Molecular Characterization of Two-Component Systems of Helicobacter pylori (original) (raw)
Abstract
Two-component systems are frequently involved in the adaptation of bacteria to changing environmental conditions at the level of transcriptional regulation. Here we report the characterization of members of the two-component systems of the gastric pathogen Helicobacter pylori deduced from the genome sequence of strain 26695. We demonstrate that the response regulators HP166, HP1043, and HP1021 have essential functions, as disruption of the corresponding genes is lethal for the bacteria, irrespective of the fact that HP1043 and HP1021 have nonconserved substitutions in crucial amino acids of their receiver domains. An analysis of the in vitro phosphorylation properties of the two-component proteins demonstrates that HP244-HP703 and HP165-HP166 are cognate histidine kinase-response regulator pairs. Furthermore, we provide evidence that the variability of the histidine kinase HP165 caused by a poly(C) tract of variable length close to the 3′ end of open reading frame 165/164 does not interfere with the kinase activity of the transmitter domain of HP165.
Helicobacter pylori is a spiral-shaped, microaerophilic, gram-negative microorganism which colonizes the human gastric mucus. H. pylori has been identified as the major cause of chronic active gastritis and peptic ulcer disease (11, 33) and is considered a risk factor for the development of gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (28, 31).
Several factors associated with the pathogenesis of H. pylori have been characterized; these include flagella (14, 37), urease (which probably enables H. pylori to survive in the acidic environment of the stomach) (10), an adhesin binding to the Lewis b histo-blood group antigen (17), and the vacuolating cytotoxin VacA (5). Furthermore, a 40-kb pathogenicity island (PAI) named cag has been identified in a subset of strains (1, 9). Based on the presence of the cag PAI, H. pylori isolates are subdivided into two types. Type I strains, containing the cag PAI, exhibit increased virulence, as they are predominantly associated with severe gastric disease; type II strains, lacking the cag PAI, are more frequently isolated from asymptomatic carriers. It has been demonstrated that some of the proteins encoded by the cag PAI trigger severe inflammatory responses of the host (9). However, the precise function of the gene products of the cag PAI and their role in virulence remain to be elucidated.
Little information is available about the mechanisms by which H. pylori regulates the expression of its virulence factors during infection. It has been reported that the expression of cagA is increased by the exposure of H. pylori to pH 6, while the expression of ureA, encoding the urease A subunit, and picB, another gene from the cag PAI, is decreased under these conditions (18). However, the molecular mechanisms of transcriptional regulation underlying these phenomena remain unclear. Recently, it has been demonstrated that the groESL, hrcA-grpE-dnaK, and cbpA-hspR-orf operons encoding the major chaperones of H. pylori are negatively regulated by the transcriptional repressor protein HspR and that the expression of several components of the flagellar apparatus is controlled by the NtrC-like two-component response regulator protein FlgR interacting with the ς54-containing RNA polymerase (35, 36).
Two-component systems are widespread prokaryotic signal transduction devices which allow the regulation of cellular functions in response to changing environmental conditions (30). Consequently, two-component systems are frequently involved in virulence gene regulation by bacterial pathogens. Usually they are composed of a sensor protein perceiving environmental stimuli via its N-terminal input domain and a cognate response regulator. In the presence of the appropriate stimulus, the sensor protein autophosphorylates at a highly conserved histidine residue in the transmitter domain. Subsequently, the phosphate group is transferred to an aspartic acid residue in the N-terminal receiver domain of the response regulator, resulting in a conformational change and the activation of its C-terminal output domain, which frequently has DNA binding capacity.
An analysis of the H. pylori genome sequence (38) revealed the presence of few regulatory genes, including four open reading frames (ORFs) with homologies to two-component sensor ORFs and six genes encoding response regulators. Based on structural and functional homologies, one sensor-response regulator pair has been assigned to be the H. pylori CheA-CheY two-component system regulating chemotaxis (6, 38), while the remaining two-component proteins probably are involved in transcriptional regulation. It has been speculated that the paucity of regulatory functions in H. pylori, which is reflected in the small number of two-component genes, compared to 29 and 32 ORFs encoding histidine kinases and response regulators, respectively, in Escherichia coli (26), is a consequence of the tight adaptation of this pathogen to the restricted ecological niche of the human stomach and the lack of competition from other microorganisms.
In this study, we report the characterization of the H. pylori two-component systems putatively involved in transcriptional regulation by construction of isogenic mutants and by an analysis of the in vitro phosphorylation properties of the purified two-component proteins.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strains G27, G25, G46, G50, and CCUG17874 are clinical isolates and have been described previously (43). When recovered from frozen stocks, the H. pylori strains were grown under microaerophilic conditions (Oxoid) on Columbia agar plates containing 5% horse blood, 0.2% cyclodextrin, and Dent's or Skirrow's antibiotic supplement at 37°C for 2 or 3 days. After passage on fresh plates, bacteria were cultured in a 5% CO2–95% air atmosphere at 37°C. When required, the blood agar plates were supplemented with kanamycin at a final concentration of 20 μg/ml. The E. coli strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in Luria-Bertani broth. When necessary, antibiotics were added to the following final concentrations: ampicillin, 100 μg/ml; and kanamycin, 25 μg/ml.
TABLE 1.
