNotch-1-Eh/XB) and religation of the blunted vectors. The constructs mNotch-1-IC+OP, mNotch-1-TKG, and mNotch-1-SPN were made by PCR. The −ΔRBP constructs correspond to the 1758WFP / LAA1760 mutation in the RAM domain of Notch-1 (47) and were made by using an in vitro mutagenesis system (Stratagene), as specified by the manufacturer's instructions, using the double-stranded oligonucleotide 5′-GCATGGCCAGCTCTTGGCCGCGGAGGGTTTCAAAGTGTC-3′. The −ΔEP in-frame deletions of mNotch-IC were constructed by_Pvu_II–EcoRV digestion and religation of the corresponding mNotch-1 expression vectors. Further details of the construction of the Notch-1 expression plasmids and the Notch-1-green fluorescent protein (GFP) fusion constructs are available on request. The expression vectors for E1A12S and for the N-terminal deletion mutant E1A12SΔ2-36 were described in references 30 and 52. The p300-specific vectors for bacterial expression of glutathiona_S_-transferase (GST) fusion proteins (p300 fragments N and A to F) were made by PCR. The PCR products were inserted into the_Bam_HI and _Eco_RI sites of the bacterial expression vector pGEX-2T (Pharmacia). The Notch-1-specific vector for bacterial expression of GST–mNotch-1-IC, pGST-TKmNotch1IC, was described previously (36). For generating the p300-expressing vector pcDNA3-p300, the p300 cDNA was digested with _Hin_dIII and_Not_I. The DNA fragment was inserted into the corresponding sites of pcDNA3. The C-terminally truncated p300 construct pcDNA3-p300ΔC was generated by digestion of pcDNA3-p300 with_Xba_I and religation of the vector. The expression vectors pCMV-RBP-VP16 and pCMV-EBNA-2 were described in reference48. The construction of pcDNA3-RBP-2N was described previously (36).
Plasmids expressing human p53 and mouse p53 (wild type and an N-terminal deletion encoding amino acids (aa) 44 to 393, referred to p53hs-wt, p53mm-wt, and p53mmΔN), originally constructed by David Lane (University of Dundee), were supplied by Neil Perkins (University of Dundee). The HES-1 promoter-specific reporter plasmid HES-1-LUC was supplied by Urban Lendahl (Karolinska Institute, Stockholm, Sweden). The luciferase reporter plasmid pGa981/6 was described previously (29).
Cell lines.
The cell lines HEK-293 (ATCC CRL 1573) and HeLa (ATCC CCL 2) were grown at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum.
Preparation of cell extracts.
Whole-cell lysates were prepared as follows. Cells were washed three times in phosphate-buffered saline (PBS) and pelleted by centrifugation at 300 × g. The pellet was resuspended in 5 volumes of ice-cold CHAPS lysis buffer consisting of 10 mM 3-[(cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM Tris-HCl (pH 7.9), 150 mM NaCl, 2 mM EDTA, 5 mM NaF, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 1 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl fluoride and incubated on ice for 40 min. The lysate was cleared by centrifugation at 80,000 × g for 30 min. Protein concentrations were determined by the Bradford method (Bio-Rad), and extracts were assayed for DNA binding activity in electrophoretic mobility shift assays (EMSA) and used for immunoprecipitation and Western blotting.
Translation of recombinant proteins.
In vitro-translated proteins were synthesized in a reticulocyte lysate-coupled transcription-translation system as specified by the manufacturer (Promega), using [35S]methionine for labeling. The quality of translation and labeling was monitored by separation of the products using the sodium dodecyl sulfate (SDS)-gel electrophoresis method. The gels were dried and exposed to X-ray films. The labeled proteins were then used for in vitro interaction assays.
In vitro interaction assay.
Purification of bacterially expressed GST fusion proteins and in vitro interaction assays were performed as described in reference 39.
EMSAs.
Approximately 5 to 10 μg of cell extract was used for electromobility gel shift assays in a binding buffer consisting of 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 0.5 mM DTT and 4% glycerol. For binding reactions, 2 μg of poly(dl-dC) (Pharmacia) and approximately 0.5 ng of 32P-labeled oligonucleotides were added. The sequence of the double-stranded oligonucleotide, SL233 (5′-CCTGGAACTATTTTCCCACGGTGCCCTTCCGCCCATTTTCCCACGAGTCG-3′), corresponds to the two RBP-Jκ binding sites within the Epstein-Barr virus TP-1 promoter. For antibody perturbation experiments, 0.5 μg of antibody K0043, directed against RBP-Jκ (14), or 5 μg of antibody, directed against the FLAG epitope (M5 [Sigma]), was added to the reaction mixture. The reaction products were separated using 5% polyacrylamide gels with 1× Tris-glycine-EDTA (TGE) at room temperature. The gels were dried and exposed to X-ray films.
DNA transfection and luciferase assay.
A total of 106 HEK-293 cells were transfected in 90-mm-diameter culture dishes with 5 to 10 μg of plasmid DNA expressing mammalian Notch-1 proteins using calcium phosphate coprecipitation, as described previously (40). At 24 h after transfection, nuclear proteins were prepared and the extracts were assayed for DNA binding activity, protein expression, and subcellular localization. HeLa cells were transfected essentially as described previously (35). Cells (2 × 105) in 35-mm-diameter culture dishes were transfected with 2 μg of reporter plasmid DNA together with various amounts of expression plasmid. To quantitate luciferase activity, cells were harvested 24 h after transfection and lysed for 10 min at room temperature in 120 μl of buffer containing 25 mM Tris-HCl (pH 7.8), 2 mM EDTA, 2 mM DTT, 10% glycerol, and 1% Triton X-100. The lysates were centrifuged at 7,000 × g for 5 min. Luciferase activity was determined from at least four independent transfections with 20 μl of cleared lysate in an LB 9501 luminometer (Berthold) using the luciferase assay system from Promega. All transfections were normalized on the level of total cellular protein.
Western blotting.
SDS-polyacrylamide gels (10 or 7.5% polyacrylamide) were transferred at room temperature to a nitrocellulose filter (BA85; Schleicher & Schuell) for 20 min at 150 mA, using a Tris-glycine buffer system. The membrane was blocked for 2 to 3 h in a solution of 5% milk powder in PBS-T (0.05% Tween 20 in PBS), incubated with the primary antibody directed against the FLAG epitope (M5) for 1 to 2 h in PBS-T containing 5% milk powder, and then washed three times for 10 min in PBS-T. The secondary antibody (1:7,000 dilution of peroxidase-conjugated sheep anti-mouse immunoglobulin G [IgG] [Sigma]) was incubated with the membrane for 30 min. To analyze immunoprecipitated proteins, membranes were sectioned horizontally such that the two proteins of interest (p300 and Notch-1) remained on separate segments of the membrane. After blocking, the sections were incubated with the corresponding primary antibodies, anti-p300 (mouse monoclonal IgG [Upstate]) or anti-Notch-1 (M20, goat polyclonal IgG [Santa Cruz]) at 4°C overnight. The secondary antibodies (1:7,000) dilution of peroxidase-conjugated sheep anti-mouse IgG [Sigma] and 1:30,000 dilution of peroxidase-conjugated rabbit anti-goat IgG [Dianova]) were incubated with membrane sections for 1 h. After washing, specific proteins were detected using an enhanced chemiluminescence system (Amersham).
Coimmunoprecipitation.
Immunoprecipitation was carried out with cell extracts from HEK-293 cells 24 h after transfection with pcDNA3-mNotch-1-IC+OP and pcDNA3-mNotch-1-IC+OP-ΔEP. The cells were lysed in 900 μl of CHAPS lysis buffer. Extracts were incubated with 40 μl of an agarose-conjugated anti-p300 antibody (N15AC [Santa Cruz]) at 4°C overnight. The mixture was divided into three aliquots, and the beads were washed three times with CHAPS lysis buffer containing 150, 500, and 1,000 mM LiCl. After a further washing step with CHAPS lysis buffer containing 150 mM LiCl, the beads were resuspended in SDS-polyacrylamide gel loading buffer and proteins were analyzed by Western blotting.
