Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana (original) (raw)
Abstract
Regulation of gene expression at the post-transcriptional level is mainly achieved by proteins containing well-defined sequence motifs involved in RNA binding. The most widely spread motifs are the RNA recognition motif (RRM) and the K homology (KH) domain. In this article, we survey the complete Arabidopsis thaliana genome for proteins containing RRM and KH RNA-binding domains. The Arabidopsis genome encodes 196 RRM-containing proteins, a more complex set than found in Caenorhabditis elegans and Drosophila melanogaster. In addition, the Arabidopsis genome contains 26 KH domain proteins. Most of the Arabidopsis RRM-containing proteins can be classified into structural and/or functional groups, based on similarity with either known metazoan or Arabidopsis proteins. Approximately 50% of Arabidopsis RRM-containing proteins do not have obvious homologues in metazoa, and for most of those that are predicted to be orthologues of metazoan proteins, no experimental data exist to confirm this. Additionally, the function of most Arabidopsis RRM proteins and of all KH proteins is unknown. Based on the data presented here, it is evident that among all eukaryotes, only those RNA-binding proteins that are involved in the most essential processes of post-transcriptional gene regulation are preserved in structure and, most probably, in function. However, the higher complexity of RNA-binding proteins in Arabidopsis, as evident in groups of SR splicing factors and poly(A)-binding proteins, may account for the observed differences in mRNA maturation between plants and metazoa. This survey provides a first systematic analysis of plant RNA-binding proteins, which may serve as a basis for functional characterisation of this important protein group in plants.
INTRODUCTION
In the last decade, it has become increasingly apparent that post-transcriptional control of gene expression in eukaryotes is very important. In particular, post-transcriptional regulatory events are of crucial importance for development. The levels of regulation include pre-mRNA splicing, polyadenylation, mRNA transport, translation and stability/decay. Regulation is mainly achieved either directly by RNA-binding proteins or indirectly, whereby RNA-binding proteins modulate the function of other regulatory factors. The large variety of possible RNA targets implies the existence of a large number of RNA-binding proteins with different binding specificities.
The most abundant nuclear RNA-binding proteins in human cells are collectively termed heterogeneous nuclear ribonucleoproteins (hnRNPs), according to their association with nascent RNA polymerase II transcripts. Molecular cloning of genes encoding hnRNPs led to the discovery of several motifs involved in RNA binding (1,2). The most widely spread is the consensus sequence RNA-binding domain (CS-RBD), also known as RNA recognition motif (RRM). The RRM contains two short consensus sequences, RNP1 (octamer) and RNP2 (hexamer) embedded in a structurally, but not sequence, conserved region of approximately 80 amino acids. RRMs are present not only in hnRNP proteins, but also in a large variety of other RNA-binding proteins involved in all post-transcriptional processes, whereby the number of RRMs per protein varies from one to four copies (1,2). The K homology (KH) motif, first identified in the human hnRNP K protein (3), is the second most frequently found RNA-binding domain (1). The KH domain is approximately 60 amino acids long with a characteristic pattern of hydrophobic residues and with the most conserved consensus sequence VIGXXGXXI mapping to the middle of the domain (1). One protein can possess up to 15 copies of the KH domain. Both RRM and KH domains seem to be ancient protein structures as they have been found in organisms ranging from bacteria to humans (1).
In this article, we surveyed the complete Arabidopsis thaliana genome for RNA-binding proteins containing RRM and KH domains. Proteins containing zinc fingers, zinc knuckles, RGG boxes or DEAD boxes as the sole possible RNA-binding motifs were not included as none of these motifs can be used to exclusively predict an RNA-binding function.
MATERIALS AND METHODS
Protein sequences of known plant or metazoan RRM-containing proteins were used to query the nucleotide sequence of the Arabidopsis genome using TBLASTN and the annotated set of predicted Arabidopsis proteins using BLASTP (4) on the TAIR server (http://www.arabidopsis.org/Blast/). All identified proteins were used for searching the non-redundant protein database at the National Center for Biotechnology Information (NCBI, Bethesda, MD) using the BLAST and PSI-BLAST programs (http://www.ncbi.nlm.nih.gov:80/BLAST/). The BLOSUM62 matrix (5) was used for scoring, and E values <0.001 were used as a threshold for inclusion into the final protein list. Final alignments presented in Figures 3–6 were generated using ClustalW (6) (http://www2.ebi.ac.uk/clustalw/) and shaded on the BoxShade server (http://www.ch.embnet.org/software/BOX\_form.html).
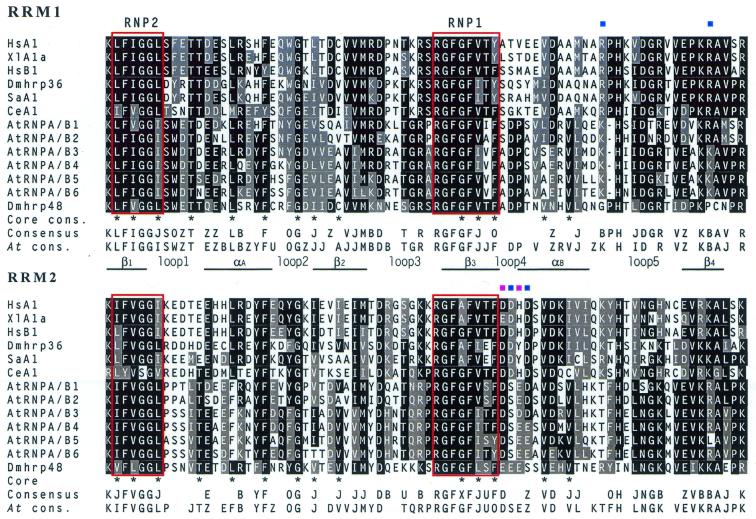
Figure 3.
Sequence analysis of Arabidopsis hnRNP A/B-like proteins. Alignment of the first (top) and the second (bottom) RRM of Arabidopsis hnRNP A/B proteins with metazoan members of the hnRNP A/B group of proteins. Sequences were aligned using the Clustal W program and shaded by the BoxShade server. Amino acids identical or similar in 50% of the sequences are shaded by black or grey background, respectively. Conserved secondary structure elements are indicated between alignments of the two RRMs. Asterisks indicate the position of residues located in the conserved hydrophobic core (52). Residues involved in formation of inter-RRM salt bridges are indicated with blue squares. The two acidic amino acids in the second RRM, possibly involved in salt bridges in Arabidopsis proteins are indicated with purple squares. The RNP1 and RNP2 motifs are indicated with red boxes. Consensus sequences at the bottom of each alignment indicate residues that are conserved in 10/13 sequences for the whole alignments or in 5/6 sequences for Arabidopsis proteins. Six groups of similar amino acids are indicated as follows: B = H, K, R; J = I, L, M, V; O = F, W, Y; U = S, T; X = A, G; Z = D, E. Hs, Homo sapiens; Xl, X.laevis; Dm, D.melanogaster; Sa, Schistocerca americana; Ce, C.elegans. Accession numbers of proteins used in alignment are as follows: HsA1, SWISS-PROT P09651; HsB1, SWISS-PROT: M29064; XlA1a, SWISS-PROT M31041; Dmhrp36, SWISS-PROT P48810; Dmhrp48, SWISS-PROT P48809; SaA1, SWISS-PROT P21522; CeA1, SWISS-PROT D10877.
Figure 6.
Alignment of RRMs of eight Arabidopsis 30K-RRM proteins. Details as in Figure 2. Consensus sequence at the bottom of alignment indicates residues that are conserved in 6/8 sequences.
The identification of KH domain-containing proteins in Arabidopsis followed the same procedure as described above for RRM-containing proteins.
A curated webpage containing results shown in Tables 1 and 2 will be available at http://www.at.embnet.org/bch/arabidopsis.htm.
