A Signal, from Human mtDNA, of Postglacial Recolonization in Europe (original) (raw)
Abstract
Mitochondrial HVS-I sequences from 10,365 subjects belonging to 56 populations/geographical regions of western Eurasia and northern Africa were first surveyed for the presence of the T→C transition at nucleotide position 16298, a mutation which has previously been shown to characterize haplogroup V mtDNAs. All mtDNAs with this mutation were then screened for a number of diagnostic RFLP sites, revealing two major subsets of mtDNAs. One is haplogroup V proper, and the other has been termed “pre*V,” since it predates V phylogenetically. The rather uncommon pre*V tends to be scattered throughout Europe (and northwestern Africa), whereas V attains two peaks of frequency: one situated in southwestern Europe and one in the Saami of northern Scandinavia. Geographical distributions and ages support the scenario that pre*V originated in Europe before the Last Glacial Maximum (LGM), whereas the more recently derived haplogroup V arose in a southwestern European refugium soon after the LGM. The arrival of V in eastern/central Europe, however, occurred much later, possibly with (post-)Neolithic contacts. The distribution of haplogroup V mtDNAs in modern European populations would thus, at least in part, reflect the pattern of postglacial human recolonization from that refugium, affecting even the Saami. Overall, the present study shows that the dissection of mtDNA variation into small and well-defined evolutionary units is an essential step in the identification of spatial frequency patterns. Mass screening of a few markers identified using complete mtDNA sequences promises to be an efficient strategy for inferring features of human prehistory.
Introduction
In the distant past, the human species was undoubtedly subject to ecological constraints similar to those of other species (Willis and Whittaker 2000). Humans, though highly adaptable to different environments, could not (for example) have lived on the vast glaciers that covered most of northern Europe during the last glaciation. Northern Europe was gradually recolonized from refugia after the Last Glacial Maximum (LGM), ∼20,000 years ago (Housley et al. 1997). The two major refugia were in southwestern France/Cantabria (Atlantic and western Mediterranean zone) and Ukraine/Central Russian Plain (Periglacial zone), but other minor refugia could have existed in between (Dolukhanov 2000). The way in which the western European refugia were interconnected after the LGM and fueled different expansion routes towards the north is, however, rather unclear. The timing of and interaction with the recolonization coming from the eastern refugia also require examination.
It is possible that the wealth of new genetic data from present-day European populations can shed some light on these questions. In particular, Semino et al. (2000) have highlighted two Y-chromosome markers, mutations M170 and M17, that may have expanded, after the LGM, from (southern) central and eastern European refugia in the northern Balkans and Ukraine, respectively. A distribution compatible with an expansion from an eastern European refugium after the LGM has also been observed, for another Y-chromosome mutation (SRY10831A), by Malaspina et al. (2000). In an earlier study (Torroni et al. 1998), it was suggested that the geographical distribution of a mtDNA marker, haplogroup V, resembles that of the second principal component of nuclear markers (Cavalli-Sforza et al. 1994) and testifies to the recolonization from western European refugia (see, however, Izagirre and de la Rúa [1999] and Simoni et al. [2000]). In the meantime, more mtDNA data (e.g., Richards et al. 2000) and additional mtDNA coding-region information (Macaulay et al. 1999) have become available, so that the variation of haplogroup V can be accurately dissected, and the geographical extent and timing of the western recolonization can now be reconsidered with more precision.
Subjects and Methods
Ethnic affiliations/geographical origins of the 10,365 subjects from 56 populations (fig. 1), whose HVS-I sequences were surveyed for the presence of 16298C, are listed in table 1. The large majority of these sequences encompassed nucleotide positions (np) 16050–16370, and, at minimum, they included np 16090–16365. Sequences harboring 16298C together with control-region motifs of haplogroups other than V—for example, C, T, U5, and Z (Torroni et al. 1993, 1996; Macaulay et al. 1999; Schurr et al. 1999)—were excluded. With the exception of 529 sequences that were extracted from the literature (table 1), all samples were collected with appropriate informed consent and were available for additional DNA typing. In addition to the HVS-I sequencing, ∼65% of these 9,836 samples have also been screened for haplogroup-diagnostic RFLP markers. In general, the remainder were not screened because they harbored an HVS-I motif that allowed them to be assigned unequivocally to a specific haplogroup (e.g., 16069T-16126C signals haplogroup J). The RFLP analysis described below was also performed on mtDNAs that lacked 16298C but harbored 16256T, as suggested by the observation of this HVS-I motif in one Basque mtDNA belonging to V (−4577 _Nla_III). This observation raises the possibility that some other small subset of V mtDNAs lacking 16298C may have been overlooked in our screening procedure.