Strains and plasmids
| Bacterial strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| Strains | ||
| H. pylori | ||
| G27 | Clinical isolate | 43 |
| G46 | Clinical isolate | 43 |
| G50 | Clinical isolate | 43 |
| G25 | Clinical isolate | 43 |
| CCUG17874 | Clinical isolate | 43 |
| G27[flgR−] | G27 with a kanamycin resistance cassette replacing the flgR (HP703) gene | 35 |
| G27/HP244::km | G27 with a kanamycin resistance cassette partially replacing the ORF for HP244 | This study |
| G27/HP165::km | G27 with a kanamycin resistance cassette replacing the ORF for HP165 | This study |
| G27/HP1364::km | G27 with a kanamycin resistance cassette replacing the ORF for HP1364 | This study |
| G27/HP1365::km | G27 with a kanamycin resistance cassette inserted into the ORF for HP1365 | This study |
| E. coli | ||
| DH5α | Strain used for high-efficiency transformation | Gibco |
| M15 | Strain used for cloning and overproducing His6 tag fusions | Qiagen |
| Plasmids | ||
| pBluescript SK | Cloning vector | Stratagene |
| pSL1180 | Cloning vector | Pharmacia |
| pGEX-3X | GST gene fusion and expression vector | Pharmacia |
| pQE-30 | Expression vector for N-terminal His6 tag cloning | Qiagen |
| pREP4 | Derivative of pACYC overexpressing LacI | Qiagen |
| pILL600 | Plasmid containing the kanamycin resistance cassette from C. coli | 21 |
| pSL-244::km | pSL1180 containing the kanamycin resistance cassette flanked by _Eco_RI/_Bam_HI and _Bam_HI/_Pst_I fragments of 541 bp and 458 bp derived from the ORFs for HP245 and HP243, respectively | This study |
| pSL-165::km | pSL1180 containing the kanamycin resistance cassette flanked by _Eco_RI/_Bam_HI and _Bam_HI/_Pst_I fragments of 463 bp and 635 bp derived from the ORFs for HP166 and HP163, respectively | This study |
| pSL-1364::km | pSL1180 containing the kanamycin resistance cassette flanked by _Eco_RI/_Bam_HI and _Bam_HI/_Pst_I fragments of 405 bp and 655 bp derived from the ORFs for HP1365 and HP1363, respectively | This study |
| pSL-166::km | pSL1180 containing the kanamycin resistance cassette flanked by _Eco_RI/_Bam_HI and _Bam_HI/_Pst_I fragments of 330 bp and 488 bp derived from the ORFs for HP166 and HP164, respectively | This study |
| pSL-1365::km | pSL1180 containing the kanamycin resistance cassette flanked by _Eco_RI/_Bam_HI and _Bam_HI/_Pst_I fragments of 304 bp and 484 bp derived from the ORFs for HP1365 and HP1364, respectively | This study |
| pSL-1043::km | pSL1180 containing the kanamycin resistance cassette flanked by _Eco_RI/_Bam_HI and _Bam_HI/_Pst_I fragments of 505 bp and 586 bp derived from noncoding DNA | This study |
| pSL-1021::km | pSL1180 containing the kanamycin resistance cassette flanked by _Eco_RI/_Bam_HI and _Bam_HI/_Pst_I fragments of 758 bp and 632 bp derived from the ORFs for HP1020 and HP1022, respectively | This study |
| pGEX-244 | pGEX-3X expressing the transmitter domain of HP244 (aa 149 to 381) fused to GST | This study |
| pGEX-165 | pGEX-3X expressing the transmitter domain of HP165 (aa 167 to 414) fused to GST | This study |
| pGEX-165(C13) | pGEX-3X expressing the transmitter domain of HP165 from H. pylori G46 fused to GST | This study |
| pGEX-165(C9) | pGEX-3X expressing the transmitter domain of HP165 from H. pylori CCUG17874 with a frameshift introduced by site-directed mutagenesis | This study |
| pGEX-1364 | pGEX-3X expressing the transmitter domain of HP1364 (aa 183 to 397) fused to GST | This study |
| pQE-166 | pQE30 expressing the His6-tagged response regulator HP166 | This study |
| pQE-1365 | pQE30 expressing the His6-tagged response regulator HP1365 | This study |
| pQE-703 | pQE30 expressing the His6-tagged response regulator HP703 | This study |
| pQE-1043 | pQE30 expressing the His6-tagged response regulator HP1043 | This study |
| pQE-1021 | pQE30 expressing the His6-tagged response regulator HP1021 | This study |
| pQE-1021R | pQE30 expressing the His6-tagged receiver domain (aa 3 to 118) of response regulator HP1021 | This study |
| pSK-165(C11) | pBluescript SK carrying a _Bam_HI/_Eco_RI fragment encoding the transmitter domain of HP165 from H. pylori CCUG17874 | This study |
General techniques.
DNA manipulations, cloning procedures, and sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) were carried out according to standard procedures. PCR amplifications were performed with a Biomed Thermocycler 60 and Deep Vent DNA polymerase (New England Biolabs). All cloned PCR products were subjected to automated sequencing (Big Dye Kit; Perkin-Elmer) to ensure proper amplification. Site-directed mutagenesis was performed with a Chameleon Double-Stranded Site-Directed Mutagenesis Kit (Stratagene).
Construction of isogenic mutants with mutations in the H. pylori ORFs encoding two-component systems by allelic exchange mutagenesis.
Transformation of H. pylori G27 was performed as described previously (6). The plasmid constructs used for transformation are derivatives of pSL1180 which carry DNA fragments flanking on the H. pylori chromosome the ORF to be inactivated, as well as a Campylobacter coli kanamycin resistance cassette inserted between these fragments. The DNA fragments that were the target sites for homologous recombination were amplified by PCR from chromosomal DNA of H. pylori G27, generating _Eco_RI/_Bam_HI and _Bam_HI/_Pst_I restriction sites at the 5′ and 3′ ends. In the resulting plasmid constructs, the coding information for the following parts of the two-component proteins has been replaced by the kanamycin resistance cassette: pSL-244::km, amino acids (aa) 108 to 381 of HP244; pSL-165::km, aa 2 to 414 of HP165; pSL-1364::km, aa 10 to 274 of HP1364; pSL-166::km, aa 110 to 181 of HP166; pSL-1365::km, aa 123 to 198 of HP1365; pSL-1043::km, aa 122 to 191 of HP1043; and pSL-1021::km, aa 9 to 296 of HP1021.
2D PAGE (pH 4 to 8) and identification of proteins by LC-mass spectrometry.
