Esc1, a Nuclear Periphery Protein Required for Sir4-Based Plasmid Anchoring and Partitioning (original) (raw)
Abstract
A targeted silencing screen was performed to identify yeast proteins that, when tethered to a telomere, suppress a telomeric silencing defect caused by truncation of Rap1. A previously uncharacterized protein, Esc1 (establishes silent chromatin), was recovered, in addition to well-characterized proteins Rap1, Sir1, and Rad7. Telomeric silencing was slightly decreased in Δ_esc1_ mutants, but silencing of the HM loci was unaffected. On the other hand, targeted silencing by various tethered proteins was greatly weakened in Δ_esc1_ mutants. Two-hybrid analysis revealed that Esc1 and Sir4 interact via a 34-amino-acid portion of Esc1 (residues 1440 to 1473) and a carboxyl-terminal domain of Sir4 known as PAD4 (residues 950 to 1262). When tethered to DNA, this Sir4 domain confers efficient partitioning to otherwise unstable plasmids and blocks the ability of bound DNA segments to rotate freely in vivo. Here, both phenomena were shown to require ESC1. Sir protein-mediated partitioning of a telomere-based plasmid also required ESC1. Fluorescence microscopy of cells expressing green fluorescent protein (GFP)-Esc1 showed that the protein localized to the nuclear periphery, a region of the nucleus known to be functionally important for silencing. GFP-Esc1 localization, however, was not entirely coincident with telomeres, the nucleolus, or nuclear pore complexes. Our data suggest that Esc1 is a component of a redundant pathway that functions to localize silencing complexes to the nuclear periphery.
The silent mating-type loci (HML and HMR) in the yeast Saccharomyces cerevisiae are maintained in a transcriptionally inactive state due to the formation of a specialized chromatin structure analogous to heterochromatin of higher eukaryotes. Silencing of the mating-type loci requires flanking sequence elements, termed silencers, which bind the transcription factors Rap1 and Abf1, as well as the multisubunit origin recognition complex (ORC) (reviewed in references 11, 15, and 27). Together, these proteins recruit the silencing proteins, Sir1, Sir2, Sir3, and Sir4, that participate in the formation of heterochromatin.
Genes placed near telomeres are also silenced. Telomeric silencing depends on Rap1, which binds to telomeric TG1-3 repeats and recruits Sir3 and Sir4 (28, 29). A Sir complex consisting of Sir2, Sir3, and Sir4 then spreads from the telomeres to nearby nucleosomes to form silent chromatin (16, 17). Although RAP1 is an essential gene, mutations that delete the 3′ end of the gene (rap1_Δ_C) are viable but lead to a complete loss of telomeric silencing (22, 26, 28). This is due to the inability of Rap1ΔC to recruit Sir3 and Sir4 to the telomeres.
Expressing Sir proteins as GAL4 DNA-binding domain (GBD) hybrids and tethering them to defective silencers in which binding sites for the ORC, Rap1, and/or Abf1 have been replaced by Gal4 binding sites can lead to silencing. This so-called targeted silencing was first demonstrated with GBD-Sir1 but has since been shown with the other Sir proteins as well as with Rap1 and Orc1 (5, 6, 38). Targeted silencing also has been achieved by forming GBD hybrids with endoplasmic reticulum or Golgi proteins and tethering them to a partially defective HMR E silencer (1). Overexpression of such membrane protein hybrids causes them to accumulate in the endoplasmic reticulum (which is contiguous with the nuclear envelope). As a consequence, the DNA-binding domain of Gal4 is in the nucleus but is anchored to the nuclear membrane. In this case it is thought that silencing occurs because the HMR locus, with Gal4 sites at the E silencer, is drawn to the periphery of the nucleus where there is a high concentration of Sir proteins (1).
Circular autonomously replicating sequence (ARS) plasmids that lack a mechanism for mitotic segregation are preferentially retained in mother cells, resulting in the generation of plasmid-free daughters. In the absence of selection, these plasmids are lost from logarithmically growing cultures (30). In contrast, ARS plasmids that contain embedded telomeric sequences or the HMR E silencer are segregated efficiently between dividing cells and are stably propagated (8, 19, 23, 24). Plasmid segregation mediated by the HMR E silencer requires the Sir proteins, and segregation mediated by telomeric sequences is improved by the silencing factors. Previously, we showed that tethering a specific domain of Sir4, the so-called partitioning and anchoring domain (PAD4), directly to ARS plasmids also confers efficient mitotic segregation (2). Using a DNA-topology assay that measures axial rotation of intracellular DNA segments, it was shown that the tethered PAD4 domain of Sir4 also immobilizes the DNA to which it is bound (2). The partitioning and DNA immobilization data suggested a model in which the Sir4 domain attaches to a nuclear component, such as a chromosome or the nuclear membrane, that divides symmetrically between cells at mitosis.
Here we describe the results of a screen for factors that, when tethered to a telomere, can reestablish silencing at a telomere in a rap1_Δ_c mutant defective in silencing. In this screen we identified three known Sir-interacting proteins, Rap1, Sir1, and Rad7, as well as a novel protein, Esc1, which we show interacts with the PAD4 domain of Sir4. We further show that Esc1 is located at the nuclear periphery and is essential for the partitioning and anchoring of plasmids by the PAD4 domain of Sir4. The results suggest that Esc1 helps recruit Sir4 to the nuclear periphery.
MATERIALS AND METHODS
Yeast strains.
