Regulation of the Forkhead Transcription Factor AFX by Ral-Dependent Phosphorylation of Threonines 447 and 451 (original) (raw)
Abstract
AFX is a Forkhead transcription factor that induces a G1 cell cycle arrest via upregulation of the cell cycle inhibitor p27Kip1. Previously we have shown that protein kinase B (PKB) phosphorylates AFX causing inhibition of AFX by nuclear exclusion. In addition, Ras, through the activation of the RalGEF-Ral pathway, induces phosphorylation of AFX. Here we show that the Ras-Ral pathway provokes phosphorylation of threonines 447 and 451 in the C terminus of AFX. A mutant protein in which both threonines are substituted for alanines (T447A/T451A) still responds to PKB-regulated nuclear-cytoplasmic shuttling, but transcriptional activity and consequent G1 cell cycle arrest are greatly impaired. Furthermore, inhibition of the Ral signaling pathway abolishes both AFX-mediated transcription and regulation of p27Kip1, while activation of Ral augments AFX activity. From these results we conclude that Ral-mediated phosphorylation of threonines 447 and 451 is required for proper activity of AFX-WT. Interestingly, the T447A/T451A mutation did not affect the induction of transcription and G1 cell cycle arrest by the PKB-insensitive AFX-A3 mutant, suggesting that Ral-mediated phosphorylation plays a role in the regulation of AFX by PKB.
AFX, together with FKHR and FKHRL1, belongs to a small subset of the Forkhead family of transcription factors (17). Chromosomal translocations in the regions of the AFX and FKHR genes are found in leukemias and rhabdomyosarcomas, respectively. For instance, translocation of the AFX gene to chromosome 11, t(X;11), results in the MLL-AFX fusion product, which may be involved in the development of certain leukemias (1). Interestingly, these transcription factors are critically involved in the regulation of cell proliferation. Overexpression of the Forkhead transcription factors causes a significant reduction of cell proliferation in a variety of cells, including a Ras-transformed cell line, by arresting the cells in the G1 phase of the cell cycle. This block in cell cycle progression is independent of functional retinoblastoma protein but dependent on the cell cycle inhibitor p27Kip1 (21). Deregulation of the Forkhead transcription factors may therefore be an important component in oncogenic transformation by promoting cell cycle progression of cells.
The Forkhead transcription factors are phosphorylated and regulated by the phosphatidylinositol 3-kinase [PI(3)K]–protein kinase B (PKB) pathway. AFX contains three putative PKB phosphorylation sites (threonine 28, serine 193, and serine 258), but AFX is phosphorylated by PKB in response to insulin only on serines 193 and 258 (18). Phosphorylation of these residues results in a transcriptional inactivation of AFX by means of nuclear exclusion, at least in part (4).
Interestingly, in addition to regulation of AFX by the PI(3)K-PKB pathway, we found that the Ras-Ral signaling route is involved in the regulation of this transcription factor (18). Ras is a small GTPase that couples growth factor signals to a variety of cellular processes, including transcription, DNA synthesis, and differentiation. RalA and RalB are very similar small GTPases that share 55% sequence identity with Ras (9). Like Ras, Ral proteins become biologically active upon exchange of bound GDP for GTP. This exchange is catalyzed in vivo by the Ral guanine nucleotide exchange factors (RalGEFs) (35). Three of the known RalGEFs—RalGDS, Rgl, and Rlf—interact with and can be activated by Ras (2). This and the observation that mitogen-dependent activation of Ral proteins requires Ras activation (37) have led to the idea that RalGEFs are Ras effector proteins. Consistent with this hypothesis is the fact that activation of Ral appears to be required for Ras-induced oncogenic growth, morphological transformation and induction of DNA synthesis (27, 31, 34, 36). In addition, overexpression of RalGEFs can cooperate with activation of other Ras effector cascades to transform cells (27, 31, 34). Together, these observations suggest that Ral may be an important mediator of Ras-induced proliferative signals.
A variety of Ral binding proteins have been identified over the past few years, yet the signal transduction pathways downstream of RalGTP and the role of Ral binding proteins in mediating Ras-induced signaling have not been resolved. The Ral binding protein 1 (RalBP1) associates with Ral in a GTP-dependent manner (6, 15). The functional importance of the interaction between Ral and RalBP1 still remains elusive. However, RalBP1 contains a GTPase activating protein (GAP) domain for Cdc42 and Rac GTPases (6, 15). This implies that Ral might negatively regulate signaling mediated by these GTPases. Also, phospholipase D (PLD) was found to interact with the N-terminal part of the Ral proteins (11, 14, 20). This interaction was, however, reported to be constitutive and independent of the nucleotide content of Ral. Although there is some evidence that PLD can contribute to oncogenic transformation (8, 19), the biological relevance of the interaction between Ral and PLD remains to be solved. Finally, filamin was reported to bind to Ral-GTP (23). The Ral-filamin interaction was described to be involved in Ral-induced filopodia formation. In contrast to a possible model of Ral being upstream of Cdc42 via RalBP1, the Ral-filamin-induced filopodia formation was reported to be downstream of Cdc42 activation.
The Ral pathway is also involved in the regulation of various transcription factors. A constitutively active mutant of the Ral exchange factor Rlf, Rlf-CAAX, stimulates transcriptional activation of the c-fos serum response element (SRE). The SRE can be activated through the regulation of the serum response factor, which forms an active complex with the ternary complex factors. Ral-mediated regulation of the SRE is most likely via activation of the ternary complex factor (36). Furthermore, Ral was found to induce phosphorylation of the c-Jun transcription factor (7). An active RalGEF induces c-Jun NH2-terminal phosphorylation similarly to the induction found after insulin treatment. Importantly, c-Jun phosphorylation in response to insulin was completely dependent on Ral activation. This pathway involves activation and phosphorylation of JNK-1 and the activation of the tyrosine kinase c-Src. Likewise Goi et al. found that c-Src, activated by EGF treatment or expression of constitutively activated Ral-GTPase, led to tyrosine phosphorylation of Stat3 and cortactin (12). Finally, expression of an activated form of Ral in quiescent rodent fibroblasts is sufficient to induce activation of NF-κB-dependent gene expression and cyclin D1 transcription (13). The regulation of cyclin D1 transcription by Ral is dependent on NF-κB activation and is mediated through an NF-κB binding site in the cyclin D1 promoter.
The observation that the Ras-Ral signaling route regulates a Forkhead transcription factor involved in regulation of cell proliferation may provide a mechanism for the effects of Ras-Ral signaling on oncogenic transformation. To further elucidate this, we searched for the sites within AFX that are phosphorylated by this pathway. We show that the Ras-Ral pathway phosphorylates AFX in its C terminus on threonines 447 and 451. A mutant protein in which both threonines are substituted for alanines (AFX-T447A/T451A) is still regulated by PKB-mediated signaling with respect to subcellular localization. However, the induction of transcription and growth suppression is greatly reduced. In agreement, inhibition of the Ral signaling pathway abolishes both AFX-mediated transcription and upregulation of the protein levels of the cell cycle inhibitor p27Kip1, while activation of Ral augments this activity. From these results we conclude that Ral-induced phosphorylation of threonines 447 and 451 of AFX is required for proper AFX activity. Interestingly, the T447A/T451A mutation had no effect on the induction of transcription and G1 cell cycle arrest induced by the PKB-insensitive, constitutively active, AFX-A3 mutant. Altogether, these results show that T447/T451 phosphorylation through the Ras-RalGEF-Ral pathway is involved in the regulation of the activity of AFX. Furthermore, our data show that T447/T451 phosphorylation is not required for AFX activity when AFX is mutated in its PKB phosphorylation sites.
MATERIALS AND METHODS
Cells and transfections.
Insulin receptor-overexpressing mouse NIH 3T3 cells (A14) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) (Gibco), penicillin (100 U/ml), streptomycin (100 μg/ml), and 0.05% l-glutamine (Imperial) as described previously (5). Jurkat-JHM1 cells, herein referred to as Jurkat cells, were maintained as described before (25) in RPMI 1640 medium supplemented with 10% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml), and 0.05% l-glutamine. Human colon carcinoma cells (DLD1) were grown in RPMI 1640 medium supplemented with 10% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml), and 0.05% l-glutamine. Insulin was added at 1 μg/ml, and LY294002 (10 μM; Sigma) was added 10 min before insulin stimulation. Transfections were carried out using the CaPO4 precipitation method for A14 cells, electroporation was used for the Jurkat cells, and Fugene 6 transfection reagent (Roche) was used to transfect the DLD1 cells.
