Isolation of a small molecule inhibitor of DNA base excision repair (original) (raw)
Abstract
The base excision repair (BER) pathway is essential for the removal of DNA bases damaged by alkylation or oxidation. A key step in BER is the processing of an apurinic/apyrimidinic (AP) site intermediate by an AP endonuclease. The major AP endonuclease in human cells (APE1, also termed HAP1 and Ref-1) accounts for >95% of the total AP endonuclease activity, and is essential for the protection of cells against the toxic effects of several classes of DNA damaging agents. Moreover, APE1 overexpression has been linked to radio- and chemo-resistance in human tumors. Using a newly developed high-throughput screen, several chemical inhibitors of APE1 have been isolated. Amongst these, CRT0044876 was identified as a potent and selective APE1 inhibitor. CRT0044876 inhibits the AP endonuclease, 3′-phosphodiesterase and 3′-phosphatase activities of APE1 at low micromolar concentrations, and is a specific inhibitor of the exonuclease III family of enzymes to which APE1 belongs. At non-cytotoxic concentrations, CRT0044876 potentiates the cytotoxicity of several DNA base-targeting compounds. This enhancement of cytotoxicity is associated with an accumulation of unrepaired AP sites. In silico modeling studies suggest that CRT0044876 binds to the active site of APE1. These studies provide both a novel reagent for probing APE1 function in human cells, and a rational basis for the development of APE1-targeting drugs for antitumor therapy.
INTRODUCTION
The DNA base excision repair (BER) pathway is required for the accurate removal of bases that have been damaged by alkylation, oxidation or ring-saturation. This pathway also handles a variety of other lesions including deaminated bases and DNA single-strand breaks (1). Although there is more than one sub-pathway of BER (2) in most cases excision of a damaged base by a DNA glycosylase enzyme leads to the formation of a potentially cytotoxic apurinic/apyrimidinic (AP) site intermediate (3,4). This is a target for an AP endonuclease, which cleaves the phosphodiester backbone on the 5′ side of the AP site via a hydrolytic mechanism (4). The major AP endonuclease in human cells, APE1 (also called previously HAP1 and Ref-1), accounts for over 95% of the total AP endonuclease activity in most cultured human cell lines (5–8). APE1 is a member of the highly conserved exonuclease III family of AP endonucleases, named after the Escherichia coli homologue of APE1 (9). A second family of AP endonucleases is found in most organisms, the prototypical member of which is E.coli endonuclease IV (10). X-ray crystallographic analysis on AP endonucleases from bacteria to human cells, have revealed that members of the exonuclease III (11,12) and endonuclease IV (13) families are structurally unrelated, despite being able to catalyze AP site cleavage reactions that generate identical products.
In common with exonuclease III (14) [and endonuclease IV (10)] APE1 performs roles in DNA repair other than AP site processing (15). APE1 exhibits a 3′-phosphodiesterase activity for removal of fragmented sugar moieties which are found at the 3′ end of DNA strand breaks induced by certain drugs, such as bleomycin, and by ionizing radiation (16). APE1 also possesses a weak 3′-phosphatase activity, a 3′–5′-exonuclease activity and an RNaseH activity; however, the functional significance of these additional activities remains obscure (15). APE1 also plays a role in the recently described nucleotide incision pathway (17). All of these activities apparently utilize a single active site in the DNA repair domain of APE1, which is the region of the protein that is conserved in exonuclease III. A separate domain in APE1, located close to the N-terminus, performs a role unrelated to the direct repair of DNA damage. This domain of APE1 performs a redox regulatory function that can maintain certain transcription factors, such as p53, c-Jun and Hif-1α, in a reduced and therefore activated state for DNA binding (18–21).
The action of APE1 on an AP site generates a strand break with a 3′-hydroxyl terminus, which can prime DNA repair synthesis, and a 5′-deoxyribose phosphate (5′dRp) terminus. The 5′dRp residue must be removed in order for the repair process to be completed. This task is accomplished by the dRp lyase domain present in DNA polymerase β, the enzyme that also performs the task of filling in the single base gap thus formed (22). Repair is then completed by ligation of the nick, which is generally catalyzed by DNA ligase III in association with its binding partner XRCC1. This pathway has been termed ‘short patch’ BER (23).
Several preclinical and clinical studies have been indicated that APE1 may be an attractive target for anticancer drug development. Although homozygous deletion of the APE1 homolog in the mouse (APEX) leads to embryonic lethality (24,25), APEX± heterozygous mice are hypersensitive to oxidative stress (26). Depletion of intracellular APE1 also sensitizes human cells to a variety of cytotoxic agents. Using either antisense oligonucleotide or RNA interference approaches, several groups have reported that APE1 downregulation confers sensitivity to mono-functional alkylating agents, such as methylmethane sulfonate (MMS), and oxidizing agents, such as hydrogen peroxide (27–31). Moreover, a recent study indicated that severe depletion of APE1 retarded cell proliferation and led to an accumulation of unrepaired AP sites (32). APE1 function may also be related to the pathogenesis of several human cancers, and its expression may have prognostic and/or predictive significance. For example, APE1 is overexpressed in several human tumors (33–40) and its expression pattern appears to have prognostic significance in cancers of the breast (33), lung (41,42) and bone (31). In cervical cancers, increased APE1 expression has been shown to be associated with radioresistance (34), and a high level of nuclear APE1 was shown to correlate with resistance to chemotherapy and poor overall survival in patients with head and neck cancer (39).
Given the strong preclinical and clinical evidence that APE1 may be a promising anticancer-drug target, a drug discovery programme was undertaken. As APE1−/− human cell lines cannot be generated, an important additional aim of this study was to generate laboratory reagents to probe the functional consequences of APE1 inactivation. A fluorescence-based, high-throughput assay was developed to screen for inhibitors of the AP endonuclease activity of APE1. A small molecule chemical library was screened and several inhibitors of APE1 were isolated. Amongst these, CRT0044876 was identified as a specific APE1 inhibitor, and this agent has been analyzed here in detail. At non-toxic concentrations, CRT0044876 potentiated the cytotoxicity of several DNA damaging agents, which generate damage that is repaired in the BER pathway, including some currently-used anticancer drugs. This potentiation of cytotoxicity was shown to correlate with an accumulation of unrepaired AP sites in CRT0044876-treated cells.
MATERIALS AND METHODS
Enzymes, chemicals and oligonucleotides
Human APE1 and uracil-DNA glycosylase were purified to homogeneity as described previously (43,44). E.coli endonuclease III and endonuclease IV were obtained from Trevigen. E.coli exonuclease III and BamHI restriction endonuclease were obtained from New England Biolabs. Topoisomerase I was obtained from Promega. Whole cell extracts were prepared by a method as described previously (45). Protein concentration was determined using the BioRad Coomassie blue assay with bovine serum albumin (BSA) as a standard.
MMS, 5-hydroxymethyl-2′-deoxyuridine (hmdUrd), camptothecin and mechlorethamine hydrochloride (nitrogen mustard) were all purchased from Sigma-Aldrich. Temozolomide was a gift from Prof. Malcolm Stevens, University of Nottingham, UK. Stock solutions of potential APE1 inhibitors and camptothecin were dissolved in DMSO. MMS, hmdUrd, temozolomide and mechlorethamine hydrochloride were dissolved in phosphate buffered saline (PBS).
