Biophysical Investigations of Engineered Polyproteins: Implications for Force Data (original) (raw)
Abstract
Dynamic force spectroscopy is rapidly becoming a standard biophysical technique. Significant advances in the methods of analysis of force data have resulted in ever more complex systems being studied. The use of cloning systems to produce homologous tandem repeats rather than the use of endogenous multidomain proteins has facilitated these developments. What is poorly addressed are the physical properties of these constructed polyproteins. Are the properties of the individual domains in the construct independent of one another or attenuated by adjacent domains? We present data for a construct of eight fibronectin type III domains from the human form of tenascin that exhibits ∼1 kcal mol−1 increase in stability compared to the monomer. This effect is salt and pH dependent, suggesting that the stabilization results from electrostatic interactions, possibly involving charged residues at the interfaces of the domains. Kinetic analysis shows that this stabilization reflects a slower unfolding rate. Clearly, if domain-domain interactions affect the unfolding force, this will have implications for the comparison of absolute forces between types of domains. Mutants of the tenascin 8-mer construct exhibit the same change in stability as that observed for the corresponding mutation in the monomer. And when Φ-values are calculated for the 8-mer construct, the pattern is similar to that observed for the monomer. Therefore, mutational analyses to resolve mechanical unfolding pathways appear valid. Importantly, we show that interactions between the domains may be masked by changes in experimental conditions.
INTRODUCTION
The field of dynamic force spectroscopy has evolved rapidly in the type of polyprotein systems that are available for investigation. There has been a shift from the use of endogenous heterogeneous polyproteins such as titin (Kellermayer et al., 1997; Rief et al., 1997; Tskhovrebova et al., 1997), spectrin (Rief et al., 1999), and tenascin (TNfn3; Oberhauser et al., 1998) to homologous repeats created using versatile cloning systems (Carrion-Vazquez et al., 1999). These developments were essential to understanding the mechanism by which proteins can resist mechanical force (Best et al., 2003; Brockwell et al., 2003; Carrion-Vazquez et al., 2003; Law et al., 2003a; Lenne et al., 2000; Rief et al., 1997; Williams et al., 2003) homologous repeats allow forces to be unambiguously assigned to domain type, and with a versatile cloning system mutational studies are possible (Best et al., 2002; Steward et al., 2002).
There is no doubt the use of homologous repeats is a powerful tool for investigating the mechanical properties of a protein. But what is rarely addressed is the effect of the multimerization of these domains. Are the domains folded in a polyprotein? Do these constructs behave as the sum of their parts? The mechanical unfolding of polyproteins assumes an uncoupled system, where the behavior of a domain in solution or under mechanical force is independent of the state of the adjacent domain.
In subsequent mutational studies, what is the effect of a specific mutation upon the ground state for unfolding? Does this mutation have the same effect in the polyprotein as the monomer? If the properties of different mutants are not known, how can the differences in the relative barrier heights to mechanical unfolding be interpreted reasonably (Li et al., 2000a; Williams et al., 2003)?
In this study, we describe the use of standard biophysical techniques, their interpretation, and the implications for the analysis of force data. We use the extensive biophysical analysis of an eight domain (8-mer) construct of the third fibronectin type III domain from the human form of TNfn3 as evidence of the importance of these initial characterizations. In light of these experimental data, we reexamine the behavior of the paradigm polyprotein, the titin 27th immunoglobulin domain (I27) construct, under alternative experimental conditions.
MATERIALS AND METHODS
Protein expression and purification
The extended form of TNfn3, residues 1-92, was used for both the monomer and 8-mer construct studies. Expression and purification of the monomer was performed as described (Clarke et al., 1997). The purification of the 8-mer construct differed in that the protein was eluted from the resin, retaining the His-tag, rather than being thrombin cleaved, using a high concentration of imidazole. Both purification methods produce protein of >95% purity as determined by SDS-PAGE. Construction of the 8-mer and mutants thereof was performed as described (Steward et al., 2002). The mutagenesis step (Quikchange, Stratagene, La Jolla, CA) for each module was undertaken in the individual T-clones, before assembly of the full-length construct. Proteins were stored at 4°C.
Equilibrium measurements
The equilibrium denaturation experiments of the TNfn3 monomer, 8-mer construct, and mutants thereof were performed as described (Clarke et al., 1997). The denaturants used were urea and GdmCl, and the buffers used were either 50 mM sodium acetate at pH 5 (35 mM sodium acetate, 15 mM acetic acid) or 50 mM MOPS at pH 7 (32 mM 3-[_N_-morpholino]propanesulfonic acid, 18 mM sodium 3-[_N_-morpholino]propanesulfonic acid). All experiments were equilibrated and performed at 25°C. The equivalent experiments for the mutant I27 monomer and 8-mer construct were performed using urea in phosphate-buffered saline (8.1 mM disodium hydrogen orthophosphate, 1.9 mM sodium dihydrogen orthophosphate, 2.7 mM KCl, 137 mM NaCl), with excitation at 280 nm and with fluorescence observed at 320 nm. All denaturations were fully reversible (the same results were obtained when starting from folded or unfolded proteins). The data obtained were the same for proteins that had been stored for several weeks as for fresh, newly purified protein.