To prepare whole-cell lysates, bacteria were harvested from plates, washed with phosphate-buffered saline (PBS), and lysed by incubation in lysis buffer {35 mM Tris (pH 7.4), 9 M urea, 65 mM dithiothreitol (DTT), 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)} for 10 min at room temperature. Two-dimensional (2D) gel electrophoresis was performed according to the method of O'Farrell (29), as modified by Hochstrasser et al. (15, 16). Isoelectric focusing was carried out with tube gels (inner diameter of capillary tubes, 1.0 mm) containing 2% ampholines (Pharmalyte; Pharmacia) for 20 h by applying a voltage gradient (200 V for 2 h, 500 V for 2 h, and 800 V for 16 h) in a Protean IIxi electrophoresis chamber (Bio-Rad). Samples contained up to 200 μg of protein. After equilibration for 5 min in 120 mM Tris-HCl (pH 6.8)–2% SDS–2% β-mercaptoethanol–10% glycerol–0.0025% bromophenol blue, the tube gel was placed on top of an SDS–12% polyacrylamide gel (1 mm) together with a small agarose gel slice containing marker proteins (SDS-PAGE standard broad range; Bio-Rad) and then covered with a thin layer of agarose. Electrophoresis was carried out at a constant current of 24 mA/gel. Gels were further processed either by silver staining or staining with colloidal Coomassie blue. Analysis of protein spots by liquid chromatography (LC)-mass spectrometry was carried out as described previously (20).
Construction of plasmids expressing H. pylori two-component sensor and regulator proteins fused to GST or His6 affinity tags.
DNA fragments encoding the transmitter domains of histidine kinases HP244 (aa 149 to 381), HP165 (aa 167 to 414), and HP1364 (aa 183 to 397) were amplified from chromosomal DNA of H. pylori G27, generating _Bam_HI and _Eco_RI restriction sites at the 5′ and 3′ termini of the fragments. The fragments were ligated into _Bam_HI/_Eco_RI-digested pGEX-3X vector DNA, creating in-frame fusions to the gene encoding glutathione _S_-transferase (GST). The resulting plasmids were named pGEX-244, pGEX-165, and pGEX-1364. pGEX-165(C13), encoding a C-terminally extended transmitter domain (aa 167 to 431; see frame c in Fig. 5A), was constructed similarly by performing PCR on chromosomal DNA of H. pylori G46. pGEX-165(C9) was obtained by site-directed mutagenesis of a PCR fragment amplified from chromosomal DNA of H. pylori CCUG17874 and cloned into pBluescript SK [pSK-165(C11)]. The mutagenized fragment was subsequently ligated into the _Bam_HI/_Eco_RI-digested pGEX-3X vector.
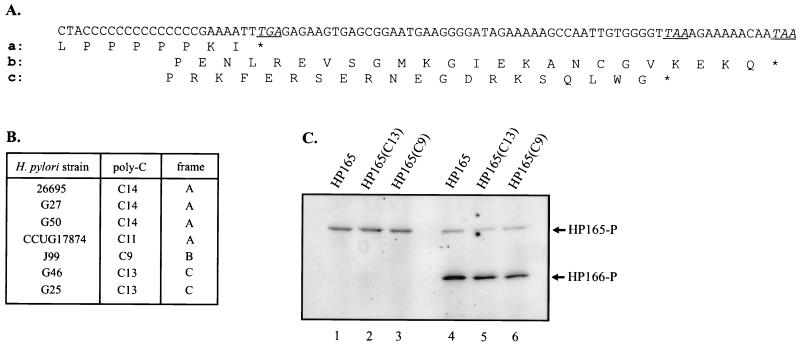
FIG. 5.
Autophosphorylation and phosphotransfer reactions of histidine kinases GST-HP165, GST-HP165(C13), and GST-HP165(C9). (A) Alignment of the different C-terminal sequences of histidine kinase HP165 due to frameshifting as a consequence of the various lengths of a poly(C) tract close to the 3′ end of the coding region. (B) Length of the poly(C) tract in ORF HP165/164 in different isolates of H. pylori. (C) Autophosphorylation of fusion proteins GST-HP165, GST-HP165(C13), and GST-HP165(C9) representing reading frames A, C, and B, respectively, in the presence of [γ-33P]ATP (lanes 1 to 3) and phosphotransfer reaction of fusion proteins GST-HP165, GST-HP165(C13), and GST-HP165(C9) in the presence of [γ-33P]ATP and the cognate response regulator HP166 (lanes 4 to 6). The positions of phosphorylated (-P) proteins are marked by arrows.
To construct plasmids pQE-166, pQE-1365, pQE-703, pQE-1021, and pQE-1043, DNA fragments encoding the response regulators HP166 (aa 3 to 225), HP1365 (aa 3 to 213), HP703 (aa 3 to 381), HP1021 (aa 3 to 298), and HP1043 (aa 3 to 223), respectively, were amplified; this process generated _Bam_HI and _Pst_I restriction sites at the 5′ and 3′ ends of the fragments to allow ligation into _Bam_HI/_Pst_I-cleaved pQE-30 vector DNA, creating an N-terminal His6 tag. For response regulator HP1021, a DNA fragment encoding the receiver domain (aa 3 to 118) also was cloned into pQE30 to yield plasmid pQE1021R.
Expression and purification of fusion proteins.
GST fusion proteins derived from the pGEX constructs were produced in E. coli DH5α. Bacteria were grown in 1 liter of Luria-Bertani broth at 37°C to an optical density at 600 nm of 0.5. Protein expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), followed by further incubation for 3 h at 30°C. The cells were harvested, washed twice with 20 ml of FP buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and frozen overnight at −20°C. The frozen bacterial pellet was resuspended in 10 ml of FP buffer, and the cells were disrupted in a French pressure cell. The lysate was cleared by centrifugation at 27,000 × g. After dilution with the same volume of PBS and the addition of Triton X-100 to a final concentration of 1%, the supernatant was loaded onto a glutathione-Sepharose 4B column (Pharmacia; bed volume, 5 ml) equilibrated with PBS. The column was washed with 75 ml of PBS, and the fusion proteins were eluted in 25 ml of 50 mM Tris-HCl (pH 8.0)–10 mM glutathione. Fractions containing the purified proteins were pooled, dialyzed against dialysis buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 20% glycerol, 1 mM DTT, 1 mM PMSF), and frozen at −80°C.
