Mitochondrial DNA–Deletion Mutations Accumulate Intracellularly to Detrimental Levels in Aged Human Skeletal Muscle Fibers (original) (raw)
Abstract
Skeletal muscle–mass loss with age has severe health consequences, yet the molecular basis of the loss remains obscure. Although mitochondrial DNA (mtDNA)–deletion mutations have been shown to accumulate with age, for these aberrant genomes to be physiologically relevant, they must accumulate to high levels intracellularly and be present in a significant number of cells. We examined mtDNA-deletion mutations in vastus lateralis (VL) muscle of human subjects aged 49–93 years, using both histologic and polymerase-chain-reaction (PCR) analyses, to determine the physiological and genomic integrity of mitochondria in aging human muscle. The number of VL muscle fibers exhibiting mitochondrial electron-transport-system (ETS) abnormalities increased from an estimated 6% at age 49 years to 31% at age 92 years. We analyzed the mitochondrial genotype of 48 single ETS-abnormal, cytochrome c oxidase–negative/succinate dehydrogenase–hyperreactive (COX−/SDH++) fibers from normal aging human subjects and identified mtDNA-deletion mutations in all abnormal fibers. Deletion mutations were clonal within a fiber and concomitant to the COX−/SDH++ region. Quantitative PCR analysis of wild-type and deletion-containing mtDNA genomes within ETS-abnormal regions of single fibers demonstrated that these deletion mutations accumulate to detrimental levels (>90% of the total mtDNA).
Aging is a complex biological process characterized by a gradual but constant decline in biochemical and physiological function of many organs. Normal aging is most evident in, and has the greatest impact on, nonreplicative tissues (i.e., heart, brain, and skeletal muscle) that rely heavily on oxidative metabolism for energy. In skeletal muscle, aging is associated with an inevitable reduction in muscle mass and strength. The process by which lean muscle, one of the most metabolically active tissues in the human body, gradually declines with age is termed “sarcopenia.”1 Sarcopenia generally begins in the early 40s but can occur as early as age 25 years.2 By ages 50–60 years, typically 10%–20% of the muscle mass is lost, and, by age 80 years, the loss increases to 40%.3 In humans and rats, the age-related loss of muscle mass results from a decrease in fiber number and/or an increase in fiber atrophy.2,4–7 In rats, the extent of sarcopenia is muscle specific, with some muscles exhibiting significant decline in weight and fiber number with age, whereas others appear resistant to sarcopenia.4,6,8–11
The role of mitochondrial abnormalities in the etiology of skeletal muscle–fiber loss with age has been extensively characterized in rats and rhesus monkeys.4,10,12 Intrafiber atrophy and fiber breakage, concomitant with electron-transport-system (ETS) abnormalities, occur along the length of muscle fibers.4,10,12 These prototypic ETS enzymatic abnormalities, also observed in mitochondrial myopathies, include a decline in the activity of the partially mitochondrial-encoded complex IV (cytochrome c oxidase [COX−]) and an increase in the nuclear-encoded complex II activity (succinate dehydrogenase [SDH++]). Fibers with longer ETS-abnormal regions are more susceptible to atrophy and breakage.4,12 Muscles that exhibit the most muscle-mass loss with age—vastus lateralis (VL) and rectus femoris—also exhibit the largest reduction in fiber number and the greatest number of ETS-abnormal fibers with associated atrophy.4,10
Age-dependent mtDNA-deletion mutations accumulate in a number of mammalian tissues. In humans, these mutations appear in the 3rd decade of life and accumulate with age in postmitotic tissues, such as skeletal muscle, heart, and brain.13–15 A 4,977-nt deletion in mtDNA (mtDNA4977) is the most studied age-associated mtDNA alteration. The mtDNA4977 deletion, located in the major arc of mtDNA, is flanked by a 13-bp direct repeat. This mtDNA-deletion mutation accumulates with age and has been detected in multiple tissues and numerous individuals.13,16–18 Many other age-dependent mtDNA deletions of various sizes have been identified in analyses of human-tissue homogenates. Although these studies have established an age-associated increase in the number of mtDNA deletions, the physiological relevance of these mutations has been questioned, since the abundance of any single mtDNA-deletion product is low (<1%) when calculated from homogenate tissue.13,17,19 Given the mosaic and segmental distribution of deletion mutations and concomitant ETS abnormalities in muscle fibers,4,20,21 analysis of homogenates is not appropriate for quantitative measurement of these mutations in single cells. The analysis of defined numbers of muscle fibers as well as in situ hybridization studies demonstrate that mtDNA-deletion mutations focally accumulate to detectable levels in single cells and are associated with abnormal ETS enzymatic activities.12,20,22,23
Previous histologic studies of human skeletal-muscle mitochondrial abnormalities examined single or undefined numbers of serial tissue sections that demonstrated an accumulation with age but provided limited information regarding the abundance of these segmental abnormalities.24–26 In rats and rhesus monkeys, ETS abnormalities are focal, mosaically distributed among cells, and segmental; they do not extend throughout the length of the affected muscle fiber but rather are localized to a small region or segment of the fiber.4,10,12,27 Thus, determination of the total number of fibers containing ETS abnormalities in a biopsy sample requires extensive histologic analyses of muscle fibers along their length. Such analysis, in addition to defining the ETS abnormalities, will also determine the length of abnormal regions and the cross-sectional area (CSA) of the fibers.
For mitochondrial enzymatic and mtDNA abnormalities to contribute to aging processes, a number of criteria must be met: (1) mtDNA abnormalities must accumulate intracellularly to high levels, (2) they must cause a phenotype (ETS abnormality) that, ultimately, has a deleterious cellular effect, and (3) there must be a sufficient number of cellular abnormalities (ETS-abnormal fibers) to affect tissue function. The analysis of the age-dependent distribution of mtDNA-deletion mutations in skeletal muscle necessitates two levels of analysis: quantification of mtDNA deletion–mutation load within ETS-abnormal regions of single skeletal-muscle fibers and determination of the number of fibers harboring mtDNA deletions that result in an abnormal phenotype. We examined the abundance of ETS abnormalities in aged human skeletal muscle and quantified the intracellular abundance of mtDNA-deletion mutations in ETS-abnormal fibers from humans of diverse ages. The number of ETS-abnormal fibers (COX−/SDH++) increased with age. mtDNA-deletion mutations were identified in all examined ETS-abnormal fibers and accumulated to >90% of the total mtDNA within ETS-abnormal regions of single fibers.
Material and Methods
Tissue Collection and Preparation
VL muscle biopsies were collected from 12 human subjects (aged 49–93 years) (table 1) with no known mitochondrial myopathies. The biopsies (∼1 g of tissue) were collected within 5 h postmortem, during the course of normal autopsy, at the University of Wisconsin Hospital and Clinics. Mitochondrial enzymes in muscle tissue retain activity for at least 5 h postmortem and for at least 10 years when frozen. The tissue was transversely cut into small pieces, was placed in optimal cutting temperature mounting media (Miles), was flash frozen in liquid nitrogen, and was stored at −80°C. Before cryosectioning, samples were brought to −20°C. Two hundred serial sections, 10 μm thick, were cut using a cryostat and were placed on labeled ProbeOn Plus microscope slides (Fisher Scientific).
Table 1. .