Kinetic measurements
Folding reactions occurring on short time scales (<2000 s) requiring rapid mixing apparatus were performed using a stopped-flow apparatus (Applied Photophysics, Leatherhead, UK). Fluorescence was measured at wavelengths >335 nm for TNfn3 and >320nm for I27, using an excitation wavelength of 280 nm. To initiate unfolding, one volume of an ∼11 _μ_M protein solution (total concentration of domains, whether in a mulitmer or as a monomer) was rapidly mixed with 10 volumes of a concentrated denaturant solution. Refolding was initiated by rapidly mixing one volume of protein, unfolded in a suitable concentration of denaturant, with 10 volumes of denaturant at different concentrations. Data collected from at least four experiments were averaged. Folding reactions requiring monitoring of longer timescales (>2000 s) necessitated a manual mixing procedure and the fluorescence, for both TNfn3 and I27 mutant L60A, was measured at 320 nm, using an excitation wavelength of 280 nm. Unfolding was initiated by manually mixing one volume of an 11 _μ_M protein solution with 10 volumes of denaturant at different concentrations. Refolding was initiated by manually mixing one volume of protein, unfolded in a suitable concentration of denaturant, with 10 volumes of a low denaturant concentration solution. All experiments contained 1× buffer and were performed at 25°C. Folding experiments performed upon the I27 L60A mutant monomer and both 8-mer constructs were all performed in the presence of 5 mM DTT. Unfolding traces were fitted to a single exponential function and refolding traces to a double exponential function. A term was included to account for baseline instability. 0 M refolding measurements made for the TNfn3 8-mer construct were performed by jumping unfolded protein at pH 12.4 to pH 5.8 and monitored using stopped-flow fluorescence with a 335-nm cut-off filter. Alkali-unfolded TNfn3 was mixed 1:1 with 50 mM sodium acetate at pH 5.5 (33.3 mM sodium acetate, 16.7 mM acetic acid). All rates determined were shown to be independent of protein concentration.
RESULTS AND DISCUSSION
TNfn3: unexpected complexity
TNfn3 is an extracellular matrix protein with an endogenous capacity to resist mechanical force (Leahy et al., 1992; Lightner and Erickson, 1990; Oberhauser et al., 1998). The saw-tooth profile of TNfn3 observed under force has been attributed to the sequential unfolding of the fibronectin type III domains (Oberhauser et al., 1998). Each domain comprises seven strands forming two sheets, which pack together to form an extensive hydrophobic core. The folding and other biophysical characteristics of the third fibronectin type III domain of human TNfn3 (Fig. 1) has previously been studied in depth (Akke et al., 1998; Carr et al., 1997; Clarke et al., 1997; Hamill et al., 1998; Leahy et al., 1992; Meekhof et al., 1998). The TNfn3 polyprotein construct studied here consists of eight of these domains in tandem.
FIGURE 1.
