The Epstein-Barr Virus BZLF1 Protein Interacts Physically and Functionally with the Histone Acetylase CREB-Binding Protein (original) (raw)
Abstract
The Epstein-Barr virus (EBV) immediate-early protein BZLF1 (Z) is a key regulator of the EBV latent-to-lytic switch. Z is a transcriptional activator which induces EBV early gene expression. We demonstrate here that Z interacts with CREB-binding protein (CBP), a histone acetylase and transcriptional coactivator. This interaction requires the amino-terminal region of CBP as well as the transactivation and leucine zipper domains of Z. We show that CBP enhances Z-mediated transactivation of EBV early promoters, in reporter gene assays and in the context of the endogenous genome. We also demonstrate that Z decreases CREB transactivation function and that this inhibitory effect is reversed by overexpression of CBP. We show that Z also interacts directly with CREB. However, mutational analysis indicates that Z inhibition of CREB activity requires the direct interaction between Z and CBP but not the direct interaction between Z and CREB. We propose that Z interacts with CBP to enhance viral early gene transcription. In addition, the Z-CBP interaction may control host cellular transcription factor activity through competition for limiting amounts of cellular CBP.
Epstein-Barr virus (EBV) is a human herpesvirus that has infected a large majority of the world’s population. EBV is responsible for the onset of infectious mononucleosis and has been found in association with a number of cancers, including Burkitt’s lymphoma and nasopharyngeal carcinoma (52, 68). EBV infects primarily two cell types: epithelial cells, where it exists in a lytic state, and B cells, where it persists in a latent state (33, 41, 52, 57). Periodically, the virus of a latently infected B cell can undergo lytic replication. This disruption of latency is characterized by a cascade of viral gene expression: immediate-early gene expression, followed by early gene expression, and then late gene expression (33).
The immediate-early gene BZLF1 encodes a transcriptional activator, Z, that plays a key role in the disruption of EBV viral latency (8, 11, 60). Z transcriptionally activates its target genes by binding to Z-responsive elements (ZREs) which are similar to AP1 sites and are present in many of the EBV early gene promoters (7, 16, 42, 51, 54), as well as in the promoters of the two immediate-early genes, BZLF1 and BRLF1 (21, 49). Z is also required for lytic replication of the virus and binds to the origin of lytic replication, oriLyt (7, 17, 18, 54).
Z is a 245-amino-acid protein that contains specific domains for transactivation, DNA binding, and protein-protein dimerization (20, 36, 49). Z is a member of the bZIP family of proteins and shares homology within the DNA-binding domain to the c-Fos and c-Jun proteins (7, 16, 21). The Z protein has been shown to interact with its cellular environment by associating with a number of cellular proteins. Our laboratory has shown that Z can interact directly with p53 and the NF-κB subunit p65 (27, 67). It is likely that Z alters the functions of these proteins in order to promote viral replication.
CREB-binding protein (CBP) is a transcriptional regulator by virtue of its histone acetylase activity (5). The association of histones and DNA becomes weakened by histone acetylation, leading to altered nucleosomal conformation and stability (48). This chromatin alteration may then facilitate transcription by the transcriptional machinery (39, 63). CBP and the highly homologous protein p300 have been shown to interact with several cellular proteins, including p53, p65, c-Jun, c-Myb, Ets-1, and NFAT1, as well as viral proteins such as adenovirus E1A, simian virus 40 (SV40) T antigen, cytomegalovirus (CMV) IE2, herpes simplex virus VP16, and human immunodeficiency virus type 1 Tat (4, 12–14, 23, 24, 28, 43, 55, 56, 62, 65).
Since Z is a potent transcriptional activator, it may target histone acetylase activity to the early viral promoters, either by interacting with CBP directly or by possessing an intrinsic histone acetylase activity. Here we demonstrate that Z and CBP physically interact both in vivo and in vitro. We have mapped the regions of this interaction, for both Z and CBP, and show that Z and CBP interact in a functional manner to activate EBV early gene expression. We have also discovered that Z inhibits the transcriptional activation function of the transcription factor CREB. We propose that Z may inhibit CREB function by competing for limiting amounts of CBP.
MATERIALS AND METHODS
Cell lines.
DG75, an EBV-negative Burkitt’s lymphoma cell line, was maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. HeLa is a cervical carcinoma cell line; D98/HE-R-1 is an EBV-positive epithelial cell line formed by the fusion of a HeLa subclone (D98) with the EBV-positive Burkitt’s lymphoma cell line P3HR/1. Epithelial cell lines were maintained in Dulbecco’s modified Eagle’s medium H supplemented with 10% fetal calf serum.
Adenovirus construction and infection.
The BZLF1 cDNA was cloned into a shuttle vector (under the control of the CMV promoter) which contains a Lox P site, the left adenovirus terminal repeat, and a packaging signal. This vector was recombined (in a cell line expressing the phage P1 Cre protein) into the Lox P site of an adenovirus lacking the E1 and E3 genes, as well as lacking a packaging sequence, to create adenovirus-Z. A control vector containing the lacZ gene (adenovirus-LacZ) was made in the same manner.
HeLa cells were plated at a cell density of 3 × 106 cells per 150-mm-diameter plate. Cells were infected with no adenovirus (mock), adenovirus-LacZ, or adenovirus-Z at a multiplicity of infection of 50. The cells were harvested 24 h postinfection.
Plasmids.
EApBS-CAT contains the early EBV BMRF1 promoter sequences from −331 to +1 linked to the chloramphenicol acetyltransferase (CAT) gene (51). BHRF1-CAT contains a 1,020-bp _Nae_I-_Hin_cII fragment of the early BHRF1 promoter (EBV positions 52800 to 53819) (27). GAL4-E1B-CAT (gift of Michael Green) contains five copies of the GAL4 DNA-binding site upstream of the E1B minimal TATA element and the CAT gene (44). GAL4-CREB (gift of Michael Green) contains the GAL4 DNA-binding domain fused to the CREB cDNA in the SG424 vector (53). Zp-CRE-CAT contains three copies of the CRE site from the Z promoter (TGACATCA) fused to the E1B minimal TATA element and the CAT gene.