The oligonucleotides; 5′-F-GCCCCCXGGGGACGTACGATATCCCGCTCC-3′ and 3′-Q-CGGGGGCCCCCTGCATGCTATAGGGCGAGG-5′ (where F is Fluorescein, Q is Dabcyl and X is Terahydrofuran, an abasic site analog) were custom-made by Eurogentec Ltd. A uracil-containing 18mer oligonucleotide 5′-CTCGCAAGUGGGTACCGA-3′ and its complementary oligonucleotide, 5′-TCGGTACCCGCTTGCGAG-3′ were synthesized by the Cancer Research UK central services laboratory (Clare Hall, UK). The 3′-phosphoglycolate (PG) substrate (5′-CAATAGAGTAACACGGpg-3′) was synthesized by Eurogentec using the method of Urata and Akagi (46), in which the 3′-phosphoglyceryl residue is oxidized sequentially by sodium periodate (NaIO4) and sodium chlorite (NaClO2). This oligonucleotide, which has been characterized previously (16), was purified by high-performance liquid chromatography (HPLC) and the presence of 3′-PG was confirmed by mass spectrometry.
Fluorescence-based, high-throughput screening
A library of 5000 ‘drug-like’ compounds was purchased from Maybridge Chemicals (Tintagel, UK). A high-throughput assay to identify potential APE1 inhibitors was developed (Figure 1). Briefly, APE1 enzyme (0.7 nM) was incubated with or without library compounds in a buffer system comprising of 50 mM Tris–HCl (pH 8.0), 1 mM MgCl2, 50 mM NaCl and 2 mM dithiothreitol (DTT) at 37°C for 10 min. The sequence 5′-F-GCCCCCXGGGGACGTACGATATCCCGCTCC-3′ and its complementary oligonucleotide 3′-Q-CGGGGGCCCCCTGCATGCTATAGGGCGAGG-5′ were annealed and the reaction was initiated by addition of the annealed substrate. Fluorescence readings were taken continuously during a 25 min incubation at 37°C (using a Molecular Devices Spectramax Gemini plate reader and Softmax Pro software package, Kinetic Mode with a 495 nM Excitation and a 530 nM Emission). If the DNA is cleaved at the abasic site at position 7 from the 5′ end by APE1, the 6mer Fluorescein-containing molecule can dissociate from its complement by thermal melting. As a result, the quenching effect of the 3′-Dabcyl molecule (which absorbs fluorescein fluorescence when in close proximity) is lost, and APE1 activity can be measured indirectly as an increase in fluorescence signal. Before the use of this assay for screening, it was shown that the 6mer Fluorescein-containing oligonucleotide did indeed fully dissociate from the complementary oligonucleotide when incubated at 37°C (data not shown).
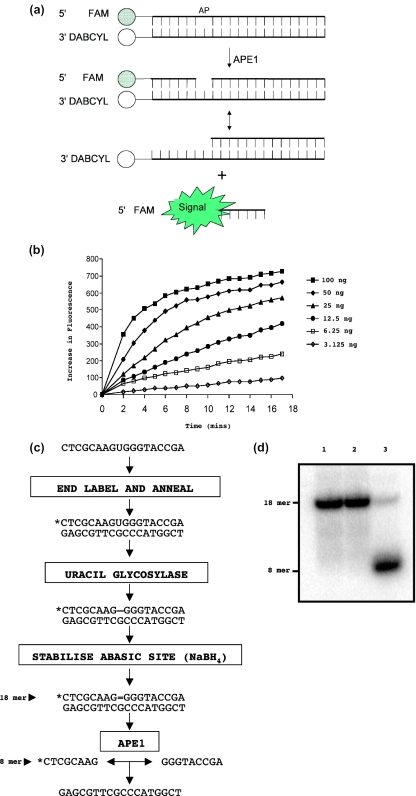
Figure 1.
Screening for APE1 inhibitors. (a) Fluorescence-based assay for APE1 activity. A DNA 30mer duplex was dual tagged with a quench (DABCYL)—fluor (FAM) pair. Separation of the quench and fluor occurs following cleavage of the single abasic site (tetrahydrofuran). (b) Fluorescence signal measurements using the APE1 assay. Increasing amounts of APE1 were mixed with 100 nM oligonucleotide substrate under conditions described in the Materials and Methods. A concentration dependent rise in fluorescence indicated AP site cleavage activity by APE1. (c) Scheme for the gel-based AP site cleavage assay. (d) Example of the assay, using a bacterial cell extract expressing recombinant APE1. The 18mer uncut substrate (lane 1) and 8mer APE1 cleavage product (lane 3) are indicated. Immunodepletion of APE1 in the extract using protein A-sepharose beads abolished the cleavage activity of the extract (lane 2) confirming that cleavage was mediated by APE1.
AP site cleavage assay
A uracil-containing oligonucleotide [5′-CTCGCAAGUGGGTACCGA-3′] was 5′ end-labeled with [γ-32P]ATP in a 20 μl reaction containing 50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 0.1 mM EDTA, 5 mM DTT, 20 μCi [γ-32P]ATP and 1 U T4-polynucleotide kinase. The mixture was incubated at 37°C for 1 h, and the reaction was terminated by the addition of 50 μl TE buffer. The DNA was then passed through a Sephadex G-25 spin column (Roche). The radiolabeled, uracil-containing oligonucleotide was then annealed to an equimolar amount of complementary oligonucleotide in TE buffer containing 100 mM KCl by incubation at 90°C for 3–5 min, followed by a slow cooling to room temperature. The annealed DNA substrate (50 pmol) was pretreated with uracil-DNA glycosylase (6.25 pmol) in 10 mM HEPES (pH 7.9), 1 mM EDTA and 100 mM KCl. The reaction mixture was incubated at 37°C for 1 h. The resulting AP site indicated was chemically reduced by the addition of sodium borohydride to 0.1 M. After incubation on ice-cold for 10 min, the reaction buffer was exchanged to TE buffer by a passage through a Sephadex G-25 spin column.
BER reaction buffer comprised 40 mM HEPES–KOH (pH 7.8), 5 mM MgCl2, 0.5 mM DTT and 0.1 mM EDTA. A 10 μl AP site cleavage reaction comprised of BER buffer mix, purified protein (3.3 nM final concentration of APE1) and 0.75 ng reduced AP site double-stranded oligonucleotide. The mixture was incubated at 37°C for 1 h. A total of 1 μl of stop buffer (50% glycerol, 10 mM Tris–HCl, 1 mM EDTA, 0.1% bromophenol blue and 0.1% Xylene cyanol) was added, and the sample mixture was denatured at 90–100°C for 2 min. The sample was then run on a 15% TBE Criterion™ Pre-Cast Gel (Bio-Rad), with electrophoresis at a constant current of 30 mA for 30 min, and the radiolabeled substrate and reaction products were visualized using a phosphorImager (Molecular Dynamics). The inhibitory activity of potential APE1-targeting compounds were analyzed at drug concentrations ranging from 0.1 to 100 μM. The resolved radiolabeled bands were quantified using ImageQuant software analysis, and IC50 values were calculated by determining the concentration of the inhibitor that reduced APE1 activity to 50% of the control values.