The strains used in this study are listed in Table 1. Deletion of ESC1 with kanMX4 was achieved by generating a targeting construct by PCR with oligonucleotides directly upstream and downstream of the ESC1 open reading frame (ORF) and with genomic DNA isolated from strain 20805 (Δ_esc1_::kanMX4) as a template (Research Genetics, Inc.). Δ_esc1_::his5+ deletions were generated by PCR-mediated targeted disruption by use of a plasmid containing the Schizosaccharomyces pombe his5+ gene (N. Dean, Stony Brook, N.Y.); this gene complements the S. cerevisiae his3 mutation. The Esc1-green fluorescent protein (GFP)-expressing strain YDZ49, strain YDZ239, and various mutant derivatives of YDZ225 were all generated using previously described plasmids and methods (25). The GAL1 promoter and the coding sequence for GFP replaced 46 nucleotides upstream of the ESC1 ORF plus the codon for the initiating methionine. W303-1a/0, YDZ13/0, and THC18/0 were derived from W303-1a, YDZ13, and THC18, respectively, by curing the strains of the 2μm plasmid using targeted DNA damage (39). The endogenous 2μm plasmid must be eliminated to study the partitioning of telomere-derived sequences that are situated on 2μm-based plasmids.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| W303-1a | MATaleu2-3112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100 | R. Rothstein |
| W303-1b | _MAT_α version of W303-1a | R. Rothstein |
| W303 | Diploid from W303-1a × W303-1b | This study |
| YDS631 | W303-1b adh4::_URA3_-(C1-3A)n | 6 |
| YDS634 | W303-1b adh4::_URA3-4xUASg_-(C1-3A)n | 6 |
| Lev8 | YDS634 rap1::LEU2 (p_RAP1-SUP4-O_) | 26 |
| L40 | MATahis3_Δ_200 trp1-901 leu2-3, 112 ade2 URA3::_(lexAop)8-lacZ LYS2::(lexAop)4_-HIS3 | 18 |
| YSB1 | W303-1b aeB::hmr::TRP1 gal4::LEU2 | 6 |
| YSB2 | YSB1 aeB::3xUASG::hmr::TRP1 | 6 |
| YSB35 | W303-1b Aeb::3xUASG::hmr::TRP1 gal4::LEU2 | 6 |
| YSB41 | YSB2 aeb::3xUASG::hmr::TRP1 | 6 |
| RS1172 | YSB2 sir1::URA3 | This laboratory |
| RS1042 | YSB2 sir2::URA3 | 6 |
| RS1061 | YSB2 sir3::URA3 | 6 |
| RS1067 | YSB2, except MATasir4::URA3 | 6 |
| YEA76 | YSB1 Aeb::UASG::hmr::URA3 | This study |
| YDZ13 | W303-1a esc1::his5+ | This study |
| YDZ239 | YDZ13 sir4::TRP1 | This study |
| YDZ20 | YDS631 esc1::his5+ | This study |
| YDZ39 | MATa/_MAT_α his3/his3 leu2/leu2 ura3/ura3 met15? lys2? | This study |
| YDZ38 | YDZ39 esc1::kanMX4/esc1::kanMX4 | This study |
| YDZ49 | W303 ESC1/GALp-GFP-ESC1::his5+ | This study |
| THC1h | MATahmr::URA3 Δ_his3 ade2-1 can1-100 leu2-3,112 trp1-1 ura3-1_ | 2 |
| YDZ61 | THC1h esc1::his5+ | This study |
| THC18 | W303-1a Δ_lys2_::rKWD5ON sir4::HIS3 | This study |
| RS927 | W303-1a hm1::TRP1 | 21 |
| YDZ62 | RS927 esc1::his5+ | This study |
| YDZ69 | YSB35 esc1::kanMX4 | This study |
| MRG6 | MATatop2-4 top1::GAL1::R his3_-Δ_200 leu2_-Δ_1 trp1_-Δ_63 ura3-52 | 2 |
| AA30 | MRG6 esc1::his5+ | This study |
| 20805 | esc1::kanMX4 | Research Genetics, Inc. |
| YDZ225 | W303-1b adh4::_URA3_-(C1-3A)n | This study |
| YDZ227 | W303-1b adh4::URA3_-(C1-3A)n_esc1::kanMX4 | This study |
| YDZ236 | W303-1b adh4::URA3_-(C1-3A)n_mlp1::his5+ | This study |
| YDZ231 | W303-1b adh4::URA3_-(C1-3A)n_mlp2::his5+ | This study |
| YDZ233 | W303-1b adh4::URA3_-(C1-3A)n_esc1::kanMX4 mlp1::his5+ | This study |
| YDZ228 | W303-1b adh4::URA3_-(C1-3A)n_esc1::kanMX4 mlp2::HIS3 | This study |
| YDZ238 | W303-1b adh4::URA3_-(C1-3A)n_mlp1::kanMX6 | This study |
| YDZ237 | W303-1b adh4::URA3_-(C1-3A)n_esc1::kanMX4 mlp1::TRP1 mlp2::HIS3 | This study |
Telomere one-hybrid screen.
The one-hybrid telomere silencing screen was performed in strain Lev8. Lev8 harbors a URA3 reporter gene with UASG sites near the left arm of chromosome VII-L; it has a RAP1 deletion on the chromosome and is kept alive by a wild-type RAP1 gene on a plasmid (26). By plasmid shuffling, the wild-type RAP1 was replaced by a C-terminal truncation allele, _rap1_Δ_670_-807, on plasmid sp18. Cells containing this rap1 allele are completely derepressed for telomeric silencing and grow slowly (26). Lev8/sp18 cells were transformed with a GBD yeast library in the vector pGBT-CYH (_CEN TRP1 CYH2 ADHp_-GBD), a gift of C. Evangelista and S. Fields, University of Washington. Transformants were selected on medium lacking tryptophan. Colonies were grown at 30°C for 4 to 5 days before being replica plated on 5-fluoroorotic acid (5-FOA) to select cells exhibiting silencing of URA3. Candidates were streaked to single colonies three times on 5-FOA plates. DNA was then isolated from these 5-FOA-resistant colonies, electroporated into Escherichia coli, and analyzed by restriction enzyme digestion. Unique GBD plasmids were retransformed into Lev8 to identify those that restored silencing. These library plasmids were subjected to sequencing.
Plasmids.
Library plasmids L4.1, L5.2, and L18.6, from the telomere one-hybrid screen described above, all encode identical GBD-Rap1 hybrids. L5.4 and L19.4 encode GBD-Sir1 and GBD-Rad7, respectively (Table 2). Library plasmid L2.4 encodes GBD-Esc1(1124-1658).
TABLE 2.
Genes identified in one-hybrid telomeric silencing screen
| GBD hybrid | Amino acids in hybrida | Comments (reference) |
|---|---|---|
| GBD-Rap1 | 354-827 | Binds to Sir3 and Sir4 (28, 29) |
| GBD-Sir1 | 163-678 | Binds to Sir4 (38) |
| GBD-Rad7 | 28-565 | Binds to Sir3 (31) |
| GBD-Esc1 | 1,124-1,658 | Binds to Sir4 (this study) |
The ESC1 insert from plasmid L2.4 was excised with _Sma_I and _Pst_I, and the fragment was subcloned into the LexA vector pBTM116-Kn-ADE2 (2μm _TRP1 ADE2 ADHp_-LexA) to create pEDA114 and into the GAD vector pGAD424 (2μm _LEU2 ADHp_-GAD) to create pEDA115. To create LexA-Esc1(1395-1551) and GAD-Esc1(1395-1551), a _Sma_I-_Pst_I fragment was cloned (from a library plasmid isolated in a similar screen to be reported elsewhere) into pBTM116-Kn-ADE2 (pEDA157) and pGAD424 (pEDA159). GBD-Esc1(1200-1448) (pEDA167) was created as follows: pEDA115 was digested with _Bam_HI and _Bgl_II and the ESC1 fragment was cloned into the _Bam_HI site of pBluescript II KS(+) to create pEDA124. pEDA124 was digested with _Nsi_I and _Sal_I and the fragment was subcloned into pKSII+ (pEDA160). A _Bam_HI-_Xho_I fragment from pEDA160 was then subcloned into pTT63, a pGBT9 derivative (2μm HIS3 ADHp-GBD) (4, 38) to make pEDA167.