The Z expression vector contains the BZLF1 cDNA, downstream of the CMV immediate-early promoter, in the pHD1013 vector (51). The ZE2ter expression vector has a deletion of the Z carboxy-terminal amino acids 228 to 245, downstream of the SV40 immediate-early promoter, in the pKSV vector (gift of Alain Sergeant). The Z311 expression vector contains a mutation in the DNA-binding domain of Z in which amino acid 185 is altered (abolishing DNA binding), downstream of the CMV immediate-early promoter, in the pHD1013 vector (25, 31). Z-CT (previously referred to as RAZΔR), contains amino acids 2 to 86 deleted (gift of Joseph Pagano) (22). ZΔ200 contains a mutation of amino acid 200 of Z from tyrosine to glutamic acid, downstream of the CMV immediate-early promoter, in the pHD1013 vector. The CBP expression vector contains the CBP cDNA, downstream of the CMV immediate-early promoter (gift of Michael Rosenfeld) (29). CBP-HAT(−) contains mutations of amino acids 1689 and 1690 of CBP from leucine-cysteine to lysine-leucine, downstream of the CMV immediate-early promoter (gift of Jenny Ting). The CREB expression vector contains the CREB gene, downstream of the Rous sarcoma virus promoter (gift of Michael Green; from Marc Montminy). The protein kinase A (PKA) expression vector contains the gene for the catalytic subunit of PKA, downstream of the mouse metallothionein 1 promoter (gift of Michael Green; from Stanley McKnight).
The Z cDNA plasmid contains the BZLF1 cDNA in the pSP64 (Promega) vector (a gift from Paul Farrell) (16). The Z cDNA was also inserted into the pBluescript (pBS; Stratagene) vector so that it could be cut with _Hin_cII to make a carboxy-terminal truncation of Z (referred to as ZΔter). ZΔLZ (from Alain Sergeant) was cut out of the pKSV vector with _Eco_RI and cloned into pBS. Z-NT contains amino acids 1 to 140 of Z. Z-CT is RAZΔR in the pBS vector (22). ZΔ200 contains a mutation at amino acid 200 of tyrosine to glutamic acid, in the pSP64 vector. ZΔ214/218 contains mutations at amino acids 214 and 218 (both leucine to serine), in the pSP64 vector.
pGEX-3X-Z contains the Z cDNA fused in frame to the glutathione _S_-transferase (GST) gene in the pGEX-3X (Pharmacia) vector (51). CBP was subcloned as five pieces into the pGEX vector (fragments 1 to 721, 706 to 1009, 1069 to 1459, 1459 to 1891, and 1892 to 2441) (gifts of Michael Rosenfeld) (29). pGEX-CREB contains the CREB cDNA fused in frame to GST in the pGEX1 vector (gift of Michael Green). pGEX-3X was used as the control vector.
DNA purification.
Plasmid DNA was purified through Qiagen columns as described by the manufacturer.
DNA transfection.
DNA (5 to 10 μg) was transfected into cells by electroporation with a Zapper electroporation unit (Medical Electronics Shop, University of Wisconsin) at 1,500 V as described elsewhere (61). All cells were resuspended in RPMI 1640 medium prior to electroporation.
CAT assays.
Cell extracts were prepared 48 h posttransfection and incubated at 37°C with [14C]chloramphenicol in the presence of acetyl coenzyme A as described elsewhere (26). The percent acetylation of chloramphenicol was quantitated by thin-layer chromatography followed by PhosphorImager screening (Molecular Dynamics).
Immunoblot analysis.
Immunoblot analysis was performed for the detection of the Z, retinoblastoma (Rb), BMRF1 (early-antigen diffuse; EAD), and CREB proteins as follows. Briefly, 10 to 100 μg of protein was loaded in each lane, and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed. The proteins were transferred overnight onto nitrocellulose (Protran), blocked in 1× phosphate-buffered saline (PBS)–5% milk–0.1% Tween 20, and incubated in primary antibody for 1 h at room temperature (BZ1 [1:40; from Alan Rickinson] [66]; anti-Rb [1:400; PharMingen]; anti-EBV [EAD; 1:100; Capricorn] anti-CREB [1:500; Santa Cruz Biotechnology]). The membrane was washed in PBS–0.1% Tween 20, incubated in secondary antibody for 1 h at room temperature (goat anti-mouse kappa-horseradish peroxidase [1:2,000; Southern Biotechnology]; goat anti-rabbit conjugated to horseradish peroxidase [1:10,000; Promega]) and washed, and the results were visualized with an ECL (enhanced chemiluminescence) kit (Amersham) according to the manufacturer’s instructions.
Z protein expression.
The Z cDNA plasmid in SP64 was linearized with _Eco_RI and transcribed with SP6 polymerase, using a Riboprobe System II kit from Promega. The resulting RNA was translated in the presence of [35S]methionine, using the rabbit reticulocyte lysate system (Promega). The Z cDNA plasmid in SP64 was also cut with _Nhe_I to make Z-NT. The Z cDNA in pBS was cut with _Hin_cII and prepared as described above to yield a carboxy-terminal deletion of Z. ZΔLZ was transcribed uncut, while Z-CT was cut with _Sal_I and prepared as for Z. ZΔ200 and ZΔ214/218 were both linearized with _Eco_RI and transcribed with SP6. Protein quantities were normalized by running aliquots of each protein on an SDS-polyacrylamide gel, followed by autoradiography.
GST, GST-Z, GST-CREB, and GST-CBP proteins were induced, sonicated, and centrifuged. Protein quantities were normalized by affinity purifying aliquots of the proteins, followed by SDS-PAGE and Coomassie staining.
Immunoprecipitation.
HeLa cells were infected with no adenovirus (mock), adenovirus-LacZ, or adenovirus-Z. Cells were harvested after 24 h and resuspended in buffer 7 (20 mM HEPES [pH 7.7], 25 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 0.05% Nonidet P-40, protease inhibitors), sonicated, and centrifuged. Anti-CBP antibody (Santa Cruz), anti-CREB antibody (Santa Cruz), or rabbit serum (Sigma) was added to 10 to 100 μg of protein and incubated for 1 h at 4°C. The reaction mixtures were then incubated with protein A-Sepharose beads for 1 h at 4°C. The beads were washed three times with buffer 7 and loaded onto an SDS-polyacrylamide gel. Ten micrograms of crude extract was loaded onto the gel for a control. Immunoblot analysis was performed with either an anti-Z antibody or an anti-CREB antibody.
Affinity chromatography.