Similar AP site cleavage assays were set up to measure exonuclease III activity [in a 10 μl reaction vol containing 10 mM Bis-Tris-Propane–HCl (pH 7.0), 10 mM MgCl2, 1 mM DTT, reduced AP site-containing double-stranded oligonucleotide (0.75 ng) and 0.1 U of Exonuclease III (New England BioLabs) and incubated for 1 h at 37°C]. Endonuclease IV was assayed in a similar manner [in a 10 μl reaction volume containing 10 mM HEPES–KOH, 100 mM KCl (pH 7.4), reduced AP site-containing double-stranded oligonucleotide (0.75 ng) and 0.01 U of Endonuclease IV (TREVIGEN®)]. Reactions were incubated for 1 h at 37°C. In assays containing HeLa whole cell extracts, a 10 μl reaction comprising of 10 mM HEPES–KOH (pH 7.4), 100 mM KCl, the reduced AP site-containing double-stranded oligonucleotide (0.75 ng) and 30 ng of HeLa whole cell extract was used, and the reaction mix was incubated for 1 h at 37°C.
3′-phosphodiesterase activity of APE1
The 3′-PG containing oligonucleotide (5′-CAATAGAGTAACACGGpg-3′) was 5′ end labeled with [γ-32P]ATP using T4-polynucleotide kinase and unincorporated label removed on a Sephadex G-25 spin column. To prepare the 3′-PG duplex substrate, the labeled 3′-PG oligonucleotide was mixed with an equimolar amount of the oligonucleotide 5′-pCGACCAGTCCCTGCCAATC-3′, and both were annealed with a 2-fold excess of the oligonucleotide 5′-GATTGGCAGGGACTGGTCGGCCGTGTTACTCTATTG-3′ at 90°C for 3–5 min followed by slow a cooling to room temperature to create an oligonucleotide with a 1 nt gap flanked by 3′-PG and 5′-phosphate ends. APE1 (0–300 fmol) was incubated with 300 fmol duplex oligonucleotide per reaction in 20 μl reaction buffer containing 50 mM HEPES–KOH (pH 7.8), 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 1.5 mM DTT, 8.5% glycerol, 2 mM ATP and 50 μg/ml BSA. For inhibitor studies, APE1 (150 fmol) was preincubated with inhibitor (0–10 μM) for 10 min on ice-cold before addition of the substrate. Reactions were incubated for 10 min at 30°C and 20 μl formamide loading dye was added (95% formamide, 0.02% Xylene cyanol and 0.02% bromophenol blue) and the samples were heated to 95°C for 3 min. Products were analyzed by 20% denaturing PAGE, and the gels were exposed to storage phosphor screens at 4°C before analysis by phosphorimaging.
3′-phosphatase activity of APE1
Through its AP-lyase activity, E.coli formamidopyrimidine [fapy]- DNA glycosylase (fpg) cleaves both 3′ and 5′ to the AP site thereby removing the AP site and leaving a one base gap between 5′ and 3′-phosphate residues. The 3′-phosphatase activity of APE1 removes the 3′-phosphate group and restores a 3′-OH priming terminus. The abasic oligonucleotide substrate was prepared as described above. One unit of fpg was added to this reaction and the mixture was incubated at 37°C for 30 min. To the above reaction mix, 3.3 nM of APE1 was added and the sample was incubated at 37°C for 1 h. A total of 1 μl of stop buffer (50% glycerol, 10 mM Tris–HCl, 1 mM EDTA, 0.1% bromophenol blue and 0.1% Xylene cyanol) was added at the end of the reaction and the sample mix was denatured at 90–100°C for 2 min. The sample was then electrophoresed in a 20% TBE-urea gel at a constant current of 30 mA for 2 h and imaged using a phosphorImager analysis (Molecular Dynamics). To test the inhibitory activity of the compounds on the 3′-phosphatase activity of APE1, a serial dilution of the potential inhibitor of APE1 was set up in the above reaction mix (after the AP lyase reaction of fpg) and the assay was perfomed as described previously.
BamHI restriction endonuclease assay
BamHI activity was assayed using conventional restriction digestion of an 8.8 Kb plasmid (β-gal-pcDNA; Invitrogen) containing two BamHI recognition sites. Reaction products were analyzed on a 0.8% agarose gel (with ethidium bromide at 0.03 ng/μl).
Topoisomerase I relaxation assay
φX174 RF I DNA (New England Biolabs) was used for measuring topoisomerase I DNA relaxation activity. Briefly, 200 ng of the DNA was added to a reaction mixture containing 50 mM Tris–HCl (pH 7.9), 5 mM MgCl2, 50 mM NaCl, 1 μg/μl BSA, 1 mM DTT and 0.25 U of wheat germ topoisomerase I and the mixture was incubated for 1 h at 37°C. A total of 1 μl of stop buffer (50% glycerol, 10 mM Tris–HCl, 1 mM EDTA, 0.1% bromophenol blue and 0.1% Xylene cyanol) was added at the end of the reaction and the samples were electrophoresed on a 1% agarose gel. The DNA was stained with 0.03 μg/ml of ethidium bromide.
Clonogenic survival assays
HT1080 fibrosarcoma cells were grown in 2% RPMI medium [supplemented with penicillin 0.06 g/l, streptomycin 0.1 g/l (pH 7.0), 10% fetal bovine serum (FBS, Life Technologies) and 4 mM glutamine]. Only cells with a plating efficiency of ≥60% were used for clonogenic survival analysis. Tissue culture dishes (10 cm) were seeded with 500 cells per dish and the culture was maintained in a humidified incubator at 37°C in an atmosphere of 5% CO2 and 95% air. To evaluate the toxicity profile of putative APE1 inhibitors, various concentrations (100–500 μM) of inhibitor were added to the medium, and cultures were incubated for 7–10 days until cell colonies were formed. Colonies were fixed [75% (v/v) methanol, 25% (v/v) acetic acid] for 30 min and stained with crystal violet (1 mg/ml in distilled water) for 4 h at room temperature. Visible colonies were counted on a colony counter (Stuart Scientific, UK).
To evaluate the potentiation of cytotoxicity of DNA damaging agents by putative APE1 inhibitors, 500 cells were plated in the absence or presence of APE1 inhibitor and allowed to adhere to the plate for 1 h. Cells were then exposed to one of the following DNA damaging agents in the concentration range indicated in parentheses: MMS for 1 h (100–500 μM), temozolomide for 1 h (200–1400 μM), mechlorethamine hydrochloride for 1 h (1–3 μM), hmdUrd for 24 h (1–4 μM), camptothecin for 24 h (2.5–50 nM), or UV light [4 Jm−2–20 Jm−2, using a UV Stratalinker (254 nM UV light). The medium was removed and 1 ml of PBS was added to each plate prior to exposure to UV light]. Analysis of colony formation and counting was done as described above. All experiments were performed in triplicate and the final concentration of DMSO was <1% when cells were treated with compounds dissolved in this solvent.
Quantification of AP sites in genomic DNA
AP sites were quantified as described by Nakamura et al. (47). Genomic DNA was extracted from a pellet of 5 × 106 cells using the guanidine/detergent lysis method. Briefly, 0.5 ml of DNAzol® (Helena Biosciences) was added to the pellet and the cell lysate was gently passed several times through a pipette. The resultant viscous solution was centrifuged at 10 000 g for 10 min at 25°C. DNA was precipitated from the supernatant using 0.25 ml of 100% ethanol by gently inverting the tube 5–8 times at room temperature for 1–3 min. The DNA was later removed by spooling with a pipette and washed twice in 0.4 ml of 75% ethanol. The DNA was then solubilized in TE buffer (pH 8.0), and the final concentration was adjusted to 100 μg/ml [using a spectrophotometer (Nanodrop® ND-1000)]. AP site determinations were performed on the genomic DNA using an aldehyde reactive probe assay kit using the protocol provided by the manufacturer (Dojindo Molecular technologies, Inc.). All experiments were performed in triplicate.