Full-length ESC1 was cloned as follows: pEDA115 was digested with _Bam_HI and _Hin_dIII to liberate the entire 3′ coding region of Esc1 and was subcloned into pRS306 (35) to make pEDA122. pEDA122 was digested at a unique _Eco_RI site within the ESC1 coding sequence, and the linear DNA was transformed into yeast. Genomic DNA was isolated from Ura+ transformants, digested with _Eag_I, incubated with T4 DNA ligase, and electroporated into E. coli strain MH3. The presence of an insert containing the entire Esc1 coding region (pEDA128) was verified by restriction enzyme analysis. A 7.0-kb _Bst_XI-_Xho_I fragment containing the full-length ORF was isolated from pEDA128 and subcloned into pKSII+ to generate pEDA129. A yeast shuttle vector containing the ESC1 gene was constructed by excising a _Sac_II-_Xho_I fragment from pEDA129 and ligating it into pRS315 (CEN/ARS LEU2) (35) cut with _Sac_II and _Sal_I to make pDZ45.
The construction of the following GAD-Sir4 constructs has been described previously: GAD-Sir4(1-1358), pCTC79; GAD-Sir4(839-1358), pCTC18; GAD-Sir4(1262-1358), pCTC24; and GAD-Sir4(839-1149), pCTC50 (6, 38). LexA-Sir4 plasmids used in two-hybrid experiments have been described previously (2). GAD-Sir4(934-1358) is clone C1-5.1, isolated in the two-hybrid screen with LexA-Esc1(1124-1658).
Plasmids LS4 (Sir4 950-1262), pAA6, and pKWD200 were described previously (2). Additional details are provided here. The 950- to 1,262-amino-acid fragment of Sir4 in LS4 terminates with a serine rather than the cysteine normally present in the Sir4 protein at position 1262. pKWD200 and pAA6 contain six copies of an oligonucleotide that has two LexA operators. Thus, the plasmids contain 12 LexA sites each. In pAA6, the _Xba_I site within the 2μm episome-derived portion of the plasmid was destroyed, thereby eliminating the Flp recognition target and errors in partitioning measurements due to interplasmid recombination with the endogenous 2μm episome (40).
Two-hybrid screens.
Two-hybrid experiments were performed in strain L40, which contains LexA operators upstream of the HIS3 and lacZ reporter genes (18). For screening with LexA-Esc1(1124-1658), cells were cotransformed with both the LexA hybrid and a yeast GAD library. For the screen with LexA-Sir4(950-1262), L40 was transformed sequentially, first with plasmid LS4(950-1262) (2) and then with the yeast GAD library. Library clones that promoted sufficient HIS3 expression for growth on SC-LEU-TRP-HIS plates (containing 5 mM 3-aminotriazole for the LexA-Sir4 screen) were subsequently tested for β-galactosidase expression. His+ β-galactosidase+ transformants were characterized as described previously to eliminate false positives (33).
Silencing assays.
Strains THC1h and YDZ61 were used to measure silencing of HMR. Strains RS927 and YDZ62 were used to measure silencing of HML. Strains YDS631 and YDZ20 were used to measure telomeric silencing. Cells were grown to saturation at 30°C in medium selecting for the GBD plasmid or, if no plasmid was involved, in yeast extract-peptone-dextrose medium. Cells were then serially diluted 10-fold and 10 μl of these dilutions was spotted onto plates containing appropriate media. For experiments with _URA3_-based reporter strains, cells were also spotted onto medium containing 0.1% 5-FOA (Angus Biochemicals, Niagara Falls, N.Y.).
Localization of GFP-Esc1.
For confocal microscopy used to detect GFP-Esc1, strain YDZ49 was grown overnight in galactose medium supplemented with adenine to reduce vacuolar autofluorescence and then centrifuged at low speed and fixed in 95% ethanol at 30°C for several hours. Cells were then washed and resuspended in SHA buffer (1 M sorbitol, 0.1 M Na-HEPES [pH 7.5], 5 mM NaN3). Spheroplasts were prepared by the addition of zymolyase and β-mercaptoethanol and the cell wall digestion was stopped by washing in SHA buffer. Spheroplasted cells were resuspended in SHA buffer and allowed to settle in the eight wells of a polylysine-coated slide. The overlaying buffer was aspirated and the cells were treated with 1 mg of RNase A per ml for 30 min at 37°C. Cells were then washed with 2 drops of SHA buffer 5 to 10 times. Propidium iodide (1 μg/ml) in SHA buffer was added to the digested cells and incubated at room temperature for 10 min. Cells were then washed 10 times with SHA buffer and preserved in mounting medium under a sealed coverslip.
Immunofluorescence.
Strains YDZ38 and YDZ39 were used for immunofluorescence, using previously described methods and antibodies (13, 14).
Plasmid stability and anchoring assays.
Plasmid stability tests and anchoring experiments were carried out as previously described (2).
RESULTS
A screen for proteins capable of restoring silencing at a telomere identifies Esc1.
A one-hybrid screen was developed to look for new proteins involved in silencing. The screen was performed in strain Lev8, which has Gal4 DNA-binding sites (UASG) and a URA3 reporter gene placed near a telomere (Fig. 1). The strain also harbors a deletion in RAP1 (Δ670-807) that removes the Sir4 interaction domain (26). Without this domain, telomeric silencing is abolished. Lev8 was transformed with a GBD hybrid plasmid library and screened for GBD fusion proteins that restored silencing of the telomeric URA3. Repression of the gene was monitored by growth on 5-FOA.
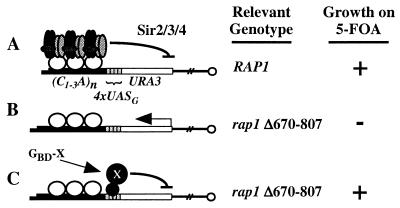
FIG. 1.
Telomeric targeted silencing screen. A telomere with a URA3 reporter gene and four adjacent Gal4 binding sites is depicted. (A) The RAP1 strain encodes a wild-type Rap1 protein that promotes the formation and spreading of a Sir2-Sir3-Sir4 complex that silences the URA3 gene. (B) The rap1 strain encodes a Rap1 protein with a deletion of amino acids 670 to 807 and hence cannot bind Sir3 and Sir4. Thus, the URA3 gene is not silenced. (C) The strain is the same as in panel B except that a GBD hybrid binds to the Gal4 binding sites and causes silencing of the URA3 gene.
Three of the four different proteins identified in this screen had been studied previously (Table 2). One of them was Rap1. Three different plasmid isolates encoded a fragment of Rap1 from amino acid 354 to the end of the protein. This portion of Rap1 includes the Sir4 interaction domain that was deleted from strain Lev8 (26) and thus was not unexpected. Also identified in the screen was Sir1, a protein known to play an essential role in the establishment of silencing at the HM loci and also shown previously to be capable of targeted silencing via an interaction with Sir4 (6, 38). Although Sir1 is dispensable for normal telomeric silencing, Sir1 enhances telomeric silencing when targeted to telomeric DNA (6). The third protein identified in the screen was Rad7, a protein implicated in the repair of DNA damage. Rad7 has been shown to interact with Sir3 (31). Thus, it is likely that the GBD-Rad7 hybrid causes silencing by recruiting Sir3 to the telomere.
