The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition (original) (raw)
Abstract
Site-specific acetylation of histone H4 by MOF is central to establishing the hyperactive male X chromosome in Drosophila. MOF belongs to the MYST family of histone acetyltransferases (HATs) characterized by an unusual C2HC-type zinc finger close to their HAT domains. The function of these rare zinc fingers is unknown. We found that this domain is essential for HAT activity, in addition to the established catalytic domain. MOF uses its zinc finger to contact the globular part of the nucleosome as well as the histone H4 N-terminal tail substrate. Point mutations that leave the zinc-finger structure intact nevertheless abolish its interaction with the nucleosome. Our data document a novel role of the C2HC-type finger in nucleosome binding and HAT activity.
INTRODUCTION
When it comes to their heteromorphic sex chromosomes, humans and fruit flies resemble each other. In both species, males are characterized by an X,Y genotype, whereas females have two X chromosomes. Accordingly, females have two copies of all X-chromosomal genes while males have only one. The process that assures that X-linked genes are expressed to a similar degree in both sexes is called dosage compensation. In Drosophila this is achieved by doubling the gene expression from the single male X chromosome (Baker et al., 1994; Lucchesi, 1998). Dosage compensation is vital for the fly; failure to adjust gene expression in males is lethal. The process is controlled by a set of male specific lethal (msl) genes coding for proteins and RNAs that associate with each other at many sites on the male, but not the female X chromosome (for reviews see Baker et al., 1994; Lucchesi, 1998). To date, five proteins, MSL1, MSL2, MSL3, MLE and MOF, as well as two non-coding RNAs, roX1 and roX2, are known to be required for dosage compensation (for review see Lucchesi, 1998, 1999; Stuckenholz et al., 1999).
Deeper insight into the functions of the dosage compensation complex (DCC) has come from the identification of MOF as a member of the MYST family of histone acetyltransferases (HATs) (Hilfiker et al., 1997, and references therein). MOF is an acetyltransferase with unusually narrow substrate specificity. MOF is targeted to the male X chromosome by interaction with other DCC subunits in a process that presumably involves interaction with non-coding roX RNA (Akhtar et al., 2000; Gu et al., 2000; Meller et al., 2000). Whether embedded in the MSL complex or isolated as a recombinant protein, MOF selectively acetylates lysine 16 of histone H4 from the pool of 14 lysine residues displayed on the histone N-termini of a nucleosome (Akhtar and Becker, 2000; Smith et al., 2000). Acetylation of histone H4 at lysine 16 is causally involved in the activation of transcription from chromatin templates in vivo and in vitro (Hilfiker et al., 1997; Akhtar and Becker, 2000). Histone acetylation is a central step involved in gene activation in other systems (for reviews see Grant and Berger, 1999; Brown et al., 2000; Strahl and Allis, 2000). Obviously, the modulation of chromatin structure, via site-specific histone acetylation, is key to the global activation of the X chromosome in male flies.
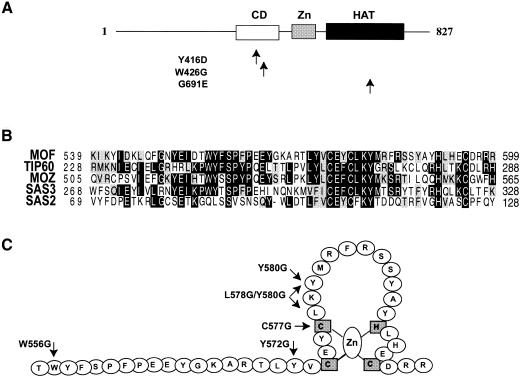
The unprecedented ability to express intact MOF in a highly active form provides a unique opportunity to understand the molecular basis for its narrow substrate specificity, which distinguishes it from other HATs (Akhtar and Becker, 2000). In addition to the HAT domain, which characterizes all HATs, so far only two other known domains have been recognized: a domain with similarity to the chromodomain (Paro and Hogness, 1991) and a putative zinc finger of the C2HC type, connected to the HAT domain by a flexible linker (Hilfiker et al., 1997). The latter feature is highly conserved among all MYST family members, with the exception of Esa1p. In order to assess the contribution of the chromo- and zinc-finger domains to chromatin binding and substrate specificity, we created MOF derivatives with point mutations in conserved residues in either the chromo- or zinc-finger domains. Surprisingly, we found that the atypical zinc finger is absolutely required for acetylation of the nucleosome.