Figure 1.
Geographical locations of the 56 populations included in the present study. The correspondence between numerical codes and populations is given in table 1.
Table 1.
Distribution of V and Pre*V mtDNAs
| V | Pre*V | ||||
|---|---|---|---|---|---|
| ID Number andRegion/Population(Code) | Sample Size | No. inSample | Estimated Frequencya(%) | No. inSample | Estimated Frequencya(%) |
| 1. Mauritania (MAU) | 30 | 0 | 0 (0–9.2) | 1 | 3.3 (.8–16.7) |
| 2. Morocco (MOR) | 92 | 1 | 1.1 (.3–5.8) | 4 | 4.3 (1.8–10.6) |
| 3. Algeria (ALG) | 49 | 3 | 6.1 (2.2–16.5) | 1 | 2.0 (.5–10.6) |
| 4. Canary Islands (CAN) | 300 | 6 | 2.0 (.9–4.3) | 0 | 0 (0–1.0) |
| 5. Spain (SPA) | 293 | 6 | 2.0 (1.0–4.4) | 8 | 2.7 (1.4–5.3) |
| 6. Portugal (POR) | 54 | 2 | 3.7 (1.1–12.5) | 0 | 0 (0–5.3) |
| 7. Basque Country (BSQ) | 97 | 12 | 12.4 (7.3–20.4) | 0 | 0 (0–3.0) |
| 8. France (FRA) | 56 | 4 | 7.1 (2.9–17.0) | 0 | 0 (0–5.1) |
| 9. Corsica (COR) | 56 | 0 | 0 (0–5.1) | 0 | 0 (0–5.1) |
| 10. North Sardinia (SAR) | 133 | 8 | 6.0 (3.1–11.4) | 1 | .8 (.2–4.1) |
| 11. Continental Italy (ITA) | 240 | 4 | 1.7 (.7–4.2) | 2 | .8 (.3–3.0) |
| 12. Sicily (SIC) | 634 | 14 | 2.2 (1.3–3.7) | 16 | 2.5 (1.6–4.1) |
| 13. Netherlands (NET) | 21 | 1 | 4.8 (1.1–22.8) | 0 | 0 (0–12.7) |
| 14. Bavaria (GER) | 33 | 1 | 3.0 (.7–15.3) | 1 | 3.0 (.7–15.3) |
| 15. Germany-Lower Saxony (GER) | 632 | 17 | 2.7 (1.7–4.3) | 9 | 1.4 (.8–2.7) |
| 16. Germany-mixed (GER)b | 157 | 7 | 4.5 (2.2–8.9) | 1 | .6 (.2–3.5) |
| 17. England (ENG) | 293 | 7 | 2.4 (1.2–4.8) | 3 | 1.0 (.4–3.0) |
| 18. Western Ireland (EIR) | 88 | 5 | 5.7 (2.5–12.6) | 1 | 1.1 (.3–6.1) |
| 19. Scotland (SCO) | 733 | 25 | 3.4 (2.3–5.0) | 1 | .1 (0–.8) |
| 20. Czech Republic (CZE) | 89 | 4 | 4.5 (1.8–11.0) | 1 | 1.1 (.3–6.0) |
| 21. Hungary (HUN) | 194 | 6 | 3.1 (1.5–6.6) | 0 | 0 (0–1.5) |
| 22. Csángó (CSA) | 68 | 0 | 0 (0–4.2) | 3 | 4.4 (1.6–12.2) |
| 23. Estonia (EST) | 148 | 1 | .7 (.2–3.7) | 0 | 0 (0–2.0) |
| 24. Finns (FIN) | 236 | 6 | 2.5 (1.2–5.4) | 0 | 0 (0–1.3) |
| 25. Skolt Saami (SSA) | 50 | 26 | 52.0 (38.5–65.2) | 0 | 0 (0–5.7) |
| 26. Inari Saami (ISA)c | 127 | 9 | 7.1 (3.8–12.9) | 0 | 0 (0–2.3) |
| 27. Erza Mordvins (ERZ) | 58 | 2 | 3.4 (1.1–11.7) | 0 | 0 (0–5.0) |
| 28. Russia (RUS) | 144 | 5 | 3.5 (1.5–7.9) | 2 | 1.4 (.4–4.9) |
| 29. Croatian Islands (CRO) | 447 | 25 | 5.6 (3.8–8.1) | 5 | 1.1 (.5–2.6) |
| 30. Bulgaria (BUL) | 81 | 0 | 0 (0–3.6) | 1 | 1.2 (.3–6.6) |
| 31. Albania (ALB) | 199 | 1 | .5 (.1–2.8) | 3 | 1.5 (.5–4.3) |
| 32. Mainland Greece (GRE) | 208 | 3 | 1.4 (.5–4.1) | 1 | .5 (.1–2.6) |
| 33. Crete (CRE) | 206 | 0 | 0 (0–1.4) | 2 | 1.0 (.3–3.4) |
| 34. Adygei (ADY) | 50 | 0 | 0 (0–5.7) | 0 | 0 (0–5.7) |
| 35. Georgia (GEO) | 137 | 1 | .7 (.2–4.0) | 0 | 0 (0–2.1) |
| 36. North Ossetia (NOS) | 106 | 0 | 0 (0–2.8) | 4 | 3.8 (1.5–9.3) |