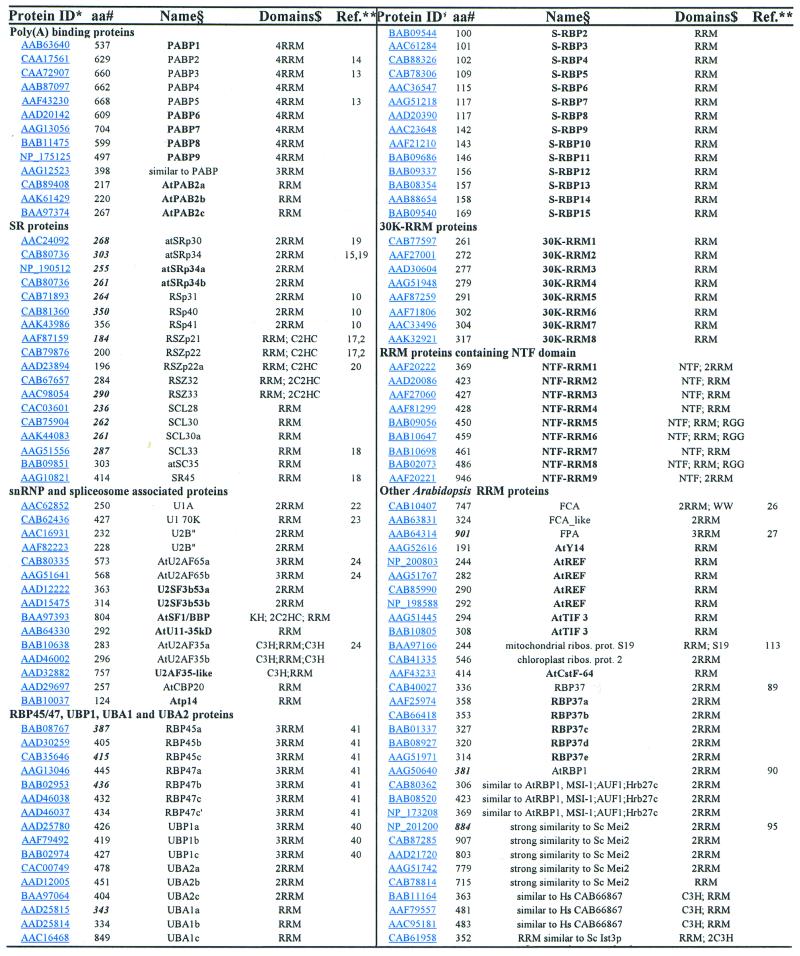
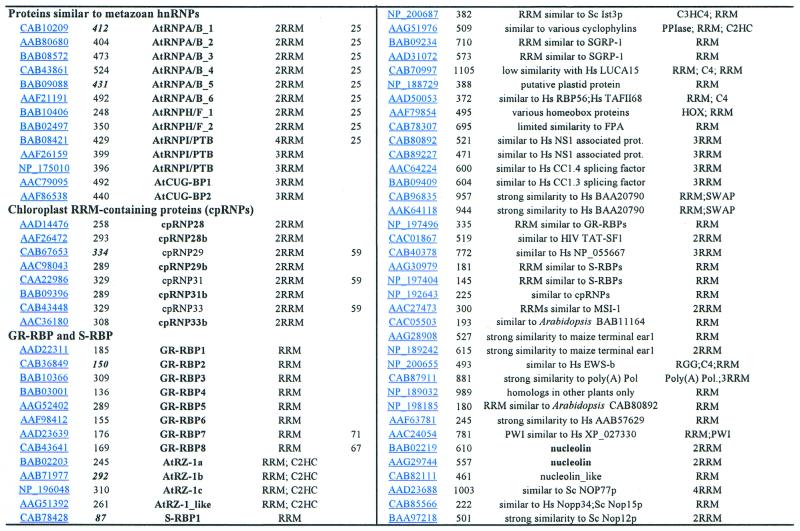
Table 1. Summary of Arabidopsis RRM-containing proteins.
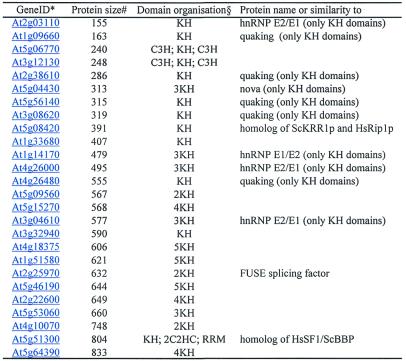
Table 2. Summary of Arabidopsis proteins containing KH domain.
RESULTS AND DISCUSSION
RNA recognition motif (RRM) proteins
The Arabidopsis genome encodes 196 different RRM containing proteins, which is more than those found in Caenorhabditis elegans [100 (7)] and Drosophila melanogaster [117 (8)]. Table 1 summarises our analysis, which comprises only RRM proteins containing both RNP1 and RNP2 submotifs. The final decision whether a protein fulfills the criteria of an RRM-containing protein was made by BLASTP and PSI-BLAST searches against the entire gene set in the GenBank. Only those proteins producing several hits with E values <0.001 were included in further analysis. This also allowed the prediction of the closest homologues in organisms other than plants (Table 1). Approximately 50% of Arabidopsis RRM proteins contain more than one RRM domain. In addition to RRM domains, 11 proteins contain C2HC-type zinc knuckles, seven proteins contain C3H-type zinc fingers and four proteins contain C4-type zinc (ring) fingers. Moreover, in nine proteins, RRMs were found in combination with the nuclear transport factor-like (NTF-like) domain (9). The Arabidopsis orthologue of SF1/BBP (BAA97393) possesses one RRM, two C2HC-type zinc knuckles and one KH domain. One protein contains, in addition to an RRM, a C2HC-type zinc knuckle and a cyclophilin domain (AAG51976). This is an unique domain combination not found in any other organism except plants. In another protein (AAF79856), an RRM was found in combination with the homeobox domain (Table 1). Domain compositions of the major types of Arabidopsis RRM-containing proteins are depicted schematically in Figure 1.
Figure 1.
Schematic representation of the modular structure of Arabidopsis RRM-containing proteins. Only major types of domain combinations are shown. Individual modules are identified by different shapes and colours. Different types of domains (RNA-binding, auxiliary domains and other distinctive regions of proteins) are listed at the bottom.
Of the 196 RRM proteins encoded by the Arabidopsis genome, ∼50% have not yet been described. However, it should not be concluded that 50% of Arabidopsis RRM proteins have an assigned function, as most of the described proteins are published or deposited in GenBank without any functional implications. In general, from our analyses it is clear that plants (Arabidopsis) express a complex set of RRM-containing proteins. At least 50% of them do not have obvious orthologues in metazoa, and among them are also some proteins that belong to protein families present in all higher eukaryotes (such as plant-specific SR proteins) (10). The Arabidopsis genome often encodes two or three very related RRM proteins. This has already been noted for other large protein groups in the Arabidopsis (11,12); indeed, ∼44% of such cases result from large intra- and inter-chromosomal duplications found in the Arabidopsis genome (12). We have not made any effort to determine how many Arabidopsis RRM proteins resulted from such events. Significantly, based on comparison with available cDNA sequences, we have noted that ∼33% (22 out of 53 analysed proteins) of Arabidopsis RRM-protein coding genes have wrongly predicted intron–exon boundaries (Table 1). Consequently, the protein sizes indicated in Table 1 should be handled with caution. In particular, this is important for cloning of Arabidopsis cDNAs based on current information in the Arabidopsis genome database.
The function for some groups of RRM proteins can be predicted based on the similarity with their metazoan counterparts. This is true for poly(A)-binding proteins (PABPs) (13,14), at least some Arabidopsis SR proteins (15–20), snRNP and spliceosome-associated RRM proteins (21–25), CstF-64 (cleavage stimulation factor of 64 kDa, a protein involved in polyadenylation), nucleolin, S19 ribosomal protein, and translation initiation factor 3 (TIF3). FCA (26) and FPA (27) are the only plant RRM proteins for which a function has been implicated based on the Arabidopsis mutant phenotype. We have identified an additional FCA-like protein (Table 1); whether this protein has a similar function in controlling flowering time in Arabidopsis remains to be established.
In the following paragraphs, we describe and discuss in more detail particular groups of Arabidopsis RRM-containing proteins.