Here we sought to study the role of the C2HC zinc finger in the context of the full-length MOF HAT protein. By site-directed mutagenesis we demonstrate that the MOF zinc-finger region is specifically required for interaction of MOF with the nucleosomes. The failure of MOF derivatives mutated in their zinc-finger domain to interact with chromatin correlates with impaired HAT activity. Chromatin binding critically involves the histone H4 N-terminal tail.
RESULTS AND DISCUSSION
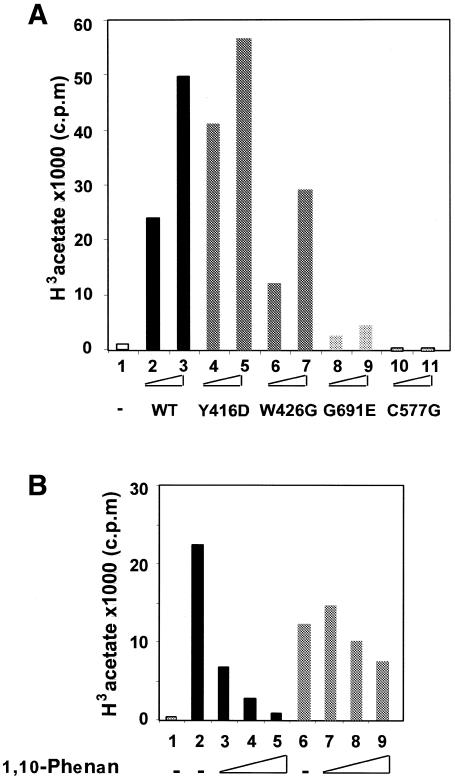
In order to assess the contribution of the chromo- and zinc-finger domains to chromatin binding and substrate specificity, we created MOF derivatives with point mutations in conserved residues in either the chromo- or zinc-finger domains. Two highly conserved residues in the chromodomain were mutated: tyrosine at position 416 was mutated to an aspartic acid (Y416D), and a tryptophan at position 426 was replaced by glycine (W426G) (Figure 1A). The corresponding residues are crucial for the silencing function of the chromodomain protein clr4 in fission yeast (Ivanova et al., 1998). In the predicted finger structure, one of the zinc-coordinating cysteines, at position 577, was replaced by a glycine (C577G; Figure 1C). Equivalent amounts of the bacterially expressed recombinant proteins, as measured by Coomassie staining (data not shown) and western blot analysis (Figure 3B), were used in a HAT assay with a mixture of Drosophila histones as substrates. As shown before, a point mutation in the HAT domain (G691E) led to the loss of HAT activity (Figure 2A, compare lanes 2 and 3 with lanes 8 and 9). Point mutations in the chromodomain had only a modest effect on acetylase activity (Figure 2A, lanes 4–7). In surprising contrast, point mutation C577G in the zinc finger abolished HAT activity entirely (Figure 2A, lanes 10 and 11). This finding is in agreement with the observation that SAS3, another HAT of the MYST family, requires the putative zinc-finger motif for acetylase activity (Takechi and Nakayama, 1999). In contrast to MOF, however, SAS3 has a relaxed specificity and is unable to acetylate nucleosomal histones (Takechi and Nakayama, 1999).