RESULTS
RBP-Jκ binding is critical for full Notch-1-IC mediated transactivation.
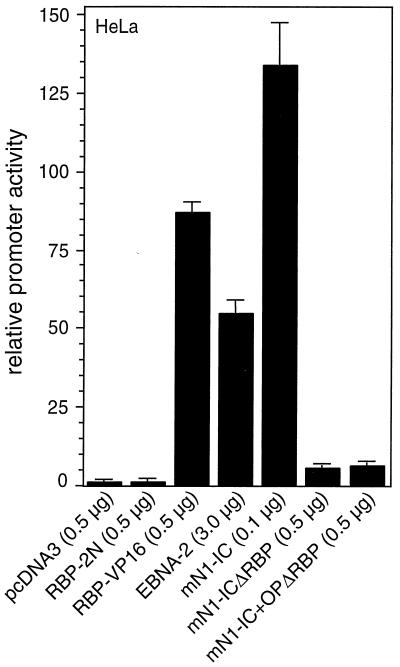
To test Notch-1-specific transcriptional activation by various Notch-1-IC mutant proteins, we used the artificial RBP-Jκ-responsive reporter construct, pGa981/6. This luciferase reporter plasmid carries six repeats of the EBNA-2-responsive element within the Epstein-Barr Virus TP-1 promoter upstream of a minimal β-globin promoter, resulting in a construct with 12 RBP-Jκ binding sites (Fig. 1A). Cotransfection of RBP-2N (9) into HeLa cells had no effect on the transcriptional activity of the promoter construct (Fig.2). In contrast, expression of RBP-VP16 resulted in a clear stimulation of promoter activity. As an additional control, we used an EBNA-2 expression plasmid (48). Overexpression resulted in approximately 50-fold stimulation. Cotransfection of the intracellular domain of the murine Notch-1 protein (mNotch-1-IC [Fig. 1B]) resulted in a dramatic increase of promoter activity (Fig. 2). This stimulation was nearly lost when we used Notch-1 mutants where the primary RBP-Jκ binding site within the RAM domain was mutated (mNotch-1-ICΔRBP and mNotch-1-IC+OPΔRBP [Fig. 1B]). These experiments demonstrate, that the artificial promoter construct pGa981/6 is a suitable tool for analyzing Notch mediated transcriptional regulation.
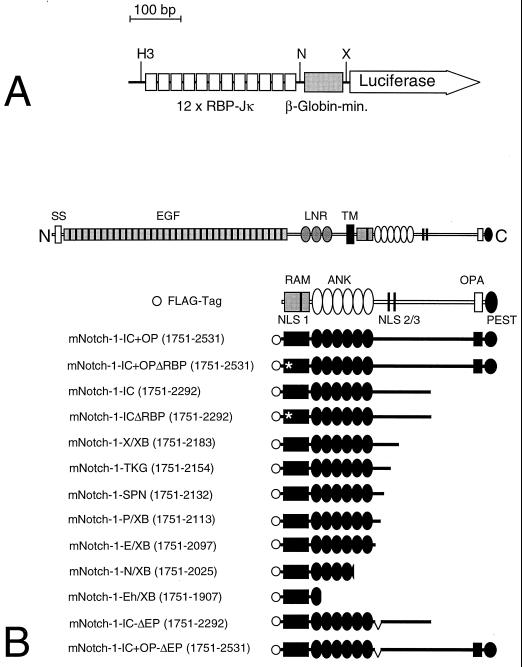
FIG. 1.
Schematic representation of reporter and mNotch-1-IC-specific expression constructs. (A) The luciferase construct pGa981/6 carries 12 RBP-Jκ binding sites upstream of a minimal β-globin promoter. Abbreviations: H3, _Hin_dIII, N, _Nsi_I, X, _Xho_I. (B) The mammalian Notch-1 receptor and the derived deletion proteins used for expression. All constructs carry an N-terminal FLAG tag (open circle). The first and last amino acids compared to the full-length protein are indicated in parentheses. The −ΔEP mutants represent in-frame deletions of aa 2098 to 2112. The asterisk designates a 1758 WFP/LAA 1760 mutation in the mRAM23 domain. Abbreviations: SS, signal sequence, EGF, epidermal growth factor-like repeats, LNR, Lin/Notch repeats, TM, transmembrane domain, RAM, RAM23 domain, ANK, ankyrin repeats, NLS, nuclear localization signal.
FIG. 2.
Notch-1-IC-mediated transcription depends on the primary RBP-Jκ binding site within the RAM domain. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells together with the indicated amounts of expression plasmids. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from at least four independent experiments are shown.
Dominant negative effect of mNotch-1-ICΔRBP in mNotch-1 mediated transcription.
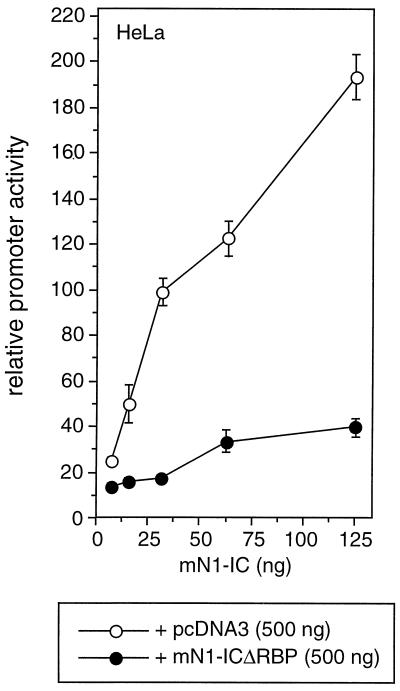
Recently, it was shown that the intracellular domain of Notch-3 represses Notch-1-mediated transcriptional activation (5). The authors presented a model in which this repression may be due to competition with Notch-1-IC for a common coactivator present in limiting amounts. If this limiting cofactor exists, a Notch-1 mutant defective in RBP-Jκ binding should compete for this factor with the wild-type Notch-1-IC protein. To address this question, we performed cotransfection experiments using the reporter constructs described above, Notch-1-IC and Notch-1-ICΔRBP. Cotransfection of increasing amounts of mNotch-1-IC together with pcDNA3 as a control resulted in a clear stimulation of promoter activity. This transactivation was significantly suppressed after coexpression of the Notch-1 mutant mNotch-1-ICΔRBP (Fig. 3). These results demonstrate that like Notch-3-IC, the mNotch-1-ICΔRBP mutant represses Notch-mediated transcription, possibly by cofactor competition.
FIG. 3.
Dominant negative effect of mNotch-1-ICΔRBP. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells together with increasing amounts of a Notch-1-IC expression plasmid and the indicated amount of pcDNA3 (open circles) or pcDNA3mN1-ICΔRBP (solid circles). Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from three independent experiments are shown.
Transcriptional activity of mNotch-1 deletion constructs.