| 37. South Ossetia (SOS) | 201 | 0 | 0 (0–1.5) | 0 | 0 (0–1.5) |
| 38. Armenia (ARM) | 192 | 0 | 0 (0–1.5) | 0 | 0 (0–1.5) |
| 39. Turkey (TUR)d | 606 | 1 | .2 (0–.9) | 2 | .3 (.1–1.2) |
| 40. Kurds (KUR) | 53 | 0 | 0 (0–5.4) | 0 | 0 (0–5.4) |
| 41. Lebanon (LEB) | 171 | 0 | 0 (0–1.7) | 0 | 0 (0–1.7) |
| 42. Druze (DRU) | 45 | 0 | 0 (0–6.3) | 0 | 0 (0–6.3) |
| 43. Jordan (JOR) | 150 | 0 | 0 (0–2.0) | 0 | 0 (0–2.0) |
| 44. Palestinians (PAL) | 117 | 0 | 0 (0–2.5) | 0 | 0 (0–2.5) |
| 45. Syria (SYR) | 36 | 1 | 2.8 (.7–14.2) | 0 | 0 (0–7.8) |
| 46. Saudi Arabia (SAB) | 194 | 0 | 0 (0–1.5) | 0 | 0 (0–1.5) |
| 47. Iraq (IRQ) | 110 | 0 | 0 (0–2.7) | 0 | 0 (0–2.7) |
| 48. Iran (IRN) | 91 | 0 | 0 (0–3.2) | 0 | 0 (0–3.2) |
| 49. Pakistan (PAK) | 100 | 0 | 0 (0–2.9) | 0 | 0 (0–2.9) |
| 50. India (IND) | 1,002 | 0 | 0 (0–.3) | 0 | 0 (0–.3) |
| 51. Oman/U.A. Emirates (OAE) | 57 | 0 | 0 (0–5.0) | 0 | 0 (0–5.0) |
| 52. Yemen (YEM) | 43 | 0 | 0 (0–6.6) | 0 | 0 (0–6.6) |
| 53. Egypt (EGY)e | 68 | 0 | 0 (0–4.2) | 0 | 0 (0–4.2) |
| 54. Nubia (NUB)e | 80 | 0 | 0 (0–3.6) | 0 | 0 (0–3.6) |
| 55. Ethiopia (ETH) | 270 | 0 | 0 (0–1.1) | 0 | 0 (0–1.1) |
| 56. Senegal (SEN)f | 240 | 0 | 0 (0–1.2) | 0 | 0 (0–1.2) |
| Total | 10,365 | 214 | 73 |
RFLP Screening
Candidate V mtDNAs underwent additional RFLP testing. As a technical prerequisite to combining the data and DNA samples acquired in the course of numerous years by a rather large number of different European laboratories, candidate V mtDNAs were screened for the presence/absence of the _Mse_I site at np 16297, a site that must be absent in the samples harboring 16298C. This allowed the identification of a number of trivial mistakes in the identification codes of DNA samples and in the aliquoting of DNAs, as well as other misassignments (Bandelt et al., in press). This preliminary step was followed by screening of the following RFLP sites: _Nla_III at np 4577, the absence of which characterizes haplogroup V (Torroni et al. 1996); _Mse_I at np 15904, the presence of which is typical of haplogroup V (Macaulay et al. 1999); _Alu_I at np 7025 the absence of which distinguishes haplogroup H (Torroni et al. 1994); and _Mse_I at np 14766, the absence of which is shared by the sister haplogroups H and V and thus defines the larger haplogroup HV (Torroni et al. 1998; Macaulay et al. 1999). The status at nps 72 and 73 in HVS-II was also checked in all samples, either through use of _Sph_I (which cuts when 72C-73A is present) and _Alw_44I (which cuts when 73G is present) (Morelli et al. 2000) or by HVS-II sequencing. 72C-73A is characteristic of haplogroup V (Torroni et al. 1998; Macaulay et al. 1999), whereas 72T-73A identifies haplogroup H (Torroni et al. 1996; Wilkinson-Herbots et al. 1996). Four mtDNAs with 16298C in HVS-I (one Italian, one Irish, one German, and one Scot) were found to lack the diagnostic haplogroup V RFLP markers. Moreover, they also lacked the _Alu_I site at np 7025 and showed 72T-73A in HVS-II; hence, they belonged to haplogroup H and were excluded.