Fig. 1. Structural features of MOF. (A) Schematic representation of the known domains of MOF. CD, chromodomain; Zn, zinc finger-containing domain; HAT, histone acetyltransferase domain. Point mutations in the CD and HAT domains are indicated. (B) Alignment of the conserved sequences of the MYST family members containing the C2HC zinc finger. (C) Summary of point mutations in the zinc-finger region generated and analyzed in this study.
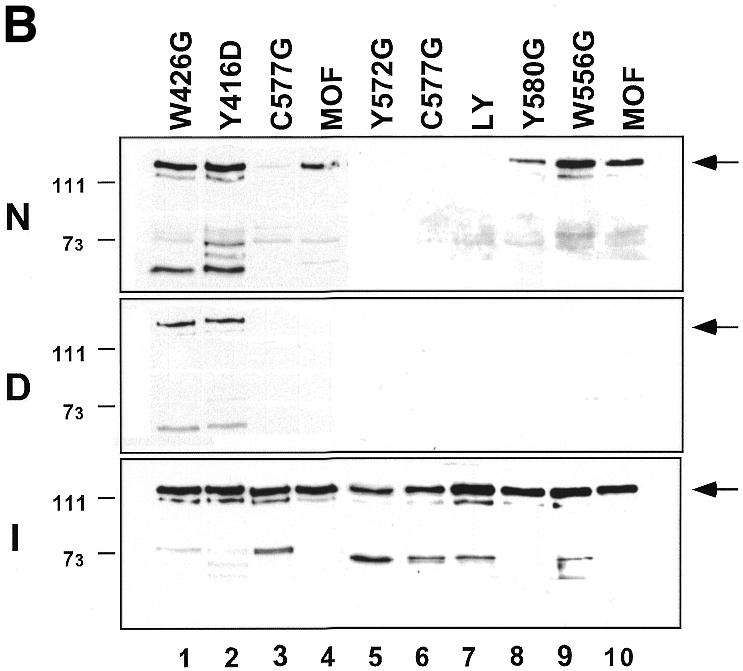
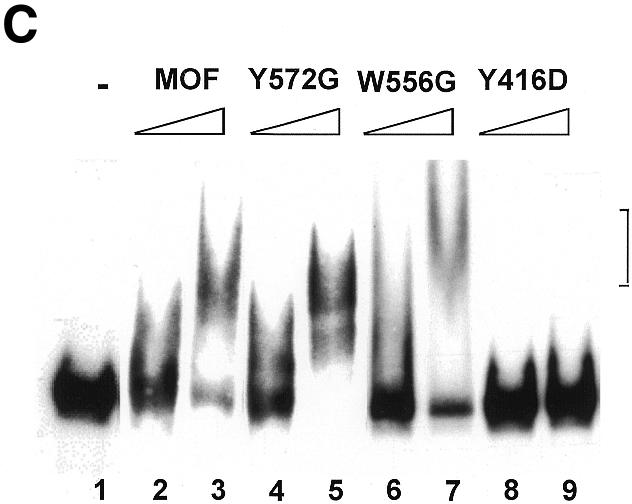
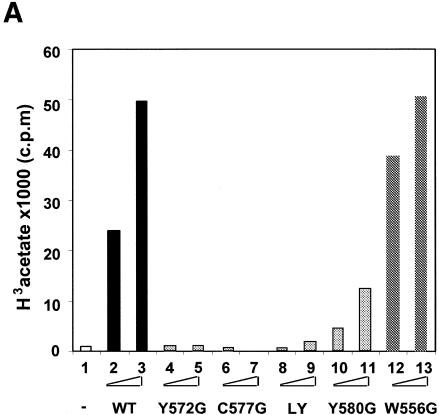
Fig. 3. The C2HC zinc finger is involved in acetyltransferase activity and chromatin binding of MOF. (A) Only mutations in or adjacent to the zinc finger affect HAT activity. See Figure 1C for an overview of the mutations. HAT assays were performed as described in Figure 2A. (B) The zinc finger, but not the chromodomain, is involved in nucleosome binding. Drosophila embryo extracts were used to assemble chromatin on linear DNA immobilized on paramagmetic beads. Three hundred nanograms of either chromatinized (N) or free DNA template (D) were used for each binding reaction with 100 ng of each MOF derivative. Stable binding was analyzed by western blot analysis. I, 20% of total input protein. Position of the molecular weight markers (in kDa) is indicated. An arrow indicates the position of full-length MOF. (C) RNA EMSA with either 150 ng (odd lanes) or 300 ng (even lanes) of MOF derivatives as indicated. Lane 1, free RNA probe.