The His6-tagged response regulator proteins derived from the various pQE constructs were overproduced in E. coli M15(pREP4) and purified with Ni2+-nitrilotriacetic acid-agarose essentially as described by Perraud et al. (32). His6-HP1043 was purified from the cell lysate supernatant on an Ni2+-nitrilotriacetic acid-agarose column equilibrated with FP buffer.
In vitro phosphorylation assays.
In vitro phosphorylation assays were carried out with a final volume of 25 μl of reaction buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2, 10 μM [γ-33P]ATP [3,000 Ci/mmol]) containing histidine kinases and response regulators in equimolar concentrations (1 μM). The phosphorylation reactions were carried out for 5 min at room temperature. After the addition of sample buffer (60 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 0.05% bromophenol blue), the reaction mixtures were separated by SDS-PAGE. The gels were washed with 45% methanol–10% acetic acid and autoradiographed.
Computational analysis.
Homology comparisons were performed using version 8 of the software package of the Genetics Computer Group. The prediction of transmembrane domains was performed using the DAS program of the protein prediction server of the University of Stockholm (www.biokemi.su.se/-server /DAS/tmdas.cgi) and the Prosite program of the ExPasy proteomics tools package (www.expasy.ch/tools/#pattern).
RESULTS
Characteristics of the H. pylori two-component proteins, as deduced from the genome sequence of H. pylori 26695.
HP1364 and HP165 are orthodox histidine kinases of 397 and 414 aa, respectively; both belong to the IIIA class of sensor proteins, according to the sequences flanking the highly conserved histidine residues in the transmitter domains (13). Two short transmembrane segments are predicted in the N-terminal halves of both sensor proteins (aa 18 to 33 and aa 157 to 177 in HP1364; aa 8 to 22 and aa 134 to 153 in HP165), suggesting that histidine kinases HP1364 and HP165 are attached to the cytoplasmic membrane, with their input domains being located in the periplasm. It should be noted that in the genome sequence of H. pylori 26695, the coding information for histidine kinase HP165 is annotated as two split ORFs (designated ORF HP165/164); this characteristic is likely to be due to a sequencing error, as the corresponding ORF is continuous in the genome of H. pylori J99 (2). Sensor protein HP244, belonging to histidine kinase subclass IIIB (13), with similarity to E. coli NtrB, is predicted to be a cytoplasmic protein of 381 aa. The regulator proteins HP1365, HP166, and HP1043 are grouped into the OmpR family of response regulators, according to sequence similarities in their output domains (Fig. 1), while HP703 is an NtrC-like protein. HP1021 is a response regulator protein of 298 aa which does not show significant sequence similarity with any other two-component regulator protein in its output domain. Surprisingly, the receiver domains of the response regulator proteins HP1043 and HP1021 show severe deviations from the consensus sequence. As shown in Fig. 2, the highly conserved aspartic acid residue corresponding to position 13 in the sequence of CheY from E. coli is replaced by lysine in the receiver domain of HP1043, while the phosphate-accepting aspartic acid residue (D57 in CheY) is shifted by one position compared to the consensus sequence. In HP1021, the phosphate-accepting aspartic acid residue is replaced by serine. Therefore, it seems questionable whether these proteins require phosphorylation to exert their function.
FIG. 1.
Relationship of response regulators HP166, HP1043, and HP1365 to the output domain of OmpR of E. coli. The residues building the hydrophobic core of the OmpR output domain (23) are marked by black shading. Gaps introduced to maximize the alignment are indicated by dots. Amino acid positions are given on the right.
FIG. 2.
Alignment of the receiver domains of the H. pylori response regulators HP166, HP1365, HP703, HP1021, and HP1043 and comparison with the receiver domain consensus sequence. Gaps introduced to maximize the alignment are indicated by dots. Positions where the indicated amino acids appear with a probability of more than 90% according to a comparison of 79 two-component response regulators (39) are highlighted by black shading. Positions considered to be invariant are indicated by arrowheads.
ORFs encoding two-component response regulator proteins HP166, HP1021, and HP1043 are essential genes.
According to the genome sequence of the H. pylori strain 26695 (38), two pairs of response regulators and histidine kinases, HP1365-HP1364 and HP166-HP165, are encoded by adjacent pairs of ORFs with the same orientation of transcription, suggesting that the corresponding proteins are cognate phosphorylation partners. The remaining ORFs, encoding histidine kinase HP244 and response regulators HP703 and HP1021, belong to different operons, while the ORF encoding response regulator protein HP1043 is predicted to be transcribed as a monocistronic mRNA.
In an attempt to generate isogenic H. pylori mutants harboring gene disruptions in the two-component protein ORFs, allelic exchange mutagenesis was performed on H. pylori strain G27 by transformation with plasmid constructs carrying a kanamycin resistance cassette flanked by _H. pylori_-specific sequences (Table 1). Colonies exhibiting normal growth on blood agar plates were obtained after transformation with plasmids generating knockout mutations in the ORFs for HP244, HP165, HP1364, and HP1365. The correct replacement of these ORFs by the kanamycin resistance cassette was confirmed by PCR experiments performed on chromosomal DNA with oligonucleotide primer pairs flanking the insertion site (data not shown). The construction of a derivative of H. pylori G27 with a gene disruption of the ORF for HP703, named G27[flgR−], has been reported previously (35). Surprisingly, in several attempts, no transformants were obtained when plasmids generating knockout mutations in ORFs for HP166 and HP1043 were used, while transformation with plasmid pSL-1021::km yielded very small colonies which could not be further passaged. The failure to construct knockout mutations in ORFs for HP166, HP1043, and HP1021 cannot be due to polar effects on downstream ORFs, as HP1021 is the last gene of its operon, HP1043 is transcribed monocistronically, and the construct used to disrupt HP166 carries the kanamycin resistance cassette in the same orientation as the construct successfully used to replace ORF HP165/164, located immediately downstream of the ORF for HP166. Therefore, we conclude that the response regulator proteins HP166, HP1043, and HP1021 provide essential functions for cell growth under in vitro conditions.