To gain further insight in the transcriptional activity of intracellular Notch-1 proteins, we used deletion constructs shown in Fig. 1B. The results of cotransfection of the reporter plasmid pGa981/6 and Notch-1-specific deletion mutants into HeLa cells are shown in Fig.4. Stimulation of promoter activity of about 200- to 250-fold was observed after cotransfection of mNotch-1-IC, mNotch-1-X/XB (aa 1751 to 2183), and mNotch-1-TKG (aa 1751 to 2154) into HeLa cells. Further C-terminal deletions in the constructs mNotch-1-SPN (aa 1751 to 2132) and mNotch-1-P/XB (aa 1751 to 2113) led to a small decrease of transcriptional activity. However, we still observed a 100-fold stimulation of promoter activity. Interestingly, after deletion of an additional 15 aa in mNotch-1-E/XB (aa 1751 to 2097), we could not detect any activation of the reporter construct in cotransfection experiments (Fig. 4). Loss of transcriptional activity was also observed after cotransfection of mNotch-1-N/XB (aa 1751 to 2025) and mNotch-1-Eh/XB (aa 1751 to 1907). Note that all of the mNotch-1 deletion constructs used in this experiment have an intact RAM domain and are capable of RBP-Jκ binding. In addition, in all deletion mutants except for mNotch-1-N/XB and mNotch-1-Eh/XB, the ankyrin domain is not affected by deletion (Fig. 1B). Therefore, we assume that a distinct domain in mNotch-1 (aa 2098 to 2113) C-terminal to the ankyrin repeats is indispensable for transactivation. We named this novel domain the EP domain.
FIG. 4.
Transcriptional activity of mNotch-1 deletion mutants. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 150 or 200 ng of plasmid expressing the indicated mNotch-1 proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from at least five independent experiments are shown.
Detection of RBP/Notch-1 complexes by EMSA.
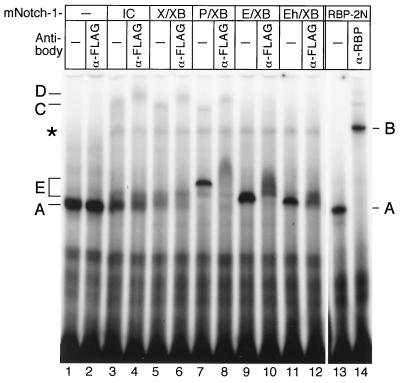
To investigate the formation of RBP complexes with selected Notch-1 deletion mutants, we performed band shift experiments (Fig.5). HEK-293 cells were transiently transfected with plasmids expressing Notch-1-IC and deletion mutants. Expression of the mutant Notch-1 proteins was verified by Western blot analysis (data not shown). As a control, we used in vitro-translated RBP-Jκ protein (Fig. 5, lanes 13 and 14). The RBP-Jκ-specific complex (band A, lane 13) was supershifted after addition of an antibody (K0043), directed against RBP (band B, lane 14). As expected, an RBP-Jκ specific complex (complex A) could already be identified in extracts of untransfected HEK-293 cells (lanes 1 and 2). Addition of an antibody directed against the FLAG epitope did not interfere with RBP-Jκ DNA binding (lane 2). The endogenous RBP-Jκ-specific DNA binding activity was markedly decreased in extracts from HEK-293 cells transfected with 5 μg of a plasmid expressing mNotch-1-IC. In these extracts, a novel, slower-migrating complex (band C) appeared (lane 3). Incubation of cell extracts with an antibody directed against the FLAG epitope supershifted complex C, resulting in complex D, which most probably contains Notch-1-IC interacting with RBP-Jκ (lane 4). Similar results were obtained with extracts from cells transfected with mNotch-1-X/XB (lanes 5 and 6) and mNotch-1-P/XB (lanes 7 and 8). Interestingly, in extracts from mNotch-1-P/XB-transfected cells, a novel complex could be detected (band E) in addition to the higher-order complex C (lane 7). This band also interferes with the anti-FLAG antibody, suggesting that this complex contains Notch-1 protein (lane 8). The novel complex could also be detected in extracts from cells transfected with mNotch-1-E/XB (lanes 9 and 10) and mNotch-1-Eh/XB (lanes 11 and 12), but no higher-order complex (band C) was generated. Note that all complexes E interfered with the anti-FLAG antibody (lanes 8, 10, and 12). The different migration properties of complexes E most probably result from different sizes of the mNotch-1 deletion mutants. Taken together, using band shift experiments, we could detect two types of Notch-1/RBP complexes: (i) a slow migration complex (C) in extracts from HEK-293 cells transfected with transcriptionally active Notch-1 proteins, and (ii) a novel complex (E) in extracts from cells transfected with transcriptionally inactive Notch-1 proteins. Only in extracts from mNotch-1-P/XB transfected cells could we detect both complexes. As shown in Fig. 4, this deletion mutant is still transactivating, although with lower activity.
FIG. 5.
Detection of RBP/Notch-1 DNA binding complexes. In cell extracts from HEK-293 cells, an RBP-Jκ-specific DNA binding complex can be detected (complex A, lanes 1 and 2). In cell extracts from HEK-293 cells transfected with expression plasmids for the indicated Notch-1 proteins, the RBP-Jκ-specific DNA binding activity was decreased and more slowly migrating complexes appeared (lanes 3 to 12). Transcriptionally active Notch-1 proteins form a slowly migrating complex (complex C, lanes 3, 5, and 7). Treatment with an anti-FLAG antibody (α-FLAG) recognized complex C and supershifted to a novel complex D (lanes 4, 6, and 8). Transcriptionally inactive Notch-1 proteins form a faster-migrating complex (complex E, lanes 9 and 11). In cell extracts from HEK-293 cells transfected with the Notch-1 mutant mNotch-1-P/XB, both complexes (C and D, lanes 7) were formed. Complex E interfered with the anti-FLAG antibody (lanes 8, 10, and 12). Cell-free synthesized RBP-2N was used as a control for RBP DNA binding. The RBP-specific DNA binding activity (A) (lane 13) was recognized by an RBP-specific antibody (α-RBP) to form a supershifted complex B (lane 14). The asterisk designates a nonspecific complex (lanes 3 to 12). The 32P-labeled oligonucleotide SL233 was used as a probe.
Transcriptional activity and subcellular localization of Notch-1-IC–GFP proteins.
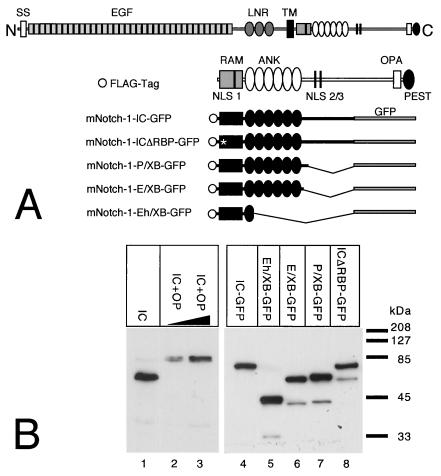
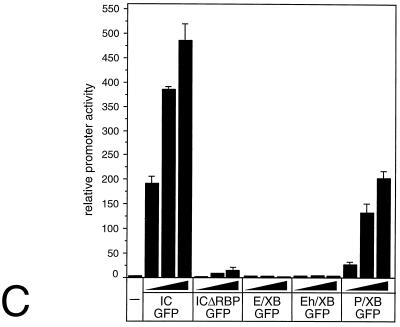
Three putative nuclear localization signals have been identified within the mammalian Notch-1 protein (3). To rule out any interference with the subcellular localization of the mNotch-1 deletion proteins, we used mNotch-1 mutants fused to GFP (Fig.6A) in transfection and cotransfection experiments. Protein expression (Fig. 6B) and transcriptional activity (Fig. 6C) were assayed 24 h after transfection. Western blot analysis revealed no significant variations in the expression of the mNotch-1–GFP proteins (Fig. 6B). Cotransfection of increasing amounts of mNotch-1-IC–GFP together with the RBP-Jκ reporter plasmid into HeLa cells resulted in a clear stimulation of promoter activity. Cotransfection of increasing amounts of mNotch-1-P/XB–GFP resulted in distinct but reduced stimulation of promoter activity compared to that in mNotch-1-IC–GFP (Fig.6C). In contrast, we could not detect any significant increase of transcriptional activity after cotransfection of mNotch-1-ICΔRBP-GFP, mNotch-1-E/XB–GFP, or mNotch-1-Eh/XB-GFP.
FIG. 6.