Fig. 2. Mutations in the zinc finger impair HAT activity of MOF. (A) Point mutants in the chromodomain (Y416D, W426G), HAT domain (G691E) and zinc finger (C577G) were tested for acetylation of Drosophila histones. (B) The HAT activity of MOF (black bars) but not HAT1 (gray bars) is sensitive to the zinc chelator 1,10-phenanthroline. HAT assays contained Drosophila histones, 3H acetyl CoA and 100 ng of either MOF (lanes 2–5) or HAT1 (lanes 6–9) in the presence of 5, 10 or 15 mM 1,10-phenanthroline (lanes 3–5 and 7–9), respectively. Lane 1, control reaction without enzyme.
In order to confirm that zinc coordination by a presumed zinc-finger structure was essential for HAT activity, we employed the zinc chelating 1,10-phenanthroline. This reagent abolishes DNA-binding activity of the GAGA factor via its zinc-finger domain (Pedone et al., 1996). The addition of increasing concentrations of 1,10-phenanthroline dramatically reduced the HAT activity of MOF (Figure 2B, lanes 2–5), whereas corresponding concentrations of the solvent, ethanol, alone did not affect HAT activity (data not shown). As a further control for non-specific effects of the chelator we established that HAT1, a yeast enzyme with no zinc fingers, was insensitive to 1,10-phenanthroline (Figure 2B, lanes 6–9).
These results imply that the zinc finger is important for HAT activity. However, mutating a cysteine residue critically involved in the coordination of the zinc ion may lead to perturbation of the neighboring HAT domain due to unfolding of the finger structure. We therefore created a series of point mutations within and adjacent to the finger structure, leaving the zinc-coordinating amino acids intact. We created MOF variants W556G, Y572G, Y580G and L578G/Y580G (LY) by individually replacing tryptophan 556 (distant to the zinc finger), tyrosine 572 (immediately adjacent to the finger) and tyrosine 580 (in the conserved loop region of the finger) with glycines, and also generated a double point mutant replacing leucine 578 and tyrosine 580 with glycines (Figure 1C). Remarkably, the mutations in the zinc finger as well as the tyrosine replacement immediately adjacent to the structure severely diminished HAT activity, whereas the W556G mutation upstream of the finger had no effect (Figure 3A). HAT assays with recombinant histones containing different complements of N-terminal tails (see below) suggest that the reduced activity of the Y580G mutant is due to a general impairment of catalysis and that the substrate preference is not affected (data not shown).
While the above results firmly establish a role for the zinc finger in HAT activity, they do not address the question of whether this structure is important for substrate recognition or catalysis. To address this issue we also tested the MOF derivatives for chromatin binding. Nucleosomal arrays were assembled on linearized templates bound to paramagnetic beads using Drosophila embryo extracts (Akhtar and Becker, 2000). Following chromatin assembly, the beads were washed and incubated with equivalent amounts (Figure 3B, I) of either wild-type MOF or of several point-mutated variants. The chromatin beads were then concentrated on a magnet, again washed in buffer, and bound proteins were analyzed by western blotting. In parallel, binding of the enzymes to free DNA was tested (Figure 3B). As shown previously, the wild-type MOF preferentially binds to nucleosomal templates (compare panels N and D of Figure 3B, lanes 4 and 10). The two point mutations in the chromodomain did not affect chromatin binding, but led to modest enhancement of binding to free DNA (Figure 3B, lanes 1 and 2). In contrast, point mutating the zinc fingers essentially abolished chromatin binding (Figure 3B, lanes 3, 5–7). Importantly, chromatin binding correlated well with HAT activity: the poor but detectable binding of Y580G mutant to chromatin (Figure 3B, lane 8) corresponded to intermediate HAT activity of this enzyme (Figure 3A, lanes 10 and 11). In contrast, the derivative W556G neither affected chromatin binding nor HAT activity.