Analysis of the protein expression patterns of the mutant H. pylori strains with disruptions of the ORFs for HP244, HP165, HP1364, HP1365, and HP703 by 2D SDS gel electrophoresis.
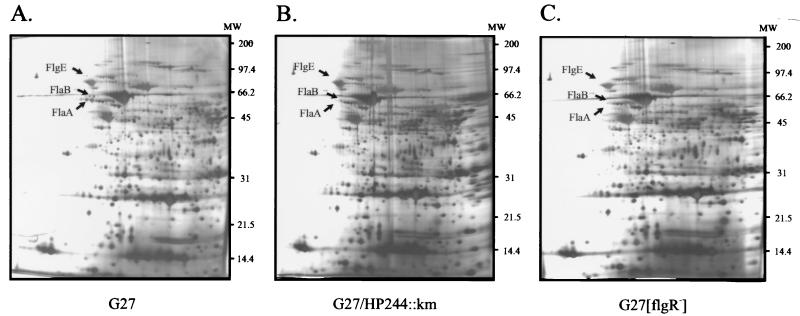
In order to identify putative target genes of the H. pylori two-component systems, whole-cell lysates of the regulatory mutants G27/HP244::km, G27/HP165::km, G27/HP1364::km, G27/HP1365::km, and G27[flgR−] were prepared, and proteins were separated by 2D gel electrophoresis using a pH gradient ranging from pH 4 to pH 8 for isoelectric focusing. For mutants G27/HP165::km, G27/HP1364::km, and G27/HP1365::km, the protein expression patterns revealed no obvious differences from the 2D protein map of the wild-type strain G27 (data not shown). However, in the 2D protein maps of G27/HP244::km and G27[flgR−], the same three protein spots were missing; these were identified by LC-mass spectrometry as the flagellin subunits FlaA and FlaB and the flagellar hook protein FlgE (Fig. 3). These data suggest that sensor protein HP244 is the cognate histidine kinase that phosphorylates response regulator HP703, named FlgR, which was previously shown to regulate the expression of several components of the H. pylori flagellar apparatus (35).
FIG. 3.
Comparison of the 2D protein maps of the H. pylori mutants G27/HP244::km (B) and G27[flgR−] (C) and the wild-type strain G27 (A). Whole-cell lysates of H. pylori were prepared as described in Materials and Methods, and 100 μg of protein extract was loaded onto the isoelectric focusing gels over a pH range from pH 4 to pH 8. The protein spots missing in the 2D maps of the mutants are indicated by arrows and were identified by LC-mass spectrometry. The positions of molecular weight (MW) standards (in thousands) are given on the right.
HP244-HP703 and HP165-HP166 are cognate histidine kinase-response regulator pairs.
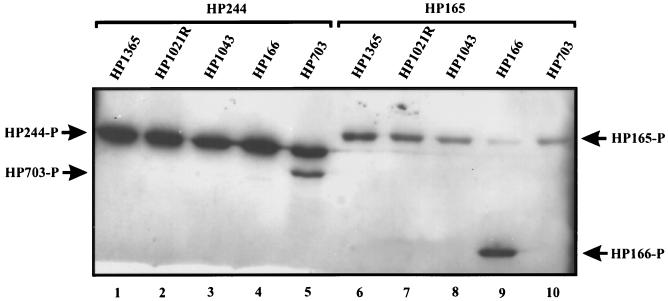
To identify cognate phosphorylation partners, the in vitro phosphorylation properties of the H. pylori two-component proteins were investigated. As deletion of the input domain generally results in a constitutively active histidine kinase in vitro, the transmitter domains of the orthodox sensor proteins HP244, HP165, and HP1364 were fused to GST, yielding fusion proteins of 52, 53, and 50 kDa, respectively. These proteins were then purified by affinity chromatography. The response regulators were overexpressed and purified as N-terminal His6 fusions (His6-HP166, 25 kDa; His6-HP1365, 24 kDa; His6-HP703, 42.5 kDa; His6-HP1034, 25 kDa; His6-HP1021, 33 kDa). As His6-HP1021 was completely insoluble, the receiver domain of HP1021 (aa 3 to 118) was separately expressed as an N-terminal His6 fusion, yielding a soluble protein of 13 kDa which was further analyzed in place of the full-length response regulator. It has been shown previously that the separated receiver domain derived from a response regulator harbors the catalytic activity required for the phosphotransfer reaction with its cognate histidine kinase (19). Incubation with [γ-33P]ATP resulted in the autophosphorylation of histidine kinases GST-HP244 and GST-HP165 (data not shown; see also Fig. 5C). Surprisingly, GST-HP1364 did not show histidine kinase activity, as no autophosphorylation of that fusion protein could be detected, irrespective of the pH of the reaction buffer (ranging from pH 6.0 to 9.0) or the presence of divalent cations other than Mg2+ (data not shown). When combined with one of the five purified response regulators in individual phosphorylation reactions, the histidine kinases GST-HP244 and GST-HP165 were able to transfer the phosphate group exclusively to a single response regulator, i.e., His6-HP703 for GST-HP244 and His6-HP166 for GST-HP165; these results demonstrate that HP244-HP703 and HP165-HP166 constitute two-component systems and show the high specificity of these sensor proteins for their cognate phosphorylation partners (Fig. 4). It should be noted that none of the response regulator proteins is phosphorylated in the presence of [γ-33P]ATP alone (data not shown).
FIG. 4.