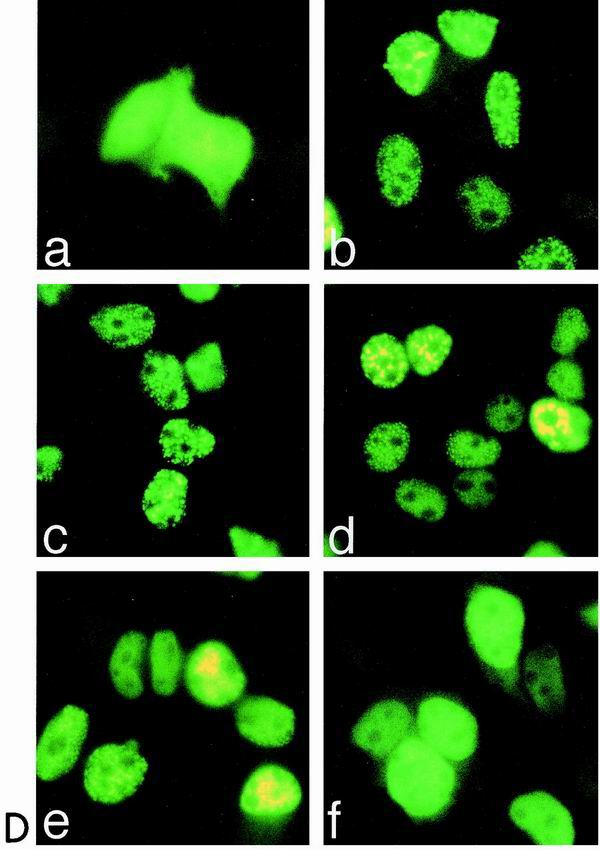
(A to C) Transcriptional activity of Notch-1–GFP fusion proteins. (A) Schematic representation of Notch-1 deletion mutants fused to GFP. For details, see Fig. 1B. (B) Cell extracts were prepared 24 h after transfection of 5-μg portions of plasmid expressing Notch-1 proteins and Notch-1-specific GFP fusion proteins. Expression was assayed by Western blotting using an anti-FLAG antibody. (C) Stimulation of the reporter construct pGa981/6 by Notch-1–GFP fusion proteins depends on the EP domain. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 150, 200, or 250 ng of plasmid expressing the indicated mNotch-1-GFP fusion proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations of four independent experiments are shown. (D) Subcellular localization of Notch-1-GFP fusion proteins in vivo. HEK-293 cells were transfected with plasmids expressing GFP (CMV-GFP) (a) or the Notch-1-IC-derived GFP fusion proteins mNotch-1-IC-GFP (b), mNotch-1-ΔRBP-GFP (c), mNotch-1-P/XB-GFP (d), mNotch-1-E/XB-GFP (e), or mNotch-1-Eh/XB-GFP (f). At 24 h after transfection, the living cells were assayed for GFP expression by fluorescence microscopy. Magnification, ×630.
Next, we investigated the subcellular localization of the mNotch-1–GFP proteins in vivo. HEK-293 cells were transiently transfected with the mNotch-1–GFP-expressing constructs shown in Fig. 6A. After 24 h the living cells were assayed for GFP expression. The microscopic observations are shown in Fig. 6D. A GFP-expressing vector (CMV-GFP) was used as a control. The GFP protein was homogeneously distributed in the nucleus as well as in the cytoplasm of the cells (Fig. 6D, panel a). The mNotch-1-IC–GFP fusion protein was found almost exclusively in the nucleus and showed a speckled pattern. We could not detect any obvious differences in subcellular localization between Notch-1-IC–GFP and the mutant Notch-1-GFP proteins, mNotch-1-ICΔRBP–GFP (panel c), mNotch-1-P/XB–GFP (panel d), and mNotch-1-E/XB–GFP (panel e), although the last two lack the nuclear localization signals 2 and 3. Only the shortest mNotch-1–GFP protein mNotch-1-Eh/XB–GFP, which lacks most of the ankyrin repeats (Fig. 6A), revealed a different localization pattern in transfected HEK-293 cells: most of the protein is found in the nucleus, but the speckled pattern is lost (panel f). Therefore, loss of transcriptional activity after deletion of the EP domain does not correlate with a change in subcellular localization of the mNotch-1 proteins. Transcriptionally, inactive mNotch-1 mutants still translocated into the nucleus.
Repression of Notch-1-mediated transactivation by E1A12S and p53.
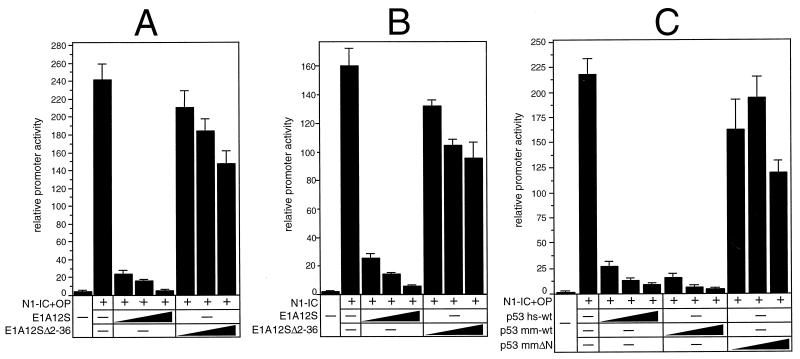
It was shown previously that the adenovirus E1A12S protein interferes with coactivator function (8, 12, 49). To investigate whether E1A interferes with Notch-1-mediated transcription, we performed cotransfection experiments using the RBP-Jκ reporter plasmid and expression plasmids for E1A12S and Notch-1 proteins. Luciferase activity of pGa981/6 was stimulated 240-fold following cotransfection of pcDNA3-mNotch-1-IC+OP. This stimulation was abrogated to basal levels by gradually increasing the amounts of E1A12S. Repression of Notch-1-mediated transcriptional activation was not observed when the N-terminal deletion mutant E1A12SΔ2-36 was used (Fig. 7A). Similar results were obtained when the C-terminal deletion mutant mNotch-1-IC, missing the OPA and PEST sequences, was used. Cotransfection resulted in 160-fold stimulation of promoter activity, which again was reduced to basal levels by gradually increasing the amount of an E1A12S expression plasmid (Fig. 7B). The inhibitory effect of E1A12S on Notch-1-mediated transcription indicated that coactivators of the p300 family might be involved in this process. For this reason, we used a second protein, p53, which was shown to be an inhibitory indicator of p300-dependent transactivation (50). The experiments were performed with murine p53 (p53 mm-wt) and human p53 (p53 hs-wt), which both strongly repressed Notch-1-mediated transcription (Fig. 7C). Deletion of the N-terminal 43 aa, which are important for p300 interaction in p53mmΔN, abolished the repressive effect of p53. These results demonstrate that E1A12S and p53 are able to suppress Notch-1-mediated transcription. This repression depends on the N-terminal sequences of E1A12S and p53.
FIG. 7.
Repression of Notch-1-mediated transactivation by E1A12S and p53. (A and B) Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 100 ng of plasmid expressing mNotch-1-IC+OP (A) or mNotch-1-IC (B), together with increasing amounts (0.2, 0.4, and 0.6 μg) of plasmids expressing the indicated E1A proteins. (C) Portions (2 μg) of reporter construct pGa981/6 were cotransfected with 100-ng portions of plasmid expressing mNotch-1-IC+OP together with increasing amounts (0.2, 0.4, and 0.6 μg) of plasmids expressing the indicated human and murine p53 proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from four independent experiments are shown.
Notch-1-IC interacts with the CH3 region of p300.