The GST fusion proteins were incubated with glutathione-agarose beads for 10 min at room temperature. The beads were washed three times with PBS, resuspended in buffer 7 or buffer CBP (25 mM HEPES [pH 7.3], 100 mM sodium chloride, 5 mM magnesium chloride, 100 mM EDTA, 0.2 mg of bovine serum albumin per ml, 0.1% Tween 20, protease inhibitors), and incubated with 2.5 to 10 μl of in vitro-translated protein for 1 h at room temperature. The beads were washed five to six times with buffer and loaded onto an SDS-polyacrylamide gel. After electrophoresis, the gel was fixed in 50% methanol plus 10% acetic acid, enhanced in 1 M sodium salicylate, and exposed to X-ray film.
RESULTS
Z interacts with CBP in vivo.
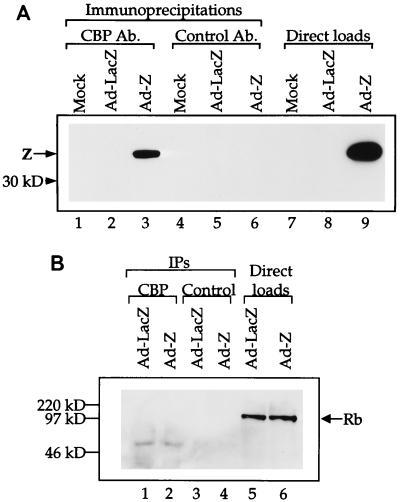
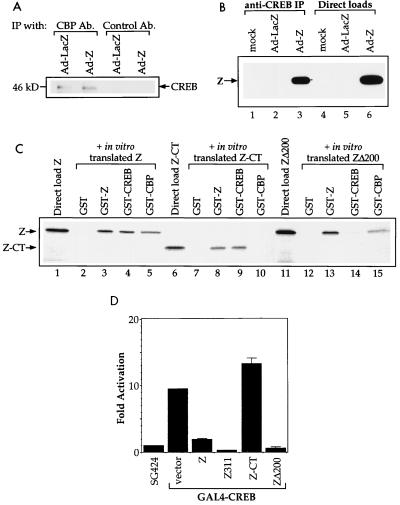
Since several transcriptional activators have recently been shown to interact with CBP and/or p300 to facilitate the opening of chromatin, we investigated whether Z, a transcriptional activator of the EBV early genes, can physically interact with CBP. CBP was immunoprecipitated from extracts of HeLa cells that had been mock infected, infected with a replication-deficient adenovirus expressing the lacZ gene (as a control), or infected with a replication-deficient adenovirus expressing the Z gene. Following the CBP immunoprecipitation, we performed immunoblot analysis with an anti-Z antibody (Fig. 1A). The results clearly demonstrate that the anti-CBP antibody coimmunoprecipitates Z from the adenovirus-Z-infected extract. A control antibody (rabbit serum) did not coimmunoprecipitate Z. Therefore, Z and CBP physically interact in vivo. As a negative control, we attempted to coimmunoprecipitate the Rb protein from the same extracts (Fig. 1B). Rb has been shown to interact with histone deacetylase activity (5a), and therefore we speculated that it would not interact with CBP. We did not coimmunoprecipitate CBP and Rb, demonstrating that our procedure does not allow for nonspecific coimmunoprecipitations.
FIG. 1.
Z and CBP associate in vivo. (A) Anti-CBP antibody and a control antibody (Ab.) (rabbit serum) were used to coimmunoprecipitate Z from mock-, adenovirus-LacZ-, or adenovirus-Z-infected HeLa cell extracts (100 μg) (lanes 1 to 6). Direct loads (10 μg) of each extract were used to confirm the presence of Z (lanes 7 to 9). Western blot analysis was performed with an anti-Z antibody. (B) Anti-CBP antibody and a control antibody (rabbit serum) were used to coimmunoprecipitate Rb from adenovirus-LacZ- or adenovirus-Z-infected HeLa cell extracts (100 μg) (lanes 1 to 4). Direct loads (20 μg) of each extract were used to confirm the presence of Rb (lanes 5 and 6). Western blot analysis was performed with an anti-Rb antibody.
Z interacts with the first 721 amino acids of CBP.
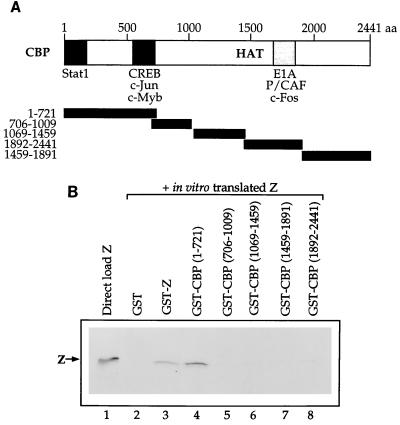
To examine which region of CBP is responsible for the CBP-Z interaction, we performed affinity chromatography experiments with five consecutive segments of CBP (as depicted in Fig. 2A) fused to GST and in vitro-translated Z protein. The in vitro-translated Z protein did not bind to GST alone but did homodimerize with GST-Z, as expected (Fig. 2B). Z bound to the GST-CBP protein containing the first 721 amino acids of CBP but not to the other regions of CBP. The amino terminus of the CBP protein has also been shown to interact with several other transcriptional activators, including CREB, c-Jun, and p65 (4, 9, 24).
FIG. 2.
Z binds to the amino-terminal portion of CBP. (A) Schematic of the CBP protein. The regions previously reported to bind various proteins are indicated. Below the CBP map are the five segments of CBP that have been fused to GST. (B) Affinity chromatography experiments were performed in which 10 μl of in vitro-translated Z was incubated with GST alone, GST-Z, or the GST-CBP segments (amino acids 1 to 721, 706 to 1009, 1069 to 1459, 1459 to 1891, and 1892 to 2441) bound to glutathione-agarose beads. Lane 1 shows the direct load of in vitro-translated Z protein (10 μl).
Several domains of Z are required for association with CBP.