We previously showed that wild-type MOF was able to interact in vivo and in vitro with RNA via its chromodomain (Akhtar et al., 2000). In order to confirm that mutations in the zinc-finger region only affected the folding of the enzyme locally, we tested selected enzymes in the established RNA bandshift assay (Akhtar et al., 2000). Mutant Y572G, which is inactive in either chromatin binding or HAT activity, is fully capable of RNA interaction in vitro (Figure 3C), suggesting that the point mutation did not affect the structure of MOF in a general way.
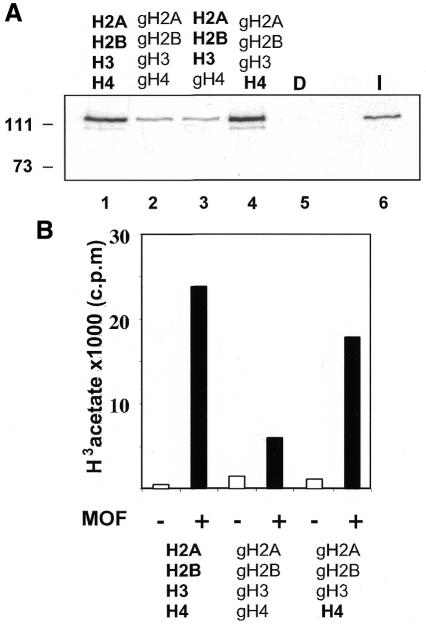
We next wished to determine the contribution of the actual acetylation target, the N-terminus of histone H4, to chromatin binding. We previously established the reconstitution of nucleosomes from recombinant histones (Luger et al., 1997) using NAP1 as a histone chaperone (Akhtar and Becker, 2000; Clapier et al., 2001). Nucleosomes were assembled on immobilized DNA from either wild-type histones (H2A/H2B/H3/H4), or only the globular histone domains (gH2A/gH2B/gH3/gH4) (Luger et al., 1997). Chimeric nucleosomes were also reconstituted, either lacking all except the H4 tail (gH2A/gH2B/gH3/H4) or containing all except the H4 N-terminus (H2A/H2B/H3/gH4). MOF was allowed to interact with these various nucleosome substrates, the chromatin beads were concentrated, washed, and interacting protein detected by western blot analysis. While MOF interacted well with intact nucleosomes, deletion of the histone N-termini, or even just of the H4 N-terminus, led to substantially reduced interaction which could nevertheless be clearly documented (Figure 4A, lanes 1–3). Remarkably, however, full binding was recovered when the nucleosomes contained just the H4 N-terminus (Figure 4A, lane 4). HAT activities measured on the histone mixes that were used for the nucleosome reconstitution corresponded well with the results of the binding assays (Figure 4B). These results, in the context of those presented in Figure 3B, point to a complex target for the zinc finger of MOF involving both the histone H4 tail as well as some other determinant on the globular part of the nucleosome. This is not surprising since lysine 16 resides rather close to the globular part of the nucleosome. The combined mutations L578G and Y580G (LY, Figure 3B) abolish both aspects of this interaction.
