Evaluation of Interactions of Human Cytomegalovirus Immediate-Early IE2 Regulatory Protein with Small Ubiquitin-Like Modifiers and Their Conjugation Enzyme Ubc9 (original) (raw)
Abstract
The human cytomegalovirus (HCMV) major immediate-early protein IE2 is a nuclear phosphoprotein that is believed to be a key regulator in both lytic and latent infections. Using yeast two-hybrid screening, small ubiquitin-like modifiers (SUMO-1, SUMO-2, and SUMO-3) and a SUMO-conjugating enzyme (Ubc9) were isolated as IE2-interacting proteins. In vitro binding assays with glutathione _S_-transferase (GST) fusion proteins provided evidence for direct protein-protein interaction. Mapping data showed that the C-terminal end of SUMO-1 is critical for interaction with IE2 in both yeast and in vitro binding assays. IE2 was efficiently modified by SUMO-1 or SUMO-2 in cotransfected cells and in cells infected with a recombinant adenovirus expressing HCMV IE2, although the level of modification was much lower in HCMV-infected cells. Two lysine residues at positions 175 and 180 were mapped as major alternative SUMO-1 conjugation sites in both cotransfected cells and an in vitro sumoylation assay and could be conjugated by SUMO-1 simultaneously. Although mutations of these lysine residues did not interfere with the POD (or ND10) targeting of IE2, overexpression of SUMO-1 enhanced IE2-mediated transactivation in a promoter-dependent manner in reporter assays. Interestingly, many other cellular proteins identified as IE2 interaction partners in yeast two-hybrid assays also interact with SUMO-1, suggesting that either directly bound or covalently conjugated SUMO moieties may act as a bridge for interactions between IE2 and other SUMO-1-modified or SUMO-1-interacting proteins. When we investigated the intracellular localization of SUMO-1 in HCMV-infected cells, the pattern changed from nuclear punctate to predominantly nuclear diffuse in an IE1-dependent manner at very early times after infection, but with some SUMO-1 protein now associated with IE2 punctate domains. However, at late times after infection, SUMO-1 was predominantly detected within viral DNA replication compartments containing IE2. Taken together, these results show that HCMV infection causes the redistribution of SUMO-1 and that IE2 both physically binds to and is covalently modified by SUMO moieties, suggesting possible modulation of both the function of SUMO-1 and protein-protein interactions of IE2 during HCMV infection.
Human cytomegalovirus (HCMV) can cause severe disease complications and pathogenesis on infection of newborns or immunocompromised individuals, whereas infection of immunocompetent individuals is typically asymptomatic (8, 44). Gene expression during the permissive lytic cycle of HCMV occurs in a three-step sequential fashion. Shortly after infection, the immediate-early (IE) genes are expressed in the absence of de novo protein synthesis. IE proteins and virion factors are required for the subsequent induction of early and late genes (36, 56). The major IE (MIE) locus of HCMV genome encodes two nuclear phosphoproteins, namely, IE1 (UL123, IE72) and IE2 (UL122, IE86), which are translated from differentially spliced mRNA species (58, 60). Some additional isoforms of IE2 can also be generated through differential splicing or through the usage of a late promoter within the IE2 coding region. Both the 72-kDa IE1 and the 86-kDa IE2 are the first and most abundantly expressed IE gene products and are also the only viral proteins detected in several nonpermissive cell types (30).
Eukaryotic cell nuclei contain several discrete domains in which different cellular processes such as DNA replication, transcription, pre-mRNA processing, and ribosome assembly take place (32). Among them, the promyelocytic leukemia protein (PML)-associated nuclear bodies known as PML oncogenic domains (PODs) or nuclear domain 10 (ND10) have been implicated as the sites for input viral DNA deposition as well as for IE transcription and initiation of viral DNA replication in a number of DNA viruses (5, 21, 23, 37, 41). We previously showed that at very early times in HCMV lytic cycle infection, the IE1 protein transiently localizes to and subsequently disrupts PODs whereas the IE2 protein localizes within or adjacent to PODs on either DNA transfection or HCMV infection (4, 5). These processes appear to be required for efficient viral gene expression and DNA replication (3). At later times after viral DNA synthesis occurs, IE2, but not IE1, accumulates within large nuclear viral DNA replication compartments (5). The IE2 protein functions as both a potent transactivator that up-regulates many viral and cellular promoters and a repressor that down-regulates its own promoter through direct DNA binding to the MIE _cis_-repression signal near the 5′ cap site in transient-cotransfection assays (35, 47, 49). IE2 has also been reported to bind to transcription factors such as TBP, TEIIB, RB, p53, Ap-1, Egr-1, CREB, and Sp1-1/Pu.1 (10, 18, 19, 28, 29, 33, 54, 55, 57, 61, 62).
Several families of ubiquitin-like proteins have recently been described, including the most prominent small ubiquitin-like modifier SUMO-1 (27, 53), which can be covalently conjugated to proteins such as RanGAP1, IκBα, PML, Sp100, and p53 through a pathway distinct from but analogous to the ubiquitin conjugation system (13, 26). SUMO-1 modification of RanGAP1 and PML is critical for their intracellular targeting as well as for their interaction with other proteins (34, 38, 40). SUMO-1 conjugation of p53 enhances its transactivation ability (16, 50), whereas sumoylation of IκBα regulates its stability (12). Two additional mammalian cDNAs encoding closely related proteins similar to SUMO-1 have been isolated, and the proteins have been designated SUMO-2/Smt3A and SUMO-3/Smt3B. Previous studies showed that both SUMO-2 and SUMO-3 could be conjugated to several target proteins via a mechanism similar to that of SUMO-1 conjugation (52).
In this study, we isolated SUMO-1, SUMO-2, and SUMO-3 as well as Ubc9 (the E2 enzyme for SUMO conjugation) as HCMV IE2-binding proteins by yeast two-hybrid screening. The results showed that IE2 interacts directly with SUMO-1, SUMO-2/3, and Ubc9 and can be covalently modified by either SUMO-1 or SUMO-2. The SUMO-1 modification takes place on either one or both of the lysine residues at positions 175 and 180. We also evaluated whether SUMO-1 modification of IE2 influences transactivation properties and examined the redistribution of SUMO-1 after infection by HCMV.
MATERIALS AND METHODS
Cell culture and virus infection.
Permissive human diploid fibroblasts (HF) cells, semipermissive U373-MG cells, 293T cells, and Vero cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Virus stocks for wild-type HCMV(Towne) and IE1-defective mutant HCMV(CR208) (17) were prepared as previously described (1, 31). Recombinant adenoviruses expressing HCMV IE1 (Ad-IE1) or IE2 (Ad-IE2) were described previously (4). For experiments involving indirect immunofluorescence assay (IFA), HF cells were seeded into four-well chamber slides at 0.4 × 105 cells per well and the subconfluent cells were infected with HCMV(Towne) at various multiplicities of infection (MOI). For experiments with immunoblot analysis, HF cells were seeded into six-well plates at 2 × 105 cells per well. Next day, the cells were mock infected or infected with wild-type HCMV(Towne) or IE1-deleted HCMV(CR208) at a MOI of 5 or with recombinant Ad at a MOI of 20. Cell lysates were then harvested at different time points after infection.
Plasmid construction.
The wild-type IE2 cDNA expression plasmid pJHA124 and its genomic version pMP18 were described previously (1). Mutant forms encoding IE2 K175R (pYX105), K180R (pYX106), K175/180R (pYX104), and S203A (pYX139) were generated from pJHA124 by the Stratagene QuikChange site-directed mutagenesis protocol. After isolation of cDNAs encoding SUMO-1, SUMO-2, and Ubc9 from a yeast two-hybrid screen, the inserts were amplified by PCR and cloned into a pJH272 expression vector (pSG5 with Flag tag) to generate pJHA312 (Flag-SUMO-1), pJHA342 (Flag-SUMO-2), and pWJ5 (Flag-Ubc9). The expression plasmid pYX107 encoding Ubc9 mutant C93S was generated from pWJ5 by the Stratagene QuikChange protocol. The _Bam_HI fragments containing wild-type and C93S Ubc9 were then subcloned into a pGEX-3X (Pharmacia)-derived vector, pGH418, to generate pYX112 for wild-type glutathione _S_-transferase (GST)–Ubc9 and pYX113 for mutant GST-Ubc9(C93S). A GST–SUMO-1 plasmid was provided by Masahiro Fujimuro (Johns Hopkins Medical Institutions, Baltimore, Md.), and GST–SUMO-2 and GST–SUMO-3 constructs were gifts from Hisato Saitoh (Picower Institute, Manhasset, N.Y.). A Flag-tagged SUMO-1(ΔGG) mutant (pJHA344) was made by a PCR-based C-terminal deletion of 6 amino acids (aa) from wild-type SUMO-1 encoded by pJHA312 and cloned into pJH272. The SUMO-1(ΔGG) insert was also subcloned into a pGEX-3X-derived background (pGH416) to generate GST-SUMO-1(ΔGG), encoded by pJHA354.
Yeast expression plasmids for GAL4 DNA-binding (DB) domain (positions 1 to 147) fusions were generated in pAS1-CYH2, a derivative of pAS1 (14). pWJ1 expressing GAL4-DB/IE2(87–542) and pJHA339 expressing GAL4-DB/IE2(87–542, K175/180R) were generated by placing the _Bgl_II-_Stu_I fragments amplified by PCR from pJHA124 and pYX104, respectively, behind the GAL4-DB domain of pAS1-CYH2. All GAL4 activation (A) domain (positions 768 to 881) fusions for expression in yeast were constructed in pACTII, a derivative of pACT (14). pJHA313 expressing GAL4-A/SUMO-1 and pJHA345 expressing GAL4-A/SUMO-1(ΔGG) were generated by inserting PCR fragments from pJHA312 and pJHA344, respectively, after the GAL4-A domain of pACTII. pCJC440 expressing GAL4-A/IE2(290–542) was described previously (2).