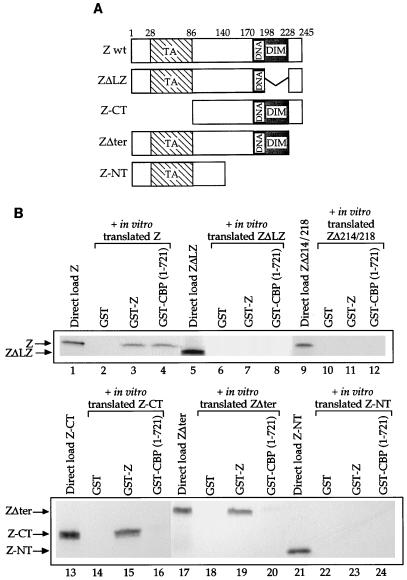
To determine which regions of Z are necessary for the Z-CBP association, we performed affinity chromatography experiments with in vitro-translated wild-type Z and Z mutants, as depicted in Fig. 3A, and GST-CBP(1-721). The results indicated that a large portion of the Z protein is necessary for the Z-CBP interaction. The leucine zipper of Z is required for interaction with CBP (as well as for Z homodimerization) (Fig. 3B, lanes 5 to 8). Furthermore, a mutant form of Z that is incapable of homodimerization (ZΔ214/218) is unable to associate with CBP (Fig. 3B, lanes 9 to 12), suggesting that the Z/CBP interaction requires Z dimerization. However, the carboxy-terminal half of Z (Z-CT) is insufficient for the CBP interaction (Fig. 3B, lanes 13 to 16). The amino-terminal half (containing the transcriptional activation domain) (Z-NT) is also required, although not in itself sufficient, for the CBP interaction (Fig. 3B, lanes 21 to 24). In addition, a mutant form of Z lacking the carboxy-terminal 17 amino acids cannot interact efficiently with CBP (Fig. 3B, lanes 17 to 20). Finally, two mutant forms of Z containing mutations in the DNA binding domain, Z311 (altering residue 185 from alanine to lysine) and Z(S186A), were still able to associate with CBP (data not shown). The finding that both the amino- and carboxy-terminal domains of Z are essential for interaction with CBP suggests that a certain conformation of Z may be required for this interaction or that Z contacts two distinct regions (within the amino terminus) of CBP.
FIG. 3.
The Z-CBP association requires several regions of the Z protein. (A) Schematics of the wild-type Z protein (Z wt) and the various Z mutants used in this study. The transactivator (TA), dimerization (DIM), and DNA binding domains are indicated. (B) Aliquots of 5 μl of in vitro-translated wild-type Z protein (lanes 2 to 4), 10 μl of in vitro-translated ZΔLZ (lanes 6 to 8), 5 μl of in vitro-translated ZΔ214/218 (lanes 10 to 12), 5 μl of in vitro-translated Z-CT (lanes 14 to 16), 5 μl of in vitro-translated ZΔter (lanes 18 to 20), or 5 μl of in vitro-translated Z-NT (lanes 22 to 24) were incubated with GST alone, GST-Z, or GST-CBP(1-721). Lanes 1, 5, 9, 13, 17, and 21 contain the direct loads for each protein (3 μl of Z, 10 μl of ZΔLZ, 5 μl of ZΔ214/218, 5 μl of Z-CT, 5 μl of ZΔter, and 3 μl of Z-NT).
Z synergizes with CBP to activate the EBV early genes BMRF1 and BHRF1.
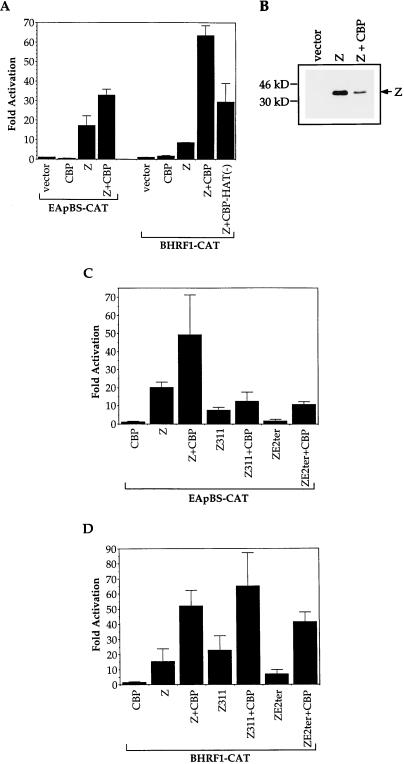
CBP enhances transcriptional activation by acting as a histone acetylase and destabilizing nucleosome structure. The disruption of chromatin organization is thought to aid in the access of the transcriptional machinery to DNA. Since we found that Z and CBP physically interact, we next examined whether CBP enhanced the ability of Z to activate EBV early gene promoters. In transient reporter gene assays, CBP enhanced the Z-mediated activation of the EBV early gene promoters, BMRF1 and BHRF1 (Fig. 4A). The increase in Z-induced activation of the BMRF1 and BHRF1 promoters in the presence of CBP was not due to an increased level of Z protein made by the CMV-Z expression construct (Fig. 4B).
FIG. 4.
CBP enhances Z-mediated activation of EBV early gene promoters. (A) Transient reporter assays were performed in which DG75 cells were transfected with 5 μg of each promoter construct (EApBS-CAT or BHRF1-CAT) and 1 μg of Z expression plasmid with or without 2 μg of CBP expression plasmid or CBP-HAT(−) expression plasmid (for a total of 8 μg DNA). CAT assays were performed as described in the text. The results are a combination of at least two separate experiments. (B) Extracts (containing either vector alone, Z, or Z plus CBP) from one of the CAT assays above were analyzed for Z protein levels. Western blot analysis was performed with an anti-Z antibody. (C and D) Transient reporter assays were performed in which DG75 cells were transfected with 5 μg of each promoter construct (EApBS-CAT [C] or BHRF1-CAT [D]) and 1 μg of Z, Z311, or ZE2ter expression plasmid with or without 2 μg of CBP expression plasmid (for a total of 8 μg DNA). CAT assays were performed as described in the text. The results are a combination of two separate experiments.
To examine whether the histone acetylase function of CBP is required for the CBP-mediated augmentation of Z transactivator function, we performed reporter gene assays with a CBP expression plasmid that contained two point mutations within the histone acetyltransferase (HAT) domain, abolishing HAT activity (35a). CBP-HAT(−) was still able to increase Z transactivator function to some extent, although it was significantly less active than wild-type CBP (Fig. 4A). This ability of CBP-HAT(−) to partially activate Z function may reflect the interaction between CBP and P/CAF, which is another histone acetylase (35a).