Molecular modeling
CRT0044876 was docked into two separate structures of APE1, crystallized in the presence and absence of DNA, using Autodock3.0.5 (48). Coordinates for the DNA bound protein were obtained from 1DE9.pdb (49). All crystallographic water molecules, DNA, protein chain B and one of the metals were removed. The Mn atom bound to chain A was retained but renamed as Zn. Missing sidechain heavy atoms were added manually using Sybyl6.9 (http://www.tripos.com). Coordinates for the unligated protein were taken from 1BIX.pdb (11). Again, water molecules and all metal ions except the one bound in the active site, which was renamed to Zn, were removed. Polar hydrogens, Kollman united atom charges, fragmental volumes and solvation parameters were added to both structures using Autodock Tools (http://www.scripps.edu/mb/olson/doc/autodock/tools.html).
All atom model of the APE1 inhibitor was built and assigned as Gasteiger-Marsili charges in Sybyl6.9. The molecule was modeled containing a carboxylate group and therefore, had an overall charge of −1. Using the Autotors utility within Autodock3, the coordinates and charges of all non-polar hydrogen atoms were merged, and the ring carbon atoms were identified as aromatic. Additionally, the bonds between the indole C2 and the carboxylate carbon, and between C7 and the nitro group were defined as rotatable.
Autorgid3 (48) was used to calculate the grid maps before docking of the inhibitor. A grid consisting of 126 points in each (x, y, z) dimension, with a grid spacing of 0.375 Å and centred on the backbone carbonyl atom of Glu96 was used. The Lamarckian genetic algorithm within Autodock3 (48) was used for the docking runs. All parameters were set to the default values, except for the number of runs, which was increased from 10 to 100. The 100 resultant docked conformations were clustered using an in-house program written by V. Lesk (Structural Bioinformatics Group, Imperial College, London, UK). A greedy algorithm was employed using the structure with the best predicted binding energy as the initial seed. A cut-off of 1.0 Å was selected, and all atoms were included in the calculation. The program was able to take an account of the symmetry in the nitro and carboxylate groups.
RESULTS
A high-throughput assay for the identification of APE1 inhibitors
A high-throughput, fluorescence-based, AP site cleavage assay was developed for use in a 96-well plate format (Figure 1a). The double-stranded DNA substrate used was engineered to contain a fluor-quench pair containing a fluorescein moiety on the 5′ end of one DNA strand adjacent to a Dabcyl quenching group on the 3′ end of the complementary strand. In this state, the fluorescence of the fluorescein moiety is quenched. The principle of the assay is that cleavage by APE1 of the phosphodiester backbone 5′ to the single tetrahydrofuran (a stable AP site analog) residue at position 7 of the fluorescein-containing oligonucleotide yields a 6mer product that cannot remain base-paired to its complement at 37°C (Figure 1a). This thermal denaturation separates the fluor-quench pair, producing a fluorescent signal that can be detected using a plate reader. Inhibitors of APE1 would be expected to block the AP endonuclease reaction (Figure 1b), and hence eliminate the fluorescent signal.
A chemical library of 5000 compounds, which represents a structurally-diverse set acquired from commercial sources and considered to be ‘drug-like’ in structure, was screened using this assay system. The compounds were initially screened at a fixed concentration of 5 μM. The primary hits (showing >50% inhibition of APE1) from this screen were re-tested in duplicate, and those that consistently inhibited APE1 activity by >50% were taken for further analysis. As a first test of target selectivity, and to eliminate compounds that non-specifically interfered with the fluorescence detection system, selected compounds were screened for their inability to inhibit HpaII restriction endonuclease in a similar fluor-quench oligonucleotide assay format. After an initial round of hit analogue reordering, 56 compounds were identified as potential APE1 inhibitors, all of which had an IC50 of <10 μM.
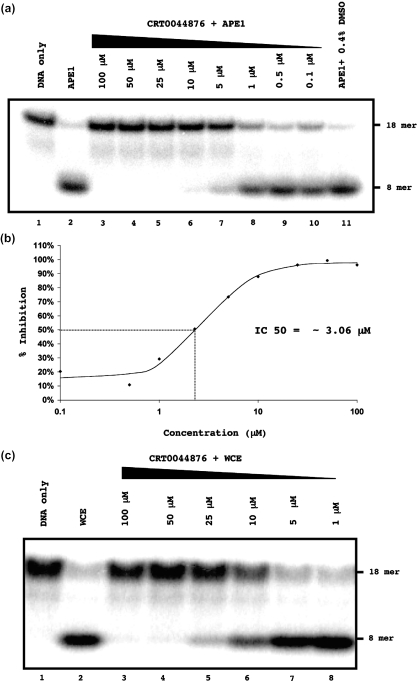
The 56 compounds from the primary screen were then tested in a more direct AP site cleavage assay in which the product of cleavage could be visualized directly. In this assay (presented schematically in Figure 1c), APE1-mediated cleavage of the AP site generates an 8mer 32P-end-labeled product that can be separated from uncleaved substrate on a polyacrylamide gel (Figure 1d). When screened at 100 μM, 15 of the selected compounds completely inhibited APE1-mediated AP site cleavage. Each of these compounds was then re-tested for inhibitory activity in this gel-based assay over a wide concentration range. From this analysis, CRT0044876 was taken for further study. CRT0044876 had an IC50 for inhibition of APE1 of ∼3 μM and not only inhibited AP site cleavage catalyzed by purified APE1, but also cleavage directed by APE1 in a HeLa whole cell extract (Figure 2).
Figure 2.
Identification of CRT0044876 as an APE1 inhibitor. (a) Inhibitory profile of CRT0044876. DNA substrate (lane 1); substrate with APE1 (lane 2); substrate with APE1 and different concentrations of CRT0044876 (lanes 3–10). Lane 11 shows that the DMSO solvent did not inhibit APE1. (b) Quantification of the data from panel A showing the percentage inhibition of APE1 as a function of CRT0044876 concentration. The IC50 value for APE1 inhibition by CRT0044876 was calculated to be 3.06 μM. (c) CRT0044876 inhibited the AP site cleavage activity in HeLa whole cell extracts (WCE).
Specificity testing on CRT0044876
It was possible that CRT0044876 was acting as a non-specific inhibitor of APE1. For example, if this compound were to bind directly to the AP site, in a manner analogous to that of methoxyamine (50), it would be expected to inhibit the action of any AP endonuclease enzyme. To address this possibility, a series of specificity analyses were undertaken. Amongst these, was a test of the ability of CRT0044876 to inhibit enzymes that interact with DNA and catalyze phosphodiester bond cleavage by mechanisms similar to or different from that used by APE1. However, no evidence for inhibition of BamHI restriction endonuclease or topoisomerase I was obtained even at CRT0044876 concentrations of 100 μM (Supplementary Figure 1).
