Open Reading Frame 50 Protein of Kaposi's Sarcoma-Associated Herpesvirus Directly Activates the Viral PAN and K12 Genes by Binding to Related Response Elements (original) (raw)
Abstract
Open reading frame (ORF) 50 protein is capable of activating the entire lytic cycle of Kaposi's sarcoma-associated herpesvirus (KSHV), but its mechanism of action is not well characterized. Here we demonstrate that ORF 50 protein activates two KSHV lytic cycle genes, PAN (polyadenylated nuclear RNA) and K12, by binding to closely related response elements located approximately 60 to 100 nucleotides (nt) upstream of the start of transcription of the two genes. The 25-nt sequence 5′ AAATGGGTGGCTAACCTGTCCAAAA from the PAN promoter (PANp) confers a response to ORF 50 protein in both epithelial cells and B cells in the absence of other KSHV proteins. The responsive region of DNA can be transferred to a heterologous minimal promoter. Extensive point mutagenesis showed that a span of at least 20 nt is essential for a response to ORF 50 protein. However, a minimum of six positions within this region were ambiguous. The related 26-nt responsive element in the K12 promoter (K12p), 5′ GGAAATGGGTGGCTAACCCCTACATA, shares 20 nt (underlined) with the comparable region of PANp. The divergence is primarily at the 3′ end. The DNA binding domain of ORF 50 protein, encompassing amino acids 1 to 490, fused to a heterologous activation domain from herpes simplex virus VP16 [ORF 50(1-490)+VP] can mediate activation of reporter constructs bearing these response elements. Most importantly, ORF 50(1-490)+VP can induce PAN RNA and K12 transcripts in transfected cells. ORF 50(1-490)+VP expressed in human cells binds specifically to duplex oligonucleotides containing the responsive regions from PANp and K12p. These DNA-protein complexes were supershifted by antibody to VP16. ORF 50(1-490) without a VP16 tag also bound to the response element. There was a strong correlation between DNA binding by ORF 50 and transcriptional activation. Mutations within PANp and K12p that impaired transactivation by ORF 50 or ORF 50(1-490)+VP also abolished DNA binding. Only one of eight related complexes formed on PANp and K12p oligonucleotides was due to ORF 50(1-490)+VP. The other complexes were due to cellular proteins. Two KSHV lytic-cycle promoters are activated by a similar mechanism that involves direct recognition of a homologous response element by the DNA binding domain of ORF 50 protein in the context of related cellular proteins.
The switch between latency and lytic-cycle gene expression of Kaposi's sarcoma-associated herpesvirus (KSHV) is initiated by a single transactivator encoded in open reading frame (ORF) 50 of the viral genome (15, 30). When plasmids that constitutively express the ORF 50 protein are transfected into cell lines derived from primary effusion lymphoma harboring KSHV in the latent stage of its life cycle they autostimulate ORF 50 expression, they activate kinetically appropriate expression of downstream early- and late-stage lytic KSHV mRNAs and polypeptides, and they induce the release of encapsidated viral DNA (7, 14, 15, 30). Thus, ORF 50 protein is sufficient to drive the viral lytic cascade to completion. Following induction of the KSHV lytic cycle by chemical stimuli such as phorbol esters and histone deacetylase inhibitors, the ORF 50 gene is rapidly expressed, within 2 to 4 h, well before many other mRNAs. Its expression is resistant to inhibitors of protein synthesis (7, 31, 40); these characteristics classify ORF 50 as an immediate-early gene.
The 691-amino-acid (aa) KSHV ORF 50 protein does not share obvious homology with cellular proteins. However, it is related to immediate-early transcriptional activator proteins of other gammaherpesviruses, such as ORF 50a encoded by Herpesvirus saimiri and Rta encoded by the BRLF1 ORF of Epstein-Barr virus (2, 11, 15, 30, 33, 35). The DNA binding and dimerization domain of KSHV ORF 50 is located in the N-terminal portion of the protein (aa 1 to 530), and an acidic activation domain is found in the C-terminal portion (aa 486 to 691) (14, 26, 34). A deletion mutant (aa 1 to 530) that removes the activation domain but retains the DNA binding domain behaves as a dominant negative for induction of the lytic cascade by ORF 50 or by chemical stimuli (14). KSHV ORF 50 protein contains two putative arginine- and lysine-rich nuclear localization signals (aa 1 to 13) and (aa 516 to 530) (4, 15). The “coactivator” CREB binding protein binds to the basic DNA binding domain and to the carboxyl-terminal activation domain of ORF 50 protein. A central proline-rich sequence (aa 385 to 600) interacts with histone deacetylase 1 (10). ORF 50 is a phosphoprotein, but its phosphorylation sites and the responsible kinases have not yet been identified (14).
In transient-transfection assays, ORF 50 protein behaves as a transcriptional activator of a number of KSHV promoters that have been fused to reporter genes. The ORF 50-responsive promoters include the following: ORF 6 (single-stranded DNA binding protein), ORF 21 (thymidine kinase [TK]), ORF 57 (posttranscriptional activator), ORF 59 (DNA polymerase-associated processivity factor), K8 (K-bZip), K9 (viral interferon response factor), K12 (kaposin), and PAN (polyadenylated nuclear RNA) (3, 4, 6, 13-15, 28). An unanswered question is whether ORF 50 protein stimulates each of these promoters by the same mechanism.
The related Epstein-Barr virus (EBV) protein Rta, which functions to disrupt EBV latency in some B-cell and epithelial-cell backgrounds, activates viral lytic-cycle promoters by several distinct mechanisms. On certain viral promoters, such as BMRF1 (the DNA polymerase processivity factor), BMLF1 (the posttranscriptional activator), and BHRF1 (the viral bc1-2 homologue), EBV Rta behaves as a sequence-specific DNA binding protein (9, 20). The binding site is large, spanning 17 to 20 nucleotides, and degenerate, with a consensus GNCCN9GGNG (9). The interaction of Rta and CREB binding protein is helpful in activating genes such as BMLF1, which are direct targets (32). However, several target EBV promoters are stimulated by Rta in an indirect manner. The promoter of the EBV DNA polymerase gene is activated via upstream stimulatory factor and E2F (12). The capacity of EBV Rta to bind to retinoblastoma protein and to displace E2F1 is related to its capacity to activate the EBV DNA polymerase promoter (36). Rta also activates Zp, the promoter of the EBV gene BZLF1, encoding ZEBRA, the other viral immediate-early transactivator that disrupts latency (22). Rta's capacity to activate Zp is linked to its ability to activate mitogen-activated protein kinases such as p38 and JNK and to activate phosphatidylinositol-3 kinase (1, 5). Activation of phosphatidylinositol-3 kinase is not required for Rta's ability to stimulate expression of genes, such as BMLF1, that are controlled by direct binding by Rta (5). The capacity of EBV Rta to autostimulate Rp, the promoter of its BRLF1 gene, to which Rta does not bind, is tightly linked to the presence of cognate binding sites for Sp1 and Sp3 (21).