Cloning and plasmids.
pMT2-HA-AFX-T447A, pMT2-HA-AFX-T451A, pMT2-HA-AFX-T454A, and pMT2-HA-AFX-T447A/T451 were generated by PCR-based site-directed mutagenesis of the pMT2-HA-AFX cDNA using the following forward primers and subsequent complementary reverse primers: T447A (5′-CCCAAGGCTCTGGGGGCTCCTGTGCTCACACC-3′), T451A (5′-GGGACTCCTGTGCTCGCACCCCCTACTGAAG-3′), T454A (5′-GCTCACACCCCCTGCTGAAGCTGCAAGC-3′), and T447A/T451A (5′-CAAGGCTCTGGGGGCTCCTGTGCTCGCACCCCCTACTG-3′). pMT2-HA-AFX1–416 was generated by creating an in-frame stop codon at position +416, using the following forward primer and subsequent complementary reverse primer: 5′-CAAGCCCCTATAGGCTCGAGGCC-3′ (the stop codon is underlined). pMT2-HA-AFXΔ419-443 was created by deleting bp +419 to +443 using PCR-based mutagenesis with primer 5′-AAGCCCCTAGAGGCTCGAGCTCTGGGGACTCCTGTG-3′. pMT2-HA-AFXΔ444–461 was created by deleting bp +444 to +461 using the following primer: 5′-AGTGCCCCCATCCCCAAGGCTATGCCTCAGGATCTAGAT-3′). pMT2-HA-AFXΔ462–501 was made by creating an in-frame stop codon at position +462 using the following forward primer and subsequent complementary reverse primer: 5′-CAAGCCAAGACAGATAGCCTCAGGATCTAG-3′ (the stop codon is underlined). The green fluorescent protein (GFP)-Pep17 constructs were made by ligating an oligo into _Sal_I/_Bam_HI cut pCMV-HA-AFX-GFP to create a hemagglutinin (HA)- and GFP-tagged construct.
Oligos used were as follows: Pep17-WT (5′-TCGACCATCCCCAAGGCTCTGGGGACTCCTGTGCTCACACCCCCTACTGAAGCTGCACCG-3′ and 3′-GGTAGGGGTTCCGAGACCCCT GAGGACACGAGTGTGGGGGATGACTTCGACGTGGCCTAG-5′), Pep17-T447S (5′-TCGACCATCCCCAAGGCTCTGGGGTCTCCTGTGCTCACACCCCCTACTGAAGCTGCACCG-3′ and 3′-GGTAGGGGTTCCGAGACCCCAGAGGACACGAGTGTGGGGGATGACTTCGACGTGGCCTAG-5′), Pep17-T451S (5′-TCGACCATCCCCAAGGC TC TGGGGAC TCC TG TGC TCTCACCCCC TAC TGAAGCTGCACCG-3′ and 3′-GGTAGGGGTTCCGAGACCCCTGAGGACACGAGAGTGGGGGATGACT TCGACGTGGCCTAG-5′), and Pep17-T447S/T451S (5′-TCGACCATCCCCAAGGCTCTGGGGTCTCCTGTGCTCTCACCCCCTACTGAAGCTGCACCG-3′ and 3′-GGTAGGGGTTCCGAGACCCCAGAGGACACGAGAGTGGGGGATGACTTCGACGTGGCCTAG-5′).
The following plasmids have been described before: pMT2-HA-AFXΔDB, pCMV-p27Kip1LUC, and pCD20 (21); pMT2-HA-AFX (18); pcDNA3-Myc-Rlf-CAAX, pMT2-HA-Rlf-CAAX, pMT2-HA-RalN28, and pSVE-RasV12 (36); pRK5-Myc-RalBP1ΔGAP (7); and pSG5-gagPKB (5); p1205LUC was a kind gift of D. Powell. The integrities of all cloning sites and point mutations were established by sequencing.
Antibodies.
The following antibodies were used: anti-p27Kip1 (Transduction Laboratories) and 12CA5 for HA-tagged proteins and 9E10 (Pharmingen).
Immunoprecipitation and Western blotting.
Cells were lysed in RIPA buffer (50 mM Tris-HCl [pH 7.5], 0.5% deoxycholate, 1% TritonX-100, 0.1% sodium dodecyl sulfate, 10 mM EDTA, 150 mM NaCl, 50 mM NaF, 1 μM leupeptin, 0.1 μM aprotinin, 0.5 mM benzamidine), and lysates were cleared for 10 min at 20,000 × g at 4°C. HA-AFX was immunoprecipitated by protein G-Sepharose beads coupled to the 12CA5 monoclonal antibody and rotation at 4°C for 2 h. Beads were washed three times in RIPA buffer and cleared of all liquid, and 20 μl of 1× Laemmli sample buffer was added. Samples were separated on a 10% acrylamide gel and transferred to a polyvinylidene difluoride membrane (Immobilon). Western blot analysis was performed under standard conditions.
[32P]orthophosphate labeling.
In vivo labeling of A14 cells transfected with HA-AFX was performed as described previously (5).
Phosphoamino acid analysis and tryptic peptide mapping.
[32P]orthophosphate-labeled HA-AFX was immunoprecipitated from A14 cells and electrophoresed and immobilized on polyvinylidene difluoride membrane. Protein was cut from the membrane and treated as described previously for phosphoamino acid analysis or peptide mapping (3). Sequence-grade trypsin was obtained from Boehringer-Mannheim.
Immunofluorescence.
Immunofluoresence studies, using the 12CA5 monoclonal antibody, were carried out as described before (32). In short, cells were cultured on coverslips, transfected with 0.2 μg of either pMT2-HA-AFX-WT or pMT2-HA-AFX-T447A/T451A either alone or in combination with 0.5 μg of pcDNA3-Myc-Rlf-CAAX or pSG5-gag-PKB, with the total amount of DNA of 2 μg equalized by empty vector, and fixed in 4% paraformaldehyde. Cells were permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS), and nonspecific binding was blocked with 0.5% bovine serum albumin (BSA) in PBS for 45 min. Incubation with the 12CA5 monoclonal antibody was for 1 h, followed by 1 h of incubation with anti-mouse-CY3 second antibody. Coverslips were washed and mounted on glass slides using Immuno-Mount (Shandon, Pittsburgh, Pa.). Subcellular localization was examined using a confocal laser scan microscope.
Isolation of transfected cells by MACS.
A14 cells were cotransfected with 2 μg of pCMV-CD20 and either 2 μg of empty vector, pMT2-HA-AFX-WT, pMT2-HA-AFX-A3 or pMT2-HA-AFX-T447A/T451A and isolated on magnetic cell sorting (MACS) separation columns type MS+ as specified by the manufacturer (Miltenyi Biotec, Bergisch Gladbach, Germany). In brief, the cells were washed with ice-cold 5 mM EDTA in PBS after stimulation and left on ice for 5 min. Then they were scraped in ice-cold 5 mM EDTA in PBS, isolated by centrifugation, washed with ice-cold buffer (1% FCS in PBS), and incubated with a monoclonal anti-CD20 antibody (DAKO). After being washed with 10 ml of wash buffer, the cells were incubated with the anti-mouse antibody coupled to iron beads. Finally, the cells were isolated by magnetic force on a separating column after being washed with wash buffer. The isolated transfected cells were lysed (in 0.5% Triton X-100, 50 mM HEPES, 100 mM NaCl, 2 mM sodium orthovanadate, 10 mM NaF), protein levels were equalized, and total cellular proteins were solubilized in Laemmli sample buffer. The samples were separated by sodium dodecyl sulfate, immunoblotted onto polyvinylidene difluoride and probed with the antibodies indicated in the figure legends.
Gene induction studies.
A14 cells were transfected with 0.1 μg of the p1205LUC reporter construct together with 2 μg of either pMT2-HA-AFX-WT, pMT2-HA-AFX-T447A/T451A, or pMT2-HA-AFX-ΔDB. Empty vector was added so that the total amount of DNA used per transfection was equal (6 μg per 5-cm-diameter dish). DLD1 cells were transfected, using Fugene 6 transfection reagent according to the manufacturers protocol, with 0.1 μg of p1205LUC reporter construct together with 0.5 μg of either pMT2-HA-AFX-WT, pMT2-HA-AFX-T447A/T451A, or pMT2-HA-AFX-ΔDB. Empty vector was added so that the total amount of DNA used per transfection was equal. The cells were washed the day after transfection, maintained in 10% FCS for 6 h, and left without serum overnight. Lysis and determination of luciferase activity were carried out 40 h after transfection and performed as described (22). The expression of cotransfected LacZ, measured by assaying β-galactosidase activity, was used as an internal control. Duplicate dishes were analyzed in all experiments, with duplicate experiments repeated at least four times. Standard deviations of fold inductions were then determined.
