Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex (original) (raw)
Abstract
BRCA1 is a breast and ovarian cancer tumor suppressor protein that associates with BARD1 to form a RING/RING heterodimer. The BRCA1/BARD1 RING complex functions as an ubiquitin (Ub) ligase with activity substantially greater than individual BRCA1 or BARD1 subunits. By using NMR spectroscopy and site-directed mutagenesis, we have mapped the binding site on the BRCA1/BARD1 heterodimer for the Ub-conjugating enzyme UbcH5c. The results demonstrate that UbcH5c binds only to the BRCA1 RING domain and not the BARD1 RING. The binding interface is formed by the first and second Zn2+-loops and central α-helix of the BRCA1 RING domain, a region disrupted by cancer-predisposing mutations. Unexpectedly, a second Ub-conjugating enzyme, UbcH7, also interacts with the BRCA1/BARD1 complex with similar affinity, although it is not active in Ub-ligase activity assays. Thus, binding alone is not sufficient for BRCA1-dependent Ub-ligase activity.
The breast and ovarian cancer tumor-suppressor protein BRCA1 has been implicated in a variety of cellular processes such as transcriptional regulation, cell-cycle control, and the cellular response to DNA damage (1–4). Consistent with such functional diversity, >30 protein partners for BRCA1 have been reported and BRCA1 has been found as a component in at least four distinct macromolecular complexes (1, 5–9). The best characterized BRCA1 interaction involves the complex formed with BARD1 (10, 11), a protein that also interacts with CstF-50, an mRNA polyadenylation factor (12, 13). In vivo, BRCA1 and BARD1 colocalize in nuclear dots during S-phase of the cell cycle and in nuclear foci that form in response to DNA damage (14, 15).
BRCA1 and BARD1 each contain a C-terminal BRCT domain and an N-terminal RING domain. The RING domains govern the tight association between the two proteins through the formation of an extensive four-helix-bundle dimerization interface (16). Formation of the BRCA1/BARD1 complex is important for their mutual stability in vivo (17, 18), their proper nuclear localization (19), and for maximal ubiquitin (Ub)-ligase (E3) activity (17, 20, 21). The demonstration that the BRCA1 RING domain functions as an E3 is of particular importance given the central role of ubiquitination for eukaryotic cell viability. Defects in ubiquitination affect a host of cellular processes including cell-cycle progression, cell differentiation, apoptosis, the response to DNA damage, DNA repair, and transcriptional regulation (22–24).
Many cellular processes rely on polyubiquitination as a signal for programmed protein destruction. Proteins tagged with a Lys-48-linked polyUb chain of four or more subunits are destined for proteosomal degradation. Ubiquitination may also serve as a regulatory signal governing protein trafficking or activity (22–24). Monoubiquitination plays a role in the regulation of histones, endocytosis, and retroviral budding (24), whereas Lys-63-linked Ub chains have been found to function in DNA repair, endocytosis, ribosome function, and the stress response (23). Recent results suggest that BRCA1 promotes the formation of non-Lys-48-linked Ub chains, implying that BRCA1-mediated ubiquitination may function as a regulatory signal (20, 21). Although the in vivo substrate(s) is not yet known, BRCA1 has been observed to undergo autoubiquitination and is capable of monoubiquitinating histones 2A and 2AX in vitro (20, 25), consistent with a role in DNA repair. In addition, a connection between BRCA1 and the monoubiquitination of the Fanconi anemia protein, FANCD2, has been suggested, based on colocalization studies and the similarity of mutant phenotypes in response to ionizing radiation (26).
All ubiquitination pathways share common steps that require the action of a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and an E3 (22). Generally, eukaryotic cells contain a single type of E1, multiple E2s, and a large number of E3s (27). BRCA1 belongs to the subset of RING-domain proteins that facilitate Ub transfer reactions and comprise the largest class of recognized Ub ligases (28, 29). The isolated BRCA1 RING domain is active in substrate-independent polyubiquitination reactions (25, 29). However, formation of the BRCA1/BARD1 complex dramatically enhances this activity (17, 20, 21). E3s are thought to facilitate ubiquitination by binding both an activated E2∼Ub complex and the target substrate. Clues to factors that govern macromolecular assembly and activity of RING E3/E2 complexes are beginning to emerge from various mutagenesis studies (17, 20, 25, 30), and structural studies on systems such as cCbl and CNOT4, in addition to BRCA1 (31, 32). BRCA1, however, is unusual among RING E3s in that a complex formed between two heterologous RING domains is required for maximal activity (17).
To define the roles each RING subunit plays in orchestrating Ub-transfer reactions, we have mapped the interactions between BRCA1/BARD1 and the E2 UbcH5c using NMR spectroscopy and mutagenesis. UbcH5c has been demonstrated to function in concert with BRCA1 in vitro (17, 29). NMR is well suited for delineating such protein interactions: protein chemical shifts are highly sensitive to environment, so chemical-shift perturbations that occur on addition of a partner protein are a sensitive indicator of residues involved in the binding interface (33). NMR titrations of BRCA1/BARD1 heterodimer and UbcH5c reveal that UbcH5c interacts only with the BRCA1 subunit of the RING E3 complex. The UbcH5c-binding surface on BRCA1 is composed of residues in the first Zn2+-loop, the second Zn2+-loop, and the central helix of the RING. The binding interface was confirmed by comparing the Ub-ligase activity of BRCA1 mutants altered specifically at the UbcH5c-binding interface. Unexpectedly, similar NMR experiments using the E2 UbcH7 revealed binding to the BRCA1 subunit with similar affinity and in the same region as UbcH5c. However, UbcH7 does not function with BRCA1 in substrate-independent ligase activity assays. Thus, binding of an E2 to an E3 protein is necessary, but is not sufficient for catalytic activity.