Age Distribution, BMI Values, and Medical History of Study Subjects
| Subject | Sex | Age at Death(years) | BMIa | Medical History and Autopsy Finding(s) |
|---|---|---|---|---|
| 1 | Fb,c | 49 | 40.3 | Cardiovascular disease, lung disease, and hyperthyroidism |
| 2 | Fb,c,d | 49 | 39.1 | Cardiovascular disease and kidney failure |
| 3 | Fd | 58 | 60.6 | Lung disease |
| 4 | Fc | 60 | 21.7 | Thyroid adenoma and brain tumor |
| 5 | Mc,d | 64 | 35.4 | Cardiovascular disease and pulmonary edema |
| 6 | Mb | 67 | 27.6 | Diabetes, nephropathy, and cardiovascular disease |
| 7 | Mb,c | 76 | 23.1 | Cardiovascular disease and nephropathy |
| 8 | Mc,d | 79 | 21.9 | Cardiovascular disease and lung disease |
| 9 | Fb,c,d | 83 | 32.0 | Cardiovascular disease |
| 10 | Mb | 92 | NA | Cardiovascular disease, lung disease, and dementia |
| 11 | Fb,c,d | 92 | 18.7 | Cardiovascular disease and leukemia |
| 12 | Fc,d | 93 | 22.0 | Cardiovascular disease and nephropathy |
Histochemical staining for COX and SDH enzyme activities was performed on serial sections, to identify fibers containing ETS-abnormal regions.28,29 Enzyme incubation media were prepared as described by Lee et al.30 The first slide was stained for COX activity, the second slide was stained for SDH activity, and the third slide was sequentially dual-stained for both COX and SDH activity. Slides 4–7 were used for the mtDNA-deletion study. Slides 8, 9, and 10 were stained again for enzyme activities (COX, SDH, and dual-stained, respectively). This staining pattern was repeated for 200 slides from each sample. After staining, slides were rinsed in distilled water and were mounted with aqueous media (Aqua Poly/Mount [Polysciences]).
Microscopy was performed on an Olympus BH2 microscope equipped with Hitachi 3-chip CCD camera (Hitachi) under bright field illumination. ImagePro Plus software (Media Cybernetics) was used to generate composite images of tissue sections by digitally overlapping images captured at 4× magnification. Each muscle fiber in the section was digitally marked using Adobe Photoshop (Adobe Systems), and fibers displaying ETS enzymatic abnormalities (COX−/SDH++) were identified. The total fiber count was determined using ImagePro Plus software.
Laser Capture Microdissection
The PixCell II laser capture microdissection (LCM) system (Arcturus Bioscience) was used, as described by Cao et al.21 and Gokey et al.,31 to isolate 10-μm thick ETS-abnormal and ETS-normal regions of single fibers. Before microdissection, the tissue sections were subjected to an ethanol and xylene dehydration series for 10–15 min at each step. Any additional noncaptured material was carefully removed using the Cap-Sure pad (Arcturus Engineering). LCM settings included a laser-spot size of 15 μm, a pulse power of 60 mW, and a pulse width of 50 ms.
DNA Isolation and Amplification
Total DNA was extracted from individual laser-captured skeletal muscle–fiber sections with use of the protocol described by Khrapko et al.32 In brief, 1 μl of digestion solution containing 2 mg/ml proteinase K, 0.5% sodium dodecyl sulfate, and 10 mM EDTA was added to each section of skeletal-muscle fiber and was incubated at 37°C for 30 min in a humidified chamber. Subsequently, 10 μml of double-distilled H2O (ddH2O) was added to the original 1 μml of digestion solution. Of the DNA solution from individual fibers, 1 μl was used for the PCR. Primers used for PCR mtDNA analyses are summarized in table 2_A_. Short-extension PCR (94°C for 5 min for “hot start” and 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 2.5 min) (primers F588–R2737) with Taq DNA polymerase (Promega) was employed to amplify small mtDNA fragments, to confirm the presence of mtDNA in the sample. The Expand Long Template PCR System (Roche) was used to amplify the whole mtDNA genome (94°C for 5 min for hot start, and 35 cycles at 94°C for 30 s, 62°C for 30 s, and 68°C for 16 min) (primers F696 and R16550). Nested PCR was used when necessary (94°C for 5 min for hot start, and 35 cycles at 94°C for 30 s, 60°C for 30 s, and 70°C for 5 min) (primers F1775 and R16424). When an amplification product was obtained, subsequent PCRs were performed using primers internal to the initial set, to define the region containing the deletion breakpoints. The amplification products were then gel-purified and directly sequenced using BigDye terminator cycle sequencing on an ABI 3700 capillary-based DNA analyzer (Applied Biosystems International) at the University of Wisconsin–Madison DNA Sequencing Facility.
Table 2. .
Primers Used to Identify mtDNA Mutations and Primers Used in Quantitative PCR[Note]
| A. Primers used to identify mtDNA deletion mutations | |||
|---|---|---|---|
| Primersa | Primer Sequence(5′→3′) | Nucleotide Location | |
| F588 | TTACCTCCTCAAAGCAATACACTG | 588–602 | |
| R2737 | AGCTCCATAGGGTCTTCTCGTCTT | 2737–2713 | |
| F696 | TGCAAGCATCCCCGTTCC | 696–714 | |
| F1775 | AGATATAGTACCGCAAGGGAAAGA | 1775–1799 | |
| R16550 | AGGGGAACGTGTGGGCTATTTAGGC | 16550–16525 | |
| R16424 | ATATTGATTTCACGGAGGATGGTG | 16424–16400 | |
| F8111 | CCGGACGTCTAAACCAAACCACTT | 8111–8135 | |
| R13765 | GGGGATGCGGGGGAAATGT | 13765–13746 | |
| B. Primers used in quantitative PCR to measure the amount of mtDNA-deletion mutations and wild-type mtDNA | |||
| Primersa | Primer Sequence(5′→3′) | Nucleotide Location | Deletion |
| F7349 | GTCCTAATAGTAGAAGAACCCTCC | 7349–7373 | 4989 |
| R12465 | AGGTGGATGCGACAATGG | 12465–12448 | |
| F6256 | GGAGGCCGGAGCAGGAAC | 6256–6274 | 7664 |
| R14144 | AGGATTGGGATGAGGATTAGTGT | 14144–14122 | |
| F8369 | CCCAACTAAATACTACCGTATGGC | 8369–8394 | 4977 |
| R13523 | CAAAGATGAGGTTTCTGGTGTAGTAG | 134523–13498 | |
| F12354 | ATAACCACCCTAACCCTGACTTCC | 12354–12378 | Wild type |
| R12573 | GTTTGTTGGGTCGAGAGGGATTCGA | 12573–12549 |
Quantitative PCR Analysis
Quantitative PCR (i.e., real-time PCR) was performed to determine the abundance of full-length and truncated mtDNA in individual sections of COX−/SDH++ abnormal fibers. Quantitative PCR was performed in an iCycler iQ Real Time Detection System instrument with 12.5 μl of iQ SYBR Green Supermix (Bio-Rad Laboratories), 0.5 μl of of each primer (10 μM), 0.5 μl of the DNA solution from individual fibers, and ddH2O to a final volume of 25 μl. Primers used for quantitative PCR are listed in table 2_B_. The primer sequences for a specific deletion-containing genome were based on the unique sequence of each mtDNA-deletion mutation determined from the breakpoint analysis. Deletion-specific primer sets flank the breakpoints such that amplification of the deletion-containing genomes yields an amplification product of ∼200 bp. The short extension time (∼20 s) in the elongation phase of the PCR emphasizes the specific amplification of deletion-containing genomes. The wild-type primer set was designed to amplify a region of the mtDNA within the deleted portion of the genome, to ensure that the wild-type primers amplify only full-length genomes and exclude deletion-containing genomes. The mtDNA from the laser-captured fibers was amplified in parallel with cloned standards that were generated using pGEM T-easy plasmid (Promega). Concentrations of the standards were determined by spectrophotometry at A260, and quantitative PCR with regression analysis was used to determine the accuracy of dilutions. A standard curve was generated for each primer set, and the starting quantities of full-length and deletion-containing mitochondrial genomes were calculated. Melting-curve analysis and gel electrophoresis were used to confirm the specificity of the amplification reactions and the absence of nonspecific products.