Jurkat T cells were transfected using electroporation. Cells (1.2 × 107) in 0.3 ml of RPMI 1640 medium were subjected to electroporation with 10 μg of either pMT2-HA-AFX-WT, pMT2-HA-AFX-T447A/T451A, or pMT2-HA-AFX-ΛDB; 4 μg of pCMV-p27Kip1-Luc reporter construct; and 4 μg of pCMV-LacZ in a total DNA amount of 50 μg equalized with empty vector plasmid, in 0.4-cm path length cuvettes using a Bio-Rad Gene Pulser set at 250 V and 960 μF. Cells were analyzed for luciferase activity 48 h after transfection. Duplicate transfections were analyzed in all experiments, with duplicate experiments repeated at least four times. Standard deviations of fold inductions were then determined.
Colony formation.
Cells were transfected with 1 μg of empty vector, pMT2-HA-AFX-WT, pMT2-HA-AFX-A3, or pMT2-HA-AFX-T447A/T451A in combination with 0.1 μg of pBabe-puro. Cells were cultured for 2 weeks in DMEM 10% FCS supplemented with 5 μg of Puromycin per ml. Puromycin-resistant colonies were scored after two weeks of selection by fixing the cells in 10% acetic acid for 10 min and subsequent staining of the cells with 0.4% crystal violet in 10% ethanol for 10 min.
Cell cycle analysis.
For DNA profiles, A14 cells were cotransfected with pEGFP (Clontech) in combination with 2 μg of either empty vector, pMT2-HA-AFX-WT, pMT2-HA-AFX-T447A/T451A or pMT2-HA-AFX-ΛDB. The amount of DNA transfected was equalized with empty vector (8 μg per 9-cm-diameter dish). Cells were grown overnight with nocodazole (250 ng/ml; Sigma). The next day, cells were harvested and fixed overnight in 70% ethanol at 4°C. After washing away the ethanol, the cells were stained with propidium iodide in a solution containing propidium iodide (10 μg/ml) and DNase-free RNase (10 μg/ml). DNA profiles of GFP-positive cells were analyzed on a fluorescence-activated cell sorter using Lysis II software flow cytometry analysis (Becton Dickinson).
RESULTS
The Ras-Ral pathway induces phosphorylation of AFX at a site different from and independent of PKB.
The Forkhead transcription factor AFX is phosphorylated upon insulin treatment of cells, and tryptic peptide mapping of in vivo-phosphorylated AFX revealed four radiolabeled peptides. Three peptides were found to be phosphorylated in a PKB-dependent manner, two of which contained PKB consensus sites. The third peptide was phosphorylated at low stoichiometry and frequently not detectable. The peptide designated peptide 4 was found to be a PKB-independent phosphorylated peptide of AFX, and instead was found to be dependent upon the Ras-Ral pathway (18).
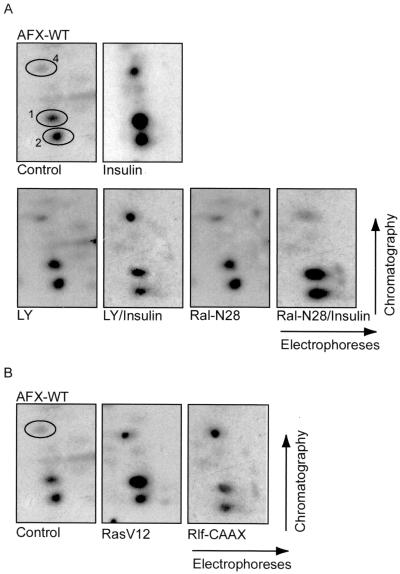
To further investigate the Ras-Ral-dependent phosphorylation of AFX, we labeled A14 cells transiently expressing hemagglutinin epitope-tagged AFX (HA-AFX) with [32P]orthophosphate. Immunoprecipitated HA-AFX was subsequently digested with trypsin and processed for peptide map analysis. Insulin stimulation of A14 cells resulted in a rapid and sustained phosphorylation of HA-AFX on the three reproducible peptides (Fig. 1A and reference 18). Pretreatment of the cells with LY294002 prior to insulin stimulation showed an inhibition of the insulin-induced phosphorylation of the PKB-dependent peptides 1 and 2. However, LY294002 did not have any effect on the insulin-induced phosphorylation of peptide 4 (Fig. 1A), confirming a PKB-independent phosphorylation of AFX. Importantly, cotransfection of the cells with the dominant negative version of Ral, Ral-N28, completely abolished the insulin-induced phosphorylation of peptide 4 (Fig. 1A). Furthermore, both RasV12 and an activated form of the Ral exchange factor Rlf, Rlf-CAAX (36), induced phosphorylation of AFX on peptide 4 (Fig. 1B). RasV12 also induced phosphorylation of the PKB peptides, in agreement with the ability of oncogenic Ras to induce PI(3)K-dependent signaling (16). Taken together, these results show that peptide 4 is phosphorylated after insulin stimulation by a pathway sensitive to Ral-N28 and independent of the PI(3)K-PKB pathway.
FIG. 1.
PKB-independent phosphorylation of AFX by the Ras-Ral pathway. (A) In vivo-phosphorylated HA-AFX was immunoprecipitated from 32P-labeled A14 cells which were left untreated, stimulated with insulin for 30 min, pretreated with LY294002 for 10 min prior to insulin stimulation, or cotransfected with 2 μg of pMT2-HA-Ral-N28. Following exposure to film, bands were cut out of the blot and processed for two-dimensional phosphopeptide mapping using trypsin as the digestive enzyme. Positions of peptides 1, 2, and 4 are indicated by number. (B) 32P-labeled HA-AFX was isolated from transfected A14 cells coexpressing either pSVE-Ras-V12 or pcDNA3-Myc-Rlf-CAAX and analyzed as described for panel A.
The Ras-Ral phosphorylation sites are threonines 447 and 451 in the C-terminal part of AFX.
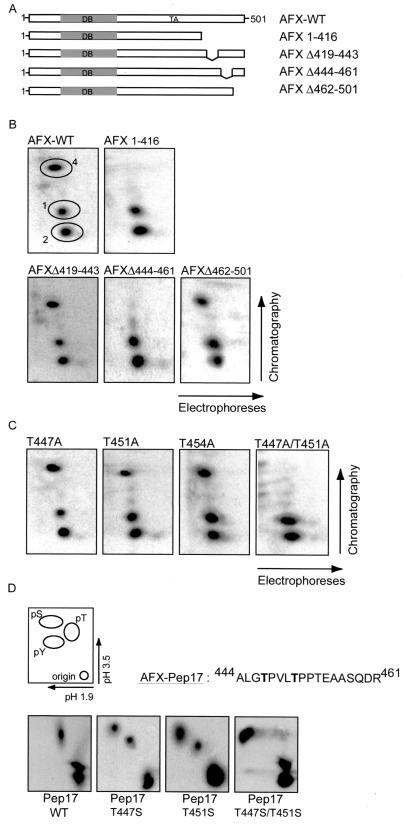
To investigate where the Ras-Ral-induced phosphorylation site was located, we made several deletion mutants of AFX. A14 cells, transiently transfected with the various deletion mutants, were labeled with [32P]orthophosphate, and AFX was immunoprecipitated and subjected to tryptic digestion. It appeared that all AFX deletion mutants missing the C terminus lost the Ras-Ral-induced phosphorylated peptide 4 (Fig. 2B, data not shown). Further deletion mapping of the C terminus revealed that the Ras-Ral-induced phosphorylation site in AFX was located within the region containing amino acids 416 to 501 (Fig. 2B). Within this region, tryptic digestion of AFX gives rise to three different peptides. Deletion mutants of AFX of each of these peptides were made (Fig. 2A). As shown in Fig. 2B, deletion of amino acids 419 to 443 (AFX-Δ419–443) still showed [32P]incorporation into peptide 4. In contrast, AFX-Δ444–461 no longer displayed a radiolabeled peptide 4, while the third deletion mutant AFX-Δ462–501 did (Fig. 2B). Within amino acid region 444 to 461 of AFX three threonines are located. Mutation of every threonine singly to an alanine resulted in an in vivo peptide map that was similar to that of AFX-WT. In all three cases, peptide 4 was still phosphorylated (Fig. 2C). However, when both threonines 447 and 451 were mutated to alanines (AFX-T447A/T451A), phosphorylation of peptide 4 was completely abolished (Fig. 2C), while combined mutation of either threonines 451 and 454 or 447 and 454 still resulted in phosphorylation of peptide 4 (data not shown). Thus, when threonine 447 is mutated to an alanine, threonine 451 will be phosphorylated while mutation of threonine 451 to an alanine will result in phosphorylation of threonine 447. These results show that AFX can be phosphorylated on both threonine 447 and threonine 451 by the Ras-Ral pathway. The preference of phosphorylation of either of these sites is currently still unclear. Importantly, simultaneous phosphorylation of both threonines 447 and 451 in AFX is most likely not the case since overlay of peptide maps of AFX-WT and AFX-T447A shows that the mobility of peptide 4 in AFX-WT is exactly the same as peptide 4 of AFX-T447A (data not shown). If indeed peptide 4 would be phosphorylated on the two threonines simultaneously, its mobility should differ from the single threonine mutant peptides in which only one phosphorylation occurs. These results are most easily explained by assuming that phosphorylation of threonines 447 and 451 is mutually exclusive.