Materials and Methods
Protein Expression and Purification.
Plasmids used for expression of wild-type proteins [myc-BRCA1(1–772), HA-BARD1, His6-BRCA1(1–304), His6-BARD1(14–189), His6-UbcH5c, and His6-UbcH7] were identical to those described (17, 34, 35). Plasmids used for the bacterial coexpression of His6-BRCA1(1–112) and BARD1(26–140) were based on PET28N and pCOT7N expression systems (generous gift of Michael Wittekind, Bristol-Myers Squibb). BRCA1 point mutations were introduced by site-directed mutagenesis (Stratagene).
Bacterially coexpressed His6-wt-BRCA1/BARD1, His6-I26A-BRCA1/BARD1, His6-UbcH5c, and His6-UbcH7 used for NMR studies were produced in BL21/DE3 bacteria grown on either rich LB medium or minimal M9 medium supplemented with [15N]ammonium chloride (Isotec). Proteins were purified by using Ni2+-affinity chromatography, according to the manufacturer's instructions (Sigma). His6-tags were removed from BRCA1/BARD1 heterodimers by thrombinolysis, according to the manufacturer's instructions (Novagen). Final purification was achieved by gel-filtration chromatography on a Sephacryl S100 column (Amersham Pharmacia) equilibrated with 25 mM sodium phosphate buffer, pH 7.0, with 0.15 M NaCl. All other proteins were purified as described (17, 34).
Ub-Ligase Activity Assays.
Details of immunoprecipitate Western blot and Ub-ligase activity assays are described in Hashizume et al. (17).
NMR Spectroscopy.
Titration of uniformly 15N-labeled wild-type (wt)- and I26A-BRCA1/BARD1 heterodimers (initial concentration 0.35 mM) with unlabeled wt-UbcH5c or wt-UbcH7 were performed at 35°C in 25 mM sodium phosphate buffer, pH 7.0, containing 0.15 M NaCl and 10% D2O. Sequential addition of 2.5 mM wt-UbcH5c or wt-UbcH7 to 0, 0.125, 0.25, 0.4, 0.5, 0.75, and 1.05 equivalents of E2 to BRCA1/BARD1 were made. 1H–15N transverse-relaxation optimized spectroscopy (TROSY) spectra were recorded on an INOVA 600 MHz spectrometer (Varian). Spectra were processed and analyzed by using nmrpipe (36) and nmrview (37).
Results
Mapping the Binding Interface for UbcH5c on the BRCA1/BARD1 RING-Domain Heterodimer.
In an NMR titration experiment, one protein component is labeled with an NMR-sensitive nucleus (i.e., 15N), whereas the titrant is unlabeled. Successive NMR spectra are recorded with each protein addition. This approach is particularly suited to study weakly interacting systems that are often difficult to study by other techniques that measure binding. To map the surface of the heterodimer that binds UbcH5c, BRCA1 and/or BARD1 were labeled with 15N in one of two ways: (i) the BRCA1 and BARD1 subunits were expressed and purified individually (34, 35) or (ii) the heterodimer complex was purified directly from a bacterial coexpression system. As previously reported, bacterial overexpression of BRCA1(1–112) or BARD1(1–115) alone results in the formation of inclusion bodies and requires refolding of the heterodimer complex. This method has the advantage that NMR-active nuclei can be incorporated selectively into only one subunit of the heterodimer, simplifying interpretation of NMR spectra. Alternatively, coexpression of subunits provides simultaneous labeling of each subunit. In this case, a soluble complex is expressed at high levels, which makes it easily purified without a refolding step. Both methods yield BRCA1/BARD1 RING domain heterodimers with identical 1H-15N TROSY spectra that are fully active in Ub-ligase activity assays (see below).