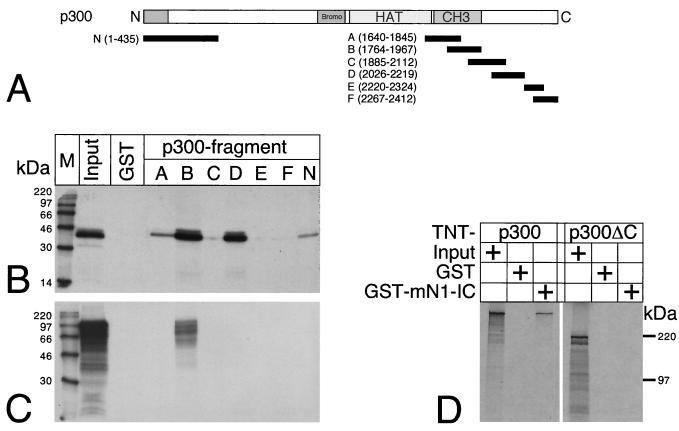
Physical interactions between mNotch-1-IC and the transcriptional coactivator p300 were analyzed in vitro by pull-down assays with GST fusion proteins. The p300-derived fragments N, A, B, C, D, E, and F shown in Fig. 8A were fused to GST and expressed in Escherichia coli. Glutathione-Sepharose beads were coated with recombinant GST or the GST fusion proteins and used as bait for cell-free synthesized and radiolabeled Notch-1-IC protein (Fig. 8C) or radiolabeled p300 protein (Fig. 8D). Radiolabeled E1A (Fig. 8B) was used as a control. As shown previously (52), strong interaction of E1A with p300 fragments B and D was observed whereas fragments A and C and the amino terminus of p300 revealed only a weak interaction with E1A. No interaction could be detected with fragments E and F and with GST protein alone (Fig. 8B). In contrast, interaction of Notch-1-IC and p300 could be detected exclusively with fragment B (Fig. 8C). This Notch-1-IC interaction domain partially overlaps the CH3 region of p300 (Fig. 8A). Interaction of Notch-1-IC with p300 was also observed when GST mNotch-1-IC was used as bait for cell-free-synthesized p300 (Fig. 8D). The interaction of p300 with GST-mNotch-IC depends on the C-terminal part of p300, since cell-free-synthesized p300ΔC (aa 1 to 1302) failed to interact with the bait (Fig. 8D).
FIG. 8.
Notch-1-IC interacts with the CH3 region of p300. (A) Schematic representation of p300-derived protein fragments fused to GST. After bacterial expression, the GST fusion proteins were used in pull-down experiments. The first and last amino acids compared to the full-length protein are indicated in parentheses. (B) Cell-free synthesized E1A protein interacts with various p300 fragments. (C) Cell-free synthesized Notch-1-IC protein interacts with p300 fragment B. (D) Cell-free synthesized p300 interacts with GSTmNotch-1-IC. C-terminally truncated p300 (aa 1 to 1302) fails to interact with GST-Notch-1-IC. GST proteins were immobilized with Sepharose beads and incubated with in vitro-translated and radiolabeled proteins. After extensive washing steps, the reaction mixtures were boiled and the proteins were separated by SDS-polyacrylamide gel electrophoresis. The positions of molecular size markers are shown.
Notch-1-IC/p300 interaction correlates with transcriptional activity.
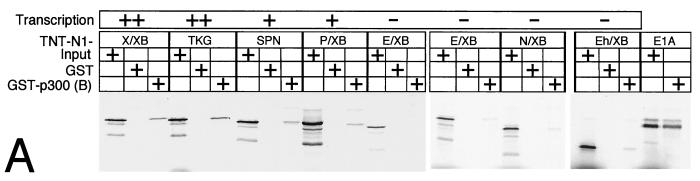
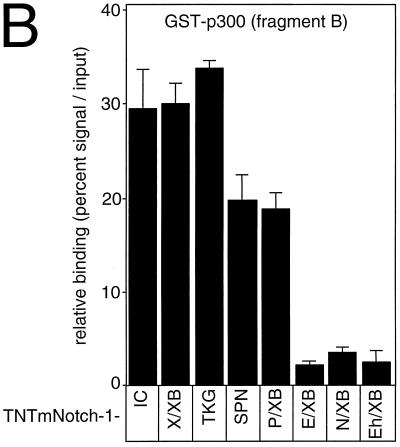
Next we asked which part of Notch-1-IC is responsible for p300 interaction. To do this we performed a set of pull-down experiments with p300 fragment B fused to GST and various C-terminal deletion mutants of Notch-1-IC. As shown in Fig.9, the C-terminal deletion mutants mNotch-1-X/XB and mNotch-1-TKG still interact with fragment B of p300. Further deletion in mNotch-1-SPN and mNotch-1-P/XB led to some loss of interaction capacity with fragment B. The interaction was further reduced using mNotch-1-E/XB, mNotch-1-N/XB, and mNotch-1-Eh/XB (Fig. 9A). A densitometric analysis of the results of six pull-down experiments is shown in Fig. 9B. Taken together, the results of these mapping experiments show that Notch-1-IC proteins are capable of interacting with p300 as long as they contain at least residues 2097 to 2113, the EP domain. Interestingly, interaction of the mNotch-1-IC-derived deletion mutants with GST-p300 fragment B correlates with their transcriptional potential in our cotransfection experiments (Fig. 4 and 6C).
FIG. 9.
(A) Notch-1-IC/p300 interaction correlates with transcriptional activity. (A) GST p300 fragment B was immobilized with Sepharose beads and incubated with in vitro-translated Notch-1-IC deletion mutants (TNT-N1-) or E1A protein as a control. After extensive washing steps, the reaction mixtures were boiled and proteins were separated by SDS-polyacrylamide gel electrophoresis. Relative transcriptional activity of the Notch-1 proteins in cotransfection experiments is shown (Fig. 4). ++, strong transcriptional activation; +, reduced activation; −, loss of activation. (B) Relative binding of in vitro-translated Notch-1-IC deletion mutants to GST-p300 fragment B obtained from densitometric analysis. Exposed films from six experiments were scanned and analyzed by the NIH Image software.
Notch-1–p300 interaction in vivo.
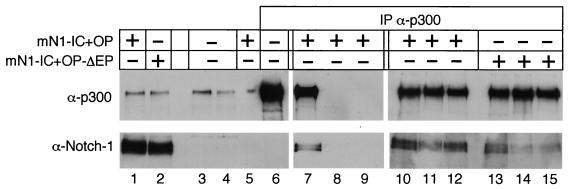
Coimmunoprecipitation experiments were performed to prove the existence of a physical interaction of p300 with mNotch-1-IC in vivo. Since it is not feasible to detect endogenous activated Notch in the nucleus by immunological methods (41), we decided to perform these experiments with transfected Notch-1 constructs. HEK-293 cells were transiently transfected with plasmids expressing mNotch-1-IC+OP or mNotch-1-IC+OPΔEP. Expression of Notch-1 proteins, as well as endogenous p300 protein, was verified by Western blotting (Fig.10, lanes 1 and 2). The quality of the p300 immunoprecipitation was monitored in untransfected HEK-293 cells. The amount of p300 protein in cell lysates (lane 3) decreased after incubation with the anti-p300 antibody (lane 4) and accumulated in the precipitate (lane 6). When the anti-p300 antibody was used for immunoprecipitation, Notch-1 protein was coimmunoprecipitated (lane 7), whereas no Notch-1 protein was found when agarose beads alone were used (lane 5). This interaction was specific, since addition of a blocking peptide (p300-N15P [Santa Cruz]) directed against the p300 antibody abolished the coimmunoprecipitation (lanes 8 and 9). Interaction of mNotch-1-IC+OP with endogenous p300 appears to be very stable, since extensive washing with 500 mM LiCl (lane 11) and 1,000 mM LiCl (lane 12) did not interfere with binding. In contrast, interaction of p300 with mNotch-1-IC+OP-ΔEP (lane 13) decreased after washing with 500 mM LiCl (lane 14) and 1,000 mM LiCl (lane 15). These results indicate that (i) Notch-1-IC binds to endogenous p300 in vivo, (ii) this interaction seems to be rather stable, and (iii) deletion of the EP domain in Notch-1-IC destabilizes this interaction.