Emerging evidence suggests that KSHV ORF 50 protein also controls its target promoters by several distinct mechanisms. Two papers published while our work was in progress suggest that the promoters of the PAN, ORF 57, and K8 genes are activated by a direct mechanism of sequence-specific DNA binding; however, the responsive region in PANp does not share obvious homology with the responsive regions of ORF 57p and K8p (13, 28). Other promoters are activated indirectly: ORF 50 activation of the KSHV TK promoter and the vIRF promoters appear to depend on Sp1 sites (4, 38). AP-1 sites were also involved in activation of the vIRF promoter (4). Autostimulation of the ORF 50 promoter is mediated by OCT 1 binding sites (25).
Our work employing extensive deletion and point mutagenesis shows that the PAN and K12 genes are activated by ORF 50 protein via a large highly conserved response element. We demonstrate a high degree of correlation between the capacity of ORF 50 protein expressed in human cells to bind to these responsive elements and its capacity to activate transcription from reporters bearing the responding DNA sequences. Moreover, we show that these two response elements are bound by similar cellular proteins, suggesting that the two promoters are regulated by a complex mechanism involving binding by ORF 50 protein and by related cellular activators or repressors.
MATERIALS AND METHODS
Cell culture.
BJAB cells are derived from a B-cell lymphoma that is not known to contain any viruses (14, 16). HH-B2 is derived from a primary effusion lymphoma infected with KSHV (7). HKB5/B5 is an EBV-negative cell line formed by fusion of HH514-16 cells with 293T cells (kindly provided by M.-S. Cho, Bayer Corporation, Berkeley, Calif.). 293T cells are human embryonic kidney cells transformed by the adenovirus E1 region and the simian virus 40 T antigen. The lymphoid cells were grown in RPMI 1640 medium plus fetal calf serum. 293T cells were cultivated in Dulbecco's modified Eagle's medium plus fetal calf serum.
Transfections.
BJAB and HH-B2 cells were transfected by electroporation (27). 293T cells were transfected by the calcium phosphate method (8). HKB5/B5 cells were transfected by using the DMRIE-C reagent (GIBCO BRL).
Plasmid construction.
pE4CAT, pRTS, and pRTS/ORF 50 (gRta) have been described (30). pCAT-Basic was from Promega. Fragments of the KSHV PAN promoter were amplified from DNA of BC-1 cells by PCR. These fragments, containing a _Sal_I site at the 5′ end and an _Xba_I site at the 3′ end, were ligated into pCAT-Basic digested with _Sal_I and _Xba_I. Double-stranded annealed oligonucleotides encompassing nucleotides −91 to −58 of the PAN promoter were cloned into pE4CAT digested with _Hin_dIII and _Xba_I. The numbering of the nucleotides in the PAN promoter (see Fig. 1) is based on the 5′ end of the PAN RNA cDNA described in reference 29. Single- and double-base-pair point mutations were introduced into PAN promoter (−90 to −1) in pCAT-Basic or PAN promoter (−91 to −58) in pE4CAT using the QuikChange site-directed mutagenesis kit (Stratagene).
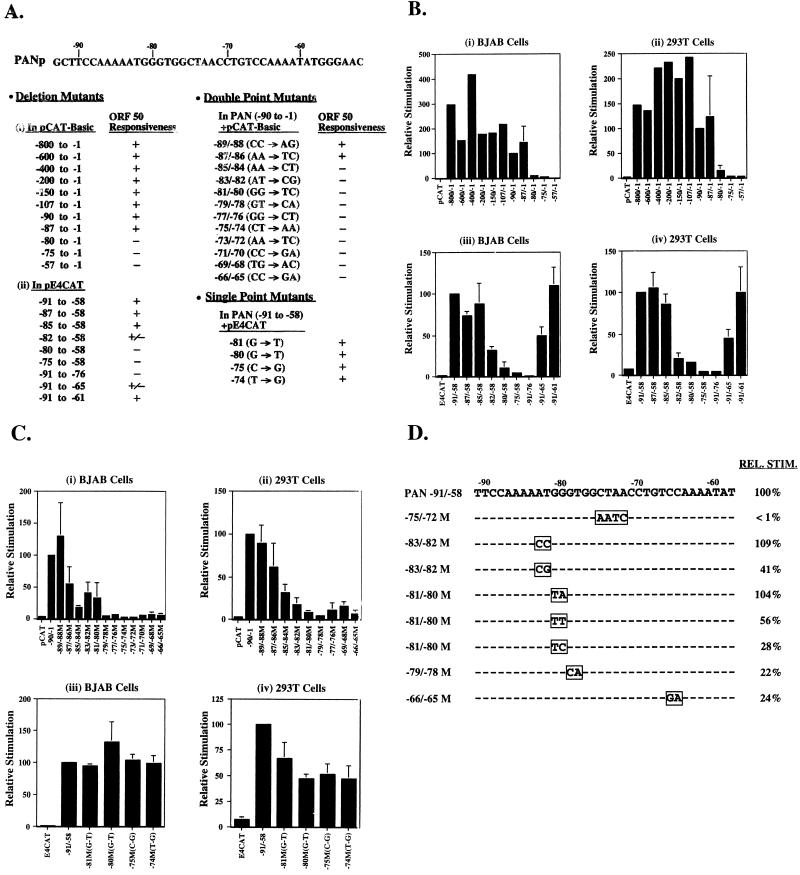
FIG. 1.
Definition of an ORF 50 protein response element in the promoter of the PAN gene. (A) Summary of mutagenesis of the PAN promoter. Deletion mutations include 5′-end deletions of the PAN promoter in pCAT-Basic (i) and fragments of PAN promoter transferred to pE4CAT (ii). Double point mutations are in the context of PAN promoter (−90 to −1) in pCAT-Basic; single point mutations are in the PANp fragment (−91 to −58) in pE4CAT. Responsiveness of each construct to ORF 50 is indicated as follows: +, wild-type activity; +/−, about 50% activity; −, markedly impaired activity. (B) Effect of deletion mutations of the PAN promoter on the response to the ORF 50 protein. The deletions were studied in two reporters, pCAT-Basic (i and ii) and pE4CAT (iii and iv). In panels i and ii, the data are expressed relative to the response of the construct (−90 to −1) pCAT. This represented 40-fold stimulation in BJAB and 62-fold stimulation in 293T cells. In panels iii and iv, the data are expressed relative to the construct −91 to −58 inserted into pE4CAT. This represented 40-fold stimulation in BJAB and 36-fold stimulation in 293T cells. (C) Effect of point mutations of the PAN promoter on response to the ORF 50 protein. The response of double point mutants was measured in pCAT (−90 to −1) (i and ii; 31-fold stimulation in BJAB and 61-fold stimulation in 293T cells) and that of single point mutants was measured in E4CAT (−91 to −58) (iii and iv; 40-fold stimulation and BJAB and 36-fold stimulation in 293T cells). (D) Activity of double or quadruple point mutants relative to the response of PANp (−91 to −58) E4CAT.