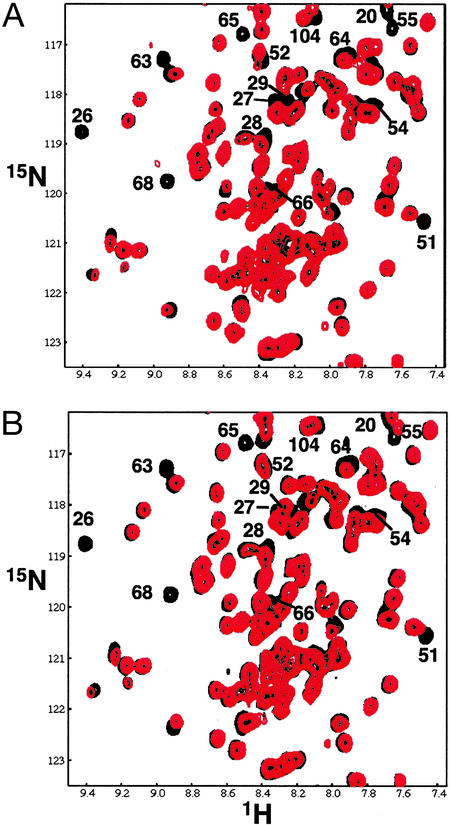
Fig. 1A shows the results of titrating unlabeled UbcH5c into a solution of [15N]BRCA1/[15N]BARD1 RING heterodimer. Regions of the spectra where BRCA1 and BARD1 resonances overlap were deciphered from titration experiments in which only the BRCA1 subunit was labeled (see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). At low concentrations of UbcH5c, a subset of resonances either broaden and shift or disappear completely. Several points regarding the interaction can be surmised from the NMR spectra: (i) Resonances that broaden and disappear are likely from residues at the binding interface. (ii) This behavior places the interaction between UbcH5c and BRCA1/BARD1 in the intermediate to fast-intermediate exchange regime on the NMR time scale. Thus, the complex formed between BRCA1/BARD1 and UbcH5c is not particularly strong (_K_d ≥ 1 μM). (iii) At higher UbcH5c concentrations there is a general broadening of all resonances, consistent with the BRCA1/BARD1-UbcH5c complex being the predominant form when excess UbcH5c is present.
Figure 1.
NMR titrations of E2 binding to the BRCA1/BARD1 heterodimer. Overlay of an expanded region of 2D 1H–15N TROSY spectra of 15N-labeled BRCA1/BARD1 heterodimer with 0 (black spectra) and 0.5 (red spectra) equivalent unlabeled UbcH5c (A) or UbcH7 (B). Resonances significantly perturbed on the addition of UbcH5c (K20, I26, C27, L28, L51, L52, Q54, K55, L63, C64, K65, N66, I68, and S104 of BRCA1) are labeled. Other perturbed resonances (C24, K45, F46, C61, and A102) lie outside this spectral region.
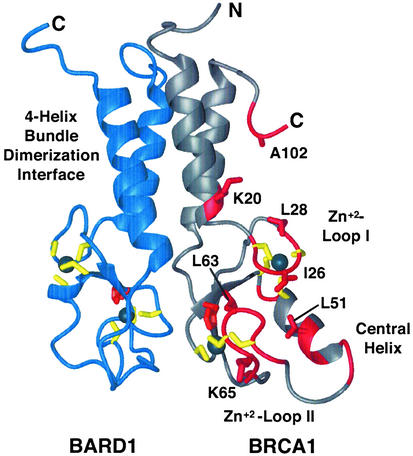
Significantly, no resonances in BARD1 are perturbed by UbcH5c. Thus, UbcH5c binds only to the BRCA1 subunit of BRCA1/BARD1 RING complex. Mapping the affected residues onto the 3D structure of BRCA1 (Figs. 2 and 3A) reveals that they delineate a contiguous region on the surface of BRCA1. These residues are located in the first Zn2+-binding loop (residues 23–29), the second Zn2+-binding loop (residues 58–68), and the central helix of BRCA1 (residues 46–53), structural elements that form a pronounced cleft on the surface of BRCA1 (16). The cleft to which UbcH5c binds is similar to surface features, which are found on the RING domains cCbl and CNOT4, RING proteins that interact with UbcH7 and UbcH5b, respectively (31, 32). In addition, several resonances from residues outside the cleft are significantly perturbed. These resonances correspond to residues in the C terminus of the BRCA1 RING domain (A102 and S104) and residue K20 at the bottom of the helical bundle.
Figure 2.
A ribbon representation of the BRCA1 (gray)/BARD1 (blue) RING heterodimer structure. The four-helix-bundle dimerization interface, Zn2+-loops I and II, and the central helix of BRCA1 are labeled. Residues whose resonances are significantly perturbed by the binding of UbcH5c (K20, C24, I26, C27, L28, K45, F46, L51, L52, Q54, K55, Q60, C61, L63, C64, K65, N66, I68, A102, and S104 of BRCA1) are shown in red. Side chains of residues discussed in the text (K20, I26, L28, L51, L63, K65, and A102) are shown in red and labeled.
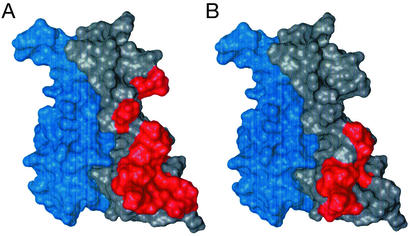
Figure 3.
Surface representation of E2 binding to the BRCA1/BARD1 heterodimer. (A) Residues in the BRCA1/BARD1 heterodimer whose resonances are significantly perturbed by the binding of UbcH5c (K20, C24, I26, C27, L28, K45, F46, L51, L52, Q54, K55, Q60, C61, L63, C64, K65, N66, I68, A102, and S104 of BRCA1) are mapped in red on a surface representation of the heterodimer. (B) Same as in A, depicting resonances significantly perturbed by the binding of UbcH7 (I26, C27, L28, L51, C61, L63, C64, K65, N66, and I68 of BRCA1).
Effect of Single-Site BRCA1 Mutants on Ub-Ligase Activity.