Autophosphorylation and phosphotransfer reactions of histidine kinases GST-HP244 and GST-HP165. GST-HP244 (lanes 1 to 5) and GST-HP165 (lanes 6 to 10) were incubated in individual reactions with [γ-33P]ATP and the respective response regulators indicated above the lanes. For HP1021, the purified receiver domain, indicated as HP1021R, was used in the phosphorylation assay. The positions of the phosphorylated (-P) proteins are marked by arrows.
Sensor protein HP165 is subject to sequence variation, which does not affect its kinase activity in vitro.
ORF HP165/164 of H. pylori 26695, encoding histidine kinase HP165, harbors a stretch of 14 C nucleotides close to its 3′ end; this stretch is also present in the corresponding gene of H. pylori G27. In 1999, Alm et al. (2) reported the complete genomic sequence of the unrelated H. pylori isolate J99 and observed that ORF JHP151, encoding the histidine kinase corresponding to HP165, contains a stretch of only nine C nucleotides, generating a frameshift which results in a sensor protein with 21 additional C-terminal amino acids (Fig. 5A). Therefore, these authors suggested that the different reading frames might represent the on or off status of the protein, which is regulated by slipped-strand mispairing. To test this hypothesis, the 3′ half of the histidine kinase gene encoding the transmitter domain was PCR amplified from chromosomal DNAs of the H. pylori strains CCUG17874, G46, G50, and G25. Sequencing of the cloned PCR fragments revealed the presence of a stretch of 11 C nucleotides in the histidine kinase gene in CCUG17874, 13 C nucleotides in G46 and G25, and 14 C nucleotides in G50, confirming the variability of the poly(C) tract and suggesting the expression of C-terminal variants of the corresponding two-component sensor proteins. To test whether histidine kinase activity is affected in the variant sensor proteins, the PCR fragment obtained from H. pylori G46 was cloned into the expression vector pGEX-3X, and the resulting fusion protein, GST-HP165(C13), was purified. As the ORF present in H. pylori J99 was not represented in the different H. pylori strains analyzed here, the corresponding histidine kinase gene fragment was constructed by site-directed mutagenesis of the cloned PCR product derived from H. pylori CCUG17874 and subsequently cloned into pGEX-3X to yield fusion protein GST-HP165(C9). As shown in Fig. 5, incubation with [γ-33P]ATP either alone or in combination with the cognate response regulator His6-HP166 resulted in similar autophosphorylation of the fusion proteins derived from the three different ORFs as well as transfer of phosphate to the response regulator. Therefore, we conclude that the C-terminal protein sequence does not interfere with the histidine kinase activity of the transmitter domain or with the phosphotransfer reaction with the cognate response regulator.
DISCUSSION
Bacterial two-component systems control a variety of physiological processes in response to certain environmental conditions. H. pylori harbors a remarkably small number of these signal transduction systems, i.e., four histidine kinases with their cognate response regulators, including the regulatory system for chemotaxis (CheA and CheY), as well as two orphan response regulators. This classification of the H. pylori two-component proteins as cognate sensor-response regulator pairs is based on convincing sequence similarities to well-known systems (HP392, CheA/HP1067, CheY), the tandem organization of the corresponding genes (HP1365-HP1364), or the direct analysis of phosphotransfer reactions between purified proteins (HP244-HP703 and HP165-HP166).
Surprisingly, the genes encoding response regulators HP166, HP1043, and HP1021 could not be inactivated by the insertion of a kanamycin resistance cassette, indicating that the corresponding regulator proteins exert some function which is essential for cell viability. Few essential two-component systems have been described so far; they include a multicomponent signal transduction pathway in the dimorphic bacterium Caulobacter crescentus, required for cell cycle regulation (34, 42), and the yycFG system in Bacillus subtilis and Staphylococcus aureus. The cellular processes regulated by these latter systems remain unknown (12, 22).
The regulator proteins HP1043 and HP1021 are unique with respect to the sequences of their receiver domains (Fig. 2). In HP1043, the conserved aspartic acid residue corresponding to position 13 in the consensus sequence is substituted by lysine. Two other response regulators have been described to harbor the Asp13→Lys mutation, i.e., FrzG from Myxococcus xanthus, which is homologous to the E. coli CheB protein (24), and FlbD from C. crescentus, an NtrC-like molecule which is involved in flagellar gene expression (41). However, both FrzG and FlbD contain additional mutations at conserved sites in the receiver domains, i.e., glycine in place of the conserved threonine or serine residue at position 87 in the consensus sequence and leucine in place of the highly conserved lysine residue at position 109; in HP1043, both positions correspond perfectly to the consensus sequence.
For the chemotaxis response regulator CheY of E. coli, an Asp13→Lys mutation renders the protein active in vivo irrespective of the presence or absence of the cognate histidine kinase CheA (8). Therefore, it was speculated that a CheY molecule harboring this mutation can adopt an active conformation in the absence of phosphorylation. In accordance with this hypothesis, it has been shown that FlbD can activate the transcription of its target promoters in the nonphosphorylated state in vitro (7). However, a cognate histidine kinase, FlbE, which is able to phosphorylate FlbD and which is responsible for the temporal and spatial regulation of FlbD-dependent genes was identified (40). At the moment, we cannot rule out the possibility that histidine kinase HP1364, belonging to the IIIA class of sensor proteins, which interact with OmpR-like response regulators, acts as a phosphor donor for HP1043, as the phosphorylation properties of this histidine kinase could not be investigated directly.