Poly(A)-binding proteins
Poly(A) tails of eukaryotic mRNAs are bound by PABPs, and this interaction was shown to be essential for stimulation of polyadenylation, control of the poly(A) tail length, translation initiation, and for mRNA degradation (28–30). In yeast, all these functions are carried out by a single protein, Pab1p, which is an essential protein present in both the nucleus and the cytoplasm. Mammalian cells contain two distinct PABPs, a cytoplasmic PABP1 which is an orthologue of the yeast Pab1p, and a nuclear PAB2 (PABP2). Consistent with their cellular localisation, PABP1 is a mammalian protein involved in translation and cytoplasmic mRNA stability, whereas PAB2 is involved in polyadenylation (28–30). In contrast to yeast and human, which possess one and two PABPs, respectively, the Arabidopsis genome encodes 12 different PABPs (Table 1). Nine of the 12 Arabidopsis PABPs are homologous to the yeast and mammalian Pab1p, and are likewise composed of four consecutive RRMs (Fig. 1 and Table 1). However, we have to mention here that PABP8 and PABP9 are highly diverged members of this protein group (Fig. 2). The other three Arabidopsis PABPs consist of an acidic N-terminal domain followed by one RRM (Fig. 1), which is reminiscent of the mammalian PAB2 protein. Also, at the primary sequence level these three proteins are highly similar to PAB2 (Fig. 2), therefore we named them AtPAB2a, AtPAB2b and AtPAB2c (Table 1). Despite the quite strong sequence divergence between individual PABPs in Arabidopsis (13,14), PABP2, PABP3 and PABP5 were capable of rescuing a Pab1p-deficient yeast strain (31–33). In a complementation assay, PABP2, which is the most diverged member of this protein family in Arabidopsis, was shown to participate in many of the same post-transcriptional processes identified for yeast Pab1p (32). As some Arabidopsis PABPs are differentially expressed, it has been hypothesised that individual PABPs regulate polyadenylation and deadenylation during different stages of plant development (13,31,34).
Figure 2.
Dendrogram of Arabidopsis PABPs, and their orthologues from yeast and human. Dendrogram and sequence alignments were generated with the PileUP program, using default parameters (Genetic Computer Group, Madison, WI).
Based on the sequence similarity, one additional protein, PABP-like (Table 1), can be assigned to this group. In contrast to PABPs which contain four RRMs (Fig. 1), this protein is predicted to have only three RRMs, and experimental data are necessary to define it as a genuine PABP. Furthermore, in Chlamydomonas reinhardtii, a nucleus-encoded PABP (RB47), is required for translational regulation of the chloroplast psbA gene (35). We were not able to unambiguously predict an Arabidopsis orthologue of RB47.
Ser/Arg (SR) and spliceosomal RRM-containing proteins
SR proteins are essential splicing factors identified in all eukaryotes except yeast. They consist of one or two N-terminally positioned RRMs and a C-terminal domain rich in SR dipeptides (Fig. 1), hence the name SR proteins. In metazoa, SR proteins play an important role in constitutive and alternative splicing by promoting interactions across intronic and exonic sequences during early steps of spliceosome assembly, thereby helping in selection of splice sites (36,37). The genes encoding most of Arabidopsis SR proteins have already been cloned (10,15,17–20), and evidence exists that at least some of them have similar activity in pre-mRNA splicing as their metazoan counterparts (15,19,20,25). In total, the Arabidopsis genome encodes 18 different SR proteins (Table 1), which is more then found in human cells (10 different human SR proteins have been characterised so far) (37). Of the 18 Arabidopsis SR proteins, clear orthologues of human SF2/ASF (atSRp34), SC35 (atSC35) and 9G8 (atRSZp21, atRSZp22 and atRSZp22a) have been identified. In addition to the previously characterised atSRp34, we have identified two novel Arabidopsis proteins having strong similarity with atSRp34 and human SF2/ASF (Table 1, atSRp34a and atSRp34b). In the TAIR database these two proteins are annotated as SF2-like, but due to the wrong prediction of the last three exons they did not contain an SR domain. The other SR proteins (RSp31, RSp40, RSp41, RSZ32 and RSZ33; Table 1) seem to be plant specific (10,18; S.Lopato and A.Barta, unpublished data). Particularly interesting are RSZ32 and RSZ33 which, in addition to one RRM, contains two consecutive C2HC-type zinc knuckles (Fig. 1 and Table 1), a situation not found in any metazoan SR protein. It is worth noting that Arabidopsis expresses three orthologues of each human SF2/ASF and 9G8 proteins. Except RSp31 and SR45, all other plant SR proteins are represented by pairs of very similar proteins. These close homologues in Arabidopsis may have partially redundant functions; however, evidence exists that pairs of homologous genes are differentially expressed (19; M.Kalyna and A.Barta, unpublished data). This indicates that they may modulate splicing during different stages of plant development, and/or that the target pre-mRNAs which are regulated by such pairs of proteins are different.
Spliceosomal proteins containing an RRM domain are easily found in Arabidopsis (Table 1). This is consistent with the observation that spliceosome composition is in general highly conserved between yeast, plants and metazoa (25,38; Z.J.Lorković and A.Barta, unpublished data). Therefore, the observed differences in intron processing between plants and metazoa must occur at the early steps of intron recognition (25,39). This is supported by the multitude of SR proteins expressed in Arabidopsis, some of which seem to be plant specific. However, additional, not yet experimentally identified, plant-specific RNA-binding proteins could also contribute to plant intron recognition (see also below).
UBP1, RBP45, RBP47, UBA1 and UBA2—oligouridylate-specific RRM proteins
A common feature of this group of nuclear RRM proteins is their specificity for oligouridylates. UBP1, RBP45 and RBP47 are also structurally related; they consist of three RRMs and a glutamine-rich N-terminus (40,41) (Fig. 1). At the primary sequence level as well as at the biological level, RBP45 and RBP47 proteins are clearly different from UBP1 (41). Protoplast transfection experiments have indicated that UBP1 from Nicotiana plumbaginifolia functions in nuclear pre-mRNA maturation by stimulating splicing efficiency of suboptimal introns and increasing the steady-state level of reporter RNAs (40). Neither RBP45 nor RBP47 from N.plumbaginifolia affected splicing and accumulation of reporter RNAs in plant protoplasts (41). The mechanism by which UBP1 increases splicing efficiency is unclear, whereas enhanced accumulation of RNA is apparently due to UBP1 interacting with the 3′-UTR and protecting mRNA from exonucleolytic degradation (40). RBP45/RBP47 and UBP1 are most similar to yeast Nam8p and metazoan TIA-1 proteins, respectively. Nam8p and TIA-1 are components of U1 snRNP (42,43), and stabilise interaction of U1 snRNP with the pre-mRNAs containing introns with suboptimal 5′-splice sites (42–44). Although direct evidence for an association of UBP1 with U1 snRNP is missing, it is possible that its effects on splicing occur in a similar way.
UBA1a and UBA2a were identified as proteins interacting with UBP1 in a yeast two-hybrid system (M.H.L.Lambermon, Y.Fu, D.A.Wieczorek Kirk, M.Dupasquier, W.Filipowicz and Z.J.Lorković manuscript submitted for publication). Like UBP1, both UBA1a and UBA2a increased the steady-state levels of reporter RNAs when overexpressed in protoplasts, but unlike UBP1 neither protein stimulated pre-mRNA splicing. It has been suggested that UBP1, UBA1 and UBA2 proteins may act as components of a complex recognising U-rich sequences in plant 3′-UTRs resulting in mRNA stabilisation in the nucleus (M.H.L.Lambermon, Y.Fu, D.A.Wieczorek Kirk, M.Dupasquier, W.Filipowicz and Z.J.Lorković manuscript submitted for publication). Neither UBA1 nor UBA2 seem to have orthologues in metazoan genomes.