A number of single-site mutations in the BRCA1 RING domain have been found in breast and ovarian cancer patients (see Breast Cancer Information Core; www.nhgri.nih.gov). The mutations genetically linked to the development of breast and ovarian cancer (i.e., C24R, C39S/Y, C44F, C47G/F, C61G, and C64G/R/Y) target residues that ligate the Zn2+ ions critical to the structure of the RING domain (16). In a previous study (38), we found that mutation of site II Zn2+-binding ligands (i.e., C39, H41, C61, and C64) alters the stability of the second Zn2+-binding loop. Residues in this loop form a major portion of the UbcH5c binding cleft (Fig. 2). Other single-site mutations have been observed in breast and ovarian cancer patients, but have not yet been directly linked to the onset of disease. Two such mutations, L52F and L63F, occur in residues whose resonances are perturbed by the binding of UbcH5c. Therefore, we introduced these mutations into the RING domain of BRCA1 to assess their impact on the Ub-ligase activity of the BRCA1/BARD1 heterodimer. Site-specific BRCA1 mutations were also introduced to corroborate the UbcH5c-binding interface delineated by the NMR chemical-shift mapping.
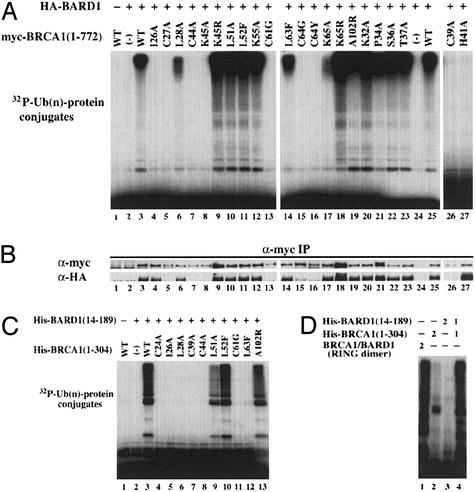
Ub-ligase assays were initially performed with BRCA1/BARD1 immunocomplexes isolated after transient transfection in 293T cells. Consistent with earlier results (17, 18), BARD1 stabilizes BRCA1 in vivo. No BRCA1 protein was isolated in the absence of BARD1 (Fig. 4 A and B, lane 1), whereas in its presence the heterodimer is readily formed and exhibits significant ligase activity (Fig. 4A, lane 3). Mutations within the first or second Zn2+-binding loops of BRCA1 result in a significant reduction or abrogation of activity (i.e., I26A, L28A, C27A, C61G, L63F, C64G/Y, and K65A), whereas mutations within the central helix (i.e., K45R, L51A, L52F, and K55A) have modest or no effect. A102R, which lies on the C-terminal end of the RING domain, has full activity, although the A102 NH resonance is significantly perturbed on binding of UbcH5c (see above). A102 may not be directly involved in the interaction with UbcH5c, but, rather, it may move to accommodate binding. Mutations outside the UbcH5c-recognition site (K32A, P34A, S36A, and T37A) did not alter ligase activity.
Figure 4.
Ub-ligase activity of wild-type and mutant BRCA1/BARD1 heterodimers. (A) Cell lysates from 293T cells transfected with 7.5 μg of plasmid encoding myc-BRCA1(1–772) and HA-BARD1(1–777) were immunoprecipitated with anti-Myc antibody followed by a substrate-independent Ub-ligase assay with UbcH5c. (B) BRCA1 and BARD1 proteins in the immunocomplex were verified by immunoblots with either anti-Myc (BRCA1) or anti-HA (BARD1) antibody. (C) Ub-ligase assays of 1 μg of purified wild-type or mutant BRCA1/BARD1 heterodimers. (D) Comparison of Ub-ligase activity of wild-type BRCA1/BARD1 heterodimers described in C with the BRCA1(1–112)/BARD1 (26–140) heterodimers used for NMR studies. Numbers at top refer to micrograms of protein added.
Mutation of BRCA1 Zn2+-liganding residues (C27, C39, H41, C44, C61, and C64) abrogate Ub-ligase activity. Inspection of Fig. 4B shows that most of these BRCA1 mutations (C27, C39, C44, C61, and C64) alter the ability of BRCA1 to form complexes with BARD1 in vivo. A notable exception is H41A, in Zn2+ site II, which clearly forms heterodimers with BARD1. This Zn2+ site is critical for maintaining the structure and stability of the second Zn2+-binding loop of BRCA1 (16, 38). Thus, this mutation is consistent with other second Zn2+ loop mutations that define this region as important in BRCA1–UbcH5c interactions.
Several BRCA1 mutations shown in Fig. 4A, including L28A and L63F, appear to have reduced activity relative to the wild-type complex. Stability of the complex formed in vivo may influence the amount of BRCA1/BARD1 heterodimer isolated and, thus, the observed activity. To control for differences in protein concentration, His-tagged BRCA1 and BARD1 proteins were purified and assayed for ligase activity (Fig. 4C). The BRCA1/BARD1 complex (Fig. 4C, lane 3) shows substantial activity, whereas the isolated subunits (Fig. 4C, lanes 1 and 2) show little or no activity. As before, mutation of critical Zn2+-liganding residues (C24A, C39A, C44A, and C61G) abrogates ligase activity. In addition, mutations in either the first (I26A and L28A) or second (L63F) Zn2+-binding loops abolish activity, whereas mutations in the central helix (L51A and L52F) have modest to no effect. The activity of L28A and L63F, which show reduced activity relative to wild type (Fig. 4A), is lost in the bacterially expressed proteins.