We also examined the ability of CBP to enhance the transactivator activity of mutant forms of Z (those which retain transactivator activity). As shown in Fig. 4C and D, in the absence of exogenous CBP, a mutant form of Z, ZE2ter (which lacks amino acids 228 to 245 and did not interact efficiently with CBP in vitro [Fig. 3]), activated the BHRF1 and BMRF1 promoters inefficiently compared to wild-type Z. However, in the presence of exogenous CBP, ZE2ter became a much more efficient transcriptional activator, suggesting that this mutant has decreased, but not absent, ability to interact with CBP in vivo. Z311, which is unable to directly bind at least some ZREs (25), activated the BMRF1 promoter to a low level compared to wild-type Z, and the addition of CBP enhanced activation by Z311 only slightly. However, Z311 unexpectedly activated the BHRF1 promoter as well as wild-type Z, and the addition of exogenous CBP greatly enhanced activation of the promoter by Z311. Z311 may directly bind to the BHRF1 promoter in vivo or, alternatively, may activate BHRF1 in an indirect manner, as it activates the Z promoter (20a).
CBP increases the ability of Z to disrupt viral latency.
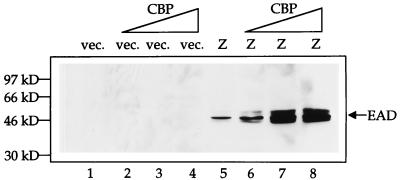
To examine the effect of CBP on Z-mediated transactivation of promoters from the endogenous viral genome, we transfected either vector DNA or Z expression plasmid with either 0, 2, 4, or 6 μg of CBP expression plasmid (Fig. 5) and then performed immunoblot analysis with an anti-BMRF1 (EAD) antibody. While CBP expression alone had no effect on viral reactivation, increasing amounts of CBP cotransfected with Z led to increasing amounts of EAD production. These data demonstrate that the Z-CBP interaction is functionally relevant in vivo, in the context of the intact viral genome, and that CBP greatly increases the efficiency of Z transactivation function.
FIG. 5.
CBP enhances Z-mediated transactivation of BMRF1 from the endogenous viral genome. D98/HE-R-1 cells were transfected with either 2 μg of vector (vec.; lanes 1 to 4) or 2 μg of Z expression plasmid (lanes 5 to 8), along with increasing amounts (0, 2, 4, and 6 μg) of CBP expression plasmid. Cells were harvested 14 h posttransfection. Western blot analysis was performed with an anti-EAD antibody.
Z inhibits CREB transactivator function.
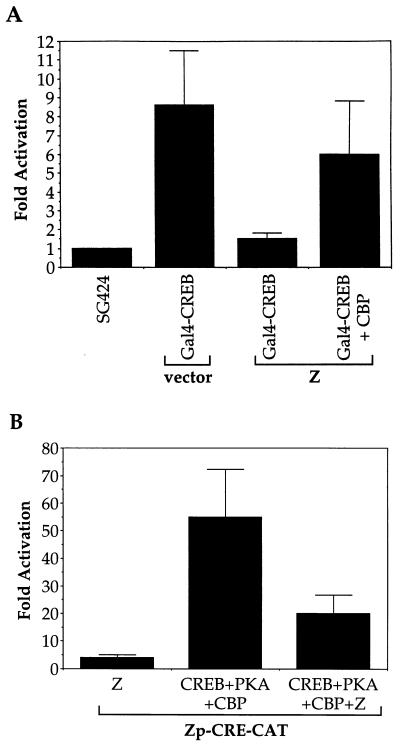
CREB is a member of the CREB/ATF family of transcriptional activators and associates with CBP in order to activate its transcriptional target promoters (9, 37). The level of CBP/p300 in cells is limiting, such that other viral proteins (including adenovirus E1A), which interact efficiently with CBP or p300, have been shown to inhibit cellular transactivator function by competing for CBP or p300 (2, 3, 40, 50, 59). Therefore, we examined the effect of Z on CREB transactivator function. As shown in Fig. 6A, Z suppressed CREB transcriptional activity in reporter gene assays. The inhibition of CREB’s activity by Z could be at least partially reversed by increasing cellular CBP levels (Fig. 6A).
FIG. 6.
Z inhibits CREB activity. (A) Transient reporter assays were performed in which DG75 cells were transfected with 5 μg of GAL4-E1B-CAT, 1 μg each of SG424 (GAL4 DNA-binding domain alone) or GAL4-CREB, and 1 μg each of control vector or Z expression plasmid with or without 1 μg of CBP expression plasmid (8 μg of total DNA). CAT assays were performed as described in the text. The results are a combination of two separate experiments. (B) DG75 cells were transfected with 5 μg of Zp-CRE-CAT and 1 μg each (or a combination) of control vector, CREB, PKA, Z, or CBP expression plasmid (9 μg in total). CAT assays were performed as described in the text. The results are a combination of two separate experiments.
The Z promoter contains a CRE/ATF motif (referred to as the ZII site) which is important for activation of Z transcription (20b) and which has been previously shown to bind CREB, ATF1, and ATF2 (6, 45, 64). CREB (which requires phosphorylation by PKA [64a] and the presence of CBP for transcriptional activation function) has been reported to activate the Z promoter through this site (reference 19, our laboratory, and unpublished data). Therefore, we examined the effect of Z on CREB’s ability to activate a promoter construct containing three copies of the ZII site linked to the minimal adenovirus E1B promoter and the CAT reporter gene. As shown in Fig. 6B, in the presence of PKA and CBP, CREB efficiently activated the construct containing the ZII motif but did not activate a control construct containing the minimal E1B promoter alone (data not shown). Furthermore, Z inhibited the ability of CREB to activate the ZII motif.
Z interacts directly with CREB.
The finding that exogenous CBP expression at least partially reversed the inhibitory effect of Z on CREB function (Fig. 6A) suggests that Z and CREB compete for limiting amounts of cellular CBP. Therefore, we examined the efficiency of the CREB-CBP interaction in the presence and absence of Z. Unexpectedly, the level of coimmunoprecipitated CREB-CBP protein complexes was not decreased in cells infected with adenovirus-Z (Fig. 7A). Nevertheless, it is possible that Z and CREB bind to different sites in CBP (although both bind to regions in the first 721 amino acids of CBP) and that both Z and CREB can bind to CBP simultaneously.
FIG. 7.