Arabidopsis proteins with homology to metazoan hnRNPs
In human cells, 20 different hnRNP proteins or groups of proteins have been characterised. hnRNP proteins have been identified in C.elegans, Xenopus laevis and D.melanogaster; in the latter organism 12 major proteins, termed hrp36 to hrp75, were identified and most of them have a strong sequence similarity to the human hnRNP A/B proteins (45–48). Metazoan hnRNP A/B proteins are composed of two adjacent N-terminally positioned RRMs and a glycine-rich C-terminal auxiliary domain (2,49,50). At the biological level, hnRNP A/B proteins are involved in alternative splicing by promoting usage of distal 5′-splice sites, thereby antagonising the alternative splicing activity of splicing factors SF2/ASF and SC35 (49–51).
Previous analysis of the Arabidopsis genome revealed six genes whose predicted protein sequences had a domain organisation typical of hnRNP A/B proteins (25). Sequence analysis of predicted Arabidopsis proteins, named AtRNP A/B_1 to 6, revealed that all of them, like metazoan proteins, are composed of two RRMs followed by a C-terminal auxiliary domain. However, the C-terminal domain in only two Arabidopsis proteins is glycine-rich, as in the metazoan hnRNP A/B proteins, whereas in the other four proteins this domain is rather equally enriched in glycine, asparagine and serine residues (Fig. 1). Comparison of the RNA-binding domains with their metazoan counterparts revealed strong sequence conservation that results in an ungapped alignment of RRM2 and one amino acid gap in loop five of the RRM1 (Fig. 3). Pairwise comparison of RRMs of AtRNP A/B proteins with metazoan hnRNP A/B RRMs resulted in identity/similarity scores ranging between 44–50% and 50–60%, which is greater than the scores obtained with RRMs from other metazoan or plant RRM-containing proteins. In addition to conserved positions within the RRMs of most RRM proteins (core consensus in Fig. 3) (52), this alignment reveals positions that are highly conserved in RRMs of both Arabidopsis and metazoan hnRNP A/B proteins, and in Arabidopsis proteins alone (consensus and At consensus lanes in Fig. 3). In terms of molecular weight, isoelectric point and amino acid composition of the C-terminal auxiliary domains, Arabidopsis proteins are most similar to D.melanogaster hrp proteins. As in D.melanogaster hrp proteins (45–47), we were unable to unambiguously identify vertebrate orthologues of the Arabidopsis hnRNP A/B proteins.
In spite of the strong similarity between Arabidopsis and metazoan hnRNP proteins, certain differences do exist. The linker region (IRL) between two RRMs, which is conserved in metazoan proteins in length (13 amino acids) and primary sequence (53), is variable in Arabidoposis proteins (11–19 amino acids). The length of the IRL is known to be important for the spatial arrangement of the two RRMs and for alternative splicing activity of the human hnRNP A1 protein (53). Another difference concerns the residues involved in formation of two salt bridges, responsible for holding the two RRMs in close contact (Fig. 3, blue squares) (53–55). The two arginines (mostly lysines in Arabidopsis proteins) in RRM1 are conserved in terms of charge, whereas only the second of the pair of acidic residues (aspartic acid 157) seems to be conserved in RRM2 of Arabidopsis proteins (Fig. 3, blue squares). Interestingly, there is another pair of acidic amino acids just one position upstream (Fig. 3, purple squares), which could potentially take over the function of Asp155 and Asp157 in making the salt bridges.
In addition to putative homologues of hnRNP A/B proteins, it seems that the Arabidopsis genome also encodes homologues of hnRNP H/F and hnRNP I proteins (Table 1) (25). hnRNP A, B, H, F and I are all involved in splicing regulation, particularly in alternative splicing (49,50). It is important to note that the splicing factors ASF/SF2 and SC35 that antagonise hnRNP A1 activity in usage of alternative 5′-splice sites are also conserved in plants (15,18,19) (Table 1; see also above). CUG-BP (or CELF) proteins that antagonise the activity of hnRNP I in 3′-splice site selection (56) are likewise conserved in Arabidopsis (Table 1). In the light of increasing evidence that alternative splicing is important in regulating gene expression in plants, it is interesting that these hnRNP proteins are conserved. Consequently, analysis of factors involved in this process, including Arabidopsis hnRNP proteins, is necessary for a better understanding of this important aspect of post-transcriptional regulation of gene expression in plants.
Database searches with the complete set of Arabidopsis RRM proteins presented in Table 1 did not reveal possible homologues of other human RRM-containing hnRNPs.
Chloroplast RRM-containing proteins (cpRNPs)
A group of nucleus-encoded, RRM-containing RNA-binding proteins has been described in the chloroplasts of higher plants (57–61). They have a characteristic domain organisation; an N-terminal transit peptide which is necessary for import into chloroplasts is followed by an acidic domain at the N-terminus of the mature protein and two consecutive RRMs at the C-terminus (Fig. 1). The Arabidopsis genome encodes eight cpRNPs and, according to sequence homology to previously described cpRNPs from different plant species (57–59), we named them cpRNP28, cpRNP29, cpRNP31 and cpRNP33. Again, as shown for SR proteins, each cpRNP is represented by two closely related proteins (Table 1). In tobacco, cpRNPs are abundant stromal proteins that exist as complexes with ribosome-free mRNAs (62). Evidence also exists that cpRNPs are involved in chloroplast mRNA 3′-end formation, RNA stabilisation (63,64); furthermore, as shown recently, some cpRNPs are involved in chloroplast RNA editing (65). A more detailed description of cpRNPs can be found in other reviews (60,61).
Glycine-rich and small RRM-containing proteins
This group comprises 27 Arabidopsis proteins. A common feature of these proteins is their similar domain organisation; they all contain one RRM at the N-terminus and a C-terminal extension. Based on differences in the C-terminal part, we divided them into two subgroups: (i) glycine-rich RNA-binding proteins (GR-RBPs) and (ii) small RNA-binding proteins (S-RBPs) (Table 1 and Fig. 1). To distinguish between cell wall-localised glycine-rich proteins (GRPs) that do not contain RRMs and RRM-containing GRPs, we propose renaming the RRM-containing GRPs as GR-RBPs (glycine-rich RNA-binding proteins). GR-RBPs are represented by eight members; all have been previously reported from different plant species (61). They have been implicated in responses to various environmental stresses (66–68) and rRNA processing (69,70), and some of them seem to be regulated by a circadian clock (61,67,71,72). Alignment of GR-RBPs revealed strong primary sequence conservation in their RRMs (Fig. 4), indicating that this is a homogenous group of proteins with similar RNA-binding specificities and maybe related functions. Furthermore, based on the sequence similarity in their RRMs, three additional proteins can be assigned to this subgroup. In contrast to GR-RBPs, the C-terminus in these three proteins is rather arginine/aspartate/glutamate-rich, with RD (BAB02203; NP_196048) or RD/RE (AAB71977) repeats. Moreover, these proteins have a C2HC-type zinc knuckle inserted between the RRM and the C-terminal domain (Fig. 1), which is not found in GR-RBPs and S-RBPs. They most probably represent orthologues of the N.silvestris RZ-1 protein, which has been found in a nucleoplasmic 60S RNP complex and in association with nuclear poly(A)+ RNA (41,73). We have found one additional protein with the same domain organisation (Table 1; RZ-1_like; AAG51392), but its C-terminal domain contains basic and acidic patches instead of RD or RD/RE repeats. Moreover, phylogenetic analysis revealed that this protein is more similar to some GR-RBPs and S-RBPs than to three AtRZ-1 proteins. Because RZ-1 and RZ-1_like proteins are encoded in genomes of different plant species, but not in metazoan genomes, we conclude that they are plant specific.
Figure 4.
Alignment of RRMs of eight Arabidopsis GR-RBPs. The three Arabidopsis RZ-1 orthologues and RZ-1_like protein are not included. Details as in Figure 2. Consensus sequence at the bottom of alignment indicates residues that are conserved in 7/8 sequences.