Fig. 4D shows that the wild-type BRCA1/BARD1 RING heterodimer used for the NMR-titration experiments has nearly the same activity as those of heterodimers that are made from larger constructs [i.e., BRCA1(1–304) and BARD1(14–189)]. Some weak Ub-ligase activity is observed for the isolated BRCA1(1–304) subunit (Fig. 4D, lane 2). Observation of such activity generally requires longer reaction times and/or autoradiograph exposures. This weak activity may be similar to that reported for other isolated BRCA1 preparations (25, 29).
Productive Versus Nonproductive E2 Binding to BRCA1.
In cases where mutations in BRCA1 do not disrupt BRCA1/BARD1 heterodimer formation, loss of ligase activity may be caused either by loss of binding or by nonproductive binding of the E2 to BRCA1. To distinguish these possibilities, an NMR-titration experiment was conducted with BRCA1/BARD1 heterodimer in which Ile-26 of BRCA1 was changed to Ala. Ile-26 was targeted because its NH resonance is the only crosspeak that disappears completely on the first addition of UbcH5c, suggesting an important role for I26 in E2 recognition and binding. The corresponding residue in cCbl (Ile-383) makes direct contact with UbcH7 in the cocrystal structure (31). Similarly, NMR titrations have identified the corresponding residue in the CNOT4 RING finger (Leu-16) as important for UbcH5b binding, and mutation to Glu or Ala abolishes Ub-ligase activity (32). The data presented herein (Figs. 1 and 4A, lane 4, and 4_C_, lane 5) demonstrate that I26 is critical to BRCA1 function as mutation to Ala abolishes Ub-ligase activity.
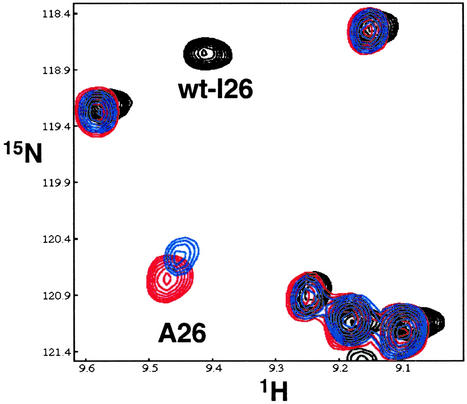
The BRCA1 I26A mutant is readily isolated as a heterodimer in our bacterial coexpression system, consistent with the coimmunoprecipitate results (Fig. 4B, lane 4). Its 1H–15N TROSY spectrum shows that the mutation results in only modest chemical shift perturbations in the BRCA1 RING motif. Resonances arising from residues in the BARD1 subunit or in the 4-helix-bundle dimerization interface show little or no change (Fig. 5). Resonances from the I26A BRCA1/BARD1 complex could be assigned by direct comparison with the wild-type spectrum. The exception (Fig. 5) is the residue 26 resonance, which is significantly shifted in the 15N dimension. Titration of UbcH5c into I26A BRCA1/BARD1 shows that even after the addition of one equivalent of UbcH5c, there is only a minor shift in the residue 26 resonance with all other resonances remaining unperturbed. Thus, the BRCA1 I26A mutation does not hinder heterodimerization with BARD1, but nearly abolishes the ability of BRCA1 to bind UbcH5c. This property makes the mutation potentially useful for distinguishing BRCA1-dependent Ub-ligase activity from other BARD1-dependent functions in vivo.
Figure 5.
NMR titration of 15N-labeled I26A BRCA1/BARD1 with UbcH5c. Shown is an overlay of expanded regions of 1H–15N TROSY spectra of 15N-labeled wild-type (black spectrum) and I26A BRCA1/BARD1 heterodimer in the presence of 0 (red spectrum) and 1.0 (blue spectrum) equivalent of unlabeled UbcH5c.
The ability of UbcH7 to interact with the BRCA1/BARD1 heterodimer was also examined. In general, Ub-conjugating enzymes share a core of ≈150 residues that are highly similar in both sequence and structure (27). UbcH5c and UbcH7 share substantial sequence similarity (37% identity, 60% similarity) particularly in the regions that mediate binding to E3 ligases. Structural and mutagenesis studies (30–32) on a number of E2s have identified these regions as the first α-helix, loop L1, and loop L2 of the E2 protein (aligned below).

Despite the sequence and structural similarity between UbcH5c and UbcH7 in these regions, only UbcH5c is functional in BRCA1-mediated substrate-independent Ub-ligase assays (ref. 17 and data not shown).
NMR-titration experiments (Fig. 1B) clearly show that UbcH7 binds to the BRCA1 subunit of the BRCA1/BARD1 heterodimer. Similar to UbcH5c (compare Fig. 1 A and B), UbcH7 causes a subset of BRCA1 subunit resonances to broaden and disappear. BARD1 resonances are not affected. The interaction between the heterodimer and UbcH7, like UbcH5c, is in intermediate exchange on the NMR time scale under the conditions used. Although only a rough measure of binding, this behavior suggests that UbcH5c and UbcH7 bind to BRCA1 with similar affinities. Substantially weaker binding would likely manifest as fast-exchange behavior in the NMR experiment, whereas substantially tighter binding may be manifest in the appearance of new NMR peaks in the so-called slow-exchange regime. These arguments assume that the UbcH5c and UbcH7 residues that interact with the BRCA1 subunit are similar with no difference in the nature or number of aromatic side chains. The sequence alignments, shown above, are consistent with this working assumption. In addition, we have recently mapped the reciprocal binding surfaces on UbcH5c and UbcH7 and have found that they use the same structural elements for binding to BRCA1 (unpublished data).