Phylogenetic Analysis and Ages
A reduced median network (Bandelt et al. 1995) of the haplotypes (defined by their HVS-I sequence plus additional RFLP information) was manually constructed and checked by the program Network 2.0d (Shareware Phylogenetic Network Software Web site). To estimate the time to the most recent common ancestor (MRCA) of subsets of mtDNAs, we used the HVS-I sequence diversity accumulated on top of the assumed ancestral sequence type within the region covered by nps 16090–16365. We used a calibration of the rate of transitional mutations in this range (μ=4.96×10-5 transitions per year; Forster et al. 1996), in conjunction with the average number of transitions on the reconstructed phylogeny from the ancestral type to each sample (ρ) (Morral et al. 1994; Forster et al. 1996; Thomson et al. 2000), to obtain an unbiased estimate, T, of the time to the MRCA (T_=ρ/μ). To summarize the uncertainty in this point estimate, we followed the approach of Saillard et al. (2000). In brief, we first used information on the relative mutability of sites (Hasegawa et al. 1993), to remove all reticulation in the reduced median network. We estimated a (multifurcating) genealogy directly from the resolved phylogeny, assigning a length to each branch that was proportional to the number of mutations that occurred on that branch. We then evaluated the variance, (Δρ)2, of ρ on this genealogy, over repeated runs of the mutation process. We use Δ_T_=Δρ/μ as the standard error of T (in the form T_±Δ_T). An approximate 95% confidence interval is given by T_-2Δ_T to T+2Δ_T. This procedure does not take into account any uncertainty in the mutation rate or in the reconstructed genealogy. We applied this approach principally to clades in the phylogeny but also to subsets of mtDNAs that are not manifestly clades, under the hypothesis that each subset may be derived from a single founder and, hence, may represent clades in the genealogy. Intrapopulation diversities of haplogroup V were computed as 1-Σ_i_ x_2_i, where x i is the relative frequency of the _i_th V haplotype (defined in the region covered by nps 16090–16365) within the set of V sequences from the respective population.
Frequency-Map Construction
Maps were obtained using Surfer, version 6 (Golden Software, Inc.), with the Kriging procedure (Delfiner 1976). A 20×12 grid was used, and estimates at each grid node were obtained by consideration of the entire data set. The advantages and disadvantages of this procedure are carefully discussed in section 1.14 of the book by Cavalli-Sforza et al. (1994).