Luciferase reporter plasmid pLA12, encoding Pol-LUC driven by the HCMV UL54 promoter, was described previously (3), and luciferase reporter plasmid pE(−207)Luc, encoding CycE-LUC driven by the human cyclin E promoter, was provided by Aubrey Thompson (University of Texas Medical Branch, Galveston, Tex.) (7).
Yeast two-hybrid interaction assays.
The yeast strain HF7C [MATa ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1-HIS3 URA3::_(GAL4 17-mers)3CYC1-lacZ_] was the host for initial library screening by the two-hybrid assay, and strain Y190 (MATa gal4Δ gal80Δ his3-200 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL1-lacZ LYS::GAL-HIS3 Cyhr) was the host for rapid assays for lacZ expression using a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) filter assay or for quantitation of interaction using a β-galactosidase assay. Media for yeast growth and the method for yeast transformation were described elsewhere (51). Both the X-Gal filter assay and the β-galactosidase assay were described previously (1).
Transient DNA transfection.
For immunoblot analysis, 293T cells or U373-MG cells were seeded into six-well plates at 4 × 105 cells per well and DNA mixtures were introduced into subconfluent cells with the _N,N_-bis-(2-hydroxyethyl)-2-aminoethanesulfonic acid-buffered saline (BBS) version of the calcium phosphate procedure, as described previously (47). For IFA, Vero cells were seeded into two-well slide chambers at 0.4 × 105 cells per well and 3 μg of total DNA was used. For luciferase (LUC) reporter assays, U373 cells were seeded into 12-well plates at 0.4 × 105 cells per well and 3 μg of total DNA was used.
Antibodies.
Mouse monoclonal antibody (MAb) 12E2 against HCMV IE2 (exon 5) was purchased from Vancouver Biotech (Vancouver, B.C., Canada). MAb CH810, which detects epitopes present in both IE1 and IE2 (exons 2 and 3), was purchased from Chemicon (Temecula, Calif.). Mouse MAb anti-Flag was purchased from Kodak. Rabbit anti-Flag polyclonal antibody (PAb) and anti-SUMO-1 PAb (FL-101) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). The rabbit antipeptide PAb referred to as PML(C), directed against amino acids 484 to 498 of PML, was described previously (1, 4). The anti-SUMO-1 MAb 21C7 was described previously (39).
In vitro binding assay with GST fusion proteins.
Plasmid DNA encoding GST fusion proteins were transformed into Escherichia coli BL21, and extracts from the bacterial cultures expressing the GST fusion proteins were prepared by standard procedures. These extracts were incubated with glutathione-Sepharose 4B beads (Pharmacia) for 3 h at 4°C. After three washes with lysis buffer, the beads were resuspended in EBC buffer (140 mM NaCl, 50 mM Tris [pH 8.0], 0.5% Triton X-100, 100 mM NaF, 200 μM Na3VO4). The [35S]Met-labeled IE2 test protein was in vitro transcribed and translated from the pJHA124 plasmid template with the Promega T7 Quick TNT kit as specified by the manufacturer. Aliquots of [35S]IE2 (5 μl) were mixed with appropriate amounts of GST fusion protein beads (containing ∼5 μg of the GST fusion protein, with unbound glutathione beads added to make a total bead volume of 20 μl) and resuspended in 500 μl of EBC buffer supplemented with bovine serum albumin at 1 mg/ml. The mixtures were then incubated for 2 h at 4°C with gentle stirring. After binding, the beads were pelleted, washed five times with 1 ml of NETT buffer (100 mM NaCl, 20 mM Tris [pH 8.0], 0.5% Triton X-100, 1 mM EDTA), resuspended in 15 μl of 2× sodium dodecyl sulfate (SDS) sample buffer, boiled for 5 min, and loaded together with the [35S]IE2 input onto SDS–8% polyacrylamide gels. After electrophoresis, the gels were stained with Coomassie blue to visualize the amount of input GST fusion proteins and the amount of [35S]IE2 bound to a given GST fusion protein was determined with an Alpha Imager (Alpha Innotech, San Leandro, Calif.).
In vitro SUMO-1 conjugation assay.
[35S]Met labeled wild-type and mutant IE2 proteins were in vitro translated from pJHA124, pYX105 (K175R), pYX106 (K180R), and pYX104 (K175/180R). A 2-μl volume of each radiolabeled protein was mixed with 2 μl of HeLa cell fraction containing E1 activity for SUMO-1 conjugation (12) and 0.5 μl of reaction mixture (2 mM ATP, 60 U of creatine kinase per ml, 15 mM creatine phosphate, 250 μg of GST-Ubc9 per ml, and 500 μg of His-tagged SUMO-1 per ml). The reaction mixtures were incubated at 37°C for 2 h, and the reactions were terminated with SDS sample buffer containing β-mercaptoethanol. After being boiled for 5 min, half of each reaction mixture was loaded onto SDS–8% polyacrylamide gels along with 1 μl of each in vitro-translated protein without the SUMO-1 conjugation reaction mixture as the control. The proteins were visualized by autoradiography on Kodak BioMax MR films.
Immunoblot analysis.
Transfected 293T or U373-MG cells and infected HF cells were washed with phosphate-buffered saline (PBS) and lysed with 200 μl of ice-cold RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitor cocktail (Santa Cruz Biotechnology). Clarified cell extracts from the equivalent of 104 cells were separated on a SDS–8% polyacrylamide gel and electroblotted onto nitrocellulose membranes. The blots were blocked by incubation for 30 min at room temperature with PBS plus 0.1% Tween 20 (PBST) containing 5% nonfat dry milk. After being washed with PBST twice, the blots were incubated with the appropriate MAb at a 1:3,000 dilution in PBST at 4°C overnight. After three 10-min washes with PBST, the blots were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibody (Amersham) for 45 min at room temperature. The blots were then washed three times with PBST, and the protein bands were visualized with the enhanced chemiluminescence system (Amersham) on Kodak XAR films.
Immunoprecipitation.
Transfected 293T cell lysates were incubated with 5 μg of MAb CH810 at 4°C overnight, and 30 μl of protein A/G-Sepharose beads was added. The remaining steps were the same as described by Buschmann et al. (9), and the immunoblot was probed with anti-SUMO-1 MAb 21C7.
IFA.
Transfected or virus-infected cells were fixed in methanol or with 1% paraformaldehyde and permeabilized with 0.2% Triton X-100 as described previously (1). The slides were then incubated with either one MAb (at a 1:200 dilution for 12E2, CH810, or anti-Flag or a 1:800 dilution for 21C7) or one PAb (at 1:1,000 dilution for PML or a 1:50 dilution for FL-101) in PBS at 37°C for 1 h and than incubated with fluorescein isothiocyanate (FITC)-labeled donkey anti-mouse IgG or with rhodamine-coupled donkey anti-rabbit IgG at a 1:100 dilution at 37°C for 45 min. For double labeling, MAb and PAb were incubated together. To stain the cell nucleus, mounting solution containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Inc., Burlingame, Calif.) was used. Slides were examined and photographed on a Leitz Dialux 20EB epifluorescence microscope with Image-Pro software (Media Cybernetics, Silver Spring, Md.).
Luciferase reporter assay.
Transfected Vero or U373-MG cells were lysed directly in 12-well plates by three freeze-thaw steps in 200 μl of 0.25 M Tris-HCl (pH 7.9) to 1 mM dithiothreitol. The extracts were clarified in a microcentrifuge, and 50-μl volumes were incubated with 350 μl of reaction buffer A (25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 5 mM ATP, 4 mM EGTA) and then mixed with 100 μl of reaction buffer B (1 mM luciferin in reaction buffer A) at 20°C in the chamber of a LUMAT LB 9501 luminometer using a 5-s assay of the photons produced (measured in relative light units).
RESULTS
Isolation of SUMO-1, SUMO-2, and SUMO-3, as well as Ubc9, as IE2-interacting proteins.
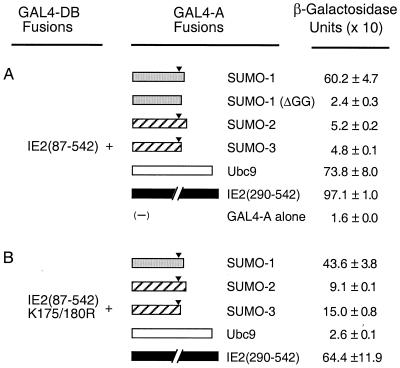
To study protein-protein interactions of HCMV IE2, cellular proteins that bound to it were isolated from a cDNA library prepared from human B lymphocytes using yeast two-hybrid screening. To generate a bait, the cDNA fragment encoding a region of IE2 from aa 87 to 542, which lacks the two distal transactivation domains of the 579-aa protein (48), was fused to GAL4-DB in the yeast two-hybrid vector pAS1-CYH2. A total of 106 transformants were screened by selection for His prototropy on yeast dropout medium lacking Trp, Leu, and His in the presence of 25 mM 3-amino-triazole. Positive colonies were confirmed by expression of β-galactosidase in X-Gal filter assays. Library plasmid DNA was isolated from yeast and amplified in E. coli, and then the cDNA inserts were sequenced. Among 80 positive clones examined, cDNAs encoding SUMO-1, SUMO-2 (hsSmt3A), and SUMO-3 (hsSmt3B) were isolated eight, one, and three times, respectively. In addition, cDNA encoding Ubc9, the E2 enzyme for SUMO conjugation, was isolated 11 times. The strength of interaction in yeast cells was quantified using a β-galactosidase activity assay. The relative strength of interactions of IE2 with SUMO-1 and Ubc9 was comparable to that of IE2 self-interaction (2, 11), whereas interactions of IE2 with SUMO-2 and SUMO-3 were less than 10% of the strength of those with SUMO-1 and Ubc9 (Fig. 1A).