The series of K12 promoter fusions to CAT (see Fig. 3C) was generated by cloning double-stranded annealed oligonucleotides into the _Hin_dIII/_Xba_I sites of pE4CAT. The series of deletion mutants of the ORF 50 gene (see Fig. 4) were derived from pRTS/gRta (30). DNA fragments encoding the VP16 activation domain (aa 413 to 490) and the simian virus 40 T antigen nuclear localization signal (KKKRK) were ligated downstream of portions of the ORF 50 gene. All constructs were confirmed by sequencing.
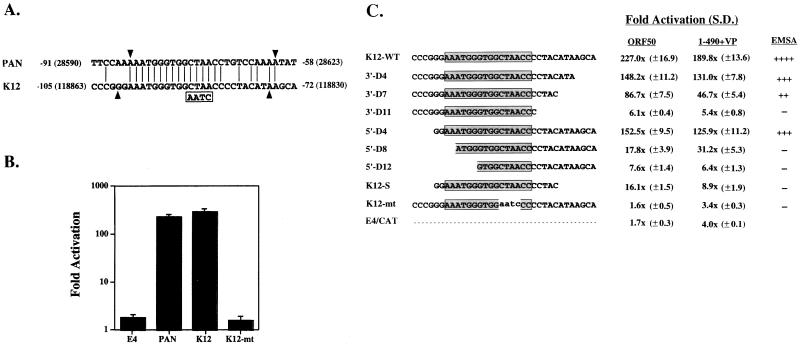
FIG. 3.
Sequence in the promoter of the K12 gene that is responsive to ORF 50 protein. (A) Comparison of sequences in the ORF 50 protein response element in the PAN promoter with a region of the K12 promoter. Identical sequences are shown by vertical lines. The sequence in the K12 mutant are boxed. Arrowheads demarcate the minimal region of the two promoters that is responsive to ORF 50 protein. (B) Response of PAN (−91 to −58), K12 (−105 to −72), and K12 mutant (K12-MT) in E4CAT to stimulation by ORF 50 protein. (C) Effect of deletions of the K12 promoter cloned in E4CAT on its response to ORF 50 protein and ORF 50(1-490)+VP. The deletions are designated by the number of nucleotides removed from the 3′ end (e.g., 3′-D4 removed 4 nt) or 5′ end. K12-S, short K12p; S.D., standard deviation. For panels B and C, the assays were performed in BJAB cells. The capacity of the duplex oligonucleotide to bind ORF 50(1-490)+VP was determined in competition EMSAs. (−, no competition; ++++, strongest competition).
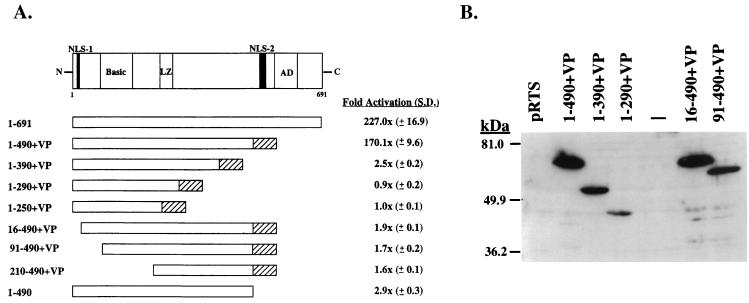
FIG. 4.
Transcriptional activation by the N-terminal 490 aa of ORF 50 protein fused to VP16. (A) Diagram of the ORF 50 protein. NLS, nuclear localization signal; LZ, leucine zipper; AD, activation domain (15). Numbers below the diagram represent amino acid residues. Results of transcriptional activation by deletion mutants of ORF 50 protein fused to VP16 are means and standard deviations (S.D.) of two experiments measuring activation of K12 promoter (−105 to −72) fused to E4CAT in BJAB cells. (B) Expression of deletion mutants of ORF 50 protein fused to VP16. A Western blot of extracts of HKB5/B5 cells was probed with antibody to VP16.
CAT assays.
BJAB or 293T cells were transfected with 5 μg of activator or vector DNA and 5 μg of reporter DNA. CAT activity was determined as described elsewhere (27). Activation was calculated as percent acetylation of chloramphenicol in the presence of activator divided by percent acetylation in the presence of control vector.
Western blot analysis.
Expression of portions of ORF 50 protein fused to VP16 was determined by immunoblotting. Protein extracts were prepared from HKB5/B5 cells that had been transfected and grown in culture for 48 h. A total of 107 cells were resuspended in 150 μl of sodium dodecyl sulfate (SDS) sample buffer and sonicated for 15 s. Samples were boiled for 5 min and electrophoresed through a 10% polyacrylamide-SDS gel before proteins were transferred to nitrocellulose. The immunoblot was reacted with a 1:150 dilution of VP16 (1-21) monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.), followed by a 1:400 dilution of a rabbit anti-mouse immunoglobulin bridge, AXL232 (Axell; Accurate Chemical and Scientific Corp.). Immunoreactive bands were detected with 125I-labeled protein A and autoradiography.
Northern blot analysis.
Expression of PAN and K12 RNA was examined in HH-B2 cells following transfection of ORF 50 and ORF 50(1-490)+VP expression plasmids. The cells were transfected with 10 μg of plasmid DNA by electroporation or were chemically induced with 3 mM sodium butyrate (17). Total cellular RNAs were prepared 48 h later with an RNeasy kit (Qiagen), fractionated on 1% formaldehyde-agarose gels, and transferred to nylon membranes (Nytran; Schleicher & Schuell). PAN and RNase P probes were labeled by the random-priming method described previously (30). To detect the K12 gene, an antisense oligonucleotide corresponding to nucleotides 118,071 to 118,103 of the KSHV genome (23) was end labeled with 32P by using T4 polynucleotide kinase (Boehringer Mannheim). Hybridization was carried out overnight in a solution containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 0.5% SDS, and 100 μg of salmon DNA per ml at 68°C for double-stranded probes and 50°C for the K12 oligonucleotide. Filters were washed in 2× SSC-0.5% SDS once for 10 min and in 0.1× SSC-0.5% SDS twice for 30 min at 68°C for double-stranded DNA probes and at 50°C for the K12 oligonucleotide probe.
Cell extracts for EMSAs.
To prepare cell extracts for electrophoretic mobility shift assays (EMSAs), aliquots of 2 × 106 HKB5/B5 cells were transfected with 4 μg of pRTS/ORF 50(1-490)+VP, pRTS/ORF 50(1-490), or pRTS. At 48 h after transfection, 107 cells were washed once with phosphate-buffered saline, and cell protein extracts were prepared by the method of Mosser et al. (18). The cell pellets were resuspended in 150 μl of lysis buffer (0.42 M NaCl, 20 mM HEPES [pH 7.5], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml). Lysates were spun at 90,000 rpm at 4°C for 15 min in a bench-top Beckman ultracentrifuge, and supernatants were aliquoted and stored at −80°C. Protein concentrations were determined by the Bradford method.