The fourth clone identified in this screen was a GBD hybrid to residues 1124 to 1658 of a previously uncharacterized yeast ORF, YMR219w, which we have named ESC1 (establishes silent chromatin). ESC1 encodes a large protein containing 1,658 amino acids. The protein is highly acidic, contains a P-loop (purine-binding motif), many predicted coiled-coils, and a putative nuclear localization sequence at the C terminus.
Targeted silencing by GBD-Esc1.
Targeted silencing assays with Esc1 are shown in Fig. 2. Three different GBD-Esc1 hybrids were used: the one found in the screen described above, a second constructed by us, and a third found in an HMR targeted silencing screen to be described elsewhere. The GBD-Esc1 hybrids gave targeted silencing, not only at telomeres, but also at various HMR alleles containing HMR E silencer deletions. The silencing required Sir2, -3, and -4 and Gal4 binding sites (UASG) (Fig. 2C), as was expected from earlier studies of targeted silencing (6). Subcloning of Esc1 demonstrated that a 54-amino-acid region from near the carboxy-terminal end of the protein (residues 1395 to 1448) was sufficient for transcriptional repression (data not shown).
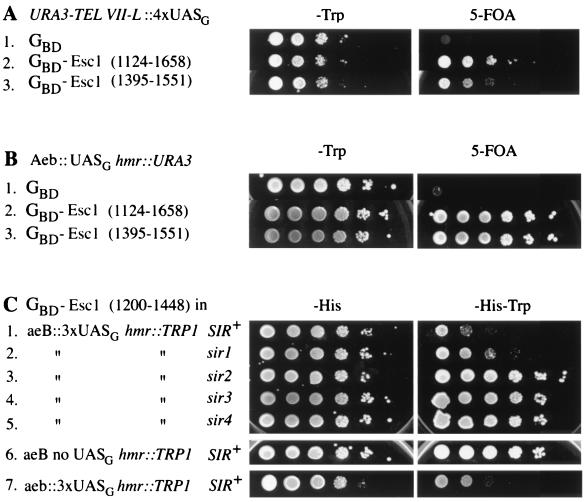
FIG. 2.
Targeted silencing by GBD-Esc1 at a telomere and at HMR. (A) Two different GBD-Esc1 hybrids silence the URA3 gene in the telomere reporter strain YDS634. Tenfold serial dilutions were plated on −Trp medium to indicate the number of cells plated and on 5-FOA medium to measure the extent of silencing. (B) GBD-Esc1 hybrids silence URA3 at an HMR locus in which the HMR E silencer has the Rap1 and Abf1 binding sites replaced by a Gal4 binding site (Aeb::UASG; strain YEA76). (C) Targeted silencing by GBD-Esc1 at an HMR locus with a TRP1 reporter gene (strain YSB2) is Sir dependent. Sir+ cells harboring GBD-Esc1(1200-1448) (row 1) grow poorly on −Trp medium due to silencing of the reporter gene, while sir mutants (rows 2 to 5) grow better. Targeted silencing by GBD-Esc1 is also dependent upon the presence of UASG sites (strain YSB1; row 6). In the absence of any silencer element at HMR E (strain YSB41), Esc1 is still capable of targeted silencing (row 7).
esc1 mutant.
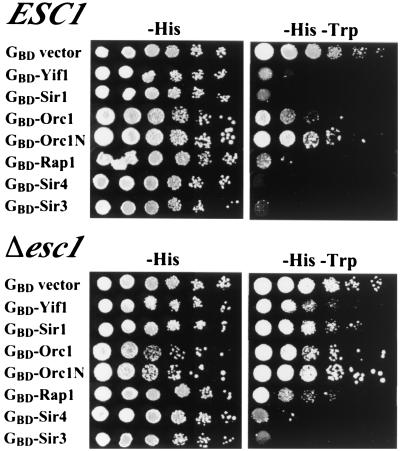
A strain with a precise deletion of the ESC1 open reading frame was constructed in a diploid. Sporulation and dissection of the heterozygous diploid yielded Δ_esc1_ haploid strains that grew normally at all temperatures tested. Strains with the Δ_esc1_ mutation were tested for silencing defects at HML, HMR, telomeres, and the ribosomal DNA (rDNA) locus. No silencing defects were observed at the HM loci or the rDNA (data not shown). A slight decrease in telomeric silencing was seen, as discussed below (see Fig. 8). In addition, the Δ_esc1_ mutant exhibited significant defects in targeted silencing. For this set of experiments, various silencing proteins were tethered to a mutated HMR E silencer via GBD and tested for the ability to silence a nearby reporter gene. The proteins examined included Rap1, Orc1, Sir1, Sir3, and Sir4. Also tested was a membrane protein, Yif1, previously shown to give efficient targeted silencing because it tethers HMR to the nuclear periphery (1). A comparison of targeted silencing by these various proteins in ESC1 versus Δ_esc1_ strains is shown in Fig. 3. Deletion of ESC1 nearly eliminates silencing by the membrane protein, Yif1, and greatly weakens silencing by Sir1, Orc1 (full length or the N-terminal fragment known to bind Sir1 [38]), and Rap1. Silencing by Sir4 is also somewhat weakened by the esc1 mutation. Introduction of a CEN plasmid containing the ESC1 gene, pDZ45, into the esc1 mutant restored silencing by these proteins (data not shown). Silencing by only one hybrid protein, GBD-Sir3, was unaffected by deleting esc1. This result will be considered further (see Discussion).
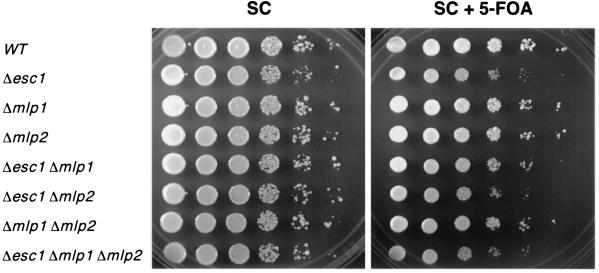
FIG. 8.
Telomeric silencing in esc1, mlp1, and mlp2 mutants. Tenfold serial dilutions are shown for each strain on complete medium (SC) and on 5-FOA medium. Growth on the latter medium indicates silencing of the URA3 reporter gene. WT, wild type.
FIG. 3.
Comparison of targeted silencing in ESC1 and Δ_esc1_ strains YSB35 and YDZ69. A TRP1 reporter gene was used and thus lack of growth on −Trp medium indicates good silencing. Note that the esc1 mutation affects silencing by all the GBD hybrids except GBD-Sir3.
Esc1 interacts with Sir4.