FIG. 1.
Interaction of IE2 with SUMO-1, SUMO-2, SUMO-3, and Ubc9 in yeast two-hybrid assays. Plasmids encoding GAL4-DB fusions and GAL4-A fusions were introduced together into Y190 cells. Transformants were selected on plates lacking Trp and Leu, and the β-galactosidase activities of the transformants were measured as described in Materials and Methods. (A) Interaction of GAL4-BD/IE2(87–542) with GAL4-A fusion proteins. The cDNAs used to make GAL4-A fusions were SUMO-1(1–101), SUMO-1(ΔGG)(1–95), SUMO-2(1–103), SUMO-3(1–95), Ubc9(1–158), and IE2(290–542). (B) Interaction of GAL4-DB/IE2(87–542, K175/180R) with GAL4-A fusion proteins. Solid arrowheads indicate the position after the double-glycine motif, which is cleaved by protease to activate the SUMO moiety for conjugation to the substrate proteins (see also Fig. 2).
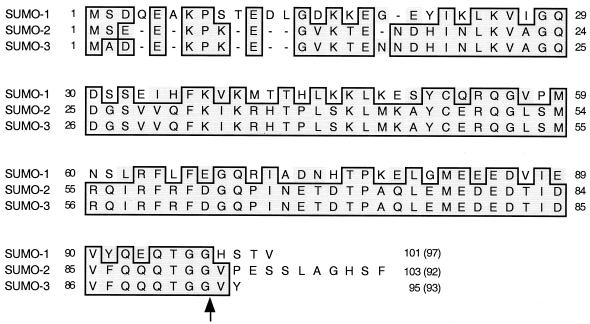
The intact cDNAs for SUMO-1, SUMO-2, and SUMO-3 isolated from the human B-lymphocyte library were sequenced and aligned with each other (Fig. 2). The resulting predicted SUMO-1 and SUMO-3 proteins were identical to those of the previously reported human SUMO-1 (GenBank no. U67122) and hsSmt3B (X99585) proteins, respectively. However, our SUMO-2 sequence differed from the hsSmt3A (X99584) sequence isolated from brain cells, with a proline instead of a serine at position 38 and a glutamate instead of an arginine at position 76. Note that our version of SUMO-2 from B lymphocytes is identical to SUMO-3 at these two positions. Furthermore, when the conjugation moieties attached to the substrates (i.e., N-terminal to the double-glycine cleavage site) were compared, there were only three amino acid differences between the SUMO-2 and SUMO-3 adducts compared to 52% amino acid difference between SUMO-1 and SUMO-2 (Fig. 2).
FIG. 2.
Comparison of amino acid sequences for SUMO-1, SUMO-2, and SUMO-3 isolated as IE2-interacting proteins from a human B-lymphocyte cDNA library. The amino acid sequences of SUMO-1, SUMO-2 (hsSmt3A), and SUMO-3 (hsSmt3B) isolated in this study were aligned. Residues that are identical in either two or three of the proteins are boxed. The arrow after the double-glycine motif indicates the position of proteolytic cleavage for conjugation. The numbers in parentheses are the numbers of amino acids presumably conjugated to the substrates.
The HCMV IE2 protein directly interacts with SUMO-1, SUMO-2/3, and Ubc9.
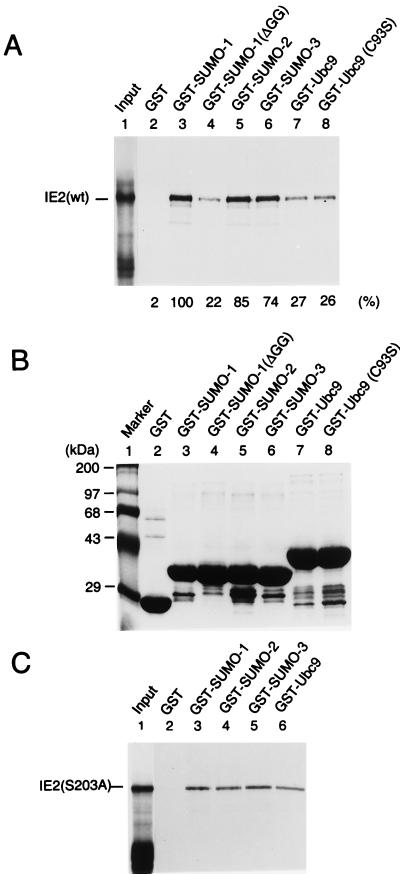
The relatively strong interaction of IE2 with SUMO-1 and Ubc9 might be attributed to either direct protein-protein interactions or covalent modification of IE2 by SUMO-1 using the Smt3 conjugation machinery available in yeast cells. For SUMO-1 conjugation to substrate proteins, the full-length SUMO-1 precursor must be activated by cleavage after the double-glycine motif near the C terminus. When a C-terminally truncated SUMO-1 referred to as SUMO-1(ΔGG), which lacks 6 amino acids including the double-glycine motif, was tested, it failed to interact with IE2 (Fig. 1A). This result suggests that the interaction of IE2 with SUMO-1 in yeast cells must either include covalent conjugation of SUMO-1 to IE2 or, alternatively, require the intact C terminus of SUMO-1 for protein-protein binding. To further test whether IE2 can interact directly with SUMO-1, SUMO-2/3, and Ubc9, an in vitro binding assay with GST fusion proteins was carried out. In the SUMO-1 conjugation pathway, Ubc9 forms a thioester bond with SUMO-1 through an active-site cysteine residue at position 93 of Ubc9 (15). The cDNAs encoding SUMO-1 (101 aa), SUMO-2 (103 aa), SUMO-3 (95 aa), truncation mutant SUMO-1(ΔGG) (95 aa), Ubc9 (158 aa), and a loss-of-function mutant Ubc9 (C93S) were each fused in frame to GST. The GST fusion proteins, as well as GST alone, were prepared from Escherichia coli (Fig. 3B), immobilized to glutathione-Sepharose beads, and used in the pulldown assays with [35S]methionine-labeled full-length IE2 protein (579 aa) generated by in vitro translation. Interestingly, SUMO-1, SUMO-2, and SUMO-3 all bound to IE2 with similar affinity, and SUMO-1(ΔGG), with the C-terminal 6-aa deletion, showed a fivefold-reduced interaction with IE2 (Fig. 3A). The latter was consistent with the result obtained from the yeast two-hybrid assay and indicated that the C-terminal end of SUMO-1 is critical for direct binding to IE2. In contrast to the results obtained with yeast, the finding that SUMO-2/3 interacted with IE2 with similar affinity to the IE2/SUMO-1 interaction in the in vitro binding assay suggests that the human SUMO-2/3 proteins are not efficiently attached to the substrate proteins in yeast cells. Wild-type Ubc9 and the C93S mutant Ubc9 showed similar levels of in vitro binding to IE2, but the affinity was 20 to 30% of that between IE2 and any of the three SUMO isoforms in this assay.
FIG. 3.
Direct interactions of IE2 with SUMO-1, SUMO-2, SUMO-3, and Ubc9. (A) In vitro binding assay of wild-type IE2 with GST fusion proteins. The same amounts of GST or GST fusion proteins immobilized to glutathione-Sepharose beads were incubated with [35S]methionine-labeled full-length IE2 proteins. One-fifth of the labeled IE2 proteins used in each binding reaction was loaded as an input control (lane 1). After in vitro binding and five washing steps, purified proteins were fractionated by electrophoresis on SDS–8% polyacrylamide gels and visualized by autoradiography. (B) Relative sizes and purity of the GST or GST fusion proteins used. The same gel as in panel A was stained with Coomassie blue before autoradiography. (C) In vitro binding assay of IE2(S203A) with GST fusion proteins. The assay conditions were exactly the same as in panel A.
Covalent modification of IE2 by SUMO-1 or SUMO-2.