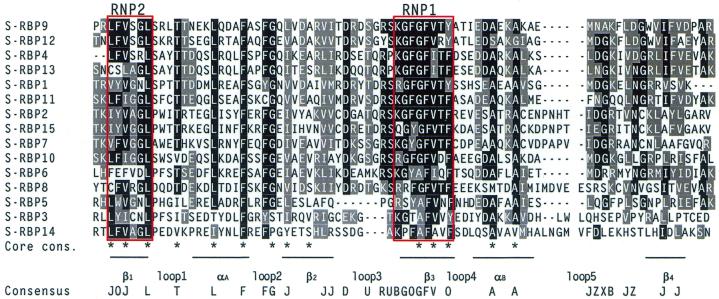
The 15 S-RBPs are grouped together based only on their predicted molecular weight. In contrast to GP-RBPs, alignment of their RRMs revealed that they are a rather heterogeneous group of proteins with low sequence homology outside the RNP1 and RNP2 submotifs (Fig. 5). BLASTP searches with individual members of this subgroup resulted in limited homology with various plant and metazoan RRM-containing proteins. The most common hits were plant GR-RBPs, human and X.laevis CIRP protein, and human RBM3 proteins which are induced by cold shock (74–77). It remains to be established whether S-RBPs also respond to cold stress or other environmental conditions.
Figure 5.
Alignment of RRMs of 15 Arabidopsis S-RBPs. Details as in Figure 2. Consensus sequence at the bottom of alignment indicates residues that are conserved in 11/15 sequences.
GR-RBPs and S-RBPs have been found in organisms ranging from Cyanobacteria to humans (61,74–78). Cyanobacterial and metazoan GR-RBPs are, like some plant GR-RBPs, induced by cold-shock (77,78 and references therein); however, the exact function of these proteins remains largely unknown. The eight GR-RBPs together with 15 S-RBPs with similarity to plant and metazoan GR-RBPs (Table 1) constitute the largest group of RRM-containing proteins in Arabidopsis. It seems that genes encoding this group of proteins became highly amplified during evolution of land plants. This may not be a surprise, because unlike metazoa, plants are sessile organisms which are constantly exposed to changes in their environment. Amplification of this gene family and subsequent acquisition of differential expression could be a way to regulate RNA metabolism under different environmental conditions.
30K-RRM proteins
This is a very homogenous group of eight proteins containing one RRM with strong sequence homology in the entire RRM region (Fig. 6), which may indicate a common ancestor or otherwise very similar functions. Another common feature of these proteins is their similar molecular weight of ∼30 kDa, hence the name 30K-RRM proteins. Like in GR-RBPs, RRMs of 30K-RRMs are located in the N-terminal half of the protein, followed by a C-terminal extension with rather unusual amino acid compositions (rich in proline, glutamine, histidine, glycine, serine and acidic amino acids) (Fig. 1). These domains could possibly play a role in protein–protein interactions. The best metazoan match found with any of these plant sequences in BLASTP searches was the human SEB4D protein whose function is not known. However, in spite of the sequence similarity with SEB4D, which is limited to the RRM regions, 30K-RRM proteins do not seem to have orthologues in metazoa. The function of all 30K-RRM proteins remains to be determined.
Arabidopsis RRM proteins containing an NTF-like domain
The NTF domain was first identified in NTF2 protein which is involved in nuclear protein import (9). Later, a related factor, p15 (or NXT1) involved in nuclear protein export was found to possess an NTF-like domain (79). Meanwhile, NTF-like domains have been found in a large variety of proteins, including nucleocytoplasmic transport factor TAP (Mex67p in yeast), some plant MAP kinases and a protein G3BP implicated in the Ras signal transduction pathway (80).
Of the nine Arabidopsis proteins containing NTF-like domain in combination with an RRM, the domain organisation of three proteins resembles that of human G3BP. An N-terminally positioned NTF domain is followed by one RRM and an RGG box at the very C-terminus (80,81) (Fig. 1). None of the NTF-RRM Arabidopsis proteins has previously been described. By analogy with metazoan proteins it is likely that they are involved in nucleocytoplasmic trafficking of RNA and/or proteins. Alternatively, they could be involved in some signal transduction pathways in plants, as is human G3BP.
Messenger RNAs are exported from the nucleus as large ribonucleoprotein complexes (82,83). A protein complex that associates with mRNA during splicing and distinguishes spliced from unspliced mRNAs has recently been identified (82,83). Among the proteins identified in this complex are two RRM proteins, REF (84) and Y14 (85), which are highly conserved in Arabidopsis (Table 1). As in mouse (84), four different REF proteins are expressed in Arabidopsis (Table 1). Only one REF homologue, Yra1p, exists in yeast (80,86), whereas Y14 does not seem not to be present in yeast at all. Other components of this protein complex (DEK, SRm160 and RNPS1) are less conserved in Arabidopsis. It is interesting to note, however, that the Arabidopsis genome does not encode a protein corresponding to TAP/Mex67p, a component of the mRNA export machinery found to interact directly with REF (84). TAP protein was shown to interact through its NTF-like domain with the p15 (NXT1) as well (80) which is likewise absent from Arabidopsis genome, and this interaction is required for efficient export of mRNA to the cytoplasm (87,88). The multitude of RRM proteins containing NTF-like domain in Arabidopsis makes it possible that at least some of these proteins take over the function of TAP in mRNA export in plants and the function of p15 could be mediated by one of the three NTF2 Arabidopsis homologues.
Other Arabidopsis RRM-containing proteins
This group consists of 69 proteins; ∼25% of these proteins have already been mentioned in the general description of Arabidopsis RRM-containing proteins, or are discussed in connection with proteins from other groups. In addition, three other members from this group, and proteins similar with them, are described below. The other proteins which are listed at the end in Table 1 show limited similarities to RRM-containing proteins from other organisms (best scores are indicated in Table 1), and are therefore not discussed further. To establish their possible functional relationships to metazoan proteins will need experimentation.
We have identified five novel proteins highly related to the previously published Arabidopsis RBP37 (Table 1) which is expressed in dividing cells during development (89). This group of proteins does not have obvious orthologues in metazoa, and their functional targets are still to be determined.
AtRBP1 protein, which consists of two N-terminal RRMs and an extension at the C-terminus, was found to be expressed in rapidly dividing tissues (90). The RRMs of this protein are most similar to those of metazoan Musashi proteins (91–93 and references therein). RRM proteins belonging to the Musashi family are specifically expressed in the nervous system, particularly in stem cells and neural progenitor cells; however, their roles are poorly understood (91–93 and references therein). We have identified three additional proteins having similarity to AtRBP1 and Musashi. Two of those proteins (BAB08520; NP_173208) contain a glycine-rich C-terminal domain which makes them similar to hnRNP A/B and D proteins (49,50). Indeed, in BLASTP searches the best scores obtained with both proteins were Musashi and hnRNP D proteins, whereas the best Arabidopsis scores were proteins designated as AthnRNP A/B proteins (Table 1). Experimental data are required to establish whether the function of these proteins is similar or equivalent to Musashi, hnRNP D or hnRNP A/B proteins.
In Schizosaccharomyces pombe, Mei2p has been shown to be required for both induction of premeiotic DNA synthesis and promotion of the first meiotic division (94). The Arabidopsis protein AML1, which is highly similar to the S.pombe Mei2p, was cloned by functional complementation of a fission yeast pheromone receptor-deficient strain (95). We have identified four novel Mei2p-like proteins in Arabidopsis. It remains to be determined whether they also participate in the regulation of meiosis.
Arabidopsis proteins containing KH domain
In metazoa, proteins containing KH domains have been implicated in transcription, mRNA stability, translational silencing and mRNA localisation (50,96,97). Mutations in KH domain proteins very often result in developmental defects. For example, the D.melanogaster how gene encodes a single KH-domain protein essential for tendon cell differentiation (98), whereas murine quaking protein is required for maturation of Schwann cells into myelin-forming cells in the peripheral nervous system (99). Quaking and two other D.melanogaster KH domain-containing proteins, FMRP and MCG10, have been shown to induce apoptosis (100–102). The FMR gene also encodes a KH domain-containing protein; transcriptional silencing of this gene or a mutation in the C-terminal KH domain leads to the fragile X syndrome (103,104). In addition, some metazoan KH proteins have been shown to be autoantigens associated with certain tumour types (105–107).