Statistical Analysis
SigmaStat 2.0 (SPSS) was used for statistical analysis. One-way analysis of variance (ANOVA) was used to determine significance between deletion sizes on the basis of the age groups. Differences were considered significant at P<.05.
Results
Extent of ETS Abnormalities and Structural Atrophy in Skeletal-Muscle Fibers of Aging Humans
VL muscle sections from six individuals (subjects 2, 6, 7, 9, 10, and 11) (table 1), with an age range of 49–92 years, were sectioned and stained for COX activity and SDH activity and were sequentially dual-stained for COX and SDH activities. ETS-abnormal fibers were identified in all analyzed human VL biopsies. Determination of abundance of COX−/SDH++ abnormalities requires extensive analyses along the length of an affected muscle fiber. Two hundred consecutive sections of tissue were analyzed from muscle biopsies for three representative ages (49, 67, and 92 years; subjects 2, 6, and 11, respectively) (table 1). The total fiber number present in each biopsy was determined, and the percentage of the COX−/SDH++ fibers within the 2,000 μm of tissue was calculated. These ETS abnormalities exhibited a mosaic and segmental distribution along the length of the fibers in the biopsy. Of the 18,367 fibers in the 49-year-old subject, 36 (0.2%) exhibited an ETS-abnormal phenotype within the 2,000 μm examined. An ETS-abnormal phenotype was exhibited in 65 (0.7%) of the 9,285 fibers in the 67-year-old individual and in 98 (0.92%) of the 10,652 fibers in the 92-year-old individual. The length of ETS-abnormal regions had a range of 170–800 μm in the 49-year-old individual, of 100–800 μm in 67-year-old individual, and of 660 to >2,000 μm in the 92-year-old individual.
To determine whether ETS-abnormal fibers exhibit intrafiber atrophy, as is the case with rat and rhesus monkey, the CSA of both COX−/SDH++ and ETS-normal fibers was measured throughout the 2,000 μm of sectioned tissue. The CSA ratio provides an estimate of the severity of intrafiber atrophy and was calculated by dividing the minimum CSA value in the COX−/SDH++ region by the average CSA value of the normal region within the same fiber. Fibers with ETS-abnormal regions that were contained within the 2,000 μm of sectioned muscle were analyzed (24 abnormal fibers from the 49-year-old subject, 20 abnormal fibers from the 67-year-old subject, and 18 fibers from the 92-year-old subject). Intrafiber atrophy (CSA ratio <0.5) was not observed in any of the analyzed ETS-abnormal fibers from the 49-year-old subject. Of the ETS-abnormal fibers from the 67-year-old and 92-year-old individuals, 5% exhibited intrafiber atrophy.
mtDNA-Deletion Mutations Colocalize with ETS-Abnormal Regions
LCM was used to isolate 10-μm thick sections of single VL fibers from both ETS-normal and ETS-abnormal regions. Forty-eight fibers exhibiting the COX−/SDH++ phenotype and 20 randomly selected ETS-normal fibers were captured. Deletion mutations were not identified in sections obtained from ETS-normal fibers. PCR analysis identified mtDNA-deletion mutations in all ETS-abnormal regions of the examined fibers (fig. 1 and table 3). Every ETS-abnormal fiber amplified a single, smaller-than-wild-type product by long-extension PCR. The size of the deletion generally differed from fiber to fiber and varied from 154 bp to 12,808 bp, which resulted in truncated mtDNA genomes of 16,415 bp to 3,761 bp (table 3). No significant difference (_P_>.05) was observed in the size of the deletion between individuals of different ages (49–92 years).
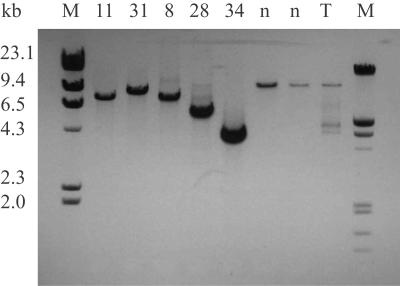
Figure 1. .
Detection of mtDNA-deletion mutations by major arc mtDNA-genome amplification. Total DNA from microdissected COX−/SDH++ fibers was subjected to long-extension PCR. mtDNA-specific primers (F4840–R766) that amplify ∼13 kb of the full-length genome, including two origins of replication, were used. Full-length amplification products were detected in ETS-normal fibers (n). Smaller amplification products were identified in ETS-abnormal fibers (8, 11, 28, 31, and 34). Amplification of total mtDNA from tissue homogenates (T) detected both full-length and smaller amplification products. M = DNA molecular-weight standards.
Table 3. .
Nucleotide-Breakpoint Location and Deletion Size[Note]
| Subjecta(Age [in years], Sex) and Fiber Number | Nucleotide Breakpoint | Deletion Size(bp) |
|---|---|---|
| 1 (49, F): | ||
| 1 | 7478–5–11439 | 3,961 |
| 2 | 11513–––13809 | 2,296 |
| 3 | 11738–––15571 | 3,833 |
| 4 | 6862–––13060 | 6,198 |
| 5 | 5786–––13923 | 8,137 |
| 6 | 6340–16–14004 | 7,664 |
| 7 | 5786–––15575 | 9,789 |
| 2 (49, F): | ||
| 8 | 9799–––15556 | 5,757 |
| 9 | 9920–––16070 | 6,149 |
| 10 | 6427–––14100 | 7,672 |
| 11 | 9665–––15182 | 5,517 |
| 12 | 8562–7–13953 | 5,385 |
| 6 (67, M): | ||
| 13 | 7181–4–14560 | 7,383 |
| 14 | 9237–4–13053 | 3,812 |
| 15 | 7722–––14109 | 6,387 |
| 16 | 7164–10–14109 | 6,935 |
| 17 | 9758–7–13748 | 3,983 |
| 7 (76, M): | ||
| 18 | 4880–––16066 | 11,185 |
| 19 | 6340–16–14004 | 7,664 |
| 20 | 10928–5–15524 | 4,591 |
| 21 | 7398–––13676 | 6,276 |
| 22 | 9342–8–13145 | 3,803 |
| 23 | 7395–11–12384 | 4,989 |
| 24 | 9577–9–13952 | 4,366 |
| 9 (83, F): | ||
| 25 | 9243–7–14421 | 5,171 |
| 26 | 8466–9–14362 | 5,905 |
| 27 | 12083–4–15853 | 3,772 |
| 28 | 7413–13–14570 | 7,170 |
| 29 | 16157–7–16318 | 154 |
| 30 | 6340–16–14004 | 7,664 |
| 31 | 7849–––13473 | 5,624 |
| 32 | 8554–6–13984 | 5,424 |
| 10 (92, M): | ||
| 33 | 6939–7–15450 | 8,311 |
| 34 | 6521–9–15049 | 8,728 |
| 35 | 10492–4–14751 | 4,263 |
| 36 | 6340–16–14004 | 7,664 |
| 37 | 6329–16–13993 | 7,664 |
| 38 | 6939–4–15450 | 8,511 |
| 39 | 6939–4–15450 | 8,511 |
| 40 | 3262–––16070 | 12,808 |
| 11 (92, F): | ||
| 41 | 10969–7–15535 | 4,559 |
| 42 | 7856–––12282 | 4,426 |
| 43 | 11046–9–15850 | 4,795 |
| 44 | 10969–7–15535 | 4,559 |
| 45 | 10931–4–15541 | 4,606 |
| 46 | 5788–––16074 | 10,286 |
| 47 | 5785–––16071 | 10,288 |
| 48 | 7097–––16071 | 8,974 |
mtDNA-Deletion Mutations Present throughout the Mitochondrial Genome
Direct mtDNA sequencing determined the breakpoints of each identified deletion mutation. The majority (96%) of the mutations were the result of deletions within the major arc of mtDNA, with both origins of replication preserved. Deletion mutations present in two fibers resulted in the removal of a portion of the minor arc and the light strand origin of replication (OL) (table 4). All mutations resulted in the partial or entire removal of the mitochondrial-protein functional genes (table 4). mtDNA deletions were clonal within an ETS-abnormal region; that is, when the same fiber was sampled from different sections within the COX−/SDH++ region, the same deletion mutation, defined by mtDNA sequence analysis, was observed. Most deletion mutations were unique to a given fiber. One mtDNA deletion mutation, with 7,664 bp (mtDNA7664) removed, was identified in the ETS-abnormal region of five fibers. Another deletion mutation, mtDNA4559, was observed in two different ETS-abnormal fibers from a 92-year-old individual. A third common deletion mutation, mtDNA8511, was observed in two fibers (tables 3 and 4).