Z interacts directly with CREB. (A) Anti-CBP antibody (Ab.) and a control antibody (rabbit serum) were used to coimmunoprecipitate (Ip) CREB from adenovirus-LacZ- or adenovirus-Z-infected HeLa cell extracts (10 μg). Western blot analysis was performed with an anti-CREB antibody. (B) Anti-CREB antibody was used to coimmunoprecipitate Z from mock-, adenovirus-LacZ-, or adenovirus-Z-infected HeLa cell extracts (100 μg) (lanes 1 to 3). Direct loads (10 μg) of each extract were used to confirm the presence of Z (lanes 4 to 6). Western blot analysis was performed with an anti-Z antibody. The results from the control antibody used in this experiment are presented in Fig. 1. (C) Aliquots of 4 μl of in vitro-translated wild-type Z protein (lanes 2 to 5), 5 μl of in vitro-translated Z-CT protein (lanes 7 to 10), or 5 μl of in vitro-translated ZΔ200 protein (lanes 12 to 15) were incubated with GST alone, GST-Z, GST-CREB, or GST-CBP(1-721). The direct loads (lanes 1, 6, and 11) contain 4 μl of Z, 5 μl of Z-CT, or 5 μl of ZΔ200. (D) DG75 cells were transfected with 5 μg of GAL4-E1B-CAT, 1 μg each of SG424 (GAL4 DNA-binding domain alone) or GAL4-CREB, and 1 μg each of vector, Z, Z311, Z-CT, or ZΔ200 expression plasmid. CAT assays were performed as described in the text. The results are a combination of two separate experiments. Fold activation was calculated by taking the ratio of GAL4-CREB activity to SG424 activity in the presence of each Z construct.
However, coimmunoprecipitation analysis of adenovirus Z-infected HeLa cells suggested that Z and CREB (both bZip proteins) also interacts directly (Fig. 7B). This interaction (as well as the Z-CBP interaction) also occurred with bacterially synthesized proteins (data not shown), excluding the possibility that the Z-CREB or Z-CBP interactions require the presence of all three proteins. To distinguish if Z inhibition of CREB transactivator function is due to the Z-CBP or the Z-CREB interaction, we compared the regions of Z required for each interaction. The amino-terminal domain of Z, which is required for interaction with CBP (Fig. 3B) but not CREB (Fig. 7C, lanes 6 to 10), is essential for Z inhibition of CREB transactivator function (Fig. 7D). Furthermore, amino acid 200, within the leucine zipper of Z, is required for interaction with CREB but not CBP (Fig. 7C, lanes 11 to 15) and is not required for Z inhibition of CREB transactivator function (Fig. 7D). These results (in addition to the finding that CBP reverses the Z inhibitory effect) suggest that the Z-CBP interaction, and not the Z-CREB interaction, causes the inhibition of CREB function.
DISCUSSION
CBP is a histone acetylase and functions as a coactivator for several cellular transcriptional activators, including p53 and p65 (24, 56). CBP is also harnessed by viral proteins, such as adenovirus E1A and SV40 T antigen, to promote viral gene transactivation (13, 14, 43). Since the level of CBP in each cell is limiting, the harboring of CBP by viral proteins can inhibit certain cellular transcription factors, including p53, MyoD, c-Fos, and c-Myb, that require CBP for their function (3, 10, 50, 59). Inhibition of cellular differentiation by E1A is mediated (in part) by titration of limiting amounts of CBP/p300 (34, 47). Therefore, competition for cellular CBP/p300 may be an important mechanism by which DNA viruses regulate the host cell environment.
Z is a critical regulatory protein of EBV which mediates the switch from latent to lytic infection. In addition, Z functions as the origin-binding protein of oriLyt (17, 18, 54) and thus is a required replicative protein. Here we have demonstrated that the EBV immediate-early Z protein engages CBP as a coactivator. Z may also associate with the closely related protein, p300, although we have not examined this interaction. Z physically associates with CBP in vivo and in vitro, as shown by coimmunoprecipitation and affinity chromatography studies, through the region comprising the first 721 amino acids of CBP. This region of CBP has also been shown to bind CREB, c-Jun, p65, and c-Myb (4, 9, 12, 24).
It appears that at least two different domains of the Z protein are required for the Z-CBP association in vitro. Previous reports indicate that many transcriptional proteins, including p53, p65, c-Jun, and c-Myb, interact with CBP through their transactivation domains (4, 12, 24, 56). Consistent with this, deletion of the Z transactivation domain (amino acids 1 to 86) completely abrogates the Z-CBP interaction. However, the leucine zipper domain of Z is also clearly required for the interaction with CBP in vitro. Z homodimerization may be required for the transactivation domain to interact with CBP. Alternatively, deletion of the leucine zipper may alter Z conformation such that the Z-CBP association cannot occur.
Although our in vitro data suggest that the carboxy terminus of Z is also required for efficient Z-CBP interaction, our in vivo data indicate that a Z mutant (ZE2ter) missing the carboxy terminus is coactivated by exogenous CBP even more efficiently than wild-type Z. It is possible that a cellular protein stabilizes the interaction between Z and CBP in vivo, such that the ZE2ter mutant can interact with CBP more efficiently in vivo. Alternatively, the low transactivation function of the ZE2ter mutant in the absence of exogenous CBP may reflect its relatively poor affinity for CBP, such that it is unable to compete for limiting amounts of CBP in the normal host cell environment but can function as well as wild-type Z when CBP is supplied exogenously.
The interaction between Z and CBP serves to increase Z-induced transactivation of lytic EBV promoters. We have demonstrated here that increased levels of CBP enhance Z-mediated transcriptional activation of two EBV early gene promoters, BMRF1 and BHRF1, in reporter gene assays and that CBP enhances Z-mediated activation of BMRF1 from the endogenous viral genome. By physically associating with CBP, Z presumably tethers CBP to EBV early gene promoters containing Z-binding sites. CBP-mediated acetylation of histones would then aid in transcriptional activation of EBV early promoters. Interestingly, the BHRF1 promoter appears to be more dependent on the Z-CBP interaction than the BMRF1 promoter. Therefore, the exact number and positioning of the ZRE sites may influence the degree Z-CBP synergy. A histone acetylase-deficient mutant of CBP partially retains the ability to enhance Z-mediated transcription. This may be explained by the fact that CBP interacts with another histone acetylase, P/CAF, and that the activity of P/CAF may contribute to the enhancement of Z function observed with CBP (35a).