In Arabidopsis we have found 26 proteins containing KH domains (Table 2). In contrast to the 27 Drosophila KH domain proteins (8), most Arabidopsis KH proteins possess more than one KH domain (Table 2). An alignment of 60 KH domains can be found on http://www.embnet.org/bch/arabidopsis.htm. In addition to KH domains, two Arabidopsis proteins possess C3H-type zinc fingers (gene IDs At5g06770; At3g12130), whereas the Arabidopsis homologue of splicing factor SF1/BBP contains two C2HC-type zinc knuckles (gene ID At5g51300). Large KH domain proteins, such as chicken vigilin, which possesses 15 KH domains were not found in the Arabidopsis genome. Vigilin homologues have been found in human, X.laevis, D.melanogaster, C.elegans, S.pombe, and Saccharomyces cerevisiae, and evidence exists for their involvement in the control of cell ploidy (108), heterochromatin structure (109), and possibly in RNA stabilisation (110). Despite the obviously highly conserved primary structure and function of vigilins in all eukaryotes, an extensive search of the Arabidopsis genome failed to reveal homologous proteins. The only Arabidopsis KH proteins that could unambiguously be predicted as orthologues of yeast or metazoan KH proteins are the splicing factor SF1/BBP (gene ID At5g51300) and the homologue of the yeast KRR1p (AtKRR1p; gene ID At5g08420). KRR1p and its metazoan orthologs (human Rip1 and Drosophila Dribble proteins) are nucleolar proteins implicated in rRNA processing (111,112 and references therein). Strong conservation of SF1/BBP and AtKRR1p is in accordance with the high conservation of pre-mRNA and rRNA processing machineries in all eukaryotes. BLASTP, PSI-BLAST and FASTA searches with Arabidopsis KH domain-containing proteins resulted in limited similarities with diverse metazoan KH proteins, but these similarities are restricted to KH domains only. For example, the KH domains of five Arabidopsis proteins (gene IDs At1g09660, At2g38610, At5g56140, At3g08620 and At2g26480) show limited similarity to respective domains of mammalian quaking proteins (Table 2), whereas the other regions of these proteins do not show significant similarity. Experimental data will be required to establish whether these proteins have similar functions. The same applies to possible Arabidopsis homologues of metazoan hnRNP K and E proteins. It seems that plants have evolved KH proteins with entirely different domain organisations, resulting most probably in different binding specificities and biological functions. Given that many metazoan KH proteins are involved in cell differentiation and development this may not be a surprise. Plant development, despite following some common themes found in metazoa in pattern formation, requires plant-specific protein functions. This is best illustrated by the existence of large number of plant-specific transcription factors (11).
It is noteworthy that none of the Arabidopsis KH-domain proteins has been characterised so far.
CONCLUSION
Plants have evolved a large number of kingdom-specific RNA-binding proteins. Only those proteins required for basic mechanisms in post-transcriptional regulation of gene expression have been preserved in all eukaryotic lineages during evolution. The function of Arabidopsis RRM and particularly KH domain proteins is largely unknown. In the years to come, the great task will be to characterise Arabidopsis RNA-binding proteins using DNA microarray technology and reverse genetics. This, together with biochemical characterisation, will aid in understanding observed differences in post-transcriptional regulation of gene expression between plants and metazoa. Such analyses will help to place individual RRM or KH proteins into a complex network regulating plant development and plant–environment interactions.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Maria Kalyna and Tim Skern for helpful and critical comments on the manuscript. We thank Joachim Seipelt for help with the webpage. This work was supported by a grant (SFB17 Nos 1710 and 1711) from the Österreichischer Fonds zur Förderung der Wissenschaftlichen Forschung to A.B.
REFERENCES
- 1.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 2.Swanson M.S. (1995) Function of nuclear pre-mRNA/mRNA binding proteins. In Lamond,A. (ed.), Pre-mRNA processing. R.G.Landes Publishers, Gergetown, TX, pp. 18–33.
- 3.Siomi H., Matunis,M.J., Michael,W.M. and Dreyfuss,G. (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res., 21, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The C.elegans Sequencing Consortium (1998) Genome sequence of the nematode C.elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 8.Lasko P. (2000) The Drosophila melanogaster genome: translation factors and RNA binding proteins. J. Cell Biol., 150, F51–F56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paschal B.M. and Gerace,L. (1995) Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell Biol., 129, 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopato S., Waigmann,E. and Barta,A. (1996) Characterisation of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell, 8, 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichmann J.L., Heard,J., Martin,G., Reuber,L., Jiang,C.-Z., Keddie,J., Adam,L., Pineda,O., Ratcliff,O.J., Samaha,R.R. et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science, 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- 12.The Arabidopsis Genome Initiative (2000) Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 13.Belostotsky D.A. and Meagher,R.B. (1993) Differential organ-specific expression of three poly(A)-binding-proteins from Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 90, 6686–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilson P., Carroll,K.L. and Masson,P.H. (1993) Molecular characterisation of PA B2, a member of the multigene family coding for poly(A)-binding proteins in _Arabidopsis thaliana._Plant Physiol., 103, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazar G., Schaal,T., Maniatis,T. and Goodman,H.M. (1995) Identification of a plant serine–arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc. Natl Acad. Sci. USA, 92, 7672–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopato S., Mayeda,A., Krainer,A.R. and Barta,A. (1996) Pre-mRNA splicing in plants: characterisation of Ser/Arg splicing factors. Proc. Natl Acad. Sci. USA, 93, 3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golovkin M. and Reddy,A.S.N. (1998) The plant U1 small nuclear ribonucleoprotein particle 70K protein interacts with two novel serine/arginine-rich proteins. Plant Cell, 10, 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golovkin M. and Reddy,A.S.N. (1999) An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1-70K protein. J. Biol. Chem., 274, 36428–36438. [DOI] [PubMed] [Google Scholar]
- 19.Lopato S., Kalyna,M., Dorner,S., Kobayashi,R., Krainer,A.R. and Barta,A. (1999) atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev., 13, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopato S., Gattoni,R., Fabiani,G., Stevenin,J. and Barta,A. (1999) A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol. Biol., 39, 761–773. [DOI] [PubMed] [Google Scholar]
- 21.Simpson G.G., Vaux,P., Clark,G.P., Waugh,R., Beggs,J.D. and Brown,J.W.S. (1991) Evolutionary conservation of the spliceosomal protein, U2B′′. Nucleic Acids Res., 19, 5213–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson G.G., Clark,G.P., Rothnie,H.M., Boelens,W., van Venrooij,W. and Brown,J.W.S. (1995) Molecular characterisation of the spliceosomal proteins U1A and U2B′′ from higher plants. EMBO J., 14, 4540–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golovkin M. and Reddy,A.S.N. (1996) Structure and expression of a plant U1 snRNP 70K gene: alternative splicing of U1 snRNP 70K gene produces two different transcripts. Plant Cell, 8, 1421–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domon C., Lorković,Z.J., Valcarcel,J. and Filipowicz,W. (1998) Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J. Biol. Chem., 273, 34603–34610. [DOI] [PubMed] [Google Scholar]
- 25.Lorkovi Z.J.M,, Wieczorek Kirk,D.A., Lambermon,M.H.L. and Filipowicz,W. (2000) Pre-mRNA splicing in higher plants. Trends Plant Sci., 5, 160–167. [DOI] [PubMed] [Google Scholar]
- 26.Macknight R., Bancroft,I., Page,T., Lister,C., Schmidt,R., Love,K., Westphal,L., Murphy,G., Sherson,S., Cobbett,C. and Dean,C. (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell, 89, 737–745. [DOI] [PubMed] [Google Scholar]