EMSA.
Annealed double-stranded oligonucleotides were end labeled with 32P using T4 polynucleotide kinase. Binding reactions contained 15 μg of cell extract protein in a solution containing 10 mM HEPES (pH 7.5), 50 mM NaCl, 2 mM MgCl2, 2.5 μM ZnSO4, 0.5 mM EDTA, 1 mM dithiothreitol, 15% glycerol, and 0.5 to 1.0 μg of poly(dI-dC) in a total volume of 20 μl. After an incubation for 5 min at room temperature, radiolabeled DNA was added. For the competition assays, increasing amounts (20, 50, and 100×) of nonradioactive competitor DNA were added in the initial reaction mix. For supershift assays, 2 μl (0.4 μg) of antibody to VP16 or antibody to ORF 50(230-250) was added 10 min following the addition of the probe. The ORF 50(230-250) antibody is an affinity-purified polyclonal serum raised against the ORF 50 peptide corresponding to aa 230 to 250 (TGSAEKRRPITTGKVTGLSYP). After a further 10 min of incubation at room temperature, the reaction mixtures were loaded onto a 4%, 0.5× Tris-borate-EDTA native polyacrylamide gel. Gels were run at 200 V, transferred to Whatman 3 MM paper, dried, and exposed to autoradiography film overnight.
RESULTS
A 25-nucleotide (nt) region in the promoter of the PAN gene is responsive to the ORF 50 protein.
PAN, the KSHV-encoded polyadenylated nuclear RNA, is highly responsive to induction by ORF 50 protein (14, 15, 30). Analysis of a series of large deletions of the PAN promoter (PANp), ranging from a deletion of positions −800 to −1 to one of positions −57 to −1, in pCAT-Basic reporter plasmids, showed that the region −87 to −1 of PAN was responsive to ORF 50 protein while the region −80 to −1 and downstream truncations were not responsive (Fig. 1A, deletion mutants, panel i, and B, panels i and ii). The response to ORF 50 protein was not dependent on the presence of KSHV. The fragment −87 to −1 of PAN was responsive to ORF 50 protein in cultured BJAB lymphoma cells and the transformed human embryonic kidney cell line 293T; both cell lines lack KSHV DNA (Fig. 1B, panels i and ii). A 34-nt region (−91 to −58) responsive to ORF 50 protein could be transferred to a heterologous minimal promoter derived from the adenovirus E4 gene (Fig. 1A, deletion mutants, panel ii, and B, panels iii and iv). This region could not be bisected; neither −91 to −76 nor −75 to −58 conferred a response. Further deletions of this fragment showed that a region encompassing position −85 at the 5′ end and −61 at the 3′ end was able to confer a near-wild-type response to ORF 50 protein in the context of the adenovirus E4 gene minimal promoter in the reporter plasmid pE4CAT. The sequence of the 25-nt response element defined by deletion mutagenesis was 5′AAATGGGTGGCTAACCTGTCCAAAA3′.
Point mutagenesis shows that the responsive region of the PAN promoter is large but ambiguous.
To determine whether most of this large region was required for the response to ORF 50 protein, a series of double point mutations were introduced into the region −89 to −65 (Fig. 1A, double point mutants, and C, panels i and ii). Double point mutations spanning the region −85 to −65 of the PAN promoter (−90 to −1) all reduced activity by more than 50% in both BJAB and 293T cells (Fig. 1C, panels i and ii). Double point mutations were also introduced into the construct PANp (−91 to −58) E4CAT. Double mutations at positions −83 and −82 (AT to CG), −81 and −80 (GG to TC), −79 and −78 (GT to CA), and −66 and −65 (CC to GA) all reduced ORF 50 protein-induced activation by 60% or more (Fig. 1D). The same double mutations that inactivated the response of PANp(−91 to −1) also inactivated the response of PANp(−91 to −58) in E4CAT. These results indicated that the region responsive to ORF 50 protein spanned at least 20 nt and that two nucleotide changes within this region both abolished the response. Furthermore, the response was independent of the core promoter.
Single point mutations in PANp in the background of E4CAT suggested that some sequence ambiguity in this responsive region was tolerated (Fig. 1A, single point mutants, 1C, panels iii and iv, and 1D). For example, single nucleotide changes at positions −81 (G to T), −80 (G to T), −75 (C to G), and −74 (T to G) permitted maximal activation in BJAB cells and 50% or greater activity in 293T cells (Fig. 1A, single point mutants, and C, panels iii and iv). Moreover, the double mutations AT to CC at positions −83 and −82 and GG to TA at positions −81 and −80 were tolerated and behaved like the wild type (Fig. 1 D). From these experiments we defined at least six positions in the ORF 50 response element (−83, −82, −81, −80, −75, and −74) that were ambiguous.
ORF 50 protein specifically activates PANp through the ORF 50 response element.
The previous results showed that the capacity of ORF 50 protein to activate PANp depended on approximately 20 to 25 bp of specific nucleotide sequences within PANp. A computer search showed that the response element in PANp was surrounded by Sp1, PTF-1β, GATA1, AP-1, and NF-κB binding sites (Fig. 2A). However, when it was transferred to E4CAT, the ORF 50 response element was upstream of binding sites for other cellular transcription factors, such as AP-2, glucocorticoid receptor, CF2, and NF-1 (Fig. 2B). Thus, the response to ORF 50 protein correlated with the ORF 50 response element and not with sites for cellular activators that could be defined by computer search.
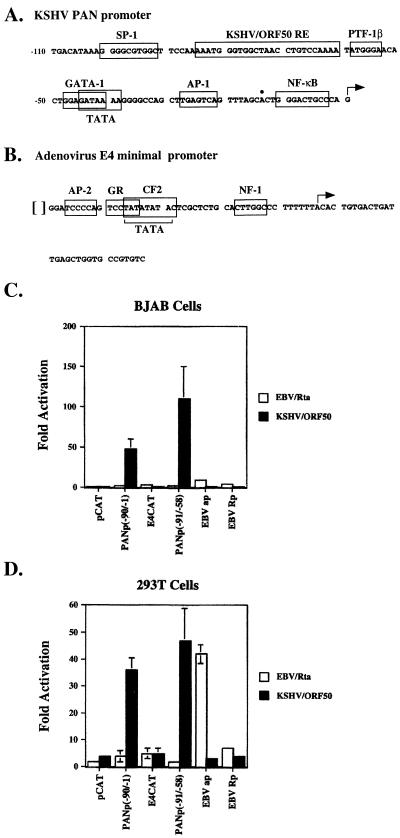
FIG. 2.