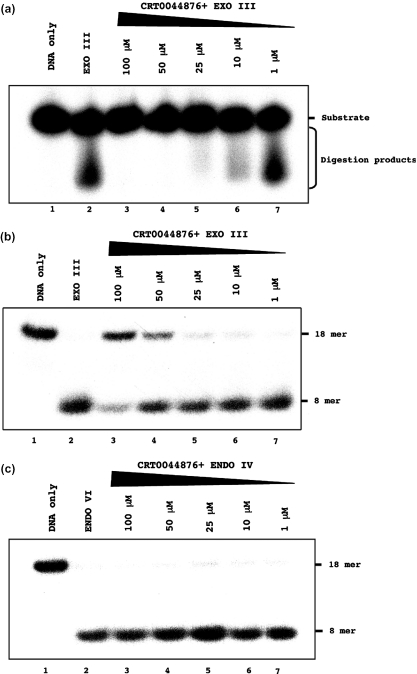
As an additional and more directed analysis of specificity, the inhibitory effect of CRT0044876 on exonuclease III and endonuclease IV was analyzed. These two bacterial AP endonucleases operate via a similar hydrolytic mechanism, but show no structural similarity to one another (11–13). Exonuclease III was chosen because it is the bacterial homologue of APE1, and, as such, has a very similar active site structure (11,12). As shown in Figure 3, CRT0044876 inhibited both the exonuclease and AP endonuclease activities of exonuclease III, but showed no inhibitory activity towards endonuclease IV. Taken together, these data argue against CRT0044876 acting via a mechanism involving direct AP site binding, in a manner analogous to methoxyamine, and suggest the possibility that CRT0044876 targets the active site of exonuclease III/APE1 family members.
Figure 3.
CRT0044876 is an inhibitor of the exonuclease III family of AP endonucleases. (a) CRT0044876 inhibits the 3′–5′-exonuclease activity of exonuclease III. DNA substrate (lane 1 as indicated on the right); exonuclease III alone, which generated digestion products from the substrate, as indicated on the right (lane 2); effects of increasing concentrations of CRT0044876 on the exonuclease activity (lanes 3–7). (b) CRT0044876 inhibits the AP endonuclease activity of exonuclease III. Lanes as per (a). (c) CRT0044876 does not inhibit the AP endonuclease activity of endonuclease IV. Lanes as per (a).
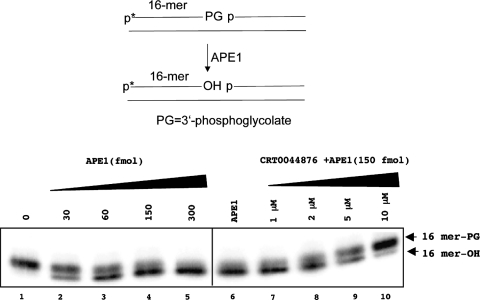
If the single DNA repair active site of APE1 were the target for CRT0044876, it might be expected that all of the enzymatic activities exhibited by APE1 would be inhibited by CRT0044876. Moreover, it might be expected that there would be a similar IC50 for inhibition of these activities to that for inhibition of the AP endonuclease activity of APE1. The data presented in Figure 4 indicate that, indeed, this is the case. CRT0044876 inhibited the 3′-phosphoglycolate diesterase activity of APE1 with an IC50 of ∼5 μM (Figure 4a and b). A similarly potent inhibition of 3′-phosphatase activity was also observed (data not shown). These data provide additional evidence that the inhibitory action of CRT0044876 on APE1 is independent of direct AP site binding.
Figure 4.
CRT0044876 inhibits the 3′-phosphodiesterase activity of APE1. (a) Assay design. The substrate contains an end-labeled 16mer oligonucleotide with a 3′-PG moiety. APE1 removes the 3′-PG residue to produce a 3′-OH residue, which has a faster mobility on a 20% polyacrylamide gel (panel b). Lanes 1–5 show an APE1 concentration-dependent increase in conversion of the 3′-PG to 3′-OH. Lanes 6–10 show that CRT00044876 inhibited the 3′-phosphodiesterase activity of APE1 (protein fixed at 150 fmol).
Effects of CRT0044876 on human cell lines
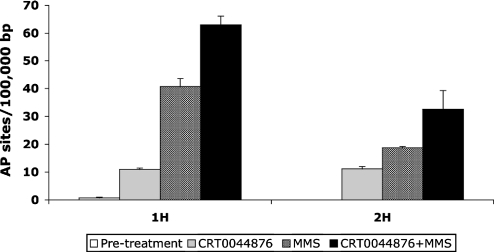
The data presented suggest that CRT0044876 can potently and selectively inhibit APE1 in vitro. However, in order to have general utility as a BER inhibitor, it was important to confirm that CRT0044876 could block APE1 function in living cells. Recent siRNA studies indicated that, in the absence of functional APE1, there is an accumulation of unrepaired AP sites in cells treated by alkylating agents (32). Using the same aldehyde reactive probe assay as utilized in that study (32), and described previously by Nakamura et al. (47), a statistically significant increase in AP site accumulation was found in HT1080 fibrosarcoma cells treated with CRT0044876 alone (Figure 5). As expected, MMS treatment of HT1080 cells also caused an elevation in the level of AP sites. However, the combination of MMS and CRT0044876 led to a synergistic increase in the level of AP sites, consistent with the notion that CRT0044876 was able to inhibit APE1 activity in vivo (Figure 5).
Figure 5.
Accumulation of AP sites in CRT0044876 treated human HT1080 cells. Quantification of the number of AP sites per 100 000 bp of nuclear DNA in untreated cells and in cells treated with CRT0044876 alone, MMS alone, or the combination of CRT0044876 and MMS (as indicated in the key below the bars). Samples were taken between 1 and 2 h after drug exposure as indicated. Values represented the mean of three independent determinations. Error bars denote Standard Errors of the mean.
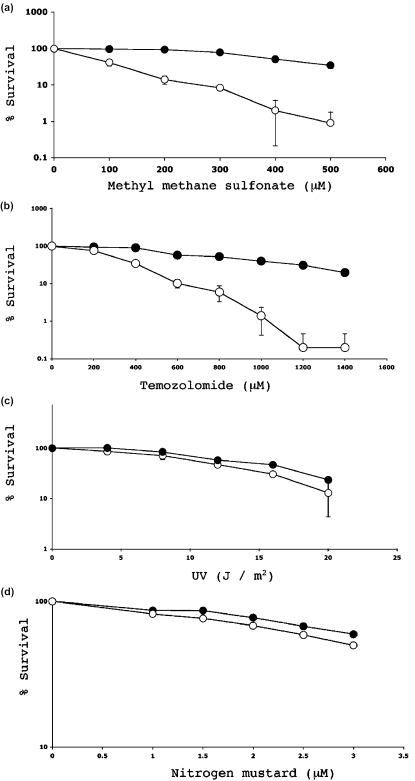
To investigate whether this apparent inhibition of APE1 by CRT0044876 had biological consequences, survival analyses were undertaken in HT1080 cells. Initially, CRT0044876 was tested for its inherent toxicity, but was shown to be non-toxic to HT1080 cells at any dose relevant to the analyses described in this study (>95% survival at concentrations up to 400 μM; data not shown). Next, the effect of combining CRT0044876 with DNA damaging agents was tested. The most commonly studied laboratory agent for analysis of AP endonuclease deficiency is the monofunctional alkylating agent, MMS. Hence this agent was tested initially. As shown in Figure 6a, whereas 500 μM MMS alone produced <10% cell killing, it was possible to kill 99% of HT1080 cells by exposure to the combination of MMS and a non-toxic concentration of CRT0044876 (200 μM). Similar results were seen in HeLa cervical carcinoma cells and in MDA-MB-231 breast carcinoma cells (data not shown). Because MMS is not used clinically, it was important to confirm this potentiation of cytotoxicity using a widely-used chemotherapeutic alkylating agent. The data in Figure 6b indicate that a similarly dramatic potentiation of temozolomide toxicity was observed in HT1080 cells treated with CRT0044876. Potentiation of cytotoxicity was also seen with hydrogen peroxide that causes oxidative DNA base damage (data not shown).