Table 4. .
Breakpoints of Deletion Mutations[Note]
| Fiber | Breakpoint Sequence | Deleted Genes |
|---|---|---|
| 1 | AGGAAGGAACCCCCCAAA(GCTGG)—(GCTGG)GTCAATAGTACTTGCCGCAG | tRNAser—ND4 |
| 2 | TATAATACGCCTCACACTCATTCTCAACCC—GCTGTCACTTTCCTAGGACTTCTAACAG | ND4 —ND5 |
| 3 | CGCCCACGGGCTTACATCCTCATTACTATT—TTCGCCTACACAATTCTCCGATCCGTCC | ND4 —Cytb |
| 4 | ATCATCGCTATCCCCACCGGCGTCAAAGT—CAGTCTCAGCCCTACTCCACTCAAGCA | COXI —ND5 |
| 5 | GAGAAGCCCCGGCAGGTTTGAAGCTGCTTC—CCTAGCATCACACACCGCACAATCCCC | tRNAcys—ND5 |
| 6 | CCTGGAG(CCTCC**G**TAGACCTAACC)—(cctcctagacctaacc) TGACTAGAA | COXI—ND5 |
| 7 | GCCCCGGCAGGTTTGAAGCTGCTTC—CCTACACAATTCTCCGATCCGTCCCTAAC | tRNAcys—Cytb |
| 8 | TTTCCGACGGCATCTACGGCTCAACATTTTT—GAATGATATTTCCTATTCGCCTACACAA | COXIII —Cytb |
| 9 | CCAAACATCACTTTGGCTTCGAAGCCGCCG—CCCATCAACAACCGCTATGTATTTCGTA | _COXIII_—HVS1 |
| 10 | ATCAATATAAAACCCCCTGCCA—CTCTCTTTCTTCTTCCCACTCATCCTA | COXI —ND5 |
| 11 | ACCTGAGCTCACCATAGTCTAATAGAA—TTACAAACTTACTATCCGCCATCCCATAC | COXIII —Cytb |
| 12 | ATTCATTGCCCCCACAATC(CTAGGCCT)—(CTAGGCCT)TCTTACGAGCCAAAACCTG | ATP6 —ND5 |
| 13 | CATCGGCGT AAATCTAACTTTCTTC(CACC)—(CACC)GCTAACAATCAATACTAAACCC | COXI —ND6 |
| 14 | CCTATCATATAGTAAAA(CCCA)—(CCCA)CCCCAGTCTCAGC | COXIII —ND5 |
| 15 | ATGCCCTTTTCCTAACACTC—TCTTCTTCCCACTCATCCTA | COXII —ND5 |
| 16 | TAAATCTAACT(TTCTTCCCAC)—(TTCTTCCCAC)TCATCCTAACCCTA | COXI —ND5 |
| 17 | AGTCTCCCTTCAC(CATTTCC)—(CATTTCC)CCCGCATCCCCCTTC | COXIII —ND5 |
| 18 | TTCTTCTCACATGACAAAAACTAGCCC—CTCACCCATCAACAACCGCTATGTATT | ND2 _—_HVS1 |
| 19 | CCTGGAG(CCTCC**G**TAGACCTAACC)—(cctcctagacctaacc) TGACTA | COXI —ND5 |
| 20 | CCCCAACCTTTTCCTCCG(ACCCC)—(ACCCC)TTAAACACCCCTCCCCAAT | ND4 —Cytb |
| 21 | AGTGACTATATGGATGCCCCCC—CCCCACCCTACTAAACCCCATTCG | COXI —ND5 |
| 22 | ACGCTCCTCATACTAGG(cc**tactaa)—(CCC**ACTAA)TCCAAACTCTAAC | COXIII —ND5 |
| 23 | CTATATGGATG(CCCCCCA**CCCT)—(ccccccat**cct)TACCACCCCGTT | COXI —ND5 |
| 24 | GGCATCACCCCGCTA(AATCCCCTA)—(AATCCCCTA)TCTAGGCCTTCTT | COXIII —ND5 |
| 25 | ATATAGTAAAACCCAGCCCA(TGACCCC)—(TGACCCC)CATGCCTCAGGATACTCCT | COXIII —ND6 |
| 26 | ATATTAAACACAAACTACCA(CCTACCTCC)—(CCTACCTCC)ATCGCTAACCCCACTAAA | ATP6 —ND6 |
| 27 | GAAAACACCCTCATGTTCATACAC(CTATC)—(CTATC)TCCCTAATTGAAAACAAAATACT | ND4 —Cytb |
| 28 | TATGGATGCCCCC(CACCC**TACCACAC)—(cacccg**accacac) CGCTAACAATCAAT | COXI —ND6 |
| 29 | TAAATACTTGACCACCTGTAGT(ACATAAA)—(ACATAAA)GCCATTTACCGTACATAGCA | _HVS_1—HVS1 |
| 30 | TGGAGCCTCC**G**(TAGACCTAACC)—(cctccTAGACCTAACC) TGACTAGAAAA | COXI —ND5 |
| 31 | CTACGCATCCTTTACATAACAGACG—TTAGCAGGAATACCTTTCCTCACAGG | COXII —ND5 |
| 32 | AATCTGTTCGCTTCATTCAT(TGCCCC)—(TGCCCC)ACTCCTCCTAGACCTAACCT | ATP6 —ND5 |
| 33 | CAGTGCTCTGAGCCCTAGGA(TTC**A**TCTT)—(ttctctt)CCTTCTCTCCTTAA | COXI —Cytb |
| 34 | AGTCCTAGCTGCTGGCATCA(ctata**ctac)—(CTATAT**TAC)GGATCATTTCTCTACTCA | COXI —Cytb |
| 35 | CCCTCATTTACATAAATATT(ATAC)—(ATAC)GCAAAACTAACCCCCTAATAA | ND4 —Cytb |
| 36 | CCTGGAG(CCTCC**G**TAGACCTAACC)—(cctcctagacctaacc) TGACTAGAA | COXI —ND5 |
| 37 | CCTGGAG(CCTCC**G**tagacctaacc)—(cctccTAGACCTAACC)TGACTAGA | COXI —ND5 |
| 38 | GCAGTGCTCTGAGCCCTAGGATTCA(TCTT)—(TCTT)CCTTCTCTCCTTAATGACATTAACA | COXI —Cytb |
| 39 | GCAGTGCTCTGAGCCCTAGGATTCA(TCTT)—(TCTT)CCTTCTCTCCTTAATGACATTAACA | COXI —Cytb |
| 40 | TAATACTAACTACCTGACTCCT(ACCCCTC)—(ACCCCTC)CCCACATCAAGCCCGAATG | tRNAleu—Cytb |
| 41 | CGCATCCTTTACATAACAGACGAGGTCAA—GCTATCCATTGGTCTTAGGCCCCAAAA | ND4 —Cytb |
| 42 | ACTCTACCTCTCTATACTA (ATCTCCCTA)—(ATCTCCCTA)ATTGAAAACAAAAT | COXII —ND4 |
| 43 | TAATACTAACTACCTGACTCCT(ACCCCTC)—(ACCCCTC)CCCACATCAAGCCCGAATG | ND4 —Cytb |
| 44 | CTGTTCCCCAACCTTTTCCTCCGAC(CCCC)—(CCCC)ACATCAAGCCCGAATGATATTTC | ND4 —Cytb |
| 45 | AGCGGCAGGTTTGAAGCTGCTTCTT—TCAACAACCGCTATGTATTTCGTACAT | ND4 —Cytb |
| 46 | AGCCCCGGCAGGTTTGAAGCTGCT—CCATCAACAACCGCTATGTATTTCGAT | OL—HVS1 |
| 47 | ATAGGAGGCTTCATTCACTGATTTCCCC—CCATCAACAACCGCTATGTATTTCGTA | OL—HVS1 |
| 48 | GATGGCAGAGCCCGGTAATCGCATAAAAC—CCCATCAACAACCGCTATGTATTTCGTAC | COXI _—_HVS1 |