FIG. 2.
The Ras-Ral-induced phosphorylation sites in AFX. (A) Deletion mutants of AFX. Abbreviations: DB, DNA binding domain; TA, transactivating domain. (B) 32P-labeled pMT2-HA-AFX-WT, pMT2-HA-AFX-1–416, pMT2-HA-AFX-Δ419–443, pMT2-HA-AFX-Δ444–461, or pMT2-HA-AFX-Δ462–501 AFX was isolated from A14 cells which were cotransfected with p-SVE-RasV12 and analyzed as described in Fig. 1A. The same results were obtained when cells were cotransfected with Rlf-CAAX (data not shown). (C) 32P-labeled pMT2-HA-AFX-T447A, pMT2-HA-AFX-T451A, pMT2-HA-AFX-T454A, or pMT2-HA-AFX-T447A/T451A was isolated from A14 cells which were cotransfected with p-SVE-RasV12 and analyzed as described in Fig. 1A. The same results were obtained when cells were cotransfected with Rlf-CAAX (data not shown). (D) A14 cells were cotransfected with either 2 μg of pCMV-GFP-Pep17-WT, pCMV-GFP-Pep17-T447S, pCMV-GFP-Pep17-T451S, or pCMV-GFP-Pep17-T447S/T451S in combination with 2 μg of p-SVE-RasV12 (or pcDNA3-Myc-Rlf-CAAX [data not shown]). Following exposure to film, bands were cut out of the blot and processed for phosphoamino acid analysis. Positions of phosphoamino acids are as indicated.
To support the idea that the phosphorylation of AFX occurs on threonines 447 and 451, we performed phosphoamino acid analysis. In order to be able to specifically look at phosphorylation of these two threonines in AFX and not any other phosphorylated residues, we used only the part of AFX that contains the sequence of peptide 4 and fused it to GFP, to make it large enough to isolate. Phosphoamino acid analysis of the wild type peptide (Pep17-WT) showed phosphorylation of threonines only, confirming the data that the Ral-dependent phosphorylation of peptide 4 is on threonines and not on serine (Fig. 2D). Importantly, when we mutated threonine 447 to a serine (Pep17-T447S), both phosphothreonine and phosphoserine were detected, showing that indeed threonine 447 is phosphorylated. Mutation of threonine 451 to serine (Pep17-T451S) gave the same result as for the threonine 447 to serine mutation, while mutating both threonines 447 and 451 to serines (Pep17-T447S/T451S) resulted in a total shift in phosphorylation from threonine to serine (Fig. 2D). Together with the data on the peptide map analysis, these results clearly show that AFX can be phosphorylated on either threonine 447 or 451 by Ras and Ral.
AFX-T447A/T451A has strongly reduced biological effects.
We next examined the effect of mutation of the Ras-Ral-induced phosphorylation sites in AFX on the biological function of this transcription factor. We mutated both threonines 447 and 451 to alanines (AFX-T447A/T451A), to abolish phosphorylation on any of these sites.
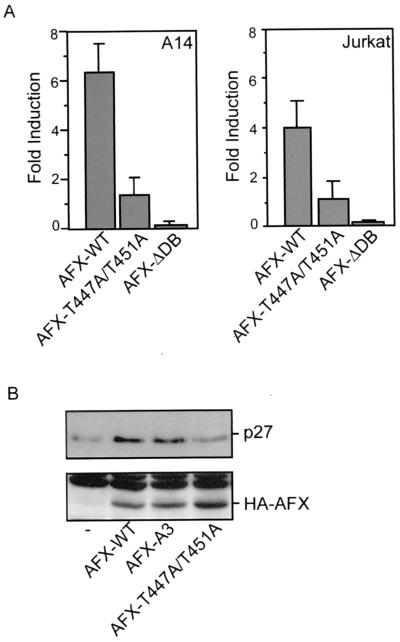
First, we investigated the transcriptional capacity of AFX-T447A/T451A compared to AFX-WT. Members of the Forkhead family regulate transcription of the insulin-like growth factor binding protein-1 (IGFBP-1) gene (18, 30). Cotransfection of AFX-WT together with a luciferase reporter construct under the control of the IGFBP-1 promoter in A14 cells resulted in a more than sixfold induction of activity of this promoter (Fig. 3A). This induction of transcriptional activity was dependent on the DNA binding capacity of AFX since AFX lacking its DNA binding domain (AFX-ΔDB) did not induce luciferase expression (Fig. 3A). Surprisingly, the AFX-T447A/T451A mutant hardly had any effect on the transcription of the IGFBP-1 promoter (Fig. 3A).
FIG. 3.
AFX-T447A/T451A has strongly reduced activity regarding both transcription and upregulation of p27Kip1 protein levels. (A) A14 cells were transfected with 2 μg of either pMT2-HA-AFX, pMT2-HA-AFX-T447A/T451A, or pMT2-HA-AFX-ΔDB in combination with 0.1 μg of p1205LUC reporter construct. The next day, the cells were allowed to recover in the presence of 10% FCS for 6 h and put on serum-free medium for 16 h. Lysates were made 40 h after transfection, and luciferase activity was determined. In each experiment, duplicate dishes were analyzed. Transfection efficiency was monitored by cotransfection of a cytomegalovirus (CMV)-LacZ construct and measuring β-galactosidase activity. Jurkat T cells were transfected by electroporation. Cells were subjected to electroporation with 10 μg of either pMT2-HA-AFX-WT, pMT2-HA-AFX-T447A/T451A, or pMT2-HA-AFX-ΛDB, with 4 μg of pCMV-p27Kip1-Luc reporter construct and 4 μg of pCMV-LacZ in a total amount of DNA of 50 μg equalized with empty vector plasmid. Cells were analyzed for luciferase activity 48 h after transfection. The increases in luciferase activity are shown as fold induction over the activity in cells transfected with control plasmid alone. The results represent the averages of at least four independent experiments. The error bars represent the standard deviations of the values. Protein expression levels were regularly controlled by immunoblotting. (B) A14 cells were transfected with 2 μg of empty vector—pMT2-HA-AFX-WT, pMT2-HA-AFX-A3, or pMT2-HA-AFX-T447A/T451A—in combination with 2 μg of pCMV-CD20. Transfected CD20-positive cells were isolated by MACS. Samples were analyzed for p27Kip1 protein levels (top panel) and expression of the different AFX constructs (bottom panel).
In addition, the cell cycle inhibitor p27Kip1 was reported to be transcriptionally activated by the different Forkhead family members (21). Transfection of Jurkat T cells with a p27Kip1-Luc reporter construct resulted in an approximately fourfold induction of activity of this promoter by AFX-WT (Fig. 3A). However, again no induction of transcriptional activity was found with AFX-T447A/T451A (Fig. 3A). Supporting these data, we found no induction of p27Kip1 protein levels by AFX-T447A/T451A compared to a significant induction by AFX-WT and AFX-A3 (Fig. 3B).