FIG. 10.
Notch-1/p300 interaction in vivo. Expression of endogenous p300, mNotch-1-IC+OP (lane 1), and mNotch-1-IC+OP-ΔEP (lane 2) in transiently transfected HEK-293 cells is shown. The amount of p300 protein in cell lysates (lane 3) decreased after incubation with the anti-p300 antibody (lane 4) and accumulated in the precipitate (lane 6). Increasing amounts of blocking peptide added to the reaction mixture (lanes 8 and 9) prevented immunoprecipitation of p300 as well as coimmunoprecipitation of mNotch-1-IC+OP (lane 7). Deletion of the EP domain destabilizes Notch-1/p300 interaction (lanes 10 to 15). Increasing amounts of LiCl in the washing buffer did not interfere with p300 binding to mNotch-1-IC+OP but destabilized p300 binding to mNotch-1-IC+OP-ΔEP (compare lanes 10 to 12 with lanes 13 to 15). Extracts were incubated with agarose beads alone (lane 5) or an agarose-conjugated anti-p300 antibody (lanes 6 to 15). The mixture was divided into three aliquots, and the beads were washed three times with CHAPS lysis buffer containing 150 mM (lanes 10 and 13), 500 mM (lanes 11 and 14), or 1000 mM LiCl (lanes 12 and 15). After a further washing step with CHAPS lysis buffer, the proteins were analyzed by Western blotting.
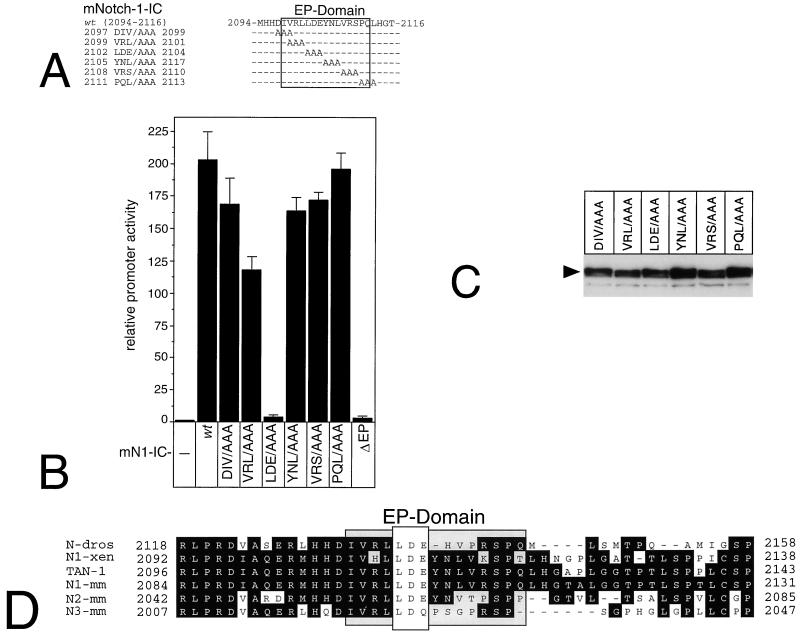
Mutational analysis of the EP domain within mNotch-1-IC.
To identify amino acids within the EP domain critical for mNotch-1-mediated transactivation, we used mNotch-1-IC-specific scanning mutants (Fig.11A). Expression of the mNotch-1-IC mutants was verified by Western analysis after transient transfection of the corresponding expression plasmids into HEK-293 cells (Fig. 11C). Cotransfection of plasmids expressing wild-type mNotch-1-IC together with the reporter plasmid pGa981/6 into HeLa cells resulted in 200-fold stimulation of luciferase activity. Interestingly, only one scanning mutation within the EP domain, mNotch-1-IC-2102 LDE/AAA 2104 resulted in complete loss of transcriptional activity, which was comparable to that of mNotch-1-ICΔEP. Cotransfection of the remaining mNotch-1-IC scanning mutants resulted in no or only a weak decrease in transactivation potential (Fig. 11B). These results demonstrate that within the EP domain of mNotch-1-IC, at least 3 aa (2102 LDE 2104) are critical for transactivation. In addition, this LDE motif within the EP domain is highly conserved in Notch proteins of various species (Fig. 11D).
FIG. 11.
Mutation analysis of the EP domain within mNotch-1-IC. (A) Schematic representation of the mNotch-1-IC specific scanning mutants used in cotransfection experiments. (B) Transcriptional activity of EP domain mutants. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 150-ng portions of plasmid expressing the indicated mNotch-1-IC proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. (C) Cell extracts from HEK-293 cells were prepared 24 h after transfection of 5 μg of plasmid expressing Notch-1-IC–EP domain mutant proteins. Expression was assayed by Western blotting using an anti-FLAG antibody. Mean values and standard deviations from four independent experiments are shown. (D) The EP domain is highly conserved within Notch proteins. Protein sequences of the indicated Notch proteins were aligned with the CLUSTAL W program. The first and last amino acids are numbered relative to the full-length proteins. The EP domain is specified. Abbreviations and accession numbers: N-dros, Notch Drosophila (P07207); N1-xen, Notch-1 Xenopus laevis (P21783); TAN-1, Notch-1 human (P46531); N1-mm, Notch-1 mouse (Q01705); N2-mm, Notch-2 mouse (BAA22094); N3-mm, Notch-3 mouse (Q61982).
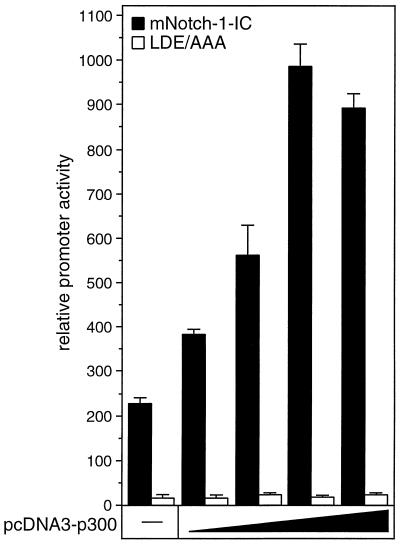
Additive effect of p300 in Notch-1-mediated transactivation.
To test the cooperation of Notch-1-IC and p300 on a functional level, we performed cotransfection experiments with HeLa cells using the RBP-Jκ reporter and expression plasmids for p300 and the intracellular domain of mNotch-1 (Fig.12). Stimulation of luciferase activity from the reporter construct by Notch-1-IC expression was further enhanced after gradually increasing the amounts of p300. Similar results were obtained when an expression plasmid for Notch-1-IC+OP was used (data not shown). In contrast, we could not detect any increase of luciferase activity when we used the Notch-1-IC-specific scanning mutant LDE/AAA in this assay (Fig.12). These results demonstrate that the additive effect of p300 in Notch-1-mediated transactivation is dependent on a functional EP domain.
FIG. 12.
Additive effect of p300 in Notch-1-mediated transactivation. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 100 ng of plasmids expressing mNotch-1-IC (black bars) or the mNotch-1 specific EP domain mutant LDE/AAA (white bars), together with increasing amounts (50, 100, 200, and 500 ng) of pcDNA3-p300. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations of three independent experiments are shown.
Transcriptional activation of the murine HES-1 promoter by Notch-1-IC depends on the EP domain.