A region of the PAN promoter is specifically responsive to ORF 50 protein. (A) Sequence of the PAN promoter from −107 to −1; •, recently mapped start of PAN RNA (28). (B) Sequence of the adenovirus E4 minimal promoter. Potential binding sites for transcription factors identified by computer search are boxed, as is the KSHV/ORF 50 response element identified by mutagenesis experiments (Fig. 1). [ ], position where fragments of PANp or K12P were inserted. (C and D) Response of CAT reporters containing the KSHV PAN promoter and the promoters of the EBV BaRF1 (ap) and BRLF1 (Rp) genes to KSHV ORF 50 protein (KSHV/ORF50) or EBV BRLF1 protein (EBV/Rta) in BJAB (C) and 293T (D) cells.
Biologic experiments previously conducted with BC-1 cells dually infected with EBV and KSHV showed that each ORF 50 homologue activated the lytic cycle of its autologous virus (30); that is, ORF 50 activated KSHV, and its homologue, the EBV BRLF1 gene product Rta, activated EBV. Figure 2C and D show that the ORF 50 response element in a PANp/CAT reporter fusion mirrored this specificity. Both CAT reporters containing the ORF 50 response element were activated by ORF 50 protein but not by the EBV BRLF1 protein (Rta). Reporters containing promoters for the EBV ribonucleotide reductase small subunit (EBVap) and EBV Rta BRLF1 (Rp) were activated by EBV Rta and not by KSHV ORF 50 protein. The KSHV ORF 50 protein was equally active in BJAB B cells and in 293T epithelial cells. However, the EBV Rta protein was significantly more active in 293T cells than in BJAB cells, consistent with the hypothesis of an epithelial-cell-enhanced function of EBV Rta (37).
A related ORF 50 response element is found in the promoter of the KSHV K12 gene.
A search of the database revealed that the response element for ORF 50 protein found in PANp was also present in a region 105 to 72 nt upstream of a 2.3-kb mRNA encompassing K12, a gene that is strongly induced during the lytic cycle (15, 24). The K12 region of KSHV is thought to encode at least three different protein products: kaposin A, a putative 60-aa hydrophobic protein encoded by a 700-nt mRNA from the K12 ORF (19, 39); kaposin B, a 48-kDa protein encoded primarily by direct repeats upstream of the K12 ORF; and possibly two proteins of 54 and 38 kDa that represent fusions between direct repeats and K12 coding sequences (24).
When transferred to pE4CAT, a 34-nt region of the K12 promoter (K12p) located from −105 to −72 upstream of the 2.3-kb mRNA was shown to be stimulated by ORF 50 protein to the same extent as PANp (Fig. 3B). This region possessed striking homology to the −91-to-−58 region of PANp: 22 of 34 nt were identical, including 16 consecutive identical nucleotides. Mutation of 4 nt (CTAA to AATC) within the sequences shared by PANp and K12p abolished the response of both promoters to ORF 50 protein (Fig. 1D and 3B).
Further deletion mutagenesis of the ORF 50 protein response element of K12p in the context of E4CAT plasmid indicated that at least three regions were required for optimal response to ORF 50 protein (Fig. 3C). These included 16 nt conserved between PANp and K12p, as well as 3′ flanking sequences and 5′ flanking sequences that were not as well conserved between PANp and K12p. The importance of the 16-nt conserved sequences can be gleaned from comparing the response of K12-WT (227-fold stimulation) with K12-MT, a mutant in which 4 nt are changed (CTAA to AATC) (1.6-fold stimulation). The role of the 11-nt 3′ flanking sequences can be surmised by the progressive decrease in response of the K12 promoter fragment as nucleotides are removed from its 3′ end. 5′ flanking sequences appeared to exert their greatest effect when accompanied by truncations of the 3′ sequences, as seen in the mutant K12-S. Thus, a minimal ORF 50 response element in K12p is 5′ GGAAATGGGTGGCTAACCCCTACATA. This 26-nt element shares 20 nt with the comparable region of PANp.
The DNA binding region of ORF 50 protein fused to a heterologous activation domain activates the K12 promoter.
The previous experiments showed that the response of the PAN and K12 promoters to activation by ORF 50 protein was mediated by a specific DNA sequence (Fig. 1 and 3) and demonstrated that these two promoters were activated efficiently by ORF 50 protein in the absence of other KSHV sequences. These results suggested that ORF 50 protein directly activated the PAN and K12 promoters by binding to DNA. To begin to test this hypothesis, we constructed a series of plasmids encompassing the putative DNA binding domain of ORF 50 fused to the transcriptional activation domain of the herpes simplex virus VP16 protein (Fig. 4). All of these constructs were expressed well in cultured human cells (Fig. 4B). A construct encoding the N-terminal 490 aa of ORF 50 protein fused to VP16 [ORF 50(1-490)+VP] activated K12p E4CAT nearly as well as the entire ORF 50 protein (Fig. 4A). Constructs which removed the putative N-terminal ORF 50 nuclear localization signal, e.g., ORF 50(16-490+VP16), which lacked an activation domain, e.g., ORF 50(1-490), or which otherwise truncated the DNA binding domain were unable to activate the K12 promoter fused to E4CAT (Fig. 4A).
The ability of ORF 50(1-490)+VP to stimulate transcription of a series of 3′ and 5′ deletions of the K12 promoter in pE4CAT was similar to the activity of intact ORF 50 protein (Fig. 3C). Promoter deletions that reduced the response to the intact ORF 50 protein containing its native activation domain also reduced the response to ORF 50(1-490)+VP with the heterologous activation domain. These results indicated that the DNA binding domain of ORF 50 was essential in mediating a response.
Activation of two lytic cycle genes by ORF 50(1-490)+VP.
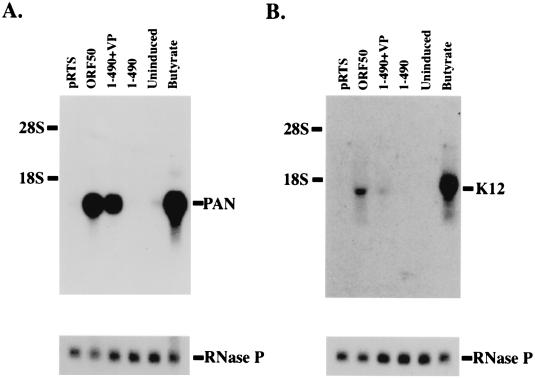
The DNA binding domain of ORF 50 fused to VP16 was competent to activate transcription of PAN and K12 from the latent KSHV genome (Fig. 5). The construct ORF 50(1-490)+VP was nearly as efficient as the entire ORF 50 protein at inducing expression of PAN RNA in the HH-B2 cell line (Fig. 6A). The K12 mRNA, which was less strongly stimulated by the intact ORF 50 protein than was PAN RNA, was weakly stimulated by transfection of ORF 50(1-490)+VP (Fig. 6B). However, these experiments indicated that the DNA binding domain of ORF 50 fused to VP16 was active on the native promoters of the KSHV genome in a latent state as well as on promoter-reporter fusions.
FIG. 5.