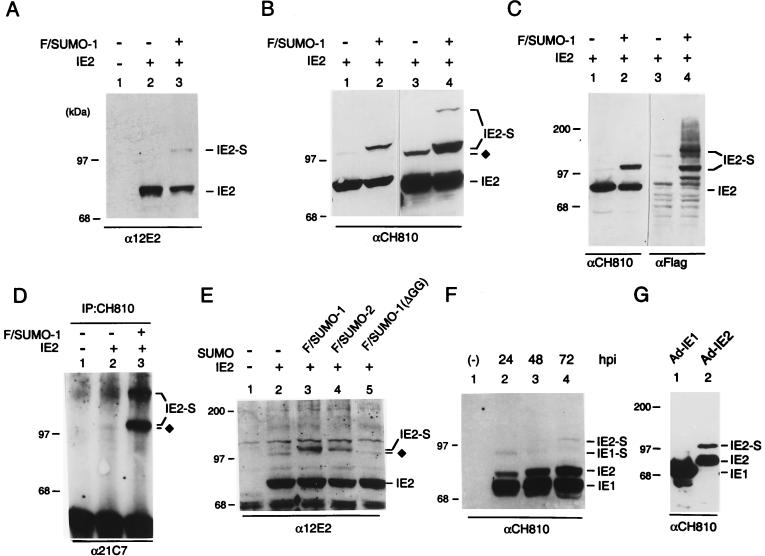
A number of known substrates for SUMO-1 modification interact with both SUMO-1 and Ubc9 in the yeast two-hybrid screen (6, 12, 16). To test whether IE2 can be covalently modified by SUMO-1 in mammalian cells, we transfected 293T cells with an IE2 expression plasmid DNA with or without cotransfection of the Flag-tagged SUMO-1 expression plasmid DNA. After lysis of cells with RIPA buffer, proteins were electrophoretically separated on an SDS–8% polyacrylamide gel and two different MAbs were used for IE2 immunoblot analysis. IE2-specific MAb 12E2 detected a minor 105-kDa band in cells cotransfected with both IE2 and Flag-SUMO-1 (Fig. 4A). MAb CH810, which is directed against epitopes shared by IE1 and IE2, was also able to detect a 105-kDa modified IE2 form even in the absence of Flag-SUMO-1 cotransfection (Fig. 4B, lane 1). In cells expressing both IE2 and Flag-SUMO-1, CH810 detected a more abundant doublet of modified IE2 species, with the upper band corresponding to IE2 conjugated by exogenous Flag-SUMO-1 and a slightly lower band that appears to represent IE2 conjugated by an endogenous SUMO moiety (Fig. 4B, lane 2). When a larger amount of the cell lysates were loaded, an additional ∼120-kDa band of modified IE2 was also detected (lane 4), suggesting that there are at least two SUMO modification sites in the IE2 protein. Control experiments with 293T cells cotransfected with UL112–113, another HCMV protein associated with PODs (5), and Flag-SUMO-1 showed that no SUMO-1 conjugation of UL112–113 was detected (data not shown).
FIG. 4.
Covalent modification of IE2 by SUMO-1 or SUMO-2. (A) Detection of SUMO-1-conjugated IE2 by Western immunoblotting in transfected 293T cells. Total cell extracts prepared from 293T cells transfected with 1 μg of empty plasmid vector DNA (lane 1) or with 1 μg of pJHA124 (encoding IE2) DNA alone (lane 2) or cotransfected with 1 μg of pJHA124 plus 2 μg of pJHA312 encoding Flag–SUMO-1 (lane 3) were fractionated on SDS–8% polyacrylamide gels, and immunoblot analysis was carried out with mouse anti-IE2 12E2 MAb. (B) Samples of the same IE2 and IE2/Flag–SUMO-1–transfected 293T cell extracts were subjected to analysis with mouse anti-IE2 CH810 MAb. Lanes 3 and 4 contained three times more loaded extract than lanes 1 and 2 and were exposed longer. (C) Samples of IE2 and IE2 plus Flag–SUMO-1–transfected 293T cell extracts were immunoblotted with mouse anti-IE2 mouse MAb CH810 (lanes 1 and 2), washed, and reprobed with mouse MAb specific for Flag epitope (lanes 3 and 4). (D) Transfected 293T cell lysates (same sample set as in panel A) were immunoprecipitated with MAb CH810, and the immunoprecipitates were fractionated by size and reacted with anti-SUMO-1 MAb 21C7. (E) U373-MG cells were transfected with empty vector (lane 1) or 1 μg of pJHA124 (encoding IE2) alone (lane 2) or cotransfected with 1 μg of pJHA124 plus 2 μg of pJHA312 (Flag–SUMO-1) (lane 3), pJHA342 (Flag–SUMO-2) (lane 4), or pJHA344 [Flag–SUMO-1(ΔGG)] (lane 5). Total cell extracts were prepared and fractionated as described above, and immunoblot analysis was carried out with anti-IE2-specific MAb 12E2. (F) HF cells were mock infected or infected with HCMV(Towne) at an MOI of 5.0 for 24, 48, or 72 h. Total cell extracts were prepared and fractionated as described above, and immunoblot analysis was performed with MAb CH810 recognizing both IE1 and IE2. (G) HF cells were infected with Ad-IE1 (lane 1) or Ad-IE2 (lane 2) at an MOI of 20 for 24 h, and immunoblot analysis of the total cell extracts with anti-IE1/IE2 CH810 MAb was carried out as in panel F. The SUMO-conjugated 90-kDa form of IE1 and 105-kDa forms of IE2 are designated IE1-S and IE2-S, and the IE2 form modified by the putative endogenous SUMO moiety is indicated by ⧫ in panels B, D and E.
To test whether the modified forms of IE2 in cotransfected cells indeed contain the SUMO-1 moiety, Western immunoblot analysis was performed with anti-Flag antibody. When the anti-IE2 CH810 immunoblot was stripped and reprobed with a rabbit anti-Flag PAb, both the 105- and 120-kDa isoforms were readily detected (Fig. 4C, lane 4). The other bands detected with the anti-Flag antibody presumably represent cellular protein substrates that were conjugated by Flag–SUMO-1 in the cotransfected cells. To provide unambiguous evidence that the 105- and 120-kDa IE2 isoforms are indeed SUMO-1 conjugates, whole-cell lysates were also immunoprecipitated with MAb CH810. The precipitated proteins were then separated by electrophoresis and visualized by Western immunoblotting with anti-SUMO-1 MAb 21C7. Both the 105-kDa doublet and the 120-kDa band were clearly detected in cells cotransfected with IE2 and Flag–SUMO-1, whereas only a small fraction of the lower band of the 105-kDa doublet was detected in cells transfected with IE2 alone (Fig. 4D, lanes 2 and 3). These results firmly established that the 105-kDa doublet corresponds to IE2 conjugated by one molecule of exogenous Flag–SUMO-1 (upper band) or of endogenous SUMO-1 (lower band) and that two molecules of Flag–SUMO-1 were conjugated to each IE2 molecule in the 120-kDa species.
Because SUMO-2 and SUMO-3 were also identified as IE2-interacting proteins in our yeast two-hybrid screening and showed similar levels of interaction with IE2 by the in vitro binding assay, we also made a Flag–SUMO-2 expression plasmid to test whether IE2 can be modified by SUMO-2 on cotransfection of semipermissive U373-MG cells (Fig. 4E). The conjugation-negative Flag–SUMO-1(ΔGG) (95-aa) mutant was used as a negative control. The results confirmed that similar levels of exogenous modified IE2 isoforms were generated by cotransfecting either Flag–SUMO-1 or Flag–SUMO-2 (lanes 3 and 4). Coexpression with the inactive Flag–SUMO-1(ΔGG) or without any SUMO-1 or SUMO-2 yielded only the minor 105-kDa modified IE2 isoform that contains an endogenous SUMO moiety (lanes 2 and 5).
After demonstrating that IE2 can be modified by both SUMO-1 or SUMO-2 in semipermissive U373-MG cells, we asked whether this type of covalent modification also occurs in fully permissive HF cells after infection with either wild-type HCMV(Towne) or recombinant Ad-IE2. We first analyzed cell lysates from HF cells infected by HCMV(Towne) for 24, 48, and 72 h by immunoblot analysis with MAb CH810. A small amount of the 105-kDa modified IE2 form accumulated at all times tested throughout the course of HCMV infection (Fig. 4F). Since MAb CH810 is also immunoreactive against IE1, the 72-kDa IE1, along with its putative 90-kDa SUMO-modified isoform, were also detected in this experiment (Y. Xu, J.-H. Ahn, M. J. Matunis, and G. S. Hayward, unpublished data). When HF cells infected for 24 h with either Ad-IE1 or Ad-IE2 were analyzed by immunoblotting with MAb CH810, much higher levels of IE2 sumoylation (30% of total IE2) was observed in the Ad-IE2-infected cells than in the Ad-IE1-infected cells (Fig. 4G). These results suggest that a substantial fraction of the IE2 protein can be covalently modified in infected cells in the absence of other viral proteins but that during a complete HCMV lytic replication cycle in permissive host cells, the levels of modification might be tightly regulated or influenced negatively by the presence of IE1.
Mapping of SUMO-1 modification sites in the IE2 protein.
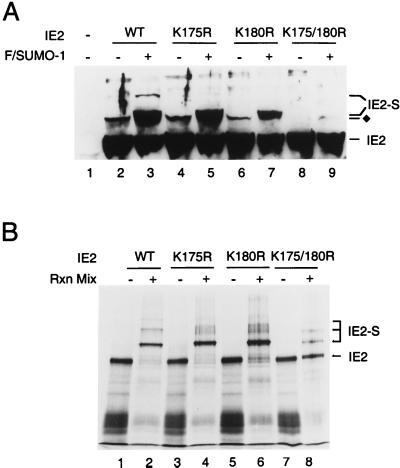
An alignment of the target sequences for SUMO-1 modification in all known SUMO-1 substrates reveals a consensus pattern of the so-called KXE motif (25). After examining the amino acid sequence of IE2, we recognized three KXE motifs at positions 175, 180, and 420. Each of these three candidate lysine residues was mutated to arginine, and the resulting IE2 proteins were tested for modification by SUMO-1 in the 293T cell cotransfection assay. The K420R IE2 mutant showed an identical Flag–SUMO-1 modification pattern to wild-type IE2 (data not shown). Cotransfection of wild-type IE2 and Flag–SUMO-1 produced both the 105- and 120-kDa modified isoforms (Fig. 5A, lane 3), but only the 105-kDa doublet could be detected with either the K175R or K180R IE2 single mutant (lanes 5 and 7). In contrast, when a K175/180R double mutant was introduced into 293T cells, it was not modified at all by exogenous Flag–SUMO-1 (lane 9). Furthermore, the endogenous SUMO modified band was also eliminated.