HES-1 was identified as a Notch target gene in mammals (17, 18, 25). To test the effect of the EP domain in Notch-1-IC-mediated transcription on a naturally existing promoter, we performed cotransfection experiments with the human HES-1 promoter fused to the luciferase gene. Cotransfection of either mNotch-1-IC or mNotch-1-IC+OP led to a clear stimulation of HES-1 promoter activity (Fig. 13). Deletion of the EP domain in mNotch-1-ICΔEP resulted in nearly complete loss of transcriptional activity, whereas the Notch-1 mutant mNotch-1-IC+OPΔEP was still able to activate transcription from the HES-1 promoter to some extent. This might be due to a transactivation domain, which was mapped to the C-terminal OPA and PEST sequences of mNotch-1 (10, 27). These results demonstrate that the identified EP domain is a prerequisite for full transcriptional activity of the intracellular domain of mNotch-1.
FIG. 13.
Transcriptional activation of the murine HES-1 promoter by Notch-1 depends on the EP domain. Portions (2 μg) of the HES-1 specific reporter construct HES-1-LUC were cotransfected into HeLa cells with 150 or 200 ng of plasmid expressing the indicated mNotch-1 proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from at least three independent experiments are shown.
DISCUSSION
One mechanism by which Notch-1-IC exerts its function is the conversion of the DNA binding protein RBP-Jκ/CBF-1 from a repressor to an activator of transcription. Three domains of Notch-1-IC are important for transactivation including, RAM, ankyrin repeats, and TAD. The RAM domain represents the primary binding site to RBP-Jκ and was shown to be important for transactivation activity of Notch-1-IC through displacement of a putative corepressor from RBP-Jκ (15,20, 47). Within this interaction domain, a stretch of 3 aa, WFP (aa 1758 to 1760), is critical for the interaction of Notch-1-IC with RBP-Jκ (47). We found that a mutant form of Notch-1-IC, mNotch-1-ICΔRBP, with a substitution of WFP to LAA, slightly transactivates a reporter gene containing multiple RBP-Jκ binding sites (Fig. 2). This is in agreement with earlier data showing that Notch-1-IC lacking the RAM domain can still activate transcription of the HES genes but does so less strongly than Notch-1-IC does (17, 21). In addition, mNotch-1-ICΔRBP, when cotransfected with Notch-1-IC, repressed the transactivation of a reporter construct in_trans_, suggesting coactivator competition (Fig. 3). Similarly, Notch-3-IC represses Notch-1-IC-mediated transcriptional activation (5). This inhibition has been suggested to rely on competition of common coactivators, which are known to be present in limiting amounts. To identify domains important for transcriptional activation of Notch-1-IC, C-terminal deletion constructs were tested for their ability to transactivate a reporter plasmid. The results show that a region C-terminal of the ankyrin repeats is important for transactivation. A construct, mNotch-1-E/XB, lacking this region did not exhibit any activity (Fig. 4). We called this putative transactivation domain the EP domain. Interestingly, this region is highly conserved among Notch proteins of various species (Fig. 11D).
An autonomous transactivation domain, TAD, has been identified in mouse Notch-1 (10, 22, 27). This domain was mapped to 200 aa containing the OPA sequence, although the OPA sequence alone was unable to mediate the transactivation function. We also found a sequential loss of transcriptional activity using the C-terminal deletion, but most of the transcriptional activity of Notch-1-IC remains after deleting TAD and is completely abolished only after C-terminal deletion of the EP-domain.
The EP domain in Notch-1 is also important for transactivation of a promoter construct derived from a well-characterized Notch target gene, HES-1. The Notch-1-specific deletion mutant mNotch-1-ICΔEP completely lost its ability to activate the HES-1 promoter (Fig. 13). Interestingly, the addition of the C-terminal OPA sequences, mNotch-1-IC+OPΔEP, rescued some of its transcriptional activity, suggesting that the function of TAD depends only in part on an intact EP domain.
In addition to impaired transcriptional function, we find a loss of protein-DNA interaction by Notch mutants lacking the EP domain. The interaction of Notch-1-IC with RBP-Jκ bound to an appropriate oligonucleotide probe can be visualized by EMSA as a slowly migrating higher-order complex (17, 36). Mutants of Notch-1-IC with C-terminal deletions in the previously described TAD still give rise to a higher-order complex containing Notch-1-IC/RBP. In contrast, this complex failed to form with transcriptional inactive mutants (Fig. 5). These data suggest the presence of a putative cofactor(s) forming a higher-order complex with transcriptionally active Notch-1 mutants.
In addition to TAD and the novel EP domain, ankyrin repeats are be important for transactivation. Missense mutations in the ankyrin repeats of mouse Notch-1 show loss-of-function phenotypes (21, 23). An autonomous function for the ankyrin domain independent of RBP-Jκ signaling has been described in_Caenorhabditis elegans_ and in mouse muscle cell differentiation (31, 38, 42). Recently, the ankyrin repeats were identified as target sequences for MAML-1 interaction. In this report, MAML-1, the human homologue of Drosophila Mastermind, acts as a transcriptional coactivator for Notch proteins (51). The question remains how these domains, ankyrin repeats, EP domain, and TAD, act together to confer strong transactivation.
The transactivation function of Notch-1-IC can be antagonized by the adenovirus protein E1A12S. This repression was not observed when the deletion mutant E1A12SΔ2-36 was used. Several studies have shown that E1A12S blocks p300/CBP coactivator functions (8, 12,45). This does not exclude the involvement of other coactivators in Notch-1-mediated transactivation, since E1A12S was also shown to interfere with P/CAF function (26). Similar to E1A12S, p53 can repress Notch-1-IC-mediated transcription, whereas the deletion mutant p53ΔN, which cannot interact with p300, does not have any repressive effect (50). Therefore, it is likely that E1A12S and p53 function in part by preventing Notch-1-IC from association with p300.
The results we obtained using several approaches support the hypothesis that the common coactivator p300 might be involved in Notch-1-dependent gene expression. (i) Notch-1-IC-mediated transcriptional activation was inhibited by E1A12S and p53, two proteins which interfere with p300 functions (Fig. 7). (ii) Notch-1-IC interacts with the CH3 region of p300 (Fig. 8). (iii) p300 interacts with a distinct domain of Notch-1-IC essential for full transcriptional activation (Fig. 9). (iv) Notch-1-IC/p300 interaction correlates with transcriptional activity. (v) Notch1-IC forms a stable complex with p300 in vivo (Fig.10). (vi) Notch-1-IC and p300 display some additive effects on transactivation, and these effects depend on a functional EP domain (Fig. 12).
Ordentlich et al. demonstrated Notch-1-IC- and Notch-2-IC-mediated repression of E47 activity (34). MyoD responds to Notch in a similar fashion (23). Although MyoD and E47 both utilize p300 as cofactors, the authors could not confirm a role for p300 in that context. The conclusion was based on the inability of Notch-1-IC to alter GAL4-p300-mediated gene expression. Moreover, GAL4-E47, E47-VP16, GAL4-MyoD, and MyoD-VP16 are all resistant to inhibition by Notch (23). This would not necessarily be contradictory to our results but points to the requirement for additional structural features.
Coactivators like p300 contribute to transcriptional regulation by modifying chromatin structure via association with members of the p160/SRC family of coactivators, as well as with P/CAF. While our work was in progress, Kurooka and Honjo reported that mouse P/CAF and GCN5 interact with Notch1-IC (26). P/CAF and GCN5 require the ankyrin repeats in addition to the previously characterized domain, TAD. The aa 2098-to-2193 sequence within Notch-1, which is involved in p300 binding is not required for interaction with P/CAF or GCN5. Therefore, two possibilities could be discussed: (i) either p300 binds primarily to Notch-1 and recruits molecules with additional HAT activity, like P/CAF, or (ii) P/CAF and GCN5 bind first and build the starting point for the formation of a larger complex. A synergism between multiple coactivator proteins has been described for the transactivation functions of hepatocyte nuclear factor 1 (HNF-1). The authors support a model in which the combined action of coactivators (here CBP and P/CAF) is recruited by HNF-1 to activate transcription (43). The interaction of a multiprotein complex including P/CAF and GCN5 with the intracellular domain of Notch-1 is an attractive model, which remains to be established.