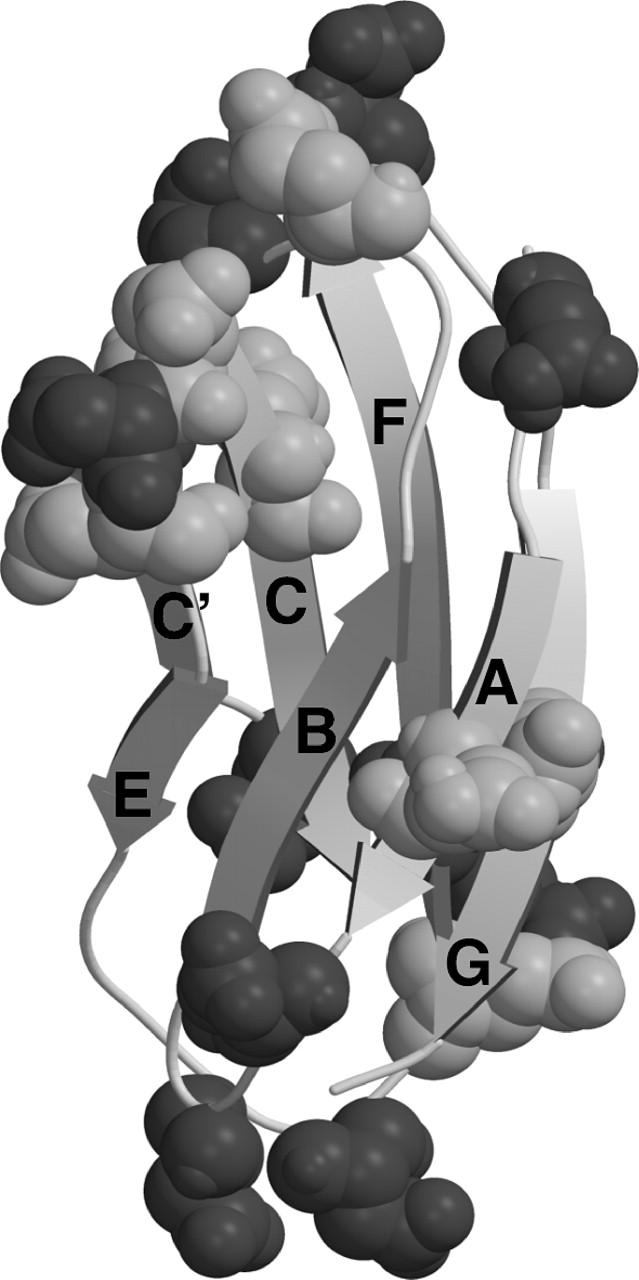
The structure of TNfn3, with Asp and Glu residues indicated in dark shaded and light shaded areas, respectively. Figure prepared using the program MolScript (Kraulis, 1991) using the file 1ten.pdb. Note that the TNfn3 module used in these constructs is extended at the C-terminus by two residues (Hamill et al., 1998).
Stability
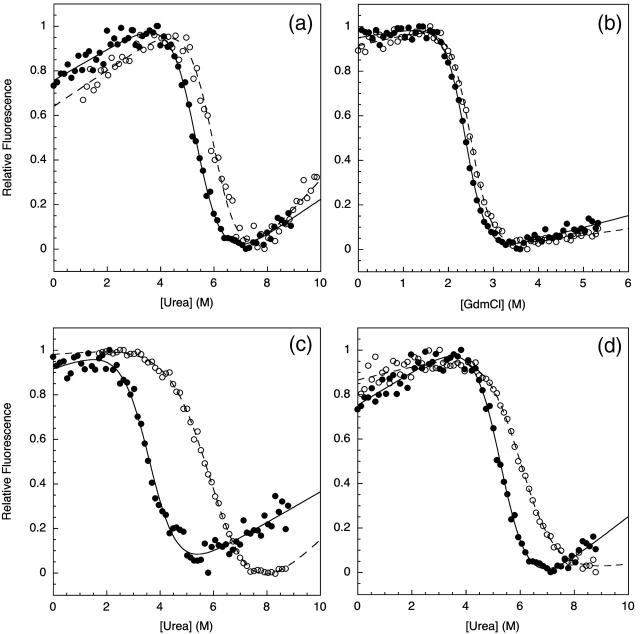
Comparative chemical denaturation measurements were performed for the TNfn3 monomer and 8-mer construct (Table 1). In urea at pH 5 an increase in stability of ∼1 kcal mol−1 for the polyprotein was observed (Fig. 2 a). Interestingly, when using guanidinium chloride (GdmCl) as a denaturant, the stabilization effect is no longer significant (Fig. 2 b). It is probable that the loss of stabilization can be attributed to the shielding of electrostatic interactions by guanidinium and chloride ions. To confirm this salt effect, the measurements were repeated in urea with the addition of 0.5 M NaCl. Under these conditions, the stabilization effect is diminished but not entirely lost (data not shown). Analysis of the structure of TNfn3 reveals acidic patches, regions rich in aspartate and glutamate residues, at either end of the molecule (Fig. 1). It has been shown that the monomer protein is more stable at pH 5 than pH 7 due to protonation of some of these acidic residues at pH 5 (Hamill et al., 1998). The stability of the monomer and 8-mer was compared at pH 7 in urea (Fig. 2 c). The monomer is ∼2.5 kcal mol−1 less stable at pH 7 than at pH 5. However, under the same conditions, the stability of the polyprotein appears less sensitive to pH; this implies shielding of the charged groups by adjacent domains. This result supports the hypothesis that electrostatic interactions may contribute to the observed stabilization of the domain upon incorporation into the polyprotein.
TABLE 1.