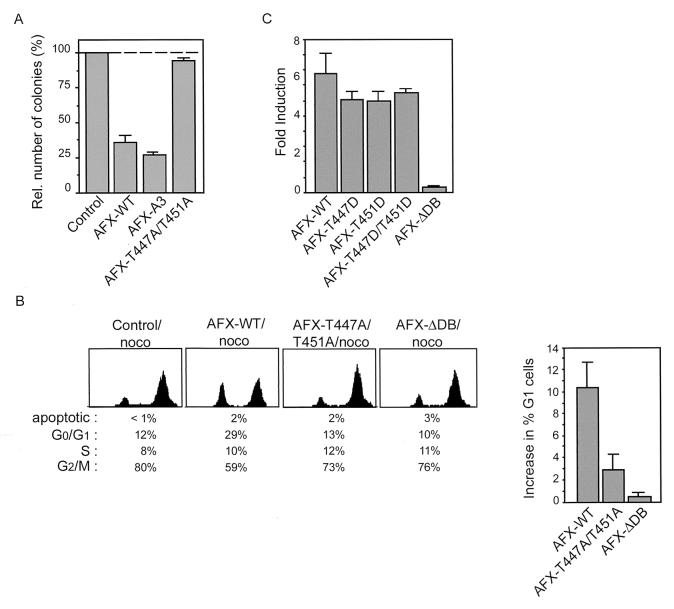
Since AFX was found to have a growth suppressive effect on cells (21), we next examined the effect of ectopic expression of AFX-WT and AFX-T447A/T451A on cell proliferation. When AFX-WT was introduced into A14 cells, it inhibited colony formation very efficiently (approximately 70%) (Fig. 4A and reference 21). When an AFX mutant lacking all three putative PKB phosphorylation sites (AFX-A3) was introduced, colony formation was inhibited even more (Fig. 4A). However, introduction of AFX-T447A/T451A hardly had any growth-suppressive effects (Fig. 4A). In addition, while introduction of AFX-WT into A14 cells resulted in accumulation of cells in G0/G1 phase of the cell cycle, AFX-T447A/T451A did not have any effect on the cell cycle progression of the cells, similar to the transcriptionally inactive AFX-ΔDB (Fig. 4B).
FIG. 4.
AFX-T447A/T451A has greatly impaired growth suppressive effects. (A) A14 cells were transfected with 1 μg of empty vector, pMT2-HA-AFX-WT, pMT2-HA-AFX-A3 or pMT2-HA-AFX-T447A/T451A in combination with 0.1 μg of pBabe-puro. Cells were cultured for two weeks in DMEM 10% FCS supplemented with 5 μg of Puromycin per ml. Puromycin-resistant colonies were scored after 2 weeks of selection. (B) A14 cells were transfected with pEGFP in combination with 2 μg of empty vector, pMT2-HA-AFX-WT, pMT2-HA-AFX-T447A/T451A, or pMT2-HA-AFX-ΛDB. Cells were grown overnight with nocodazole (250 ng/ml). DNA profiles of GFP-positive cells were analyzed on a fluorescence-activated cell sorter. The absolute increase in percentage of A14 cells in the G0/G1 phase upon expression of the indicated proteins is presented in a graph. The error bars represent the standard deviations of the values. Protein expression levels were controlled by immunoblotting. (C) A14 cells were transfected with 2 μg of either pMT2-HA-AFX, pMT2-HA-AFX-T447D, pMT2-HA-AFX-T451D, pMT2-HA-AFX-T447D/T451D, or pMT2-HA-AFX-ΔDB in combination with 0.1 μg of p1205LUC reporter construct and analyzed as described in the legend of Fig. 3A. The increases in luciferase activity are shown as fold inductions over the activity in cells transfected with control plasmid alone. The results represent the averages of at least four independent experiments. The error bars represent the standard deviations of the values. Protein expression levels were regularly controlled by immunoblotting.
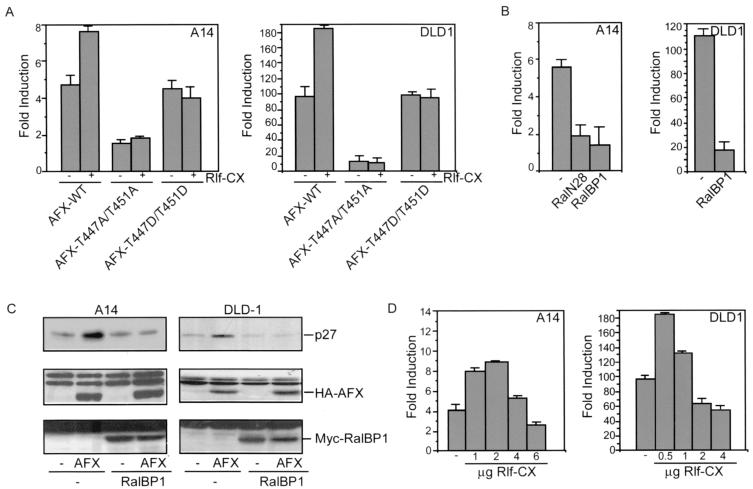
To further support that phosphorylation on threonines 447 or 451 is required for proper functioning of AFX, we made a threonine-to-aspartate mutation in these sites, which is reported to mimic phosphorylation. Expression of AFX-T447D, AFX-T451D, or AFX-T447D/T451D all resulted in a reversion of the inactivity of the AFX-T447A/T451A mutant. They all had transcriptional activity similar to that of AFX-WT (Fig. 4C), indicating that indeed phosphorylation of these sites is required for the activity of AFX. Taken together, these results show that mutation of both threonine 447 and threonine 451 to alanines in AFX abolishes AFX activity, indicating the importance of these sites for proper functioning of AFX with regard to important biological effects of this transcription factor on transcriptional activation and growth suppression.
Inhibition of the Ral pathway inhibits AFX activity.
To further investigate the role of Ral in the regulation of AFX, we examined the effect of activation or inactivation of the Ral pathway on the transcriptional activity of this transcription factor. Therefore, we cotransfected either A14 cells or DLD1 cells, which are human colon carcinoma cells, with AFX and the luciferase reporter construct under the control of the IGFBP-1 promoter in combination with Rlf-CAAX. As shown in Fig. 5A, cotransfection of Rlf-CAAX in both A14 and DLD1 cells resulted in an induction of AFX transcriptional activity. No effect of Rlf-CAAX on either the inactive AFX-T447A/T451A mutant or the AFX-T447D/T451D mutant was found (Fig. 5A). Moreover, expression of either RalN28 or the minimal Ral-binding domain of RalBP1 resulted in a significant decrease of both AFX transcriptional activity (Fig. 5B) and AFX-induced upregulation of p27Kip1 protein levels (Fig. 5C). These results show that Ral-induced phosphorylation is involved in the activation of AFX, in agreement with the mutational analysis.
FIG. 5.
Inhibition of the Ral pathway inhibits AFX activity. (A) A14 cells were transfected with 4 μg of pMT2-HA-AFX, pMT2-HA-AFX-T447A/T451A, or pMT2-HA-AFX-T447D/T451D in combination with 1 μg of pMT2-HA-Rlf-CAAX and 0.1 μg of p1205LUC reporter construct. DLD1 cells were transfected with 0.5 μg of either pMT2-HA-AFX, pMT2-HA-AFX-T447A/T451A, or pMT2-HA-AFX-T447D/T451D in combination with 0.5 μg of pMT2-HA-Rlf-CAAX and 0.1 μg of p1205LUC reporter construct. Samples were analyzed as described in the legend to Fig. 3A. The increases in luciferase activity are shown as fold induction over the activity in cells transfected with control plasmid alone. In each experiment, duplicate dishes were analyzed. The results represent the averages of at least three independent experiments. The error bars represent the standard deviations of the values. Protein expression levels were regularly controlled by immunoblotting. (B) A14 cells were transfected with 4 μg of pMT2-HA-AFX in combination with 2 μg of either pMT2-HA-RalN28 or pRK5-Myc-RalBP1ΔGAP and 0.1 μg of p1205LUC reporter construct. DLD1 cells were transfected with 0.5 μg of pMT2-HA-AFX in combination with 1 μg of pRK5-Myc-RalBP1ΔGAP and 0.1 μg of p1205LUC reporter construct. Samples were analyzed as described in the legend to Fig. 3A. The increases in luciferase activity are shown as fold induction over the activity in cells transfected with control plasmid alone. In each experiment, duplicate dishes were analyzed. The results represent the averages of at least three independent experiments. The error bars represent the standard deviations of the values. Protein expression levels were regularly controlled by immunoblotting. (C) A14 cells were transfected with 2 μg of empty vector or pMT2-HA-AFX in combination with 2 μg of pRK5-Myc-RalBP1ΔGAP and 2 μg of pCMV-CD20. DLD1 cells were transfected with 0.5 μg of empty vector (−) or pMT2-HA-AFX in combination with 1 μg of pRK5-Myc-RalBP1ΔGAP and 1 μg of pCMV-CD20. Transfected CD20-positive cells were isolated by MACS. Samples were analyzed for p27Kip1 protein levels (top panel), expression of AFX (middle panel), and expression of Myc-RalBP1ΔGAP (bottom panel). (D) A14 cells were transfected with 4 μg of pMT2-HA-AFX in combination with 1,2, 4, or 6 μg of pMT2-HA-Rlf-CAAX and 0.1 μg of p1205LUC reporter construct. DLD1 cells were transfected with 0.5 μg of pMT2-HA-AFX in combination with 0.5, 1, 2, or 4 μg of pMT2-HA-Rlf-CAAX and 0.1 μg of p1205LUC reporter construct. Samples were analyzed as described in the legend to Fig. 3A. The increases in luciferase activity are shown as fold induction over the activity in cells transfected with control plasmid alone. In each experiment, duplicate dishes were analyzed. The result depicted in this figure is a representative example of at least three independent experiments. The error bars represent the standard deviations of the duplicate values in this particular experiment. Protein expression levels were regularly controlled by immunoblotting.