Results
RFLP Screening
The survey of 10,365 HVS-I sequences identified 291 candidate V mtDNAs (with 16298C or 16256T). The RFLP screening revealed that they all belonged to haplogroup HV (–14766 _Mse_I), but only 210 turned out to be regular members of haplogroup V in having −4577 _Nla_III and +15904 _Mse_I (15904T). Four mtDNAs (one Italian, one Irish, one German, and one Scot) showing +4577 _Nla_III, −7025 _Alu_I, and 72T-73A belonged to haplogroup H. Four Sardinian mtDNAs had −4577 _Nla_III and −15904 _Mse_I; however, sequence analysis of the region around np 15900 showed that they bore the mutation 15905C in addition to 15904T, so that the _Mse_I site generated by the mutation at 15904 has experienced a subsequent loss because of the mutation at 15905. Thus, the total number of V mtDNAs is 214 (table 1). Results of HVS-II analyses showed that these mtDNAs generally harbored 72C-73A, but a number of exceptions were observed: 2 with 73G and 17 with 72T (fig. 2).
Figure 2.
Network of pre*V and V mtDNAs. Areas of circles are proportional to the number of sampled individuals, and population codes correspond to those in table 1. The listed nucleotide positions are those differing from the Cambridge Reference Sequence (Anderson et al. 1981). Mutations are transitions (T↔C, A↔G), unless the base change is specified explicitly. Recurrent mutations are underlined, and arrows point to the presence of the restriction site. Squares indicate the phylogenetic position of the roots of the nested haplogroups H, HV, and R (Macaulay et al. 1999).
The remaining 73 mtDNAs all had +4577 _Nla_III but were polymorphic at the other V site: 13 bore +15904 _Mse_I, whereas 60 had −15904 _Mse_I and thus were indistinguishable from the root of haplogroup HV when coding-region RFLP markers were used. This observation reveals the temporal order of mutations leading to V (first, the gain of the 15904 _Mse_I site; then, the loss of the 4577 _Nla_III site), and identifies a slightly larger haplogroup, pre-V, which is solely defined by the two control-region mutations 16298C and 72C. Haplogroup pre-V encompasses the previously defined V plus additional mtDNAs which we refer to as “pre*V” (fig. 2). Pre*V as a paraphyletic grouping comprises at least two major subsets separated by 15904 _Mse_I. Also, in the case of pre*V, considerable variation at np 72 was observed in 15 subjects characterized by 72T and in 1 subject with 72A (fig. 2). Full details of the HVS-I sequence and the 72-73 status of all pre-V mtDNAs are available at V.M.'s Web site.
It should be emphasized that published complete sequences of pre-V mtDNAs (Ingman et al. 2000; Finnilä et al. 2001) confirm that the four transitions at nps 72, 16298, 15904, and 4580 are the only mutations that separate the roots of V and HV. The ultimate level of resolution has thus been reached for this part of the phylogeny.
Population Distribution
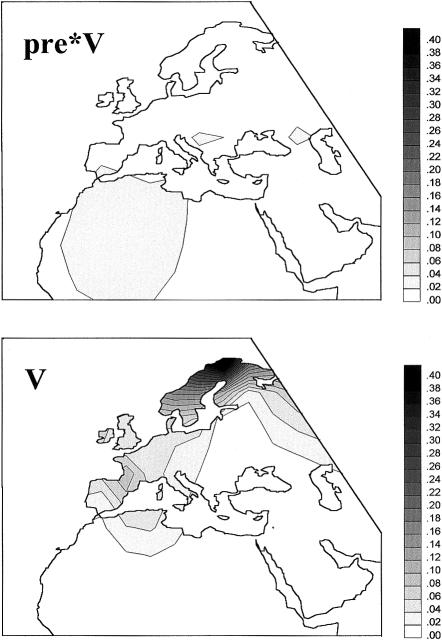
Table 1 reports the frequencies of pre*V and V in the 56 populations studied. Pre*V mtDNAs are generally rare and apparently scattered throughout Europe, although they are less uncommon in the Mediterranean area (including northwestern Africa). In contrast, V mtDNAs are more frequent and tend to be restricted to western, central, and northern Europe, with the highest population frequencies in the Skolt Saami (52%) and the Basques (12%). The difference in the frequency patterns of pre*V and V is strikingly clear in the geographical frequency maps (fig. 3).
Figure 3.
Spatial frequency distributions. Distributions of pre*V (top)and V (bottom) mtDNAs are shown.