Thermodynamic data of monomeric protein compared to 8-mer constructs
| Urea | GdmCl | ||||
|---|---|---|---|---|---|
| Construct | pH | [Urea]50%* (M) | Δ_G_D − N† kcal mol−1 | [GdmCl]50%* (M) | Δ_G_D − N† kcal mol−1 |
| TNfn3 monomer | 5‡ | 5.33 | 6.50 | 2.38¶ | 6.19 |
| 7§ | 3.80†† | 3.80 | |||
| TNfn3 8-mer | 5 | 6.00 | 7.32 | 2.49¶ | 6.47 |
| 7 | 5.76 | 5.76 | |||
| TNfn3 in TNfn3-I27 8-mer | 5 | 6.00 | 7.32 | ||
| I27 (L60A) monomer | 7.4‖ | 3.03 | 3.33 | ||
| I27 (L60A) 8-mer | 7.4 | 3.43 | 3.74 |
FIGURE 2.
Chemical denaturation measurements comparing the relative stabilities of the TNfn3 monomer (•) and 8-mer construct (○) performed in (a) urea at pH 5, (b) GdmCl at pH 5, and (c) urea at pH 7. Chemical denaturation was monitored by fluorescence at 320 nm at 25°C. (d) The stability of the TNfn3 domain is the same in the TNfn3-I27 8-mer where each TNfn3 domain has an adjacent I27 domain. The apparent change in _m_-value is due to the contribution of I27 modules in the construct beginning to unfold at the higher denaturant concentrations.
Kinetics
For both pH 5 and pH 7 in urea, the stabilization of the polyprotein can be predominantly ascribed to a slower unfolding rate (Fig. 3, a and c) compared to the monomer under the same conditions. When the chevrons are repeated using GdmCl as a denaturant at pH 5, the unfolding rates are comparable for the polyprotein and monomer (Fig. 3 b). The refolding arms of the monomer and polyprotein overlay under all conditions, indicating that the component domains of the polyprotein fold independently of each other, unaffected by the presence of their neighbors. The previous folding studies of TNfn3 have shown that the peripheral regions (the A and G strands and the loops) are not involved in early folding, but mutations in these regions can affect the unfolding rate significantly. An interaction between the domains of the protein that stabilized the terminal strands or peripheral loop regions would be consistent with the stabilization effect observed in the 8-mer construct. These results support the idea of a domain-domain interaction between the folded TNfn3 modules involving the peripheral strands and loops of the proteins.
FIGURE 3.
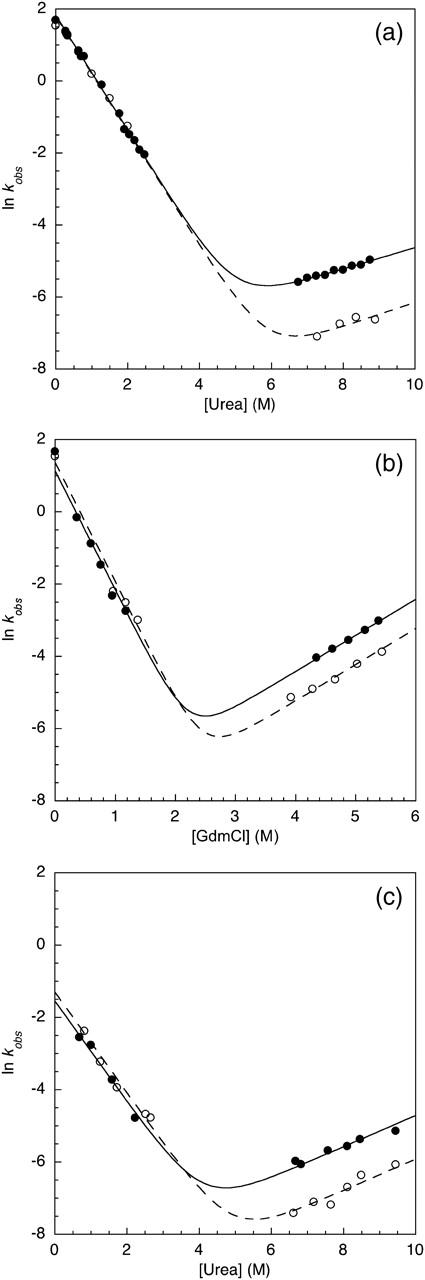
Kinetic data for the unfolding and refolding of the TNfn3 monomer (•) and 8-mer construct (○) performed in (a) urea at pH 5, (b) GdmCl at pH 5, and (c) urea at pH 7. The Chevrons display the major rates determined from the kinetic measurements.
The interaction is not specific
To test the specificity of this interaction between domains, TNfn3 was studied in a polyprotein where every alternate domain was titin I27. The stability of the TNfn3 domains could be monitored independently of the I27 since I27 is much more stable than TNfn3. The TNfn3 domain was stabilized to the same extent in the TNfn3-I27 8-mer as in the TNfn3 8-mer (Fig. 2 d). Interestingly, there are also charged patches on the surface of I27.