The deletion breakpoints occurred throughout the genome, with the highest concentration observed in the ND5 (∼41%) (GenBank accession number 2052366), Cytb (∼33%) (GenBank accession number AAB58955), COXI (∼33%) (GenBank accession number AAB58945), and COXIII (∼16%) (GenBank accession number 2052365) genes (fig. 2). The majority of the mutations contained direct repeat sequences at the breakpoints. The size of direct-repeat sequences typically had a range of 4–16 bp in length, with one repeat deleted and the other maintained. In two fibers, the direct repeats were partially deleted (table 4). Of the repeats, ∼65% were perfect (identical sequences flanking the deletion breakpoints), whereas imperfect direct repeats (repeats differing in sequence by 1 or 2 nt) were present in other deletions (table 4).
Figure 2. .
Schematic representation of the human mtDNA-deletion mutations obtained from 48 COX−/SDH++ fibers. mtDNA-deletion mutations were detected in all 48 ETS-abnormal fibers, and breakpoints were determined by DNA sequence analysis. Arcs represent the deleted regions of the genome. The location of the light-strand (OL) and the heavy-strand (OH) origins of replication are indicated. Cyt = cytochrome.
mtDNA-Deletion Mutations Accrue to High Levels in the ETS-Abnormal Region of Human Skeletal-Muscle Fibers
The mtDNA-deletion load present in 10-μm thick sections from single LCM-dissected fibers was measured by quantitative PCR. The absolute level of deletion-containing and full-length mitochondrial genomes was determined along the length of four muscle fibers exhibiting the COX−/SDH++ phenotype. Three of the fibers (19, 36, and 37) contained the deletion mutation mtDNA7664, removing nucleotides 6340–14004, and one fiber (23) contained the deletion mutation mtDNA4989, removing nucleotides 7395–12384 of the mitochondrial genome. Total mtDNA increased in the COX−/SDH++ region, an increase resulting from the accumulation of the specific mtDNA-deletion mutation. The amount of full-length DNA did not increase in the ETS-abnormal region. Full-length genomes were detected in both ETS-normal and -abnormal regions, with the abundance typically declining in the COX−/SDH++ regions (see fig. 3).
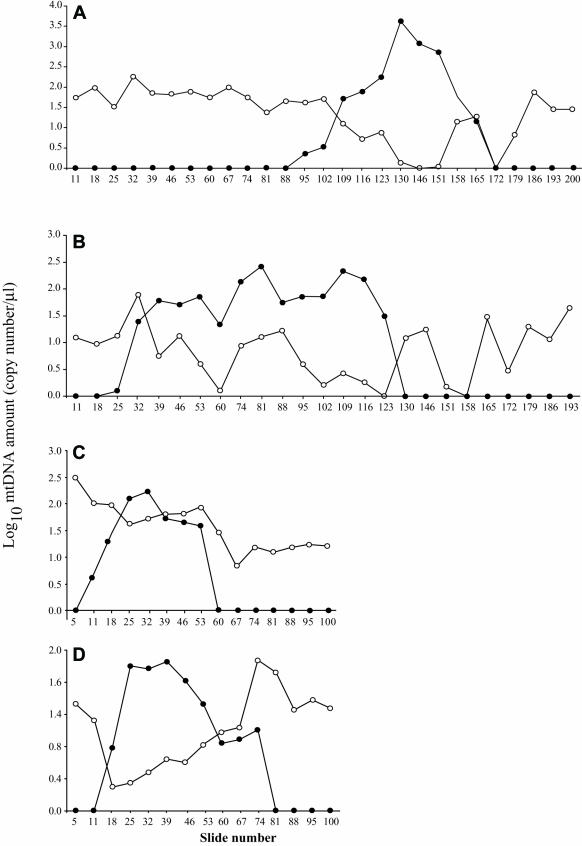
Figure 3. .
Log10 of mtDNA wild-type and partially deleted genomes along the length of fibers 19 (A), 36 (B), 37 (C), and 23 (D). Unblackened circles represent wild-type mtDNA, and blackened circles represent deletion-containing genomes.
The mtDNA deletion–mutation load approached 99% of the mtDNA within each fiber’s COX−/SDH++ abnormal region (fig. 4). An intermediate ETS phenotype (COXlow/SDHhigh) was observed in sections adjacent to the COX−/SDH++ regions. Deletion mutations were present in these transition regions, albeit at a lower abundance (30%–40%) compared with COX−/SDH++ regions. mtDNA-deletion mutations were present at a very low abundance (<20%) in the ETS-normal segments immediately adjacent to the abnormal regions, whereas, in the ETS-normal regions distant from the ETS abnormality, only wild-type genomes were observed.
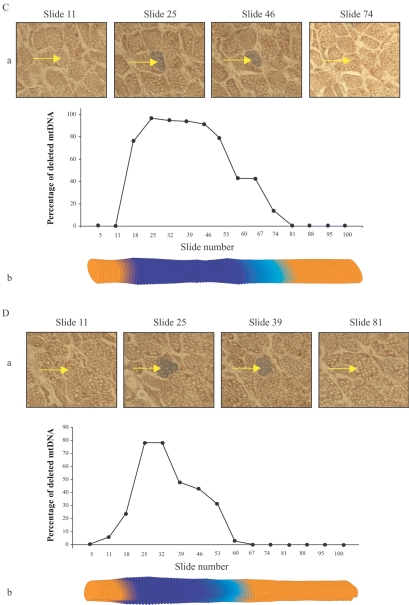
Figure 4. .
mtDNA deletion–mutation abundance along the length of COX−/SDH++ fibers. LCM was used to isolate 10-μm thick tissue sections at numerous sites along the length of single muscle fibers. Quantitative PCR defined the abundance of full-length and deleted mtDNA. A, B, and C, Percentage of deleted mtDNA7664 in the abnormal region of fibers 19, 36, and 37, respectively. D, Percentage of deleted mtDNA4989 in the abnormal region of fiber 23. a, Photomicrographs of tissue cross-sections sequentially double stained for COX and SDH activity. The individual fiber (arrow) displays an ETS-abnormal phenotype. b, Digital reconstruction of CSA silhouettes. The COX−/SDH++ region is shown in dark blue, the COXlow/SDHhigh region in light blue, and the COXnormal/SDHnormal in orange.