Relatively recent founder events in specific populations, however, could have strongly affected the observed population frequencies. To address this issue, the diversity of V mtDNAs was measured in the six populations (Sicilians, Scots, Germans, Basques, Croatians, and Saami) in which >10 individual V mtDNAs were found (each population sample contributing >5% to the total V data set) (table 2). Diversity values <.5 were observed in the Saami (.074), the Croatians (.349), and the Basques (.486). The exclusion of the Saami from the frequency map had a major impact on the overall distribution of V, by completely erasing the frequency peak centered in Scandinavia (fig. 4_A_). The exclusion of the Croatians did not have any major impact (fig. 4_B_), and the exclusion of the Basques lowered, but did not erase, the frequency peak centered in southwestern Europe (fig. 4_C_).
Table 2.
Diversity of Haplogroup V in Samples Contributing >5% to the Total Set of V Sequences
| Samplea | Haplogroup V Diversity |
|---|---|
| SSA | .074 |
| CRO | .349 |
| BSQ | .486 |
| GER | .666 |
| SCO | .701 |
| SIC | .755 |
Figure 4.
Spatial frequency distributions of V mtDNAs, excluding possible outlier populations. A, without Saami; B, without Saami and Croatians; C, without Saami, Croatians, and Basques.
Time Depths
Age estimates were calculated for the different subsets of pre-V. The two subsets of pre*V, with and without the _Mse_I site at np 15904, gave age estimates of 26,400 and 14,800 years, respectively, when treated as potential clades, albeit with rather large standard errors (table 3).
Table 3.
Age Estimates of Different Subsets of Haplogroup Pre-V
| Set of SequencesConsidered | No. ofSubjects | ρa | T (± Δ_T_)b(years) |
|---|---|---|---|
| Pre-V (all populations) | 278 | 175/278 = .629 | 12,700 (±2,400) |
| V (all populations) | 205 | 114/205 = .556 | 11,200 (±2,700) |
| V (without SSA) | 179 | 113/179 = .631 | 12,700 (±3,100) |
| V (without SSA and CRO) | 154 | 108/154 = .701 | 14,200 (±3,400) |
| V (without SSA, CRO, and BSQ) | 142 | 103/142 = .725 | 14,600 (±3,600) |
| V (western) | 98 | 79/98 = .806 | 16,300 (±4,800) |
| V (eastern, without Saami) | 81 | 34/81 = .420 | 8,500 (±2,300) |
| Pre*V (+15904_Mse_I; all populations)c | 13 | 17/13 = 1.308 | 26,400 (±11,100) |
| Pre*V (–15904_Mse_I; all populations)c | 60 | 44/60 = .733 | 14,800 (±5,700) |
For V, a somewhat younger time depth of 11,200±2,700 years was estimated when all populations were included. This value, however, progressively increased when the potential outlier populations—first the Saami, then the Croatians, and, finally, the Basques—were excluded, eventually reaching 14,600±3,600 years. Moreover, there is a contrast in V diversities between west and east. We dissected the total sample along a NW-SE axis, so that northwest Africa, Iberia, France, Italy, the Netherlands, and the British Isles (a total of 98 V mtDNAs) were on the western side, and the rest of Europe (excluding the Saami because of their extreme V homogeneity), the Caucasus, and the Near East (a total of 81 V mtDNAs) were on the eastern side. The V age in the west is then 16,300±4,800 years, and that in the east is 8,500±2,300 years.
Discussion
The geographical distribution of haplogroup V is much more clear-cut than that of the pooled pre-V distribution considered in our earlier study (Torroni et al. 1998). Except for rare, isolated occurrences, V is virtually absent in the southern Balkans, Turkey, the Caucasus, and the Near East. Attested migrations from the north(west) into these regions in historic times are certainly sufficient to explain these few “erratics” (Richards et al. 2000, p. 1269). With regard to age and frequency, there is a clear cline from west to east (fig. 4); the age for V in the west (∼16,000 years) is almost twice that in the east, indicating the direction of settlement. We interpret this pattern in the following way: the older age reflects the onset of the recolonization of Europe from western refugia (Housley et al. 1997), whereas later founder events are responsible for the limited occurrences and reduced diversity of V mtDNAs in the east.