Recently, domains of Notch-1 which are important for transformation of E1A-immortalized baby rat kidney cells in vitro have been characterized (6). These mapped sequences overlap with the EP domain. Neoplastic transformation by Notch is likely to be RBP-Jκ independent, although this process requires nuclear translocation of Notch (10, 19). One possible explanation for the role of the EP domain in this transformation process might be a transcriptional cross talk between Notch-1 and the tumor suppressor protein p53. Notch-1 and other p300-associated transcription factors compete for limiting quantities of complexes containing p300 and other coactivator proteins. Notch might block p300 function, and the effect on cell transformation might occur by inhibiting the actions of p53. In this model, Notch-1 could be viewed as having an effect of promoting resistance to apoptosis. A similar regulation between RelAp65 and p53 has been previously suggested (50).
On a functional level, a 3-aa substitution within the EP domain (2102 LDE/AAA 2104) leads to a clear reduction of Notch-1-mediated transactivation, which was comparable to that of the ΔEP mutant. In addition, the additive effect of p300 in Notch-mediated transactivation is completely lost by the LDE mutant protein (Fig. 12). On the other hand, the in vivo interaction of the Notch-1-ΔEP mutant with p300 is not completely abolished but appears to be sensitive to high salt concentrations. Therefore, we found some discrepancies between our functional and biochemical data. From our pull-down experiments, one could speculate that in addition to the EP domain, C-terminal sequences might be necessary for full p300 binding to Notch-1 in vivo. However, a deletion construct lacking these C-terminal sequences (mNotch-1-P/XB) is still transcriptionally active. Only a further deletion of an additional 15 aa (mNotch-1-E/XB) leads to a transcriptionally inactive Notch-1 protein, and this mutant fails to interact with p300 in our GST pull-down assays. Taken together, these results raise the question whether the function of p300 as a coactivator for Notch-1-mediated transcription depends mainly on the strength of interaction or whether the situation is more complex. In this context, it is important to note that very recently, a novel, more dynamic role for DNA binding proteins was considered when another transcription factor, HNF-1α, was studied. Two naturally existing mutant forms of this transcription factor are still able to recruit coactivators; however, the interaction is nonproductive (44). The authors support a model in which transcription factors not only recruit coactivators but also modulate their enzymatic activity. This idea might explain the fact that a transcriptionally inactive Notch-1 mutant protein still interacts with p300 to some extent.
In this study, we have identified and characterized a novel domain in Notch-1-IC, the EP domain, which is important for a Notch-1-IC/p300 interaction. The interaction of this common coactivator with Notch-1 adds another level of complexity to this signaling pathway. Other p300 binding proteins may directly influence Notch-mediated transactivation and thus explain at least in part the pleiotropic involvement of Notch in the complex biological processes that affect cell growth, transformation, and differentiation.
ACKNOWLEDGMENTS
We thank U. Wegenka, G. Schneider, U. Zechner, and H. Häcker for critically reading the manuscript. We also thank T. Honjo for providing the RBPJκ-specific antibody, K0043, and N. Perkins for providing the p53-expressing plasmids. For excellent technical assistance we thank J. Koehler, R. Rittelmann, and C. Heber.
This study was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 322,C4, to S.L. and R.M.S.
REFERENCES
- 1.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 4.Bailey A M, Posakony J W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 5.Beatus P, Lundkvist J, Oberg C, Lendahl U. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126:3925–3935. doi: 10.1242/dev.126.17.3925. [DOI] [PubMed] [Google Scholar]
- 6.Capobianco A J, Zagouras P, Blaumueller C M, Artavanis-Tsakonas S, Bishop J M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 8.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 9.Dou S, Zeng X, Cortes P, Erdjument-Bromage H, Tempst P, Honjo T, Vales L D. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol Cell Biol. 1994;14:3310–3319. doi: 10.1128/mcb.14.5.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumont E, Fuchs K P, Bommer G, Christoph B, Kremmer E, Kempkes B. Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J signalling. Oncogene. 2000;19:556–561. doi: 10.1038/sj.onc.1203352. [DOI] [PubMed] [Google Scholar]
- 11.Egan S E, St. Pierre B, Leow C C. Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr Top Microbiol Immunol. 1998;228:273–324. doi: 10.1007/978-3-642-80481-6_11. [DOI] [PubMed] [Google Scholar]
- 12.Feng X H, Zhang Y, Wu R Y, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortini M E, Artavanis-Tsakonas S. The suppressor of hairless protein participates in Notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 14.Hamaguchi Y, Yamamoto Y, Iwanari H, Maruyama S, Furukawa T, Matsunami N, Honjo T. Biochemical and immunological characterization of the DNA binding protein. (RBP-J κ) to mouse J κ recombination signal sequence. J Biochem (Tokyo) 1992;112:314–320. doi: 10.1093/oxfordjournals.jbchem.a123898. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh J J, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh J J, Zhou S, Chen L, Young D B, Hayward S D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 18.Jarriault S, Le B O, Hirsinger E, Pourquie O, Logeat F, Strong C F, Brou C, Seidah N G, Israel A. Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffries S, Capobianco A J. Neoplastic transformation by Notch requires nuclear localization. Mol Cell Biol. 2000;20:3928–3941. doi: 10.1128/mcb.20.11.3928-3941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao H Y, Ordentlich P, Koyano N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 22.Kidd S, Lieber T, Young M W. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogasterembryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 26.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 27.Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 29.Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl L J, ZimberStrobl U, Bornkamm G W, Honjo T. RBP-L, a transcription factor related to RBP-Jκ. Mol Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevels M, Täuber B, Kremmer E, Spruss T, Wolf H, Dobner T. Transforming potential of the adenovirus type 5 E4orf3 protein. J Virol. 1999;73:1591–1600. doi: 10.1128/jvi.73.2.1591-1600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 32.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olave I, Reinberg D, Vales L D. The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev. 1998;12:1621–1637. doi: 10.1101/gad.12.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ordentlich P, Lin A, Shen C P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oswald F, Dobner T, Lipp M. The E2F transcription factor activates a replication-dependent human H2A gene in early S-phase of the cell cycle. Mol Cell Biol. 1996;16:1889–1895. doi: 10.1128/mcb.16.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oswald F, Liptay S, Adler G, Schmid R M. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol Cell Biol. 1998;18:2077–2088. doi: 10.1128/mcb.18.4.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 38.Roehl H, Kimble J. Control of cell fate in C. elegansby a GLP-1 peptide consisting primarily of ankyrin repeats. Nature. 1993;364:632–635. doi: 10.1038/364632a0. [DOI] [PubMed] [Google Scholar]
- 39.Rubenwolf S, Schutt H, Nevels M, Wolf H, Dobner T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol. 1997;71:1115–1123. doi: 10.1128/jvi.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid R M, Liptay S, Betts J C, Nabel G J. Structural and functional analysis of NF-κ B. Determinants of DNA binding specificity and protein interaction. J Biol Chem. 1994;269:32162–32167. [PubMed] [Google Scholar]
- 41.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 42.Shawber C, Nofziger D, Hsieh J J, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 43.Soutoglou E, Papafotiou G, Katrakili N, Talianidis I. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J Biol Chem. 2000;275:12515–12520. doi: 10.1074/jbc.275.17.12515. [DOI] [PubMed] [Google Scholar]
- 44.Soutoglou E, Viollet B, Vaxillaire M, Yaniv M, Pontoglio M, Talianidis I. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 2001;20:1984–1992. doi: 10.1093/emboj/20.8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J κ/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 48.Waltzer L, Bourillot P Y, Sergeant A, Manet E. RBP-J κ repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H G, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webster G A, Perkins N D. Transcriptional cross talk between NF-κB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu L, Aster J C, Blacklow S C, Lake R, Artavanis-Tsakonas S, Griffin J D. MAML1, a human homologue of Drosophilamastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 52.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 53.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]