FIG. 5.
Mapping of the lysine residues in IE2 that are conjugated by SUMO-1. (A) Total cell extracts were prepared from 293T cells transfected with 1 μg of vector alone (lane 1), or 1 μg of pJHA124 encoding wild-type IE2 (lane 2), pYX105 encoding IE2(K175R) (lane 4), pYX106 encoding IE2(K180R) (lane 6), or pYX104 encoding IE2(K175/180R) (lane 8) or the same four plasmids plus 2 μg of pJHA312 encoding Flag–SUMO-1 (lanes 3, 5, 7, and 9). Total cell extracts were prepared and subjected to electrophoresis on SDS–8% polyacrylamide gels, and immunoblot analysis was carried out with MAb CH810. (B) In vitro SUMO-1 conjugation assays. [35S]methionine-labeled wild-type and mutant IE2 proteins were incubated at 37°C in the presence or absence of SUMO-1 modification reaction mixtures (Rxn Mix) as described in Materials and Methods. The reaction products were visualized by autoradiography. IE2-S, Flag–SUMO-1 conjugated IE2; ⧫, IE2 forms conjugated by an endogenous SUMO moiety.
To confirm these mapping data, we also tested wild-type and mutant IE2 proteins by in vitro SUMO-1 conjugation assays. Surprisingly, wild-type IE2 was almost completely sumoylated into three higher-molecular-mass isoforms (Fig. 5B, lane 2) while the K175R and K180R single mutants were primarily monosumoylated into the 105-kDa isoform (lanes 4 and 6). About 80% of the K175/180R double-mutant protein remained unmodified, although a small fraction of monosumoylated isoform was detected (lane 8). These observations suggest that a third lysine residue in IE2 has the potential to be a minor SUMO-1 modification site in the presence of large amounts of recombinant SUMO-1 and Ubc9 proteins used for the in vitro conjugation assays.
Taken together, these results provide compelling evidence that two of the three KXE motifs in IE2 are utilized as alternative SUMO-1 modification sites in vivo, with a third lysine as a minor SUMO-1 conjugation site in vitro.
Evaluation of protein interactions of IE2 in yeast two-hybrid system: a possible role for SUMO moieties as a bridge for protein-protein interaction.
We also examined the effect of the K175/180R double mutation on the interaction of IE2 with SUMO-1, SUMO-2/3, or Ubc9 in yeast cells (Fig. 1B). Interestingly, the IE2 bait fragment with the K175/180R double mutations still gave a strong interaction with SUMO-1 (retaining 70% of the wild-type IE2/SUMO-1 interaction level). Furthermore, SUMO-2 and SUMO-3 each showed a two- to threefold-stronger interaction with this double mutant IE2 than with the wild-type IE2 bait fragment. There was little effect on IE2 self-interaction (dimerization). However, Ubc9 failed to show a significant level of interaction with this mutant form of IE2. In light of our in vitro binding data presented above, these results suggest that the yeast two-hybrid readout of the IE2–SUMO-1 interaction includes a direct protein-protein binding component as well as covalent conjugation whereas the IE2-Ubc9 interaction might be primarily indirect through recognition of SUMO-1-modified forms of IE2.
Very recently, Minty et al. proposed that hhXSXS/Taaa (h, hydrophobic residues; a, acidic residues) represents a putative motif for physical interaction with SUMO-1 (43). Interestingly, this consensus sequence is well conserved in HCMV IE2 as IVISDSEEE between positions 200 and 208 near the two lysine residues at positions 175 and 180 that are modified by SUMO-1. Therefore, we wished to test whether this consensus sequence motif in IE2 might account for our evidence for a direct interaction with SUMO moieties. Because S and S/T residues with a single amino acid spacing between them were shown to be critical to the interaction of this motif with SUMO-1 (43), we generated a mutant IE2 protein in which the serine at position 203 was changed to alanine (S203A). GST pulldown assays with 35S-labeled IE2(S203A) revealed only relatively weak interactions with all three SUMO isoforms, similar to the level obtained with Ubc9 (Fig. 3C). These results with IE2(S203A) contrasted with those obtained with wild-type IE2, whose interactions with all three SUMO isoforms were three to fivefold stronger than that with Ubc9 (Fig. 3A), suggesting that the IVISDSEEE motif in IE2 may indeed be involved in efficient direct binding to SUMO moieties.
We also investigated whether SUMO-1 acts as a bridge for interaction of IE2 with cellular proteins by testing the interactions of other IE2-interacting proteins with SUMO-1 in yeast two-hybrid assays. Interestingly, the results showed that 6 of the 22 independent cellular proteins (GAL4-A fusions) identified as IE2 partners in our yeast two-hybrid study also gave significant interactions with GAL4-DB/SUMO-1 (data not shown). These included PIAS-1 and thymidine DNA glycosylase, which were also identified as interaction partners of the SUMO-1-interacting protein p73α, as well as being found to interact independently with SUMO-1 (43). These observations suggest that SUMO moieties directly bound or covalently conjugated to IE2 might act as a bridge for interaction between IE2 and other SUMO-1-interacting proteins.
SUMO-1 modification is not required for the POD targeting of IE2 but may enhance IE2-mediated transactivation.
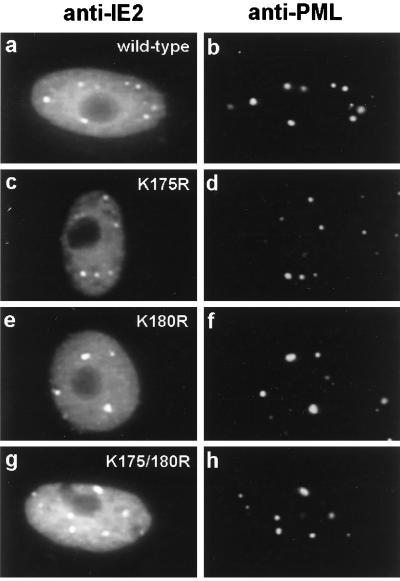
Previous studies have shown that PODs are targeted by both IE1 and IE2 within a few hours after HCMV infection (4). IE1 only transiently colocalizes with PML in the PODs, and subsequently both IE1 and PML become distributed as a nuclear diffuse pattern whereas some IE2 remains in punctate forms. In DNA-transfected cells, IE2 localizes as a mixed pattern of nuclear diffuse and PML-associated punctate forms (5). For the PML protein, modification by SUMO-1 has been suggested to be required for its localization in PODs (46, 63). After generating IE2 mutants that are not modified by SUMO-1, we asked whether their targeting to PODs has been affected in DNA-transfected Vero cells. Double-label IFA analysis was carried out with PML(C) PAb to detect endogenous punctate POD structures and with MAb 12E2 to detect the IE2 mutants. However, both single IE2 K-to-R mutants and the double K175/180R IE2 mutant still exhibited the typical wild-type IE2 mixed punctate and diffuse pattern (Fig. 6). Therefore, SUMO-1 modification is not required for POD targeting by IE2 in DNA-transfected cells.
FIG. 6.
Intracellular localization patterns of mutant IE2 proteins unable to be modified by SUMO-1. Vero cells were transfected with pJHA124 encoding wild-type IE2 (a and b), pYX105 encoding IE2(K175R) (c and d), pYX106 encoding IE2(K180R) (e and f), or pYX104 encoding IE2(K175/180R) (g and h). At 48 h after transfection, the cells were fixed by the paraformaldehyde procedure and double-label IFA was carried out with MAb 12E2 for IE2 (a, c, e, and g) or PAb PML(C) for endogenous PML (b, d, f, and h). FITC-labeled anti-mouse IgG and rhodamine-coupled anti-rabbit IgG were used for visualization.
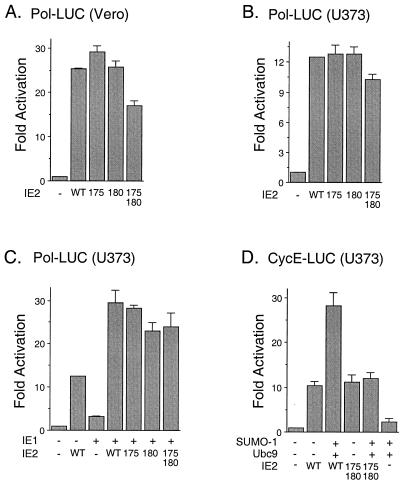
Recent results have shown that SUMO-1 modification of p53 enhances its transactivation activity (16, 50), whereas SUMO-1 modification of c-Jun down-regulates its transactivation (45). Therefore, we were interested in testing whether covalent modification by SUMO-1 has any effect on the transactivation activity of IE2. We first examined the basal transactivation of the HCMV early-gene UL54 (Pol) promoter by mutant IE2 proteins in comparison to wild-type IE2 in the absence of SUMO-1 cotransfection. Vero cells and semipermissive U373-MG cells were cotransfected with a reporter plasmid (Pol-LUC) expressing luciferase under the control of the viral delayed-early Pol promoter and plasmids expressing either wild-type or mutant IE2 (K175R, K180R, or K175/180R). The results of reporter gene readout showed that the activity of the mutant IE2 protein was similar to that of wild-type IE2 in both cell types (Fig. 7A and B). Transactivation of the Pol promoter by IE2 was enhanced by coexpression with IE1. However, there was also little effect of the lysine-to-arginine mutations on this synergistic effect by IE1 in U373-MG cells (Fig. 7C). When SUMO-1 was coexpressed with the wild-type IE2 protein in U373-MG cells, the transactivation of the viral Pol promoter was also similar to that by IE2 alone (data not shown). During HCMV infection, the protein levels of cyclin E and cyclin B are increased (24). Recently, IE2 was shown to activate the exogenous cyclin E promoter through direct binding to the promoter in a transient-transfection assay (7), although in another study the endogenous cyclin E promoter was not activated by IE2 (42). We found that transactivation of the exogenous cellular cyclin E promoter by IE2 was enhanced two- to threefold by coexpression with either SUMO-1 (data not shown) or SUMO-1 plus Ubc9, but this enhancement was not detected when the K175/180R mutant IE2 was used (Fig. 7D). Therefore, as with p53 sumoylation, SUMO-1 appears to show a small positive cooperative effect on the transactivation of some but not all target promoters mediated by IE2.