- 27.Schomburg F.M., Patton,D.A., Meinke,D.W. and Amasino,R.M. (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell, 13, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller W. and Minvielle-Sebastia,L. (1997) A comparison of a mammalian and yeast pre-mRNA 3′-end processing. Curr. Opin. Cell Biol., 9, 329–336. [DOI] [PubMed] [Google Scholar]
- 29.Minvielle-Sebastia L. and Keller,W. (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol., 11, 352–357. [DOI] [PubMed] [Google Scholar]
- 30.Wahle E. and Ruegsegger,U. (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- 31.Belostotsky D.A. and Meagher,R.B. (1996) A pollen-, ovule- and early embryo-specific poly(A) binding protein from Arabidopsis complements essential functions in yeast. Plant Cell., 8, 1261–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palanivelu R., Belostotsky,D.A. and Meagher,R.B. (2000) Arabidopsis thaliana poly(A) binding protein 2 (PAB2) functions in yeast translational and mRNA decay processes. Plant J., 22, 187–198. [DOI] [PubMed] [Google Scholar]
- 33.Chekanova J.A., Shaw,R.J. and Belostotsky,D.A. (2001) Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr. Biol., 11, 1207–1214. [DOI] [PubMed] [Google Scholar]
- 34.Palanivelu R., Belostotsky,D.A. and Meagher,R.B. (2000) Conserved expression of Arabidopsis thaliana poly(A) binding protein 2 (PAB2) in distinct vegetative and reproductive tissues. Plant J., 22, 199–210. [DOI] [PubMed] [Google Scholar]
- 35.Yohn C.B., Cohen,A., Rosch,C., Kuchka,M.R. and Mayfield,S.P. (1998) Translation of the chloroplast psbA mRNA requires the nuclear-encoded poly(A)-binding protein, RB47. J. Cell Biol., 142, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu X.-D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- 37.Graveley B.R. (2000) Sorting out the comlexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mount S.M. and Salz,H.K. (2000) Pre-messenger RNA processing factors in the Drosophila genome. J. Cell Biol., 150, F37–F43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown J.W.S. and Simpson,C.G. (1998) Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Physiol. Plant Mol. Biol., 49, 77–95. [DOI] [PubMed] [Google Scholar]
- 40.Lambermon M.H.L., Simpson,G.G., Wieczorek Kirk,D.A., Hemmings-Mieszczak,M., Klahre,U. and Filipowicz,W. (2000) UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J., 19, 1638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorković Z.J., Wieczorek Kirk,D.A., Klahre,U., Hemmings-Mieszczak,M. and Filipowicz,W. (2000) RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA, 6, 1610–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puig O., Gottschalk,A., Fabrizio,P. and Seraphin,B. (1999) Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev., 13, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Gatto-Konczak F., Bourgeois,C.F., Le Guiner,C., Kister,L., Gesnel,M.C., Stevenin,J. and Brethnach,R. (2000) The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol., 20, 6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Förch P., Puig,O., Kedersha,N., Martinez,C., Granneman,S., Seraphin,B., Anderson,P. and Valcarcel,J. (2000) The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell, 6, 1089–1098. [DOI] [PubMed] [Google Scholar]
- 45.Haynes S.R., Raychaudhuri,G. and Beyer,A.L. (1990) The Drosophila Hrb98DE locus encodes four protein isoforms homologous to the A1 protein of mammalian heterogeneous nuclear ribonucleoprotein complexes. Mol. Cell. Biol., 10, 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haynes S.R., Johnson,D., Raychaudhuri,G. and Beyer,A.L. (1991) The Drosophila Hrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res., 19, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matunis E.L., Matunis,M.J. and Dreyfuss,G. (1992) Characterisation of the major hnRNP proteins from Drosophila melanogaster. J. Cell Biol., 116, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matunis M.J., Matunis,E.L. and Dreyfuss,G. (1992) Isolation of hnRNP complexes from Drosophila melanogaster. J. Cell Biol., 116, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weighardt F., Biamonti,G. and Riva,S. (1996) The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays, 18, 747–756. [DOI] [PubMed] [Google Scholar]
- 50.Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 51.Mayeda A., Helfman,D.M. and Krainer,A.R. (1993) Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell. Biol., 13, 2993–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Birney E., Kumar,S. and Krainer,A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayeda A., Munroe,S.H., Xu,R.-M. and Krainer,A.R. (1998) Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA, 4, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shamoo Y., Krueger,U., Rice,L.M., Williams,K.R. and Steitz,T.A. (1997) Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 Å resolution. Nature Struct. Biol., 4, 215–222. [DOI] [PubMed] [Google Scholar]
- 55.Xu R.M., Jokhan,L., Cheng,X., Mayeda,A. and Krainer,A.R. (1997) Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure, 5, 559–570. [DOI] [PubMed] [Google Scholar]
- 56.Ladd A.N., Charlet,N. and Cooper,T.A. (2001) The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol., 21, 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y. and Sugiura,M. (1990) Three distinct ribonucleoproteins from tobacco chloroplasts: each contains a unique amino terminal acidic domain and two ribonucleoprotein consensus motifs. EMBO J., 9, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mieszczak M., Klahre,U., Levy,J.H., Goodall,G.J. and Filipowicz,W. (1992) Multiple plant RNA binding proteins identified by PCR: expression of cDNAs encoding RNA binding proteins targeted to chloroplasts in Nicotiana plumbaginifolia. Mol. Gen. Genet., 234, 390–400. [DOI] [PubMed] [Google Scholar]
- 59.Ohta M., Sugita,M. and Sugiura,M. (1995) Three types of nuclear genes encoding chloroplast RNA-binding proteins (cp29, cp31 and cp33) are present in Arabidopsis thaliana: presence of cp31 in chloroplsts and its homologue in nuclei/cytoplasm. Plant Mol. Biol., 27, 529–539. [DOI] [PubMed] [Google Scholar]
- 60.Sugita M. and Sugiura,M. (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol., 32, 315–326. [DOI] [PubMed] [Google Scholar]
- 61.Mar Alba M. and Pages,M. (1998) Plant proteins containing the RNA-recognition motif. Trends Plant Sci., 3, 15–21. [Google Scholar]
- 62.Nakamura T., Ohta,M., Sugiura,M. and Sugita,M. (2001) Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem., 276, 147–152. [DOI] [PubMed] [Google Scholar]
- 63.Schuster G. and Gruissem,W. (1991) Chloroplst mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J., 8, 4163–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayes R., Kudla,J., Schuster,G., Gabay,L., Maliga,P. and Gruissem,W. (1996) Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J., 15, 1132–1141. [PMC free article] [PubMed] [Google Scholar]
- 65.Hirose T. and Sugiura,M. (2001) Involvement of a site-specific _trans_-acting factors and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J., 20, 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Didierjean L., Frendo,P. and Burkard,G. (1992) Stress responses in maize: sequence analysis of cDNAs encoding glycine-rich proteins. Plant Mol. Biol., 18, 847–849. [DOI] [PubMed] [Google Scholar]
- 67.Carpenter C.D., Kreps,J.A. and Simon,A.E. (1994) Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol., 104, 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunn M.A., Brown,K., Lightowlers,R. and Hughes,M.A. (1996) A low-temperature-responsive gene from barley encodes a protein with single-stranded nucleic acid-binding activity which is phosphorylated in vitro. Plant Mol. Biol., 30, 947–959. [DOI] [PubMed] [Google Scholar]
- 69.Mar Alba M., Culianez-Macia,F.A., Goday,A., Freire,M.A., Nadal,B. and Pages,M. (1994) The maize RNA-binding protein, MA16, is a nucleolar protein located in the dense fibrillar component. Plant J., 6, 825–834. [Google Scholar]
- 70.Moriguchi K., Sugita,M. and Sugiura,M. (1997) Structure and subcellular localisation of a small RNA-binding protein from tobacco. Plant J., 12, 215–221. [DOI] [PubMed] [Google Scholar]