Discussion
For mtDNA-deletion mutations and their associated ETS abnormalities to be physiologically relevant, they must be present in a significant number of cells and accumulate to high levels intracellularly. In this study of human muscle, ETS-abnormal (COX−/SDH++) fibers were identified in all biopsy samples, three of which were extensively analyzed (200 consecutive 10-μm sections) and provided an estimate of the abundance of fibers exhibiting ETS abnormalities. Extrapolating the percentage of ETS abnormalities found in the analyzed 2,000-μm region to the entire length of VL fibers (∼6.5 cm),33 we estimate that ∼6% of muscle fibers from the 49-year-old individual, ∼22% of muscle fibers from the 67-year-old individual, and ∼31% from the 92-year-old individual contain ETS abnormalities somewhere along their length. These calculations suggest that a large number of fibers in the muscles of older subjects contain ETS abnormalities and, thus, initiate a process that will eventually lead to fiber breakage and, ultimately, fiber loss. Similar calculations in aged rats and rhesus monkeys indicate that 5%–15% and 28%–60% of the fibers, respectively, contain an ETS-abnormal phenotype somewhere along the length.4,12
The mosaic distribution and segmental nature of ETS abnormalities necessitated the analysis of mtDNA mutations from single-fiber sections. LCM was combined with PCR and direct mtDNA sequencing to define the mtDNA genotype of fibers exhibiting a COX−/SDH++ abnormal phenotype. mtDNA-deletion mutations, with removal of 1%–72% of the genome, were observed in every analyzed COX−/SDH++ region. These results contrast with those of Fayet et al.,34 which showed mtDNA mutations in 10% of the COX− fibers. However, Fayet et al.34 examined COX− fibers, whereas our study focused on fibers with a dual ETS-abnormal phenotype, COX−/SDH++.
Although mtDNA-deletion mutations accrue and accumulate with age in many species, our studies have identified a number of subtle differences in the nature of the deletion breakpoints. Direct repeat sequences are present at many of the mtDNA-deletion breakpoints in human and rhesus monkey COX−/SDH++ skeletal-muscle fibers but are rarely observed at rat mtDNA-deletion breakpoints.21,31 Interestingly, in humans, most of the mtDNA-deletion mutations with direct repeat sequences were identified in fibers from older individuals (aged 60–90 years). The majority of the deletion breakpoints in human VL muscle were located in the major arc, with both origins of replication preserved and with only two deletion mutations missing the OL. A similar loss of the OL region was observed in 28% of the mtDNA-deletion mutations present in ETS-abnormal fibers from aged rhesus monkey skeletal muscle.12,31 Deletion mutations are, however, exclusively confined within the major arc in single ETS-abnormal fibers from aged rats.4,21 Patterns of mtDNA deletions in humans, rats, and rhesus monkeys suggest species-specific differences that could be due to different life spans or different processes associated with deletion formation.
mtDNA-deletion mutations located in the hypervariable site 1 (HVS1 [nucleotide position 16024–16383, GenBank accession 1944628) of the genome were observed in four fibers. This region of the genome is thought to be a mutational hotspot region for both germline and somatic mutations of mtDNA.35 Deletions of this site were described elsewhere in patients with CPEO (MIM 530000) as well as in healthy older individuals.34,36
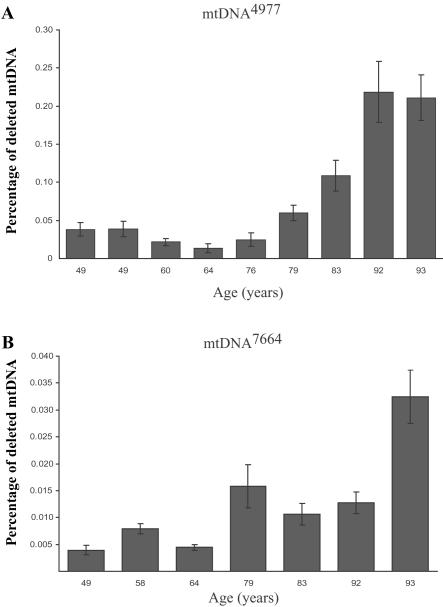
Considerable attention has been focused on the mtDNA4977 deletion mutation that causes many sporadic mitochondrial myopathies. Initial mtDNA-aging studies focused solely on this mutation and found that it accumulates with age in a variety of tissues.16,17 We did not identify mtDNA4977 in any of the analyzed individual COX−/SDH++ fibers. The deletion was, however, detected in every sample (from subjects aged 49–93 years) when tissue homogenates (tens of thousands of fibers) were analyzed (see fig. 5). Quantitative PCR analysis of tissue homogenates demonstrated that the abundance of mtDNA4977 increased with age (see fig. 6_a_). The mtDNA7664 deletion mutation, which is also linked to sporadic myopathies, was identified in five COX−/SDH++ fibers. Interestingly, in the same tissue homogenates, the abundance of the mtDNA7664 deletion mutation—which, in COX−/SDH++ fibers, was found to accumulate to high levels—was ∼10-fold lower than mtDNA4977 (see fig. 6_b_). Our inability to identify ETS-abnormal fibers containing the mtDNA4977 mutation may reflect the number of other deletion mutations that can also accumulate and trigger the COX−/SDH++ phenotype. In support of this argument, other studies have reported the presence of mtDNA4977 in a very small proportion of COX− fibers.34,37 Another possibility is that mtDNA4977 rarely accumulates to sufficiently high levels to result in an observable phenotype. Alternatively, the generation of deletion mutations could be tissue specific, since tissue homogenates contain—in addition to muscle fibers—nervous and connective tissue, blood vessels, and adipose cells.
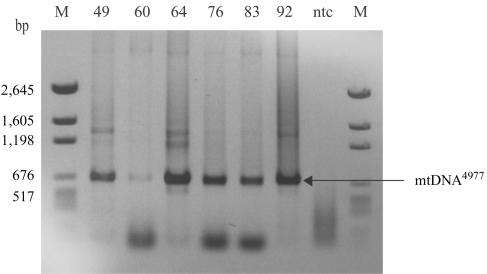
Figure 5. .
mtDNA4977 deletion mutation in tissue homogenate from aged human skeletal muscle. Total DNA was isolated from skeletal muscle–tissue homogenates of 49-, 60-, 64-, 76-, 83-, and 92-year-old human subjects. To investigate the presence of mtDNA4977 mutations in skeletal muscle–tissue homogenate at different ages, total DNA was subjected to PCR with primers (F8111–R13756) (table 2_A_) immediately flanking the deleted region. This deletion mutation was present in all analyzed tissue homogenates. In addition, 57 ETS-normal fibers (fibers that exhibited the COXnormal/SDHnormal phenotype throughout 2,000 μm of analyzed tissue) were randomly selected and analyzed for the presence of mtDNA4977 from a 49-year-old subject (20 fibers) and from a 92-year-old subject (37 fibers). mtDNA4977 was not detected in any of these fibers. M = DNA molecular-weight standards; ntc = no template control.