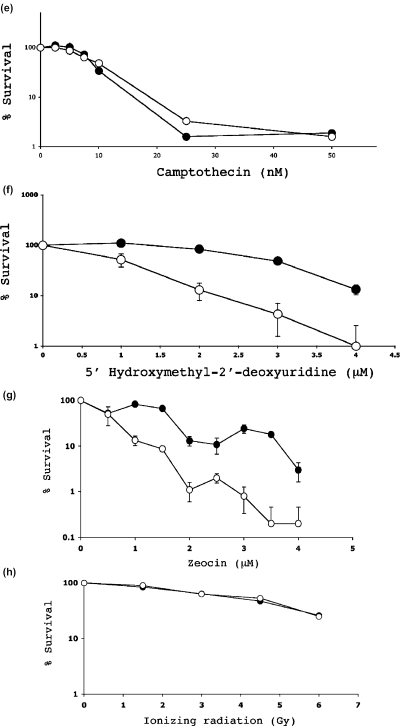
Figure 6.
Clonogenic survival analysis. The percentage survival of HT1080 cells as a function of drug dose or radiation dose is indicated in (a–h). In each case the filled circles indicate the survival for cells exposed to the DNA damaging agent alone, and the open circles indicate the survival for cells exposed to the damaging agent in the presence of 200 μM CRT0044876.
It was possible that these effects of CRT0044876 reflected a non-specific response to any form of DNA damage, and not a specific inhibition of the BER pathway. To address this, additional survival analyses were conducted using agents that generate DNA damage that is not repaired by BER. Consistent with CRT0044876 being a specific BER inhibitor, CRT0044876 did not potentiate the cytotoxicity of UV light (Figure 6c), nitrogen mustard (Figure 6d), or camptothecin (Figure 6e). To provide further evidence to substantiate this point, cytotoxicity studies were also conducted with hmdUrd. This compound is incorporated into DNA, and can be excised by the SMUG1 glycosylase to generate an AP site. Nevertheless, hmdUrd is generally regarded as innocuous unless faulty repair is invoked. For example, previous studies have shown that hmdUrd is highly toxic in mouse cells lacking functional DNA polymerase β, but not in wild-type cells, because of the build up of toxic BER intermediates that are not processed adequately due to the polymerase β deficiency (51). Consistent with CRT0044876 being a specific BER inhibitor, a strong potentiation of hmdUrd cytotoxicity was seen in CRT0044876-treated cells (Figure 6f).
Zeocin™ is a formulation of Phleomycin D1, a copper–chelated glycopeptide that belongs to the bleomycin family of anti-tumor antibiotics. One lesion characteristic of the bleomycins is a strand break with a 3′-PG terminus. Apart from processing AP sites, APE1 has been shown to be critically involved in the removal of 3′-PG termini (16). A strong potentiation of Zeocin cytotoxicity was seen in CRT0044876-treated cells (Figure 6g). This is entirely consistent with the biochemical data presented above showing that CRT0044876 inhibited the 3′-PG diesterase activity of APE1. Ionizing radiation induces single-strand DNA breaks, double-strand DNA breaks, oxidative DNA base damage and many other toxic lesions. BER, non-homologous end rejoining (NHEJ) and homologous recombination (HR) are some of the DNA repair processes involved in the repair of such damage (1). CRT0044876 did not potentiate the cytoxicity of ionizing radiation (Figure 6h). This may be because BER is only one of the many pathways that are involved in the repair of ionizing radiation-induced lesions.
In silico modeling of CRT0044876 binding to APE1
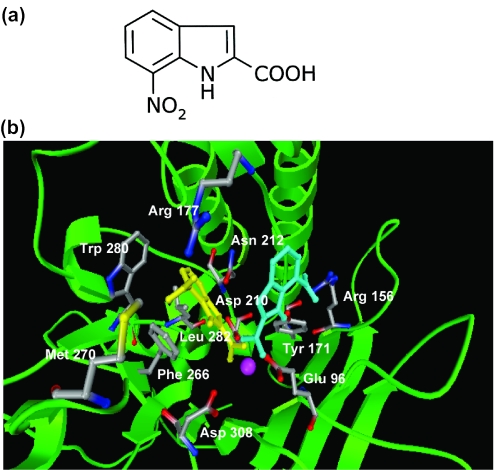
The finding that CRT0044876 can inhibit two exonuclease III family members, but not endonuclease IV or a number of other DNA cleaving enzymes, strongly suggested that this compound targets the APE1 active site. To investigate how this might occur, an in silico modeling approach was undertaken (Figure 7). CRT0044876 was docked to two X-ray crystal structures of APE1 using Autodock3 (Materials and Methods). In the first structure, 1DE9.pdb, the protein was crystallized bound to DNA that had been cleaved at the phosphodiester bond 5′ to the abasic site. As the resolution of this complex was relatively poor, a second structure, 1BIX.pdb, with only metal ions bound was also selected from the PDB. After removal of all HETAM groups, except the catalytic metal, the two proteins were prepared for the Autodock calculations as described in the Materials and Methods. CRT0044876 was modeled with the carboxylic group in the ionized form, as would be expected at neutral pH. Both the bonds from the indole ring to the carboxylate group, and the nitro group of CRT0044876, were free to rotate in the calculations.
Figure 7.
Molecular Modeling. CRT0044876 is a chemically known as 7-nitro-1H-indole-2-carboxylic acid (a). Through molecular modeling studies, CRT0044876 was found to dock on to the active site of APE1 in two different low energy conformations (b) designated cluster I (in yellow) and cluster II (in blue). APE1 is shown as a ribbon structure. Selected active site amino acid residues are indicated. See text for details.
Using Autodock3, 100 complexes of CRT0044876 docked to APE1 from both 1DE9.pdb and 1BIX.pdb were generated and the conformations were clustered using an in-house program (written by V. Lesk) and a 1.0 Å cutoff. The solution with the best predicted binding energy from each cluster was selected as a representative conformation for the cluster. The clusters were ranked by the binding energy of the representative molecules and by their population size.
For both protein structures, the two major drug binding modes identified were in the active site. Binding mode 1 (Figure 7b) resulted in the indole ring of CRT0044876 being located in the abasic deoxyribose binding pocket, bordered by residues Phe-266, Trp-280 and Leu-282. The nitro group was orientated away from the bulk of the protein towards the sidechain of Arg-177 and the carboxylate group was coordinated to the metal ion. This binding mode was the major one (top ranked energy and 25 out of 100 runs) when the X-ray structure 1DE9.pdb was used. The second ranked cluster by both energy and population defined binding mode 2. Again, the carboxylate group coordinated with the metal ion, but the nitro group was seen to hydrogen bond to the sidechain of Arg-156. A hydrogen bond could also be formed from the indole NH to the hydroxyl oxygen of Tyr-171 (Figure 7b). Both binding modes 1 and 2 were also located when 1BIX.pdb was used for protein coordinates.