Activation of viral transcripts by ORF 50(1-490)+VP. HH-B2 cells were transfected with plasmids expressing empty vector (pRTS), full-length ORF 50 protein (ORF 50), ORF 50(1-490)+VP, or ORF 50(1-490). Uninduced and _n_-butyrate-treated cells were negative and positive controls for lytic gene expression. Northern blots were probed with the PAN RNA gene (A) or an oligonucleotide from the K12 gene (B).
FIG. 6.
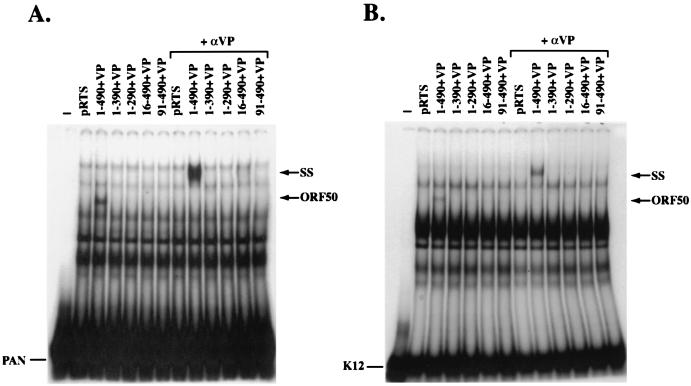
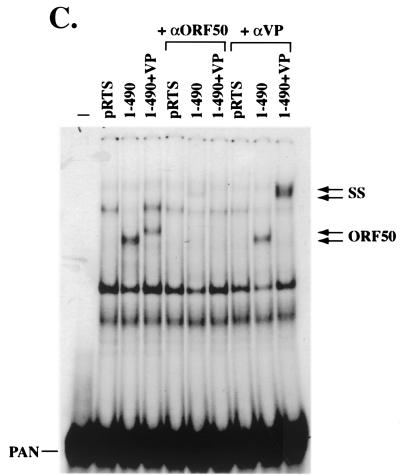
The N-terminal 490 aa of ORF 50 protein fused to VP16 binds to the promoters of the KSHV PAN and K12 genes. Extracts of HKB5/B5 cells transfected with deletion mutants of ORF 50 protein fused to VP16 were used in EMSAs. Antibody to VP16 (αVP) was used to supershift the complexes formed. The probes were the PAN promoter (−91 to −58) (A) and the K12 promoter (−105 to −72) (B). The ORF 50-specific complexes and the supershifted (SS) complex are indicated. (C) Comparison of DNA binding by ORF 50(1-490) and ORF 50(1-490)+VP to PANp. The binding reactions were supershifted with antibody to ORF 50(230-250) or with antibody to VP16.
Direct evidence for DNA binding by ORF 50(1-490)+VP.
The family of constructs encoding N terminal portions of ORF 50 protein fused to VP16 was examined for their ability to bind DNA by EMSA that employed extracts of human cells that expressed the proteins (Fig. 6). ORF 50(1-490)+VP expressed a protein that bound specifically to DNA (Fig. 6). The same plasmid encoded a protein that was competent to activate transcription from a K12p/E4CAT reporter (Fig. 4A) and to strongly activate PAN RNA transcription from the latent KSHV genome (Fig. 5). ORF 50(1-490)+VP bound to duplex oligonucleotides derived from both the PAN (Fig. 6A) and K12 (Fig. 6B) promoters. Other constructs encoding ORF 50 aa 1 to 390, 1 to 290, 16 to 490, or 91 to 490 fused to VP16 did not bind DNA, even though these proteins were expressed adequately (Fig. 4B and 6). The VP16 tag was not required for ORF 50 to bind DNA. A construct encoding ORF 50(1-490) specifically bound to PAN promoter (Fig. 6C). As expected, the complex between ORF 50(1-490) and PANp was supershifted by antibody to ORF 50 protein but not by antibody to VP16 (Fig. 6C).
There were at least eight comparable complexes formed on the two promoters by cell extracts containing ORF 50(1-490)+VP; only one of these complexes (complex 3) was specific to the ORF 50 protein (1-490)+VP (Fig. 7). Complex 3 was observed only in cells transfected with ORF 50(1-490)+VP and not in cells transfected with vector pRTS. Complex 3 was supershifted by antibody to VP16 (Fig. 6 and 7). The abundance of the other complexes formed by cellular proteins was not altered by the presence of ORF 50(1-490)+VP, nor were they affected by antibody to VP16 (Fig. 7).
FIG. 7.
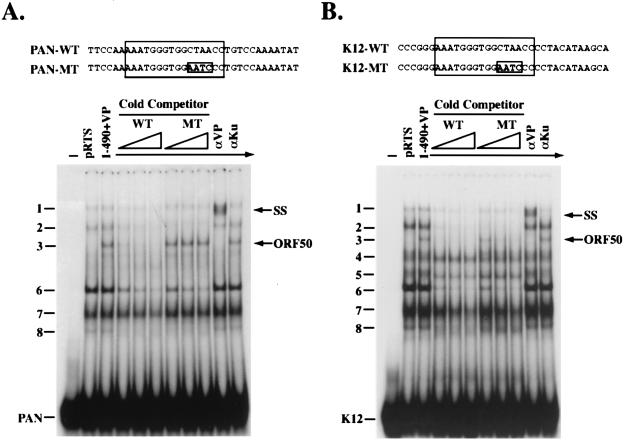
Specificity of DNA binding by the N-terminal 490 aa of ORF 50 fused to VP16. Extracts for EMSA were prepared from HKB5/B5 cells transfected with empty vector (pRTS) or a vector expressing ORF 50(1-490)+VP16. DNA binding reactions were competed with wild-type (WT) or mutant (MT) duplex oligonucleotides. (A) Binding to a duplex oligonucleotide from the PAN promoter; (B) binding to a duplex oligonucleotide from the K12 promoter.
Specificity of binding of ORF 50(1-490)+VP demonstrated by competition with unlabeled duplex oligonucleotides.
The specific binding of ORF 50 protein to the PAN promoter was competed by a 20- to 100-fold molar excess of wild-type oligonucleotide (Fig. 7A). This oligonucleotide competed for binding by ORF 50 (complex 3) as well as for binding by the cellular proteins responsible for complexes 2 and 6. Comparable amounts of an unlabeled duplex oligonucleotide containing four mutations (CTAA to AATC) at positions −75 to −72 failed to compete for binding of ORF 50(1-490)+VP. However, this mutant oligonucleotide was able to compete for formation of complexes 2 and 6.
Similarly, binding of ORF 50(1-490)+VP to the K12 promoter was competed by excess cold wild-type K12 oligonucleotide (Fig. 7B). The wild-type oligonucleotide competitor also reduced the formation of complexes 2 and 6. Competition with a mutant oligonucleotide with four point mutations (CTAA to AATC) produced a reduction in complexes 2 and 6 formed by cellular proteins that was similar in magnitude to the reduction of the complexes by wild-type K12 oligonucleotide. However, this mutant oligonucleotide had little effect on the ORF 50-specific complex 3.