FIG. 7.
Effect of sumoylation on transactivation by IE2. (A and B) Comparison of transactivation by wild-type and sumoylation mutant forms of IE2 on the HCMV Pol promoter. Vero (A) and U373-MG (B) cells were cotransfected with 1 μg of plasmid DNA containing a reporter gene driven by the HCMV Pol promoter (Pol-LUC) and 2 μg of pJHA124 (encoding intact IE2) (WT), pYX105(K175R), pYX106(K180R), or pYX104(K175/180R). At 48 h after transfection, total-cell extracts were prepared and assayed for luciferase activity. (C) Comparison of cooperative transactivation with IE1 by wild-type and sumoylation mutant forms of IE2 on the HCMV Pol promoter. U373-MG cells were cotransfected with 0.5 μg of reporter plasmid (Pol-LUC) and with 1 μg of either pJHA303 (encoding IE1) or pJHA124 (IE2) alone or with pJHA303 plus wild-type or mutant IE2 plasmid. (D) Comparison of transactivation of the cyclin E promoter by wild-type and sumoylation mutant IE2. U373-MG cells were cotransfected with 0.5 μg of plasmid DNA containing a reporter gene driven by the cellular cyclin E promoter (CycE-LUC) and with various combinations of plasmid pJHA312 encoding Flag–SUMO-1, pWJ5 encoding Flag-Ubc9, and plasmids for wild-type or mutant IE2. Luciferase activities are indicated as fold activation over the basal level of each reporter gene and shown as an average of duplicated experiments.
Redistribution of SUMO-1 during HCMV infection in an IE1-dependent manner.
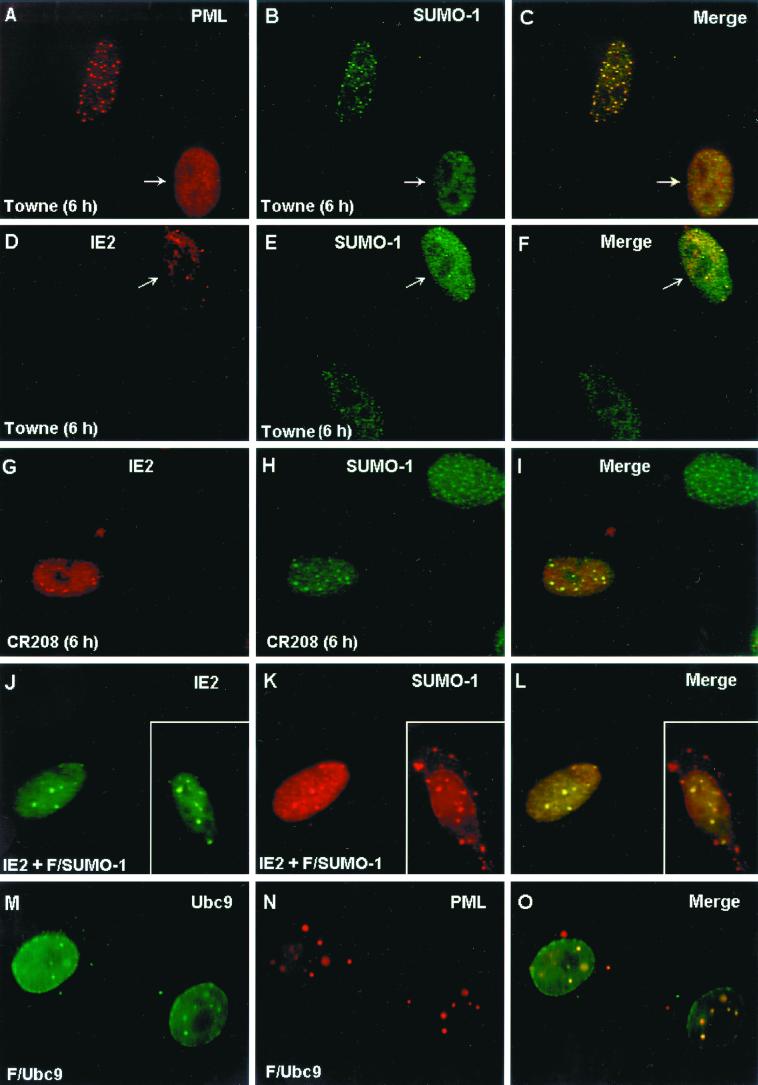
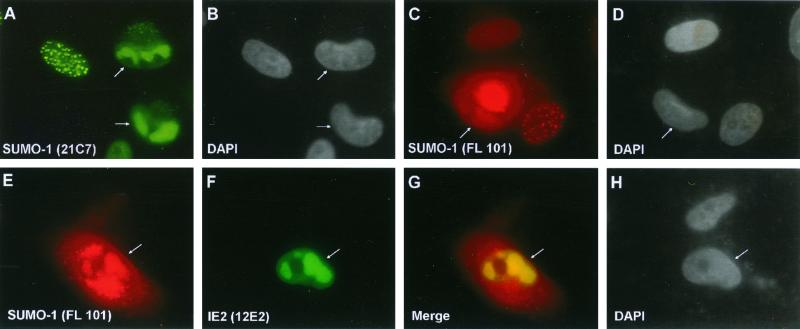
The localization pattern of SUMO-1 was investigated in permissive HF cells infected with wildtype HCMV(Towne) or with the IE1-defective mutant HCMV(CR208). Staining of uninfected HF cells with mouse MAb 21C7 against SUMO-1 showed a mixed IFA pattern including both a nuclear punctate form and a nuclear diffuse form (Fig. 8B). As previously reported, the punctate form of SUMO-1 colocalized with PML in PODs in uninfected cells when observed by double-label IFA with anti-PML PAb (Fig. 8A to C). As expected, at 6 h after HCMV(Towne) infection in HF cells, PODs in infected cells were disrupted and PML was displaced from the PODs into a nuclear diffuse form (Fig. 8A). The SUMO-1 pattern was also changed into a predominantly nuclear diffuse pattern with a small number of residual punctate spots in the HCMV(Towne)-infected cells (Fig. 8B and E). Staining of IE2 with anti-peptide rabbit PAb P3 at 6 h after infection showed a nuclear punctate IFA pattern with nuclear diffuse background (Fig. 8D). Interestingly, when double-label IFA for both IE2 and SUMO-1 was carried out, the punctate forms of both proteins proved to be colocalized in POD-like structures in infected cells (Fig. 8D to F). Therefore, this residual punctate SUMO-1 form appears to be IE2 associated, but not PML associated, because all of the PML protein is already displaced in HCMV(Towne)-infected cells (4). Our previous studies have shown that the disruption of PODs by HCMV infection is due to the function of the virus-encoded IE1 protein (1, 4). To confirm that the change of SUMO-1 localization pattern in HCMV infection requires the presence of IE1 protein, HF cells were also infected with HCMV(CR208) and double labeled for IE2 and SUMO-1. As expected, the localization of SUMO-1 in punctate domains at 6 h was not affected by infection in this case (Fig. 8G to H). However, some of the punctate SUMO-1 signals now colocalized with punctate IE2 (Fig. 8I), although there were more punctate SUMO-1 domains than IE2 domains (compare Fig. 8G and H). These IE2-associated SUMO-1 domains are presumably also PML-associated forms, because the PODs are not disrupted by HCMV(CR208) infection.
FIG. 8.
Displacement of SUMO-1 in HCMV-infected cells and colocalization of IE2 with SUMO-1. (A to I) HF cells were infected with HCMV(Towne) at a low MOI (<1.0 PFU/cell) or with HCMV(CR208) at an MOI of 5.0. At 6 h after infection, the cells were fixed in methanol and double-label IFA was carried out with anti-peptide rabbit PAb PML(C) against PML and mouse MAb 21C7 against SUMO-1 (A to C) or with anti-peptide rabbit PAb P3 against IE2 and MAb 21C7 (D to I). (J to L) Vero cells were cotransfected with pMP18 plasmid DNA encoding wild-type intact IE2 and pJHA312 encoding Flag–SUMO-1 (F/SUMO-1) and then stained for double-labeled IFA for IE2 and Flag–SUMO-1 with mouse MAb and anti-Flag rabbit PAb. (M to O) Vero cells were transfected with plasmid pWJ5 encoding Flag-Ubc9 (F/Ubc9) and double-labeled for Flag-Ubc9 and the endogenous PML with anti-Flag mouse MAb and PAb PML(C). For color IFA visualization, FITC-labeled anti-mouse IgG (green) and rhodamine-coupled anti-rabbit IgG (red) were used.