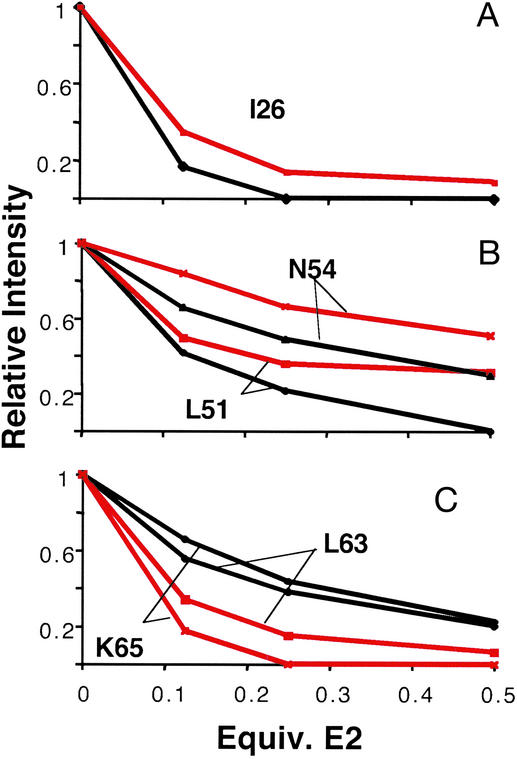
Mapping the UbcH7-induced chemical shift perturbations onto BRCA1 (Fig. 3B) shows that UbcH7 binds to the same surface cleft as UbcH5c. However, close comparison of the UbcH7- and UbcH5c-induced intensity and chemical-shift changes reveal some clear differences (Figs. 1 A and B and 6). Fig. 6 highlights intensity changes from representative residues (I26 in Zn2+-loop I, L51 and N54 in the central helix, and L63 and K65 in Zn2+-loop II). In general, UbcH5c causes larger perturbations in Zn2+-loop I, the central helix, and in the C-terminal residues (A102 and S104) of residues in the BRCA1 subunit. UbcH7 causes larger changes in Zn2+-loop II of BRCA1, whereas little or no effects are observed for resonances of C-terminal residues (A102 and S104) and the four-helix-bundle (i.e., K20). A comparison of all measurable resonances can be seen in Fig. 8, which is published as supporting information on the PNAS web site.
Figure 6.
Comparison of E2-induced intensity changes in BRCA1 resonances. Normalized resonance intensity changes in I26 in Zn2+-loop I (A), L51 and N54 in the central helix (B), and L63 and K65 in Zn2+-loop II (C) induced by UbcH5c (black traces) and UbcH7 (red traces) binding are shown. Chemical shift perturbations are not shown.
Discussion
RING finger proteins that function as E3 Ub ligases tend to be classified into two broad categories: complex and simple. Complex RING E3s exist as small subunits in large macromolecular complexes, for example, the complex of ROC1 or Rbx1 with Skp-1-Cul1-F-box (SCF) (39, 40). Simple RING E3s are generally one domain within a larger protein, such as is found in cCbl (31). BRCA1 does not easily fit into either category. As demonstrated herein and elsewhere, the substrate-independent polyubiquitination activity is markedly dependent on complex formation with the RING domain of BARD1. This RING E3 complex is unusual in that it contains two RING fingers instead of only one, raising the question of the role of each RING in Ub-chain formation.
The NMR data presented clearly demonstrate that only the BRCA1 RING interacts with E2 proteins. The binding interface lies within a surface cleft formed by the first Zn2+-loop, the second Zn2+-loop, and the central helix. A similar surface cleft, formed by the same RING structural elements, is recognized by UbcH7 on binding to the cCbl RING finger. Across the family of RING finger proteins, the segment between the third and fourth pairs of Zn2+ ligands varies considerably, containing from 5 to 48 residues. The sequence and structure of this segment is undoubtedly a critical determinant for Ub-ligase activity. In this segment, BARD1 has five fewer residues than BRCA1, no central helix, and no surface cleft, and does not bind UbcH5c or UbcH7. Thus, mutation of Zn2+-liganding residues in BARD1 has a lesser effect on the Ub-ligase activity of the heterodimer (refs. 17 and 21, and Fig. 9, which is published as supporting information on the PNAS web site) than the corresponding mutations in BRCA1, which abolish activity. In addition, changes in activity caused by BARD1 mutations can generally be correlated with a reduced ability to heterodimerize with BRCA1 (21).