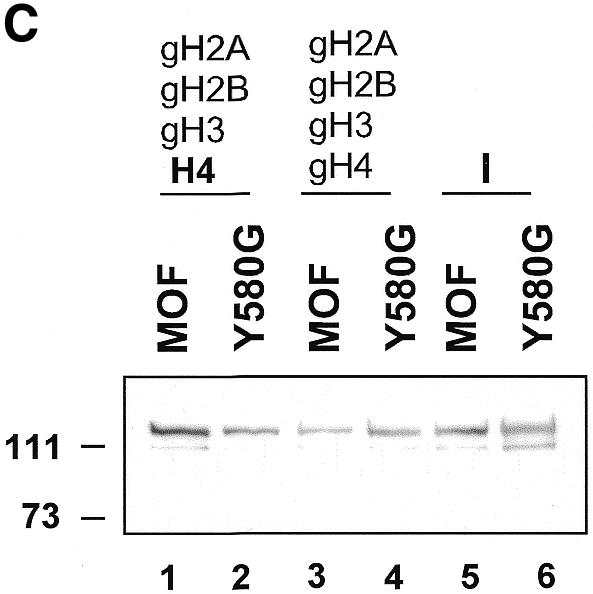
Fig. 4. Substrate recognition by MOF requires the histone H4 tail. (A) Interaction assay. Chromatin was assembled from recombinant Xenopus histones using the NAP1 histone chaperone. The histone mixes that were used to reconstitute chromatin are indicated above the lanes. Histones from which the N-terminus has been deleted (i.e. the globular domains) are indicated with the prefix ‘g’. D, free DNA; I, 20% input protein. Bound proteins were resolved by SDS–PAGE and detected by western blot analysis. (B) HAT activity in the absence (–) or presence (+) of MOF using different recombinant histone substrates, as indicated. (C) Tail-dependence of the interaction of the Y580G derivative. Binding of wild-type MOF and Y580G derivative to nucleosomes either lacking all histones tails (lanes 3 and 4) or displaying just the H4 N-terminus (lanes 1 and 2) was analyzed as in (A).
Interestingly, mutating only tyrosine 580 resulted in an intermediate binding affinity (Figure 3B). In order to explore the possibility that the H4-tail interaction had been selectively disrupted under these circumstances, we assayed the interaction of the Y580G derivative as a function of the H4 tail (Figure 4C). Nucleosomes lacking all histone tails, or containing just the H4 tails, were reconstituted on immobilized DNA and assayed for interaction of wild-type MOF or the Y580G derivative. As before, the Y580G mutation led to a reduced interaction with H4 tail-containing nucleosomes (Figure 4C, lanes 1 and 2). This effect may be underestimated due to a somewhat higher input of mutant enzyme in these assays (Figure 4C, lanes 5 and 6). Remarkably, both enzymes interacted with a similar, reduced affinity with a nucleosome lacking the H4 tail (Figure 4C, lanes 3 and 4), supporting the idea that the zinc finger in the vicinity of tyrosine 580 directly interacts with the H4-tail substrate.
Our results demonstrate that the histone H4 tail is a crucial determinant of nucleosome binding by MOF. On the other hand, we document a hitherto unknown contribution of the C2HC-type zinc finger, a hallmark of MYST family acetylases, to chromatin binding, a prerequisite for HAT activity. Significantly, the minimal catalytic domain of the HAT Gcn5p, which is unrelated to the MYST family, does not contain a zinc finger (Candau et al., 1997).
Zinc fingers of the C2HC-type are atypical and rare. So far, roles for C2HC fingers, particularly in the context of LIM domains, in contacting various other protein domains have been documented (Schmeichel and Beckerle, 1997, and references therein). In contrast to the majority of classical zinc fingers, C2HC fingers are unlikely to be involved in DNA contacts. However, interactions with RNA remain a possibility since several C2HC finger proteins are RNA-binding proteins (Arrizabalaga and Lehmann, 1999; Gorelick et al., 1999; Urbaneja et al., 1999). From their careful mutagenesis study on the C2HC finger of the MMLV nucleocapsid protein, Gorelick et al. (1999) concluded that replacing the CCHC structure with corresponding CCCC or CCHH fingers led to defects in the production of full-length reverse transcripts of the viral RNA, emphasizing the distinct functions of the CCHC structure beyond simply providing a zinc-coordinating scaffold.