The essential protein HP1021, to our knowledge, is the first response regulator with a serine residue substituted for the phosphate-accepting aspartic acid residue (position 57 in the consensus sequence). Recently, it was reported that an Asp57→Asn mutant of E. coli CheY can use Ser56 as an alternative phosphorylation site (4). Although the rate of phosphotransfer is much lower than that in the wild-type CheY protein, this finding demonstrates that a two-component histidine kinase providing a high-energy phosphoramidate can serve as a phosphate source to generate serine phosphate in the response regulator. Another prokaryotic protein exhibiting serine protein kinase activity is the anti-sigma factor SpoIIAB of B. subtilis, which shows some homology to histidine kinases. This protein contains subdomains N, G1, F, and G2 of the transmitter domain, lacks the H domain (harboring the phosphorylated histidine residue), and catalyzes the phosphorylation of the anti-anti-sigma factor SpoIIAA on a serine residue (25). However, SpoIIAA does not bear any homology to two-component response regulator proteins. It should be noted that the recently published genome sequence of the related pathogen Campylobacter jejuni (Sanger Centre) encodes orphan response regulators exhibiting 72 and 54% similarities to HP1043 and HP1021, respectively. While the ortholog of HP1021 also contains a serine residue in place of the phosphate-accepting aspartic acid residue, the protein homologous to HP1043 corresponds perfectly to the receiver consensus sequence.
Although deletion of the input domain generally results in the constitutive active phenotype of a histidine kinase in vitro, fusion protein GST-HP1364 did not autophosphorylate under the applied experimental conditions. In HP1364, the phosphorylated histidine residue is located at a distance of only 22 aa from the predicted second membrane-spanning segment confining the putative input domain, indicating the presence of a very short linker region. Therefore, it is conceivable that the lack of autophosphorylation of GST-HP1364 is not due to the indispensability of the input domain but to a misfolding of the transmitter domain as a consequence of the choice of an improper fusion to GST. However, this notion could not be tested experimentally, as fusion proteins harboring C-terminal parts of the input domain including the putative transmembrane segment could not be overexpressed in E. coli (data not shown). Although the phosphotransfer reactions of HP1364 could not be analyzed directly, considering the tandem organization of the ORFs for HP1365 and HP1364 and the fact that HP1365 could not be phosphorylated by the other histidine kinases, we suppose that HP1364 is the cognate histidine kinase that phosphorylates the response regulator HP1365. For histidine kinases HP244 and HP165, it could be demonstrated that these proteins exclusively phosphorylate their cognate response regulators, i.e., HP703 and HP166, respectively (Fig. 4).
A comparison of the 2D protein expression patterns of regulatory H. pylori mutants with the wild-type 2D map is a suitable approach for the identification of regulated target genes but is hampered by several technical limitations: (i) proteins which are not abundant in the cell cannot easily be detected by this technique, and (ii) regulated proteins with a pI beyond pH 4 to 8 are beyond the resolution capacity of the isoelectric focusing gels. Furthermore, it must be considered that in vitro growth conditions may not provide the environmental stimuli necessary for the activation of the sensor proteins under investigation.
So far, regulated target genes could be identified only for the HP244-HP703 two-component system. It has been reported previously that HP703, named FlgR, by its interaction with the ς54-containing RNA polymerase, positively regulates the expression of several components of the flagellar apparatus (35). This report has been confirmed by an analysis of the protein expression patterns of the H. pylori mutants G27/HP244::km and G27[flgR−], which demonstrated the lack of expression of flagellins FlaA and FlaB and the flagellar hook protein FlgE in these mutants (Fig. 3). Although the expression of the flaA gene is derepressed in an flgR mutant at the transcriptional level (35), the FlaA protein cannot be detected in this mutant, suggesting the rapid degradation of FlaA when the other components of the flagella are not produced. It should be noted that the gene encoding histidine kinase HP244 is cotranscribed with flgI, encoding a component of the flagellar basal body; however, this operon is not preceded by a ς54-dependent promoter, arguing against an autoregulatory mechanism in the expression of HP244. As HP244 is supposed to be a cytoplasmic protein, it is tempting to speculate that additional sensory transmembrane proteins interacting with HP244 might be involved in signal perception. It will be interesting to identify the signals triggering flagellar gene expression, as motility is considered essential for H. pylori virulence.
Due to a stretch of C nucleotides of variable lengths, histidine kinase HP165 is expressed with various C-terminal sequences in different H. pylori strains (Fig. 5A). Introduction of translational frameshifts due to changes in the lengths of such polynucleotide tracts, generating inactive truncated gene products, and restoration of the reading frames and the activities of the gene products in subsequent cycles of replication represent a mechanism well known as a cause of phase variation in pathogenic bacteria. Recently, phase variation affecting the structure of lipopolysaccharide due to the changing lengths of a poly(C) tract in the α3-fucosyltransferase genes has also been demonstrated to occur in H. pylori (3). However, in histidine kinase HP165, only the C terminus of the protein is affected by a frameshift due to the variable lengths of the poly(C) tract in the corresponding gene; in vitro phosphorylation experiments have demonstrated that neither the kinase activity of HP165 nor its ability to serve as the phosphate donor for response regulator HP166 is altered by the different C-terminal sequences (Fig. 5C). If HP165 also harbors phosphatase activity for the phosphorylated form of its cognate response regulator, which is frequently the case for two-component sensor proteins, then that activity also should be unaffected in the three different gene products of ORF HP165/164, as the ratio of phosphorylated histidine kinase to response regulator remains unchanged in all cases (Fig. 5C). We cannot rule out the possibility that in vivo, the C-terminal part of HP165 interacts with the linker sequence connecting the input and transmitter domains and thereby interferes negatively with the signal transduction process, causing an altered phenotype of the sequence variants as a consequence of the lack of regulation of target genes. An example of phase variation affecting a two-component histidine kinase is provided by the BvgS protein, which regulates virulence gene expression in Bordetella spp. However, in this case, inactivation of the protein is an irreversible process due to the spontaneous occurrence of small deletions in the coding sequence (27).
ACKNOWLEDGMENTS
G. Spohn and V. Scarlato are acknowledged for providing the H. pylori mutant G27[flgR−]. We thank R. Gross, B. Kimmel, and V. Scarlato for critically reading the manuscript.
D.B. is the recipient of a fellowship from the Deutsche Krebsforschungszentrum. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (BE 1543/2-1).