TNfn3: mutational analysis
Φ-Value analysis uses mutational probes to determine the structure of the transition state for (un)folding (Fersht et al., 1992). Originally performed using bulk solution techniques, the same principles have recently been applied to probe the mechanical transition state using force data (Best et al., 2002, 2003). Φ-value analysis can resolve the difference between solution and mechanical transition states, which may not be obvious when solely comparing unfolding rates at 0 M denaturant and at zero force. In a Φ-value analysis, the effect of the mutation on the unfolding kinetics is normalized against the effect of the same mutation on the native state (Fersht et al., 1992). This has previously been estimated using the monomeric form of the protein, the assumption being that a mutation will have the same destabilizing effect whether the domain is a monomer or an 8-mer. To test this, we compared the effect of a number of mutations in the monomer and the 8-mer forms of TNfn3.
Stability
Nine conservative mutations were made, equating to at least one probe per strand. All mutant polyproteins, with the same single mutation in each of the domains, exhibited the same change in Δ_G_D − N as that observed for the monomer (Table 2). This is an important result: the relative change in free energy is unchanged by the domain-domain interaction of TNfn3. This validates the comparative studies of the mutant constructs and therefore the conclusions drawn from them. However, this reflects the nature of the mutations made (buried, conservative, hydrophobic deletions). Had surface, polar residue mutations (particularly at the termini) been made, the results may not have been the same. Clearly ΔΔ_G_D − N values must be determined for the mutants in the 8-mer construct before mechanical Φ-values of any meaning may be calculated.
TABLE 2.
Thermodynamic and kinetic data for mutants of TNfn3
| Monomer | 8-mer | ||||
|---|---|---|---|---|---|
| Mutation (strand) | [Urea]50% (M)* | ΔΔ_G_D − N* kcal mol−1 | [urea]50% (M) | ΔΔ_G_D − N† kcal mol−1 | _k_f (_s_−1)‡ |
| Wild-type | 5.33 (0.03) | — | 6.00 (0.05) | — | 1.20 (0.03) |
| L2A (A) | 3.59 (0.1) | 2.1 (0.1) | 4.35 (0.07) | 2.0 (0.1) | 1.43 (0.13) |
| I8A (A) | 2.98 (0.1) | 2.9 (0.1) | 3.99 (0.06) | 2.5 (0.1) | 1.34 (0.11) |
| I20A (B) | 2.31 (0.1) | 3.7 (0.1) | 3.48 (0.08) | 3.1 (0.1) | 0.20 (0.01) |
| Y36A (C) | 1.86 (0.1) | 4.2 (0.1) | 2.86 (0.03) | 3.8 (0.1) | 0.07 (0.01) |
| I48A (C′) | 3.52 (0.1) | 2.2 (0.1) | 4.35 (0.09) | 2.0 (0.1) | 0.13 (0.01) |
| I59A (E) | 3.63 (0.1) | 2.1 (0.1) | 4.30 (0.07) | 2.1 (0.1) | 0.26 (0.03) |
| Y68F (F) | 2.75 (0.1) | 3.1 (0.1) | 3.53 (0.03) | 3.0 (0.1) | 0.04 (0.01) |
| V70A (F) | 3.01 (0.1) | 2.8 (0.1) | 3.73 (0.05) | 2.8 (0.1) | 0.25 (0.01) |
| T90A (G) | 2.50 (0.1) | 3.4 (0.1) | 3.52 (0.10) | 3.0 (0.1) | 0.37 (0.01) |
Kinetics
One of the aims of comparing mechanical and denaturant induced unfolding transition states is to determine how the folding pathway changes when the protein experiences mechanical stress. The tethering effect of the termini attributed to the domain-domain interaction observed for the TNfn3 polyprotein made it necessary to determine whether the Φ-values of the polyprotein were comparable to those previously determined for the monomer. These Φ-values were obtained by determining the 0 M refolding rate of the wild-type polyprotein and mutants, by stopped-flow fluorescence, using pH-jump techniques. All the published folding studies have been performed at the pH where the monomer is most stable, pH 5. However, aggregation of the unfolded polyprotein at 0 M denaturant at this pH prevented reasonable rates being obtained, so these experiments were performed at pH 5.8. Although the refolding rates obtained were consistently slower at pH 5.8, the relative differences in the wild-type and mutant rates could be determined, and Φ-values calculated. The aggregation effects observed at pH 5 may still be affecting the data obtained at pH 5.8 (the local concentration of protein is high in a polyprotein and cannot be reduced by dilution); therefore, these Φ-values are associated with a relatively large error.