While C2HC-type fingers fold as independent units (Hammarstrom et al., 1996), they frequently perform their functions in combination with other structural elements, such as C2H2-type fingers in LIM-domain proteins (Schwabe and Klug, 1994). Comparison of the features of MYST HATs to other more distantly related HATs reveals structural elements that may cooperate with the C2HC finger in substrate recognition. At the current level of understanding, the HAT domain itself may provide a complementary structural element, since conserved residues distinguish MYST family HAT domains from others (Hilfiker et al., 1997). On the other hand, the zinc finger itself is embedded in a larger element of sequence similarity between MYST HATs (see Figure 1C; Hilfiker et al., 1997). Resolution of the structural context within which this zinc finger exerts its function may significantly enhance our understanding of substrate recognition and specificity among histone acetyltransferases.
METHODS
Mutagenesis and expression of mutant proteins
A site-directed mutagenesis kit (Stratagene) was used for mutagenesis. The identity of all mutants was verified by sequencing. Proteins were expressed in bacteria as described (Akhtar and Becker, 2000).
Chromatin assembly and binding experiments
Nucleosomal arrays were assembled using Drosophila embryo extracts for 6 h on 300 ng linearized templates containing the Hsp26 gene immobilized on paramagnetic beads, as described (Akhtar and Becker, 2000). Following assembly, beads were washed twice in 500 µl of wash buffer [25 mM HEPES pH 7.6, 100 mM KCl, 1.5 mM MgCl2, 10 % glycerol, 0.025 % NP40, 0.1 mM dithiothreitol (DTT)]. For assembly of recombinant nucleosomes, 300 ng of immobilized DNA (Akhtar and Becker, 2000) were assembled into nucleosomes from recombinant Xenopus histones (Luger et al., 1997) using NAP1 as a histone chaperone (Clapier et al., 2001). Assembly was allowed to proceed at 26°C for 4 h, and followed by two washes with 500 µl of wash buffer as described above. The nucleosomal arrays used in Figure 4C were assembled by a salt step protocol (Godde et al., 1995).
Binding of MOF to nucleosomes was performed in binding buffer [50 mM Tris pH 8.0, 50 mM NaCl, 0.05 % NP40, 0.1 mM EDTA, 1 mM MgCl2, 0.1 mg/ml bovine serum albumin (BSA)] for 30 min at 26°C. This was followed by two washes with wash buffer. Beads were then resuspended in SDS loading buffer and proteins resolved on an 8% SDS–polyacrylamide gel. The proteins were then blotted onto a PVDF membrane (0.2 µM), probed with anti MOF antibody (gift from M. Kuroda) and detected with an ECL kit (Amersham).
HAT and RNA binding assays were as described (Akhtar et al., 2000).
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. Imhof for HAT1, K. Nightingale for recombinant histones, C. Clapier for NAP1 and M. Kuroda for antibody against MOF. We are also grateful to S. Kass, T. Gibson, G. Vriend and members of the laboratory for helpful discussions and critical reading of the manuscript. A.A. gratefully acknowledges the receipt of a fellowship from HFSPO.