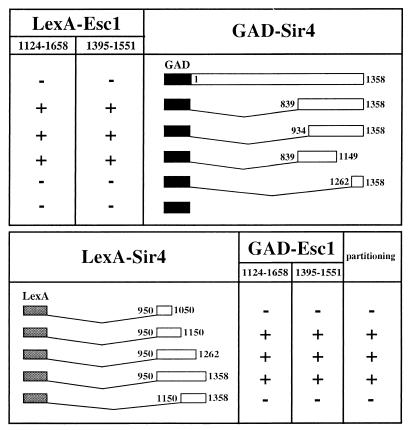
Since Esc1 had targeted silencing activity, it seemed likely that it was recruiting a known silencing protein. To identify Esc1-interacting proteins, a two-hybrid screen was undertaken using the domain of Esc1 recovered in the targeted silencing screen (amino acids 1124 to 1658) as a bait. A C-terminal fragment of Sir4 (amino acids 934 to 1358) was found as a specific interacting partner. The two-hybrid system was used to delineate the domains required for the Esc1-Sir4 interaction. This directed approach showed that GAD hybrids containing amino acids 839 to 1149 of Sir4 were sufficient for interaction with LexA-Esc1 (amino acids 1124 to 1658), whereas a GAD hybrid containing the extreme C-terminal lamin-like heptad repeats of Sir4 (amino acids 1262 to 1358) was not (Fig. 4). An identical Sir4 interaction profile was observed using a smaller Esc1 hybrid, LexA-Esc1 (amino acids 1395 to 1551), indicating that this 157-amino-acid domain of Esc1 contained a Sir4-interacting motif. Neither Esc1 hybrid interacted with full-length GAD-Sir4.
FIG. 4.
Summary of two-hybrid data showing the interaction between Esc1 and Sir4. +, significant two-hybrid interaction in strain L40; −, no interaction. The various LexA-Sir4 hybrids were also tested for improved partitioning of plasmid pAA6 as described previously (2). Symbols for partitioning column: +, improved partitioning; −, no improvement.
The region of Sir4 found to interact with Esc1 coincided with a previously described domain of Sir4, the partitioning and anchoring domain (PAD4). This Sir4 domain, when bound to plasmids via LexA, greatly increases mitotic stability of plasmids, and furthermore, appears to anchor them as judged by a topological assay (2). Interestingly, a two-hybrid search using the PAD4 domain of Sir4 (amino acids 950 to 1262) as bait identified four different Esc1 fragments. Three of them encompassed the C terminus of Esc1, from amino acids 944, 1013, and 1227 to the end of the protein. The fourth PAD4-interacting clone was a small Esc1 fragment spanning amino acids 1440 to 1473, indicating that this 34-amino-acid fragment was sufficient for interaction with Sir4.
Further evidence that the PAD4 domain of Sir4 coincided with the Esc1-interacting domain is shown in Fig. 4. Five different LexA hybrids containing different regions of Sir4, encompassing part or all of the operationally defined PAD4 domain, were tested against two GAD-Esc1 hybrids in the two-hybrid system. A perfect correlation could be seen between those LexA-Sir4 hybrids that interacted with Esc1 and those that were positive in the plasmid partitioning assay (2), which we describe below.
Plasmid partitioning by targeted Sir4 requires Esc1.
An association between Esc1 and the PAD4 domain of Sir4 suggested that Esc1 might be required for Sir4-mediated partitioning of plasmids. To test this possibility, strain W303-1a and the isogenic Δ_esc1_ mutant were cotransformed with a LexA-PAD4 expression vector and an unstable circular ARS plasmid, pAA6, that contains 12 LexA binding sites. Partitioning of pAA6 was evaluated by measuring the mitotic stability and the loss rate of the plasmid. The former parameter corresponds to the fraction of cells that contain pAA6 under selective growth conditions whereas the latter corresponds to the rate of pAA6 loss during nonselective growth conditions. In agreement with earlier results, partitioning of pAA6 in W303-1a was enhanced by the expression of LexA-PAD4 but not by LexA alone (2). Strikingly, PAD4-mediated partitioning of pAA6 was abolished in the Δ_esc1_ strain. Mitotic stability dropped nearly fivefold and plasmid loss rate increased more than sixfold (Table 3). Introduction of an ESC1 plasmid, pDZ45, into the esc1 mutant restored PAD4-mediated partitioning (data not shown). These results indicate that ESC1 is required for PAD4-mediated plasmid stabilization.
TABLE 3.
Partitioning of plasmid pAA6 by a LexA-Sir4p hybrid requires ESC1
| LexA hybrid | Strain | Relevant genotype | % Mitotic stabilitya | Plasmid loss ratea |
|---|---|---|---|---|
| LS4 (950-1262) | W303-1a | ESC1 | 57.0 ± 5.0 | 0.08 ± 0.03 |
| LS4 (950-1262) | YDZ13 | Δ_esc1_::his5+ | 12.0 ± 3.0 | 0.51 ± 0.02 |
| LexA only | W303-1a | ESC1 | 20.2 ± 2.1 | 0.39 ± 0.10 |
One explanation for the partitioning defect in the Δ_esc1_ strain is that the gene plays a role in DNA replication. This possibility is not supported by the finding that a CEN/ARS plasmid exhibited similar mitotic stability and plasmid loss rate in both the ESC1 and Δ_esc1_ strains (Table 4). Decreased plasmid persistence, therefore, does not result from an inability to replicate plasmids properly. An alternative explanation for the partitioning defect is that ESC1 is required for the LexA chimera to bind DNA. In a Δ_esc1_ strain, however, LexA-PAD4 can bind the promoter of a lacZ reporter gene and block activation by an upstream UASG, indicating that expression, nuclear transport, and DNA binding of LexA-PAD4 can occur in the absence of ESC1 (data not shown).
TABLE 4.
CEN/ARS plasmid partitioning does not require ESC1a
| Strain | Relevant genotype | % Mitotic stabilityb | Plasmid loss rateb |
|---|---|---|---|
| W303-1A | ESC1 | 84.8 ± 2.4 | 0.08 ± 0.01 |
| YDZ13 | Δ_esc1_::his5+ | 96.3 ± 2.4 | 0.09 ± 0.02 |
Sir4-mediated partitioning of telomere-based plasmids requires Esc1.
The finding that Esc1 promotes plasmid partitioning by artificially tethered Sir4 suggested that ESC1 might also contribute to segregation of plasmids that recruit Sir4 naturally. We therefore measured the role of ESC1 in segregating two circular plasmids that possess telomeric sequences. The first plasmid, pYET1, contains a telomeric TG1-3 tract and a subtelomeric X repeat (sometimes referred to as an X-TAS, for X-telomere-associated sequence), whereas the second plasmid, pETACC, contains only the TG1-3 tract (24). Previous work showed that both plasmids partitioned well. However, the X repeat of pYET1 provided additional stabilization and this added effect required Sir2, Sir3, and Sir4 (8, 23). Accordingly, deletion of SIR4 from the strain used here increased the pYET1 loss rate from 0.10 to 0.15 (Table 5). Deletion of ESC1 resulted in a comparable increase in the pYET1 loss rate, suggesting that ESC1 is required for the enhanced partitioning of telomeric sequences by Sir4. The esc1 sir4 double mutant had the same loss rate as each single mutant, showing that both genes act in the same pathway. The loss rate of pETACC, on the other hand, was not changed significantly by deletion of either SIR4 or ESC1. The combined evidence supports a model in which telomeric sequences recruit Sir4 as part of the Sir silencing complex and then Sir4 in turn associates with Esc1 to confer partitioning.