What then is the function of BARD1? One possibility is that BARD1 stabilizes the conformation of BRCA1 required for E3-ligase activity. There are few contacts between the two RING motifs in the structure of the BRCA1/BARD1 heterodimer (16). Thus, heterodimerization is unlikely to alter either RING motif, as the structure of this region is primarily dictated by folding around the Zn2+ ions. Instead, interaction between the two subunits occurs primarily by means of flanking helices, which combine to form the four-helix-bundle dimerization interface (Fig. 2). Therefore, it is likely that BARD1 stabilizes an active conformation of BRCA1 by properly positioning the BRCA1 RING relative to its dimerization helices.§ Consistent with this proposal, mutations in BARD1 that affect dimerization with BRCA1 significantly reduce or abolish BRCA1-mediated Ub-ligase activity (21). Alternatively, BARD1 could be involved in the recognition and binding of appropriate protein targets for ubiquitination. Although target candidates for BRCA1-mediated ubiquitination have been suggested, the in vivo substrates are not known. Potentially, binding of target proteins could occur anywhere along the full-length proteins in the BRCA1/BARD1 complex. In any case, the minimal BRCA1/BARD1 RING domain is sufficient for substrate-independent polyUb-chain formation (Fig. 4D). Thus, individual molecules of Ub can be considered as substrates in this reaction. In this context, it is surprising that we see no interaction of free Ub with BARD1 or BRCA1 in NMR experiments (unpublished data).
Several mammalian E2 proteins have been screened for activity in BRCA1-dependent ligase reactions, including UbcH2, Cdc34, UbcH5b, UbcH5c, UbcH7 (17, 29), and UbcH6 (data not shown). Only UbcH5b and UbcH5c exhibit substrate-independent activity in conjunction with BRCA1. Remarkably, we find that UbcH5c and UbcH7 both bind to BRCA1, on the same surface cleft, and with roughly similar affinities. We have also mapped the reciprocal binding surfaces on UbcH5c and UbcH7 and find they use similar structural elements, but the perturbations are more extensive on UbcH5c (data not shown). The observations raise the question of what determines an active E2/E3 pair. The patterns of NMR perturbations observed, although similar, are not identical. On the first addition of UbcH5c (0.125 eq) the NH resonance of BRCA1 Ile-26 in the first Zn2+-binding loop nearly disappears. In contrast, the crosspeak for Lys-65 in the second Zn2+-binding loop disappears on addition of an identical amount of UbcH7. Furthermore, UbcH5c binding perturbs more resonances corresponding to residues in the BRCA1 central helix and in the C terminus of the RING domain (residues 102 and 104); UbcH7 is nearly silent with respect to the same BRCA1 C-terminal residues. The role of these residues in UbcH5c binding is not clear. Although close to the C terminus in our minimal construct, these residues are structured in solution and interact with residues on the exposed face of the four-helix bundle. It is possible that they are directly involved in binding to UbcH5c, whereas they do not recognize UbcH7. However, mutation of Ala-102 to Arg has essentially no effect on the Ub-ligase activity of BRCA1. An alternative possibility is that these residues must move to accommodate the binding of UbcH5c. These observations suggest that proper orientation between E2 and E3 may be a critical element for Ub-ligase activity.
Our results show that a single residue change on the UbcH5c-recognition surface of BRCA1 is sufficient to severely decrease or abrogate the Ub-ligase activity of BRCA1/BARD1. The most effective mutations involve residues in the two Zn2+-binding loops; mutation of residues in the central helix of the BRCA1 RING retain activity. This finding is in contrast to the E2/E3 pair of UbcH5/cCbl, where mutation of Trp-408 to Ala in the central helix of cCbl abrogates activity (41). In the cocrystal structure of cCbl-UbcH7, Trp-408 contacts UbcH7 loop L1 residue Phe-63 (31). As shown above, loop L1 is highly conserved between UbcH5c and UbcH7, including Phe-63. However, mutation of the BRCA1 residue that is structurally analogous to Trp-408 in cCbl (i.e., L51A) does not abolish the activity of the BRCA1/BARD1 complex. L51A had a more pronounced effect on the much weaker activity of the isolated BRCA1 RING domain (25). On the other hand, both proteins have a structurally equivalent Ile residue in the first Zn2+-binding loop, a feature shared by many RING finger proteins. In BRCA1 and other RING fingers, the integrity of this residue is critical for Ub-ligase activity and may represent a general determinant for E2 recognition and binding. This finding has important implications for functional studies, at least for BRCA1, as mutation of Ile-26, unlike mutations of Zn2+-liganding residues, does not disrupt the overall structure of the BRCA1 RING domain, or prevent interactions with other protein partners such as BARD1.
Given the high degree of similarity in both sequence and structure of the E2s UbcH5 and UbcH7, it is not surprising that they both bind to BRCA1/BARD1. The interaction between BRCA1/BARD1 and UbcH5c, as well as the cocrystal of cCbl/UbcH7, may be thought to represent an E2/E3 product complex present after Ub has been transferred and the target protein has diffused away. In this respect, the two systems differ substantially. The interaction between cCbl and UbcH7 is very strong, as demonstrated by the ability to cocrystallize the complex. This tight binding pair, however, is inactive in Ub-transfer reactions (N. Zheng and C. Joazeiro, personal communication). In contrast, UbcH5c interacts weakly with the BRCA1 RING domain and such interactions are unlikely to be detected by binding assays such as coimmunoprecipitation. Weak interactions in the product complex may be necessary for turnover and Ub-chain elongation. It is possible that appropriate target proteins and/or the presence of conjugated Ub on the E2 increase the affinity and specificity of this macromolecular enzyme complex. Further characterizations that include these additional molecular components will surely reveal additional insights into the determinants of BRCA/BARD1-mediated ubiquitination.