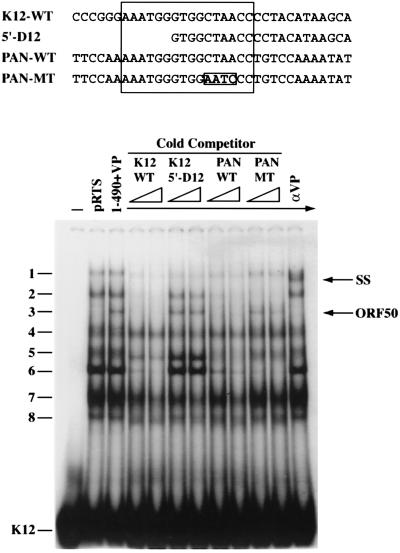
A PAN or K12 promoter fragment with the 4-nt mutation (CTAA to AATC) was not activated by full-length ORF 50 protein or ORF 50(1-490)+VP (Fig. 1D and 3B and C). Thus, there was a correlation between the ability of a fragment of K12 promoter DNA to bind ORF 50 protein and its capacity to confer activation in a reporter assay. To explore this correlation between DNA binding and activation further, all the K12 promoter deletions illustrated in Fig. 3C were examined for their ability to compete for binding by ORF 50 protein in EMSAs. K12p deletions which markedly impaired the transcriptional response, such as 3′-D11, 5′-D8, and 5′-D12 (Fig. 3C), also failed to compete for ORF 50 binding (Fig. 8 and data not shown). Conversely, K12p mutations which permitted activation, such as 3′-D4, also competed for binding to the K12p probe (Fig. 3C and data not shown).
FIG. 8.
Cross competition of ORF 50 protein binding to the K12 and PAN promoters. The probe was a duplex oligonucleotide derived from the K12 promoter. The competitors were wild-type (WT) K12, K12 5′-D12 (Fig. 3C), wild-type PAN, and a mutant oligonucleotide from the PAN promoter with the same changes as K12-MT (Fig. 7B). The sequences of the competitors are above the EMSA gel. The sequence shared between PAN promoter and K12 promoter is boxed.
Cross-competition experiments demonstrate that related complexes form on the promoters of the PAN and K12 genes.
Figure 8 compares the effect of addition of 50- to 100-fold molar excess of unlabeled oligonucleotide from the K12 promoter or PAN promoter used as a competitor for binding of ORF 50(1-490)+VP and cellular proteins to radiolabeled K12p. Competition with cold wild-type K12p and PANp produced similar results: the sharp complex 3 due to ORF 50(1-490+VP) was markedly reduced, as were complexes 2 and 6 due to cellular products. The effect of a mutant of PANp with the 4-nt changes CTAA to AATC as a cold competitor was, like WT PANp, to reduce cellular complexes 2 and 6. However, this mutant did not compete for binding by ORF 50(1-490)+VP. The K12 promoter mutant 5′-D12, which was not activated by ORF 50 protein, did not compete for binding by ORF 50(1-490)+VP. This mutant oligonucleotide also failed to bind cellular proteins responsible for complexes 2 and 6. These results indicated that the region of PANp and K12p responsive to ORF 50 bound similar cellular proteins. The same mutation (CTAA to AATC) destroyed binding of ORF 50 to both promoters. Promoter mutations that impaired binding by ORF 50 also impaired transcriptional activation by ORF 50(1-490)+VP.
DISCUSSION
Our work employed deletion and point mutagenesis to define two related DNA elements upstream of the PAN and K12 genes that are critical for conferring a response to the ORF 50 protein. These DNA elements conferred activation in the context of the native minimal promoter and could be transferred to a heterologous promoter. They responded to ORF 50 in epithelial cells and in B cells; other KSHV gene products were not required. The ORF 50 DNA binding domain was responsible for activating transcription from these response elements. Those deletion and point mutations within the response elements that abolished transcriptional activation by ORF 50 protein also blocked the ability of ORF 50 protein to bind to the response elements. Moreover, the DNA binding domain of ORF 50 protein fused to a heterologous activation domain activated expression of PAN RNA and K12 mRNA from a latent KSHV genome. Taken together, our experiments indicate that the PAN and K12 genes are activated by ORF 50 protein binding to these related response elements in their promoters.
ORF 50 response elements.
Two papers published while our work was in progress contribute to the identification of ORF 50 response elements. Using deletion mutagenesis, Song et al. defined a 32-nt region upstream of the PAN gene that conferred a response to ORF 50 protein (28). This region overlaps but is not identical to the region that we defined. Using primer extension, Song et al. mapped a start to PAN RNA that is 14 nt upstream of the start of transcription that was identified by cDNA cloning in earlier experiments (28). They found that the response element in the PAN promoter is located from −38 to −69. By using their numbering system, our data, in close agreement, would map the response element from −46 to −70. Our system of double point mutagenesis provides the novel information that at least 20 nt of this region are essential (from −50 to −70). We discovered that an ORF 50 response element closely related to that in the PAN promoter was present in the promoter of the K12 gene.
Lukac et al. identified a 25-nt ORF 50 response element in the promoter of the KSHV ORF 57 gene which they designated 50RE57 (13). The sequence 5′AGTGTAACAATAATGTTCCCACGGC shows no obvious similarity to the ORF 50 response elements that we have defined in the promoters of the PAN and K12 genes. Within 50RE57 was a partial 12-bp palindrome, 5′ AACAATAATGTT, which proved essential for the ORF 50 response and which was also present in the promoter of the K8 (K-bZIP) gene. The palindrome in the ORF 57 and K8 response elements is not found in the ORF 50 response elements of the PAN or K12 genes. Conversely, the essential 16-nt conserved element in the PAN and K12 promoters that we have identified does not appear to be present in the promoters of the ORF 57 and K8 genes.
The available data therefore suggest either that ORF 50 protein can bind to several unrelated sequences or that the sequence recognized by ORF 50 protein is highly degenerate. That the ORF 50 response element may be ambiguous is illustrated by our point mutagenesis in the PAN promoter, which shows that a response may still be obtained when six positions are altered (Fig. 1A and D). However, it would be distinctly unusual for a DNA binding transcriptional activator to recognize totally unrelated sequences.
The ORF 50 response elements in PANp and K12p consist of two components, a conserved core element and less highly conserved flanking sequences. Both groups of sequences are essential for activation and DNA binding. The core element of 16 identical consecutive nucleotides is shared by PANp and K12p. Invasion of this core element by deletion or installation of the 4-nt mutation CTAA to AATC within the core element destroys the response of either promoter. An optimal response also required 8 nt (K12) or 9 nt (PAN) of DNA downstream of the conserved core element. In this region five of nine nucleotides are identical. Deletion of sequences at the 3′ end of the core element as in the K12p mutant 3′-D11 (Fig. 3C) reduces ORF 50-mediated activation by 97% and eliminates DNA binding. Furthermore, the K12 promoter also appears to require 2 nt upstream of the conserved core element which are not essential for a response by PANp.