REFERENCES
- Akhtar A. and Becker, P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyl transferase essential for dosage compensation in Drosophila. Mol. Cell, 5, 367–375. [DOI] [PubMed] [Google Scholar]
- Akhtar A., Zink, D. and Becker, P.B. (2000) Chromodomains as RNA interaction modules. Nature, 407, 405–409. [DOI] [PubMed] [Google Scholar]
- Arrizabalaga G. and Lehmann, R. (1999) A selective screen reveals discrete functional domains in Drosophila Nanos. Genetics, 153, 1825–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.S., Gorman, M. and Marin, I. (1994) Dosage compensation in Drosophila. Annu. Rev. Genet., 28, 491–521. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Lechner, T., Howe, L. and Workman, J.L. (2000). The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Candau R., Zhou, J.X., Allis, C.D. and Berger, S.L. (1997). Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C., Längst, G., Corona, D.F.V., Becker, P.B. and Nightingale, K.P. (2001). A key role for the histone H4 N-terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde J.S., Nakatani, Y. and Wolffe, A.P. (1995). The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res., 23, 4557–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R.J., Fu, W., Gagliardi, T.D., Bosche, W.J., Rein, A., Henderson, L.E. and Arthur, L.O. (1999) Characterization of the block in replication of nucleocapsid protein zinc finger mutants from moloney murine leukemia virus. J. Virol., 73, 8185–8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.A. and Berger, S.L. (1999) Histone acetyltransferase complexes. Semin. Cell Dev. Biol., 10, 169–177. [DOI] [PubMed] [Google Scholar]
- Gu W., Wei, X., Pannuti, A. and Lucchesi, J.C. (2000) Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J., 19, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarstrom A., Berndt, K.D., Sillard, R., Adermann, K. and Otting, G. (1996) Solution structure of a naturally-occurring zinc-peptide complex demonstrates that the N-terminal zinc-binding module of the Lasp-1 LIM domain is an independent folding unit. Biochemistry, 35, 12723–12732. [DOI] [PubMed] [Google Scholar]
- Hilfiker A., Hilfiker-Kleiner, D., Pannuti, A. and Lucchesi, J.C. (1997) mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J., 16, 2054–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A.V., Bonaduce, M.J., Ivanov, S.V. and Klar, A.J. (1998) The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nature Genet., 19, 192–195. [DOI] [PubMed] [Google Scholar]
- Lucchesi J.C. (1998) Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr. Opin. Genet. Dev., 8, 179–184. [DOI] [PubMed] [Google Scholar]
- Lucchesi J.C. (1999) Dosage compensation: roX marks the spot. Curr. Biol., 9, R807–R808. [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner, T.J., Flaus, A.J., Waye, M.M. and Richmond, T.J. (1997) Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol., 272, 301–311. [DOI] [PubMed] [Google Scholar]
- Meller V.H., Gordadze, P.R., Park, Y., Chu, X., Stuckenholz, C., Kelley, R.L. and Kuroda, M.I. (2000) Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr. Biol., 10, 136–143. [DOI] [PubMed] [Google Scholar]
- Paro R. and Hogness, D.S. (1991) The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedone P.V., Ghirlando, R., Clore, G.M., Gronenborn, A.M., Felsenfeld, G. and Omichinski, J.G. (1996) The single Cys2-His2 zinc finger domain of the GAGA protein flanked by basic residues is sufficient for high-affinity specific DNA binding Proc. Natl Acad. Sci. USA, 93, 2822–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeichel K.L. and Beckerle, M.C. (1997) Molecular dissection of a LIM domain. Mol. Biol. Cell, 8, 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe J.W. and Klug, A. (1994) Zinc mining for protein domains. Nature Struct. Biol., 1, 345–349. [DOI] [PubMed] [Google Scholar]
- Smith E.R., Pannuti, A., Gu, W., Steurnagel, A., Cook, R.G., Allis, C.D. and Lucchesi, J.C. (2000) The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol., 20, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis, C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Stuckenholz C., Kageyama, Y. and Kuroda, M.I. (1999) Guilt by association: non-coding RNAs, chromosome-specific proteins and dosage compensation in Drosophila. Trends Genet., 15, 454–458. [DOI] [PubMed] [Google Scholar]
- Takechi S. and Nakayama, T. (1999) Sas3 is a histone acetyltransferase and requires a zinc finger motif. Biochem. Biophys. Res. Commun., 266, 405–410. [DOI] [PubMed] [Google Scholar]
- Urbaneja M.A., Kane, B.P., Johnson, D.G., Gorelick, R.J., Henderson, L.E. and Casas-Finet, J.R. (1999) Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol., 287, 59–75. [DOI] [PubMed] [Google Scholar]