Supplementary Material
Supporting Figures
Acknowledgments
We thank Dr. David Hoyt for assistance with the NMR experiments, which were performed in the Environmental Molecular Sciences Laboratory (a national scientific user facility sponsored by Department of Energy Biological and Environmental Research) located at Pacific Northwest National Laboratory (Richland, WA) and operated by Battelle, and Dr. Kei Arima for technical support. This work was supported by National Cancer Institute Grant CA79953 (to R.K.), grants from the Japan Society for the Promotion of Science, and the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to T.O.).
Abbreviations
Ub
ubiquitin
E2
Ub-conjugating enzyme
E3
Ub ligase
TROSY
transverse-relaxation optimized spectroscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
§
Ideally, we would like to compare the interaction of E2s with BRCA1 in the absence and presence of BARD1; however, the properties of the isolated BRCA1 RING domain are not amenable to NMR studies.
References
- 1.Welcsh P L, King M C. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 2.Welcsh P L, Owens K N, King M C. Trends Genet. 2000;16:69–74. doi: 10.1016/s0168-9525(99)01930-7. [DOI] [PubMed] [Google Scholar]
- 3.Yu V. Breast Cancer Res. 2000;2:82–85. doi: 10.1186/bcr37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scully R, Puget N. Biochimie. 2002;84:95–102. doi: 10.1016/s0300-9084(01)01359-1. [DOI] [PubMed] [Google Scholar]
- 5.Bochar D A, Wang L, Beniya H, Kinev A, Xue Y, Lane W S, Wang W, Kashanchi F, Shiekhattar R. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 6.Chiba N, Parvin J D. J Biol Chem. 2001;276:38549–38554. doi: 10.1074/jbc.M105227200. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong Q, Chen C F, Li S, Chen Y, Wang C C, Xiao J, Chen P L, Sharp Z D, Lee W H. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 9.Deng C X, Brodie S G. BioEssays. 2000;22:728–737. doi: 10.1002/1521-1878(200008)22:8<728::AID-BIES6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 10.Wu L C, Wang Z W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M C, Hwang L Y, Bowcock A M, Baer R. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 11.Irminger-Finger I, Leung W C. Int J Biochem Cell Biol. 2002;34:582–587. doi: 10.1016/s1357-2725(01)00161-3. [DOI] [PubMed] [Google Scholar]
- 12.Kleiman F E, Manley J L. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 13.Kleiman F E, Manley J L. Science. 1999;285:1576–1579. doi: 10.1126/science.285.5433.1576. [DOI] [PubMed] [Google Scholar]
- 14.Jin Y, Xu X L, Yang M C, Wei F, Ayi T C, Bowcock A M, Baer R. Proc Natl Acad Sci USA. 1997;94:12075–12080. doi: 10.1073/pnas.94.22.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 16.Brzovic P S, Rajagopal P, Hoyt D W, King M C, Klevit R E. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 17.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 18.Joukov V, Chen J, Fox E A, Green J B, Livingston D M. Proc Natl Acad Sci USA. 2001;98:12078–12083. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabbro M, Rodriguez J A, Baer R, Henderson B R. J Biol Chem. 2002;277:21315–21324. doi: 10.1074/jbc.M200769200. [DOI] [PubMed] [Google Scholar]
- 20.Chen A, Kleiman F E, Manley J L, Ouchi T, Pan Z Q. J Biol Chem. 2002;277:22085–22092. doi: 10.1074/jbc.M201252200. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y, Pao G, Chen H W, Verma I M, Hunter T. J Biol Chem. 2003;278:5255–5263. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- 22.Weissman A M. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 23.Pickart C M. Trends Biochem Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 24.Hicke L. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 25.Ruffner H, Joazeiro C A, Hemmati D, Hunter T, Verma I M. Proc Natl Acad Sci USA. 2001;98:5134–5139. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Andrea A D, Grompe M. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 27.Pickart C M. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 28.Joazeiro C A, Weissman A M. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 29.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Noel G, Muller U, Harbers K. Eur J Biochem. 2001;268:5912–5919. doi: 10.1046/j.0014-2956.2001.02541.x. [DOI] [PubMed] [Google Scholar]
- 31.Zheng N, Wang P, Jeffrey P D, Pavletich N P. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 32.Albert T K, Hanzawa H, Legtenberg Y I, de Ruwe M J, van den Heuvel F A, Collart M A, Boelens R, Timmers H T. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuiderweg E R. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 34.Meza J E, Brzovic P S, King M C, Klevit R E. J Biol Chem. 1999;274:5659–5665. doi: 10.1074/jbc.274.9.5659. [DOI] [PubMed] [Google Scholar]
- 35.Brzovic P S, Meza J, King M C, Klevit R E. J Biol Chem. 1998;273:7795–7799. doi: 10.1074/jbc.273.14.7795. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Johnson B A, Blevins R A. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 38.Brzovic P S, Meza J E, King M C, Klevit R E. J Biol Chem. 2001;276:41399–41406. doi: 10.1074/jbc.M106551200. [DOI] [PubMed] [Google Scholar]
- 39.Zheng N, Schulman B A, Song L, Miller J J, Jeffrey P D, Wang P, Chu C, Koepp D M, Elledge S J, Pagano M, et al. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 40.Ohta T, Michel J J, Schottelius A J, Xiong Y. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 41.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figures