Figure 6. .
Accumulation, with age, of mtDNA4977 and mtDNA7664 deletion mutation in tissue homogenate. A, Primers specific for mtDNA4977 (F8369–R13498) were used to measure the amount of deletion mutation. The wild-type primer set (F12354–R12549) was designed to amplify a region of the genome within the breakpoints of the deleted mtDNA, to ensure that the primers amplified full-length wild-type genomes. The amount of mtDNA4977 deletion mutation had a range of 0.02%–0.06% of the wild-type genomes for ages 49-79 years. The deletion-mutation level significantly increased to 0.1%–0.225% of the wild-type genomes for ages 83–93 years. B, Primers specific for mtDNA7664 (F6256–R14122) were used to measure the amount of deletion mutation. Primer set F12354–R12549 was used to measure the amount of wild-type mtDNA present. The mtDNA7664 deletion mutation load fluctuated between ages 49 and 92 years. A significant (P<.05) increase was found at age 93 years.
Quantitative analysis of the abundance of mtDNA-deletion mutations along the length of individual COX−/SDH++ skeletal-muscle fibers facilitates a better understanding of the relationship between mtDNA-deletion mutations and their concomitant phenotypes: ETS abnormalities and intrafiber atrophy. Studies of mitochondrial myopathies show that ∼80%–100% of the mtDNA must be of the mutant type for there to be an observable ETS phenotype.23,38,39 We found mtDNA-deletion mutations to clonally accumulate to high levels within COX−/SDH++ regions (>80% of total mtDNA). Declining levels of the deletion-containing genomes in the transition regions and in ETS-normal regions immediately adjacent to the transition regions suggest that the accumulation of mtDNA-deletion mutations precedes the ETS-abnormal phenotype. These observations strongly support the hypothesis that mtDNA-deletion mutations play a causal role in mitochondrial and cellular dysfunction of skeletal-muscle fibers with age, a process that ultimately leads to fiber loss.
The data presented in this work further delineate the involvement of mitochondria in skeletal-muscle aging and suggest a specific order of molecular and cellular events that occur in response to the accumulation of mtDNA mutations. Sporadic mtDNA-replication errors or mtDNA-replication errors resulting from oxidative damage may cause large-scale deletions that result in the loss or truncation of mtDNA genes that encode subunits of the respiratory chain, ATP synthase, rRNA, and tRNA genes.40–44 Within a fiber, a given genome with deletion replicates to high levels, impairing the enzymatic activity of the ETS complex and altering the mitochondrial metabolism. Whereas wild-type mtDNA is present in both ETS-normal and -abnormal regions at a relatively constant level, mtDNA-deletion mutations accumulate to very high levels in COX−/SDH++ regions of muscle fibers. Our data indicate that there is a threshold of mtDNA deletion–mutation abundance necessary for the expression of an ETS-abnormal phenotype. An ETS-normal phenotype is observed when only full-length mtDNA or low levels of mtDNA-deletion mutations are present (<20%). When 20%–80% of the genomes have deletions, the fiber exhibits an intermediate ETS-abnormal phenotype, the COX−/SDHnormal phenotype. As the ratio of deletion-containing genomes increases (to >80%), the fibers manifest the COX−/SDH++ phenotype. With time, the ETS-abnormal region extends throughout the length of the fiber and results in fiber atrophy, fiber breakage, and thus fiber loss.
Acknowledgments
We thank Susan McKiernan and Chris Johnson for their critical reviews of this manuscript. This work was supported by National Institutes of Health grant AG11604.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for ND5 [accession number 2052366], Cytb [accession number AAB58955], COXI [accession number AAB58945], COXIII [accession number 2052365], and HVS-1 [accession number 1944628])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CPEO) [PubMed]
References
- 1.Rosenberg IH (1997) Sarcopenia: origins and clinical relevance. J Nutr 127:990S–991S [DOI] [PubMed] [Google Scholar]
- 2.Lexell J, Taylor CC, Sjostrom M (1988) What is the cause of the ageing atrophy? Total number, size and proportion of different fiber types studied in whole vastus lateralis muscle from 15- to 83-year-old men. J Neurol Sci 84:275–294 10.1016/0022-510X(88)90132-3 [DOI] [PubMed] [Google Scholar]
- 3.Baumgartner RN, Koehler KM, Gallanger D (1998) Epidemiology of sarcopenia among the elderly in New Mexico. Am J Epidemiol 147:755–763 [DOI] [PubMed] [Google Scholar]
- 4.Wanagat J, Cao Z, Pathare P, Aiken JM (2001) Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting and oxidative damage in sarcopenia. FASEB J 15:322–332 10.1096/fj.00-0320com [DOI] [PubMed] [Google Scholar]
- 5.Reimers CD, Harder T, Saxe H (1998) Age-related muscle atrophy does not affect all muscles and can partly be compensated by physical activity: an ultrasound study. J Neurol Sci 159:60–66 10.1016/S0022-510X(98)00134-8 [DOI] [PubMed] [Google Scholar]
- 6.Brown M, Hasser EM (1996) Complexity of age-related change in skeletal muscle. J Gerontol A Biol Sci Med Sci 51:B117–B123 [DOI] [PubMed] [Google Scholar]
- 7.Overend TJ, Cunningham DA, Paterson DH, Lefcoe MS (1992) Thigh composition in young and elderly men determined by computed tomography. Clin Physiol 12:629–640 [DOI] [PubMed] [Google Scholar]
- 8.Ishihara A, Naitoh H, Katsuta S (1987) Effects of ageing on the total number of muscle fibers and motoneurons of the tibialis anterior and soleus muscles in the rat. Brain Res 435:355–358 10.1016/0006-8993(87)91624-6 [DOI] [PubMed] [Google Scholar]
- 9.Cartee GD, Bohn EE, Gibson BT, Farrar RP (1996) Growth hormone supplementation increases skeletal muscle mass of old male Fischer 344/Brown Norway rats. J Gerontol A Biol Sci Med Sci 51:B214–B219 [DOI] [PubMed] [Google Scholar]
- 10.Bua EA, McKiernan SH, Wanagat J, McKenzie D, Aiken JM (2002) Mitochondrial abnormalities are more frequent in muscles undergoing sarcopenia. J Appl Physiol 92:2617–2624 [DOI] [PubMed] [Google Scholar]
- 11.McKiernan SH, Bua E, McGorray J, Aiken JM (2004) Early-onset caloric restriction conserves fiber number in aging skeletal muscle. FASEB J 18:580–581 [DOI] [PubMed] [Google Scholar]
- 12.Lopez ME, VanZeeland NL, Dahl DB, Weindruch R, Aiken JM (2000) Cellular phenotypes of age-associated skeletal muscle mitochondrial abnormalities in rhesus monkeys. Mutation Res 452:123–138 [DOI] [PubMed] [Google Scholar]
- 13.Cortopassi GA, Shibata D, Soong NW, Arnheim NA (1992) Pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc Natl Acad Sci USA 89:7370–7374 10.1073/pnas.89.16.7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Baumer A, Maxwell RJ, Linnane AW, Nagley P (1992) Multiple mitochondrial DNA deletions in the elderly human individual. FEBS Lett 297:34–38 10.1016/0014-5793(92)80321-7 [DOI] [PubMed] [Google Scholar]