In contrast, the distribution of pre*V—that is, the non-V mtDNAs of the more ancient haplogroup, pre-V—suggests that these rather rare mtDNAs must have been present in more than one refuge area. The estimated age and the conspicuous, though scattered, occurrence of potential pre*V clades in the Mediterranean suggests that pre-V could have originated before the LGM (like its larger sister haplogroup H), perhaps in eastern Europe, whence it spread along an east-west axis with Gravettian contacts.
The present study demonstrates the potential of a phylogeographic approach based on the screening of markers that are now being identified from complete mtDNA sequences. A haplogroup-focused sequencing strategy (Finnilä et al. 2000, 2001) allows the systematic detection of numerous mutations that define subhaplogroups (i.e., clades of the phylogeny), which may have interesting geographical distributions. In the same way that we have assayed several thousand samples for a small set of specific mutations (viz., those relevant for assessment of V and pre*V status), one can perform a mass screening of a few sites characteristic of other clades of the mtDNA phylogeny (Richards and Macaulay 2001). Inasmuch as there is evidence that ⩾3/4 of the present-day European genes are descended from indigenous Paleolithic ancestors (Richards et al. 2000; Semino et al. 2000), a full understanding of the genetic landscape in Europe requires the (partial) reconstruction and distinction of the gene pools in the various glacial refugia.
Acknowledgments
This research received support from Italian Consiglio Nazionale delle Ricerche grant 99.02620.CT04 and Telethon-Italy grant E.0890 (both to A.T.), as well as from Fondo d’Ateneo per la Ricerca 2001 dell'Università di Pavia (to A.T. and A.S.S.-B.); Progetto Finalizzato C.N.R. “Beni Culturali” (Cultural Heritage, Italy) (to R.S., A.S.S.-B., O.R., and A. C.); Grandi Progetti Ateneo Università di Roma “La Sapienza” (to R.S.); the Italian Ministry of the University, Progetti Ricerca Interesse Nazionale 1999 and 2001 (to A.T., R.S., A.S.S.-B., and A. C.); the “Istituto Pasteur Fondazione Cenci Bolognetti,” Università di Roma “La Sapienza” (to R.S.); a Research Career Development Fellowship from the Wellcome Trust (to V.M.); Ministry of Science and Technology of the Republic of Croatia grant 019601 (to P.R.); Ministerio de Educación y Ciencia grant PB93-00558 (to V.M.C.); and Academy of Finland grants 38826 and 42183 (to M.-L.S.).
Electronic-Database Information
The URLs for data in this article are as follows:
- Author's Web site, http://www.stats.ox.ac.uk/~macaulay/preV2001/index.html (for HVS-I data)
- Shareware Phylogenetic Network Software, http://www.fluxus-engineering.com/sharenet.htm (for Network 2.0d)
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organisation of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Lahermo P, Richards M, Macaulay V. Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med (in press) [DOI] [PubMed] [Google Scholar]
- Calafell F, Underhill P, Tolun A, Angelicheva D, Kalaydjieva L (1996) From Asia to Europe: mitochondrial DNA sequence variability in Bulgarians and Turks. Ann Hum Genet 60:35–49 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Comas D, Calafell F, Mateu E, Perez-Lezaun A, Bertranpetit J (1996) Geographic variation in human mitochondrial DNA control region sequence: the population history of Turkey and its relationship to the European populations. Mol Biol Evol 13:1067–1077 [DOI] [PubMed] [Google Scholar]
- Delfiner P (1976) Linear estimation of non-stationary spatial phenomena. In: Guarasio M, David M, Haijbegts C (eds) Advanced geostatistics in the mining industry. Dordrecht, Reidel, pp 49–68 [Google Scholar]
- Dolukhanov PM (2000) “Prehistoric revolutions” and languages in Europe. In: Künnap A (ed) The roots of peoples and languages of northern Eurasia II and III. University of Tartu, Tartu, pp 71–78 [Google Scholar]
- Finnilä S, Hassinen IE, Ala-Kokko L, Majamaa K (2000) Phylogenetic network of the mtDNA haplogroup U in northern Finland based on sequence analysis of the complete coding region by conformation-sensitive gel electrophoresis. Am J Hum Genet 66:1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt H-J (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Graven L, Passarino G, Semino O, Boursot P, Santachiara-Benerecetti S, Langaney A, Excoffier L (1995) Evolutionary correlation between control region sequence and restriction polymorphisms in the mitochondrial genome of a large Senegalese Mandenka sample. Mol Biol Evol 12:334–345 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Di Rienzo A, Kocher TD, Wilson AC (1993) Toward a more accurate time scale for the human mitochondrial DNA tree. J Mol Evol 37:347–354 [DOI] [PubMed] [Google Scholar]