TABLE 5.
ESC1 and SIR4 stabilize telomeric plasmids containing a subtelomeric X repeat
| Telomere-based plasmid | Straina | Relevant genotype | Plasmid loss rateb |
|---|---|---|---|
| pYET1 | W303-1a/0 | Wild type | 0.10 ± 0.008 |
| YDZ13/0 | Δ_esc1_::his5+ | 0.17 ± 0.013 | |
| THC18/0 | Δ_sir4_::HIS3 | 0.15 ± 0.013 | |
| YDZ239/0 | Δ_esc1_-Δ_sir4_ | 0.15 ± 0.005 | |
| pETACC | W303-1a/0 | Wild type | 0.20 ± 0.036 |
| YDZ13/0 | Δ_esc1_::his5+ | 0.28 ± 0.06 | |
| THC18/0 | Δ_sir4_::HIS3 | 0.25 ± 0.086 | |
| YDZ239/0 | Δ_esc1_-Δ_sir4_ | 0.28 ± 0.057 |
Anchoring of DNA by targeted Sir4 requires Esc1.
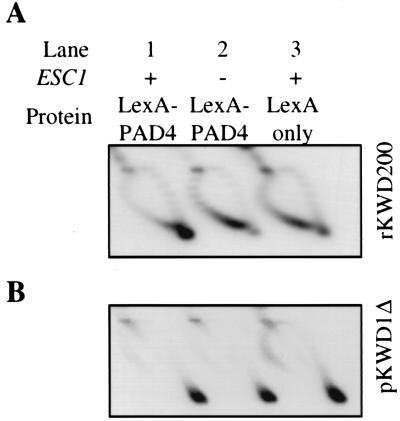
The domain of Sir4 that confers plasmid partitioning also hinders the axial rotation (swiveling) of DNA fragments to which it is bound (2). A role for ESC1 in this form of DNA immobilization, termed DNA anchoring, was tested with a DNA-topology-based assay (for a detailed description, see reference 2). Briefly, twin domains of positive and negative supercoils are generated in a DNA ring formed by site-specific recombination. Proteins that block DNA rotation prevent cancellation of the two domains. In top1 top2 yeast mutants expressing the negative supercoil-specific E. coli topoisomerase I, selective relaxation of negative supercoils results in positive supercoil accumulation. Under these conditions, DNA rings containing LexA sites became highly positively supercoiled when LexA-PAD4 was expressed (2) (Fig. 5A, lane 1). The result indicates that the LexA-PAD4-DNA complex is anchored in vivo. Substantial positive supercoiling did not occur, however, if the experiment was performed in an isogenic Δ_esc1_ strain. The level of supercoiling in this mutant was comparable to the level in an ESC1 strain that expressed LexA alone (Fig. 5A, lanes 2 and 3). These data demonstrate that ESC1 is required for Sir4-mediated DNA anchoring.
FIG. 5.
DNA anchoring by targeted Sir4 requires ESC1. Assays were performed in strains MRG6 (ESC1) and AA30 (Δ_esc1_) transformed with an E. coli topoisomerase I expression plasmid [YEp-topA(PGPD)], a LexA-PAD4 expression plasmid [LS4(950-1262)], and an excision substrate vector (pKWD200) that produces a ring (rKWD200) with 12 LexA sites. (A) Analysis of ring rWKD200 supercoiling by two-dimensional gel electrophoresis in buffer containing chloroquine. DNA topoisomers were resolved into an arc with positively supercoiled rings coalescing in a spot at the extreme clockwise end (lane 1) and negatively supercoiled rings occupying a broad region at the counterclockwise end (lane 2). The minor population of positive supercoils seen in lanes 2 and 3 appears occasionally with rings that lack DNA anchors. (B) Analysis of plasmid pKWD1Δ supercoiling. rKWD200 and pKWD1Δ were visualized sequentially by hybridization with randomly primed radiolabeled probes.
To test whether ESC1 plays a general role in DNA anchoring, the blot in Fig. 5A was stripped and rehybridized with a probe to a second plasmid that contains the partitioning locus of the 2μm plasmid, REP3, which was previously shown to form a DNA anchor (12). The plasmid, pKWD1Δ, accumulates positive supercoils efficiently in both the ESC1 and Δ_esc1_ strains, irrespective of whether LexA or LexA-PAD4 is expressed (Fig. 5B). Thus, the role for ESC1 in the DNA anchoring assay is specific for the anchor formed by LexA-PAD4.
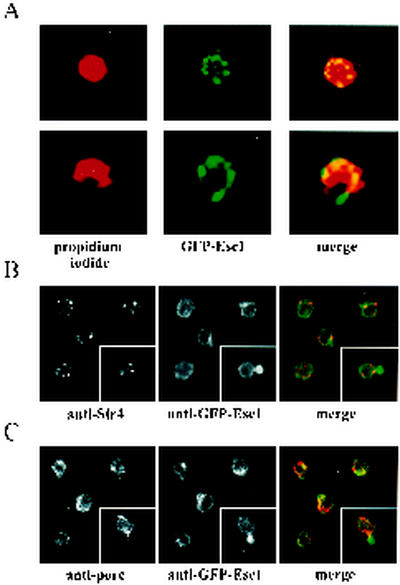
Subcellular localization of Esc1.
To identify the subcellular location of Esc1, a strain was generated that expresses a GFP-Esc1 fusion protein under control of the GAL1 promoter (25). This construct complemented a Δ_esc1_ mutant in the partitioning assay described above (data not shown). Confocal microscopy of galactose-induced cells showed that the expressed GFP fusion protein localized to the periphery of the nucleus in a punctate pattern (Fig. 6A). Our finding that Esc1 and Sir4 interact prompted us to examine the relative locations of the two proteins. Indirect immunofluorescence showed that the two proteins colocalized at many positions along the nuclear periphery (Fig. 6B). GFP-Esc1, however, was also found at other peripheral positions that lacked Sir4. Colocalization studies with the nucleolar marker Nop1 demonstrated that Esc1 did not localize to the nucleolus (data not shown).
FIG. 6.
Esc1 localizes to the nuclear periphery. (A) Confocal images of GFP-Esc1 and propidium iodide fluorescence; the latter stains the nuclear DNA. (B) Esc1 and Sir4 are both at the nuclear periphery but they do not colocalize. Both proteins were visualized by immunofluorescence. (C) Esc1 does not colocalize with nuclear pores. The insets in panels B and C show examples of cells with very strong GFP-Esc1 fluorescence at the bud neck. This was seen for 5 to 10% of cells.