The colocalization of IE2 with SUMO-1 in a nuclear punctate form was further investigated in Vero cells after cotransfection with IE2 and Flag–SUMO-1 expression plasmids. The overexpressed Flag–SUMO-1 detected with a PAb against the Flag epitope efficiently localized either in a nuclear punctate form (presumably the POD-associated form) or in a nuclear envelope-associated form (presumably the nuclear pore RanGAP1-associated form) (Fig. 8K; the nuclear membrane-associated form is shown in the inset). Double labeling with anti-IE2 MAb 12E2 and anti-Flag PAb confirmed that the two proteins were colocalized in the punctate form but not in the nuclear membrane-associated form (Fig. 8J to L). These IFA results demonstrate that the localization pattern of endogenous SUMO-1 at early stages of the lytic cycle in HCMV-infected cells is changed from a predominantly nuclear punctate PML-associated pattern to a predominantly nuclear diffuse pattern by the IE1 protein, but with some SUMO-1 now associated with the punctate IE2 “immediate-early” transcription domains and prereplication compartments.
The intranuclear localization pattern of Ubc9 has been controversial. When we addressed the question by transfecting Vero cells with Flag-tagged Ubc9 (Flag-Ubc9), we found that the protein was predominantly localized both to nuclear envelope patches and in a nuclear punctate form (Fig. 8M). Double labeling of Flag-Ubc9 and PML showed complete colocalization of the punctate forms of Ubc9 with PML in PODs (Fig. 8M to O). When Vero cells were cotransfected with both IE2 and Flag-Ubc9, the proteins were colocalized in a nuclear punctate form (data not shown).
SUMO-1 accumulates in viral DNA replication compartments at late times after infection.
In an effort to obtain some insights into the role(s) of SUMO-1 redistribution and sumoylation of IE2 during HCMV replication, we further investigated the localization pattern of SUMO-1 at late times after HCMV infection. When the HCMV(Towne)-infected HF cells were fixed at 60 h after infection and stained with mouse MAb 21C7 for SUMO-1, most infected cells showed globular nuclear subdomains that resembled mature viral DNA replication compartments (Fig. 9A and B). A similar pattern of SUMO-1 localization was observed with rabbit PAb FL-101 for SUMO-1, although this rabbit PAb, like most other rabbit PAbs at late times, also gave a nonspecific cytoplasmic staining pattern (Fig. 9C and D). IE2 also accumulates in viral DNA replication compartments at late times after infection (5). Double labeling with rabbit PAb FL-101 (for SUMO-1) and mouse MAb 12E2 (for IE2) showed that the SUMO-1 and IE2 IFA signals were colocalized in the same nuclear subdomains, indicating that the SUMO-1-positive nuclear structures do indeed correspond to viral DNA replication compartments (Fig. 9E to H). These results indicate that SUMO-1 (possibly in the form of sumoylated cellular proteins) and IE2 are both very efficiently incorporated into viral DNA replication compartments and suggest that sumoylation of proteins including IE2 may participate in or affect viral DNA replication processes.
FIG. 9.
Localization of SUMO-1 within viral DNA replication compartments in HCMV-infected cells. HF cells were infected with HCMV(Towne) at an MOI of 3.0. At 60 h after infection, the cells were fixed by the paraformaldehyde procedure. (A to D) Single-label IFA for SUMO-1 was carried out either with mouse MAb 21C7 (A) or with rabbit PAb FL-101 (C). (E to H) Double-label IFA for both SUMO-1 and IE2 was performed with both rabbit PAb FL-101 and mouse MAb 12E2 (for IE2). SUMO-1, IE2, and the merge images from a single microscopic field are shown in panels E, F, and G, respectively. For color IFA visualization, FITC-labeled anti-mouse IgG (green) and rhodamine-coupled anti-rabbit IgG (red) were used. DAPI staining for the same fields of panels A, C, and E to G are shown in panels B, D, and H, respectively. HCMV-infected cells showing viral DNA replication compartments are indicated by arrows. The rabbit PAb gives nonspecific cytoplasmic staining in addition to specific nuclear staining in late-stage HCMV-infected cells.
DISCUSSION
The data presented here demonstrate that both covalent conjugation and a direct protein-protein interaction occur between the HCMV pleotrophic regulatory protein IE2 and ubiquitin-like SUMO proteins. Two lysine residues were identified as the major alternative SUMO-1 attachment sites, which are surrounded by a short consensus sequence similar to that present in all other known SUMO-1 substrates. Either one or two molecules of SUMO-1 could be covalently attached to a single 86-kDa IE2 polypeptide in vivo, giving rise to two distinct isoforms with apparent molecular masses of 105 and 120 kDa (Fig. 4B, lane 4, and Fig. 5A, lane 3). These minor isoforms of IE2 (representing up to 30% of the protein in some cases) were detected directly in both U373 and HF cells infected with HCMV, as well as in cells infected with Ad-IE2. Similarly, some of the IE2 protein synthesized in DNA-transfected Vero cells was modified by SUMO, with the levels being greatly enhanced by cotransfection with either Flag–SUMO-1 or Flag–SUMO-2.
Recently, Hofmann et al. reported the covalent modification of IE2 by SUMO-1 or SUMO-3 on either one or other of the same two lysine residues that we have mapped in this study (20). However, unlike their results, we were able to detect an additional 120-kDa IE2 isoform either with IE2 MAb (CH810) in direct immunoblot analysis of transfected cell lysates or with anti-SUMO-1 MAb in IE2 immunoprecipitates. This form presumably has two molecules of Flag–SUMO-1 attached to it. The existence of the 120-kDa isoform, along with our site-directed mutagenesis mapping data, provided compelling evidence that both lysine 175 and 180 can be modified by SUMO-1 simultaneously, in contrast to the “mutually exclusive” scenario proposed by Hofmann et al. (20). Furthermore, we found that the IE2 protein could be almost completely sumoylated in vitro, with a third lysine being used as a minor SUMO-1 conjugation site, and that IE2 also contains a motif involved in direct physical interaction with SUMO-1.
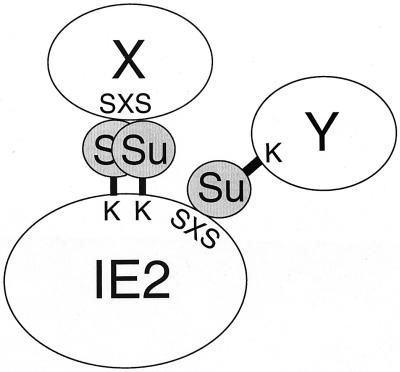
As far as we are aware, the HCMV IE1 and IE2 proteins are the first virus-encoded proteins demonstrated to be subjected to modification by the novel cellular mechanism that conjugates SUMO to an important subset of cellular nuclear proteins. Some of the previously characterized cellular SUMO-1 substrates, including PML, RanGAP1, and IκBα, are also known to interact with SUMO-1 and/or Ubc9 in yeast two-hybrid analysis. Such interactions were merely considered preludes to potential covalent conjugation. However, the recent identification of a SUMO-1 interaction motif among several cellular proteins (43) revealed the previously ignored importance of noncovalent interaction between certain proteins and SUMO-1. Our results established that the IE2–SUMO-1 interaction includes both covalent conjugation and noncovalent interaction. Intriguingly, IE2 contains a potential SUMO-1 interaction motif almost identical to the reported consensus sequence (43) and the in vitro interaction between IE2 and SUMO-1 was reduced fivefold by a point mutation in this motif. The finding that IE2 is able to interact physically with SUMO-1 and that some IE2-interacting partners identified in this study also interact directly with SUMO-1 in yeast two-hybrid assays led to our working hypothesis that IE2 may associate with several cellular proteins through binding to covalently conjugated SUMO moieties (Fig. 10).
FIG. 10.
Possible role for SUMO-1 as a bridge for the interaction of IE2 with cellular proteins. SUMO-1 can be either covalently attached to lysine residues (K) or physically bound to the putative SUMO-1-binding consensus sequence (IVISDSEEE, designated SXS) within IE2. Both cellular proteins containing SUMO-1-binding consensus sequences (X) or those conjugated by SUMO-1 (Y) appear capable of interacting with IE2 via the SUMO-1 bridging molecules.
Our yeast two-hybrid screen detected not only SUMO-1 but also SUMO-2 and SUMO-3 as IE2-interacting partners, although the IE2–SUMO-1 interaction was 30 times stronger than the IE2–SUMO-2 or IE2–SUMO-3 interaction in yeast cells. However, SUMO-2 could be conjugated to IE2 with similar efficiency to SUMO-1 in our mammalian cell cotransfection assay. Furthermore, our in vitro binding data showed no difference in direct binding affinity between IE2 and any of the three SUMO isoforms. The difference in the β-galactosidase readout in yeast two-hybrid assays could be explained by preferential conjugation of SUMO-1 to IE2 or by differential expression efficiency or protein stability of SUMO-1 and SUMO-2 in yeast cells. Human SUMO-1 (but not SUMO-2) restores growth to the yeast smt3Δ mutant (27), suggesting that yeast Ubc9 can distinguish between human SUMO-1 and SUMO-2/3. Most recently, Saitoh and Hinchey (52) found that protein-damaging stimuli rapidly induce the accumulation of high-molecular-mass SUMO-2/3 conjugates, while SUMO-2/3 conjugated poorly to one of the major SUMO-1 substrates, RanGAP1. It will be intriguing to investigate whether there is a functional distinction between SUMO-1 and SUMO-2/3 conjugation of the IE2 protein.