The ORF 50 protein DNA binding domain.
Song et al. showed that FLAG-tagged full-length ORF 50 protein expressed in Escherichia coli bound to duplex oligonucleotides from the PAN promoter (28), while Lukac et al. carried out DNA binding assays on the ORF 57 and K-bZIP promoters with a His-tagged version of full-length ORF 50 protein expressed from baculovirus-infected Sf9 insect cells (13). Although the specificity of binding to DNA of these partially purified proteins was demonstrated by competition with wild-type and mutant oligonucleotides neither of the previous studies provided the gold standard of direct DNA binding, i.e., a supershift with specific antibody to the DNA binding protein. Lukac et al. also showed that the N-terminal 272 aa of ORF 50 protein expressed in E. coli represented a minimal region of the protein that could bind to the ORF 57-responsive sequence (13).
Our experimental results extend these findings in several ways. We show that the N-terminal 490 aa of ORF 50 protein expressed in human cells represents a domain that is competent to recognize the PAN and K12 response element (Fig. 6). The VP16 epitope tag facilitated antibody supershift experiments (Fig. 6). However, the VP16 epitope tag was not required for binding to DNA, as the construct ORF 50(1-490) without VP16 also bound to DNA and was supershifted by antibody to ORF 50 (Fig. 6C). The VP16 activation domain tag also permitted the demonstration of a tight correlation between portions of the ORF 50 protein expressed in human cells which function to activate the PAN and K12 promoters (Fig. 4A) and those portions of the protein which were able to bind DNA (Fig. 6). N-terminal fragments of ORF 50 protein that ranged from 250 to 390 aa were expressed well in human cells (Fig. 4B) but did not bind to DNA (Fig. 6) or activate transcription (Fig. 4A). The variance of our results from those which identify the N-terminal 272 aa expressed in E. coli as a functional DNA binding domain may be related to the sources of protein or the DNA sequences being studied.
Cellular proteins that may affect the ORF 50 response.
It is likely that ORF 50-responsive promoters are regulated by complex interactions between ORF 50 protein and cellular proteins. The response of the ORF 50 gene itself to autostimulation appears to be mediated by the cellular Oct 1 protein (25), while the response of the viral TK promoter, and perhaps other promoters, to ORF 50 protein seems to require Sp1 binding sites (4, 38). Neither the ORF 50 promoter nor the TK promoter has been shown to bind the ORF 50 protein itself. Our results show that the ORF 50-responsive regions of PANp and K12p bind several cellular proteins as well as ORF 50 protein (Fig. 7 and 8). Binding of ORF50 protein might be facilitated by one of these cellular proteins that bind to the PAN and K12 promoters. The binding sites for some cellular proteins overlap but are distinct from the binding site for ORF 50 protein. For example, in competition EMSA studies the CTAA-to-AATC mutant oligonucleotides from the PAN and K12 promoter fail to bind ORF 50 protein but still specifically bind at least two cellular proteins (Fig. 7). However, other mutants, e.g., 5′-D12 of the K12 promoter, eliminate binding of both ORF 50 and conserved cellular proteins (Fig. 8). The identification and elucidation of the function of the cellular proteins that bind to PANp and K12p together with ORF 50 is a topic for fruitful further study.
Acknowledgments
This work was supported by NIH grants CA70036 and CA16038 to G.M.
We thank Jill Countryman and Ayman El-Guindy for helpful critiques of the manuscript.
REFERENCES
- 1.Adamson, A. L., D. Darr, E. Holley-Guthrie, R. A. Johnson, A. Mauser, J. Swenson, and S. Kenney. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74**:**1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66**:**5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, S. R., C. Bloomer, and B. Chandran. 1998. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology 240**:**118-126. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74**:**8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darr, C. D., A. Mauser, and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BRLF1 induces the lytic form of viral replication through a mechanism involving phosphatidylinositol-3 kinase activation. J. Virol. 75**:**6135-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146**:**403-413. [DOI] [PubMed] [Google Scholar]
- 7.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74**:**6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52**:**456-467. [DOI] [PubMed] [Google Scholar]
- 9.Gruffat, H., and A. Sergeant. 1994. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 22**:**1172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75**:**1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62**:**2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, C., N. D. Sista, and J. S. Pagano. 1996. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J. Virol. 70**:**2545-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75**:**6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73**:**9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252**:**304-312. [DOI] [PubMed] [Google Scholar]
- 16.Menezes, J., W. Leibold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22**:**276-284. [PubMed] [Google Scholar]
- 17.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71**:**314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosser, D. D., N. G. Theodorakis, and R. I. Morimoto. 1988. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol. 8**:**4736-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muralidhar, S., A. M. Pumfery, M. Hassani, M. R. Sadaie, M. Kishishita, J. N. Brady, J. Doniger, P. Medveczky, and L. J. Rosenthal. 1998. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J. Virol. 72**:**4980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21**:**1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ragoczy, T., and G. Miller. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 75**:**5240-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragoczy, T., and G. Miller. 1999. Role of the Epstein-Barr virus Rta protein in activation of distinct classes of viral lytic cycle genes. J. Virol. 73**:**9858-9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93**:**14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadler, R., L. Wu, B. Forghani, R. Renne, W. Zhong, B. Herndier, and D. Ganem. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73**:**5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75**:**6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seaman, W. T., D. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology 263**:**436-449. [DOI] [PubMed] [Google Scholar]
- 27.Serio, T. R., J. L. Kolman, and G. Miller. 1997. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J. Virol. 71**:**8726-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75**:**3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, R., S. F. Lin, L. Gradoville, and G. Miller. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 93**:**11883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95**:**10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73**:**2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swenson, J. J., E. Holley-Guthrie, and S. C. Kenney. 2001. Epstein-Barr virus immediate-early protein BRLF1 interacts with CBP, promoting enhanced BRLF1 transactivation. J. Virol. 75**:**6228-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurau, M., A. Whitehouse, S. Wittmann, D. Meredith, and H. Fickenscher. 2000. Distinct transcriptional and functional properties of the R transactivator gene orf50 of the transforming herpesvirus saimiri strain C488. Virology 268**:**167-177. [DOI] [PubMed] [Google Scholar]
- 34.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146**:**1415-1426. [DOI] [PubMed] [Google Scholar]
- 35.Whitehouse, A., M. Cooper, K. T. Hall, and D. M. Meredith. 1998. The open reading frame (ORF) 50a gene product regulates ORF 57 gene expression in herpesvirus saimiri. J. Virol. 72**:**1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zacny, V. L., J. Wilson, and J. S. Pagano. 1998. The Epstein-Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J. Virol. 72**:**8043-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93**:**9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, L., J. Chiu, and J. C. Lin. 1998. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 17**:**735-742. [DOI] [PubMed] [Google Scholar]
- 39.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93**:**6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73**:**5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]