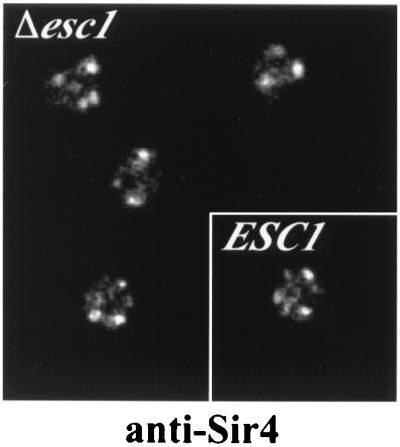
Previously Sir4 and other telomere binding proteins were shown to colocalize with telomeres in discrete perinuclear foci (13, 32). Disruption of a single telomere binding factor often leads to dispersion of others and sometimes leads to unclustering of the telomeric DNA sequences (reviewed in reference 7). Indirect immunofluorescence of Sir4, however, showed that telomeric foci were not disrupted in a Δ_esc1_ strain (Fig. 7).
FIG. 7.
Telomere clustering, visualized with an antibody to Sir4, is not altered in Δ_esc1_ mutants. For comparison, a wild-type ESC1 strain is shown in the inset.
GFP-Esc1 localization somewhat resembled the distribution of nucleoporins. The relative distribution of these two entities was examined using an antibody to nuclear pore proteins (3). Esc1 and nuclear pores were only occasionally found to be coincident (Fig. 6C). Thus, Esc1 does not appear to be a component of the nuclear pore complex. This is consistent with the result that Esc1 was not found in a comprehensive study of yeast nuclear pore proteins (34).
Interestingly, in cells in which GFP-Esc1 was most highly expressed, the Esc1 signal was also detected in a single projection from the nucleus (Fig. 6B and C, inset panels). This projection did not appear to contain Sir4, although nuclear pores could be detected at the base of the projection, suggesting that the projection may be an extension of the nuclear membrane.
Telomeric silencing in esc1 and mlp1 and -2 mutants.
Two related yeast proteins, called Mlp1 and Mlp2 (myosin-like protein), have been shown to localize to the nuclear periphery and bind to nuclear pore proteins (36). An mlp1 mlp2 double mutant has been reported to have telomeric silencing defects (9, 10). Since Esc1, Mlp1, and Mlp2 are all large coiled-coil proteins that localize to the nuclear periphery, we wondered whether they played overlapping roles in silencing. We therefore constructed all combinations of single, double, and triple mutants and tested them for telomeric silencing. As mentioned above, esc1 single mutants have a slight but reproducible silencing defect (Fig. 8). To our surprise, mlp1 and mlp2 single and double mutants show no silencing defect and do not exacerbate the esc1 silencing defect (Fig. 8). However, we did observe an unusual colony morphology for mlp1 mlp2 double mutants, just as had been reported previously (10). This confirms that we had indeed mutated the MLP1 and MLP2 genes. We have no explanation for why our silencing results differ from those previously published for the mlp1 mlp2 mutant (9, 10). It seems unlikely that strain differences could account for such a drastic difference.
DISCUSSION
Esc1 was identified in a screen for proteins that confer silencing when targeted to silencing-defective loci. The protein was shown to silence both a derepressed telomere and a mutated HMR silencer when tethered to these sites (Fig. 2). Paradoxically, strains lacking Esc1 have no obvious silencing defects at the native HM loci or rDNA and only minor defects at telomeres (Fig. 8). Nevertheless, several lines of evidence suggest that the protein plays a role in Sir-mediated repression; silencing by targeted Esc1 requires the other Sir proteins (Fig. 2), and Esc1 is required for silencing by other targeted proteins (Fig. 3). Esc1 also interacts with Sir4 (Fig. 4). This interaction may explain why the protein was recovered in the targeted silencing screen: a tethered fragment of Esc1 complexes with Sir4, which in turn associates with Sir2 and Sir3 to nucleate silent chromatin. While the genetic evidence presented here makes it likely that Esc1 binds directly to Sir4, we have no in vitro evidence to support this. It is conceivable that the interaction is bridged by another yeast protein.
Esc1 localizes to the nuclear periphery (Fig. 6) and a potential role for the protein in silencing may be to tether silent chromatin complexes to this subnuclear compartment. Though Sir proteins are limiting in the nucleus, the factors concentrate at telomeres, which cluster at the nuclear periphery (7). Silencing of the mating-type loci is thought to benefit from their proximity to telomeres and the associated pools of Sir proteins. Indeed, tethering a defective silencer to the telomere-rich nuclear periphery imparts Sir-dependent transcriptional repression (1). Thus, Esc1 may function in silencing because it colocalizes the silent mating-type loci with perinuclear pools of Sir proteins.
A role for Esc1 as a localization factor is supported by the finding that Esc1 influences the intracellular behavior of Sir4. We show here that partitioning and anchoring of plasmids by a tethered fragment of Sir4 requires Esc1 (Table 3; Fig. 5). Moreover, Esc1 is required for Sir4-enhanced partitioning of a telomere-based plasmid, an extrachromosomal element that recruits Sir4 as part of a silent chromatin complex (Table 5). It seems likely that Esc1 facilitates the segregation process by recruiting Sir4-bound plasmids to the nuclear membrane or some other structure at the nuclear periphery that divides between progeny at mitosis.
Why does Esc1 not play a greater role in silencing at native silent loci? One explanation is that silencing is a process with built-in redundancies. Native silencers, for example, bind redundant combinations of proteins that cooperate to recruit Sir proteins for remarkably efficient silencing. Elimination of one silencer-bound protein is often not sufficient to disrupt silencing. Targeted silencing, on the other hand, is always measured in situations in which some component of silencing has been weakened by mutation. According to this view, some redundant feature of native silencing must be eliminated before the deleterious effects of an esc1 deletion can be observed. Redundancy might not be limited to silencers and the proteins that bind them. There might also be overlapping mechanisms for targeting silent chromatin to the nuclear periphery. The Ku protein and Mlp1 and Mlp2 have been implicated in tethering telomeres to the nuclear periphery and these proteins may contribute to or overlap with Esc1-mediated targeting of silent chromatin (10, 20). But, as shown in Fig. 8, we see no evidence that Mlp1 and Mlp2 play any role in telomeric silencing. Nevertheless, additional factors are likely to contribute to telomere localization since the left telomere of chromosome VII remains at the periphery in Ku and silencing-deficient strains (37).
Why is targeted silencing by GBD-Sir3 ESC1 independent while silencing by all the other hybrid proteins tested is weakened by deleting ESC1 (Fig. 3)? A simple explanation is that tethering Sir3 to a silencer bypasses the requirement for ESC1 in silencing. According to this view, ESC1 acts upstream of Sir3 recruitment. Conversely, targeted silencing by GBD-Sir4 is facilitated by ESC1. Thus, the data indicate that ESC1 plays a role subsequent to Sir4 recruitment.