Previously we reported that Rlf-CAAX inhibited AFX-mediated transcription (18), a result at variance with the results in the present work. However, we noticed that the effect of Rlf-CAAX on AFX-mediated transcription was sensitive to the amount of Rlf-CAAX used (Fig. 5D). Consistently, at relatively low concentrations of Rlf-CAAX transcriptional activity of AFX was induced, whereas at higher levels of Rlf-CAAX the activity was inhibited. No effect of Rlf-CAAX at either concentration was found on the inactive AFX-T447A/T451A mutant or the active AFX-T447D/T451D mutant (data not shown). Apparently, at higher concentrations of Rlf-CAAX additional signaling occurs that results in the inactivation of AFX. The mechanism of this inactivation is currently unknown.
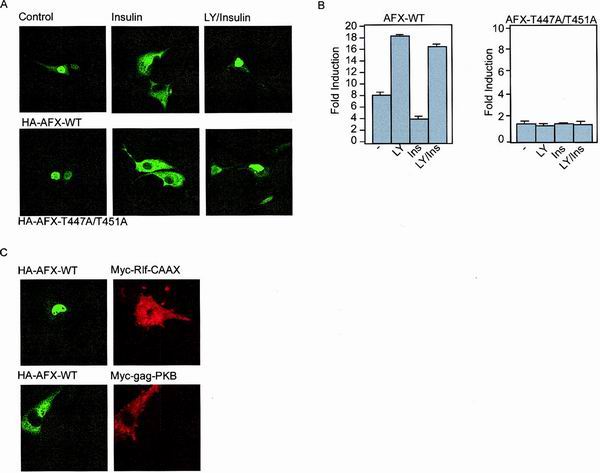
AFX-T447A/T451A is normally regulated by PKB with respect to subcellular relocalization.
In normal serum-starved cells, AFX, as well as other members of the Forkhead transcription family, is mainly localized to the nucleus. Upon stimulation with insulin or specific activation of the PI(3)K-PKB pathway, AFX is retained in the cytoplasm (4) and thereby inactivated. We examined whether the Ras-Ral-mediated phosphorylation of AFX affects its subcellular localization. Therefore, we examined the localization of AFX-T447A/T451A in cells either unstimulated or stimulated with insulin. As shown in Fig. 6A, both AFX-WT and AFX-T447A/T451A are located in the nucleus in serum-starved cells. Stimulation of the cells with insulin led to cytoplasmic retention of both AFX-WT and AFX-T447A/T451A within 30 min. Pretreatment of the cells with LY294002 prior to insulin stimulation blocked this relocalization of AFX, showing the PI(3)K-dependency (Fig. 6A).
FIG. 6.
The Ras-Ral signaling pathway does not induce a change in the steady-state distribution of AFX from the nucleus to the cytoplasm. (A) A14 cells were transfected with 0.2 μg of either pMT2-HA-AFX-WT or pMT2-HA-AFX-T447A/T451A. The next day, the cells were allowed to recover in the presence of 10% FCS for 6 h and put on serum-free medium for 16 h. Cells were stimulated with insulin (30 min) and LY294002 (10 min prior to insulin treatment) as indicated. Cells were fixed and stained for AFX using the 12CA5 monoclonal antibody. (B) A14 cells were transfected with 2 μg of either pMT2-HA-AFX or pMT2-HA-AFX-T447A/T451A in combination with 0.1 μg of p1205LUC reporter construct. The next day, the cells were allowed to recover in the presence of 10% FCS for 6 h and put on serum-free medium for 16 h. Cells were stimulated with insulin (30 min) and LY294002 (10 min prior to insulin treatment) as indicated. Lysates were made 40 h after transfection, and luciferase activity was determined. In each experiment, duplicate dishes were analyzed. Transfection efficiency was monitored by cotransfection of a CMV-LacZ construct and measuring β-galactosidase activity. (C) A14 cells were transfected with 0.2 μg of pMT2-HA-AFX-WT in combination with 0.5 μg of pcDNA3-Myc-Rlf-CAAX or pSG5-Myc-gag-PKB. The next day, the cells were allowed to recover in the presence of 10% FCS for 6 h and put on serum-free medium for 16 h. Cells were fixed and stained for AFX using the 12CA5 monoclonal antibody and 9E10 antibody to stain Rlf-CAAX- and gag-PKB-expressing cells.
As shown previously by Kops et al. for AFX-WT, this nuclear-cytoplasmic location nicely correlated with activation of the IGFBP-1 promoter luciferase construct (18). Addition of LY294002 resulted in a more active AFX, whereas insulin inhibited AFX. This inhibition could be abolished by inhibition of PI3K (Fig. 6B and reference 18). In contrast, AFX-T447A/T451A was completely inactive even though it was located predominantly in the nucleus after LY294002 treatment (Fig. 6B). All together, these results show that the subcellular localization of AFX-T447A/T451A is not altered compared to AFX-WT and that the PI(3)K-PKB-mediated induction of relocalization of AFX from the nucleus to the cytoplasm is not influenced by the T447A/T451 mutation. Importantly, these results also show that AFX-T447A/T451A is a normally processed protein considering that the nuclear import and export are not altered, indicating that the inactivity of this mutant is not the result of incorrect processing or folding.
Ras-Ral signaling does not influence the nuclear localization of AFX.
We next investigated the effect of activation of the Ral pathway on the localization of AFX. Expression of Rlf-CAAX did not induce a shift in the steady-state localization of AFX from the nucleus to the cytoplasm (Fig. 6C). Also, introduction of RalN28 did not affect nuclear localization of AFX (data not shown), whereas, as expected, transfection of active gag-PKB inhibits nuclear import of AFX resulting in cytoplasmic retention (Fig. 6C and reference 4). Together, these results show that the Ras-Ral pathway does not seem to impinge on nuclear localization of AFX.
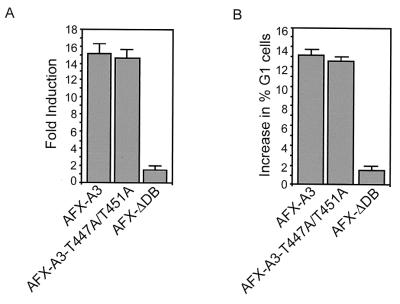
The T447A/T451A mutation does not affect the activity of an AFX mutant that lacks intact PKB sites.
AFX-WT can be inhibited by phosphorylation of two PKB sites resulting in, among others, the inhibition of nuclear import. Since nuclear translocation is not affected by T447A/T451A mutation of AFX, we investigated the effect of the T447A/T451A mutation on the background of an AFX mutated in its PKB phosphorylation sites (AFX-A3). Therefore, we made an AFX mutant that has mutations of both threonines 447 and 451 to alanines and mutations of the three putative PKB sites (threonine 28, serine 193 and 258) to alanines (AFX-A3-T447A/T451A). First, we examined the transcriptional capacity of this particular mutant using the p27Kip1-Luc reporter construct. We cotransfected this reporter construct together with AFX-A3 in Jurkat T cells, which resulted in a 15-fold induction of promoter activity (Fig. 7A). Surprisingly, expression of AFX-A3-T447A/T451A resulted in a very similar induction of transcriptional activity of the p27Kip-1 promoter (Fig. 7A). We also investigated the cell cycle progression of cells expressing either mutant. In support of the results obtained in the transcription assay, no difference in increase of cells in the G1 phase of the cell cycle induced by either AFX-A3 or AFX-A3-T447A/T451A could be observed (Fig. 7B). Together, these results clearly show that mutation of threonines 447 and 451 to alanines does not affect the activity of AFX-A3. Similar results were obtained with AFX-SASA (serine 193 and 258 to alanine mutation only [data not shown and reference 18]). From these results we conclude that mutation of the PKB sites in AFX is sufficient to make AFX insensitive to mutation of threonines 447 and 451.
FIG. 7.