- Hofmann, S, Jaksch M, Bezold R, Mertens S, Aholt S, Paprotta A, Gerbitz KD (1997) Population genetics and disease susceptibility: characterization of central European haplogroups by mtDNA gene mutations, correlations with D loop variants and association with disease. Hum Mol Genet 6:1835–1846 [DOI] [PubMed] [Google Scholar]
- Housley RA, Gamble CS, Street M, Pettitt P (1997) Radiocarbon evidence for the late glacial human recolonisation of northern Europe. Proc Prehist Soc 63:25–54 [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Izagirre N, de la Rúa C (1999) A mtDNA analysis in ancient Basque populations: implications for haplogroup V as a marker for a major paleolithic expansion from southwestern Europe. Am J Hum Genet 65:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings M, Salem AE, Bauer K, Geisert H, Malek AK, Chaix L, Simon C, Welsby D, Di Rienzo A, Utermann G, Sajantila A, Pääbo S, Stoneking M (1999) mtDNA analysis of Nile River Valley populations: a genetic corridor or a barrier to migration? Am J Hum Genet 64:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A (1999) The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspina P, Cruciani F, Santolamazza P, Torroni A, Pangrazio A, Akar N, Bakalli V, Brdicka R, Jaruzelska J, Kozlov A, Malyarchuck B, Mehdi SQ, Michalodimitrakis E, Varesi L, Memmi MM, Vona G, Villems R, Parik J, Romano V, Stefan M, Stenico M, Terrenato L, Novelletto A, Scozzari R (2000) Patterns of male-specific inter-population divergence in Europe, West Asia and North Africa. Ann Hum Genet 64:395–412 [DOI] [PubMed] [Google Scholar]
- Morelli L, Grosso MG, Vona G, Varesi L, Torroni A, Francalacci P (2000) Frequency distribution of mitochondrial DNA haplogroups in Corsica and Sardinia. Hum Biol 72:585–595 [PubMed] [Google Scholar]
- Morral N, Bertranpetit J, Estivill X, Nunes V, Casals T, Giménez J, Reis A, et al (1994) The origin of the major cystic fibrosis mutation (ΔF508) in European populations. Nat Genet 7:169–175 [DOI] [PubMed] [Google Scholar]
- Rando JC, Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM, Bandelt H-J (1998) Mitochondrial DNA analysis of Northwest African populations reveals genetic exchanges with European, Near-Eastern, and sub-Saharan populations. Ann Hum Genet 62:531–550 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V (2001) The mitochondrial gene tree comes of age. Am J Hum Genet 68:1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S (2000) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr TG, Sukernik RI, Starikovskaya YB, Wallace DC (1999) Mitochondrial DNA variation in Koryaks and Itel’men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am J Phys Anthropol 108:1–39 [DOI] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 [DOI] [PubMed] [Google Scholar]
- Simoni L, Calafell F, Pettener D, Bertranpetit J, Barbujani G (2000) Geographic patterns of mtDNA diversity in Europe. Am J Hum Genet 66:262–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R, Pritchard JK, Shen P, Oefner PJ, Feldman MW (2000) Recent common ancestry of human Y chromosomes: evidence from DNA sequence data. Proc Natl Acad Sci USA 97:7360–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, D'Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savontaus M-L, Bonné-Tamir B, Scozzari R (1998) MtDNA analysis reveals a major late Palaeolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet 62: 1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Lott MT, Cabell MF, Chen Y-S, Lavergne L, Wallace DC (1994) MtDNA and the origin of Caucasians. Identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am J Hum Genet 55:760–776 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993) Asian affinities and continental radiation of the four founding Native American mitochondrial DNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-Herbots H, Richards M, Forster P, Sykes B (1996) Site 73 in hypervariable region II of the human mitochondrial genome and the origin of European populations. Ann Hum Genet 60: 499–508 [DOI] [PubMed] [Google Scholar]
- Willis KJ, Whittaker RJ (2000) The refugial debate. Science 287:1406–1407 [DOI] [PubMed] [Google Scholar]