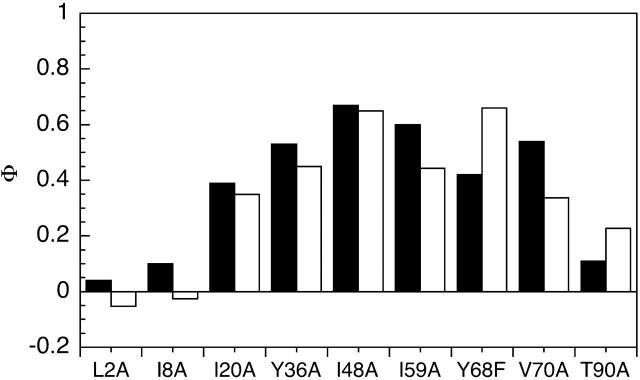
The Φ-values obtained were compared to the Φ-values previously reported for the monomer (Fig. 4). A high Φ-value indicates the residue is in a region that is structured, and a low Φ-value indicates that the residue is in a region that is generally unstructured in the transition state for (un)folding. Within the limits of our confidence in the 8-mer folding data, we infer that the Φ-values of our mutants have not been significantly altered by inclusion of the domain into the 8-mer, and the pattern of Φ-values in the 8-mer is similar to that in the monomer (Hamill et al., 2000). We infer that the folding pathway is unchanged by inclusion of the domain into an 8-mer construct.
FIGURE 4.
Comparison of the Φ-values calculated for the TNfn3 monomer (black) and polyprotein (white) from 0 M refolding rates obtained at pH 5 and pH 5.8, respectively. The Φ-values are the same, within the error of our measurements (± 0.1). 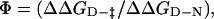 where
where 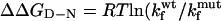 and
and 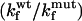 are the folding rates of wild-type and mutant, respectively, with the values for the monomer taken from Hamill et al. (2000).
are the folding rates of wild-type and mutant, respectively, with the values for the monomer taken from Hamill et al. (2000).
I27: a simple case?
I27 from the muscle protein titin has been studied extensively, with both the different transition state structures for the solution and mechanical unfolding pathways resolved using Φ-value analysis (Best et al., 2003; Fowler and Clarke, 2001; Wright et al., 2003). The stabilities of the I27 monomer and the 8-mer construct, determined by equilibrium denaturation, were demonstrated to be the same (Best et al., 2001). The kinetics of the I27 monomer and 8-mer construct also proved to be comparable under these same conditions. However, these studies used GdmCl as a denaturant. In light of the salt and pH dependent stabilization effect observed for the TNfn3 construct, we decided to repeat these experiments in urea. However, wild-type I27 is significantly more stable than TNfn3; in fact too stable to be studied in urea. Therefore, the destabilized mutant L60A (with a deeply buried core mutation) was chosen, which allowed the study of I27 to be performed in urea without influencing the domain interfaces.
Stability
In contrast to what had been observed previously, a small stabilization effect of ∼0.6 kcal/mol (Table 1) was observed for the I27 mutant construct when compared to the monomeric form (Fig. 5 a). This suggests that there are some small stabilizing electrostatic interactions between I27 domains that were previously missed when GdmCl was used as a denaturant. However, this is less than the increase in stability observed for the TNfn3 construct.
FIGURE 5.
Biophysical studies of a titin mutant. (a) Chemical denaturation measurements comparing the relative stabilities of the I27 monomer (•) and 8-mer construct (○) performed in urea at pH 7.4. Chemical denaturation was monitored at 320 nm. (b) Kinetic data for the unfolding and refolding of the I27 L60A mutant monomer (•) and 8-mer construct (○) performed in urea at pH 7.4. The chevrons display the major rates determined from kinetic measurements.
Kinetics
The stabilization effect observed for the I27 mutant construct in urea can also be attributed to a decrease in the unfolding rate (Fig. 5 b), as the refolding arms of the monomer and polyprotein overlay. As for TNfn3, the comparable refolding rates observed for the I27 mutant monomer and polyprotein indicate that the component domains of the construct fold independently of each other.
CONCLUSIONS
Domain-domain interactions: implications for force data?
Both the TNfn3 and the mutant I27 domains exhibit a stabilization effect associated with incorporation into their respective homologous 8-mer constructs. For both TNfn3 and the I27 mutant, this stabilization effect has been attributed to a decrease in the unfolding rate of the component domains of the construct. The force at which a protein unfolds depends upon the height of the barrier between the ground state and the transition state. In titin I27, although the unfolding transition state is not the same as that probed in denaturant unfolding experiments, the unfolding force has been shown to be related to the unfolding rate of the monomer (Li et al., 2000a; Scott et al., 2002). This is because the same region of the protein unfolds early in both events (Best et al., 2003). Therefore, it is important to consider that when in a multimer, tethering may affect the unfolding force. The folding Φ-values determined for the TNfn3 polyprotein were comparable to those observed for the monomer. Therefore, it is reasonable to assume that the domain-domain interaction is lost comparatively early in unfolding. However, it must be noted that a property observed in solution may not necessarily be influential upon the measured force to unfold. The low force regime of tethering the molecule between the atomic force microscope (AFM) tip and the surface may be sufficient to “melt” out any interaction that may be formed in solution. If this is the case, comparison of absolute forces between domain types is possible. Fortunately, this will be testable. Does salt, or pH, for example, affect the unfolding forces? If domain-domain interactions do make a contribution to the forces measured for a domain unfolding, comparisons may only safely be made between mutants of the same domain type, not between proteins.