In addition to the role of the Z-CBP interaction for inducing early gene transcription, this interaction could potentially be important for Z’s role as the oriLyt origin-binding protein. Origin-binding proteins may function to open chromatin, allowing formation of the replication complex. The interaction between Z and CBP may thus be required for histone acetylation near oriLyt ZRE sites, thereby allowing access of the replication machinery. In addition, CBP has been recently shown to localize within promyelocytic leukemia protein (PML)-containing nuclear bodies (38). The ability of herpes simplex virus and CMV immediate-early proteins to localize within and subsequently disperse PML bodies appears to be important for lytic replication (1, 15, 30, 35, 46). Assuming that lytic EBV replication likewise requires modulation of PML-associated nuclear bodies, the Z-CBP interaction could potentially provide a mechanism for disrupting the PML-CBP interaction.
Although the Z-CBP interaction is likely required for efficient transactivation of EBV early promoters, it may also be important for regulating host cell transcription factors. Our results clearly indicate that Z inhibits CREB transactivation function in GAL4 reporter gene assays. This inhibition is alleviated by the overexpression of CBP; therefore it is likely that Z competes with CREB for CBP. These results are in contrast with the effect of the CMV immediate-early IE2 protein, which utilizes its interaction with CBP to enhance CREB-mediated transactivation (55), but are in accordance with the effect of the adenovirus E1A protein, which has been shown to decrease CREB activity (2). The inhibition of CREB by Z requires the amino terminus of Z, consistent with our in vitro data showing that the CBP-Z interaction involves this region.
The interpretation that Z inhibits CREB function by competing for limiting amounts of CBP may be overly simplistic, in that the total number of CREB-CBP complexes was not decreased in the presence of Z (Fig. 7A). Furthermore, Z and CREB also directly interact. Nevertheless, our mutational analysis suggests that Z inhibition of CREB function requires its ability to interact directly with CBP but not CREB. One explanation for these results is that Z also interacts with p300 through the same domains required for its interaction with CBP and that the Z-p300 interaction competitively inhibits the CREB-p300 interaction. At this point, the functional role of the Z-CREB interaction remains unknown, but it could potentially be important for regulating early EBV transcription through either ZRE/AP1- or CREB-binding motifs.
CREB-binding sites have been identified in two promoters within the EBV genome, the Z promoter (6, 45, 64) and the LMP1 promoter (58). CREB has been shown to activate the Z promoter through the ZII regulatory element (reference 19, our laboratory, and unpublished data). Z inhibition of CREB function could serve to autoregulate the Z promoter and turn off Z expression after critical amounts of the protein are produced. In addition, we have previously shown that Z inhibits the activity of the LMP1 promoter (32).
E1A, by interacting with CBP, decreases the activities of other transcription factors, including p53, by competing for limiting amounts of cellular CBP (59). Although we have specifically shown that Z inhibits CREB transactivator function, the Z-CBP interaction could likewise inhibit the functions of other transcription factors regulated by CBP. Interestingly, we have previously shown that two of the transcription factors known to interact with CBP (p53 and p65) can inhibit BZLF1 function (and vice versa) (27, 67). Interaction with CBP/p300 may prove to be a common mechanism by which viruses regulate the host cell environment.
ACKNOWLEDGMENTS
This work was supported by grants RO1-CA58853, RO1-CA66519, and PO1-CA19014 from the National Institutes of Health.
We thank Brian Ashburner from Al Baldwin’s laboratory and Jonathon Harton from Jenny Ting’s laboratory for providing CBP constructs and for helpful discussions, Amy Mauser for the adenovirus-Z, and the UNC Gene Therapy Core (R. Jude Samulski and Douglas McCarty) for preparing the adenovirus-LacZ and adenovirus-Z.
REFERENCES
- 1.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetylase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5a.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone acetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 6.Cen H, McKnight J L C. EBV-immortalized isogenic human B-cell clones exhibit differences in DNA-protein complex formation on the BZLF1 and BRLF1 promoter regions among latent, lytic and TPA-activated cell lines. Virus Res. 1994;31:89–107. doi: 10.1016/0168-1702(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y, Dong D, Hayward G, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV) encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an early EBV promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Colgin M A, Nyborg J K. The human T-cell leukemia virus type 1 oncoprotein Tax inhibits the transcriptional activity of c-Myb through competition for the CREB binding protein. J Virol. 1998;72:9396–9399. doi: 10.1128/jvi.72.11.9396-9399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Countryman J, Miller G. Activation of expression of latent Epstein-Barr virus after gene transfer with a small cloned fragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 13.Dorsman J C, Teunisse A F, Zantema A, van der Eb A J. The adenovirus 12 E1A proteins can bind directly to proteins of the p300 transcription co-activator family, including the CREB-binding protein CBP and p300. J Gen Virol. 1997;78:423–426. doi: 10.1099/0022-1317-78-2-423. [DOI] [PubMed] [Google Scholar]
- 14.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrell P, Rowe D, Rooney C, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to consensus Ap1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fixman E, Hayward G, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fixman E, Hayward G, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus oriLyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flamand L, Menezes J. Cyclic AMP-responsive element-dependent activation of Epstein-Barr virus Zebra promoter by human herpesvirus 6. J Virol. 1996;70:1784–1791. doi: 10.1128/jvi.70.3.1784-1791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flemington E K, Borras A M, Lytle J P, Speck S H. Characterization of the Epstein-Barr virus BZLF1 protein transactivation domain. J Virol. 1992;66:922–929. doi: 10.1128/jvi.66.2.922-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Flemington E K, Lytle J P, Cayrol C, Borras A M, Speck S H. DNA-binding-defective mutants of the Epstein-Barr virus lytic switch activator Zta transactivate with altered specificities. Mol Cell Biol. 1994;14:3041–3052. doi: 10.1128/mcb.14.5.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20b.Flemington E, Speck S. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1217–1226. doi: 10.1128/jvi.64.3.1217-1226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemington E, Speck S. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc Natl Acad Sci USA. 1991;87:9459–9463. doi: 10.1073/pnas.87.23.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furnari F B, Zacny V, Quinlivin E B, Kenney S, Pagano J S. RAZ, an Epstein-Barr virus transdominant repressor that modulates the viral reactivation mechanism. J Virol. 