- 71.Heintzen Ch., Nater,M., Apel,K. and Staiger,D. (1997) AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 94, 8515–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Staiger D. and Apel,K. (1999) Circadian clock-regulated expression of an RNA-binding protein in Arabidopsis: characterisation of a minimal promoter element. Mol. Gen. Genet., 261, 811–819. [DOI] [PubMed] [Google Scholar]
- 73.Hanano S., Sugita,M. and Sugiura,M. (1996) Isolation of a novel RNA-binding protein and its association with a large ribonucleoprotein particle present in the nucleoplasm of tobacco cells. Plant Mol. Biol., 31, 57–68. [DOI] [PubMed] [Google Scholar]
- 74.Derry J.M., Kerns,J.A. and Francke,U. (1995) RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum. Mol. Genet., 4, 2307–2311. [DOI] [PubMed] [Google Scholar]
- 75.Danno S., Nishiyama,H., Higashitsuji,H., Yokoi,H., Xue,J.H., Itoh,K., Matsuda,T. and Fujita,J. (1997) Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem. Biophys. Res. Commun., 236, 804–807. [DOI] [PubMed] [Google Scholar]
- 76.Nishiyama H., Itoh,K., Kaneko,Y., Kishishita,M., Yoshida,O. and Fujita,J. (1997) A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J. Cell Biol., 137, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsumoto K., Aoki,K., Dohmae,N., Takio,K. and Tsujimoto,M. (2000) CIRP2, a major cytoplasmic RNA-binding protein in Xenopus oocytes. Nucleic Acids Res., 28, 4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maruyama K., Sato,N. and Ohta,N. (1999) Conservation of structure and cold-regulation of RNA-binding proteins in cyanobacteria: probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nucleic Acids Res., 27, 2029–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Black B.E., Levesque,L., Holaska,J.M., Wood,T.C. and Paschal,B.M. (1999) Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein import. Mol. Cell. Biol., 19, 8616–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suyama M., Doerks,T., Braun,C.C., Sattler,M., Izaurralde,E. and Bork,P. (2000) Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep., 1, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parker F., Maurier,F., Delumeou,I., Duchense,M., Faucher,D., Debussche,L., Digie,A., Schweighoffer,F. and Tocque,B. (1996) A Ras-GTPase-activating protein SH3-domain-binding protein. Mol. Cell. Biol., 16, 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conti E. and Izaurralde,E. (2001) Nucleocytoplsmic transport enters the atomic age. Curr. Opin. Cell Biol., 13, 310–319. [DOI] [PubMed] [Google Scholar]
- 83.Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- 84.Stutz F., Bachi,A., Doerks,T., Braun,I.C., Seraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kataoka N., Yong,J., Kim,V.N., Velasquez,F., Perkinson,R.A., Wang,F. and Dreyfuss,G. (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell, 6, 673–682. [DOI] [PubMed] [Google Scholar]
- 86.Portman D.S., O’Connor,P. and Dreyfuss,G. (1997) YRA1, an essential Saccharomyces cerevisiae gene, encodes a novel nuclear protein with RNA annealing activity. RNA, 3, 527–537. [PMC free article] [PubMed] [Google Scholar]
- 87.Braun I.C., Herold,A., Rode,M., Conti,E. and Izaurralde,E. (2001) Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem., 276, 20536–20543. [DOI] [PubMed] [Google Scholar]
- 88.Strasser K., Bassler,J. and Hurt,E. (2000) Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol., 150, 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hecht V., Stiefel,V., Delseny,M. and Gallois,P. (1997) A new Arabidopsis nucleic-acid-binding protein gene is highly expressed in dividing cells during development. Plant Mol. Biol., 34, 119–124. [DOI] [PubMed] [Google Scholar]
- 90.Suzuki M., Kato,A. and Komeda,Y. (2000) An RNA-binding protein, AtRBP1, is expressed in actively proliferative regions in Arabidopsis thaliana. Plant Cell Physiol., 41, 282–288. [DOI] [PubMed] [Google Scholar]
- 91.Nakamura M., Okano,H., Blendy,J.A. and Montell,C. (1994) Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron, 13, 67–81. [DOI] [PubMed] [Google Scholar]
- 92.Good P., Yoda,A., Sakakibara,S., Yamamoto,A., Imai,T., Sawa,H., Ikeuchi,T., Tsuji,S., Satoh,H. and Okano,H. (1998) The human Musashi homolog 1 (MSL1) gene encoding the homolgue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cells and neural progenitor cells. Genomics, 52, 382–384. [DOI] [PubMed] [Google Scholar]
- 93.Yoda A., Sawa,H. and Okano,H. (2000) MSI-1, a neural RNA-binding protein, is involved in male mating behaviour in Caenorhabditis elegans. Genes Cells, 5, 885–895. [DOI] [PubMed] [Google Scholar]
- 94.Watanabe Y. and Yamamoto,M. (1994) S.pombe mei2 encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell, 78, 487–498. [DOI] [PubMed] [Google Scholar]
- 95.Hirayama T., Ishida,C., Kuromori,T., Obata,S., Shimoda,C., Yamamoto,M., Shinozaki,K. and Ohto,C. (1997) Functional cloning of a cDNA encoding Mei2-like protein from Arabidopsis thaliana using a fission yeast pheromone receptor deficient mutant. FEBS Lett., 413, 16–20. [DOI] [PubMed] [Google Scholar]
- 96.Ostareck-Lederer A., Ostareck,D.H. and Hentze,M.W. (1998) Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci., 23, 409–411. [DOI] [PubMed] [Google Scholar]
- 97.Gibson T.J., Thompson,J.D. and Heringa,J. (1993) The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acids. FEBS Lett., 324, 361–366. [DOI] [PubMed] [Google Scholar]
- 98.Nabel-Rosen H., Dorevitch,N., Reuveny,A. and Volk,T. (1999) The balance between two isoforms of the Drosophila RNA-binding protein how controls tendon cell differentiation. Mol. Cell, 4, 573–584. [DOI] [PubMed] [Google Scholar]
- 99.Ebersole T.A., Chen,Q., Justice,M.J. and Artzt,K. (1996) The quaking gene product necessery in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nature Genet., 12, 260–265. [DOI] [PubMed] [Google Scholar]
- 100.Wan L., Dockendorf,T.C., Jongens,T.A. and Dreyfuss,G. (2000) Characterisation of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell. Biol., 20, 8536–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu J. and Chen,X. (2000) MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G2-M. Mol. Cell. Biol., 20, 5601–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pilotte J., Larocque,D. and Richard,S. (2001) Nuclear translocation controlled by alternatively spliced isoforms inactivates the QUAKING apoptotic inducer. Genes Dev., 15, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gibson T.J., Rice,P.M., Thompson,J.D. and Heringa,J. (1993) KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem. Sci., 18, 331–333. [DOI] [PubMed] [Google Scholar]
- 104.Siomi H., Choi,M., Siomi,M.C., Nussbaum,R.L. and Dreyfuss,G. (1994) Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell, 77, 33–39. [DOI] [PubMed] [Google Scholar]
- 105.Buckanovich R.J., Posner,J.B. and Darnell,R.B. (1993) Nova, the paraneoplastic Ri antigene, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron, 11, 657–672. [DOI] [PubMed] [Google Scholar]
- 106.Buckanovich R.J., Yang,Y.Y. and Darnell,R.B. (1996) The onconeural antigene Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J. Neurosci., 16, 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Darnell R.B. (1996) Onconeural antigenes and the paraneoplastic neurologic disorders: at the intersection of cancer, immunity and the brain. Proc. Natl Acad. Sci. USA, 93, 4529–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wintersberger U., Kühne,C. and Karwan,A. (1995) Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast, 11, 929–944. [DOI] [PubMed] [Google Scholar]
- 109.Cortes A., Huertas,D., Fanti,L., Pimpinelli,S., Marsellach,F.X., Pina,B. and Azorin,F. (1999) DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J., 18, 3820–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dodson R.E. and Shapiro,D.J. (1997) Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J. Biol. Chem., 272, 12249–12252. [DOI] [PubMed] [Google Scholar]
- 111.Sasaki T., Toh,E.A. and Kikuchi,Y. (2000) Yeast krr1p physically and functionally interacts with a novel essential kri1p and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol., 20, 7971–7979.11027267 [Google Scholar]
- 112.Chan H.Y., Brogna,S. and O’Kane,C.J. (2001) Dribble, the Drosophila KRR1p homologue, is involved in rRNA processing. Mol. Biol. Cell., 12, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sanchez H., Fester,T., Kloska,S., Schroder,W. and Schuster,W. (1996) Transfer of rps19 to the nucleus involves the gain of an RNP-binding motif which may functionally replace RPS13 in Arabidopsis mitochondria. EMBO J., 15, 2138–2149. [PMC free article] [PubMed] [Google Scholar]