- 15.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K (2006) Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet 38:518–520 10.1038/ng1778 [DOI] [PubMed] [Google Scholar]
- 16.Linnane AW, Baumer A, Maxwell RJ, Preston H, Zhang CF, Marzuki S (1990) Mitochondrial gene mutation: the ageing process and degenerative diseases. Biochem Int 22:1067–1076 [PubMed] [Google Scholar]
- 17.Cortopassi GA, Arnheim N (1990) Detection of specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res 18:6927–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melov S, Shoffner JM, Kaufman A, Wallace DC (1995) Marked increase in the number and variety of mitochondrial DNA rearrangements in aging human skeletal muscle. Nucleic Acids Res 23:4122–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper JM, Mann VM, Schapira AHV (1992) Analyses of mitochondrial respiratory chain function and mitochondrial DNA deletion in human skeletal muscle: effect of aging. J Neurol Sci 113:91–98 10.1016/0022-510X(92)90270-U [DOI] [PubMed] [Google Scholar]
- 20.Müller-Höcker J, Pongratz D, Hubner G (1983) Focal deficiency of cytochrome-c-oxidase in skeletal muscle of patients with progressive external ophthalmoplegia: cytochemical-fine-structural study. Virchows Arch A Pathol Anat Histopathol 402:61–71 10.1007/BF00695049 [DOI] [PubMed] [Google Scholar]
- 21.Cao Z, Wanagat J, McKiernan SH, Aiken JM (2001) Mitochondrial DNA deletion mutations are concomitant with ragged red regions of individual, aged muscle fibers: analysis by laser-capture microdissection. Nucleic Acids Res 29:4502–4508 10.1093/nar/29.21.4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarze SR, Lee CM, Chung SS, Roecker EB, Weindruch R, Aiken JM (1995) High levels of mitochondrial DNA deletions in skeletal muscle of old rhesus monkeys. Mech Ageing Dev 83:91–101 10.1016/0047-6374(95)01611-3 [DOI] [PubMed] [Google Scholar]
- 23.Shoubridge EA, Karpati G, Hastings KE (1990) Deletion mutants are functionally dominant over wild-type mitochondrial genomes in skeletal muscle fiber segments in mitochondrial disease. Cell 62:43–49 10.1016/0092-8674(90)90238-A [DOI] [PubMed] [Google Scholar]
- 24.Müller-Höcker J, Schneiderbanger K, Stefani FH, Kadenbach B (1992) Progressive loss of cytochrome c oxidase in the human extraocular muscles in ageing—a cytochemical-immunohistochemical study. Mutat Res 275:115–124 [DOI] [PubMed] [Google Scholar]
- 25.Müller-Höcker J, Seibel P, Schneiderbanger K, Kadenbach B (1993) Different in situ hybridization patterns of mitochondrial DNA in cytochrome c oxidase-deficient extraocular muscle fibres in the elderly. Virchows Arch A Pathol Anat Histopathol 422:7–15 10.1007/BF01605127 [DOI] [PubMed] [Google Scholar]
- 26.Rifai Z, Welle S, Kamp C, Thornton C (1995) Ragged red fibers in normal aging and inflammatory myopathy. Ann Neurol 37:24–29 10.1002/ana.410370107 [DOI] [PubMed] [Google Scholar]
- 27.Bua E, McKiernan SH, Aiken JM (2004) Calorie restriction limits the generation but not the progression of mitochondrial abnormalities in aging skeletal muscle. FASEB J 18:582–584 [DOI] [PubMed] [Google Scholar]
- 28.Seligman AM, Karnovsky MJ, Wasserkrug HL, Hanker JS (1968) Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J Cell Biol 38:1–14 10.1083/jcb.38.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubowitz V (1985) Muscle biopsy: a practical approach. Bailliere Tindall, London, pp 19–40 [Google Scholar]
- 30.Lee CM, Lopez ME, Weindruch R, Aiken JM (1998) Association of age-related mitochondrial abnormalities with skeletal muscle fiber atrophy. Free Radic Biol Med 25:964–972 10.1016/S0891-5849(98)00185-3 [DOI] [PubMed] [Google Scholar]
- 31.Gokey NG, Cao Z, Pak JW, Lee D, McKiernan SH, McKenzie D, Weindruch R, Aiken JM (2004) Molecular analyses of mtDNA deletion mutations in microdissected skeletal muscle fibers from aged rhesus monkeys. Aging Cell 3:319–326 10.1111/j.1474-9728.2004.00122.x [DOI] [PubMed] [Google Scholar]
- 32.Khrapko K, Bodyak N, Thilly WG, Van Orsouw NJ, Zhang X, Coller HA, Perls TT, Upton M, Vijg J, Wei JY (1999) Cell-by-cell scanning of whole mitochondrial genomes in aged human heart reveals a significant fraction of myocytes with clonally expanded deletions. Nucleic Acids Res 27:2434–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickiewicz TL, Roy RR, Powell PL, Edgerton VR (1983) Muscle architecture of the human lower limb. Clin Orthop Relat Res 179:275–283 [PubMed] [Google Scholar]
- 34.Fayet G, Jansson M, Sternberg D, Moslemi AR, Blondy P, Lombes A, Fardeau M, Oldfors A (2002) Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul Disord 12:484–493 10.1016/S0960-8966(01)00332-7 [DOI] [PubMed] [Google Scholar]
- 35.Stoneking M (2000) Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet 67:1029–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moraes CT, Andreetta F, Bonilla E, Shanske S, DiMauro S, Schon EA (1991) Replication-competent human mitochondrial DNA lacking the heavy-strand promoter region. Mol Cell Biol 11:1631–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brierley EJ, Johnson MA, Lightowlers RN, James OF, Turnbull DM (1998) Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann Neurol 43:217–223 10.1002/ana.410430212 [DOI] [PubMed] [Google Scholar]
- 38.Schon EA, Hirano M, DiMauro S (1994) Mitochondrial encephalomyopathies: clinical and molecular analysis. J Bioenerg Biomembr 26:291–299 10.1007/BF00763100 [DOI] [PubMed] [Google Scholar]
- 39.He L, Chinnery PF, Durham SE, Blakely EL, Wardell TM, Borthwick GM, Taylor RW, Turnbull DM (2002) Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res 30:e68 10.1093/nar/gnf067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung SS, Weindruch R, Schwarze SR, McKenzie D, Aiken JM (1994) Multiple age-associated mitochondrial DNA deletions in skeletal muscle of mice. Aging 6:193–200 [DOI] [PubMed] [Google Scholar]
- 41.Copeland WC, Ponamarev MV, Nguyen D, Kunkel TA, Longley MJ (2003) Mutations in DNA polymerase gamma cause error prone DNA synthesis in human mitochondrial disorders. Acta Biochim Pol 50:155–167 [PubMed] [Google Scholar]
- 42.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly M, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429:417–423 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- 43.Adachi K, Fujiura Y, Mayumi F, Nozuhara A, Sugiu Y, Sakanashi T, Hidaka T, Toshima H (1993) A deletion of mitochondrial DNA in murine doxorubicin-induced cardiotoxicity. Biochem Biophys Res Commun 195:945–951 10.1006/bbrc.1993.2135 [DOI] [PubMed] [Google Scholar]
- 44.Prithivirajsingh M, Story S, Bergh F, Geara K, Kian Ang S, Ismail C, Stevens T, Buchholz W (2004) Accumulation of the common mitochondrial DNA deletion induced by ionizing radiation. FEBS Lett 571:227–232 10.1016/j.febslet.2004.06.078 [DOI] [PubMed] [Google Scholar]