Mutation of threonines 447 and 451 does not affect the activity of AFX-A3. (A) Jurkat T cells were transfected by electroporation. Cells were subjected to electroporation with 10 μg of either pMT2-HA-AFX-A3, pMT2-HA-AFX-A3-T447A/T451A, or pMT2-HA-AFX-ΛDB; 4 μg of pCMV-p27Kip1-Luc reporter construct; and 4 μg of pCMV-LacZ in a total of 50 μg of DNA equalized with empty vector plasmid. Cells were analyzed for luciferase activity 48 h after transfection. The increases in luciferase activity are shown as fold induction over the activity in cells transfected with control plasmid alone. The results represent the averages of at least four independent experiments. The error bars represent the standard deviations of the values. Protein expression levels were regularly controlled by immunoblotting. (B) A14 cells were transfected with pEGFP in combination with 2 μg of either empty vector, pMT2-HA-AFX-A3, pMT2-HA-AFX-A3-T447A/T451A, or pMT2-HA-AFX-ΛDB. Cells were grown overnight with nocodazole (250 ng/ml). DNA profiles of GFP-positive cells were analyzed on a fluorescence-activated cell sorter. The absolute increase in percentage of A14 cells in the G0/G1 phase upon expression of the indicated proteins is presented. The error bars represent the standard deviations of the values. Protein expression levels were controlled by immunoblotting.
DISCUSSION
The Ras-Ral pathway phosphorylates AFX on threonines 447 and 451.
We have previously shown that, in addition to the PI(3)K-PKB pathway, the Ras-Ral signaling pathway regulates the Forkhead transcription factor AFX. Both the PI(3)K-PKB pathway and the Ras-Ral pathway induce phosphorylation of AFX. To investigate the link between Ras-Ral signaling and AFX in further detail, we have determined the site of phosphorylation induced by the Ras-Ral pathway. This phosphorylation site was clearly located in peptide 4, which resides in the C-terminal transcriptional activation domain of AFX. Further analysis showed that peptide 4 contains three threonines at positions 447, 451 and 454. Single mutation of threonine 447, threonine 451 or threonine 454 to alanine did not abolish phosphorylation, but replacing both threonines 447 and 451 with alanines resulted in a loss of phosphorylation of peptide 4. Phosphorylation of threonine 447 and threonine 451 appears to be mutually exclusive, since mutation of one of the two sites to alanine did not result in the predicted mobility shift of the peptide. Apparently, the kinase responsible for this phosphorylation can switch between these two identical threonine-proline sites. This kinase does not have a clear preference for one of the sites since changing one of the threonines to serine, results in an apparently equal usage of the sites (Fig. 2D).
Ral-mediated phosphorylation results in the activation of AFX.
Using an AFX mutant with both threonines 447 and 451 mutated to alanines (AFX-T447A/T451A), we observed that the activity of this mutant is greatly reduced with respect to both activation of transcription and proliferation. However, this mutant is still regulated by PKB-mediated signaling with respect to subcellular localization, indicating that the mutant protein is still properly responsive to other signals. In addition, AFX-T447D/T451D, in which both threonines 447 and 451 are mutated to phosphorylation-mimicking aspartate residues, is as active as AFX-WT and regulatable by PKB. From these results we conclude that phosphorylation of threonines 447 and 451 is required for proper AFX activity. The role of Ral-mediated phosphorylation in the regulation of the activity of AFX was further supported by the observation that inhibition of Ral, using either the dominant negative mutant RalN28, or the Ral-binding domain of the effector RalBP1, also inhibits AFX-mediated transcription. Furthermore, activation of Ral signaling at low concentrations of Rlf-CAAX resulted in the activation of AFX-mediated transcription. Previously, we reported that introduction of Rlf-CAAX resulted in the inhibition of AFX-mediated transcription. For this effect higher concentrations of Rlf-CAAX are required. Apparently at these concentrations additional effects of Rlf-CAAX on the cell occur, independently of the phosphorylation on threonines 447 and 451, that result in the inactivation of transcription.
Since activation of AFX results in a cell cycle arrest by, among others, the activation of the transcription of the cell cycle inhibitor p27Kip1, the results predict that inhibition of Ral would also inhibit transcription of the p27Kip1 gene. Indeed, introduction of a minimal Ral-binding domain of RalBP1 inhibits p27Kip1 transcription. However, previously it was shown that Rlf-CAAX induces cell proliferation and RalGDS, another RalGEF, complements other Ras effectors, like Raf1, in oncogenic transformation. It may well be that in these cases selection for high expression levels of Rlf-CAAX and RalGDS caused the growth supporting effects (35). Concentration-dependent effects on cell proliferation of Ras effectors is not without precedent. Previously, it was found that low levels of Raf1 result in the induction of cell proliferation, whereas high levels result in cessation of cell proliferation (29, 38). It should be noted that the Ras-Ral signaling route is linked not only to cell proliferation but also to the induction of cell differentiation and G1 arrest (24, 26, 28, 33). The consequence of our finding may be that we have to revise our view on the Ral pathway being a mediator of Ras-induced cell proliferation. Perhaps this pathway operates as a negative control in preventing Ras-induced cell cycle progression through activation of AFX. With respect to tumorigenesis, these results mean that the expression of a constitutively active Ras affects signaling in a different way from activation of Ras by growth factors. Thus, in tumors containing mutant Ras, AFX may be inactivated, leading to cell cycle progression, while in normal cells endogenous Ras activation through growth factors may result in activation of AFX, possibly acting as a negative feedback mechanism for the activation of growth stimulatory pathways.
Phosphorylation of threonines 447 and 451 is required for PKB-mediated regulation of AFX.
Previously we have shown that AFX-A3, a mutant in which all three putative PKB phosphorylated sites are mutated to alanines, is an active mutant with respect to the induction of G1 cell cycle arrest and the induction of transcription (18). Interestingly, when the T447A/T451A mutation was made in the A3 background, the activity of AFX was not affected. This contrasted with the inhibition of AFX by the T447A/T451A mutation in the WT background. Since the only difference between AFX-T447A/T451A and AFX-A3-T447A/T451A is the ability to become phosphorylated and inhibited by phosphorylation on the PKB sites, our results indicate that Ras-Ral-induced phosphorylation of threonines 447 and 451 is only functional in the presence of AFX phosphorylated on its PKB sites. This implies that T447/451 phosphorylation is required to relieve the inhibition imposed by phosphorylation of the PKB sites. These results predict that inhibition of AFX phosphorylation by PKB, for instance by using an inhibitor of the upstream PI(3)-kinase, would result in a constitutively active T447A/T451A mutant like the AFX-A3-T447A/T451A. However, after LY294002 treatment the T447A/T451A mutant is still inactive (Fig. 6B). This apparent discrepancy can be explained by the observation that at the time point of starting LY294002 treatment, basal phosphorylation of AFX on the PKB sites is high (Fig. 1A); therefore, AFX is retained in the cytoplasm and thus inactive. Another possible explanation is that T447/451 phosphorylation is required to dephosphorylate AFX at the PKB sites. This would predict that basal phosphorylation of the PKB sites in AFX-T447A/T451A is higher than in AFX-WT. However, we never found a consistent increase in phosphorylation of the PKB peptides in our peptide map analysis. Also, no increase in phosphorylation was observed using a phosphospecific antibody against PKB phosphorylated serine 193. Thus, even though inhibition of PKB signaling results in retention of AFX in the nucleus, phosphorylation of the Ral sites is required for proper activation of AFX.
How does Ras-Ral induce the phosphorylation of AFX?
The components downstream of Ral responsible for the phosphorylation of AFX described in this work are currently unknown. Since in the absence of signaling, threonines 447 and 451 are already phosphorylated, indicating the presence of Ras-Ral-independent kinase activity, it may be that the Ras-Ral route either further activates this kinase or that it inhibits a phosphatase. Previously, Ral was reported to phosphorylate c-Jun, which is mediated both by JNK and the tyrosine kinase c-Src (7). However, the JNK-Src pathway that was suggested to be downstream of Ral, leading to c-Jun phosphorylation, is not involved in the phosphorylation of AFX by the Ras-Ral signaling pathway. First, PP1, a Src tyrosine kinase inhibitor, does not affect the phosphorylation of AFX by Ras or Ral. Second, in JNK−/− cells AFX is still phosphorylated by Ras and Ral, indicating that JNK activity is not responsible for the phosphorylation of AFX (N. D. de Ruiter, unpublished data). Also, filamin was reported to bind to Ral. Filamin efficiently cross-links actin filaments and is a docking site for various cell surface receptors and certain intracellular proteins involved in signal transduction (10). Therefore, filamin could mediate complex formation between, for instance, Ral and the kinase that is responsible for the phosphorylation of AFX. However, the kinase that was found to interact with filamin, SEK1, is the upstream kinase in the JNK signaling pathway. Since there is no apparent involvement of the JNK route in AFX regulation, it is not very likely that filamin plays a role in Ral-mediated regulation of AFX. Finally, MAP kinase, p90rsk, p70s6k, protein kinase A, protein kinase C enzymes sensitive to 12-d-tetradecanoyl phorbol-13-acetate (TPA), calmodulin kinase II, p38/HOG1, and glycogen synthase kinase-3 do not seem to be involved in the phosphorylation of AFX by the Ras-Ral route based on the use of different inhibitors and activators of these kinases (data not shown and reference 18).