Our results comparing wild-type IE2 and the IE2 sumoylation-negative double mutant imply that IE2-dependent transactivation can be modulated somewhat by sumoylation, although only after inclusion of exogenous SUMO-1 in a transient-cotransfection assay for enhancement of cyclin E expression. However, there was no effect on the viral Pol promoter target even under conditions of IE1 plus IE2 synergistic activation. Hofmann et al. (20) reported a five- to sixfold-reduced stimulation of the viral UL112-113 or UL84 promoter targets by sumoylation-negative IE2. Based on previous studies on IE2, together with insights from other SUMO-1 substrates such as p53, PML, and RanGAP1, we propose two possible mechanisms for these effects. First, sumoylation of IE2 might regulate its association with proteins in the basal transcriptional machinery such as TBP and TFIIB or with proteins involved in coactivator-corepressor complexes. SUMO-1 modification of RanGAP1 was hypothesized to induce a conformational change and expose its RanBP2-binding domain (40). This scenario of sumoylation regulating protein-protein interactions appears to be one of the general themes of SUMO-1 conjugation pathways. Second, the oligomerization of IE2 was shown to be critical for specific DNA binding by IE2 to CRS-like target sequences and probably also plays a role in transcriptional regulation (60a). Although we found that the sumoylation-negative mutant IE2 still down-regulated the MIE promoter to normal levels in a dosage-dependent manner (data not shown), the possibility remains that SUMO-1 conjugation to IE2 might increase (or decrease) its oligomerization and/or protein stability.
Recent studies have established that SUMO-1 modification of PML is critical for both formation of PODs and recruitment of other POD-targeting proteins such as Sp100, Daxx, and CREB-binding protein (22, 46, 63). However, SUMO-1 conjugation of another POD component protein, Sp100, is not required for its targeting to PODs (59). Similarly, we found that SUMO-1 modification of IE2 is not required for its localization to PODs in DNA-transfected cells. Our finding that the intranuclear punctate form of Ubc9 completely colocalizes with PML in PODs (Fig. 8M to O) raises the possibility that sumoylation of certain POD proteins could occur after their targeting to PODs. Our preliminary results also suggest that the nuclear punctate IE2 domains still form after transient expression in PML−/− MEF cells (Y. Xu, J.-H. Ahn, P. Salomoni, P. P. Pandolfi and G. S. Hayward, unpublished data). These observations led to the hypothesis that the intranuclear punctate localization of IE2 does not involve direct recruitment by PML protein, in contrast to the PML-dependent transient targeting of IE1 to PODs (1).
The subnuclear localization pattern of SUMO-1 in early-stage HCMV-infected cells showed that in addition to PML and Sp100, most SUMO-1 was rapidly dispersed into a nuclear diffuse form in an IE1-dependent fashion. However, some residual SUMO-1 (but not PML) was retained within or relocated to the IE2 punctate “immediate-early” subnuclear domains, which are adjacent to but distinct from PML punctate domains (5, 23). Most dramatically, SUMO-1 (but, again, not PML) was very efficiently recruited together with IE2 into viral DNA replication compartments at later stages of the HCMV lytic cycle. We do not know at present whether the dispersed SUMO-1 in HCMV-infected cells is still associated with its cellular substrates or released from them. Potentially, the SUMO-1 recruited to both IE2 punctate domains and viral replication compartments could also include forms that are bound or conjugated to other proteins.
Overall, these observations suggest that in addition to IE2, the sumoylation pattern and the associated functions of some key cellular proteins may be affected by HCMV infection. Although the changes in cellular localization of PML and SUMO-1 effected by IE1 are not essential for the HCMV lytic cycle at high MOI in cell culture, the absence of IE1 has profound effects under the presumably more physiological conditions of low MOI (3, 17). At low MOI, in the absence of IE1 in a U373 cell line that overexpresses PML, the IE2 immediate-early punctate domains persist for much longer than normal and progression through to the late stages of infection typified by replication compartment formation is either highly delayed or absent in many IE2-positive cells (3). We expect that displacement of PML and SUMO-1 from PODs and finally recruitment of both IE2 and SUMO-1 (but not IE1 and PML) into mature replication compartments are events whose efficiency and timing may be controlled or greatly affected by the levels of sumoylation of both IE1 and IE2 and by the interaction of IE2 with sumoylated cellular proteins.
ACKNOWLEDGMENTS
J.-H. Ahn and Y. Xu contributed equally to this work.
This study was funded by Public Health Service research grants RO1 AI24576 to G.S.H. from the National Institute of Allergy and Infectious Diseases and PO1 HL56091 to G.S.H. from the National Heart, Lung, and Blood Institute.
We thank Dolores Ciufo for the PML(C) polyclonal antibody and Jianchao Zong for DNA sequencing. We also thank Aubrey Thompson, Hisato Saitoh, and Masahiro Fujimuro for gifts of plasmids. We are grateful to Edward S. Mocarski for samples of the pair of IE1-deleted CR208 virus and its parent HCMV(Towne) virus. We also thank Cecile M. Pickart for helpful suggestions.
REFERENCES
- 1.Ahn J H, Brignole III E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J H, Chiou C J, Hayward G S. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1998;210:25–36. doi: 10.1016/s0378-1119(98)00056-0. [DOI] [PubMed] [Google Scholar]
- 3.Ahn J H, Hayward G S. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology. 2000;274:39–55. doi: 10.1006/viro.2000.0448. [DOI] [PubMed] [Google Scholar]
- 4.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn J H, Jang W J, Hayward G S. The human cytomegalovirus IE2 and UL112–113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73:10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 7.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 8.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 9.Buschmann T, Fuchs S Y, Lee C G, Pan Z Q, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 10.Caswell R, Hagemeier C, Chiou C J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 11.Chiou C J, Zong J, Waheed I, Hayward G S. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol. 1993;67:6201–6214. doi: 10.1128/jvi.67.10.6201-6214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desterro J M, Rodriguez M S, Hay R T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 13.Desterro J M, Thomson J, Hay R T. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 14.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 15.Gong L, Kamitani T, Fujise K, Caskey L S, Yeh E T. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J Biol Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 16.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz S E, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann H, Floss S, Stamminger T. Covalent modification of the transactivator protein IE2–p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J Virol. 2000;74:2510–2524. doi: 10.1128/jvi.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishov A M, Sotnikov A G, Negorev D, Vladimirova O V, Neff N, Kamitani T, Yeh E T, Strauss III J F, Maul G G. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson E S, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson E S, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 27.Johnson P R, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 28.Jupp R, Hoffmann S, Depto A, Stenberg R M, Ghazal P, Nelson J A. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J Virol. 1993;67:5595–5604. doi: 10.1128/jvi.67.9.5595-5604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jupp R, Hoffmann S, Stenberg R M, Nelson J A, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–7546. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaFemina R-L, Hayward G S. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J Gen Virol. 1988;69:355–374. doi: 10.1099/0022-1317-69-2-355. [DOI] [PubMed] [Google Scholar]
- 31.LaFemina R L, Pizzorno M C, Mosca J D, Hayward G S. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology. 1989;172:584–600. doi: 10.1016/0042-6822(89)90201-8. [DOI] [PubMed] [Google Scholar]
- 32.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 33.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Leo C, Zhu J, Wu X, O'Neil J, Park E J, Chen J D. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1. cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukonis C J, Weller S K. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J Virol. 1997;71:2390–2399. doi: 10.1128/jvi.71.3.2390-2399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 39.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matunis M J, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. [Google Scholar]
- 42.McElroy A K, Dwarakanath R S, Spector D H. Dysregulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130. J Virol. 2000;74:4192–4206. doi: 10.1128/jvi.74.9.4192-4206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 44.Mocarski E S. Cytomegalovirus and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 45.Muller S, Berger M, Lehembre F, Seeler J S, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 46.Muller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pizzorno M C, Mullen M A, Chang Y N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pizzorno M C, O'Hare P, Sha L, LaFemina R L, Hayward G S. trans-Activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez M S, Desterro J M, Lain S, Midgley C A, Lane D P, Hay R T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose M D, Winston F, Hieter P. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 52.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 53.Saitoh H, Pu R T, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 54.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spaete R R, Mocarski E S. Insertion and deletion mutagenesis of the human cytomegalovirus genome. Proc Natl Acad Sci USA. 1987;84:7213–7217. doi: 10.1073/pnas.84.20.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Speir E, Modali R, Huang E S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 58.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sternsdorf T, Jensen K, Reich B, Will H. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J Biol Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- 60.Stinski M F, Thomsen D R, Stenberg R M, Goldstein L C. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60a.Waheed I, Chiou C-J, Ahn J-H, Hayward G S. Binding of the human cytomegalovirus 80-kD immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology. 1998;252:235–257. doi: 10.1006/viro.1998.9448. [DOI] [PubMed] [Google Scholar]
- 61.Wara-aswapati N, Yang Z, Waterman W R, Koyama Y, Tetradis S, Choy B K, Webb A C, Auron P E. Cytomegalovirus IE2 protein stimulates interleukin-1β gene transcription via tethering to Spi-1/PU.1. Mol Cell Biol. 1999;19:6803–6814. doi: 10.1128/mcb.19.10.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoo Y D, Chiou C J, Choi K S, Yi Y, Michelson S, Kim S, Hayward G S, Kim S J. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–7070. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong S, Muller S, Ronchetti S, Freemont P S, Dejean A, Pandolfi P P. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]