1994;68:1827–1836. doi: 10.1128/jvi.68.3.1827-1836.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Rodriguez C, Rao C. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP) J Exp Med. 1998;187:2031–2036. doi: 10.1084/jem.187.12.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giot J-F, Mikaelian I, Buisson M, Manet E, Joab I, Nicolas J-C, Sergeant A. Transcriptional synergy and interference between the EBV transcription factors EB1 and R require both the basic region and the activation domains of EB1. Nucleic Acids Res. 1991;19:1251–1258. doi: 10.1093/nar/19.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorman C, Moffat L, Howard B. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutsch D, Holley-Guthrie E, Zhang Q, Stein B, Blanar M, Baldwin A, Kenney S. The bZIP transactivator, BZLF-1, of Epstein-Barr virus functionally and physically interacts with the p65 subunit of NFκB. Mol Cell Biol. 1994;14:139–149. doi: 10.1128/mcb.14.3.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hottinger M O, Nabel G J. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–8256. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 30.Kelly C, Van Driel R, Wilkinson G W. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 31.Kenney S, Holley-Guthrie E, Quinlivan E, Gutsch D, Zhang Q, Bender T, Giot J, Sergeant A. The cellular oncogene c-myb can interact synergistically with the Epstein-Barr virus BZLF1 transactivator in lymphoid cells. Mol Cell Biol. 1992;12:136–146. doi: 10.1128/mcb.12.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenney S, Kamine J, Holley-Guthrie E, Lin J-C, Mar E-C, Pagano J. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J Virol. 1989;63:1729–1736. doi: 10.1128/jvi.63.4.1729-1736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 34.Kirshenbaum L, Schneider M D. Adenovirus E1A represses cardiac gene transcription and reactivates DNA synthesis in ventricular myocytes, via alternative pocket protein- and p300-binding proteins. J Biol Chem. 1995;270:7791–7794. doi: 10.1074/jbc.270.14.7791. [DOI] [PubMed] [Google Scholar]
- 35.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 35a.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 36.Kouzarides T, Packham G, Cook A, Farrell P. The BZLF1 protein of EBV has a coiled coil dimerization domain without a heptad leucine repeat but homology to the C/EBP leucine zipper. Oncogene. 1991;6:195–204. [PubMed] [Google Scholar]
- 37.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 38.LaMorte V J, Dyck J A, Ochs R L, Evans R M. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 40.Lee J-S, See R H, Deng T, Shi Y. Adenovirus E1A downregulates c-Jun- and JunB-mediated transcription by targeting their coactivator p300. Mol Cell Biol. 1996;16:4312–4326. doi: 10.1128/mcb.16.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q X, Young L S, Niedobitek G, Dawson C W, Birkenbach M, Wang F, Rickinson A B. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature. 1992;356:347–350. doi: 10.1038/356347a0. [DOI] [PubMed] [Google Scholar]
- 42.Lieberman P, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lill N L, Tevethia M J, Eckner R, Livingston D M, Modjtahedi N. p300 family members associate with the carboxyl terminus of simian virus 40 large tumor antigen. J Virol. 1997;71:129–137. doi: 10.1128/jvi.71.1.129-137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lillie J, Green M. Transcriptional activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 45.Liu P, Liu S, Speck S H. Identification of a negative cis element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J Virol. 1998;72:8230–8239. doi: 10.1128/jvi.72.10.8230-8239.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-finger domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 47.Mymryk J S, Lee R W H, Bayley S T. Ability of adenovirus 5 E1A proteins to suppress differentiation of BC3H1 myoblasts correlates with their binding to a 300 kDa cellular protein. Mol Biol Cell. 1992;3:1107–1115. doi: 10.1091/mbc.3.10.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norton V G, Imai B S, Yau P, Bradbury E M. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989;57:449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- 49.Packham G, Economou A, Rooney C M, Rowe D T, Farrel P J. Structure and function of the Epstein-Barr virus BZLF1 protein. J Virol. 1990;64:2110–2116. doi: 10.1128/jvi.64.5.2110-2116.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlivan E, Holley-Guthrie E, Norris M, Gutsch D, Bachenheimer S, Kenney S. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 1993;21:1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 53.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schepers A, Pich D, Hammerschmidt W. A transcription factor with homology to the Ap1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 1993;12:3921–3929. doi: 10.1002/j.1460-2075.1993.tb06070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz R, Helmich B, Spector D H. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J Virol. 1996;70:6955–6966. doi: 10.1128/jvi.70.10.6955-6966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scolnick D M, Chehab N H, Stavridi E S, Lien M C, Caruso L, Moran E, Berger S L, Halazonetis T D. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 57.Sixby J W, Nedrud J G, Raab-Traub N, Hanes R A, Pagano J S. Epstein-Barr virus replication in oropharyngeal epithelial cells. N Engl J Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 58.Sjoblom A, Yang W, Palmqvist L, Jansson A, Rymo L. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J Virol. 1998;72:1365–1376. doi: 10.1128/jvi.72.2.1365-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somasundaram K, El-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 60.Takada K, Shimizu N, Sakuma S, Ono Y. Transactivation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J Virol. 1986;57:1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tonneguzzo F, Hayday A C, Keating A. Electric field-mediated DNA transfer: transient and stable gene expression in human and mouse lymphoid cells. Mol Cell Biol. 1986;6:703–706. doi: 10.1128/mcb.6.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Ebernarter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 63.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y-C J, Huang J-M, Montalvo E A. Characterization of proteins binding to the ZII element in the Epstein-Barr virus BZLF1 promoter: transactivation by ATF1. Virology. 1997;227:323–330. doi: 10.1006/viro.1996.8326. [DOI] [PubMed] [Google Scholar]
- 64a.Yamamoto K K, Gonzalez G A, Biggs III W H, Montimony M R. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 65.Yang C, Shapiro L H, Rivera M, Kumar A, Brindle P K. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young L, Lau R, Rowe M, Niedobitek G, Packham G, Shanahan F, Rowe D, Greenspan D, Greenspan J, Rickinson A, Farrell P. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator in oral hairy leukoplakia. J Virol. 1991;65:2868–2874. doi: 10.1128/jvi.65.6.2868-2874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zur Hausen H, Schulte-Holthauzen H, Klein G, Henle G, Henle W, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1956–1958. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]