An interesting corollary to this work lies in the following question: do natural domain pairs with natural linkers influence the unfolding of each other? The answer appears to be both yes and no. Spectrin domains are both stabilized by each other in equilibrium experiments (MacDonald and Pozharski, 2001) and can be observed to unfold cooperatively in AFM experiments (Law et al., 2003a). (Interestingly, this forced unfolding cooperativity is lost at higher temperatures (Law et al., 2003b).) On the other hand, titin domains have been shown to fold and unfold independently in both equilibrium and kinetic experiments (Scott et al., 2002). But, in apparent contradiction, in AFM experiments, the force to unfold I28 was shown to be higher when I28 was in tandem array (linked naturally) with I27 than in an I28 polyprotein. This was interpreted as the mechanical properties of I28 being modified by the presence of I27 (Li et al., 2000b). However, intriguingly, since the forces required to unfold I28 are significantly higher than those observed for I27, all of the I27 domains were unfolded before I28. That is, I28 has greater mechanical stability in the presence of unfolded I27. The biophysical data from Scott et al. (2002) shed some light on this—they suggest that the domain boundaries of I28 in the AFM study were “too short”, resulting in a protein that is less stable (of course, with I27 attached, this problem is solved at the N-terminus). The data we have presented here suggest another contributing factor—could the tandem I28 domains in close proximity be destabilizing each other? In either case, the force observed to unfold I28 in the homologous construct would be an underestimate of the true force to unfold I28 observed in the heterologous construct including I27. Again, this emphasizes the need to characterize the AFM protein substrates thoroughly before carrying out the AFM experiments.
Acknowledgments
This work was supported by the Medical Research Council (R.W.S.R.) and the Wellcome Trust (J.C. and A.S.). J.C. is a Wellcome Trust Senior Research Fellow.
References
- Akke, M., J. Liu, J. Cavanagh, H. P. Erickson, and A. G. Palmer. 1998. Pervasive conformational fluctuations on microsecond time scales in a fibronectin type III domain. Nat. Struct. Biol. 5:55–59. [DOI] [PubMed] [Google Scholar]
- Best, R. B., S. B. Fowler, J. L. Toca-Herrera, and J. Clarke. 2002. A simple method for probing the mechanical unfolding pathway of proteins by protein engineering phi-value analysis. Proc. Natl. Acad. Sci. USA. 99:12143–12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best, R. B., S. B. Fowler, J. L. Toca-Herrera, A. Steward, E. Paci, and J. Clarke. 2003. Mechanical unfolding of a titin Ig domain: structure of transition state revealed by combining atomic force microscopy, protein engineering and molecular dynamics simulations. J. Mol. Biol. 330:867–877. [DOI] [PubMed] [Google Scholar]
- Best, R. B., H. B. Li, A. Steward, V. Daggett, and J. Clarke. 2001. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 81:2344–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockwell, D. J., E. Paci, R. C. Zinober, G. S. Beddard, P. D. Olmsted, D. A. Smith, R. N. Perham, and S. E. Radford. 2003. Pulling geometry defines the mechanical resistance of a _β_-sheet protein. Nat. Struct. Biol. 10:731–737. [DOI] [PubMed] [Google Scholar]
- Carr, P. A., H. P. Erickson, and A. G. Palmer. 1997. Backbone dynamics of homologous fibronectin type III cell adhesion domains from fibronectin and tenascin. Structure. 5:949–959. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez, M., H. B. Li, H. Lu, P. E. Marszalek, A. F. Oberhauser, and J. M. Fernandez. 2003. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 10:738–743. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez, M., A. F. Oberhauser, S. B. Fowler, P. E. Marszalek, S. E. Broedel, J. Clarke, and J. M. Fernandez. 1999. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 96:3694–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J., S. J. Hamill, and C. M. Johnson. 1997. Folding and stability of a fibronectin type III domain of human tenascin. J. Mol. Biol. 270:771–778. [DOI] [PubMed] [Google Scholar]
- Cota, E., S. J. Hamill, S. B. Fowler, and J. Clarke. 2000. Two proteins with the same structure respond very differently to mutation—the role of plasticity in protein stability. J. Mol. Biol. 302:713–725. [DOI] [PubMed] [Google Scholar]