In summary, we have identified a second regulatory mechanism of the Forkhead transcription factor AFX involving phosphorylation induced by the Ras-Ral pathway that impinges on the inhibitory constraint of AFX induced by phosphorylation of PKB sites. We propose that a balance between Ras-Ral-mediated activating or inactivating and PI(3)K-PKB-induced inactivating signals determines the response of AFX on transcription.
ACKNOWLEDGMENTS
We thank Geert Kops for continuous discussions and help during this project, Lydia de Vries and René Medema for help with certain experiments, Kris Reedquist for critically reading of the manuscript, and other members of our laboratory for support.
This work was supported by the Dutch Cancer Society.
REFERENCES
- 1.Borkhardt A, Repp R, Haas O A, Leis T, Harbott J, Kreuder J, Hammermann J, Henn T, Lampert F. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23) Oncogene. 1997;14:195–202. doi: 10.1038/sj.onc.1200814. [DOI] [PubMed] [Google Scholar]
- 2.Bos J L. All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 1998;17:6776–6782. doi: 10.1093/emboj/17.23.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 4.Brownawell A M, Kops G J, Macara I G, Burgering B M. Inhibition of nuclear import by protein kinase B (AKT) regulates the subcellular distribution and activity of the Forkhead transcription factor AFX. Mol Cell Biol. 2001;21:3534–3546. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgering B M, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 6.Cantor S B, Urano T, Feig L A. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Ruiter N D, Wolthuis R M, van Dam H, Burgering B M, Bos J L. Ras-dependent regulation of c-Jun phosphorylation is mediated by the Ral guanine nucleotide exchange factor-Ral pathway. Mol Cell Biol. 2000;20:8480–8488. doi: 10.1128/mcb.20.22.8480-8488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Exton J H. New developments in phospholipase D. J Biol Chem. 1997;272:15579–15582. doi: 10.1074/jbc.272.25.15579. [DOI] [PubMed] [Google Scholar]
- 9.Feig L A, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 10.Fox J W, Lamperti E D, Eksioglu Y Z, Hong S E, Feng Y, Graham D A, Scheffer I E, Dobyns W B, Hirsch B A, Radtke R A, Berkovic S F, Huttenlocher P R, Walsh C A. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 11.Frankel P, Ramos M, Flom J, Bychenok S, Joseph T, Kerkhoff E, Rapp U R, Feig L A, Foster D A. Ral and Rho-dependent activation of phospholipase D in v-Raf-transformed cells. Biochem Biophys Res Commun. 1999;255:502–507. doi: 10.1006/bbrc.1999.0234. [DOI] [PubMed] [Google Scholar]
- 12.Goi T, Shipitsin M, Lu Z, Foster D A, Klinz S G, Feig L A. An EGF receptor/Ral-GTPase signaling cascade regulates c-Src activity and substrate specificity. EMBO J. 2000;19:623–630. doi: 10.1093/emboj/19.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry D O, Moskalenko S A, Kaur K J, Fu M, Pestell R G, Camonis J H, White M A. Ral GTPases contribute to regulation of cyclin D1 through activation of NF-κB. Mol Cell Biol. 2000;20:8084–8092. doi: 10.1128/mcb.20.21.8084-8092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H, Luo J Q, Urano T, Frankel P, Lu Z, Foster D A, Feig L A. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature. 1995;378:409–412. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- 15.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis J H. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 16.Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 17.Kops G J, Burgering B M. Forkhead transcription factors: new insights into protein kinase B (c-akt) signaling. J Mol Med. 1999;77:656–665. doi: 10.1007/s001099900050. [DOI] [PubMed] [Google Scholar]
- 18.Kops G J, de Ruiter N D, De Vries-Smits A M, Powell D R, Bos J L, Burgering B M. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 19.Lu Z, Hornia A, Joseph T, Sukezane T, Frankel P, Zhong M, Bychenok S, Xu L, Feig L A, Foster D A. Phospholipase D and RalA cooperate with the epidermal growth factor receptor to transform 3Y1 rat fibroblasts. Mol Cell Biol. 2000;20:462–467. doi: 10.1128/mcb.20.2.462-467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo J Q, Liu X, Hammond S M, Colley W C, Feig L A, Frohman M A, Morris A J, Foster D A. RalA interacts directly with the Arf-responsive, PIP2-dependent phospholipase D1. Biochem Biophys Res Commun. 1997;235:854–859. doi: 10.1006/bbrc.1997.6793. [DOI] [PubMed] [Google Scholar]
- 21.Medema R H, Kops G J, Bos J L, Burgering B M. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 22.Medema R H, Wubbolts R, Bos J L. Two dominant inhibitory mutants of p21ras interfere with insulin-induced gene expression. Mol Cell Biol. 1991;11:5963–5967. doi: 10.1128/mcb.11.12.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta Y, Suzuki N, Nakamura S, Hartwig J H, Stossel T P. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci USA. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson M F, Paterson H F, Marshall C J. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature. 1998;394:295–299. doi: 10.1038/28425. [DOI] [PubMed] [Google Scholar]
- 25.Reedquist K A, Fukazawa T, Panchamoorthy G, Langdon W Y, Shoelson S E, Druker B J, Band H. Stimulation through the T cell receptor induces Cbl association with Crk proteins and the guanine nucleotide exchange protein C3G. J Biol Chem. 1996;271:8435–8442. doi: 10.1074/jbc.271.14.8435. [DOI] [PubMed] [Google Scholar]
- 26.Ridley A J, Paterson H F, Noble M, Land H. Ras-mediated cell cycle arrest is altered by nuclear oncogenes to induce Schwann cell transformation. EMBO J. 1988;7:1635–1645. doi: 10.1002/j.1460-2075.1988.tb02990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 28.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 29.Sewing A, Wiseman B, Lloyd A C, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unterman T G, Fareeduddin A, Harris M A, Goswami R G, Porcella A, Costa R H, Lacson R G. Hepatocyte nuclear factor-3 (HNF-3) binds to the insulin response sequence in the IGF binding protein-1 (IGFBP-1) promoter and enhances promoter function. Biochem Biophys Res Commun. 1994;203:1835–1841. doi: 10.1006/bbrc.1994.2401. [DOI] [PubMed] [Google Scholar]
- 31.Urano T, Emkey R, Feig L A. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- 32.van Weering D H, Moen T C, Braakman I, Baas P D, Bos J L. Expression of the receptor tyrosine kinase Ret on the plasma membrane is dependent on calcium. J Biol Chem. 1998;273:12077–12081. doi: 10.1074/jbc.273.20.12077. [DOI] [PubMed] [Google Scholar]
- 33.Verheijen M H, Wolthuis R M, Defize L H, den Hertog J, Bos J L. Interdependent action of RalGEF and Erk in Ras-induced primitive endoderm differentiation of F9 embryonal carcinoma cells. Oncogene. 1999;18:4435–4439. doi: 10.1038/sj.onc.1202834. [DOI] [PubMed] [Google Scholar]
- 34.White M A, Vale T, Camonis J H, Schaefer E, Wigler M H. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J Biol Chem. 1996;271:16439–16442. doi: 10.1074/jbc.271.28.16439. [DOI] [PubMed] [Google Scholar]
- 35.Wolthuis R M, Bos J L. Ras caught in another affair: the exchange factors for Ral. Curr Opin Genet Dev. 1999;9:112–117. doi: 10.1016/s0959-437x(99)80016-1. [DOI] [PubMed] [Google Scholar]
- 36.Wolthuis R M, de Ruiter N D, Cool R H, Bos J L. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 1997;16:6748–6761. doi: 10.1093/emboj/16.22.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolthuis R M, Zwartkruis F, Moen T C, Bos J L. Ras-dependent activation of the small GTPase Ral. Curr Biol. 1998;8:471–474. doi: 10.1016/s0960-9822(98)70183-6. [DOI] [PubMed] [Google Scholar]
- 38.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]