Localization of Esc1 at the nuclear periphery does not coincide exactly with Sir4 or with nuclear pores, although there is some overlap (Fig. 6). Esc1 is a large protein with predicted coiled-coil domains throughout its length. It has always been unclear why yeasts do not have nuclear lamins of the type found in larger eukaryotes. Perhaps Esc1 plays that role in yeast, along with other coiled-coil proteins. Identification of factors that maintain Esc1 at the periphery may address this question. In summary, we think that Esc1 is a component of the nuclear periphery that helps attract silent chromatin complexes containing Sir4 to the periphery. Of course, it may have other roles yet to be uncovered.
Acknowledgments
E.D.A., D.C.Z., and A.A. contributed equally to this work.
We thank C. Evangelista and S. Fields for the GBD library, J. Berman and D. Gottschling for plasmids, S. Gasser for advice and encouragement, and J. Speh and G. Mandel for help with confocal microscopy.
This work was supported by National Institutes of Health grants GM51402 to M.R.G. and GM28220 to R.S.
REFERENCES
- 1.Andrulis, E. D., A. M. Neiman, D. C. Zappulla, and R. Sternglanz. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394**:**592-595. (Erratum, **395:**525.) [DOI] [PubMed] [Google Scholar]
- 2.Ansari, A., and M. R. Gartenberg. 1997. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol. Cell. Biol. 17**:**7061-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aris, J. P., and G. Blobel. 1989. Yeast nuclear envelope proteins cross react with an antibody against mammalian pore complex proteins. J. Cell Biol. 108**:**2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel, P. L., J. A. Roecklein, D. SenGupta, and S. Fields. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12**:**72-77. [DOI] [PubMed] [Google Scholar]
- 5.Buck, S. W., and D. Shore. 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9**:**370-384. [DOI] [PubMed] [Google Scholar]
- 6.Chien, C. T., S. Buck, R. Sternglanz, and D. Shore. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75**:**531-541. [DOI] [PubMed] [Google Scholar]
- 7.Cockell, M., and S. M. Gasser. 1999. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9**:**199-205. [DOI] [PubMed] [Google Scholar]
- 8.Enomoto, S., M. S. Longtine, and J. Berman. 1994. Enhancement of telomere-plasmid segregation by the X-telomere associated sequence in Saccharomyces cerevisiae involves SIR2, SIR3, SIR4 and ABF1. Genetics 136**:**757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feuerbach, F., V. Galy, E. Trelles-Sticken, M. Fromont-Racine, A. Jacquier, E. Gilson, J. C. Olivo-Marin, H. Scherthan, and U. Nehrbass. 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4**:**214-221. [DOI] [PubMed] [Google Scholar]
- 10.Galy, V., J. C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403**:**108-112. [DOI] [PubMed] [Google Scholar]
- 11.Gartenberg, M. R. 2000. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr. Opin. Microbiol. 3**:**132-137. [DOI] [PubMed] [Google Scholar]
- 12.Gartenberg, M. R., and J. C. Wang. 1993. Identification of barriers to rotation of DNA segments in yeast from the topology of DNA rings excised by an inducible site-specific recombinase. Proc. Natl. Acad. Sci. USA 90**:**10514-10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan, and S. M. Gasser. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134**:**1349-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotta, M., S. Strahl-Bolsinger, H. Renauld, T. Laroche, B. K. Kennedy, M. Grunstein, and S. M. Gasser. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16**:**3243-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunstein, M. 1998. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell 93**:**325-328. [DOI] [PubMed] [Google Scholar]
- 16.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80**:**583-592. [DOI] [PubMed] [Google Scholar]
- 17.Hecht, A., S. Strahl-Bolsinger, and M. Grunstein. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383**:**92-96. [DOI] [PubMed] [Google Scholar]
- 18.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15**:**3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimmerly, W. J., and J. Rine. 1987. Replication and segregation of plasmids containing _cis_-acting regulatory sites of silent mating-type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol. Cell. Biol. 7**:**4225-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde, E. J. Louis, and S. M. Gasser. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8**:**653-656. [DOI] [PubMed] [Google Scholar]
- 21.Le, S., C. Davis, J. B. Konopka, and R. Sternglanz. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13**:**1029-1042. [DOI] [PubMed] [Google Scholar]
- 22.Liu, C., X. Mao, and A. J. Lustig. 1994. Mutational analysis defines a C-terminal tail domain of RAP1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics 138**:**1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longtine, M. S., S. Enomoto, S. L. Finstad, and J. Berman. 1993. Telomere-mediated plasmid segregation in Saccharomyces cerevisiae involves gene products required for transcriptional repression at silencers and telomeres. Genetics 133**:**171-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longtine, M. S., S. Enomoto, S. L. Finstad, and J. Berman. 1992. Yeast telomere repeat sequence (TRS) improves circular plasmid segregation, and TRS plasmid segregation involves the RAP1 gene product. Mol. Cell. Biol. 12**:**1997-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14**:**953-961. [DOI] [PubMed] [Google Scholar]
- 26.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275**:**986-990. [DOI] [PubMed] [Google Scholar]
- 27.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8**:**489-498. [DOI] [PubMed] [Google Scholar]
- 28.Moretti, P., K. Freeman, L. Coodly, and D. Shore. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8**:**2257-2269. [DOI] [PubMed] [Google Scholar]
- 29.Moretti, P., and D. Shore. 2001. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 21**:**8082-8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray, A. W., and J. W. Szostak. 1983. Pedigree analysis of plasmid segregation in yeast. Cell 34**:**961-970. [DOI] [PubMed] [Google Scholar]
- 31.Paetkau, D. W., J. A. Riese, W. S. MacMorran, R. A. Woods, and R. D. Gietz. 1994. Interaction of the yeast RAD7 and SIR3 proteins: implications for DNA repair and chromatin structure. Genes Dev. 8**:**2035-2045. [DOI] [PubMed] [Google Scholar]
- 32.Palladino, F., T. Laroche, E. Gilson, A. Axelrod, L. Pillus, and S. M. Gasser. 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75**:**543-555. [DOI] [PubMed] [Google Scholar]
- 33.Park, H., and R. Sternglanz. 1998. Two separate conserved domains of eukaryotic DNA topoisomerase I bind to each other and reconstitute enzymatic activity. Chromosoma 107**:**211-215. [DOI] [PubMed] [Google Scholar]
- 34.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148**:**635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122**:**19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strambio-de-Castillia, C., G. Blobel, and M. P. Rout. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144**:**839-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tham, W. H., J. S. Wyithe, P. K. Ferrigno, P. A. Silver, and V. A. Zakian. 2001. Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol. Cell 8**:**189-199. [DOI] [PubMed] [Google Scholar]
- 38.Triolo, T., and R. Sternglanz. 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381**:**251-253. [DOI] [PubMed] [Google Scholar]
- 39.Tsalik, E. L., and M. R. Gartenberg. 1998. Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast 14**:**847-852. [DOI] [PubMed] [Google Scholar]
- 40.Tschumper, G., and J. Carbon. 1983. Copy number control by a yeast centromere. Gene 23**:**221-232. [DOI] [PubMed] [Google Scholar]