- Fersht, A. R., A. Matouschek, and L. Serrano. 1992. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J. Mol. Biol. 224:771–782. [DOI] [PubMed] [Google Scholar]
- Fowler, S. B., and J. Clarke. 2001. Mapping the folding pathway of an immunoglobulin domain: structural detail from phi-value analysis and movement of the transition state. Structure. 9:355–366. [DOI] [PubMed] [Google Scholar]
- Hamill, S. J., A. E. Meekhof, and J. Clarke. 1998. The effect of boundary selection on the stability and folding of the third fibronectin type III domain from human tenascin. Biochemistry. 37:8071–8079. [DOI] [PubMed] [Google Scholar]
- Hamill, S. J., A. Steward, and J. Clarke. 2000. The folding of an immunoglobulin-like Greek key protein is defined by a common-core nucleus and regions constrained by topology. J. Mol. Biol. 297:165–178. [DOI] [PubMed] [Google Scholar]
- Kellermayer, M. S. Z., S. B. Smith, H. L. Granzier, and C. Bustamante. 1997. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 276:1112–1116. [DOI] [PubMed] [Google Scholar]
- Kraulis, P. 1991. MolScript, a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946–950. [Google Scholar]
- Law, R., P. Carl, S. Harper, P. Dalhaimer, D. W. Speicher, and D. E. Discher. 2003a. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys. J. 84:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, R., G. Liao, S. Harper, G. L. Yang, D. W. Speicher, and D. E. Discher. 2003b. Pathway shifts and the thermal softening in temperature-coupled forced unfolding of spectrin domains. Biophys. J. 85:3286–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy, D. J., W. A. Hendrickson, I. Aukhil, and H. P. Erickson. 1992. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science. 258:987–991. [DOI] [PubMed] [Google Scholar]
- Lenne, P. F., A. J. Raae, S. M. Altmann, M. Saraste, and J. K. H. Horber. 2000. States and transitions during forced unfolding of a single spectrin repeat. FEBS Lett. 476:124–128. [DOI] [PubMed] [Google Scholar]
- Li, H. B., M. Carrion-Vazquez, A. F. Oberhauser, P. E. Marszalek, and J. M. Fernandez. 2000a. Point mutations alter the mechanical stability of immunoglobulin modules. Nat. Struct. Biol. 7:1117–1120. [DOI] [PubMed] [Google Scholar]
- Li, H. B., A. F. Oberhauser, S. B. Fowler, J. Clarke, and J. M. Fernandez. 2000b. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. USA. 92:6527–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner, V. A., and H. P. Erickson. 1990. Binding of hexabrachion (tenascin) to the extracellular matrix and substratum and its effect on cell adhesion. J. Cell Sci. 95:263–277. [DOI] [PubMed] [Google Scholar]
- MacDonald, R. I., and E. V. Pozharski. 2001. Free energies of urea and of thermal unfolding show that two tandem repeats of spectrin are thermodynamically more stable than a single repeat. Biochemistry. 40:3974–3984. [DOI] [PubMed] [Google Scholar]
- Meekhof, A. E., S. J. Hamill, V. L. Arcus, J. Clarke, and S. M. V. Freund. 1998. The dependence of chemical exchange on boundary selection in a fibronectin type III domain from human tenascin. J. Mol. Biol. 282:181–194. [DOI] [PubMed] [Google Scholar]
- Oberhauser, A. F., P. E. Marszalek, H. P. Erickson, and J. M. Fernandez. 1998. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 393:181–185. [DOI] [PubMed] [Google Scholar]
- Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- Rief, M., J. Pascual, M. Saraste, and H. E. Gaub. 1999. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286:553–561. [DOI] [PubMed] [Google Scholar]
- Scott, K. A., A. Steward, S. B. Fowler, and J. Clarke. 2002. Titin; a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 315:819–829. [DOI] [PubMed] [Google Scholar]
- Steward, A., J. L. Toca-Herrera, and J. Clarke. 2002. Versatile cloning system for construction of multimeric proteins for use in atomic force microscopy. Protein Sci. 11:2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tskhovrebova, L., J. Trinick, J. A. Sleep, and R. M. Simmons. 1997. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 387:308–312. [DOI] [PubMed] [Google Scholar]
- Williams, P. M., S. B. Fowler, R. B. Best, J. L. Toca-Herrera, K. A. Scott, A. Steward, and J. Clarke. 2003. Hidden complexity in the mechanical properties of titin. Nature. 422:446–449. [DOI] [PubMed] [Google Scholar]
- Wright, C. F., K. Lindorff-Larsen, L. G. Randles, and J. Clarke. 2003. Parallel protein-unfolding pathways revealed and mapped. Nat. Struct. Biol. 10:658–662. [DOI] [PubMed] [Google Scholar]