DISCUSSION
We have developed a robust and facile high-throughput screening assay for the identification of inhibitors of the APE1 protein. This screening methodology is suitable for analysis of inhibitors of any AP site-processing enzyme that cleaves the phosphodiester backbone. Even enzymes that do not directly break the DNA backbone such as some DNA glycosylases, can be analyzed using the assay, because the AP site generated by their action can easily be chemically or enzymatically cleaved in a modified 2-stage assay procedure. Indeed, we would propose that this assay system has far wider utility as a drug screening methodology in which it can readily be adapted for use in the analysis of other DNA modifying enzymes that cleave or unwind the duplex. Amongst several putative APE1 inhibitors identified using this assay, we chose to focus on CRT0044876 because of its potency, selectivity and lack of inherent toxicity to human cell lines. Other than exonuclease III, the APE1 homologue in bacteria, we have yet to identify any DNA modifying enzyme that is inhibited by CRT0044876 at concentrations of up to 100 μM.
Currently available inhibitors of AP endonuclease function, such as methoxyamine (which is in phase I clinical trials for solid tumors) directly target the AP site in the DNA (50), not the APE1 enzyme. We would argue that targeting DNA repair enzymes in human cells is a more promising strategy for the inhibition of DNA repair during cancer chemotherapy because APE1 performs DNA repair roles in addition to removal of AP sites. In particular, it is likely that the 3′-phosphodiesterase activity of APE1 is important in the protection of cells against the toxic effects of certain agents, such as bleomycin, that generate DNA strand breaks associated with a fragmented sugar group at 3′ terminus. These fragments prevent the priming of DNA repair synthesis and must be removed. Indeed, APE1 has been shown to be the major 3′-PG diesterase activity in human cells (16).
This study provides a clear preclinical proof-of-principle that targeting the DNA base excision repair pathway can potentiate the cytotoxic effects of certain DNA damaging agents, including clinically-used alkylating agents. Clearly, these findings have significant translational implications. While we would propose that the current findings provide important validation of APE1 as an anticancer drug target, it is likely that further development will be required before a genuine clinical lead compound can emerge. One approach to the development of the next generation of APE1 inhibitors will be to utilize structure-aided design. The modeling data reported here provides the objective evidence that CRT0044876 targets the APE1 active site, which is entirely consistent with specific targeting of exonuclease III family members by this compound. Future analyses will include co-crystallization of APE1 with this and/or related compounds in order to drive a synthetic chemistry programme aimed at improving the potency and selectivity of compounds still further.
Structural features of APE1 make this protein an attractive target for small molecule inhibitors. A comparison of the architecture of the catalytic active site in the absence and presence of bound DNA substrate indicates that there is little or no remodeling of the active site upon AP site binding (11,12). This point is reinforced by the molecular docking study reported here in which the protein structures were rigid. Using X-ray structures determined in the presence and absence of DNA, CRT0044876 was seen to favor similar binding modes in the active site in both cases. Having a ‘pre-formed’ active site pocket of this sort opens the way to targeting the catalytic centre of the enzyme prior to the introduction of cellular DNA damage, which should increase the likelihood of ablating APE1 function in tumors during chemotherapy.
It is interesting to note that there are many parallels between our findings and those presented recently by Wang et al. (31) and Fung and Demple (32) using an siRNA approach to disable APE1 function. Wang et al. showed that down regulation of APE1 was associated with enhanced cytotoxicity and induction of apoptosis following exposure to alkylating agents, hydrogen peroxide and other DNA damaging agents (31). This potentiation of cytotoxicity was very similar to the effects of CRT0044876 seen in our study. Recently, Fung and Demple (32) observed that siRNA-mediated APE1 depletion led to an accumulation of AP sites, a reduction in cell proliferation and an induction of apoptosis in the absence of any other treatments. The results of that study suggest that the primary essential function of APE1 is likely to be in DNA repair (32). However, in the present study, CRT0044876 was shown to be non-toxic to several human cell lines at doses well in excess of those that potentiated cell killing by alkylating agents. Taken together, these data would imply that the inherent toxicity associated with APE1 depletion by siRNA may be a result of disabling both the BER functions of APE1 and functions distinct from DNA repair, such as its recently described and apparently essential role in acetylation-mediated gene regulation (52). It should be emphasized, however, that this conclusion is tentative at this stage and additional studies are required to address this point in detail.
In summary, we have identified small molecule inhibitors of the APE1 DNA repair enzyme. Amongst these, CRT0044876 was identified as a promising lead compound for future development. CRT0044876 represents the first generation of APE1-targeting lead compounds for blocking BER during cancer chemotherapy. Numerous factors, including the pharmacokinetics of BER inhibitors in vivo and their potency, will have to be considered in the design of the next generation of compounds. Nevertheless, CRT0044876 represents a valuable reagent for probing APE1 function and the BER process in general in vitro and in vivo.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
[Supplementary Material]
Acknowledgments
We thank members of the CR-UK Genome Integrity and DNA Repair Groups for helpful discussions, and Dr P. McHugh for critical reading of the manuscript. This work was supported by Cancer Research UK and Medical Research Council. Funding to pay the Open Access publication charges for this article was provided by JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Boiteux S., Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Dianov G.L., Sleeth K.M., Dianova II, Allinson S.L. Repair of abasic sites in DNA. Mutat. Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Robson C.N., Hickson I.D. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E.coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demple B., Herman T., Chen D.S. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl Acad. Sci. USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D.S., Herman T., Demple B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robson C.N., Hochhauser D., Craig R., Rack K., Buckle V.J., Hickson I.D. Structure of the human DNA repair gene HAP1 and its localisation to chromosome 14q 11.2-12. Nucleic Acids Res. 1992;20:4417–4421. doi: 10.1093/nar/20.17.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barzilay G., Hickson I.D. Structure and function of apurinic/apyrimidinic endonucleases. Bioessays. 1995;17:713–719. doi: 10.1002/bies.950170808. [DOI] [PubMed] [Google Scholar]
- 10.Ramotar D. The apurinic-apyrimidinic endonuclease IV family of DNA repair enzymes. Biochem. Cell. Biol. 1997;75:327–336. [PubMed] [Google Scholar]