REFERENCES
- 1.Akopyants N S, Clifton S W, Kersulyte D, Crabtree J E, Youree B E, Reece C A, Bukanov N O, Drazek E S, Roe B A, Berg D E. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Appelmelk B J, Martin S L, Monteiro M A, Clayton C A, McColm A A, Zheng P, Verboom T, Maaskant J J, van den Eijnden D H, Hokke C H, Perry M B, Vandenbroucke-Grauls C M J E, Kusters J G. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in α3-fucosyltransferase genes. Infect Immun. 1999;67:5361–5366. doi: 10.1128/iai.67.10.5361-5366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appleby J L, Bourret R B. Activation of CheY mutant D57N by phosphorylation at an alternative site, Ser-56. Mol Microbiol. 1999;34:915–925. doi: 10.1046/j.1365-2958.1999.01653.x. [DOI] [PubMed] [Google Scholar]
- 5.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 6.Beier D, Spohn G, Rappuoli R, Scarlato V. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J Bacteriol. 1997;179:4676–4683. doi: 10.1128/jb.179.15.4676-4683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson A K, Ramakrishnan G, Ohta N, Feng J, Ninfa A, Newton A. The Caulobacter crescentus FlbD protein acts at ftr sequence elements both to activate and repress transcription of cell cycle regulated flagellar genes. Proc Natl Acad Sci USA. 1994;91:4989–4993. doi: 10.1073/pnas.91.11.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourret R B, Drake S K, Chervitz S A, Simon M I, Falke J J. Activation of the phosphosignaling protein CheY. Analysis of the activated mutants by 19F NMR and protein engineering. J Biol Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- 9.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cussac V, Ferrero R L, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick J D. Helicobacter (Campylobacter) pylori: a twist on an old disease. Annu Rev Microbiol. 1990;108:70–90. doi: 10.1146/annurev.mi.44.100190.001341. [DOI] [PubMed] [Google Scholar]
- 12.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabret C, Feher V A, Hoch J A. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas R, Meyer T F, van Putten J P M. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using shuttle mutagenesis. Mol Microbiol. 1993;8:753–760. doi: 10.1111/j.1365-2958.1993.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 15.Hochstrasser D F, Augsburger V, Funk M, Appel R, Pelegrini C, Müller A F. Immobilized pH gradients in capillary tubes and two-dimensional gel electrophoresis. Electrophoresis. 1986;7:505–511. [Google Scholar]
- 16.Hochstrasser D F, Harrington M G, Hochstrasser A C, Miller M J, Merril C R. Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal Biochem. 1988;173:424–435. doi: 10.1016/0003-2697(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 17.Ilver D, Arnqvist A, Ögren J, Frick I-M, Kersulyte D, Incecik E T, Berg D E, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 18.Karita M, Tummuru M R, Wirth H-P, Blaser M J. Effect of growth phase and acid shock on Helicobacter pylori cagA expression. Infect Immun. 1996;64:4501–4507. doi: 10.1128/iai.64.11.4501-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimmel B, Bosserhoff A, Frank R, Gross R, Goebel W, Beier D. Identification of immunodominant antigens from Helicobacter pylori and evaluation of their reactivity with different patient sera. Infect Immun. 2000;68:915–920. doi: 10.1128/iai.68.2.915-920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labigne-Roussel A, Courcoux P, Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988;170:1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin P K, Li T, Sun D, Biek D P, Schmid M B. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J Bacteriol. 1999;181:3666–3673. doi: 10.1128/jb.181.12.3666-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 24.McCleary W, McBride M, Zusman D. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990;172:4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Min K T, Hilditch C M, Diederich B, Errington J, Yudkin M D. Sigma F, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 26.Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 27.Monack D M, Arico B, Rappuoli R, Falkow S. Phase variants of Bordetella bronchiseptica arise by spontaneous deletions in the vir locus. Mol Microbiol. 1989;3:1719–1728. doi: 10.1111/j.1365-2958.1989.tb00157.x. [DOI] [PubMed] [Google Scholar]
- 28.Nomura A, Stemmermann G N, Chyou P-H, Kato I, Perez-Perez G I, Blaser M J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 29.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 30.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 31.Parsonnet J, Hansen S, Rodriguez L, Gelb A B, Warnke R A, Jellum E, Orentreich N, Vogelman J H, Friedman G D. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 32.Perraud A-L, Kimmel B, Weiss V, Gross R. Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol Microbiol. 1998;27:875–887. doi: 10.1046/j.1365-2958.1998.00716.x. [DOI] [PubMed] [Google Scholar]
- 33.Peterson W L. Helicobacter pylori and peptic ulcer disease. N Engl J Med. 1991;324:1043–1048. doi: 10.1056/NEJM199104113241507. [DOI] [PubMed] [Google Scholar]
- 34.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 35.Spohn G, Scarlato V. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J Bacteriol. 1999;181:593–599. doi: 10.1128/jb.181.2.593-599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spohn G, Scarlato V. The autoregulatory HspR repressor protein governs chaperone gene transcription in Helicobacter pylori. Mol Microbiol. 1999;34:663–674. doi: 10.1046/j.1365-2958.1999.01625.x. [DOI] [PubMed] [Google Scholar]
- 37.Suerbaum S, Josenhans C, Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol. 1993;175:3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 39.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 40.Wingrove J A, Gober J W. Identification of an asymmetrically localized sensor histidine kinase responsible for temporally and spatially regulated transcription. Science. 1996;274:597–601. doi: 10.1126/science.274.5287.597. [DOI] [PubMed] [Google Scholar]
- 41.Wu J, Benson A K, Newton A. Global regulation of a ς54-dependent flagellar gene family in Caulobacter crescentus by the transcriptional activator FlbD. J Bacteriol. 1995;177:3241–3250. doi: 10.1128/jb.177.11.3241-3250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Ohta N, Newton A. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiang Z, Censini S, Bayelli P F, Telford J L, Figura N, Rappuoli R, Covacci A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63:94–98. doi: 10.1128/iai.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]