- 11.Gorman M.A., Morera S., Rothwell D.G., de La Fortelle E., Mol C.D., Tainer J.A., Hickson I.D., Freemont P.S. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. Embo J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mol C.D., Kuo C.F., Thayer M.M., Cunningham R.P., Tainer J.A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 13.Hosfield D.J., Guan Y., Haas B.J., Cunningham R.P., Tainer J.A. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 14.Kuo C.F., Mol C.D., Thayer M.M., Cunningham R.P., Tainer J.A. Structure and function of the DNA repair enzyme exonuclease III from E.coli. Ann. N Y Acad. Sci. 1994;726:223–234. doi: 10.1111/j.1749-6632.1994.tb52820.x. discussion 234–225. [DOI] [PubMed] [Google Scholar]
- 15.Hickson I.D., Gorman Michael A., Freemont Paul S. Structure and Functions of the Major Human AP Endonuclease HAP1/Ref-1. Totowa, NJ: Humana Press Inc.; 2000. [Google Scholar]
- 16.Parsons J.L., Dianova II, Dianov G.L. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gros L., Ishchenko A.A., Ide H., Elder R.H., Saparbaev M.K. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32:73–81. doi: 10.1093/nar/gkh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xanthoudakis S., Miao G., Wang F., Pan Y.C., Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. Embo J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xanthoudakis S., Miao G.G., Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc. Natl Acad. Sci. USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker L.J., Robson C.N., Black E., Gillespie D., Hickson I.D. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell. Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tell G., Damante G., Caldwell D., Kelley M.R. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 22.Prasad R., Beard W.A., Strauss P.R., Wilson S.H. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J. Biol. Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 23.Dianov G., Price A., Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol. 1992;12:1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xanthoudakis S., Smeyne R.J., Wallace J.D., Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig D.L., MacInnes M.A., Takiguchi Y., Purtymun P.E., Henrie M., Flannery M., Meneses J., Pedersen R.A., Chen D.J. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res. 1998;409:17–29. doi: 10.1016/s0921-8777(98)00039-1. [DOI] [PubMed] [Google Scholar]
- 26.Meira L.B., Devaraj S., Kisby G.E., Burns D.K., Daniel R.L., Hammer R.E., Grundy S., Jialal I., Friedberg E.C. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–5557. [PubMed] [Google Scholar]
- 27.Walker L.J., Craig R.B., Harris A.L., Hickson I.D. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D.S., Olkowski Z.L. Biological responses of human apurinic endonuclease to radiation-induced DNA damage. Ann. NY Acad. Sci. 1994;726:306–308. doi: 10.1111/j.1749-6632.1994.tb52834.x. [DOI] [PubMed] [Google Scholar]
- 29.Silber J.R., Bobola M.S., Blank A., Schoeler K.D., Haroldson P.D., Huynh M.B., Kolstoe D.D. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin. Cancer Res. 2002;8:3008–3018. [PubMed] [Google Scholar]
- 30.Lau J.P., Weatherdon K.L., Skalski V., Hedley D.W. Effects of gemcitabine on APE/ref-1 endonuclease activity in pancreatic cancer cells, and the therapeutic potential of antisense oligonucleotides. Br. J. Cancer. 2004;91:1166–1173. doi: 10.1038/sj.bjc.6602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D., Luo M., Kelley M.R. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol. Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 32.Fung H., Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 33.Kakolyris S., Kaklamanis L., Engels K., Fox S.B., Taylor M., Hickson I.D., Gatter K.C., Harris A.L. Human AP endonuclease 1 (HAP1) protein expression in breast cancer correlates with lymph node status and angiogenesis. Br. J. Cancer. 1998;77:1169–1173. doi: 10.1038/bjc.1998.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herring C.J., West C.M., Wilks D.P., Davidson S.E., Hunter R.D., Berry P., Forster G., MacKinnon J., Rafferty J.A., Elder R.H., et al. Levels of the DNA repair enzyme human apurinic/apyrimidinic endonuclease (APE1, APEX, Ref-1) are associated with the intrinsic radiosensitivity of cervical cancers. Br. J. Cancer. 1998;78:1128–1133. doi: 10.1038/bjc.1998.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y., Moore D.H., Broshears J., Liu L., Wilson T.M., Kelley M.R. The apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme is elevated in premalignant and malignant cervical cancer. Anticancer Res. 1997;17:3713–3719. [PubMed] [Google Scholar]
- 36.Moore D.H., Michael H., Tritt R., Parsons S.H., Kelley M.R. Alterations in the expression of the DNA repair/redox enzyme APE/ref-1 in epithelial ovarian cancers. Clin. Cancer Res. 2000;6:602–609. [PubMed] [Google Scholar]
- 37.Thomson B., Tritt R., Davis M., Kelley M.R. Histology-specific expression of a DNA repair protein in pediatric rhabdomyosarcomas. J. Pediatr. Hematol. Oncol. 2001;23:234–239. doi: 10.1097/00043426-200105000-00011. [DOI] [PubMed] [Google Scholar]
- 38.Bobola M.S., Blank A., Berger M.S., Stevens B.A., Silber J.R. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin. Cancer Res. 2001;7:3510–3518. [PubMed] [Google Scholar]
- 39.Koukourakis M.I., Giatromanolaki A., Kakolyris S., Sivridis E., Georgoulias V., Funtzilas G., Hickson I.D., Gatter K.C., Harris A.L. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int. J. Radiat. Oncol. Biol. Phys. 2001;50:27–36. doi: 10.1016/s0360-3016(00)01561-3. [DOI] [PubMed] [Google Scholar]
- 40.Fritz G., Grosch S., Tomicic M., Kaina B. APE/Ref-1 and the mammalian response to genotoxic stress. Toxicology. 2003;193:67–78. doi: 10.1016/s0300-483x(03)00290-7. [DOI] [PubMed] [Google Scholar]
- 41.Kakolyris S., Giatromanolaki A., Koukourakis M., Kaklamanis L., Kanavaros P., Hickson I.D., Barzilay G., Georgoulias V., Gatter K.C., Harris A.L. Nuclear localization of human AP endonuclease 1 (HAP1/Ref-1) associates with prognosis in early operable non-small cell lung cancer (NSCLC) J. Pathol. 1999;189:351–357. doi: 10.1002/(SICI)1096-9896(199911)189:3<351::AID-PATH435>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 42.Puglisi F., Aprile G., Minisini A.M., Barbone F., Cataldi P., Tell G., Kelley M.R., Damante G., Beltrami C.A., Di Loreto C. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer Res. 2001;21:4041–4049. [PubMed] [Google Scholar]
- 43.Barzilay G., Walker L.J., Robson C.N., Hickson I.D. Site-directed mutagenesis of the human DNA repair enzyme HAP1: identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res. 1995;23:1544–1550. doi: 10.1093/nar/23.9.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slupphaug G., Eftedal I., Kavli B., Bharati S., Helle N.M., Haug T., Levine D.W., Krokan H.E. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka M., Lai J.S., Herr W. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992;68:755–767. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- 46.Urata H., Akagi M. A convenient synthesis of oligonucleotides with a 3′-phosphoglycolate and 3′-phosphoglycaldehyde terminus. tetrahedron Lett. 1993;34:4015–4018. [Google Scholar]
- 47.Nakamura J., Swenberg J.A. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 48.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a lamarckian genetic algorithm and and empirical binding free energy function. J. Computational Chemistry. 1998;19:1639. [Google Scholar]
- 49.Mol C.D., Izumi T., Mitra S., Tainer J.A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 50.Liu L., Gerson S.L. Therapeutic impact of methoxyamine: blocking repair of abasic sites in the base excision repair pathway. Curr. Opin. Investig. Drugs. 2004;5:623–627. [PubMed] [Google Scholar]
- 51.Horton J.K., Joyce-Gray D.F., Pachkowski B.F., Swenberg J.A., Wilson S.H. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair (Amst). 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 52.Izumi T., Brown D.B., Naidu C.V., Bhakat K.K., Macinnes M.A., Saito H., Chen D.J., Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl Acad. Sci. USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]